Abstract
Background & Aims
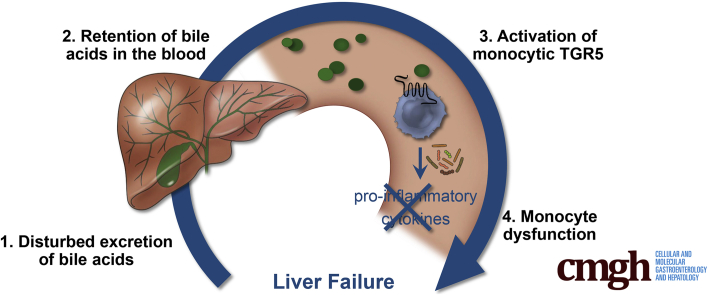
Retention of bile acids in the blood is a hallmark of liver failure. Recent studies have shown that increased serum bile acid levels correlate with bacterial infection and increased mortality. However, the mechanisms by which circulating bile acids influence patient outcomes still are elusive.
Methods
Serum bile acid profiles in 33 critically ill patients with liver failure and their effects on Takeda G-protein–coupled receptor 5 (TGR5), an immunomodulatory receptor that is highly expressed in monocytes, were analyzed using tandem mass spectrometry, novel highly sensitive TGR5 bioluminescence resonance energy transfer using nanoluciferase (NanoBRET, Promega Corp, Madison, WI) technology, and in vitro assays with human monocytes.
Results
Twenty-two patients (67%) had serum bile acids that led to distinct TGR5 activation. These TGR5-activating serum bile acids severely compromised monocyte function. The release of proinflammatory cytokines (eg, tumor necrosis factor α or interleukin 6) in response to bacterial challenge was reduced significantly if monocytes were incubated with TGR5-activating serum bile acids from patients with liver failure. By contrast, serum bile acids from healthy volunteers did not influence cytokine release. Monocytes that did not express TGR5 were protected from the bile acid effects. TGR5-activating serum bile acids were a risk factor for a fatal outcome in patients with liver failure, independent of disease severity.
Conclusions
Depending on their composition and quantity, serum bile acids in liver failure activate TGR5. TGR5 activation leads to monocyte dysfunction and correlates with mortality, independent of disease activity. This indicates an active role of TGR5 in liver failure. Therefore, TGR5 and bile acid metabolism might be promising targets for the treatment of immune dysfunction in liver failure.
Keywords: Liver Failure, Bile Acids, TGR5, GPBAR1
Abbreviations used in this paper: ACLF, acute-on-chronic liver failure; ADRB2, β2 adrenergic receptor; DMSO, dimethyl sulfoxide; EV, empty vector; IFN, interferon; LPS, lipopolysaccharide; NanoBRET, bioluminescence resonance energy transfer using nanoluciferase; Nluc, nanoluciferase; TGR5, Takeda G-protein–coupled receptor 5; TLCA, taurolithocholic acid; UDCA, ursodeoxycholic acid
Graphical abstract
Summary.
Serum bile acids accumulate in liver failure. We show that, depending on their composition and quantity, these bile acids activate Takeda G-protein–coupled receptor 5, an immunomodulatory receptor that is highly expressed in monocytes. Takeda G-protein–coupled receptor 5 activation leads to monocyte dysfunction and correlates with mortality.
Cholestasis is a condition that complicates not only liver but also nonhepatic diseases such as critical illness, and is related to increased mortality.1, 2, 3 Recent studies have shown that the disturbed excretion of bile acids and their accumulation in the blood are associated especially with infections and fatal outcomes in these diseases.4, 5, 6, 7 The mechanisms by which circulating bile acids influence patient outcomes still are elusive.
Bile acids, secreted by the liver into the bile, have long been believed to be simple emulsifiers of dietary fats.8 However, increasing evidence has indicated that bile acids are complex signaling molecules that activate various nuclear and membrane-bound receptors.9,10 One of these receptors, the Takeda G-protein–coupled receptor 5 (TGR5, also known as G-protein–coupled bile acid receptor 1), is an immunomodulatory receptor that fine-tunes innate immune function and has been suggested to mediate immune tolerance in the gut.11,12 Monocytes and macrophages express TGR5.9,13,14 In general, these myeloid cells produce high amounts of proinflammatory cytokines in response to bacterial pathogens. The same cells also may be involved in tolerance toward the plethora of bacteria colonizing the human gut. This tolerance is supposed to be mediated, at least in part, by intestinal bile acids, that activate TGR5 on myeloid cells, thereby preventing the production of proinflammatory cytokines and subsequent inflammation.11,12
In various liver diseases, the bile acid flow from the liver into the bile is disturbed, leading to an accumulation of bile acids in the blood.15,16 To date, massively increased serum bile acid levels in liver diseases were considered mainly as a surrogate parameter for excretory liver failure. We hypothesized that the quantity and composition of circulating bile acids may impact myeloid cell functionality via TGR5. Therefore, we investigated the bile acid profiles of patients with liver failure requiring admission to the intensive care unit and analyzed the effect of the complex bile acid profiles on monocyte function. Furthermore, our study determined the relative potency of several bile acids to activate TGR5 and analyzed how complex bile acid mixtures affect TGR5 activity.
Results
Patients With Liver Failure Show Serum Bile Acids Activating TGR5
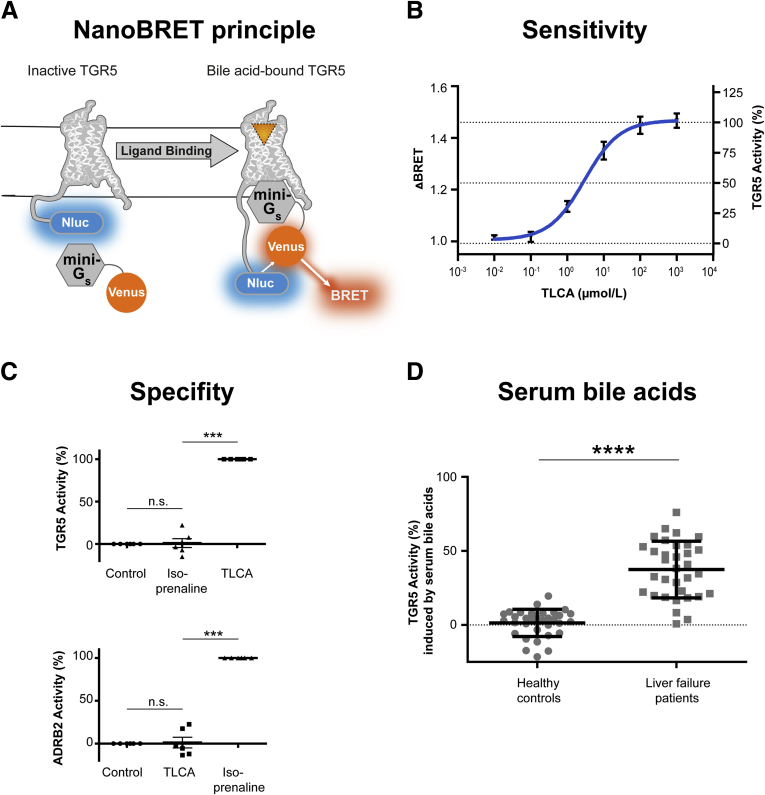
To investigate TGR5 activity in response to circulating bile acids, we established a TGR5 reporter assay using a recently described highly selective and specific nanoluciferase bioluminescence resonance energy transfer assay (NanoBRET, Promega Corp, Madison, WI) in combination with a mini-Gs-protein.17 Mini-Gs-protein recruitment by the G-protein–coupled receptor TGR5 was measured via NanoBRET (Figure 1A). TGR5, C-terminally tagged with a nanoluciferase (Nluc), served as the donor, and the mini-Gs-Venus fusion protein was the acceptor. Human embryonic kidney (HEK) 293T cells were transiently transfected with both vectors and NanoBRET change was monitored before and after ligand addition. As shown in Figure 1B, stimulation with taurolithocholic acid (TLCA), the most potent naturally occurring TGR5 agonist,9 showed a dose-dependent recruitment of the mini-Gs-protein. The NanoBRET change induced by 100 μmol/L TLCA was set to 100%, reflecting maximal TGR5 activity. To control the specificity of our assay, the β2 adrenergic receptor (ADRB2), another G-protein–coupled receptor recruiting Gs proteins, also was fused genetically to a nanoluciferase. The TGR5 agonist did not induce an ADRB2 NanoBRET change, and the ADRB2 agonist isoprenaline did not activate TGR5 NanoBRET (Figure 1C), indicating specificity of the NanoBRET assay. Using this method, we evaluated whether serum bile acids would activate TGR5. We assessed the individual serum bile acid profiles of 33 critically ill patients with liver failure and 33 healthy controls by tandem mass spectrometry. To exclude unspecific serum effects, the pure serum bile acid profile from each subject was remixed in buffer and measured for their potential to activate TGR5. Serum bile acids from healthy human volunteers hardly induced any TGR5 activation (means ± SD, 1.3% of maximal activation ± 9.0%). By contrast, serum bile acids from patients with liver failure activated TGR5 significantly (means ± SD, 37.4% ± 19.1%) (Figure 1D).
Figure 1.
Patients with liver failure show serum bile acids activating TGR5. (A and B) The TGR5 NanoBRET assay measured dose-dependent Gs recruitment by bile acids. N = 10. Data are means ± SEM. (C) TLCA selectively activated TGR5 NanoBRET. The ADRB2 agonist isoprenaline selectively activated ADRB2 NanoBRET. N = 6. Data are means ± SEM, 1-way repeated-measures analysis of variance with Bonferroni correction. (D) Patients with liver failure show serum bile acids significantly activating TGR5. n = 33/group. Data are means ± SD, Student t test. ∗∗∗P < .001, ∗∗∗∗P < .0001.
In Acute-on-Chronic Liver Failure Patients, 95% Show Serum Bile Acids Activating TGR5
Serum bile acids from our patients induced TGR5 activity ranging from 0.8% to 76.1% (Table 1). Some patients showed levels that were comparable with healthy controls, while others had a serum bile acid composition potently activating TGR5. To identify patients with serum bile acids that activate TGR5 relevantly, we defined the mean value of healthy individuals ± 3× SD as the reference range. Consequently, serum bile acid profiles inducing a TGR5 activity of ≥28.3% were defined as pathologic and were classified as TGR5-activating serum bile acids. By using this cut-off value, none of the healthy volunteers had TGR5-activating serum bile acids. By contrast, 67% of our critically ill patients with liver failure had serum bile acids that activated TGR5 relevantly (22 of 33 patients) (Table 1).
Table 1.
TGR5 Activity Induced by Serum Bile Acids From Patients With Liver Failure
| Patient ID | TGR5 activity,a% | Patient ID | TGR5 activity,a% | Patient ID | TGR5 activity,a% |
|---|---|---|---|---|---|
| 01 | 3.7 | 12 | 76.1 | 23 | 54.2 |
| 02 | 30.7 | 13 | 16.5 | 24 | 45.9 |
| 03 | 57.2 | 14 | 8.3 | 25 | 38.2 |
| 04 | 48.3 | 15 | 19.9 | 26 | 28.7 |
| 05 | 65.0 | 16 | 23.0 | 27 | 53.7 |
| 06 | 34.6 | 17 | 19.1 | 28 | 18.3 |
| 07 | 59.5 | 18 | 59.7 | 29 | 40.9 |
| 08 | 44.1 | 19 | 18.7 | 30 | 0.8 |
| 09 | 31.8 | 20 | 52.9 | 31 | 49.6 |
| 10 | 50.7 | 21 | 32.7 | 32 | 22.2 |
| 11 | 62.3 | 22 | 21.2 | 33 | 47.2 |
Pathologically increased values are shown in bold (ie, bile acids that mediate TGR5 activity ≥28.3%).
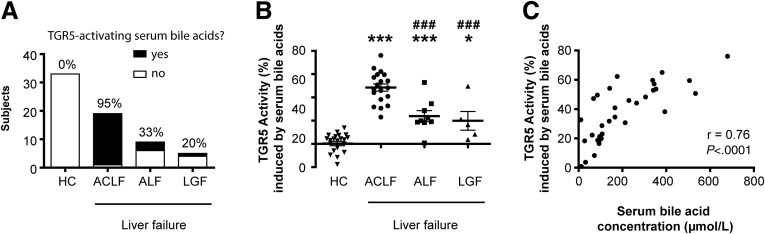
Liver failure in critically ill patients is a heterogeneous condition with different etiologies and characteristics. Interestingly, almost all patients with acute-on-chronic liver failure (ACLF) showed bile acid profiles activating TGR5 (18 of 19 patients; 95%) (Figure 2A and Table 2). Although pathologically increased TGR5 activity also was induced by bile acid profiles of patients with acute liver failure (33% with TGR5-activating serum bile acids) or liver graft failure (20% with TGR5-activating serum bile acids), the proportion was significantly lower. Accordingly, the mean TGR5 activity induced by serum bile acids was highest in patients with ACLF (Figure 2B).
Figure 2.
In acute-on-chronic liver failure patients, 95% show serum bile acids activating TGR5. (A) Proportion of subjects with serum bile acids activating TGR5. (B) TGR5 activity induced by serum bile acid profiles. Data are means ± SD. One-way repeated-measures analysis of variance with Bonferroni correction. ∗P < .05 compared with HC, ∗∗∗P < .001 compared with HC, and ###P < .001 compared with ACLF. (C) Correlation between serum bile acid concentration and TGR5 activity induced by serum bile acids, Spearman correlation rank test. ACLF, acute-on-chronic liver failure; ALF, acute liver failure; HC, healthy control; LGF, liver graft failure.
Table 2.
Characteristics of Liver Failure Patients With and Without TGR5-Activating Serum Bile Acids
| Characteristics | TGR5-activating serum bile acids? |
P | |
|---|---|---|---|
| Yes |
No |
||
| n = 22 | n = 11 | ||
| Age, y | 55 ± 12 | 56 ± 13 | NS |
| Male sex, % (n) | 50 (11) | 64 (7) | NS |
| Etiology of liver failure, % (n) | |||
| Acute-on-chronic | 81.8 (18) | 9.1 (1) | |
| Alcoholic cirrhosis | 50.0 (11) | ||
| Viral cirrhosis | 4.5 (1) | ||
| Viral and alcoholic cirrhosis | 4.5 (1) | ||
| Cryptogenic cirrhosis | 9.0 (2) | 4.5 (1) | |
| Other cirrhosisa | 13.6 (3) | ||
| Acute | 13.6 (3) | 54.5 (6) | |
| Liver graft failure | 4.5 (1) | 36.4 (4) | |
| Hemodialysis, % (n) | 55 (12) | 45 (5) | NS |
| Serum levels | |||
| ALT, U/L | 42 (27–99) | 54 (40–88) | NS |
| AST, U/L | 86 (64–353) | 78 (57–126) | NS |
| AST/ALT | 2.2 (1.4–3.2) | 1.6 (1.3–2.1) | NS |
| ALP, U/L | 118 (79–161) | 147 (70–444) | NS |
| Total bilirubin, μmol/L | 357 ± 120 | 350 ± 81 | NS |
| Albumin, g/L | 18 (17–25) | 19 (16–23) | NS |
| Bile acid serum parameter | |||
| Total serum bile acids, μmol/L | 285 (166–476) | 72 (46–96) | <.001 |
| TGR5 activity by bile acids, % | 48.4 ± 12.4 | 15.6 ± 7.7 | <.001 |
NOTE. Values are means ± SD or median with 95% CI.
ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
Other causes of cirrhosis included the following: 1 patient with primary sclerosing cholangitis, 1 patient with primary biliary cholangitis, and 1 patient with Alagille–Watson syndrome.
Increased serum bile acids are commonly understood as an indicator for excretory liver failure.15,16 Consequently, TGR5 activation by serum bile acids might correlate with bilirubin, alkaline phosphatase, or other liver function tests indicative of cholestasis. We therefore compared blood tests of patients with and without TGR5-activating serum bile acids (Table 2). Blood tests indicated severe liver failure, but there was no difference between both groups. Consequently, none of the routine blood tests we used could predict TGR5 activation induced by serum bile acids. This suggests that the TGR5 activation by complex serum bile acid profiles is an independent factor in the pathogenesis of liver failure.
Only the total serum bile acid concentration assessed by a colorimetric assay was correlated to TGR5 activity (Figure 2C and Table 2).
TGR5-Activating Serum Bile Acids Induce Monocyte Dysfunction
TGR5 activation has been linked closely to immune suppression in monocytes.9,18 TGR5-activating serum bile acids therefore might modulate monocyte function in patients. To investigate this hypothesis, we treated monocytes from healthy human volunteers producing proinflammatory cytokines in response to bacterial lipopolysaccharide (LPS) with bile acids. We remixed the mean bile acid profile from the patients with TGR5-activating serum bile acids. We compared its effect with the mean bile acid profile of matched healthy controls (both mean bile acid profiles are shown in Table 3). Monocytes were preincubated with bile acid mixtures and subsequently stimulated with LPS. Cells left untreated served as a negative control and the TGR5 agonist TLCA as a positive control.
Table 3.
Serum Bile Acid Profile of Healthy Controls vs Liver Failure Patients With TGR5-Activating Serum Bile Acids
| Bile acid profile, μmol/L | Healthy controls (n = 22) | Patients (n = 22) | P | TGR5 activity,a% |
|---|---|---|---|---|
| Primary bile acids | ||||
| Cholic acid | 0.3 ± 0.5 | 0.1 ± 0.1 | NS | 0.0 |
| Taurocholic acid | 0.0 ± 0.1 | 7.8 ± 9.9 | NS | 0.9 |
| Glycocholic acid | 0.2 ± 0.2 | 15.4 ± 15.8 | < .01 | 1.6 |
| Chenodeoxycholic acid | 0.4 ± 0.7 | 0.2 ± 0.2 | NS | 0.1 |
| Taurochenodeoxycholic acid | 0.1 ± 0.1 | 23.7 ± 12.3 | < .001 | 16.5 |
| Glycochenodeoxycholic acid | 0.9 ± 0.6 | 29.2 ± 21.1 | < .001 | 17.4 |
| Secondary bile acids | ||||
| Lithocholic acid | 0.0 ± 0.0 | 0.0 ± 0.0 | NS | 0 |
| Taurolithocholic acid | 0.0 ± 0.0 | 0.0 ± 0.1 | NS | 0 |
| Deoxycholic acid | 0.5 ± 0.8 | 0.0 ± 0.0 | NS | 0 |
| Taurodeoxycholic acid | 0.1 ± 0.1 | 0.2 ± 0.8 | NS | 0.4 |
| Glycodeoxycholic acid | 0.3 ± 0.3 | 0.4 ± 1.3 | NS | 0.6 |
| Tertiary bile acids | ||||
| Ursodeoxycholic acid | 0.1 ± 0.3 | 6.0 ± 6.4 | NS | 1.0 |
| Tauroursodeoxycholic acid | 0.0 ± 0.0 | 21.0 ± 14.3 | < .001 | 3.2 |
| Glycoursodeoxycholic acid | 0.1 ± 0.2 | 81.2 ± 56.9 | < .001 | 8.3 |
| Total | 50.0 |
NOTE. Values are means ± SD, 1-way analysis of variance with Bonferroni correction.
TGR5 activity caused by single serum bile acids from patients; calculated according to the formula shown in Figure 6C (eg, glycocholic acid in serum from patients was increased significantly and caused a TGR5 activity of 1.6%).
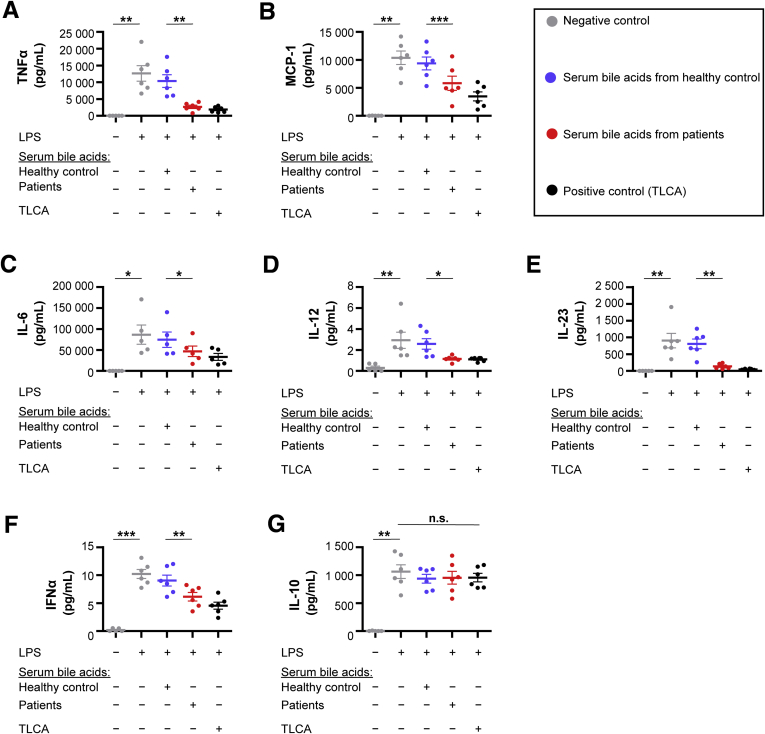
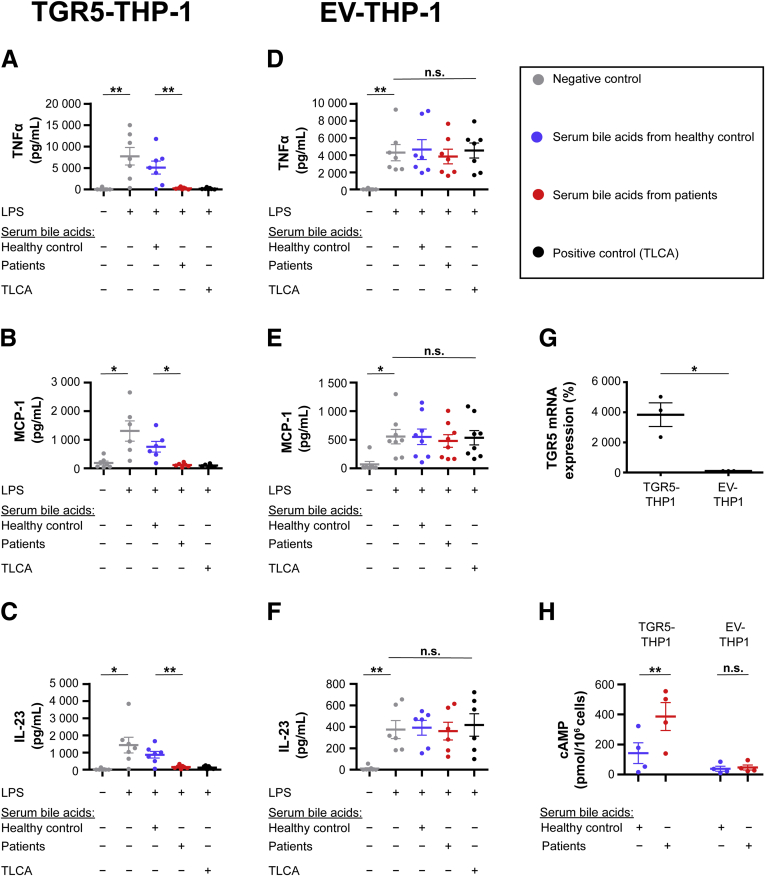
Monocytes stimulated with TGR5-activating serum bile acids from patients showed a significant immune dysfunction (Figure 3A–F). The release of proinflammatory cytokines and chemokines, such as tumor necrosis factor α, interleukin 6, and monocyte chemoattractant protein-1, was reduced significantly. Interestingly, the anti-inflammatory cytokine interleukin 10 was not affected (Figure 3G).
Figure 3.
TGR5-activating serum bile acids induce monocyte dysfunction. (A–G) Monocytes were incubated with the mean bile acid profile from either healthy controls or liver failure patients with TGR5-activating serum bile acids. TGR5-activating serum bile acids significantly inhibited the release of proinflammatory cytokines induced by LPS. Anti-inflammatory interleukin (IL)10 was not altered. Data are means ± SEM, N = 5–6. Two-way repeated-measures analysis of variance with post hoc paired t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. IFN, interferon; MCP-1, monocyte chemoattractant protein-1; TLCA, taurolithocholic acid; TNFα, tumor necrosis factor α.
To assure that the effects of the serum bile acids on monocytes were mediated by TGR5, we repeated our experiments using the human monocytic cell line THP-1, which does not express TGR5 endogenously.9,19 THP-1 monocytes were transduced with TGR5 or empty vector (EV). TGR5-activating serum bile acids from liver failure patients triggered adenosine 3′,5′-cyclic monophosphate production (Figure 4H) and immune suppression (Figure 4A–C) in TGR5-expressing THP-1. By contrast, EV–THP-1 did not show any bile acid effects (Figure 4D–F and H), indicating that the immunosuppressive effects of serum bile acids were mediated by TGR5.
Figure 4.
Effects of serum bile acids are mediated by TGR5. THP-1 monocytes were transduced with TGR5 or EV and incubated with the mean bile acid profile from either healthy controls or liver failure patients with TGR5-activating serum bile acids. Serum bile acids from patients significantly inhibited the secretion of proinflammatory cytokines in (A–C) TGR5–THP-1, but not in (D–F) EV–THP-1. Two-way repeated-measures analysis of variance with post hoc paired t test, N = 6–8. (G) TGR5–THP-1 showed a significantly higher TGR5 expression compared with EV–THP-1, paired t test, N = 3. (H) Serum bile acids from patients induced significant adenosine 3′,5′-cyclic monophosphate (cAMP) production in TGR5-THP-1, but not in EV-THP-1. Two-way repeated-measures analysis of variance with the post hoc Holm–Šidák test, N = 4. All data are means ± SEM. ∗P < .05, ∗∗P < .01. MCP-1, monocyte chemoattractant protein-1; mRNA, messenger RNA; TNFα, tumor necrosis factor α.
TGR5-Activating Serum Bile Acids Are Associated With Increased Mortality
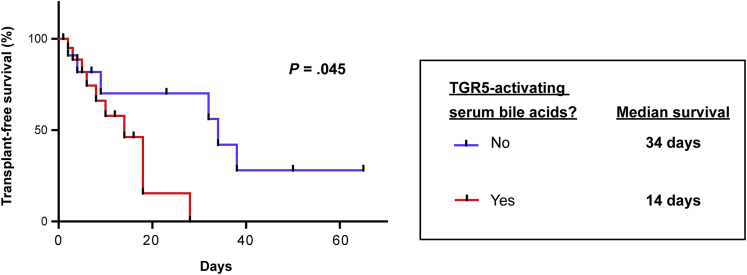
Monocyte dysfunction in liver failure is associated with a poor outcome. In particular, an inhibited ex vivo tumor necrosis factor α secretion after LPS challenge has been linked to high mortality.20 From our 33 liver failure patients investigated, 22 showed serum bile acids activating TGR5. We therefore compared the outcome of these 22 patients with the other 11 patients. Indeed, TGR5-activating serum bile acids were associated with significantly increased mortality (Figure 5). Serum bile acids already have been shown to be a risk factor for a fatal outcome in other studies, but were considered mainly as a surrogate parameter for liver failure.4, 5, 6 To exclude that the increased mortality of the 22 patients with TGR5-activating serum bile acids was the result of confounding factors, we assessed severity scores that have been described to predict mortality in liver failure and intensive care patients. As indicated by multivariable Cox proportional hazards regression, severity scores and TGR5 activation by serum bile acids predicted mortality independently (Table 4). Furthermore, TGR5-activating serum bile acids were a risk factor for fatal outcome independent of the patients’ total serum bile acid concentration (Table 5), suggesting an active role of TGR5 in the pathogenesis of liver failure.
Figure 5.
TGR5-activating serum bile acids are associated with increased mortality. Critically ill patients with liver failure were grouped according to their serum bile acid profiles. Patients with TGR5-activating serum bile acids had a significantly shorter survival (n = 22, red line) as compared with patients without (n = 11, blue line).
Table 4.
Mortality Hazard Ratios for TGR5-Activating Serum Bile Acids and Severity Scores
| Predictors | HR | 95% CI | P value |
|---|---|---|---|
| TGR5-activating serum bile acids | 7.785 | 1.294–46.853 | .025 |
| MELD-Na | 1.300 | 1.075–1.570 | .007 |
| SAPS II | 0.887 | 0.808–0.974 | .012 |
| APACHE II | 1.279 | 1.054–1.552 | .013 |
| SOFA | 1.386 | 1.052–1.827 | .020 |
NOTE. Cox proportional hazard regression.
APACHE II, Acute Physiology And Chronic Health Evaluation II; HR, hazard ratio; MELD-Na, Model for End-Stage Liver Disease-Sodium; SAPS II, Simplified Acute Physiology Score II; SOFA, Sequential Organ Failure Assessment Score.
Table 5.
Mortality Hazard Ratios for TGR5-Activating Serum Bile Acids and Total Serum Bile Acid Concentration
| Predictors | HR | 95% CI | P value |
|---|---|---|---|
| TGR5-activating serum bile acids | 10.600 | 1.801–62.392 | .009 |
| Total serum bile acid concentration | 0.995 | 0.990–1.001 | .078 |
NOTE. Cox proportional hazard regression.
TGR5 Activation Is Caused by the Additive Effect of Conjugated Primary and Tertiary Serum Bile Acids
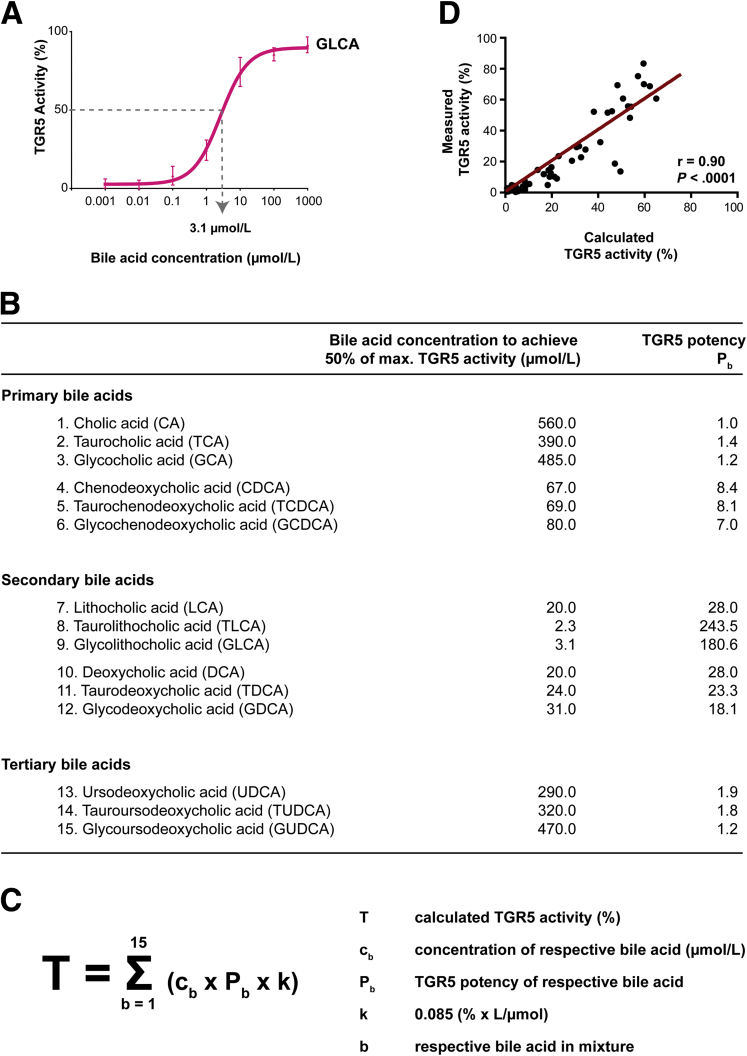
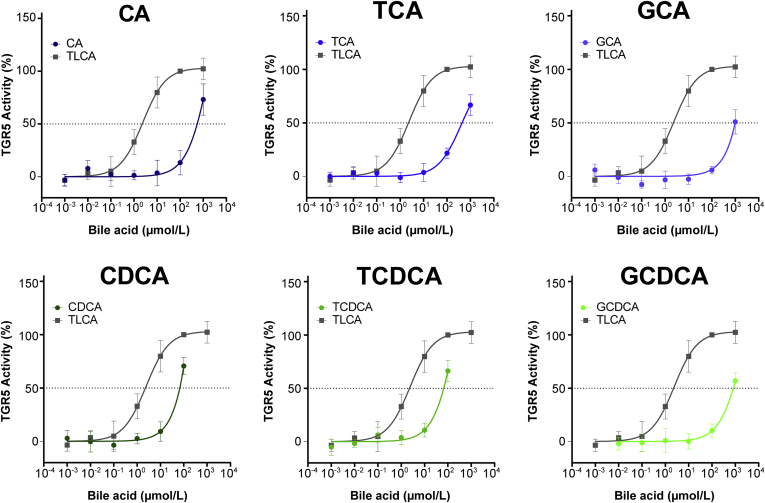
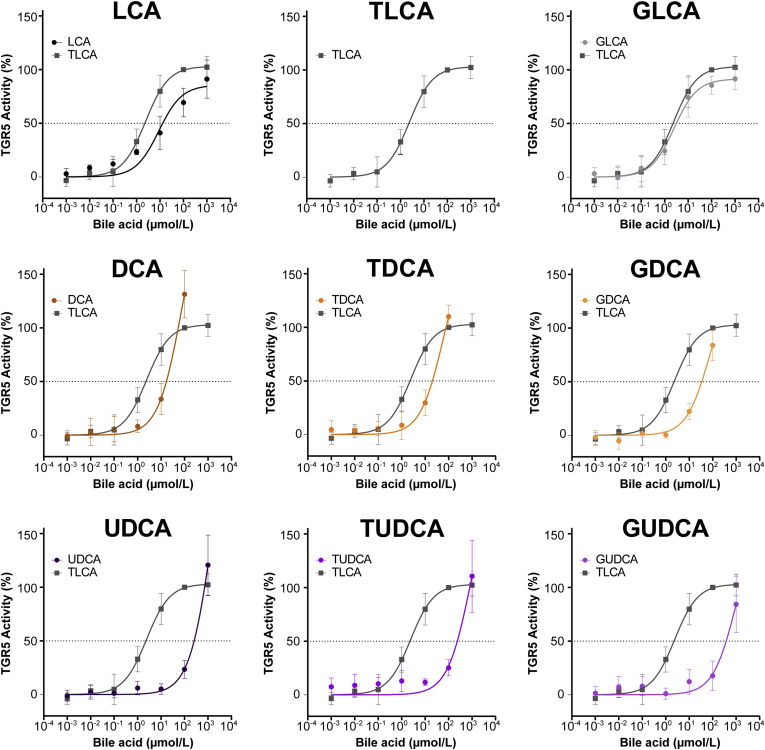
The human serum bile acid profile consists of circulating primary bile acids (bile acids, which are produced in the liver), secondary bile acids (primary bile acids, which have been modified by gut microbiota), and tertiary bile acids (ursodeoxycholic acid derivatives, ie, secondary bile acids that have been metabolized further by the liver or were administered directly as a drug). To assess which serum bile acids contribute to TGR5 activation, we used the TGR5 NanoBRET assay to determine the TGR5 potency of the 15 most abundant human serum bile acids. Therefore, we quantified the concentration of each bile acid that was required to achieve the half-maximal effect of TLCA (the most potent naturally occurring bile acid).9 One exemplary calculation is shown in Figure 6A. The curves for all bile acids are shown in Figure 7 and Figure 8. The potency of each bile acid to activate TGR5 was expressed as equivalent of cholic acid (Figure 6B).
Figure 6.
TGR5 activation is caused by the additive effect of conjugated primary and tertiary bile acids. (A) TGR5 potency was measured for each bile acid (eg, glycolithocholic acid [GLCA]) at increasing concentrations. The concentration that was required to achieve the half-maximal effect of TLCA was calculated. (B) TGR5 potencies of bile acids were expressed as a multiple of cholic acid potency. (C) Formula to calculate the TGR5 activity induced by serum bile acids using the bile acid concentrations and TGR5 potencies (potencies of bile acids). (D) Calculated and measured TGR5 activity correlated closely, N = 66, Spearman correlation rank test. max., maximum.
Figure 7.
TGR5 activation by primary bile acids. TGR5 activity was measured for each bile acid at increasing concentrations. The concentration that was required to achieve the half-maximal effect of TLCA was calculated. CA, cholic acid; CDCA, chenodeoxycholic acid; GCA, glycocholic acid; GCDCA, glycochenodeoxycholic acid; TCA, taurocholic acid; TCDCA, taurocholic acid.
Figure 8.
TGR5 activation by secondary and tertiary bile acids. TGR5 activity was measured for each bile acid at increasing concentrations. The concentration that was required to achieve the half-maximal effect of TLCA was calculated. DCA, deoxycholic acid; GDCA, glycodeoxycholic acid; GLCA, glycolithocholic acid; GUDCA, glycoursodeoxycholic acid; LCA, lithocholic acid; TDCA, taurodeoxycholic acid; TUDCA, tauroursodeoxycholic acid.
Secondary bile acids were the most potent activators of TGR5 (up to 243.5 times as potent as cholic acid). Primary and tertiary bile acids only showed mild to moderate TGR5 potency (1.0–8.4 times as potent as cholic acid). This is in line with previous studies comparing the effects of primary, secondary, and tertiary bile acids.9,21,22
We hypothesized that the effect of different bile acids might be additive in mixtures. Consequently, the total TGR5 activity induced by bile acid mixtures might be calculated by the sum of effects of the single bile acids. To address this question, we calculated the TGR5 activity induced by each single serum bile acid using the product of (1) its bile acid concentration, (2) its respective TGR5 potency, and (3) k, a constant determined by previous measurements. The products of all serum bile acids were added for the total TGR5 activity (Figure 6C).
By using this formula, we calculated the TGR5 activity induced by the serum bile acids for each patient and healthy volunteer. Each calculated TGR5 activity was compared with the TGR5 activity measured by TGR5 NanoBRET. Interestingly, there was a significant and close correlation of calculated and measured TGR5 activity (Figure 6D). This indicates that bile acids found in serum indeed had an additive effect.
We now compared the serum bile acid profiles of the 22 patients with TGR5-activating serum bile acids with their age- and sex-matched controls (Table 3). The bile acid profiles showed significantly enhanced levels of conjugated cholic acid, chenodeoxycholic acid, and ursodeoxycholic acid. Using our formula (Figure 6C), we calculated the TGR5 activity induced by each single bile acid (Table 3, last column). Chenodeoxycholic acid conjugates, although only moderate activators of TGR5, caused most of the serum TGR5 activation in our patients (16.5% and 17.4% of a total of 50% serum TGR5 activity). Furthermore, the weak TGR5 activators glycocholic acid, glycoursodeoxycholic acid, and tauroursodeoxycholic acid contributed to TGR5 activation.
Discussion
The immunomodulatory effects of the bile acid receptor TGR5 were first described in 2003.9 Since then, many studies have confirmed immunosuppressive actions of single bile acids on monocytes and macrophages mediated by TGR5.9,14,23 However, studies investigating the effects of complex bile acid profiles are lacking. To overcome the challenge of multiple TGR5-activating bile acids in human serum, we established a highly specific and sensitive TGR5 NanoBRET assay. This assay showed that none of the tested healthy individuals, but 67% of our patients, had serum bile acid profiles activating TGR5. These bile acid profiles inhibited monocyte function significantly (Figure 3). In line with previous studies investigating single bile acids in macrophages, we showed that the proinflammatory cytokine secretion upon LPS stimulation was reduced significantly, while the anti-inflammatory functions were preserved.14,23
Functionally impaired monocytes are an abundant hallmark of liver failure, which makes patients highly susceptible to infections and fatal outcome.24 Impaired monocytes in liver failure are characterized by their inability to produce proinflammatory cytokines upon LPS challenge,25,26 a feature that can be induced in healthy monocytes by incubating them with plasma from liver failure patients.27,28 Our study showed a link between increased serum bile acids, which frequently are seen in liver failure, and immune dysfunction. Bile acids that accumulate in the blood of patients with liver failure activate TGR5 in monocytes and thus impair monocyte function. Because monocyte dysfunction leading to infection is a major complication of liver dysfunction and an important driver of mortality,24 TGR5 and bile acid metabolism might be interesting targets for pharmacologic approaches. Diets, changes of the microbiome, certain medications, and treatments alter the bile acid composition in the intestine and serum.29, 30, 31, 32 Consequently, therapeutic bile acids (ursodeoxycholic acid, obeticholic acid), inhibitors of reabsorption, and liver dialysis might be evaluated for their potential to reduce TGR5 activity and improve immune function.
Our study showed that TGR5-activating serum bile acids were associated with increased mortality in liver failure. Strikingly, these serum bile acids were a risk factor for a fatal outcome independent of disease severity and commonly used prognostic scores (Table 4). Predicting the mortality in liver failure is of utmost importance because it is the base for allocation of donor livers worldwide.33 A detailed knowledge of individual risk factors is pivotal for an equitable allocation policy. In most countries, deceased donor livers are allocated to the patients with the Model for End-Stage Liver Disease score.33 TGR5 activation by serum bile acids might reflect the increased waitlist mortality seen in some cholestatic liver diseases (eg, primary biliary cirrhosis).34,35 Nevertheless, our observation on mortality was conducted in only 33 patients. Whether studies with larger numbers of subjects will confirm this correlation remains to be investigated.
In our patients, TGR5 activation was not owing to the occurrence of a single bile acid, but was the sum of effects of several bile acids (Table 3). (Patho-)Physiological conditions, in which mixtures of ligands bind to the same receptor with different affinities, frequently are observed in clinical practice. One strategy to handle the pharmacologic complexity is to define the potencies of the ligands. Prominent examples are glucocorticoid potencies and the relative strength of opioids. These potencies allow an estimate of the effect of drug combinations. We assessed the potency of each bile acid relative to cholic acid. In line with the literature, secondary bile acids, that is, lithocholic acid, deoxycholic acid, and their conjugates, had the highest potency to activate TGR5.9,22 Using these potencies, we calculated the TGR5 activity induced by bile acid mixtures as the sum of TGR5 activity of single bile acids (Figure 6C). Calculated TGR5 activity in 66 individuals correlated closely with the measured TGR5 activity (Figure 6D). The formula therefore might be a useful tool to understand the complex bile acid profiles seen in patients. It allows calculation of the effect of bile acids on TGR5 directly from serum bile acid profiles and could be used to compare bile acid effects before and after distinct therapies.
This study had several limitations: first and foremost, our study was limited to 33 patients and further studies will be needed to ascertain the TGR5-mediated effects of bile acid profiles in liver failure. Second, according to departmental standard operating procedures, all patients with liver failure received UDCA therapy (3 × 250 mg/d). Although the weak TGR5 activator UDCA and its conjugates accumulate in serum, UDCA therapy lowers serum concentration of bile acids that are potent TGR5 activators.29,36 The effect of bile acid profiles therefore might be underestimated in our study.
Taken together, TGR5-activating serum bile acids were abundant in critically ill patients with liver failure and associated with a fatal outcome. In addition to other recently identified mechanisms,25,37 serum bile acids may contribute to immune dysfunction in liver diseases. Therefore, circulating bile acids and TGR5 are interesting targets for new therapeutic approaches.
Materials and Methods
Patients and Healthy Volunteers
Patients with liver failure treated in the surgical intensive care unit of Jena University Hospital were screened for study inclusion. Inclusion criteria were as follows: (1) age ≥18 years; (2) plasma disappearance rate of indocyanine green <10%/minute; (3) plasma bilirubin level >170 μmol/L; and (4) international normalized ratio >1.5 and/or overt hepatic encephalopathy (grades II–IV according to West Haven Criteria). Liver failure included ACLF, acute liver failure, and liver graft failure. According to our departmental standard operating procedures, all patients with liver failure received UDCA therapy (3 × 250 mg/d). All clinical data including scores (Acute Physiology And Chronic Health Evaluation II, Simplified Acute Physiology Score II, and Sequential Organ Failure Assessment Score) and laboratory data were obtained on the day of blood sampling. Model for End-Stage Liver Disease including serum sodium score was calculated from these data. Survival was monitored until death, discharge from the hospital, or liver transplantation. Follow-up time was censored at the date of discharge from the hospital or liver transplantation. For each patient, an age- and sex-matched healthy volunteer was selected. Healthy volunteers had no past medical history of cholestatic or liver diseases and were classified according to American Society of Anesthesiology (ASA) physical status classification system as ASA I or II.
Study Approval
The protocol was approved by the Ethical Committee of the Jena University Hospital. All patients or their legal representatives and healthy volunteers provided written informed consent before inclusion in the study.
Bile Acids
Total bile acid levels in serum were quantified by an enzymatic colorimetric kit (Abbott Laboratories, Abbott Park, IL). Bile acid profiles were measured after protein precipitation and filtration from serum samples. Samples were loaded on an Agilent 1200 high-performance liquid chromatography system (Agilent Technologies, Santa Clara, CA) with a CTC-PAL autosampler coupled to an API 4000 Triple Quadrupole mass spectrometer with electrospray ionization source (AB SCIEX, Framingham, MA). All chromatographic separations were performed with a reverse-phase analytical column.
Pure bile acids and bile acid profiles for NanoBRET assays or cell culture experiments were remixed in the respective assay buffer or medium. Tauroursodeoxycholic acid was purchased from VWR International (Darmstadt, Germany), glycolithocholic acid was purchased from Santa Cruz Biotechnology (Dallas, TX), and all other bile acids were obtained from Sigma-Aldrich (Taufkirchen, Germany). Bile acid stock solutions were dissolved in dimethyl sulfoxide (DMSO).
NanoBRET
TGR5 plasmid was purchased from Sino Biological (Beijing, China). ADRB2 plasmid already has been used before by our group.38 Nanoluciferase (Promega Corp, Madison, WI) was appended directly to the respective C terminus of the receptors via an isothermal assembly reaction (New England Biolabs, Ipswich, MA) using polymerase chain reaction products, which introduced 20– to 40-base pair overlap. Plasmid sequences were controlled by restriction enzyme digest and sequencing (Eurofins, Ebersberg, Germany). The plasmid encoding a fragment of Gsα subunit (mini-Gs) tagged with Venus fluorophore was generated as described previously.17 HEK293T cells were transiently transfected with mini-Gs-Venus and either TGR5-Nluc or ADRB2-Nluc in a 7:3 ratio using Effectene Transfection Reagent (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions. After incubation for 24 hours at 37°C and 5% CO2, transfected cells were seeded into white opaque 96-well plates. The following day, medium was replaced with BRET buffer supplemented with Nano-Glo luciferase substrate (Promega Corp). Pure bile acids or bile acid profiles were remixed in BRET buffer. NanoBRET measurements were conducted using the following: (1) 0.5% DMSO solvent control (negative control); (2) 100 μmol/L TLCA (maximal stimulation; later set to 100%); and (3) bile acid or bile acid profile. All samples contained 0.5% DMSO. The emission of the NanoBRET donor and acceptor was measured on a Synergy Neo2 plate reader (BioTek, Winooski, VT), using a custom-made filter (excitation bandwidth, 541–550 nm; emission, 560–595 nm; fluorescence filter, 620/15 nm). BRET ratios were calculated by the division of the acceptor emission by the donor emission, individually corrected for baseline measurements and buffer conditions and normalized to the maximal stimulation. Maximal stimulation induced by 100 μmol/L TLCA for TGR5 and 100 μmol/L isoprenaline for ADRB2 was set to 100%. Each measurement was performed in triplicate and repeated at least 2 times.
Cell Culture
Primary human monocytes were isolated from healthy donors by negative selection (Thermo Fisher Scientific, Waltham, MA) as described previously.39 Monocyte purity was 92.5% ± 0.9%, and viability was 99.1% ± 0.5%. Monocytes suspended in RPMI-1640 (Dutch modification) including 10% fetal calf serum, 100 U/mL penicillin, 100 U/mL streptomycin (all Sigma-Aldrich), 1 mmol/L pyruvate and 2 mmol/L GlutaMax (both Thermo Fisher Scientific), were seeded into a 96-well plate (5 × 105 cells/well). After 60 minutes, cells were stimulated with the following: (1) 0.1% DMSO solvent control (negative control); (2) 100 μmol/L TLCA (positive control); (3) mean bile acid profile of the 22 patients with TGR5-activating serum bile acids; or (4) mean bile acid profile of 22 respective healthy controls. All samples contained 0.1% DMSO. After 60 minutes of incubation, 100 ng/mL LPS from Escherichia coli (Sigma-Aldrich) was added to the medium for 6 hours. Supernatants were frozen at -80°C for further analysis. Cytokine levels were assessed by a bead-based immunoassay (BioLegend, San Diego, CA). Each experiment was performed in duplicate.
pLV-C-GFPSpark (EV, LVCV-01) and pLV–G-protein–coupled bile acid receptor 1–GFPSpark (TGR5 vector, HG11472-ACGLN) were purchased from Sino Biological, Inc. THP-1 cells (TIB-202; ATCC, Manassas, VA) were stably transduced with both vectors and sorted for equal GFP expression. TGR5 expression was verified by quantitative real-time polymerase chain reaction. Total RNA was isolated using the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany). Primer sequences are listed in Table 6. Cytokine experiments were performed as described earlier. Adenosine 3′,5′-cyclic monophosphate was measured using an enzyme-linked immunosorbent assay kit (Cayman, Ann Arbor, MI) after incubation of cells in serum-free medium including 500 μmol/L 3-isobutyl-1-methylxanthine (Sigma-Aldrich).
Table 6.
Primers Used for Quantitative Real-Time Polymerase Chain Reaction
| Gene name | Gene symbol | Gene ID | Forward primer, 5’ to 3’ | Reverse primer, 5’ to 3’ |
|---|---|---|---|---|
| Target gene | ||||
| TGR5 | GPBAR1 | 151306 | CTCCCAGGCTATCTTCCCAG | ACAGAGAGGAAGGCAGCAG |
| Housekeeping genes | ||||
| Calnexin | CANX | 821 | AAGGTTACTTACAAAGCTCC | ACTCCTTCATTTCCTCTACC |
| Ubiquitin C | UBC | 7316 | TCGCAGCCGGGATTTGG | TCACGAAGATCTGCATTGTCAAG |
| Proteasome 20S subunit β3 | PSMB3 | 5691 | GTCCACATCATCGAGAAGGACA | GGCTCTGGGAACAGGGTTAG |
Statistics
Statistical analysis was conducted as indicated in the respective figure legends using GraphPad Prism (La Jolla, CA) or SPSS (IBM, Chicago, IL).
Acknowledgment
The authors thank our patients and healthy volunteers for their participation in this study. The authors gratefully acknowledge the support of registered study nurses. The authors thank Julia Gantner, from the Institute of Medical Statistics, Computer and Data Sciences (University Hospital Jena), for supporting our statistical analyses.
CRediT Authorship Contributions
Julia Leonhardt, MD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Raphael Silvanus Haider (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Visualization: Supporting; Writing – review & editing: Supporting)
Christoph Sponholz, PD, MD (Conceptualization: Supporting; Investigation: Supporting; Project administration: Supporting; Writing – review & editing: Supporting)
Silke Leonhardt, MD (Conceptualization: Supporting; Formal analysis: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Julia Drube, PhD (Methodology: Supporting; Writing – review & editing: Supporting)
Katrin Spengler, PhD (Investigation: Supporting; Writing – review & editing: Supporting)
Diana Mihaylov, MD (Investigation: Supporting; Writing – review & editing: Supporting)
Sophie Neugebauer, PhD (Investigation: Supporting; Writing – review & editing: Supporting)
Michael Kiehntopf, PD, MD, PhD (Methodology: Supporting; Writing – review & editing: Supporting)
Nevin A. Lambert, PhD (Resources: Supporting; Writing – review & editing: Supporting)
Andreas Kortgen, MD (Conceptualization: Supporting; Project administration: Supporting; Writing – review & editing: Supporting)
Tony Bruns, MD (Writing – review & editing: Supporting)
Frank Tacke, MD (Writing – review & editing: Supporting)
Carsten Hoffmann, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Methodology: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Michael Bauer, MD (Conceptualization: Supporting; Funding acquisition: Supporting; Project administration: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Regine Heller, MD (Conceptualization: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Methodology: Supporting; Project administration: Supporting; Resources: Supporting; Supervision: Lead; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the Federal Ministry of Education and Research, Germany, FKZ 01EO1502 (J.L.), by the Department of Anesthesiology and Critical Care Medicine, Jena University Hospital, and the Institute for Molecular Cell Biology, Jena University Hospital.
References
- 1.Horvatits T., Drolz A., Trauner M., Fuhrmann V. Liver injury and failure in critical illness. Hepatology. 2019;70:2204–2215. doi: 10.1002/hep.30824. [DOI] [PubMed] [Google Scholar]
- 2.Cordoba J., Ventura-Cots M., Simon-Talero M., Amoros A., Pavesi M., Vilstrup H., Angeli P., Domenicali M., Gines P., Bernardi M., Arroyo V., Consortium Canonic Study Investigators of EASL-CLIF Consortium Characteristics, risk factors, and mortality of cirrhotic patients hospitalized for hepatic encephalopathy with and without acute-on-chronic liver failure (ACLF) J Hepatol. 2014;60:275–281. doi: 10.1016/j.jhep.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Malinchoc M., Kamath P.S., Gordon F.D., Peine C.J., Rank J., ter Borg P.C. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology. 2000;31:864–871. doi: 10.1053/he.2000.5852. [DOI] [PubMed] [Google Scholar]
- 4.Horvatits T., Drolz A., Roedl K., Rutter K., Ferlitsch A., Fauler G., Trauner M., Fuhrmann V. Serum bile acids as marker for acute decompensation and acute-on-chronic liver failure in patients with non-cholestatic cirrhosis. Liver Int. 2017;37:224–231. doi: 10.1111/liv.13201. [DOI] [PubMed] [Google Scholar]
- 5.Horvatits T., Drolz A., Rutter K., Roedl K., Langouche L., Van den Berghe G., Fauler G., Meyer B., Hulsmann M., Heinz G., Trauner M., Fuhrmann V. Circulating bile acids predict outcome in critically ill patients. Ann Intensive Care. 2017;7:48. doi: 10.1186/s13613-017-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Recknagel P., Gonnert F.A., Westermann M., Lambeck S., Lupp A., Rudiger A., Dyson A., Carre J.E., Kortgen A., Krafft C., Popp J., Sponholz C., Fuhrmann V., Hilger I., Claus R.A., Riedemann N.C., Wetzker R., Singer M., Trauner M., Bauer M. Liver dysfunction and phosphatidylinositol-3-kinase signalling in early sepsis: experimental studies in rodent models of peritonitis. PLoS Med. 2012;9 doi: 10.1371/journal.pmed.1001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mannes G.A., Thieme C., Stellaard F., Wang T., Sauerbruch T., Paumgartner G. Prognostic significance of serum bile acids in cirrhosis. Hepatology. 1986;6:50–53. doi: 10.1002/hep.1840060110. [DOI] [PubMed] [Google Scholar]
- 8.Copple B.L., Li T. Pharmacology of bile acid receptors: evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol Res. 2016;104:9–21. doi: 10.1016/j.phrs.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., Hinuma S., Fujisawa Y., Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 10.Sipe L.M., Chaib M., Pingili A.K., Pierre J.F., Makowski L. Microbiome, bile acids, and obesity: how microbially modified metabolites shape anti-tumor immunity. Immunol Rev. 2020;295:220–239. doi: 10.1111/imr.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiorucci S., Biagioli M., Zampella A., Distrutti E. Bile acids activated receptors regulate innate immunity. Front Immunol. 2018;9:1853. doi: 10.3389/fimmu.2018.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biagioli M., Carino A., Cipriani S., Francisci D., Marchiano S., Scarpelli P., Sorcini D., Zampella A., Fiorucci S. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol. 2017;199:718–733. doi: 10.4049/jimmunol.1700183. [DOI] [PubMed] [Google Scholar]
- 13.Keitel V., Spomer L., Marin J.J., Williamson C., Geenes V., Kubitz R., Haussinger D., Macias R.I. Effect of maternal cholestasis on TGR5 expression in human and rat placenta at term. Placenta. 2013;34:810–816. doi: 10.1016/j.placenta.2013.06.302. [DOI] [PubMed] [Google Scholar]
- 14.Yoneno K., Hisamatsu T., Shimamura K., Kamada N., Ichikawa R., Kitazume M.T., Mori M., Uo M., Namikawa Y., Matsuoka K., Sato T., Koganei K., Sugita A., Kanai T., Hibi T. TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn’s disease. Immunology. 2013;139:19–29. doi: 10.1111/imm.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makino I., Nakagawa S., Mashimo K. Conjugated and unconjugated serum bile acid levels n patients with hepatobiliary diseases. Gastroenterology. 1969;56:1033–1039. [PubMed] [Google Scholar]
- 16.Trottier J., Bialek A., Caron P., Straka R.J., Milkiewicz P., Barbier O. Profiling circulating and urinary bile acids in patients with biliary obstruction before and after biliary stenting. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan Q., Okashah N., Inoue A., Nehme R., Carpenter B., Tate C.G., Lambert N.A. Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J Biol Chem. 2018;293:7466–7473. doi: 10.1074/jbc.RA118.001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis N.D., Patnaude L.A., Pelletier J., Souza D.J., Lukas S.M., King F.J., Hill J.D., Stefanopoulos D.E., Ryan K., Desai S., Skow D., Kauschke S.G., Broermann A., Kuzmich D., Harcken C., Hickey E.R., Modis L.K. A GPBAR1 (TGR5) small molecule agonist shows specific inhibitory effects on myeloid cell activation in vitro and reduces experimental autoimmune encephalitis (EAE) in vivo. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki-Anzai S., Masuda M., Levi M., Keenan A.L., Miyazaki M. Dual activation of the bile acid nuclear receptor FXR and G-protein-coupled receptor TGR5 protects mice against atherosclerosis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berres M.L., Schnyder B., Yagmur E., Inglis B., Stanzel S., Tischendorf J.J., Koch A., Winograd R., Trautwein C., Wasmuth H.E. Longitudinal monocyte human leukocyte antigen-DR expression is a prognostic marker in critically ill patients with decompensated liver cirrhosis. Liver Int. 2009;29:536–543. doi: 10.1111/j.1478-3231.2008.01870.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen G., Wang X., Ge Y., Ma L., Chen Q., Liu H., Du Y., Ye R.D., Hu H., Ren R. Cryo-EM structure of activated bile acids receptor TGR5 in complex with stimulatory G protein. Signal Transduct Target Ther. 2020;5:142. doi: 10.1038/s41392-020-00262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 23.Haselow K., Bode J.G., Wammers M., Ehlting C., Keitel V., Kleinebrecht L., Schupp A.K., Haussinger D., Graf D. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J Leukoc Biol. 2013;94:1253–1264. doi: 10.1189/jlb.0812396. [DOI] [PubMed] [Google Scholar]
- 24.Triantafyllou E., Woollard K.J., McPhail M.J.W., Antoniades C.G., Possamai L.A. The role of monocytes and macrophages in acute and acute-on-chronic liver failure. Front Immunol. 2018;9:2948. doi: 10.3389/fimmu.2018.02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triantafyllou E., Pop O.T., Possamai L.A., Wilhelm A., Liaskou E., Singanayagam A., Bernsmeier C., Khamri W., Petts G., Dargue R., Davies S.P., Tickle J., Yuksel M., Patel V.C., Abeles R.D., Stamataki Z., Curbishley S.M., Ma Y., Wilson I.D., Coen M., Woollard K.J., Quaglia A., Wendon J., Thursz M.R., Adams D.H., Weston C.J., Antoniades C.G. MerTK expressing hepatic macrophages promote the resolution of inflammation in acute liver failure. Gut. 2018;67:333–347. doi: 10.1136/gutjnl-2016-313615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry P.A., Antoniades C.G., Carey I., McPhail M.J., Hussain M.J., Davies E.T., Wendon J.A., Vergani D. Severity of the compensatory anti-inflammatory response determined by monocyte HLA-DR expression may assist outcome prediction in cirrhosis. Intensive Care Med. 2011;37:453–460. doi: 10.1007/s00134-010-2099-7. [DOI] [PubMed] [Google Scholar]
- 27.Antoniades C.G., Khamri W., Abeles R.D., Taams L.S., Triantafyllou E., Possamai L.A., Bernsmeier C., Mitry R.R., O’Brien A., Gilroy D., Goldin R., Heneghan M., Heaton N., Jassem W., Bernal W., Vergani D., Ma Y., Quaglia A., Wendon J., Thursz M. Secretory leukocyte protease inhibitor: a pivotal mediator of anti-inflammatory responses in acetaminophen-induced acute liver failure. Hepatology. 2014;59:1564–1576. doi: 10.1002/hep.26933. [DOI] [PubMed] [Google Scholar]
- 28.Bernsmeier C., Pop O.T., Singanayagam A., Triantafyllou E., Patel V.C., Weston C.J., Curbishley S., Sadiq F., Vergis N., Khamri W., Bernal W., Auzinger G., Heneghan M., Ma Y., Jassem W., Heaton N.D., Adams D.H., Quaglia A., Thursz M.R., Wendon J., Antoniades C.G. Patients with acute-on-chronic liver failure have increased numbers of regulatory immune cells expressing the receptor tyrosine kinase MERTK. Gastroenterology. 2015;148:603–615 e14. doi: 10.1053/j.gastro.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 29.Manna L.B., Ovadia C., Lovgren-Sandblom A., Chambers J., Begum S., Seed P., Walker I., Chappell L.C., Marschall H.U., Williamson C. Enzymatic quantification of total serum bile acids as a monitoring strategy for women with intrahepatic cholestasis of pregnancy receiving ursodeoxycholic acid treatment: a cohort study. BJOG. 2019;126:1633–1640. doi: 10.1111/1471-0528.15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y., Li F., Zalzala M., Xu J., Gonzalez F.J., Adorini L., Lee Y.K., Yin L., Zhang Y. Farnesoid X receptor activation increases reverse cholesterol transport by modulating bile acid composition and cholesterol absorption in mice. Hepatology. 2016;64:1072–1085. doi: 10.1002/hep.28712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegade V.S., Kendrick S.F., Dobbins R.L., Miller S.R., Thompson D., Richards D., Storey J., Dukes G.E., Corrigan M., Oude Elferink R.P., Beuers U., Hirschfield G.M., Jones D.E. Effect of ileal bile acid transporter inhibitor GSK2330672 on pruritus in primary biliary cholangitis: a double-blind, randomised, placebo-controlled, crossover, phase 2a study. Lancet. 2017;389:1114–1123. doi: 10.1016/S0140-6736(17)30319-7. [DOI] [PubMed] [Google Scholar]
- 32.Sponholz C., Matthes K., Rupp D., Backaus W., Klammt S., Karailieva D., Bauschke A., Settmacher U., Kohl M., Clemens M.G., Mitzner S., Bauer M., Kortgen A. Molecular adsorbent recirculating system and single-pass albumin dialysis in liver failure--a prospective, randomised crossover study. Crit Care. 2016;20:2. doi: 10.1186/s13054-015-1159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tschuor C., Ferrarese A., Kuemmerli C., Dutkowski P., Burra P., Clavien P.A. Liver Allocation Study G. Allocation of liver grafts worldwide - is there a best system? J Hepatol. 2019;71:707–718. doi: 10.1016/j.jhep.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 34.Staufer K., Kivaranovic D., Rasoul-Rockenschaub S., Soliman T., Trauner M., Berlakovich G. Waitlist mortality and post-transplant survival in patients with cholestatic liver disease - Impact of changes in allocation policy. HPB (Oxford) 2018;20:916–924. doi: 10.1016/j.hpb.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Singal A.K., Fang X., Kaif M., Hasanin M., McGuire B.M., Kuo Y.F., Wiesner R.H. Primary biliary cirrhosis has high wait-list mortality among patients listed for liver transplantation. Transpl Int. 2017;30:454–462. doi: 10.1111/tri.12877. [DOI] [PubMed] [Google Scholar]
- 36.Batta A.K., Arora R., Salen G., Tint G.S., Eskreis D., Katz S. Characterization of serum and urinary bile acids in patients with primary biliary cirrhosis by gas-liquid chromatography-mass spectrometry: effect of ursodeoxycholic acid treatment. J Lipid Res. 1989;30:1953–1962. [PubMed] [Google Scholar]
- 37.O’Brien A.J., Fullerton J.N., Massey K.A., Auld G., Sewell G., James S., Newson J., Karra E., Winstanley A., Alazawi W., Garcia-Martinez R., Cordoba J., Nicolaou A., Gilroy D.W. Immunosuppression in acutely decompensated cirrhosis is mediated by prostaglandin E2. Nat Med. 2014;20:518–523. doi: 10.1038/nm.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann C., Leitz M.R., Oberdorf-Maass S., Lohse M.J., Klotz K.N. Comparative pharmacology of human beta-adrenergic receptor subtypes--characterization of stably transfected receptors in CHO cells. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:151–159. doi: 10.1007/s00210-003-0860-y. [DOI] [PubMed] [Google Scholar]
- 39.Leonhardt J., Grosse S., Marx C., Siwczak F., Stengel S., Bruns T., Bauer R., Kiehntopf M., Williams D.L., Wang Z.Q., Mosig A.S., Weis S., Bauer M., Heller R. Candida albicans beta-glucan differentiates human monocytes into a specific subset of macrophages. Front Immunol. 2018;9:2818. doi: 10.3389/fimmu.2018.02818. [DOI] [PMC free article] [PubMed] [Google Scholar]