Abstract
Background
Despite an impressive effort in clinical research, no standard therapeutic approach for coronavirus disease 2019 (COVID-19) patients has been established, highlighting the need to identify early biomarkers for predicting disease progression and new therapeutic interventions for patient management. The present study aimed to evaluate the involvement of the human endogenous retrovirus -W envelope (HERV-W ENV) in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection considering recent findings that HERVs are activated in response to infectious agents and lead to various immunopathological effects. We analysed HERV-W ENV expression in blood cells of COVID-19 patients in correlation with clinical characteristics and have discussed its potential role in the outcome of the disease.
Methods
We analysed HERV-W ENV expression in blood samples of COVID-19 patients and healthy donors by flow cytometry and quantitative reverse transcriptase PCR analysis, and evaluated its correlation with clinical signs, inflammatory markers, cytokine expression, and disease progression.
Findings
HERV-W ENV was highly expressed in the leukocytes of COVID-19 patients but not in those of healthy donors. Its expression correlated with the markers of T-cell differentiation and exhaustion and blood cytokine levels. The percentage of HERV-W ENV-positive lymphocytes correlated with inflammatory markers and pneumonia severity in COVID-19 patients. Notably, HERV-W ENV expression reflects the respiratory outcome of patients during hospitalization.
Interpretation
Given the known immuno- and neuro-pathogenicity of HERV-W ENV protein, it could promote certain pathogenic features of COVID-19 and therefore serve as a biomarker to predict clinical progression of disease and open to further studies for therapeutic intervention.
Funding
Information available at the end of the manuscript.
Keywords: Cytokine storm, COVID-19, HERV-W. human endogenous retroviruses, Inflammation, Respiratory outcome, T-cell exhaustion
Abbreviation: AS, Asymptomatic; BiP, Bilateral interstitial pneumonia; BiP/Bact, BiP with bacterial co-infection; CM, Central memory; COV, SARS-CoV-2-positive individuals; COVID-19, Human coronavirus disease 2019; C-PAP, Continuous Positive Airway Pressure; CT, Computed tomography; Ct, Threshold cycle; CXCL6, C-X-C Motif chemokine ligand 6; CXCR1, C-X-C chemokine receptor type 1; DTT, Dithiothreitol; EM, Effector memory; GUSβ, Beta-glucuronidase; HD, Healthy donors; HERVs, Human Endogenous Retroviruses; HERV-W ENV, Human Endogenous Retrovirus -W envelope; IL-10, Interleukin-10; IL-17, Interleukin-17; IL-17RA, Interleukin-17 receptor A; IL-6, Interleukin-16; INFγ, Interferon γ; LDH, Lactate dehydrogenase; MCP1, monocyte chemoattractant protein-1; MiP, Monolateral or minimal interstitial pneumonia; Mod, Moderate; NC/VMKs, Nasal cannula or Venturi masks; NIV, Non-invasive ventilation; NK, Natural killer; OTI, Orotracheal intubation; P No, interstitial pneumonia; PBS, Phosphate-buffered saline; PCR, Polymerase chain reaction; PS, Paucisymptomatic; PT%, Prothrombin activity percentage; PT, Prothrombin time; PT-INR, Prothrombin time international normalized ratio; PTV, Policlinico Tor Vergata; qRT-PCR, Quantitative reverse-transcriptase PCR; Rho, Spearman correlation coefficient; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; Sev, Severe; TEM, Terminal effector memory; TNFα, Tumor necrosis factor α
Research in context.
Evidence before this study
Although the exact pathophysiology of Coronavirus Disease 2019 (COVID-19) has not been fully elucidated, it has been established that the various clinical manifestations of the disease are caused by the dysregulation of innate immunity associated with hyper-inflammation induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Addressing the severe form of the disease, currently, few treatment options are available for controlling the host's immune response against SARS-CoV-2 and the associated hyper-inflammation. Furthermore, these methods revealed to be mostly ineffective in reducing the mortality rate.
Human Endogenous Retroviruses (HERVs) have been described as genetic elements present in the human genome. Their activation has increasingly been found to be associated with various diseases including cancer, multiple sclerosis, type 1 diabetes, and rheumatoid arthritis, which are all underlying diseases in patients developing severe forms of COVID-19. Several studies confirmed that the immunopathogenic envelope protein of HERV family W (HERV-W ENV) is involved in inflammatory and neurological diseases. Moreover, HERV-W was shown to be directly activated by exogenous viruses with various outcomes depending on triggering viruses and susceptible cells or tissues.
Added value of this study
We demonstrate for the first time an elevated expression of the HERV-W ENV protein in the blood cells of COVID-19 patients compared to that of healthy donors and its correlation with markers of inflammation including cytokines expression, and T cell differentiation and exhaustion, thus proposing its role in predicting the outcome of the disease. Based on the potential early reactivation by SARS-CoV-2, we propose HERV-W ENV protein as a contributing factor to the immunopathology of COVID-19.
Implications of all the available evidence
The identification of the association between HERV-W ENV expression and inflammatory and immune dysfunction in COVID-19 opens an avenue for further investigation of its role as a trigger of detrimental immune response.
Alt-text: Unlabelled box
1. Introduction
The human coronavirus disease 2019 (COVID-19) caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently associated with high morbidity and mortality, and hospitals worldwide are facing the increasing demand of patients requiring oxygen supply, ventilators, and intensive care. Despite dominant respiratory tract tropism, both pulmonary and extra-pulmonary SARS-CoV-2-related forms have been recognized [1], which predominantly involve the impairment of immune functions [2]. Innate immune hyper-activation combined with adaptive immune dysregulation has been recognized to play a critical role in the progression of the disease and thus in the clinical outcome of COVID-19 patients [3,4], suggesting the critical evolution driven by exacerbated innate immunity and associated inflammation and lymphopenia [5,6]. The immunological involvement includes hyper-immune reactions such as the “cytokine storm” syndrome (mostly occurring in pulmonary forms), the pediatric multisystem inflammatory syndrome, and immune-mediated cutaneous or neurological diseases along with various autoimmune manifestations such as dysregulation of coagulation mechanisms [7], [8], [9], [10].
With the aim to identify new factors contributing to dysregulated inflammation in COVID-19, as well as targets for therapeutic intervention and patient management, we studied the human endogenous retrovirus family W (HERV-W) that encodes an immunopathogenic and neurotoxic envelope protein HERV-W ENV, which has been associated with human inflammatory and/or autoimmune diseases [11,12].
HERVs are genetic elements resulting from ancestral infection of germ line cells mediated by exogenous retroviruses and constitute up to 8% of the human genome. In the general population, their co-opting in physiological functions[13] and inter-individual variations such as copy number variations, unfixed copies and polymorphisms have been demonstrated [14]. Activation of HERVs has been demonstrated in cancer, multiple sclerosis, type 1 diabetes and rheumatoid arthritis [11,12,15], which represent the underlying conditions frequently detected in patients developing severe forms of COVID-19. Recent studies have confirmed the involvement of HERVs in inflammatory and neurological diseases [14,16]. Specifically, the immunopathogenic protein HERV-W ENV can be activated by exogenous viruses [17,18] and induces proinflammatory responses through the toll-like receptor 4 pathway (TLR4) [19,20]. Furthermore, a humanized monoclonal antibody targeting the HERV-W ENV protein has been proposed as an innovative therapeutic approach for multiple sclerosis [21]. The present study aimed to evaluate the expression of HERV-W ENV in the peripheral blood cells of COVID-19 patients, its correlation with clinical characteristics, and its potential relationship with predicting the outcome of the disease.
2. Methods
2.1. Patients and healthy donors
Thirty SARS-CoV-2-positive individuals (COV) were enrolled in an open study by the Infectious Diseases Clinic, Department of Systems Medicine, University of Rome “Tor Vergata” and Policlinico Tor Vergata (PTV) Foundation. Ethical approval for the collection and use of human samples was obtained from the ethical board of the Hospital, COronaVIrus Disease: Safety and Efficacy of Experimental Treatment (COVID_SEET prot.7562/2020, 9th April 2020, experimental register 46.20). Blood cells from healthy donors (n = 17, HD) were obtained from individuals attending the local blood transfusion center and referred to the Virology Unit for screening. The donors have been matched for age and sex to the best possible extent with patients and provided written informed consent. SARS-CoV-2 infection was diagnosed by the Virology Unit of PTV using Allplex™ 2019-nCoV multiplex Real-time polymerase chain reaction (PCR) assay, according to the manufacturer's instructions. All clinical data of COVID-19 patients are reported in Table 1.
Table 1.
Demographics and clinical classification of COVID-19 patients at enrolment and respiratory outcome
| Demographics and Clinical severity at enrolment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Asymptomatic (AS) | Paucisymptomatic (PS) | Mild (Mild) | Moderate (Mod) | Severe (Sev) | Tot | ||||
| Number | 5 | 10 | 9 | 3 | 3 | 30 | |||
| Age (Mean ± SD) | 61 ± 12.1 | 57 ± 15 | 60 ± 6.6 | 56 ± 14 | 48 ± 1.2 | 58 ± 12 | |||
| Sex (M/F) | 4/1 | 6/4 | 7/0 | 2/1 | 3/0 | 24/6 | |||
| Hospitalization (days+) | 2 ± 2 | 1 ± 1.1 | 1 ± 0.9 | 1 ± 1.6 | 1.6 ± 0.1 | 2 ± 1.45 | |||
| +days between hospitalization and sampling | |||||||||
| Radiological and clinical characteristics, treatments and outcome | |||||||||
| AS | PS | Mild | Mod | Sev | Tot | ||||
| Pneumonia* | None | 1 | 2 | 0 | 0 | 0 | 3 (10%) | ||
| P | 1 | 1 | 1 | 0 | 0 | 3 (10%) | |||
| MiP | 1 | 5 | 0 | 0 | 0 | 6 (20%) | |||
| BiP | 2 | 1 | 8 | 3 | 3 | 17 (56.6%) | |||
| Comorbidities | Obesity | 0 | 2 | 3 | 1 | 2 | 8 (26.6%) | ||
| Diabetes mellitus | 1 | 2 | 5 | 1 | 0 | 9 (30%) | |||
| Cardiovascular | 6 | 4 | 5 | 3 | 1 | 19 (63.3%) | |||
| Others** | 2 | 7 | 4 | 0 | 2 | 15 (66.6%) | |||
| Mortality | 0 | 0 | 1 | 1 | 1 | 3/30 (10%) | |||
| Treatment | Antiviral | 0 | 1 | 3 | 3 | 2 | 9 | ||
| Corticosteroids | - | 1 | 4 | 3 | 3 | 11 | |||
| * none =no pneumonia, P= no interstitial pneumonia, MiP= Monolateral or minimal interstitial pneumonia, BiP= bilateral interstitial pneumonia ** other comorbidities or habits: solid organ replacement, gastrointestinal, smoke | |||||||||
| Blood Count | |||||||||
| AS | PS | Mild | Mod | Sev | Tot | ||||
| Red Blood cells (4.40-6.00) 106 /μl | 4 ± 0.13 | 5 ± 0.042 | 5 ± 0.72 | 5 ± 1.02 | 5 ± 0.43 | 4.32 ± 0.5 | |||
| Haemoglobin (13.00–18.00) g/dl | 11 ± 1.47 | 13 ± 1.69 | 13 ± 1.78 | 13 ± 2.43 | 15 ± 2.2 | 12.40 ± 1.7 | |||
| White Blood Cells (4.30–10.80) 105/μl | 3 ± 4.13 | 5 ± 0.19 | 6 ± 2.99 | 5 ± 1.93 | 6 ± 3.28 | 7.28 ± 3.2 | |||
| Neutrophils | Abs count | 2.0 ± 1.16 | 3.0 ± 0.03 | 5.0 ± 2.68 | 5.0 ± 2.31 | 9.0 ± 2.55 | 5.27 ± 3.0 | ||
| % (40–75) | 64 ± 16 | 55.5 ± 1.31 | 71.2 ± 14.7 | 61.49 ± 16.59 | 68 ± 1.83 | 67.8 ± 1.0 | |||
| Lymphocytes | Abs count | 1.0 ± 0.55 | 2.0 ± 0.14 | 2.0 ± 0.9 | 2.0 ± 0.95 | 1.0 ± 0.57 | 1.45 ± 0.70 | ||
| % (20–45) | 22.2 ± 12.70 | 32.91 ± 1.6 | 22.5 ± 11.9 | 30.62 ± 13.29 | 24.46 ± 2 | 23.04 ± 1.0 | |||
| Monocytes | Abs count | 0.14 ± 0.10 | 0.46 ± 0.07 | 0.38 ± 0.17 | 0.33 ± 0.2 | 0.40 ± 0.40 | 0.45 ± 0.17 | ||
| % (3.4–11) | 4 ± 2.93 | 10 ± 1.83 | 5.19 ± 1.8 | 5.72 ± 2.97 | 6.16 ± 2.6 | 6.89 ± 2.60 | |||
| Eosinophils | Abs count | 0.15 ± 0.2 | 0.17 ± 0.05 | 0.09 ± 0.07 | 0.05 ± 0.02 | 0.07 ± 0.3 | 0.10 ± 0.15 | ||
| % (0–7) | 3.3 ± 5.49 | 4.04 ± 1.06 | 1.96 ± 1.67 | 0.87 ± 0.35 | 1.0 ± 3.60 | 1.90 ± 3.62 | |||
| Basophils | Abs count | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.001 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.00 | ||
| % (0–1.5) | 0.3 ± 0.2 | 0.34 ± 0.28 | 0.27 ± 0.07 | 0.16 ± 0.10 | 0.13 ± 0.07 | 0.20 ± 0.17 | |||
| Respiratory Outcome*** | |||||||||
| AS | PS | Mild | Mod | Sev | Tot | ||||
| None | 4 | 10 | 2 | 0 | 0 | 16 (53.3%) | |||
| NC/VMK | 0 | 0 | 6 | 1 | 1 | 8 (26.6%) | |||
| NIV/C-PAP/OTI | 1 | 0 | 1 | 2 | 2 | 6 (20.0%) | |||
| *** None = No oxygen support, NC/VMK = Nasal Cannula/Venturi Mask, NIV/C-PAP = Non-invasive ventilation or OTI = invasive ventilation | |||||||||
| Biochemicals | |||||||||
| AS | PS | Mild | Mod | Sev | Tot | ||||
| PT % (70-130) | 93 ± 8.77 | 91 ± 6.5 | 89 ± 8.27 | 83.10 ± 4.54 | 83.06 ± 0.1 | 89.09 ± 7.83 | |||
| PT-INR (0.80-1.20) | 1.09 ± 0.06 | 1.99 ± 0.045 | 1.1 ± 0.067 | 1.15 ± 0.042 | 1.91 ± 0.01 | 1.10 ± 0.06 | |||
| PT sec | 12 ± 0.8 | 13 ± 0.55 | 13.69 ± 0.78 | 13.92 ± 0.49 | 13.80 ± 0.1 | 13.16 ± 0.72 | |||
| Fibrinogen (200-400) mg/dl | 341 ± 144 | 460 ± 171 | 495 ± 53 | 520 ± 122 | 527 ± 1.3 | 474 ± 124 | |||
| Antitrombin III % (75-128) | 91 ± 15 | 100 ± 3 | 104 ± 17 | 106 ± 14 | 99 ± 1.1 | 101 ± 14 | |||
| D-Dmers (0-500) ng/ml | 1133 ± 618 | 670 ± 966 | 574 ± 106 | 648 ± 182 | 558 ± 532 | 662 ± 511 | |||
| Glucose (70-100) mg/dl | 93 ± 27 | 94 ± 73 | 152 ± 77 | 94 ± 30 | 100 ± 35 | 104 ± 51 | |||
| BUN (15-40) mg/dl | 143 ± 66 | 30 ± 8.73 | 38 ± 8.8 | 32 ± 11 | 45 ± 28 | 53 ± 49 | |||
| AST (5-34) U/liter | 24 ± 61 | 21 ± 10 | 59 ± 10 | 50 ± 127 | 195 ± 1 | 54 ± 67 | |||
| ALT (0-55) U/liter | 28 ± 31 | 28 ± 7.5 | 44 ± 46 | 71 ± 183 | 34 ± 68 | 54 ± 83 | |||
| LDH (125-220) U/liter | 311 ± 136 | 212 ± 91 | 269 ± 100 | 321 ± 97 | 459 ± 90 | 305 ± 121 | |||
| Reactive C Protein (CRP) (0-5) mg/liter | 35 ± 40 | 25 ± 11 | 38 ± 44 | 54 ± 55 | 40 ± 32 | 39 ± 40 | |||
2.2. Sample size
This was an exploratory study based on the number of patients available fitting the inclusion/exclusion criteria. No outliers were excluded.
2.3. RNA extraction from blood samples
Blood samples (250 μL) were centrifuged at 800 × g for 8 min, and the obtained pellets were treated with 150 μL of red blood cell lysing buffer (Sigma Aldrich, St. Louis, Missouri, US) for 5 min at room temperature (20 °C), twice to remove the red blood cells. After washing with Dulbecco's phosphate-buffered saline (PBS, PAN-Biotech, Aidenbach, Bavaria), the pellets were resuspended in 400 μL of R1 buffer and 1 mM DTT, incubated at room temperature for 5 min, mixed with 70% ethanol, and transferred to RNA mini spin column, according to the manufacturer's instructions (Total RNA Kit – Blood & Cultured cells – GriSP, Porto Portugal). Treatment with DNase I ‘in column’ at room temperature for 15 min ensured removal of contaminating DNA. RNA samples were evaluated by Nanodrop DS 11 (DeNovix), showing a ratio of 260/280 of about 2.0 and a concentration ranging from 10 ng/μL to 100 ng/μL.
2.4. Quantitative reverse-transcriptase PCR (qRT-PCR)
DNase-treated RNA (100 ng) was reverse-transcribed into cDNA using ImProm-II Reverse Transcription System (Promega, Fitchburg, Wisconsin, USA) and oligo dT according to the manufacturer's protocol; controls without template and another without enzyme were included in each RT reaction. Initial RNA (2.5 ng) in RT reaction was used for quantitative evaluation of the transcriptional levels of HERV-W ENV genes and the gene expression of IL-6, IL-10, IL-17 as well as its receptor IL-17RA, TNF-α, INF-γ, MCP-1, CXCR1, and CXCL6 by qRT-PCR (primer pairs used are listed in Table S1). The assays were performed in a Bio-rad instrument (CFX96 Real-Time System, Biorad, Hercules, California, USA) using SYBR Green chemistry (Fast QPCR Master Mix, SmoBio, Taiwan). Each sample was analysed in triplicate, and a negative control (no template reaction) was included to check for any possible contamination. The housekeeping gene beta-glucuronidase was used to normalize the results. Each experiment was completed with a melting curve analysis to confirm the specificity of amplification and the lack of any non-specific product and primer dimers. Quantification was performed using the threshold cycle (Ct) comparative method, and the relative expression was calculated as follows: 2 −[∆Ct (sample)−∆Ct (calibrator) = 2−∆∆Ct] where ∆Ct (sample) = [Ct (target gene) − Ct (housekeeping gene)] and ∆Ct (calibrator) was the mean of ∆Ct of all HD samples.
2.5. Flow cytometry for HERV ENV expression and immunophenotyping analysis
Blood cells (100 μL) were incubated for 15 min in the dark with VersaLyse Lysing Solution (Beckman Coulter, BC) to lyse red blood cells and then permeabilized with IntraPrep Permeabilization Reagent kit (BC), followed by staining with anti HERV-W ENV monoclonal antibody (ENV-W01; GeNeuro) and a secondary FITC-labeled goat anti-mouse antibody (Beckton Dickinson, BD). For the analysis of leukocyte subpopulations, before intracellular staining, the cells were incubated with monoclonal antibodies of interest: CD4-APC and CD56-PE (BD), CD8-APC700, CD3-APC750, CD14-PE, CD19 ECD or PC7 (BC). For certain analyses, the DuraClone IM T-cell subsets Tube (B53328, BC) was used. Stained cells were then washed with PBS analysed via CytoFLEX and CytExpert 2.2 software (BC), at least 4.5 × 105 cells in the gate of Leukocytes were recorded. The gating strategy is reported in Figure S1. Results are expressed as the percentage of HERV-W ENV-positive cells in COV with respect to HD used as a control group.
2.6. in vitro stimulation with IL-6 and HERV-W ENV proteins in PBMCs from Healthy donors
Peripheral blood mononuclear cells (PBMCs) from heparinized blood samples from 4 Healthy Donors were isolated by density gradient centrifugation (Pancoll human, PAN-Biotech, Aidenbach, Bavaria) and cultured at density of 0.25×106/mL in RPMI 1640 (PAN-Biotech, Aidenbach, Bavaria) enriched with 2 mM of l-glutamine, 100 U/mL of Penicillin, 0.1 mg/ml of Streptomycin, 12% fetal bovine serum in presence of human recombinant interleukin-2 (IL-2), 20 U/ml (all from Sigma, MO, USA). PBMCs were exposed to SARS-CoV-2 Spike protein active trimer (1 and 5 nM BIOSYSTEM Acro, Bay Area, CA) or IL-6 10 ng/ml (rh-IL6, Immuno Tolls, Gladiolenweg, Germany) or HERV-W ENV protein 10 and 100 ng/ml (GeNeuro, Geneva, Switzerland) for 3 h, 24 h and 5 days at 37 °C in 5% CO2. At the end of incubation, PBMCs were harvested and analyzed by flow cytometry and Real time PCR for HERV-W ENV and IL-6 expression, depending on culture condition. All cultures were performed in duplicate.
2.7. Quantification of plasma IL-6 concentration
IL-6 was measured in plasma using an automated ELISA assay (ELLA microfluidic analyzer, Protein Simple, Bio-Techne, USA) using the “COVID-19″ kit. “COVID-19 SIMPLE PLEX COVID-19 CYTOKINE STORM PANEL” cartridges are supplied as “ready to use” format with preloaded antibodies. Wells were filled with 25µL of plasma or wash buffer following manufacturer's instructions.
2.8. Ethics
Ethical approval “COronaVIrus Disease: Safety and Efficacy of Experimental Treatment” was obtained from the independent ethical board of the Tor Vergata Hospital in Rome, for the collection and use of human samples (COVID_SEET prot.7562/2020, 9th April 2020, experimental register 46.20).
Blood cells from healthy donors were obtained from individuals attending the local blood transfusion center that referred to the Virology Unit for screening at Tor Vergata Hospital in Rome. All study subjects provided written informed consent.
2.9. Statistical analysis
Statistical analysis of group-wise expression levels was performed through nonparametric Mann Whitney test in case of 2-independent samples or Kruskall Wallis test and Bonferroni's correction in case of n-independent samples. Pairwise associations between continuous variables were tested through the Spearman correlation coefficient. Statistically significant comparisons were considered at p < 0.05. The Bonferroni's post-hoc multiple comparison ANOVA test was utilized for in vitro experiments analysis. Data analyses were performed using SPSS statistical software system (version 24.0 for Windows, USA).
2.10. Role of the funding source
No specific funding was received for this work. Geneuro innovation provided materials and reagents. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. Demographics and clinical classification of COVID-19 patients
We analysed the clinical status at enrolment and the respiratory outcome of 30 COVID-19 patients who were hospitalized at the Policlinico Tor Vergata hospital in Rome during September–October 2020 (Table 1). The study cohort was divided into five groups based on patients’ clinical characteristics and physical examination at hospital admission or study enrolment: 5 (16.66%) patients who did not show any symptoms related to COVID-19 (referred to as asymptomatic or AS), although they were already admitted at the hospital for pre-existing morbid conditions; 10 (33.33%) who were symptomatic with few clinical manifestations (pauci symptomatic or PS) including at least one COVID-19-related symptom such as cough or fever not showing signs of pneumonia on physical examination. In PS group on a chest CT scan performed during the course of the hospitalization, minimal pneumonia was evidenced in 7 out of 9 individuals. None of them needed oxygen support during the whole hospitalization; 9 (30%) who were considered mild (Mild) with typical symptoms of COVID-19 and clinical evidence of pneumonia on physical examination, without showing shortness of breath or dyspnea on enrolment; 3 (10%) who were considered moderate (Mod) with typical symptoms of COVID-19 and clinical evidence of pneumonia on physical examination, shortness of breath or dyspnea on enrolment and a saturation of oxygen ≥94% on room air; and 3 (10%) who were considered severe (Sev) with typical symptoms of COVID-19, clinical evidence of pneumonia on physical examination, shortness of breath or dyspnea on enrolment and a saturation of oxygen <94% on room air, or a ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2) <300 mm Hg, or respiratory frequency >30 breaths/min. Overall, among the 30 COVID-19 patients enrolled, 23 (76.7%) showed radiological signs of SARS-CoV-2-related pneumonia defined as monolateral interstitial pneumonia (MiP) in 6 cases (20%) and bilateral (BiP) in 17 cases (56.6%). Considering comorbidities, 26 (86%) patients showed the presence of at least one pre-existing chronic disease, with cardiovascular conditions being the most common comorbidity present in 19 (63.3%) patients. Upon sampling, the patients had been hospitalized for about 2 days, with nine (30%) of them being treated with antiviral and corticosteroid therapies and two (7%) with only corticosteroid therapy; unfortunately, three of these patients died (fatality rate 10%). Patients did not show a reduction in the absolute lymphocytes count at the time of sampling, although the relative counts tended to be within the lower limit of the normal count. Considering coagulopathy markers and other biochemical parameters, the levels of C-reactive protein, fibrinogen, D-Dimers, blood urea nitrogen, aspartate transaminase, alanine transaminase, and lactate dehydrogenase (LDH) were increased on average, considering the overall cohort, although no differences were found among the five COVID-19 groups. The 30 COVID-19 patients were evaluated for respiratory outcomes during the hospitalization period and divided into three groups: 16 (53.3%) patients who did not need oxygen therapy; 8 (26,6%) who were supported by a nasal cannula or Venturi masks (VMKs); and 6 (20%) who were supported by non-invasive ventilation (NIV), continuous Positive Airway Pressure (C-PAP), and oro-tracheal intubation (OTI) or mechanical invasive ventilation.
The healthy donors were best matched for age and sex with COVID-19 patients: the cohort included 30 COVID-19 patients (median age 61+/−12, 24 males and 6 females) and 17 healthy donors (median age 55+/−13, 11 males and 6 females); all clinical parameters analysed in the HD were within normal reference values.
3.2. HERV-W envelope mRNA and protein were highly expressed in leukocytes of COVID-19 patients and correlated with markers of differentiation and exhaustion of T cells
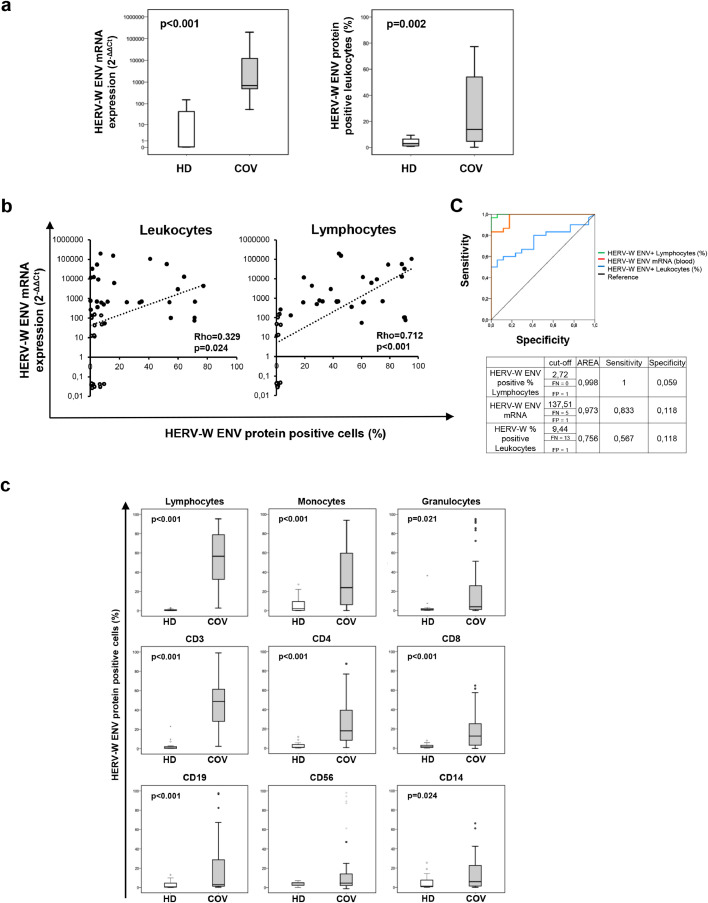
The transcriptional level of the HERV-W ENV gene was evaluated by qRT PCR, whereas the expression of HERV-W ENV protein was analysed by flow cytometry in permeabilised leukocytes after staining with a mouse monoclonal antibody. Highly significant differences were noted in the levels of HERV-W ENV mRNA and protein between COV and HD groups (p<0.001 for mRNA and p = 0.002 for protein; Fig. 1a and Table 2). Of note, the Spearman analysis performed on COV and HD samples demonstrated a significant positive correlation between the HERV-W ENV mRNA and the percentage of HERV-W ENV-positive cells in both total leukocytes and lymphocytes (Fig. 1b). Interestingly, this correlation was much stronger for HERV-W protein detection in lymphocytes (Rho=0.712, p<0.001) than in all leukocytes (Rho=0.329, p = 0.024), suggesting that lymphocytes are the major source of this mRNA. ROC curve analysis confirmed the percentage of lymphocytes HERV-W ENV positive cells as a more accurate marker than HERV-W ENV mRNA and leukocytes HERV-W ENV positive cells in COV group respect to HD (Fig. 1c).
Fig. 1.
Pathogenic HERV-W ENV mRNA expression and the percentage of HERV-W ENV protein positive cells are significantly expressed in blood cells of COVID-19 patients versus healthy donors.
HERV-W expression in blood samples and the percentage of HERV-W ENV-positive cells were analysed in thirty hospitalized COVID-19 patients (COV, gray box plots) and seventeen healthy donors (HD, white box plots). (a) HERV-W ENV expression (mRNA) and HERV-W ENV protein were analysed in leukocytes by qRT-PCR and flow cytometry, respectively. (b) Scatter Plots of the HERV-W ENV mRNA levels (Y-axis) and the percentage HERV-W ENV-positive leukocytes (left) and lymphocytes (right) (X-axis). Pairwise associations between continuous variables were tested through the Spearman correlation coefficient (Rho). Statistically significant values were considered when p <0.050. (c) ROC Curve of the% HERV-W ENV in Lymphocytes (green line), leukocytes (blue line) and HERV-W ENV mRNA levels (orange line). (d) Different subpopulations of leukocytes were analysed according to the morphology, and cell phenotype using specific phenotypic markers (see the gating strategy in supplementary data Fig.S1). The percentage of HERV-W ENV-positive cells has been evaluated in lymphocytes, monocytes, granulocytes, CD3+ (T Lymphocytes), CD4+ (T Helper cells), CD8+ (T Cytotoxic cells), CD19+ (B Lymphocytes), CD56+ (Natural Killer cells) and CD14+ (monocytes) of COVID-19 patients and healthy donors. Non-parametric Mann–Whitney test was used to compare the two groups. Statistically significant values were considered when p <0.050.
Table 2.
Median values, interquartile range and Mann Whitney U test of HERV-W env transcriptional activity and cytokines and receptors expression (mRNA) in blood samples from COVID19 and HD.
| HERV-W ENV | IL-6 | IL-10 | IL-17 | IL-17RA | TNFα | INFγ | MCP1 | CXCR1 | CXCL6 | ||
| COVID-19 PATIENTS number | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | |
| Median | 691.787 | 125.985 | 11.998 | 31.951 | 12.618 | 3.506 | 3.333 | 7.003 | 3.615 | 2.492 | |
| Interquartile Range (IQR) | 25 | 463.330 | 65.769 | 1.309 | 0.664 | 2.649 | 1.574 | 0.491 | 2.418 | 2.350 | 0.175 |
| 50 | 691.787 | 125.986 | 11.998 | 31.951 | 12.618 | 3.506 | 3.333 | 7.003 | 3.615 | 2.492 | |
| 75 | 12,460.919 | 375.029 | 126.432 | 93.247 | 123.115 | 10.866 | 14.101 | 31.884 | 16.205 | 20.118 | |
| HEALTHY DONORS number | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | |
| Median | 0.045 | 0.0403 | 0.269 | 0.594 | 0.225 | 0.490 | 0.354 | 1.717 | 0.274 | 2.474 | |
| Interquartile Range (IQR) | 25 | 0.038 | 0.0284 | 0.111 | 0.044 | 0.164 | 0.159 | 0.176 | 0.100 | 0.188 | 0.020 |
| 50 | 0.045 | 0.0403 | 0.269 | 0.594 | 0.225 | 0.490 | 0.354 | 1.717 | 0.274 | 2.747 | |
| 75 | 45.339 | 92.546 | 11.544 | 25.985 | 35.813 | 4.614 | 12.233 | 13.349 | 7.589 | 26.096 | |
| Healthy donors vs COVID19 patients | <0.001* | <0.001 | 0.004 | 0.004 | 0.002 | 0.132 | 0.298 | 0.016 | 0.024 | 0.507 | |
*Mann Witney U test.
HERV-W ENV protein expression was further analysed by flow cytometry in leukocyte subpopulations (Fig. 1d). The lymphocytes, monocytes, and granulocytes showed a significantly higher percentage of HERV-W ENV protein-positive cells in the COV group than in HD (Table 3) (p<0.001, p<0.001, and p = 0.021, respectively). Lymphocytes displayed the highest values among all leukocytes, and in particular, T-lymphocytes (CD3+) showed the highest percentage of HERV-W ENV-positive cells, clearly differentiating between COV and HD groups. CD4+ and CD8+ T-lymphocytes showed similar median percentages of HERV-W ENV-positive cells in the COV group but showed significantly higher percentages of positivity, compared to HD. HERV-W ENV protein was expressed at a lower percentage in B cells (CD19+) and monocytes (CD14+), although it remained significantly higher in the COV group than in HD (p<0.001 and p = 0.024, respectively).
Table 4.
Median values, interquartile range and Kruskal-Wallis test of HERV-W ENV, cytokines and receptors expression (mRNA) in blood samples of COVID19 and HD according to clinical status.
| HERV-W ENV | IL-6 | IL-10 | IL-17 | IL-17RA | TNFα | INFγ | MCP1 | CXCR1 | CXCL6 | ||
| Healthy donors number | 17 | ||||||||||
| Median | 0.045 | 0.040 | 0.269 | 0.594 | 0.225 | 0.490 | 0.354 | 1.717 | 0.274 | 2.474 | |
| Interquartile Range (IQR) | 25 | 0.039 | 0.028 | 0.111 | 0.044 | 0.164 | 0.159 | 0.177 | 0.100 | 0.188 | 0.021 |
| 50 | 0.045 | 0.040 | 0.269 | 0.594 | 0.225 | 0.490 | 0.354 | 1.717 | 0.274 | 2.474 | |
| 75 | 45.339 | 92.546 | 11.544 | 25.985 | 35.813 | 4.614 | 12.233 | 13.349 | 7.589 | 26.096 | |
| Asymptomatic and | |||||||||||
| Paucisympromatic patients number | 15 | ||||||||||
| Median | 684.997 | 103.630 | 2.000 | 1292 | 3.598 | 3.104 | 2.287 | 6.382 | 3.158 | 0.323 | |
| Interquartile Range (IQR) | 25 | 498.451 | 63.360 | 1.255 | 0.260 | 2.342 | 1.396 | 0.744 | 1.198 | 2.221 | 0.115 |
| 50 | 684.997 | 103.630 | 2.000 | 1.292 | 3.598 | 3.104 | 2.287 | 6.382 | 3.158 | 0.323 | |
| 75 | 9421.437 | 325.770 | 45.422 | 39.068 | 36.705 | 3.842 | 10.522 | 17.867 | 11.029 | 6.956 | |
| Mild, moderate and severe | |||||||||||
| patients number | 15 | ||||||||||
| Median | 4376.194 | 240.628 | 34.432 | 82.683 | 92.885 | 7.019 | 11.163 | 16.268 | 12.542 | 11.773 | |
| Interquartile Range (IQR) | 25 | 357.968 | 100.991 | 5.916 | 26.721 | 7.401 | 2.001 | 0.147 | 2.827 | 3.132 | 0.268 |
| 50 | 4376.194 | 240.628 | 34.432 | 82.683 | 92.885 | 7.019 | 11.163 | 16.268 | 12.542 | 11.773 | |
| 75 | 56,563.819 | 469.597 | 198.821 | 124.940 | 171.080 | 44.286 | 38.627 | 50.997 | 45.354 | 43.816 | |
Table 3.
Median values, interquartile range and Mann Whitney U test of the percentage of HERV-W ENV protein positive cells in leukocyte and leukocyte subpopulations in blood samples from COVID19 and HD.
| Leuko cytes | Lympho cytes | Mono cytes | Granulo cytes | CD3 | CD4 | CD8 | CD19 | CD56 | CD14 | ||
| COVID-19 PATIENTS number | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | |
| Median | 13.800 | 56.535 | 23.935 | 3.925 | 48.920 | 17.990 | 12.510 | 3.045 | 4.600 | 5.985 | |
| Interquartile Range (IQR) | 25 | 4.687 | 32.017 | 6.040 | 0.940 | 27.672 | 8.1275 | 3.165 | 1.242 | 2.097 | 1.262 |
| 50 | 13.800 | 56.535 | 23.935 | 3.925 | 48.920 | 17.990 | 12.510 | 3.045 | 4.600 | 5.985 | |
| 75 | 54.325 | 81.235 | 59.757 | 32.080 | 61.527 | 43.307 | 26.597 | 28.985 | 16.797 | 24.325 | |
| HEALTHY DONORS number | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | 17 | |
| Median | 2.700 | 0.630 | 2.390 | 0.920 | 1.020 | 1.090 | 1.540 | 0.760 | 3.770 | 1.290 | |
| Interquartile Range (IQR) | 25 | 1.400 | 0.265 | 0.410 | 0.345 | 0.285 | 0.740 | 0.600 | 0.350 | 2.215 | 0.470 |
| 50 | 2.700 | 0.630 | 2.390 | 0.920 | 1.020 | 1.090 | 1.540 | 0.760 | 3.770 | 1.290 | |
| 75 | 7.745 | 1.200 | 10.975 | 2.038 | 2.580 | 4.145 | 3.725 | 6.370 | 5.370 | 8.150 | |
| Healthy donors vs COVID19 patients | 0.004 | <0.001* | <0.001* | 0.021 | <0.001 | <0.001 | <0.001* | 0.002 | 0.111 | 0.024 | |
* Mann Witney U test.
No significant difference was found in natural killer cells (NK; CD3-CD56+) between COV and HD groups. Nonetheless, elevated HERV-W ENV positivity in NK cells was found in a few COV samples.
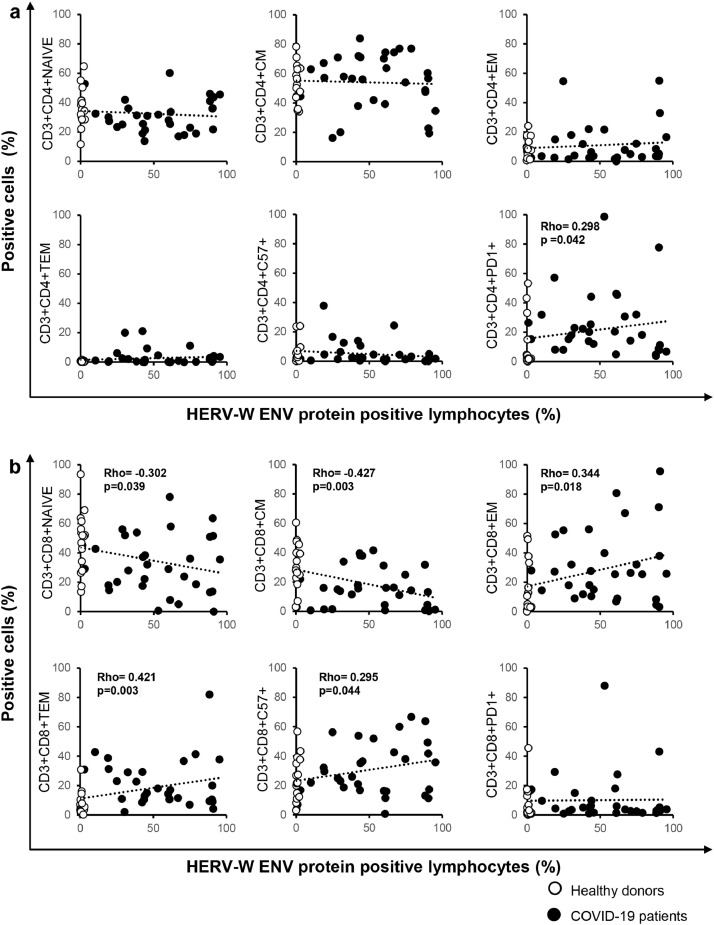
Further analysis of T-lymphocyte cell phenotype revealed a significant correlation between the important markers of T cell differentiation and exhaustion and the percentage of HERV-W ENV-positive lymphocytes. In CD3+CD4+ T cells (Fig. 2a), HERV-W ENV positively correlated with the percentage of cells expressing PD1 exhaustion marker (p = 0.042) but not with other CD3+CD4+ T cell subtypes. In CD3+CD8+ T cells (Fig. 2b), no correlation was found between HERV-W ENV expression and PD1 expression, but HERV-W ENV expression was associated with a decrease in naive (p = 0.039) and central memory (p = 0.003) cells but positively correlated with effector memory (p = 0.018) and terminal effector memory (p = 0.003) cells. More interestingly, in CD8+ T cells, HERV-W ENV positively correlated with the expression of the senescence marker CD57.
Fig. 2.
The percentage of HERV-W ENV-positive lymphocytes correlates with markers of T cell differentiation and exhaustion.
Scatter plot of the percentage of HERV-W ENV-positive lymphocytes (X axis) and the expression of markers of differentiation and exhaustion in CD3+CD4+ T cells (a) and CD3+CD8+ T cells (b) (Y axis) in COVID-19 patients (n = 30, black dots) and healthy donors (n = 17, white dots). The gating strategy to analyze markers related to differentiation, activation status, senescence, and exhaustion in T cells was provided by Beckman Coulter (Duraclone), specifically, naïve (NAIVE) (CCR7+CD45RA+CD28+CD27+), central memory (CM) (CCR7−CD45RA+CD28+CD27+/−), effector memory (EM) (CCR7−CD45RA−CD28+/−CD27+/−), terminal effector memory (TEM) (CCR7−CD45RA+CD28−CD27+/−), PD1+ exhausted and CD57+ senescent T cells. Pairwise associations between continuous variables were tested through the Spearman correlation coefficient. Statistically significant values were considered when p <0.050.
No statistically significant differences of HERV-W ENV expression depending on sex and age (both mRNA and protein) were found in patients and healthy controls, as well as no changes in HERV-W ENV expression between treated patients (both with antiviral and/or corticosteroids) and untreated patients at the time of sampling were found.
3.3. HERV-W ENV mRNA correlated with the expression of pro-inflammatory cytokines involved in COVID-19 and was early activated in PBMCs upon SARS-CoV-2 Spike stimulation
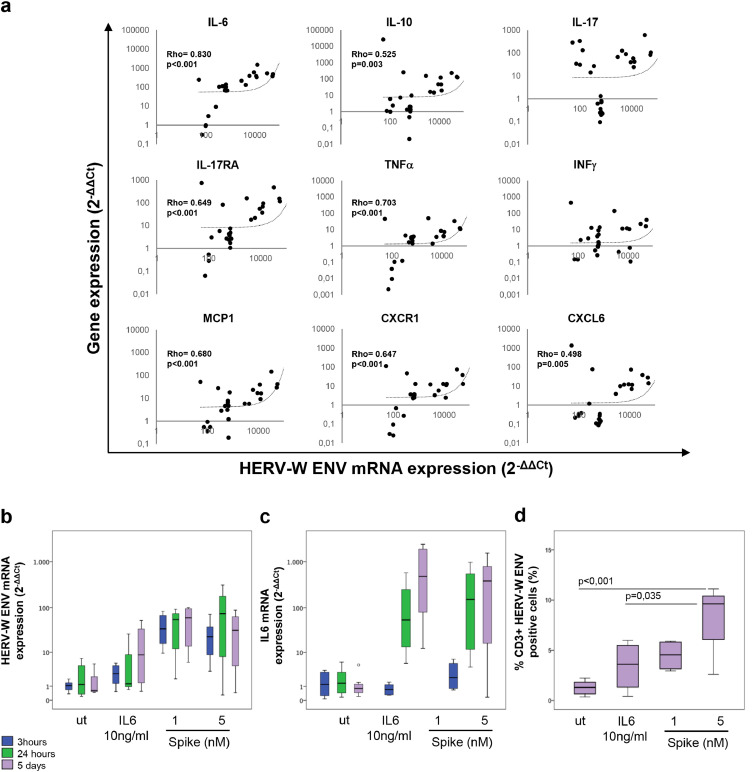
The expression of a selected group of cytokines and cytokine receptors was analysed in blood samples of COV and HD patients by qRT-PCR (Table 2). Higher mRNA expression levels of IL-6 (p<0.001), IL-10 (p = 0.004), IL-17 (p = 0.004), IL-17RA (p = 0.002), MCP1 (p = 0.016), and CXCR1 (p = 0.024) were found in COV patients than in HD, whereas no statistically significant differences were detected in the case of TNFα and INFγ. Interestingly, Spearman analysis revealed a positive correlation between HERV-W ENV mRNA levels and the levels of proinflammatory mediators (Fig. 3), such as IL-6 (Rho=0.830, p<0.001), IL-10 (Rho=0.525, p = 0.003), IL-17RA (Rho=0.649, p<0.001), TNFα (Rho=0.703, p<0.001), MCP1 (Rho=0.680, p<0.001), CXCL6 (Rho=0.498, p = 0.005) and CXCR1 (Rho=0.647, p<0.001) in the leukocytes of the COV group.
Fig. 3.
HERV-W ENV mRNA expression correlates with the expression of several pro-inflammatory cytokines and innate immunity mediators in blood samples of COVID-19 patients.
Scatter plots of the HERV-W ENV mRNA levels (X-axis) and cytokine/cytokine receptor expression (Y-axis: IL-6, IL-10, IL-17, IL-17RA, TNFα, INFγ, MCP1, CXCR1, CXCL6), obtained by qRT-PCR analysis. Pairwise associations between continuous variables were tested through the Spearman correlation coefficient (Rho). Statistically significant values were considered when p <0.050 (a). PBMCs from Healthy donors(n = 4) stimulated with SARS-CoV2 spike protein (1 or 5 nM) and IL-6 protein (10 ng) for 3,24 h and 5 days(Box plots of HERV-W ENV (b) and IL-6 (c) mRNA levels, obtained by qRT-PCR analysis, d) Box plot of the percentage of HERV-W ENV-positive in CD3+ T lymphocytes after 5 days of stimulation with SARS-CoV-2 spike protein and IL-6 protein. For comparison the Bonferroni's post-hoc multiple comparison ANOVA test was utilized.
At the protein level, the analysis of plasma cytokines revealed elevated levels of IL-6, TNFα and IFNγ in COV patients compared to those of HDs (Figure S2 and Table S2), although they did not directly correlate with HERV-W ENV protein levels.
To address the contribution of SARS-CoV-2 on the activation of HERV-W ENV, in vitro experiments were performed stimulating PBMCs from healthy donors with SARS-CoV-2 Spike protein and monitoring the induction of HERV-W ENV mRNA expression after 3, 24 h and 5 days. The exposure to SARS-CoV-2 Spike protein (Fig. 3b and Table S3) induced an early expression of HERV-W ENV, which was already significantly increased compared to the untreated control at 3 h, and ahead of the induction of IL6 which was significantly induced from 24 h onwards (Fig. 3c). After 5 days of stimulation with SARS-CoV-2 Spike protein, a significant increase in the percentage of CD3+ HERV-W ENV positive cells was observed by flow cytometry (Fig. 3d and Table S3).
3.4. HERV-W ENV protein expression in leukocyte subpopulations was associated with COVID-19 severity
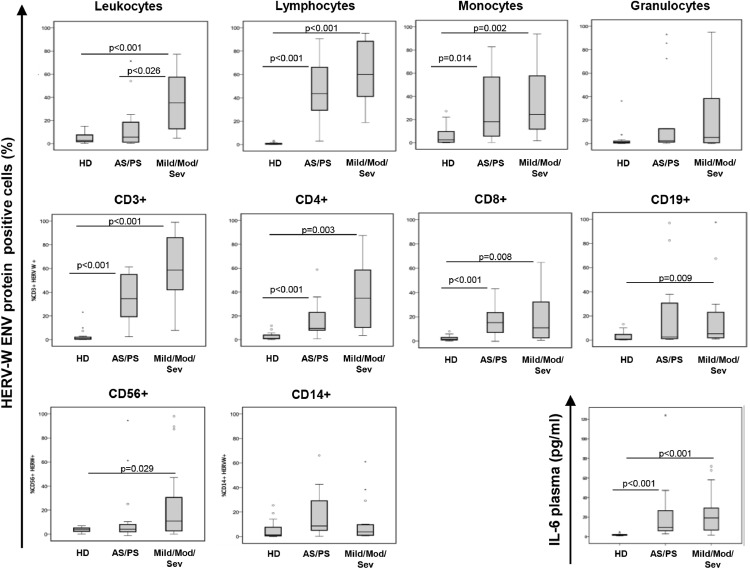
To evaluate the expression of HERV-W ENV in relation to clinical status, we stratified the patients based on the clinical score and analysed the expression of HERV-W ENV in leukocyte subpopulations. As shown in Fig. 4 and according to statistical analysis (Table 3), a significantly higher percentage of HERV-W ENV-positive lymphocytes was observed in COV patients compared to HD (p ≤ 0.007), with a high percentage of HERV-W ENV-positive lymphocytes and monocytes, but not granulocytes, being correlated with an increase in the severity score. This percentage progressively increased in asymptomatic/pauci-symptomatic and symptomatic patients compared to HD in CD3+ T-lymphocytes, mainly in CD4+ subpopulation. HERV-W ENV mRNA levels were found to be significantly elevated in the COV group irrespective to clinical status when compared to those of HD group (p ≤ 0.001; Figure S5 and Table S4). Moreover, IL-6 expression was found to be elevated in all the clinical groups when compared to that in HD (p ≤ 0.0010). The mRNA levels of pro-inflammatory cytokines, except INFγ and CXCL6, were significantly higher in the mild/moderate/severe group than in HD (Figure S5 and Table S5).
Fig. 4.
HERV-W ENV protein expression in the leukocytes of COVID-19 patients was associated with disease severity.
The COVID-19 patients have been stratified based on clinical status as asymptomatic and pauci-symptomatic (AS/PS, n = 15), mild, moderate and severe (Mild/Mod/Sev, n = 15).The percentage of HERV-W ENV-positive cells has been analysed in leukocytes, lymphocytes, monocytes, granulocytes and in T cell, B cell and NK lymphocyte subtypes. IL-6 plasma concentration was also evaluated. Data are represented as box plots (white box: healthy donors, HD; gray box: all patients positive for SARS-CoV-2). Non-parametric Kruskall Wallis test and Bonferroni's correction was used to compare groups (Table 4).
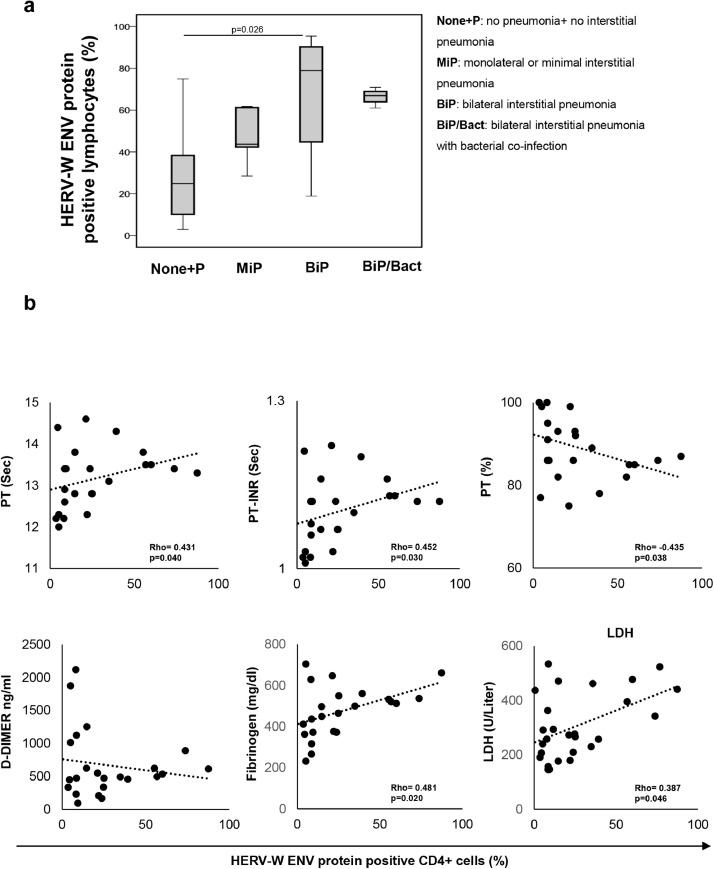
To analyze the association between HERV-W ENV expression in lymphocytes and pneumonia status, the COV group was sub-grouped according to chest computed tomography (CT) images at the time of hospitalization and sampling. As shown in Fig. 5a and Table S6, a significant increase in the percentage of HERV-W ENV-positive lymphocytes across the four groups with different pulmonary involvement was observed, with statistically significant variation in the group with bilateral interstitial pneumonia (BiP; p = 0.026) when compared to the group without or no interstitial pneumonia (None+ P). In addition, the percentage of HERV-W ENV-positive cells in CD4+ T cells significantly correlated with coagulopathy markers and biochemical parameters associated with COVID-19 severity (Fig. 5b).
Fig. 5.
Elevated percentage of HERV-W ENV-positive lymphocytes is associated with pulmonary involvement and correlates with biochemical markers in COVID-19 patients. Patients were stratified into five groups based on pulmonary status: no pneumonia or non-interstitial pneumonia (None+P, n = 7), monolateral or minimal interstitial pneumonia (MiP, n = 6), bilateral or severe pneumonia (BiP, n = 14) and pneumonia with bacterial co-infection (BiP+Bact, n = 3). (a) The percentage of HERV-W ENV-positive lymphocytes was represented as a box plot in all the groups examined and statistical difference was shown. (b) Scatter plot of the percentage of HERV-W ENV-positive CD4+ T cells (X axis) and the biochemical markers (Y axis) in COVID-19 patients. The biochemical markers examined were prothrombin time (PT, sec), prothrombin time internationl normalized ratio (PT-INR, sec), prothrombin activity percentage (PT%), d-DIMERs (ng/ml), fibrinogen (mg/dl) and lactate dehydrogenase (LDH, U/liter). Non-parametric Kruskall Wallis and Bonferroni's correction were used to compare groups; pairwise associations between the continuous variables were tested through the Spearman correlation coefficient. Statistically significant values were considered when p <0.050.
3.5. The percentage of HERV-W ENV-positive lymphocytes reflected the respiratory outcome of COVID-19 patients during hospitalization
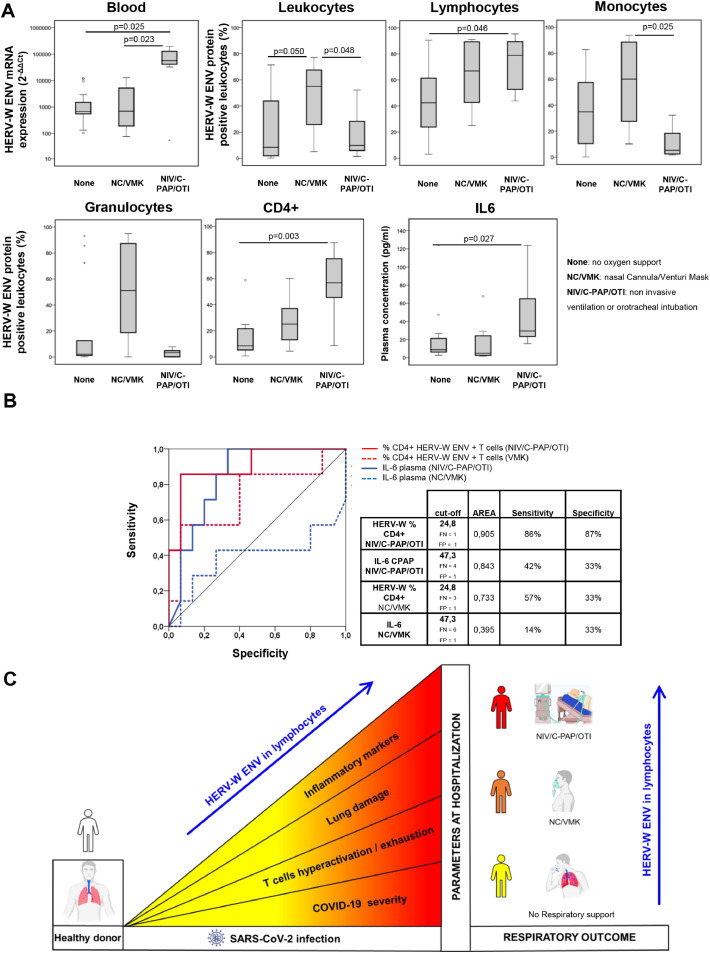
Using information on required oxygen supply during the course of the disease, the COVID-19 patients were divided into three groups representing their respiratory outcome: no oxygen support needed (none), oxygen support with NC/VMKs, and oxygen support by NIV/C-PAP/OTI. The mRNA levels of HERV-W ENV were found to be elevated in the NIV/C-PAP/OTI group compared to the ‘none’ group (p = 0.023) and were significantly higher in the NIV/C-PAP/OTI group than in the NC/VMK group (p = 0.025; Fig. 6a). Surprisingly, the percentage of HERV-W ENV protein-positive cells in the total leukocytes did not reflect the mRNA expression, with a significant increase only in the NC/VMK group (p = 0.050). Nevertheless, an increase in the percentage of HERV-W ENV-positive lymphocytes, mainly in CD4+ T cells, across the three groups was observed with a significant increase in the NIV/C-PAP/OTI group compared to the ‘none’ group (Lymphocytes p = 0.019, CD4+ T cells p = 0.03). This increase was not found in monocytes or granulocytes, suggesting that the percentage of HERV-W ENV-positive lymphocytes reflects the respiratory outcome of COVID-19 patients. Interestingly, analysis of the inflammatory and coagulation markers across the three groups demonstrated that plasma level of IL-6 and LDH were able to discriminate the NIV/C-PAP/OTI group from the ‘none’ group (IL-6 p = 0.027, Fig. 6a; LDH p = 0.002 Figure S6 and Tables S7–9), whereas a significant increase in the transcriptional levels of several pro-inflammatory cytokines was found across the three groups (p ≤ 0.035, Figure S6). Of note, ROC curve analysis revealed the percentage of CD4+ HERV-W ENV positive cells evidenced a more specific marker than the IL6 concentration in plasma for patients needing respiratory support (Fig. 6b).
Fig. 6.
Levels of HERV-W ENV mRNA expression and the percentage of positive lymphocytes reflect respiratory outcome of COVID-19 patients. The COVID-19 patients were stratified according to respiratory needs during hospitalization: no oxygen support needed (None; n = 16), oxygen support with nasal cannula or ventimask (NC/VMK; n = 8), oxygen support by non-invasive ventilation, continuous positive airway pressure or orotracheal intubation (NIV/C-PAP/OTI; n = 6). (a) The expression of HERV-W ENV mRNA in leukocytes and the percentage of HERV-W ENV-positive cells in leukocytes, lymphocytes, monocytes, granulocytes, CD4+ T cells and IL-6 in plasma were represented as box plots. Non-parametric Kruskall Wallis test and Bonferroni's correction were used to compare groups and statistically significant values were considered when p <0.050. (b) ROC Curve of the% HERV-W ENV Lymphocytes and CD3+ T cells, and IL6 plasma concentration in COV (n = 30) respect to HD (n = 17) left panel; ROC Curve of the% HERV-W ENV in Lymphocytes and in CD4+ T cells and IL-6 plasma concentration respect to the respiratory outcome. (c) Association of high expression of HERV-W ENV protein in the blood cells of COVID-19 patients, and several clinical and biological parameters linked to patient status at hospitalization and disease progression.
4. Discussion
The present pilot study analysed the potential involvement of the pathogenic HERV-W ENV protein as a contributing factor in COVID-19 immunopathology, based on previous reports of its activation induced by certain infectious agents [17,18]. The study demonstrates for the first time a high expression of HERV-W ENV protein in the blood cells of COVID-19 patients, which is associated with several clinical and biological parameters linked to patient status at hospitalization and disease progression (Fig. 6c). Analysis of leukocytes on admission revealed elevated levels of HERV-W ENV mRNA and protein in the COVID-19 patients compared to healthy donors. The detection of HERV-W ENV protein was most significantly associated with lymphocytes and, importantly, the correlation between values from orthogonal assays, i.e., RT-qPCR mRNA quantification and those from protein expression analysis by flow cytometry, was highly significant and thereby validated the initially observed results in COVID-19 patients. Of note, a positive correlation was found between HERV-W ENV mRNA levels and the expression of several pro-inflammatory mediators involved in COVID-19 and the associated cytokine storm. Particularly, in COVID-19 patients, HERV-W ENV mRNA levels strongly correlated with the levels of IL-6, which is a well-known factor for indicating a worsening prognosis of disease [22], but also with IL-17, TNF-α and chemokines such as CXCL6 and MCP1 known to play a role in the pathogenesis of airway inflammation and found elevated in patients [23].
The involvement of the immune response in the pathogenesis of SARS CoV-2 infection and in the inflammatory complications associated with COVID-19 has been well described [24,25]. In our patients, a dysregulation of the immune response was confirmed and found associated with an elevated expression of HERV-W ENV in cell subpopulations that mediate this response. Immuno-phenotyping analysis revealed a significant positive correlation between the percentage of HERV-W ENV-positive lymphocytes and hyper-activated/exhausted CD4+PD1+ T-lymphocytes in COVID-19 patients, which was not observed in HD. Interestingly, the percentage of HERV-W ENV-positive lymphocytes varied among the CD3+CD8+ T-cytotoxic cell subsets, which correlated with a decrease in naive and central memory cells and, conversely, an increase in effector memory as well as in terminal effector memory cells. Moreover, it is well known that PD1 is highly expressed both alone or in combination with CD57 in COVID-19 disease. At the beginning it acts as a marker of hyperactivation, however, the subsequent step would be to exhaust its function to protect the cell or drive the senescence by expressing the CD57 marker [24,26]. In the CD8 compartment we evidenced the expression of HERV-W correlates with CD57 expression. These results suggest that HERV-W ENV expression may affect the activation and differentiation states of lymphocytes, thereby potentially being involved in adaptive immune dysfunctions. An altered adaptive immune response, particularly via CD3+CD8+ T-cells subsets, has already been characterized in COVID-19 [27] patients; and therapies aiming at restoring T-cell function [28] or neutralizing the involved co-factors/cytokines have been proposed. No correlation of disease severity with the absolute number of lymphocyte counts was found in the present study; however, as samples were collected early in the course of the disease, this correlation requires longitudinal analysis, in particular to verify a possible role of HERV-W ENV in delayed lymphopenia.
Interestingly, the percentage of HERV-W ENV positive CD3+ T-lymphocytes progressively increased in asymptomatic/pauci-symptomatic and symptomatic patients. Alongside, an elevated transcriptional expression of HERV-W ENV was found in blood cells of the symptomatic group. A similar profile was found for IL-6 mRNA levels suggesting early HERV activation consistent with a potential role of HERV ENV in further immune hyperstimulation and lymphocyte deregulation. Accordingly, the in vitro stimulation of PBMCs from healthy donors with HERV-W ENV protein induced IL-6 expression. HERV-W ENV protein expression in lymphocytes was also associated with lung involvement as assessed by radiological imaging, and found increased in patients with extensive interstitial pneumonia. Moreover, HERV-W ENV protein expression was significantly correlated with biochemical markers such as LDH, which is an important indicator of poor prognosis [29].
Most importantly, ENV expression in lymphocytes at sampling reflected the respiratory outcome during the global course of hospitalization, which suggested its involvement in disease pathogenesis. Interestingly, the percentage of CD4+ HERV-W ENV positive cells was found to be a more specific marker predicting a need for respiratory support than the IL6 concentration in plasma.
These findings provide consistent avenues for further in-depth and large-scale studies that are required to decipher the cellular and molecular mechanism of HERV-W transactivation by SARS-CoV-2 and the immunopathogenic impact of HERV-W ENV on the clinical evolution of COVID-19. The immunopathogenic effects of HERV-W ENV have been demonstrated both via in vitro experiments and in a few chronic inflammatory human diseases [12,20,[30], [31], [32]]. Detection of high expression of HERV-W ENV in the blood cells of COVID-19 patients, particularly T-lymphocytes, has not been observed in any other condition studied to date. Moreover, this elevated expression in blood cells should be studied in relation with its neurotoxic effects [31], [32], [33], considering the recently identified neurological symptoms in COVID-19 [10]. A limitation of the study was the impossibility to characterize the kinetics of HERV-W ENV expression and the relationship with inflammation markers and clinical progression of COVID-19 over time, but considering the rapid evolution of the clinical severity of COVID-19 and the need to move patients to ICU facilities or to discharge non-severe patients, a cross-sectional evaluation at the moment of hospital admission was performed. Further longitudinal studies specifically designed for this purpose are needed. Moreover, further analysis of HERV-ENV protein expression in lymphocytes in association with inflammatory markers should be investigated in other infectious and chronic inflammatory diseases. Another limitation of the study is that the in vitro stimulation with the Spike protein, although significantly induced HERV-W ENV expression, showed a variable response among donors, suggesting that the individual and genetic variability that can be found in the population of COVID-19 patients may already reflect selected characteristics. Further in vitro studies must also address factors underlying this individual susceptibility, which could not be addressed in the present study,
Because HERV-W ENV was identified quite early in COVID-19 patients, it may significantly contribute to a delayed onset of immunopathological and/or neurological syndromes. Thus, studying mechanisms underlying immune hyperstimulation associated with lymphocyte dysfunction and lymphopenia may also help identify relevant targets for therapeutic intervention preventing severe clinical evolutions of COVID-19.
Contributors
Conceptualization, C.M., E.B., A.M, H.P.; Lab work, V.P., M.F, and A.M.; Flow cytometry C.M., A.M.; Real time analysis E.B., V.P., and M.F.; Clinical patients and sample management M.A, S.G., L.S., M.I., V.M., M.Z., P.V., S.B., B.C.; Statistical analysis C.M., A.M. and E.B.; Writing-Original Draft, C.M., E.B., A.M., H.P., P.S.V.; revising manuscript critically for important intellectual content B.H, E.G, P.D.F., P.S.V.; all authors contributed to data interpretation and critical revision of the manuscript and approved the final version of the manuscript.
Declaration of Competing Interests
C.M. reports grant from Gilead, outside the submitted work; M.I. reports personal fees from Biogen srl, personal fees from Becton, Dickinson and Company, outside the submitted work; HP and BC receive compensation for their work by Geneuro-Innovation. The other authors have nothing to disclose.
Acknowledgments
Acknowledgments
No specific funding was received for this work. The study was supported by Geneuro innovation who provided materials and reagents for HERV analyses. We acknowledge Martina Giudice for the technical assistance.
Data Sharing Statement
Data from this manuscript is available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103341.
Appendix. Supplementary materials
References
- 1.Gupta A., Madhavan M.V., Sehgal K. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 2.Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J Exp Med. 2020;217 doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Jiang M., Chen X., Montaner L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Tan Y., Ling Y. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 6.Huang I., Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J Intensive Care. 2020;8:36. doi: 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta P., McAuley D.F., Brown M. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020;8:E46–E47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diorio C., Henrickson S.E., Vella L.A. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J Clin Invest. 2020;130:5967–5975. doi: 10.1172/JCI140970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nath A., Smith B. Neurological issues during COVID-19: an overview. Neurosci Lett. 2021;742 doi: 10.1016/j.neulet.2020.135533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Küry P., Nath A., Créange A. Human endogenous retroviruses in neurological diseases. Trends Mol Med. 2018;24:379–394. doi: 10.1016/j.molmed.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levet S., Medina J., Joanou J. An ancestral retroviral protein identified as a therapeutic target in type-1 diabetes. JCI Insight. 2017;2:e94387. doi: 10.1172/jci.insight.94387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank J.A., Feschotte C. Co-option of endogenous viral sequences for host cell function. Curr Opin Virol. 2017;25:81–89. doi: 10.1016/j.coviro.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas J., Perron H., Feschotte C. Variation in proviral content among human genomes mediated by LTR recombination. Mob DNA. 2018;9:36. doi: 10.1186/s13100-018-0142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matteucci C., Balestrieri E., Argaw-Denboba A., Sinibaldi-Vallebona P. Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol. 2018;53:17–30. doi: 10.1016/j.semcancer.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Kazazian H.H., Jr, Moran J.V. Mobile DNA in Health and Disease. N Engl J Med. 2017;377:361–370. doi: 10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charvet B., Reynaud J.M., Gourru-Lesimple G., Perron H., Marche P.N., Horvat B. Induction of pro-inflammatory multiple sclerosis-associated retrovirus envelope protein by human herpesvirus-6A and CD46 receptor engagement. Front Immunol. 2018;9:2803. doi: 10.3389/fimmu.2018.02803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F., Nellåker C., Sabunciyan S. Transcriptional derepression of the ERVWE1 locus following influenza A virus infection. J Virol. 2014;88:4328–4337. doi: 10.1128/JVI.03628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madeira A., Burgelin I., Perron H., Curtin F., Lang A.B., Faucard R. MSRV envelope protein is a potent, endogenous and pathogenic agonist of human toll-like receptor 4: relevance of GNbAC1 in multiple sclerosis treatment. J Neuroimmunol. 2016;291:29–38. doi: 10.1016/j.jneuroim.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Duperray A., Barbe D., Raguenez G. Inflammatory response of endothelial cells to a human endogenous retrovirus associated with multiple sclerosis is mediated by TLR4. Int Immunol. 2015;27:545–553. doi: 10.1093/intimm/dxv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtin F., Perron H., Kromminga A., Porchet H., Lang A.B. Preclinical and early clinical development of GNbAC1, a humanized IgG4 monoclonal antibody targeting endogenous retroviral MSRV-Env protein. MAbs. 2015;7:265–275. doi: 10.4161/19420862.2014.985021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han H., Ma Q., Li C. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z., Wherry E.J. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vabret N., Britton G.J., Gruber C. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Candia P., Prattichizzo F., Garavelli S., Cells Matarese G.T. Warriors of SARS-CoV-2 infection. Trends Immunol. 2021;42:18–30. doi: 10.1016/j.it.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Biasi S., Meschiari M., Gibellini L. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matteucci C., Minutolo A., Balestrieri E. Thymosin alpha 1 mitigates cytokine storm in blood cells from COVID-19 patients. Open Forum Infect Dis. 2020;8:ofaa588. doi: 10.1093/ofid/ofaa588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang S.M., Zhu H.Q., Xie Y.N. Temporal changes in laboratory markers of survivors and non-survivors of adult inpatients with COVID-19. BMC Infect Dis. 2020;20:952. doi: 10.1186/s12879-020-05678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kremer D., Gruchot J., Weyers V. pHERV-W envelope protein fuels microglial cell-dependent damage of myelinated axons in multiple sclerosis. Proc Natl Acad Sci U S A. 2019;116:15216–15225. doi: 10.1073/pnas.1901283116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faucard R., Madeira A., Gehin N. Human endogenous retrovirus and neuroinflammation in chronic inflammatory demyelinating polyradiculoneuropathy. EBioMedicine. 2016;6:190–198. doi: 10.1016/j.ebiom.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Horssen J., van der Pol S., Nijland P. Human endogenous retrovirus W in brain lesions: rationale for targeted therapy in multiple sclerosis. Mult Scler Relat Disord. 2016;8:11–18. doi: 10.1016/j.msard.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Johansson E.M., Bouchet D., Tamouza R. Human endogenous retroviral protein triggers deficit in glutamate synapse maturation and behaviors associated with psychosis. Sci Adv. 2020;6 doi: 10.1126/sciadv.abc0708. eabc0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.