Abstract
Background
The aryl hydrocarbon receptor (AHR) and autophagy are both important to maintain skin homeostasis. However, they are also involved in skin disorders. So far, their roles in psoriasis pathogenesis are unknown.
Objective
We studied the immunohistochemical and gene expression of AHR, CYP1A1, and microtubule-associated protein light chain 3 (LC3) in lesional skin of psoriasis patients to determine correlations among them.
Methods
We included 24 psoriasis patients and ten healthy volunteers. Skin biopsies were collected. AHR, CYP1A1, and LC3 protein expression was examined by immunohistochemistry, immunofluorescence, and western blotting. AHR, CYP1A1, LC3, ATG5, BECN1 and Nrf2 mRNA levels were measured by quantitative polymerase chain reaction.
Results
AHR and CYP1A1 protein expression were higher in psoriasis lesional skin than in normal skin. LC3 protein expression was lower in psoriasis lesions than in normal controls. AHR and CYP1A1 protein expression in psoriasis lesions showed significant positive correlations with mean epidermal thickness and inflammatory cell density. Significant negative correlations were noted between LC3 protein expression in psoriasis lesions and the mean epidermal thickness or inflammatory cell density. A significant negative correlation was found between AHR and LC3 expression in psoriatic skin. AHR, CYP1A1 and Nrf2 mRNA expression were upregulated while LC3, ATG5, and BECN1 mRNA were down-regulated, in psoriatic lesional skin compared with normal controls.
Conclusion
AHR and autophagy could play a role in psoriasis pathogenesis by modifying epidermal hyperproliferation and inflammation. AHR and autophagy regulation are potential therapeutic targets in chronic inflammatory skin diseases.
Keywords: Autophagy, Cytochrome P-450 CYP1A1, Psoriasis, Receptors, aryl hydrocarbon
INTRODUCTION
Psoriasis is a chronic, immune-mediated, inflammatory skin disorder affected by both genetic and environmental factors. Epidemiologic reports confirm correlations between psoriasis and exposure to environmental pollutants, such as polycyclic aromatic hydrocarbons (PAHs) including benzo [a]pyrene as an aryl hydrocarbon receptor (AHR) ligand1,2. AHR is a ligand-activated transcription factor expressed in various human tissues including skin, lung, liver, thymus, and kidney. AHR in the cytoplasm can be activated by environmental factors and migrate into the nuclei of skin cells3. In the nucleus, AHR dimerizes with the AHR nuclear translocator (ARNT) and participates in canonical signaling. The ligand–AHR–ARNT complex binds the xenobiotic-responsive element of its specific DNA recognition site causing target gene transcription, such as CYP1A1, which is a xenobiotic-metabolizing enzyme4. Environmental toxicants, dioxins including 2,3,7,8,-tetrachlorodibenzo-p-dioxin (TCDD), activate AHR by functioning as high-affinity AHR ligands, subsequently upregulating CYP1A1 expression. The AHR-CYP1A1 pathway also stimulates oxidative stress generation, which in turn induces inflammatory cytokine release5. AHR can also interact in a non-canonical manner with other signaling pathways, such as inflammatory and immune regulatory cascades, including the NF-κB6, mitogen–activated protein kinases7, and signal transducer and activator of transcription (STATs) pathways8. AHR was also recently suggested to be a critical skin integrity and immunity modulator, which are crucially involved in the pathogenesis of multiple skin diseases9.
Autophagy is the endogenous housekeeping process responsible for degrading dysfunctional and redundant cellular organelles and proteins10. Autophagy not only serves a physiological function in maintaining skin homeostasis, but is also related to pathological processes in multiple skin disorders11. Cellular microtubule-associated protein light chain 3 (LC3) is an autophagy marker that indicates autophagosome quantity as the lipidated form of LC3-II accumulates in autophagosomal membranes12. Evidence shows that dysfunctional autophagy leads to inflammatory immune responses13,14. In cultured keratinocytes, autophagy defects result in increased inflammatory cytokine production and cell proliferation, suggesting a role in promoting psoriasis development15.
Previously, we reported that crosstalk between the AHR pathway and autophagy may be linked to psoriasis pathogenesis in human keratinocytes. Specifically, AhR activation reduced autophagy, promoting skin inflammation through the p65NF-κB/p38MAPK pathways16. To validate the potential roles of the AHR pathway and autophagy in human psoriasis, we examined the expression of AHR, its downstream molecule CYP1A1, and autophagy marker LC3 in skin biopsies from psoriasis and normal controls, and evaluated the correlation among these markers in psoriasis skin tissues.
MATERIALS AND METHODS
Patients
This study included 24 patients with psoriasis vulgaris, and 10 normal control subjects from the Outpatient Clinic of the Department of Dermatology, Hallym University Kangnam Sacred Heart Hospital. Psoriasis was diagnosed by 2 dermatologists based on clinical and pathological features.
No subjects received topical or systemic treatment at least 3 months before skin biopsy. Patients with any other dermatological or uncontrolled systemic diseases and pregnant or lactating woman were excluded. A complete history was obtained from each patient, and all patients underwent general and dermatological examinations of disease severity using PASI scoring for psoriasis17.
Ethics statement
Written informed consent was provided by all subjects before their participation. The protocol was approved by the Institutional Review Board of Hallym University Kangnam Sacred Heart Hospital (IRB no. 2018-05-024).
Skin biopsies
Four-millimeter punch skin biopsies were taken under local anesthesia from lesional skin of psoriasis and control subjects. Specimens embedded in paraffin blocks were divided into 4 sections (each 4-µm thick); one was stained with hematoxylin and eosin to evaluate pathological changes and the other three were placed onto poly-L-lysine-coated slides for immunostaining. Skin tissues were also preserved in liquid nitrogen to prepare total RNA and total protein lysate.
Immunohistochemistry
Immunohistochemistry was conducted in 10% formalin-fixed, paraffin-embedded tissues. Dissected tissues were cleansed twice with distilled water, and residual fixatives were removed by 1 hour treatment with 1% sodium. The tissues were pre-treated with 3% hydrogen peroxide for 10 minutes, cleansed with distilled water; and cultured for 5 minutes with 1 TBST (Tris-buffered 5 saline with 0.1% Tween 20). To prevent nonspecific reactions, tissues were treated with normal goat serum (Vector Laboratories, Burlingame, CA, USA) at room temperature for 1 hour, and then cultured overnight with rabbit anti-AHR (1:300; Abcam, Cambridge, MA, USA), rabbit anti-CYP1A1 (1:300; Abcam), and rabbit anti-LC3 (1:300; Novus Biologicals, Littleton, CO, USA). Tissues were cleansed with 1 TBST, and cultured with biotinylated secondary antibody solution from the Dako REAL EnVision Detection System (Dako, Glostrup, Denmark) for 30 minutes at room temperature. The tissues then then cleansed with distilled water; counterstained with hematoxylin (Sigma-Aldrich, St. Louis, MO, USA); dehydrated and clarified based on a conventional method; and then prepared for Leica microsystem DFi8 LASX software light microscopy (Leica, Wetzlar, Germany).
Immunohistochemical analysis
The staining pattern was evaluated using 4 separate fields in the epidermal cell layers. According to previous studies, brown cytoplasmic and/or nuclear staining for AHR, brown cytoplasmic staining for CYP1A1, and brown cytoplasmic and/or nuclear staining for LC3 were considered positive18,19.
AHR, CYP1A1, and LC3 intensity scoring was performed via quantitative evaluation of the immunostained slides. Positive keratinocyte counts were described as the percentage of total keratinocyte counts from 0 to +3 as follows: 0 (negative expression)=no staining, +1 (mild expression) <25%, +2 (moderate expression)=25%~50%, and +3 (strong expression) >50%20. A positive percentage was given in epidermis after counting 500 cells in each section21.
Quantitative morphometric analysis was conducted with LASX computerized digital image analysis software (Leica). Using a calibrated ruler, epidermis thickness was measured from the bottom of the rete ridge to the bottom of the stratum corneum; the mean was calculated from the measurements in 4 fields at ×100 magnification. Inflammatory cell density was calculated by identifying the area with the maximum density of inflammatory cells at low magnification (×100), after which inflammatory cells were counted in 4 fields at ×400. The mean density of inflammatory cells was calculated as the inflammatory cell count/dermal area. The results are described as the mean±standard deviation of the optical density of five different digital images20. All immunohistochemical analyses were determined independently by a single pathologist (LHJ) and single dermatologist (KJE) blinded to patient identity or clinical outcome. Disagreements were resolved through a consensus conference. The rate of initial concurrences was >90% in our study.
Immunofluorescence staining
Formalin-fixed, paraffin-embedded human tissue sections were deparaffinized and rehydrated in xylene and ethanol. More details are included in the Supplementary Materials.
Western blotting
Human skin tissues were harvested in pro-prep lysis buffer (Intron, Seoul, Korea) with a protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). The copper (II) sulfate solution in bicinchoninic acid solution (Sigma-Aldrich) was used to measure protein concentrations. The same amount of protein (20 µg) was separated by 10% SDS-PAGE and transferred to enhanced chemiluminescence (ECL) nitrocellulose membranes (GE Healthcare, Buckinghamshire, UK), and blocked for 1 hour with 5% skim milk in TBST. The membranes were incubated overnight at 4℃ with rabbit anti-LC3 (1:1,000; Abcam) antibodies. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit, 1:1,000; Abcam) and chemiluminescent luminol (LUMINOGRAPH II; Atto, Tokyo, Japan). Immunocomplexes were detected using an enhanced horseradish peroxidase/luminol chemiluminescence system (ECL Plus; Amersham International PLC, Little Chalfont, UK). Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control.
Quantitative reverse transcriptase-polymerase chain reaction
Based on manufacturer instructions, the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) was used to extract total RNA. The transcriptor First Strand cDNA synthesis kit (Roche Applied Science, Mannheim, Germany) was used to synthesize cDNA from 1 µg of total RNA. Quantitative reverse transcriptase-polymerase chain reaction (PCR) was performed three times using the TaqMan master mix (Applied Biosystems, Foster City, CA, USA) and the real-time PCR System (Applied Biosystems). The primers for mRNA detection are included in the Supplementary Materials. mRNA levels of AHR, CYP1A1, LC3, ATG5, BECN1 and NRF2 were normalized to that of GAPDH. Relative quantification was performed using a Light Cycler® 96 Instrument (Roche Diagnostics).
Statistical analyses
Qualitative data were expressed as numerical values and percentages; quantitative data were expressed as the range, mean, standard deviation, and median. Comparisons between different groups of categorical variables were performed using Fisher's exact test. For continuous variables, Student's t-test wasused. By using Spearman's correlation coefficient, correlations between variables were determined. All statistical analyses were performed using IBM SPSS Statistics 24.0 (IBM Corp., Armonk, NY, USA). p-values <0.05 were considered statistically significant.
RESULTS
Clinicopathologic data
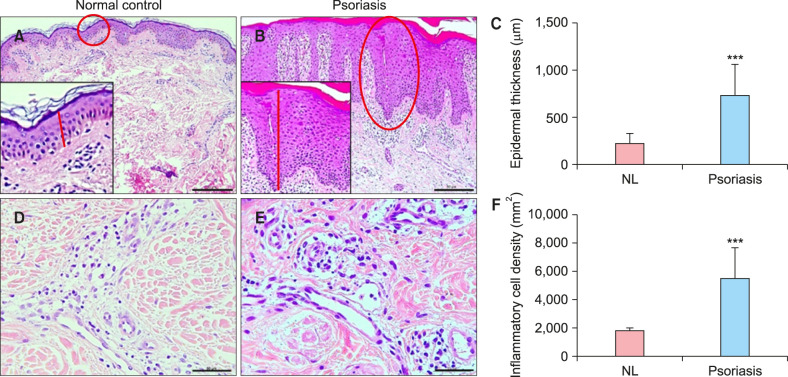
Demographic and clinical patient data are presented in Table 1. Histopathologic data are shown in Table 2. No significant differences were observed in age and sex between groups. Epidermal thickness in the psoriasis group (729.19±331.37 µm) was higher than that in the control group (215.28±109.21 µm, p<0.001; Fig. 1). Inflammatory cell density/mm2 in the psoriasis group (5,489.4±2,149.1) was higher than that in the control group (1,773.34±235.79, p<0.001; Fig. 1).
Table 1. Clinical characteristics of psoriasis and control groups.
| Variable | Psoriasis group (n=24) | Control group (n=10) | p-value |
|---|---|---|---|
| Age (yr) | 0.38 | ||
| Mean±SD | 42.1±15.47 | 36.78±13.65 | |
| Median | 41 | 42 | |
| Range | 10~67 | 13~55 | |
| Sex, n (%) | 0.704 | ||
| Male | 14 (58.3) | 6 (60.0) | |
| Female | 10 (41.7) | 4 (40.0) | |
| Duration (mo) | - | - | |
| Mean±SD | 98.9±128.32 | ||
| Median | 24 | ||
| Range | 1∼480 | - | - |
| PASI score | |||
| Mean±SD | 10.74±7.14 | ||
| Median | 10.2 | ||
| Range | 1.4∼30.6 |
SD: standard deviation, PASI: psoriasis area and severity index, -: not available.
Table 2. Epidermal thickness and inflammatory cell density in psoriasis and control groups.
| Variable | Psoriasis group (n=24) | Control group (n=10) | p-value |
|---|---|---|---|
| Epidermal thickness (μm) | <0.001* | ||
| Mean±SD | 729.19±331.37 | 215.28±109.21 | |
| Median | 690.23 | 214.45 | |
| Range | 210.34∼1,663.75 | 71.31∼434.76 | |
| Inflammatory cell (density/mm2) | <0.001* | ||
| Mean±SD | 5,489.4±2,149.1 | 1,773.34±235.79 | |
| Median | 4,963.2 | 1,768.6 | |
| Range | 1,683.5∼9,683.5 | 1,368.9∼2,065.1 | |
SD: standard deviation. *p<0.05.
Fig. 1. Epidermal thickness and inflammatory cell density of psoriasis and control groups. (A~C) Epidermal thickness measurement from the bottom of the rete ridge to the bottom of the stratum corneum (red line). (A) Normal control specimen (H&E , ×200; bar=75 µm). (B) Lesional skin of a psoriasis patient (H&E, ×200; bar=75 µm). (C) Epidermal thickness qualification. The epidermal thickness in lesional skin of psoriasis was thicker significantly compared to that in controls. (D~F) Morphometric counting of inflammatory cell density. (D) Normal control specimen (H&E, ×400; bar=50 µm). (E) Lesional skin of a psoriasis patient (H&E, ×400; bar=50 µm). (F) Inflammatory cell density qualification. Inflammatory cell density in lesional skin of psoriasis was higher significantly compared to that in controls. NL: normal control. Statistical significance was determined by Student's t-test (***p<0.001).
AHR and CYP1A1 immunohistochemical expression
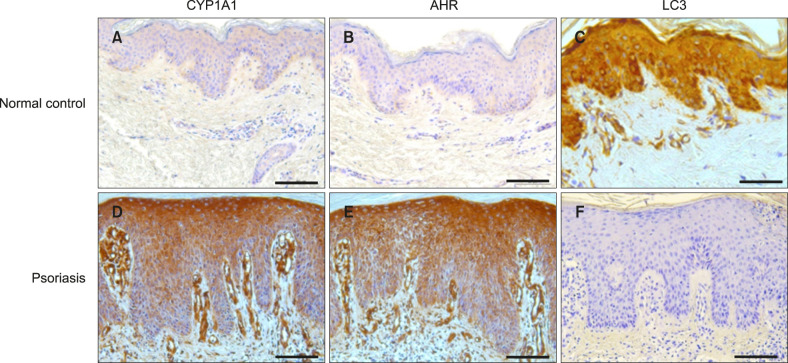
Positive AHR staining in epidermal keratinocytes was found in 100% of psoriasis patients and 30% of healthy controls. CYP1A1 was expressed in epidermal keratinocytes in 100% of psoriasis patients and 40% of controls. AHR and CYP1A1 expression intensities in psoriasis patients were stronger than those in controls (p<0.001, p<0.001, respectively; Table 3, Fig. 2). In immunofluorescence staining, AHR expression was significantly higher in lesional psoriatic skin than in normal controls (Supplementary Fig. 1).
Table 3. Comparison between psoriasis and control groups according to AHR and CYP1A1 expression.
| Variable | Psoriasis group (n=24) | Control group (n=10) | p-value |
|---|---|---|---|
| AHR expression | |||
| Intensity | <0.001* | ||
| None | 0 (0) | 7 (70.0) | |
| Mild | 3 (12.5) | 3 (30.0) | |
| Moderate | 9 (37.5) | 0 (0) | |
| Strong | 12 (50.0) | 0 (0) | |
| Distribution | <0.001* | ||
| Negative | 0 (0) | 7 (70.0) | |
| Basal | 1 (4.2) | 0 (0) | |
| Both basal and suprabasal | 21 (87.5) | 3 (30.0) | |
| Full thickness | 2 (8.3) | 0 (0) | |
| CYP1A1 expression | |||
| Intensity | <0.001* | ||
| None | 0 (0) | 6 (60.0) | |
| Mild | 0 (0) | 4 (40.0) | |
| Moderate | 7 (29.2) | 0 (0) | |
| Strong | 17 (70.8) | 0 (0) | |
| Distribution | <0.001* | ||
| Negative | 0 (0) | 6 (60.0) | |
| Basal | 0 (0) | 0 (0) | |
| Both basal and suprabasal | 17 (70.8) | 4 (40.0) | |
| Full thickness | 7 (29.2) | 0 (0) |
Values are presented as number (%). AHR: aryl hydrocarbon receptor. *p<0.05.
Fig. 2. AHR, CYP1A1, and LC3 immunohistochemical expression in psoriasis lesional skin and normal skin. (A) Normal control skin sample with mild (+1) basal pattern CYP1A1 expression (×200). (B) Normal control skin sample with negative. (0) AHR expression (×200). (C) Normal control skin sample with negative. (0) LC3 expression (×200). (D) Psoriasis specimen (lesional skin) showing a moderate (+2) basal and suprabasal pattern of CYP1A1 expression (×200). (E) Psoriasis specimen (lesional skin) showing a strong (+3) basal and suprabasal pattern of AHR expression (×200). (F) Psoriasis specimen (lesional skin) showing a mild (+1) basal and suprabasal pattern of LC3 expression (×200). Bar=75 µm. AHR: aryl hydrocarbon receptor, LC3: microtubule-associated protein light chain 3.
LC3 protein expression
Epidermal keratinocytes showed positive LC3 staining in 33.3% of psoriasis patients and 90% of controls. LC3 expression intensity was significantly lower in psoriasis patients than in controls (p<0.001; Table 4, Fig. 2). Immunofluorescence analyses showed significantly lower LC3 protein expression in lesional psoriatic skin than in normal controls (Supplementary Fig. 3). There was a decrease in LC3-II protein in lesional skin tissue of psoriasis by western blot analysis (Supplementary Fig. 2).
Table 4. Comparison of LC3 expression between psoriasis and control groups.
| LC3 expression | Psoriasis group (n=24) | Control group (n=10) | p-value |
|---|---|---|---|
| Intensity | <0.001* | ||
| None | 16 (66.7) | 1 (10.0) | |
| Mild | 8 (33.3) | 0 (0) | |
| Moderate | 0 (0) | 5 (50.0) | |
| Strong | 0 (0) | 4 (40.0) | |
| Distribution | 0.001* | ||
| Negative | 16 (66.7) | 1 (10.0) | |
| Basal | 0 (0) | 0 (0) | |
| Both basal and suprabasal | 8 (33.3) | 4 (40.0) | |
| Full thickness | 0 (0) | 5 (50.0) |
Values are presented as number (%). LC3: microtubule-associated protein light chain 3. *p<0.05.
Relationships among the lesional AHR, CYP1A1, and LC3 immunohistochemical expression, clinical data, and histopathological parameters in psoriasis patients
AHR expression in psoriasis lesions was significantly positively correlated with mean epidermal thickness (R=0.57, p=0.002) and mean inflammatory cell density (R=0.56, p=0.002). CYP1A1 expression also showed significant positive correlations with mean epidermal thickness (R=0.59, p=0.001) and mean inflammatory cell density (R=0.57, p=0.006). LC3 expression in psoriasis lesions was significantly negatively correlated with mean epidermal thickness (R=−0.59, p=0.001) and mean inflammatory cell density (R=−0.47, p=0.01). No significant correlation was observed between psoriasis severity and AHR, CYP1A1, and LC3 intensities (Supplementary Table 1).
Correlation among AHR, CYP1A1, and LC3 protein expression in psoriasis patients
There was a significant negative correlation between AHR and LC3 expression in psoriatic skin (R=−0.63, p=0.006). In addition, a positive, significant correlation between AHR and CYP1A1 expression was found in skin lesions of psoriasis patients (R=0.67, p=0.004; Supplementary Table 1).
AHR, autophagy-related molecules and Nrf2 gene expression in psoriatic skin tissue
To further validate the significance of AHR and autophagy-related factors in psoriasis, we measured mRNA expression of AHR, CYP1A1, LC3, BECN1, and ATG5 in psoriasis lesional skin tissue. These samples expressed relatively higher AHR and CYP1A1 mRNA compared to that in normal controls. However, the autophagy-related factors LC3, BECN1, and ATG5 were decreased in lesional skin of psoriasis compared with normal controls (Supplementary Fig. 3).
AHR signaling and autophagy both act upon the oxidative-stress response22,23. Thus, we examined the expression of NRF2 gene as a redox-sensitive transcription factor in psoriatic skin tissue and found that psoriasis lesional skin had higher NRF2 mRNA expression than normal controls (Supplementary Fig. 3).
DISCUSSION
Previously, we performed in vitro and ex vivo studies to confirm the effects of AHR activation on autophagy in human keratinocytes, as well as the relevance of AHR and autophagy in psoriasis pathogenesis. Results from in vitro polycytokine-stimulated human keratinocytes and ex vivo psoriasis skin tissues revealed that AHR activation decreased autophagy, inducing inflammation through p65NF-κB/p38MAPK pathways16. Here, we examined the relevance of both AHR (a ligand-dependent transcription factor with various ligands including environmental pollutants such as PAHs) and LC3 (autophagy marker) in skin lesions of psoriasis patients. Immunohistochemical staining showed that AHR and CYP1A1 (the downstream target of AHR) were upregulated in psoriasis lesions compared to expression in control samples. However, expression of the representative autophagy marker, LC3, was downregulated in psoriasis lesions compared to controls. Furthermore, AHR and CYP1A1 expression in psoriasis lesions showed significant positive correlations with mean epidermal thickness and mean inflammatory cell density. Alternatively, significant negative correlations were observed between LC3 expression in psoriasis lesions and mean epidermal thickness and mean inflammatory cell density. There was also a significant negative correlation between AHR and LC3 expression in psoriatic skin. Furthermore, AHR and CYP1A1 mRNA expression were increased in psoriasis lesional skin compared to normal controls. The mRNA expression of autophagy-related molecules was decreased in psoriasis lesional skin compared with normal controls. Together, these findings suggest the important roles of AHR and autophagy in psoriasis development.
AHR is a member of the bHLH/PAS family that is widely expressed in many human tissues including skin. Evidence shows that AHR is involved in various pathways important to cell proliferation and differentiation and immune reactions24,25. Epidemiological studies report a relationship between psoriasis and exposure to AHR ligands in hazardous environmental pollutants (such as PAHs)1,26. Moreover, AHR activation was reported to affect inflammatory profiles in both an imiquimod-induced mouse model of psoriasiform inflammation and skin tissues of psoriasis patients27. Corroborating findings of the current study, researchers found that AHR and CYP1A1 proteins and mRNA were overexpressed in psoriasis skin lesions, and that a significant positive correlation existed between AHR and CYP1A1 protein expression and mean epidermal thickness and mean inflammatory cell density. Laboratory studies reported that AHR activation via PAHs such as TCDD or endogenous ligands affect the proliferation and differentiation of Th17 cells, which have critical roles in psoriasis pathogenesis28,29. Together, AHR overexpression and activation might contribute to psoriasis pathogenesis. It remains unclear whether AHR activation in skin disease is beneficial or harmful. Regarding this, it was speculated that AHR functions in ligand-specific, tissue-specific, and cell-specific manners30,31,32,33. In the context of psoriasis, Di Meglio et al.27 found that transient AhR activation by endogenous, rapid-metabolizing AHR ligand, FICZ results in a decreased inflammatory reaction in a psoriasis mouse model and human psoriatic skin. In support of these findings, topical application of an AHR agonist, tapinarof, was reported to be efficacious in psoriasis clinical trials34. Tapinarof (5-[(E)-2-phenylethenyl]-2-[propan-2-yl] benzene-1, 3-diol) is a hydroxylated stilbene generated by bacterial symbionts of entomopathogenic nematodes. Tapinarof is a high-affinity AHR ligand that exhibits antioxidative activity via NRF2 activation. Tapinarof also repairs the skin barrier dysfunction by promoting FLG and IVL expression, while suppressing interleukin (IL)-17A production and enhancing IL-22 production by affecting T-helper 17 (Th17) and Treg cell differentiation35. However, we previously reported that slow-metabolizing dioxin, TCDD-mediated AHR activation induces inflammation in human keratinocytes and ex vivo skin biopsies of psoriasis16.
Few studies have evaluated the expression of autophagy-related proteins, including LC3, in psoriasis. Here, LC3 expression in psoriasis lesions was downregulated. A previous study showed that while LC3 expression was absent in all layers of psoriasis lesional skin epidermis, LC3 was expressed throughout the full thickness of the epidermis in normal sample36. Varshney and Saini13 also found evidence of decreased LC3 expression in lesional psoriatic skin tissue compared to that in non-lesional tissue. These reports are consistent with our study. In addition, a previous report indicating that a lack of LC3 expression in psoriatic skin lesions is correlated with parakeratosis consistent with our results36. We also found significant negative correlations between LC3 expression in psoriasis lesions the mean epidermal thickness and mean inflammatory cell density. Notably, we found a significant negative correlation between AHR and LC3 expression in psoriatic skin. Based on these results, we speculate that the inverse functional correlation between AHR activation and autophagy could affect psoriasis development by modulating epidermal hyperproliferation and inflammation.
Moreover, NRF2 is a key factor involved in regulating antioxidant signaling. The relationship between NRF2 and AHR signaling and autophagy is plausible as AHR and autophagy also act on the oxidative/antioxidant axis22,23. We, therefore, measured the expression of NRF2 mRNA in psoriatic and healthy control skin tissues and found that it was increased in psoriatic skin lesions compared to that in normal controls. Similarly, a previous study reported that NRF2 mRNA and protein expression was increased in epidermis of psoriasis lesion compared to that in normal control skin. Furthermore, using human keratinocytes and an imiquimod-induced psoriasis mouse model, Yang et al.37 demonstrated that IL-17 and IL-22 stimulated NRF2 translocation to the epidermis of psoriasis lesions, promoting upregulation of keratin 6, keratin 16, and keratin 17 and inducing keratinocyte proliferation, which is associated with psoriasis pathogenesis.
In this study, we found a significant difference in AHR, CYP1A1, and LC3 expression levels in lesional psoriatic skin in comparison to normal skin. However, no significant differences were observed in AHR, CYP1A1, and LC3 intensity according to the PASI score. These results might be explained by the fact that the local inflammation severity of psoriasis lesional skin tissue, which evaluated the expression levels of AHR, CYP1A1, and LC3, could be different to the PASI score in assessing the severity of the psoriasis in the whole body. In other words, even if the PASI score is high, it is possible that the local skin lesion severity of inflammation examined by biopsy is not severe, or vice versa. Furthermore, it is possible that AHR or autophagy could be related to psoriasis development (disease initiation stage), but not to disease severity. Further large-scale studies are needed to confirm this result.
Several limitations were noted. First, group sample sizes were small. Therefore, a large-scale study will be needed to confirm our results. Second, the temporal relationship between AHR and LC3 remains unclear as this study only demonstrated the overall expression of AHR and LC3 in skin tissues. Currently, only a limited number of studies have sought to unravel the complicated roles of AHR and autophagy in psoriasis pathogenesis; to our knowledge, this study is the first to demonstrate the negative correlation between AHR and the expression of the autophagy marker LC3 in lesional skin of psoriasis patients. We anticipate that this finding will aid in the discovery of more detailed mechanisms regarding psoriasis pathogenesis.
In conclusion, AHR and autophagy affected psoriasis pathogenesis through the regulation of epidermal hyperproliferation and skin inflammation. Therefore, AHR and autophagy modification may prove useful as novel therapeutic targets in psoriasis.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING SOURCE: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2018R1C1B6007998, 2020M3E5D1A02030067), Hallym University Research Fund (HURF-2019-72), and Soonchunhyang University Research Fund.
DATA SHARING STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
SUPPLEMENTARY MATERIALS
Supplementary data can be found via http://anndermatol.org/src/sm/ad-33-138-s001.pdf.
The correlation co-efficients between clinicopathologic parameters in patients with psoriasis
Immunofluorescence analysis of AHR and LC3 expression in psoriatic skin lesion. (A) The level of AHR (red), LC3 (green) and nuclei (blue) expression was determined by immunofluorescence. In merged images, areas within the red circle are shown as a magnified image on the left lower square. AHR expression was increased and LC3 expression was decreased in psoriasis skin compared to controls. Data are representative of psoriasis (n=5) and healthy volunteers (n=5). Original magnification ×200, Bar=75 µm, Bar=50 µm (×400) on the left lower square of the merged image. Quantification of AHR (B) and LC3 (C) expression on psoriatic skin lesion. The level of staining was semi-quantitatively analyzed using LASX software (Leica, Wetzlar, Germany). HC: healthy control, AHR: aryl hydrocarbon receptor, LC3: microtubule-associated protein light chain 3. Statistical significance was determined by Student's t-test (***p<0.001) compared to normal controls.
LC3-II protein expression in psoriasis of human skin tissues. (A) Western blot of LC3-I and LC3-II in human skin tissues of HC, and psoriasis lesions. Data normalization was based on GAPDH. The results are representative of 3 independent experiments. (B) The densitometric analysis (LC3-II/GAPDH). Data represent the mean±standard deviation of 3 independent experiments. LC3: microtubule-associated protein light chain 3, HC: healthy control. Statistical significance was determined by student's t-test (***p<0.001).
AHR, CYP1A1, LC3, ATG5, BECN1, NRF2 mRNA expression in psoriatic lesional skin. Quantitative polymerase chain reaction analysis of (A) AHR, (B) CYP1A1, (C) LC3, (D) BECN1, (E) ATG5, and (F) NRF2 expression in human psoriasis lesion (n=5) and HC tissues (n=5). Data represent the mean±standard deviation of three independent experiments (each performed in duplicate). LC3: microtubule-associated protein light chain 3, HC: healthy control. Statistical significance was determined by Student's t-test (*p<0.05, **p<0.01, ***p<0.001) in comparison to controls.
References
- 1.Naldi L, Peli L, Parazzini F. Association of early-stage psoriasis with smoking and male alcohol consumption: evidence from an Italian case-control study. Arch Dermatol. 1999;135:1479–1484. doi: 10.1001/archderm.135.12.1479. [DOI] [PubMed] [Google Scholar]
- 2.Gupta MA, Gupta AK, Watteel GN. Cigarette smoking in men may be a risk factor for increased severity of psoriasis of the extremities. Br J Dermatol. 1996;135:859–860. doi: 10.1111/j.1365-2133.1996.tb03909.x. [DOI] [PubMed] [Google Scholar]
- 3.Ikuta T, Namiki T, Fujii-Kuriyama Y, Kawajiri K. AhR protein trafficking and function in the skin. Biochem Pharmacol. 2009;77:588–596. doi: 10.1016/j.bcp.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Miao W, Hu L, Scrivens PJ, Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 5.Kopf PG, Walker MK. 2,3,7,8-tetrachlorodibenzo-p-dioxin increases reactive oxygen species production in human endothelial cells via induction of cytochrome P4501A1. Toxicol Appl Pharmacol. 2010;245:91–99. doi: 10.1016/j.taap.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritsche E, Schäfer C, Calles C, Bernsmann T, Bernshausen T, Wurm M, et al. Lightening up the UV response by identification of the arylhydrocarbon receptor as a cytoplasmatic target for ultraviolet B radiation. Proc Natl Acad Sci U S A. 2007;104:8851–8856. doi: 10.1073/pnas.0701764104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev. 2015;67:259–279. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sukseree S, Eckhart L, Tschachler E, Watanapokasin R. Autophagy in epithelial homeostasis and defense. Front Biosci (Elite Ed) 2013;5:1000–1010. doi: 10.2741/e679. [DOI] [PubMed] [Google Scholar]
- 12.Zheng HY, Zhang XY, Wang XF, Sun BC. Autophagy enhances the aggressiveness of human colorectal cancer cells and their ability to adapt to apoptotic stimulus. Cancer Biol Med. 2012;9:105–110. doi: 10.3969/j.issn.2095-3941.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varshney P, Saini N. PI3K/AKT/mTOR activation and autophagy inhibition plays a key role in increased cholesterol during IL-17A mediated inflammatory response in psoriasis. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5 Pt A):1795–1803. doi: 10.1016/j.bbadis.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Park MJ, Lee SY, Moon SJ, Son HJ, Lee SH, Kim EK, et al. Metformin attenuates graft-versus-host disease via restricting mammalian target of rapamycin/signal transducer and activator of transcription 3 and promoting adenosine monophosphate-activated protein kinase-autophagy for the balance between T helper 17 and Tregs. Transl Res. 2016;173:115–130. doi: 10.1016/j.trsl.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Lee HM, Shin DM, Yuk JM, Shi G, Choi DK, Lee SH, et al. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J Immunol. 2011;186:1248–1258. doi: 10.4049/jimmunol.1001954. [DOI] [PubMed] [Google Scholar]
- 16.Kim HR, Kang SY, Kim HO, Park CW, Chung BY. Role of aryl hydrocarbon receptor activation and autophagy in psoriasis-related inflammation. Int J Mol Sci. 2020;21:2195. doi: 10.3390/ijms21062195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt J, Wozel G. The psoriasis area and severity index is the adequate criterion to define severity in chronic plaque-type psoriasis. Dermatology. 2005;210:194–199. doi: 10.1159/000083509. [DOI] [PubMed] [Google Scholar]
- 18.Pan ZY, Chen J, Wu Q, Hu TT, Lu L, Ju Q. Activation and overexpression of the aryl hydrocarbon receptor contribute to cutaneous squamous cell carcinomas: an immunohistochemical study. Diagn Pathol. 2018;13:59. doi: 10.1186/s13000-018-0740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshihara N, Takagi A, Ueno T, Ikeda S. Inverse correlation between microtubule-associated protein 1A/1B-light chain 3 and p62/sequestosome-1 expression in the progression of cutaneous squamous cell carcinoma. J Dermatol. 2014;41:311–315. doi: 10.1111/1346-8138.12439. [DOI] [PubMed] [Google Scholar]
- 20.Hodeib AA, Neinaa YME, Zakaria SS, Alshenawy HA. Glucose transporter-1 (GLUT-1) expression in psoriasis: correlation with disease severity. Int J Dermatol. 2018;57:943–951. doi: 10.1111/ijd.14037. [DOI] [PubMed] [Google Scholar]
- 21.Seleit I, Bakry OA, Al Sharaky D, Ragheb E. Evaluation of aquaporin-3 role in nonmelanoma skin cancer: an immunohistochemical study. Ultrastruct Pathol. 2015;39:306–317. doi: 10.3109/01913123.2015.1022241. [DOI] [PubMed] [Google Scholar]
- 22.Furue M, Uchi H, Mitoma C, Hashimoto-Hachiya A, Chiba T, Ito T, et al. Antioxidants for healthy skin: the emerging role of aryl hydrocarbon receptors and nuclear factor-erythroid 2-related factor-2. Nutrients. 2017;9:223. doi: 10.3390/nu9030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng S, Cao Z, Wang X. Role of aryl hydrocarbon receptor in cancer. Biochim Biophys Acta. 2013;1836:197–210. doi: 10.1016/j.bbcan.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 26.Kavli G, Førde OH, Arnesen E, Stenvold SE. Psoriasis: familial predisposition and environmental factors. Br Med J (Clin Res Ed) 1985;291:999–1000. doi: 10.1136/bmj.291.6501.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Meglio P, Duarte JH, Ahlfors H, Owens ND, Li Y, Villanova F, et al. Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity. 2014;40:989–1001. doi: 10.1016/j.immuni.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 29.Schiering C, Vonk A, Das S, Stockinger B, Wincent E. Cytochrome P4501-inhibiting chemicals amplify aryl hydrocarbon receptor activation and IL-22 production in T helper 17 cells. Biochem Pharmacol. 2018;151:47–58. doi: 10.1016/j.bcp.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 30.van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, et al. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J Clin Invest. 2013;123:917–927. doi: 10.1172/JCI65642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuji G, Hashimoto-Hachiya A, Kiyomatsu-Oda M, Takemura M, Ohno F, Ito T, et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. 2017;8:e2931. doi: 10.1038/cddis.2017.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hidaka T, Ogawa E, Kobayashi EH, Suzuki T, Funayama R, Nagashima T, et al. The aryl hydrocarbon receptor AhR links atopic dermatitis and air pollution via induction of the neurotrophic factor artemin. Nat Immunol. 2017;18:64–73. doi: 10.1038/ni.3614. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy LH, Sutter CH, Leon Carrion S, Tran QT, Bodreddigari S, Kensicki E, et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated production of reactive oxygen species is an essential step in the mechanism of action to accelerate human keratinocyte differentiation. Toxicol Sci. 2013;132:235–249. doi: 10.1093/toxsci/kfs325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins K, Bissonnette R, Maeda-Chubachi T, Ye L, Peppers J, Gallagher K, et al. Phase 2, randomized dose-finding study of tapinarof (GSK2894512 cream) for the treatment of plaque psoriasis. J Am Acad Dermatol. 2019;80:714–721. doi: 10.1016/j.jaad.2018.10.037. [DOI] [PubMed] [Google Scholar]
- 35.Smith SH, Jayawickreme C, Rickard DJ, Nicodeme E, Bui T, Simmons C, et al. Tapinarof is a natural AhR agonist that resolves skin inflammation in mice and humans. J Invest Dermatol. 2017;137:2110–2119. doi: 10.1016/j.jid.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Akinduro O, Sully K, Patel A, Robinson DJ, Chikh A, McPhail G, et al. Constitutive autophagy and nucleophagy during epidermal differentiation. J Invest Dermatol. 2016;136:1460–1470. doi: 10.1016/j.jid.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Fan X, Cui T, Dang E, Wang G. Nrf2 promotes keratinocyte proliferation in psoriasis through up-regulation of keratin 6, keratin 16, and keratin 17. J Invest Dermatol. 2017;137:2168–2176. doi: 10.1016/j.jid.2017.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The correlation co-efficients between clinicopathologic parameters in patients with psoriasis
Immunofluorescence analysis of AHR and LC3 expression in psoriatic skin lesion. (A) The level of AHR (red), LC3 (green) and nuclei (blue) expression was determined by immunofluorescence. In merged images, areas within the red circle are shown as a magnified image on the left lower square. AHR expression was increased and LC3 expression was decreased in psoriasis skin compared to controls. Data are representative of psoriasis (n=5) and healthy volunteers (n=5). Original magnification ×200, Bar=75 µm, Bar=50 µm (×400) on the left lower square of the merged image. Quantification of AHR (B) and LC3 (C) expression on psoriatic skin lesion. The level of staining was semi-quantitatively analyzed using LASX software (Leica, Wetzlar, Germany). HC: healthy control, AHR: aryl hydrocarbon receptor, LC3: microtubule-associated protein light chain 3. Statistical significance was determined by Student's t-test (***p<0.001) compared to normal controls.
LC3-II protein expression in psoriasis of human skin tissues. (A) Western blot of LC3-I and LC3-II in human skin tissues of HC, and psoriasis lesions. Data normalization was based on GAPDH. The results are representative of 3 independent experiments. (B) The densitometric analysis (LC3-II/GAPDH). Data represent the mean±standard deviation of 3 independent experiments. LC3: microtubule-associated protein light chain 3, HC: healthy control. Statistical significance was determined by student's t-test (***p<0.001).
AHR, CYP1A1, LC3, ATG5, BECN1, NRF2 mRNA expression in psoriatic lesional skin. Quantitative polymerase chain reaction analysis of (A) AHR, (B) CYP1A1, (C) LC3, (D) BECN1, (E) ATG5, and (F) NRF2 expression in human psoriasis lesion (n=5) and HC tissues (n=5). Data represent the mean±standard deviation of three independent experiments (each performed in duplicate). LC3: microtubule-associated protein light chain 3, HC: healthy control. Statistical significance was determined by Student's t-test (*p<0.05, **p<0.01, ***p<0.001) in comparison to controls.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.