Abstract
Background
Chronic gastroesophageal reflux disease (GERD) is a major risk factor for the development of metaplastic Barrett's esophagus (BE) and its progression to esophageal adenocarcinoma (EAC). Uncontrolled accumulation of reactive oxygen species (ROS) in response to acidic bile salts (ABS) in reflux conditions can be lethal to cells. In this study, we investigated the role of APE1/REF1 in regulating nuclear erythroid factor-like 2 (NRF2), the master antioxidant transcription factor, in response to reflux conditions.
Results
We found that APE1 protein was critical for protecting against cellular ROS levels, oxidative DNA damage, double strand DNA breaks, and cell death in response to conditions that mimic reflux. Analysis of cell lines and de-identified tissues from patients with EAC demonstrated overexpression of both APE1 and NRF2 in EAC cells, as compared to non-neoplastic esophageal cells. Using reflux conditions, we detected concordant and prolonged increases of APE1 and NRF2 protein levels for several hours, following transient short exposure to ABS (20 min). NRF2 transcription activity, as measured by ARE luciferase reporter, and expression of its target genes (HO-1 and TRXND1) were similarly increased in response to ABS. Using genetic knockdown of APE1, we found that APE1 was required for the increase in NRF2 protein stability, nuclear localization, and transcription activation in EAC. Using knockdown of APE1 with reconstitution of wild-type and a redox-deficient mutant (C65A) of APE1, as well as pharmacologic APE1 redox inhibitor (E3330), we demonstrated that APE1 regulated NRF2 in a redox-dependent manner. Mechanistically, we found that APE1 is required for phosphorylation and inactivation of GSK-3β, an important player in the NRF2 degradation pathway.
Conclusion
APE1 redox function was required for ABS-induced activation of NRF2 by regulating phosphorylation and inactivation of GSK-3β. The APE1-NRF2 network played a critical role in protecting esophageal cells against ROS and promoting cell survival under oxidative reflux conditions.
Keywords: Acidic bile salt, GERD, NRF2, APE1, GSK-3β, ROS, Oxidative stress
Highlights
-
•
APE1 protected esophageal neoplastic cells from acidic bile salts (ABS)-induced oxidative stress, DNA damage and cell death.
-
•
APE1 regulated oxidative stress through mediating the activity of master antioxidant transcription factor, NRF2.
-
•
APE1 was required to maintain NRF2 stability in the nucleus and suppressed NRF2 protein degradation, mainly through GSK-3β.
-
•
APE1 redox function was required for proper phosphorylation and inactivation of GSK-3β, and subsequent activation of NRF2.
1. Introduction
Esophageal cancer is the sixth most common cause of cancer-related deaths, with a five-year survival rate of less than 25% worldwide [1]. Over the past few decades, epidemiological trends have shown a massive shift in esophageal cancer histology. There are more esophageal adenocarcinoma (EAC) cases than esophageal squamous cell carcinoma (ESCC), in the Western world, with a five-year survival rate of around 18% [[2], [3], [4], [5]]. Chronic gastroesophageal reflux disease (GERD) is the main risk factor for the development of a metaplastic glandular condition in the lower esophagus, known as Barrett's esophagus (BE), and its progression to EAC [6]. Exposure of esophageal cells to gastric acid and bile salts in patients with GERD generates high levels of reactive oxygen species (ROS) and oxidative DNA damage [7,8]. Uncontrolled accumulation of ROS and DNA damage would be lethal to cells. Therefore, esophageal cells must develop a protective antioxidant capacity to protect against the lethal effects of ROS.
Nuclear erythroid factor 2, like 2 (NRF2), is an important antioxidant transcription factor that regulates the expression of a large panel of genes through the antioxidant response element (ARE) binding sites in their promoter regions [9]. Targets of NRF2 include genes that encode antioxidant enzymes and proteins involving in xenobiotic metabolism and protection against heavy metal toxicity [10]. Under normal physiological conditions, NRF2 activity and levels are controlled by Kelch-like ECH -associated protein 1 (KEAP1) in the cytosol by KEAP1-CUL3-mediated ubiquitination and degradation mechanism. Under stress conditions, an increase in ROS levels mediates transient and rapid oxidation of KEAP1 with immediate release of NRF2 from the KEAP1-CUL3 complex. This leads to transient accumulation and nuclear translocation of NRF2, where it binds to the conserved ARE region to activate a battery of antioxidative and cellular defense targets [11]. NRF2 functions can safeguard against carcinogenesis via quick activation of several antioxidant genes with potent enzymatic activity for quenching ROS [12]. Its high constitutive levels have been detected in several cancer types such as lung [13], gall bladder [14], epithelial ovarian [15], cervical [16], and squamous cell carcinoma of esophagus and skin [17]. These findings raised questions about its double-edged sword activity where it can act as an anti-tumorigenic or pro-tumorigenic factor in a context dependent fashion. It has been reported that activation of NRF2 in cancer cells promoted metastasis [18] and conferred resistance to chemo and radiotherapy [19]. It is, therefore, important to understand the etiological risk factors and context-dependent regulation of NRF2 to develop preventive and therapeutic approaches.
Apurinic/apyrimidinic endonuclease 1 (APE1), also known as reduction-oxidation factor-1 (REF1), is a multifunctional enzyme with 36.5 KDa molecular weight. APE1 plays a key role in base excision repair pathway and its redox function has been associated with activation of several redox-dependent transcription factors such as activator protein-1 (AP-1), nuclear factor kappa B (NFκB), hypoxia-inducible factor 1-α (HIF1-α), and signal transducer activator of transcription 3 (STAT3) [[20], [21], [22]]. Recent studies have shown that APE1 promotes cancer cell invasion by activating ARF6-mediated MMP-14 [23] and cell survival and proliferation by activating EGFR–STAT3 through its redox-dependent activity [22]. APE1 is also shown to suppress JNK and p38 MAP-kinases [21] upon exposure to acidic bile salts.
The connection between APE1 and NRF2 remains controversial. While one study suggested that APE1 might negatively regulate NRF2 in PaCa-2 cells and cancer-associated fibroblast lines [24], another study suggested that APE1 has an opposite function in lung cancer cell lines [25]. These findings raised the possibility of a cell context-dependent regulation of APE1 and NRF2 in carcinogenesis. In this study, we investigated the role of APE1 and NRF2 in oxidative stress signaling induced by reflux conditions in EAC tumorigenesis. The results suggest that NRF2 activation in response to reflux conditions is dependent on APE1 redox function.
2. Experimental details and methods
2.1. Cell culture and chemicals
CPB (dysplastic BE) and FLO1 (EAC) cell lines were obtained from American Tissue culture collection (ATCC, Manassas, VA). OE33 was a kind gift from Dr. David Beer (University of Michigan). CPB cells were maintained in DMEM/F12 media with 5% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin-streptomycin (P/S). Additional nutrients include bovine pituitary extract, hydrocortisone, recombinant epidermal growth factor (EGF), 1X insulin transferrin selenium supplement and l-adenine. FLO1 and OE33 cell lines were maintained in DMEM and RPMI 1640 medium, respectively, supplemented with 10% FBS and 1% P/S. All cells were grown at 37 °C in a humidified incubator with 5% CO2. All cell lines were routinely authenticated and tested for mycoplasma contamination.
2.2. Acidic bile salts treatment
A cocktail of bile salts (BS) was prepared with an equimolar concentration of a mixture of bile salts (20 mM each) that included glycocholic acid (GCA), taurocholic acid (TCA), glycodeoxycholic acid (GDCA), glycochenodeoxycholic acid (GCDCA) and deoxycholic acid (DCA). Cells were treated with 100 μM of BS cocktail for 20 min in corresponding acidic culture media (pH 4.0, ABS), followed by recovery in regular media. The ABS cocktail mimicked the bile acids mixture in the distal esophagus in patients with GERD [26].
2.3. Antibodies and reagents
The following antibodies were purchased from commercial sources; NRF2 antibody from ABCAM (ab62352, Cambridge, MA, USA), APE1 antibody from Thermo Fisher Scientific (MA5-31586; Waltham, MA, USA), KEAP 1 antibody from Proteintech (10503-2-AP, Rosemont, IL, USA), β-actin antibody from Sigma-Aldrich (A5441), GSK-3-β (Total) (12456S), p-GSK-3-β (S9) (5558S), PARP, c-PARP and β-tubulin antibodies from Cell Signaling Technology (Danvers, MS, USA) and p84 from Genetex (Irvine, CA, USA). The E3330, an APE1 redox inhibitor, was obtained from Novus Biologicals (NBP1-49581, Centennial, CO, USA) and GSK-3β inhibitors; lithium chloride (Calbiochem) and CHIR-98014 (Selleck chemicals). Control siRNA and APE1siRNA were purchased from Dharmacon (ON-TARGET plus Human APEX1 siRNA, L-010237-00-0005). Transfection reagents, lipojet (for siRNAs) and polyjet (for plasmid DNAs), were purchased from SignaGen laboratories (Rockville, MD, USA).
2.4. Small hairpin RNA (shRNA) and APE1 expression vectors
APE1 shRNA and control shRNA lentiviral plasmids were obtained from Vector Builder Inc. (Santa Clara, CA, USA). Plasmids were co-transfected with second generation packaging mix (Abm) into 293LTV cells (CellBiolabs, San Diego, CA), following the manufacturer's protocol. After 72 h transfection, the media supernatants were collected and stored at −80 °C. CPB, FLO1 and OE33 cells were infected with control or APE1shRNA media supernatants in the presence of polybrene (4 μg/mL) for 72 h. Stable cell lines expressing control or APE1shRNA were selected using puromycin and used for further experiment. The FLAG-tagged coding sequence of APE1 and its mutant C65A were cloned into pcDNA3.1 mammalian expression plasmid (Invitrogen, Carlsbad, CA USA).
2.5. Cell fractionation
Cells were treated with 100 μM ABS for 20 min, followed by recovery in regular media at different time points. Cells were trypsinized and processed for cytosol and nuclear separation as per the protocol provided in the cell fractionation kit (Cell Signaling Technologies, USA). Briefly, after the treatment, the cells were trypsinized, washed with 1X PBS, and centrifuged for 500 g for 5 min. Then, the resulted pellet was mixed with 500 μL of cytoplasmic isolation buffer (CIB), vortexed for 5 s, incubated in ice for 5 min, and centrifuged at 500 g for 5 min. The supernatant was collected as the cytosolic fraction. Then, the pellet was remixed with 500 μL of membrane isolation buffer (MIB), vortexed for 15 s, incubated for 5 min, and centrifuged at 8000 g for 5 min. The resulted supernatant was collected as a membrane fraction. Then the resulted pellet was mixed with Cytoskeletal/Nuclear isolation buffer (CyNIB) and sonicated twice at 20X speed. The isolated cytosolic and nuclear fractions were mixed with the 4X Laemmli sample buffer and electrophoresed using SDS-PAGE.
2.6. Real-time RT-PCR
Total RNA was extracted from the cells using Trizol manually using chloroform, isopropanol method. Total 1 μg/sample RNA was subjected to cDNA synthesis using the TaqMan reverse transcription reagents kit (Applied Biosystems, Thermofisher Scientific, Waltham, MA, USA). The primers for NRF2 target genes such as HO-1, NQO1, TRXND1 and GR were designed using primer 3 online tools (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) and were obtained from Integrated DNA Technologies (Coralville, Iowa). Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out using an iCycler (Biorad laboratories) with the threshold cycle number determined by iCycler software version 3.0. All the reactions were performed in triplicates. The threshold numbers were averaged. The fold expression was calculated and normalized to the average CT value of HPRT1 using the method reported previously [27].
2.7. Luciferase reporter assay
Briefly, the cells were seeded in 12 well plates. The next day, the cells were co-transfected with PGL 4.37 [luc2P/ARE/Hygro] reporter (Promega, Madison, WI), as a measure of NRF2 transcription activity, along with renilla as the internal control using polyjet DNA transfecting agent. 24 h after transfection, the cells were treated with a 200 μM mixture of bile salts in pH7 medium or PBS for another 24 h. The cells were harvested and lysed with 1X luciferase passive lysis buffer. Luciferase activity was measured after adding the luciferase reagent and renilla after adding the stop solution using a dual-luciferase reporter assay system (Promega) in a FLUOstar OPTIMA microplate reader (BMG LABTECH, Cary, NC). Luciferase activity was calculated by normalizing the luciferase with the corresponding renilla value and represented as relative luciferase activity.
2.8. Western blot assay
Cells were lysed using the RIPA buffer (Santa Cruz), sonicated and centrifuged at 13,000 rpm for 10 min at 4 °C. The supernatant was collected and mixed with a 4X LDS sample buffer, heated at 70 °C for 10 min, electrophoresed at 100 V for 90 min, and transferred into the nitrocellulose membrane. The membrane was then blocked with 5% BSA for 1 h at room temperature and incubated with the primary antibodies against NRF2 (1:1000; ABCAM), APE1 (1: 7000, Thermofisher Scientific), KEAP-1 (1:1000; Proteintech), GSK-3β (1:1000, Cell Signaling), p-GSK-3β (S9) (1:1000, Cell Signaling) and β-actin (1:10,000; Sigma-Aldrich) for overnight. The membrane was then washed thrice with the 1X TBST and then incubated with their corresponding secondary antibodies (1:5000 dilution) for 2 h. The membranes were again washed thrice. After the washing, the immune complex was detected by the ECL kit. The amount of protein relative to the loading control was quantified by Quantity One software (BioRad Laboratories, USA).
2.9. 3D organotypic culture
3D organotypic cultures (OTC) were performed using CPB and OE33 cells, following the method previously described [28]. Briefly, human esophageal fibroblasts (ScienCell, Carlsbad, CA, USA) were seeded into a 3D matrix (75,000 cells/well) containing collagen I (High concentration rat-tail collagen, Corning) and Matrigel (BD Biosciences, Franklin Lakes, NJ USA) and incubated for 7 days at 37 °C. Following incubation, epithelial cells were seeded (500,000 cells/well) on the top of the fibroblast matrix. Cultures were then allowed to grow for an additional 7 days and treated with ABS (100 μM, pH4) or PBS for 30 min, followed by recovery in complete media for 3 h. OTC cells were then harvested, fixed in 70% ethanol, and sent to histology core laboratory for paraffin embedding, H&E staining, and slide sectioning for immunocytochemistry.
2.10. Immunocytochemistry of 3D organotypic cell culture
Paraffin-embedded 3D OTC slides were deparaffinized and rehydrated following standard protocol. Antigen retrieval was performed by boiling the slides in 1 M Tris EDTA, pH 9.0 for 10 min. Slides were allowed to cool down to room temperature before incubation in 5% BSA in PBS for 1 h. Primary antibodies anti-APE1 (Thermo Fisher Scientific, mouse monoclonal, #13B 8E5C2) and anti-NRF2 (Abcam) were incubated with slides overnight at 4 °C in a humidified chamber. The next day following incubation, the slides were washed with PBS and incubated with Alexa Fluor-conjugated anti-mouse or anti-rabbit secondary antibody (Fluor-488 or Fluor-586) for 1 h at room temperature, protected from light. The slides were washed again with PBS and mounted with a Vectashield mounting medium with DAPI (Vector Laboratories). Images were captured with the BZ-X710 KEYENCE All-in-one fluorescence microscope (Atlanta, GA).
2.11. Immunofluorescence
1.2 × 104 cells were seeded in an eight well slide chamber. The cells were washed with PBS and treated with ABS (100 μM/20 min), washed again with PBS, added complete media for the recovery for 1, 3, 6 h. Cells were fixed with 4% paraformaldehyde for 45 min, washed once with 1X ice-cold PBS and added permeabilization buffer for 10 min in ice. The cells were then washed twice and blocked using the goat antiserum for 20 min at room temperature. Then the cells were incubated with primary antibody anti-NRF2 (1:200 dilution), anti-APE1 (1:500 dilution), anti-KEAP1 (1:200 dilution), anti-8-oxoguanine (1:100) (MAB3560, Sigma-Aldrich, St. Louis, MO, USA), anti-phosphor histone H2A.X (Ser 139) (1:250) for overnight. The next day, the cells were washed with 1X PBS thrice, followed by incubation with Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 568 goat anti-mouse (1:500) secondary antibodies for 45 min. After 45 min, the cells were washed thrice, and the slides were mounted with Vectashield mounting medium with DAPI and sealed with a coverslip. The images were captured by using the BZ-X710 KEYENCE All-in-one fluorescence microscope (Atlanta, GA).
2.12. Detection of intracellular ROS levels
The intracellular ROS levels were determined using flow cytometry for CM-H2DCFDA dye staining [29]. Upon stress, CM-H2DCFDA passively diffuses into cells, where its acetate groups are cleaved by intracellular esterase and its thiol-reactive chloromethyl group reacts with intracellular glutathione and other thiols. Subsequent oxidation yields a fluorescent adduct that is trapped inside the cell. Oxidation of these probes can be detected by monitoring the increase in fluorescence with flow cytometry at Ex/Em: ~492–495/517–527 nm. Briefly, 1.5 × 105 counts of OE33 control and APE1shRNA cells were seeded into 12 well plates. On the next day, cells were treated with 100 μM ABS for 10 min, washed with PBS, and incubated with 5 μM CM-H2DCFDA for 30 min. Then the cells were washed with PBS, trypsinized and resuspended with 500 μL phenol red-free media. Cells were then subjected to flow cytometry analysis in Flow Cytometry Shared Resource and a total of 10, 000 cells were analyzed per sample.
2.13. Annexin V staining
Apoptosis analysis was performed using PE Annexin V Apoptosis Detection Kit I (BD Pharmingen™, San Jose, CA, USA) following the manufactory's protocol. 2.5 × 105 cells were seeded in duplicate in 6-well plates and on the next day, the cells were transfected with 60 nM Control and APE1 siRNA. On the following evening, the cells were split again into 12 well plates (1.5 × 105 cells/well) as triplicates. 48 h after transfection, cells were treated with ABS (100 μM, pH 4.0) or PBS for 20 min, followed by recovery in complete media for 3 h. Cells were then harvested and stained with Annexin-V and propidium iodide (PI). The cells were washed with PBS and re-suspended in a binding buffer (HEPES buffered saline solution supplemented with 2.5 mM CaCl2) and then subjected to fluorescence-activated cell sorting (FACS) analysis using a flow cytometer (Becton Dickinson). Apoptotic cell death was determined by counting the cells that stained positive for Annexin-V.
2.14. Statistical analysis
Biochemical experiments were repeated 3 times in at least two independent cell lines and conditions. Quantified results were expressed as mean ± SD. All the statistical analyses were performed using GraphPad Prism, version 8.0 (GraphPad Software). P < 0.05 was considered statistically significant.
3. Results
3.1. APE1 protected esophageal neoplastic cells against ABS-induced oxidative stress, DNA damage and cell death
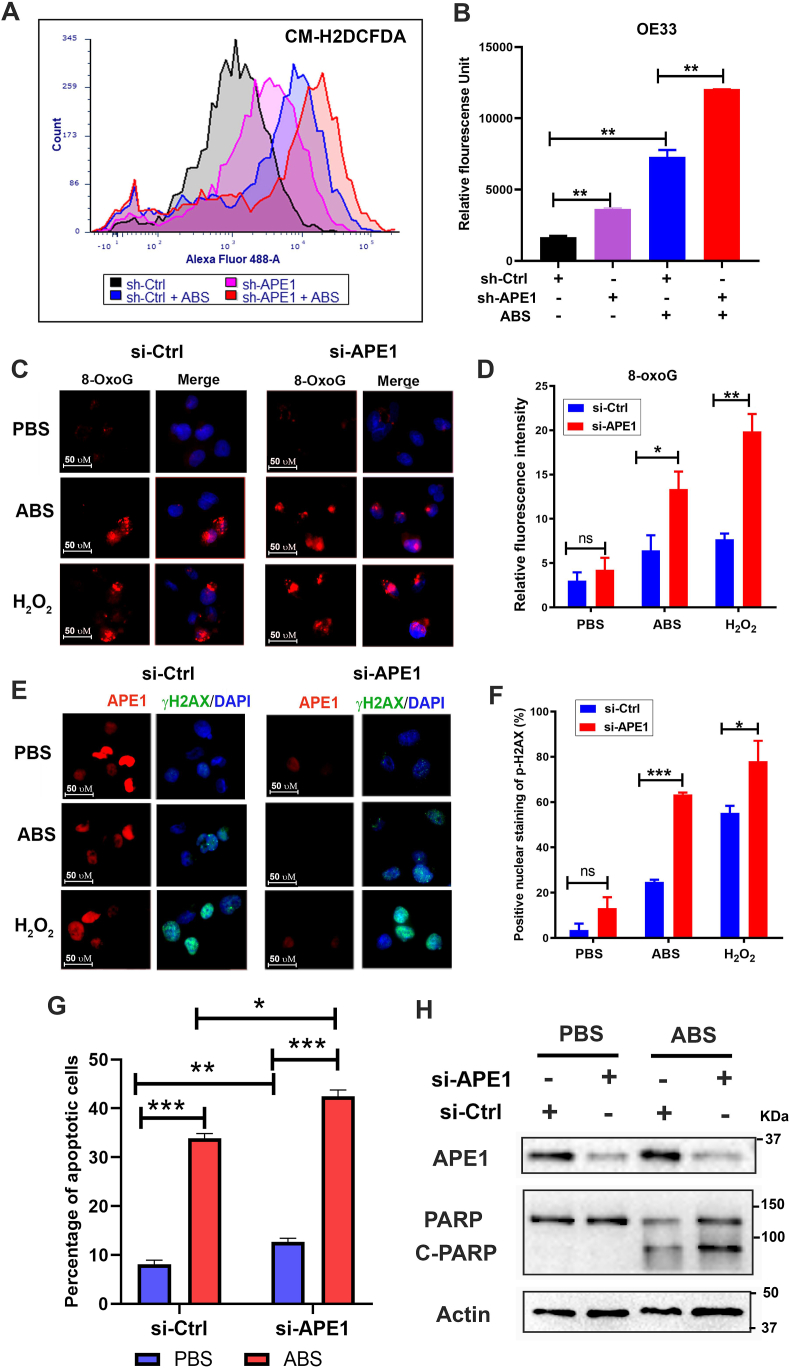
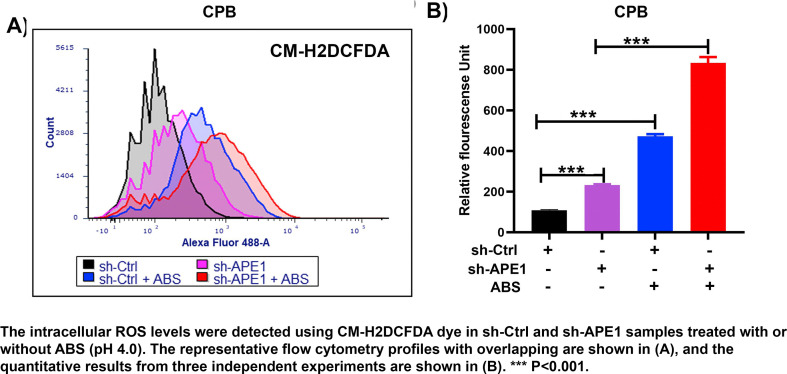
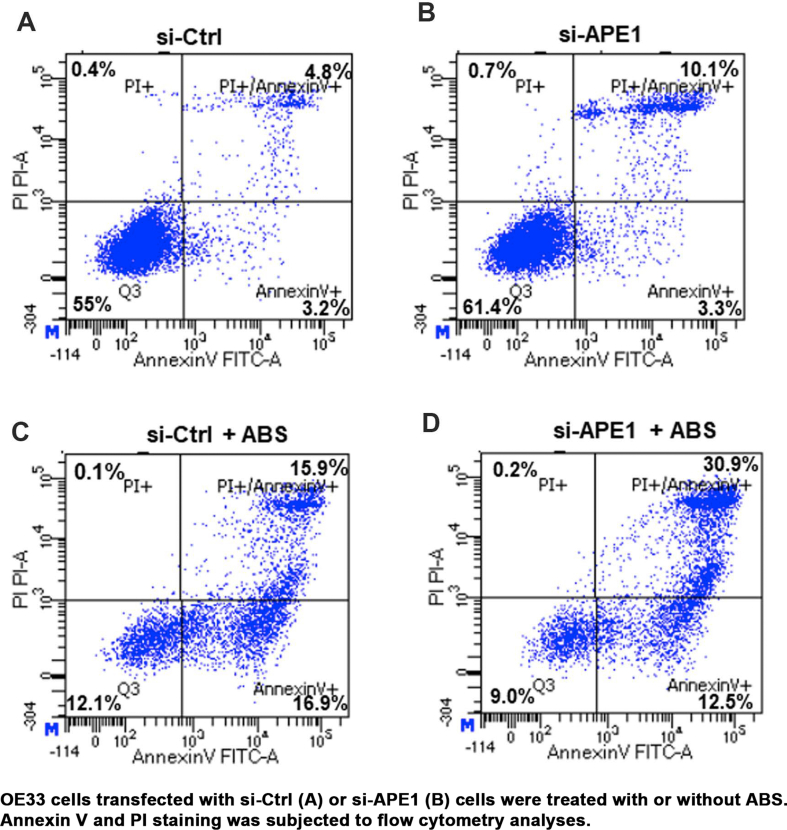
We and others have reported that exposure of esophageal cells to acidic bile salts (ABS), mimicking GERD episodes, induced significant ROS and oxidative stress [[30], [31], [32], [33]]. Therefore, neoplastic esophageal cells must develop mechanisms to survive in this harsh oxidative environment. To determine if APE1 could regulate oxidative stress in esophageal neoplastic cells under reflux conditions, we treated these cells with an ABS cocktail to mimic GERD episodes. By using CM-H2DCFDA, an intracellular ROS indicator, we found that silencing of APE1 significantly increased the ROS production in OE33 cells and promoted ROS generation in response to ABS treatment (P < 0.05) (Fig. 1 A and B). Similar results were obtained in CPB cells (P < 0.001) (Supplementary Fig. S1 A and B). Next, we investigated the levels of 8-OxoG following APE1 knockdown (si-APE1) and exposure to ABS, or H2O2, as a positive control. The 8-oxoguanine (8-OxoG) is a biomarker for oxidative DNA damage [34]. We detected a significant increase in 8-OxoG immunostaining following exposure to ABS or H2O2 in APE1 knockdown conditions, as compared with controls (P < 0.05) (Fig. 1C and D). By using gamma-H2AX (γ-H2AX, p-H2AX (s139)), a maker of DNA double-strand DNA breaks, we detected a significant increase in γ-H2AX immunostaining following exposure to ABS or H2O2 in conditions of APE1 knockdown, as compared to controls (P < 0.05) (Fig. 1 E and F). Uncontrolled accumulation of oxidative stress and DNA damage is lethal to cells. In line with this, the knockdown of APE1 significantly sensitized esophageal neoplastic cells to ABS-induced genotoxic stresses. APE1 knockdown promoted cell death, as evidenced by flow cytometry of Annexin V and Western blotting of cleaved PARP (Fig. 1G and H and Supplementary Fig. S2). In addition to apoptosis, bile acid exposure may also induce other cell death types, such as necroptosis, and ferroptosis, which require further investigations in the future. These results demonstrated that APE1 is an important antioxidant pro-survival factor that protected neoplastic esophageal cells against harsh oxidative reflux conditions.
Fig. 1.
Loss of APE1 resulted in increased oxidative stress, DNA damage, and promoted cell death in response to reflux conditions. (A–B) Flow cytometry analyses of OE33 cells to detect the intracellular ROS levels using CM-H2DCFDA dye in APE1 knockdown (sh-APE1) and control (sh-Ctrl) cells treated with or without ABS (pH 4.0). Representative flow cytometry profiles are shown in (A). The quantitative results from 3 independent experiments are shown in (B). (C–F) OE33 cells were transfected with si-Ctrl and si-APE1 for 48 h, followed by acidic bile salts (ABS) treatment for 20 min and recovery in full medium for 3 h. Cells treated with 100 μM H2O2 were used as a positive control. 8-Oxoguanine (8-OxoG), an oxidative DNA damage marker, was used for immunofluorescence. Representative immunofluorescence images are shown in (C) and the quantitative data, using ImageJ, is shown in panel (D). p-H2AX (S139, γH2AX, green), a DNA double-strand breaks marker, and APE1 (red) were used for immunofluorescence. The representative images are shown in panels, (E) and the quantitative data is shown in panel (F). DAPI (blue) was used as a nuclear counterstain. (G) Flow cytometry analysis of annexin V staining in si-Ctrl and si-APE1 cells with or without ABS treatment. The quantitative analysis data from three independent experiments are shown. (H) Western blotting analysis of cleaved PARP in si-Ctrl and si-APE1 cells with ABS or control PBS treatment. *P < 0.05; **P < 0.01; ***P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. ABS exposure induced ROS-dependent APE1 upregulation and activation of NRF2
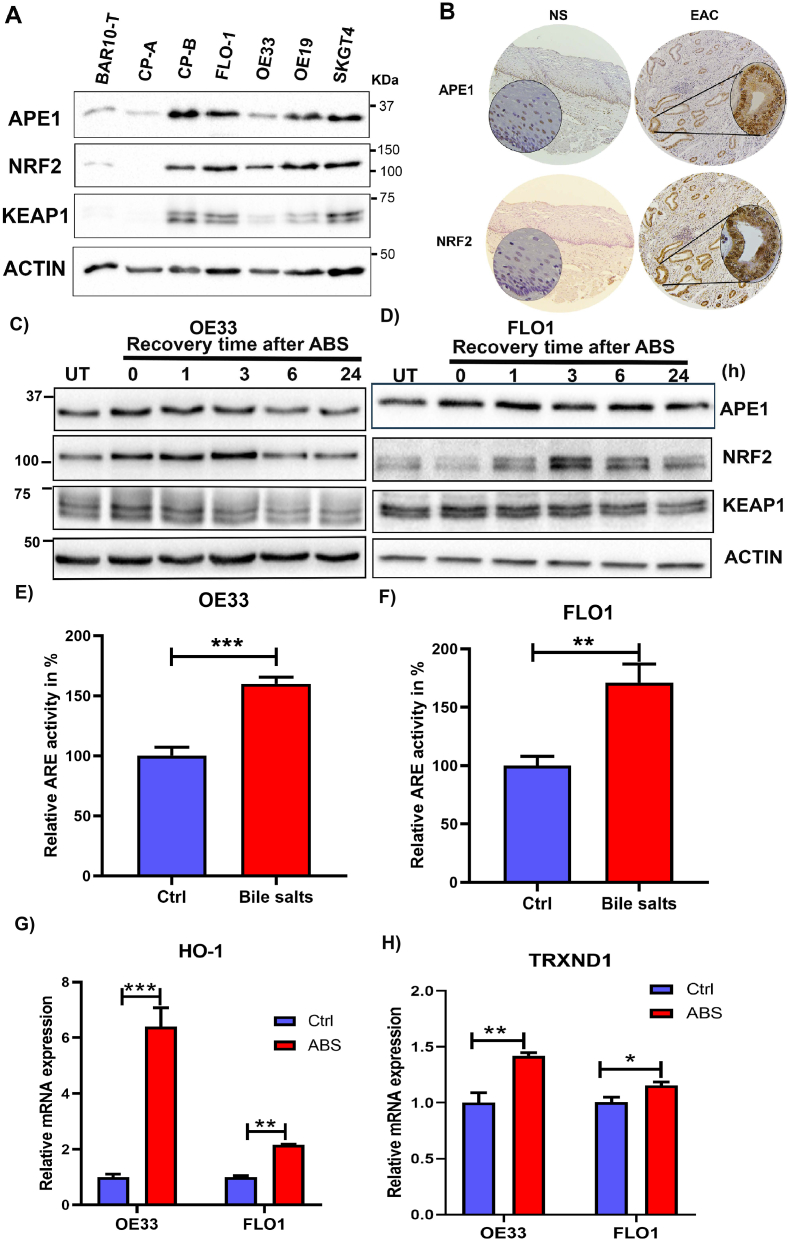
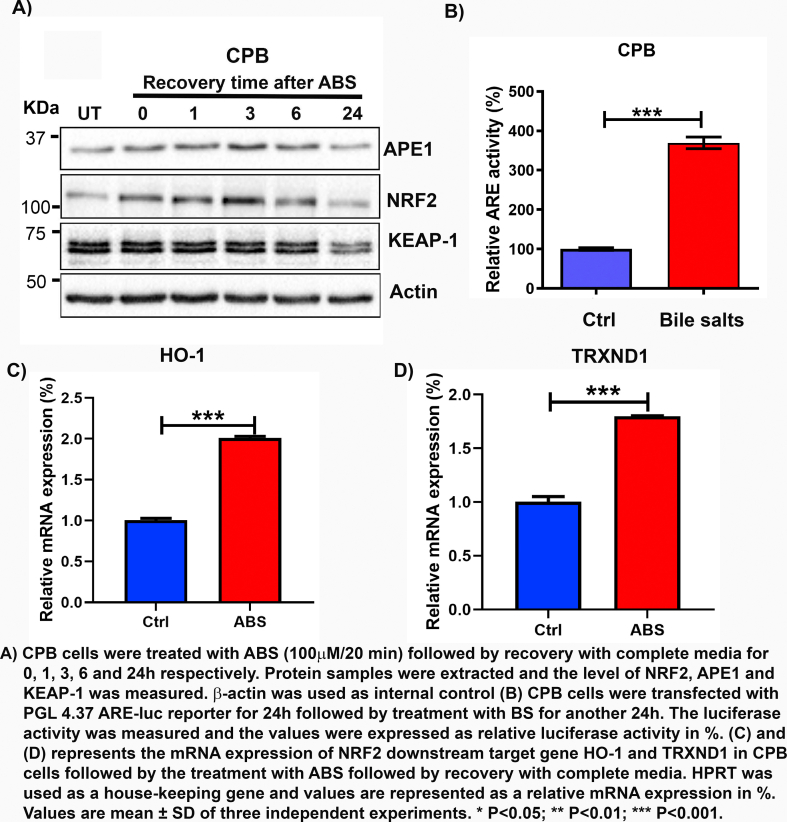
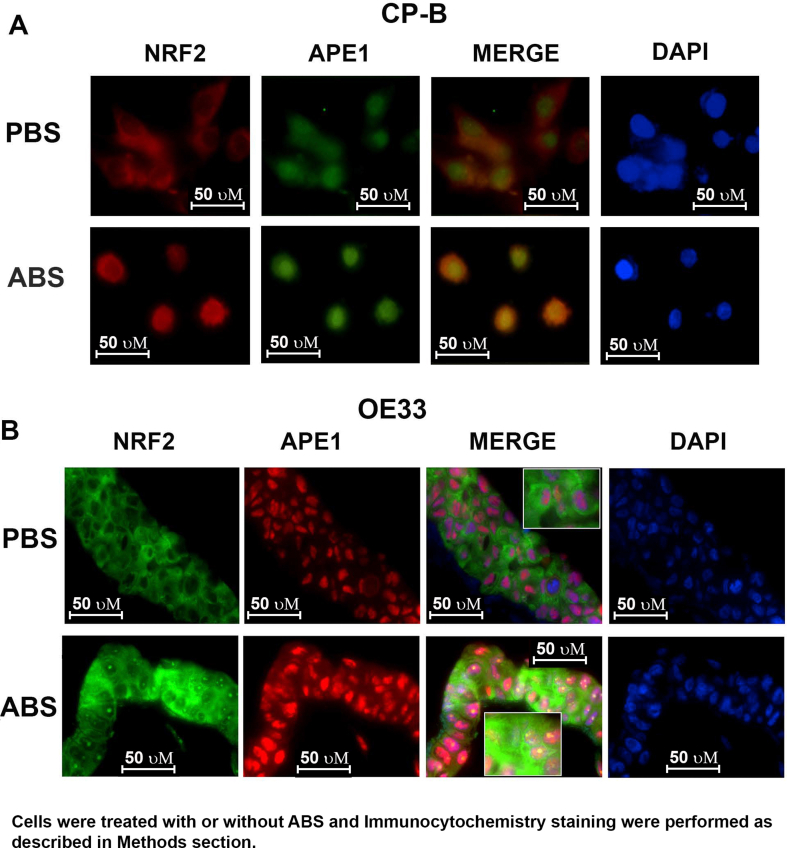
We previously reported that APE1 is overexpressed in dysplastic BE and EACs [21,22]. Our above findings of the role of APE1 in regulating ROS levels prompted us to explore the underlying mechanisms. NRF2 is a known master regulator of intracellular redox homeostasis and transactivates a wide spectrum of antioxidant genes under various stress conditions [35]. Interestingly, we found that both APE1 and NRF2 were significantly upregulated in neoplastic cell lines, compared to Barrett's cell lines (BAR10-T and CP-A) (Fig. 2A). We also observed high levels of APE1 and NRF2 protein in EAC tissue samples compared to the normal esophagus (Fig. 2B). To mimic the pathophysiological condition of GERD, we treated the high-grade dysplastic BE (CPB) and EAC (OE33 and FLO1) cells with ABS (100 μM or 200 μM, pH 4.0) for 20 min followed by recovery in complete media for 0, 1, 3, 6 and 24 h post-treatment. Immunoblot analyses revealed that the levels of both APE1 and NRF2 were increased simultaneously at 1 h and 3 h recovery time points and returned to basal level after 24 h (Fig. 2C and D, Supplementary Fig. S3A). Using an ARE luciferase reporter assay as a measure of NRF2 transcription activity, we detected a significant increase in transcriptional activity of NRF2 (P < 0.01) in cells treated with bile salts, as compared with control cells (Fig. 2 E and F, Supplementary Fig. S3B). Consequently, we found a significant increase in the expression levels of NRF2 target genes, HO-1 and TRXND1, in cells exposed to ABS, compared to control cells (Fig. 2 G and H, Supplementary Fig. S3 C and D, P < 0.05). Immunofluorescence staining of NRF2 and APE1 in CPB (in 2D culture) and OE33 (in 3D organotypic culture) further confirmed the increased protein expression of NRF2 and APE1 in a similar pattern (Supplementary Fig. S4 A and B).
Fig. 2.
APE1 and NRF2 were induced by acidic bile salts. (A) Western blot analysis of APE1, NRF2 and KEAP1 protein levels in esophageal cell lines from Barrett's (BAR10-T, CPA), dysplastic Barrett's (CPB) and EAC (FLO1, OE33, OE19 and SKGT4). (B) Immunohistochemistry staining of APE1 and NRF2 in a representative normal esophagus and an esophageal adenocarcinoma tissue samples (x10) with insets of higher magnification (×40). (C–D) Western blot analysis of APE1, NRF2 and KEAP-1 levels in OE33, and FLO1 cells. Cells were exposed to 100 μM or 200 μM (FLO1) ABS for 20 min with recovery in the complete media for the indicated time points. Whole-cell lysates were used for western blotting. (E, F) The ARE luciferase reporter assay for the NRF2 transcriptional activity in OE33 and FLO1 cells, following treatment with a mixture of bile salts (200 μM) or PBS (Ctrl). The luciferase reporter activity values were normalized to β-gal expression levels and are represented as percentage luciferase activity relative to control (set as 100%). (G, H) qRT-PCR analyses of NRF2 downstream target genes, HO-1 and TRXND1 in FLO1 and OE33 cells treated with ABS. Values are mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
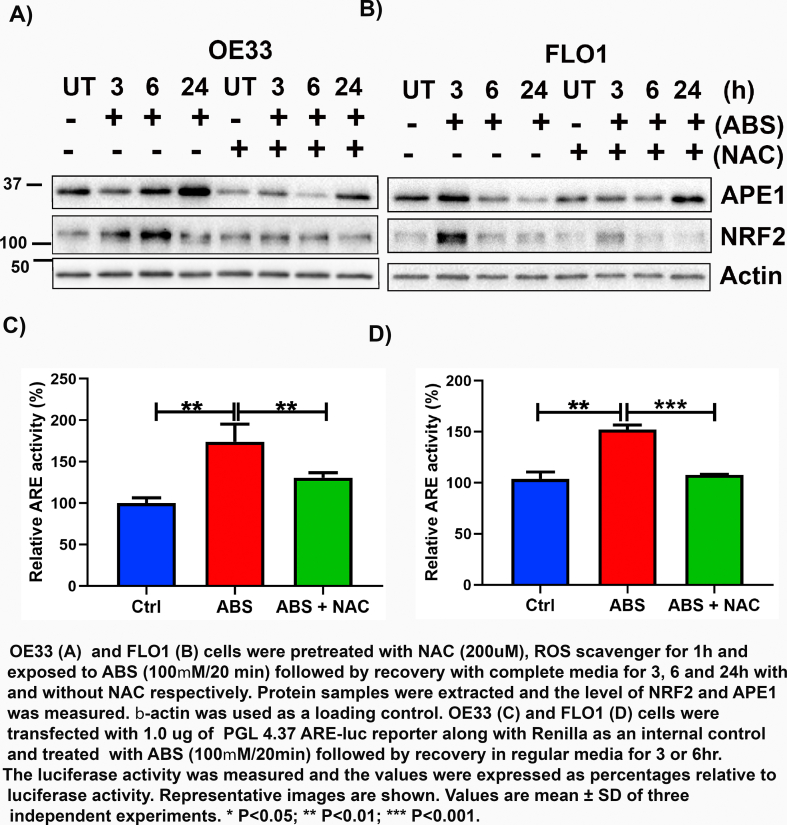
Next, we investigated if the induction of APE1 and NRF2 by ABS is mediated through ROS. OE33 and FLO1 cells were pretreated with N-acetyl cysteine (200 μM), a known ROS scavenger, for 1 h and then treated with ABS (100 μM/20 min) followed by recovery with corresponding media for 3, 6 and 24 h with and without NAC, respectively. We found that pretreatment with NAC abolished the ABS-mediated increase in both APE1 and NRF2 levels in Western blot analyses (Supplementary Fig. S5 A-B), as well as NRF2 activity in ARE reporter assay (Supplementary Fig. S5C-D). These results suggest that APE1 regulation of NRF2 in conditions of ABS was largely ROS-dependent.
3.3. Induction and regulation of NRF2 were dependent on APE1 in EAC cells
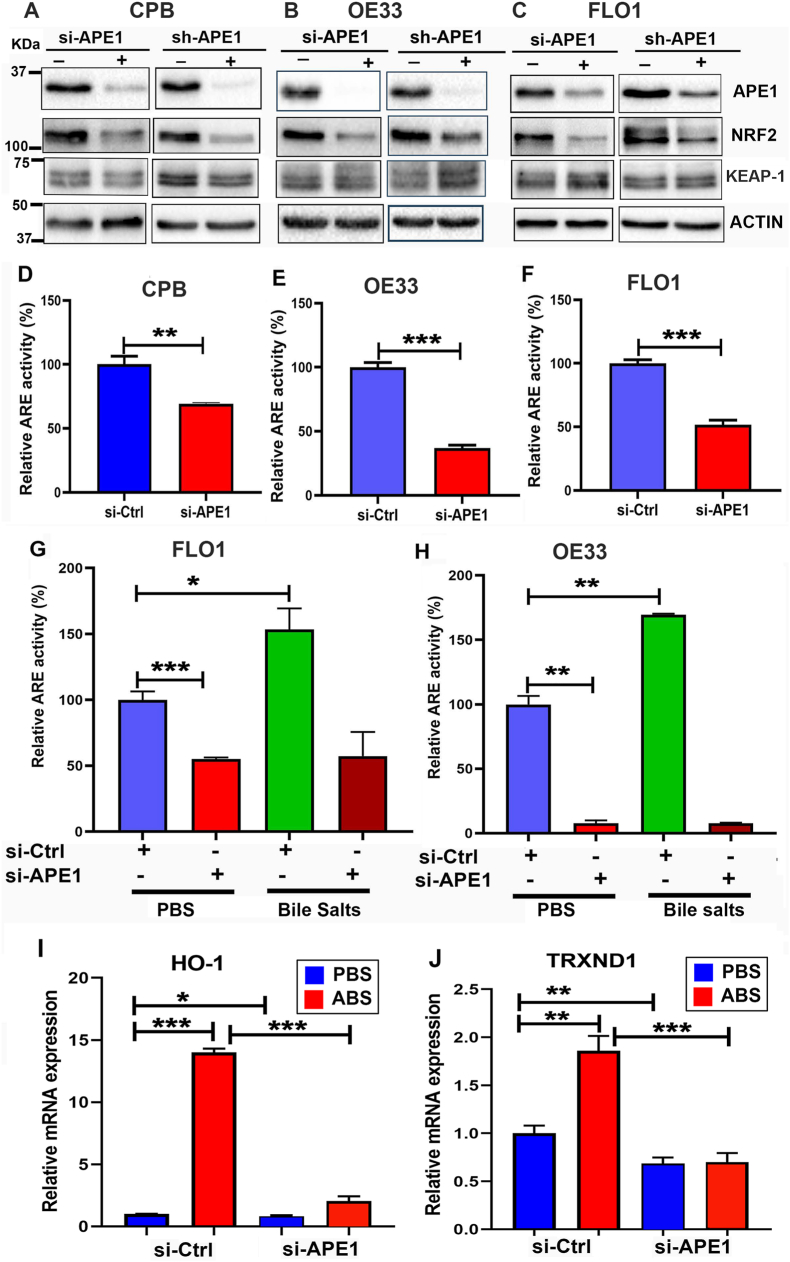
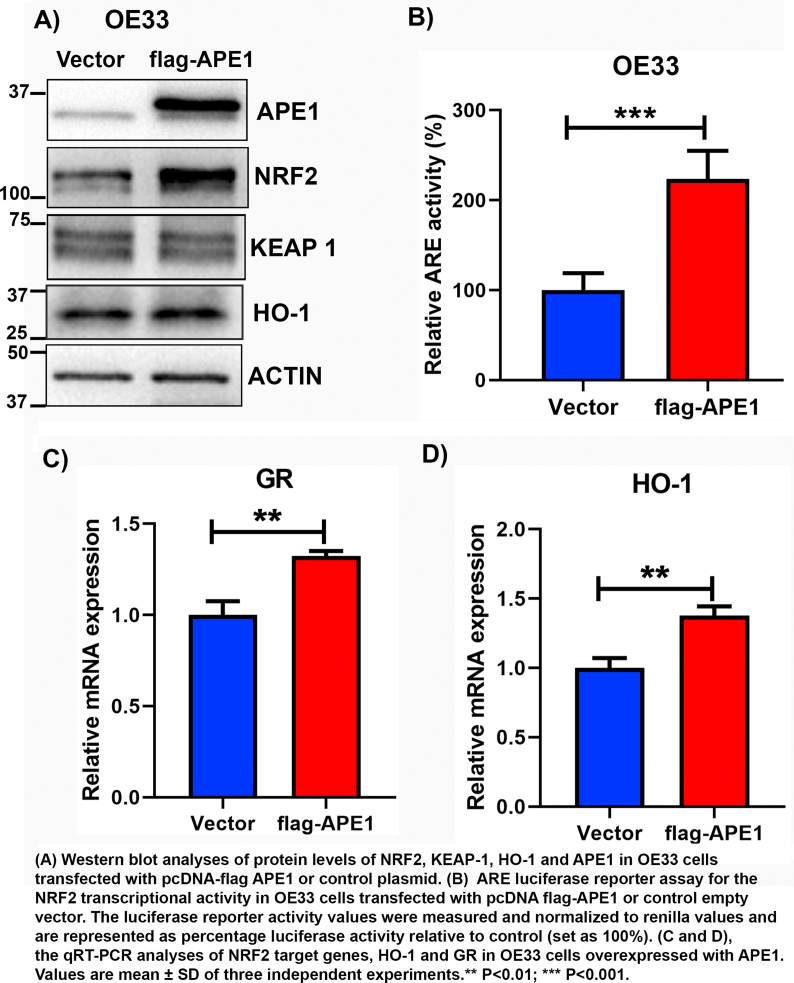
To investigate the mechanistic relationship between APE1 and NRF2, we knocked down APE1 using transient siRNA (si-APE1) and stable lentiviral shRNA (sh-APE1). Western blot analyses revealed that knockdown of APE1 decreased the level of NRF2 in CPB, OE33, and FLO1 cells, compared to scrambled controls (si-Ctrl and sh-Ctrl) (Fig. 3A–C). However, the levels of KEAP-1 were not significantly changed in APE1 knockdown cells as compared with controls. We then determined the impact of APE1 knockdown on the NRF2 transcriptional activity using the ARE luciferase reporter assay. Indeed, the knockdown of APE1 significantly reduced NRF2 transcriptional activity in CPB, OE33, and FLO1 cells (P < 0.001), as compared with control cells (si-Ctrl) (Fig. 3D–F). Moreover, the knockdown of APE1 blocked bile salts-induced increase in ARE transcriptional activity (Fig. 3G and H). Consequently, ABS-induced upregulation of NRF2 downstream target genes, HO-1 and TRXND1, were blocked in APE1 knockdown cells as compared to that in control cells (Fig. 3I and J). To confirm these results, we performed overexpression of flag-APE1 in OE33 cells because OE33 cells have a relatively lower level of APE1, as compared with other EAC cells. Overexpression of APE1 led to an increase in NRF2 protein level and its downstream HO-1 level (Supplementary Fig. S6A). Similarly, the ARE luciferase activity level and its downstream target genes expression levels were upregulated as compared with the empty vector (Supplementary Fig. S6 B-D). Taken together, these experiments confirmed the role of APE1 in mediating the protein level and activity of NRF2.
Fig. 3.
Induction and regulation of NRF2 are APE1-dependent. (A–C) Knockdown of APE1 downregulated NRF2 protein levels in CPB, OE33, and FLO1 cells. Cells were transfected with si-APE1 or sh-APE1 and controls (si-Ctrl or sh-Ctrl). Whole-cell lysates were collected for western blotting analysis of APE1, NRF2, and KEAP-1. (D–F) Relative ARE luciferase activity was determined in CPB, OE33, and FLO1 cells with or without APE1 knockdown followed by transfection with PGL3- NRF2-ARE-luc and renilla for 48 h. (G–H) FLO1 and OE33 cells were knockdown of APE1 followed by transfection with PGL3- NRF2-ARE-luc and renilla for 24 h and treatment with 200 μM bile salts cocktails for another 24 h, ARE luciferase activity was determined as previously described. (I–J) mRNA expression of NRF2 downstream genes HO-1 and TRXND1 in OE33 cells treated with and without ABS. Values are mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
3.4. APE1 is required for NRF2 nuclear accumulation
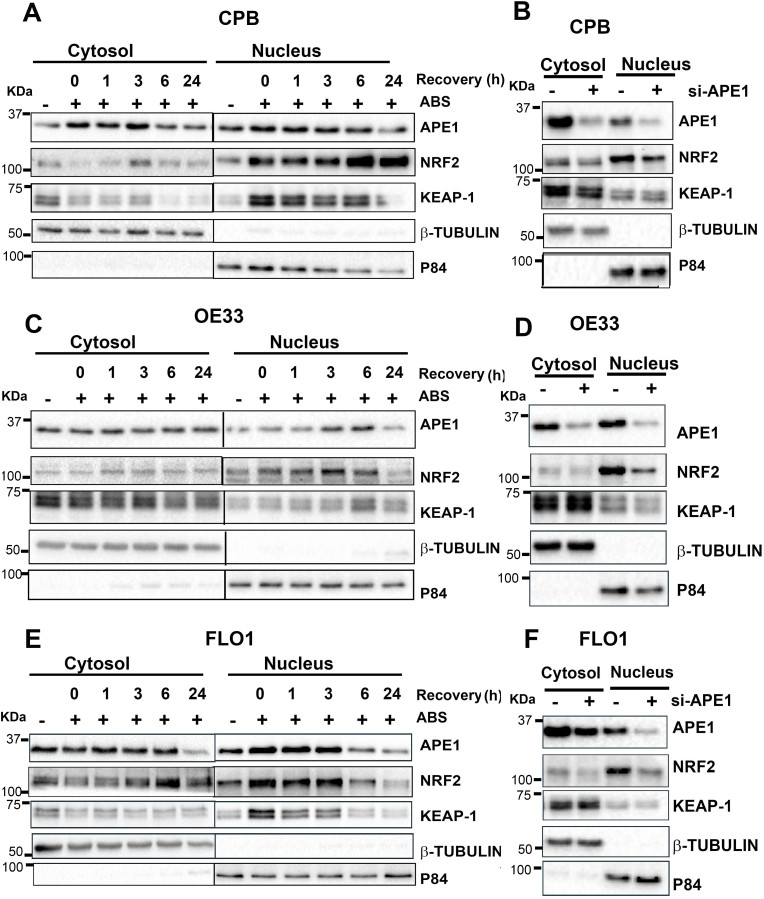
When the NRF2 pathway is activated, NRF2 accumulates and translocates into the nucleus to induce expression of its target genes. Using immunofluorescence staining, we observed that exposure to ABS increased the nuclear accumulation of NRF2 and APE1 (Supplementary Fig. S4). We carried out a nuclear and cytosolic fractionation assay. We detected a significant increase in the NRF2 protein levels in the nucleus after ABS treatment in CPB, OE33 and FLO1 cells (Fig. 4A, C and E). To determine the role of APE1 in regulating nuclear NRF2, cells were transfected with si-APE1 for 48 h. Immunoblot analysis displayed a significant decrease in NRF2 protein levels in a nuclear fraction in APE1 knockdown cells (Fig. 4B, D and F), suggesting that APE1 is required for nuclear accumulation and activation of NRF2.
Fig. 4.
Acidic bile salts induced NRF2 nuclear accumulation where APE1 is required for NRF2 nuclear retention. CPB (A), OE33 (C) and FLO1 (E) cells were treated with ABS for 20 min, followed by recovery in complete media. The samples were collected at 0, 1, 3, 6, and 24 h time points post treatment. Cytosolic and nuclear fractions were isolated and evaluated by western blotting for the levels of NRF2, APE1 and KEAP1. CPB (B), OE33 (D) and FLO1 (F) cells were transfected with si-Ctrl and si-APE1 for 48 h. Cytosolic and nuclear fractions were isolated and evaluated by western blotting for the levels of NRF2, APE1 and KEAP1. β-tubulin and p84 were used as a loading control for cytosolic and nuclear fractions, respectively.
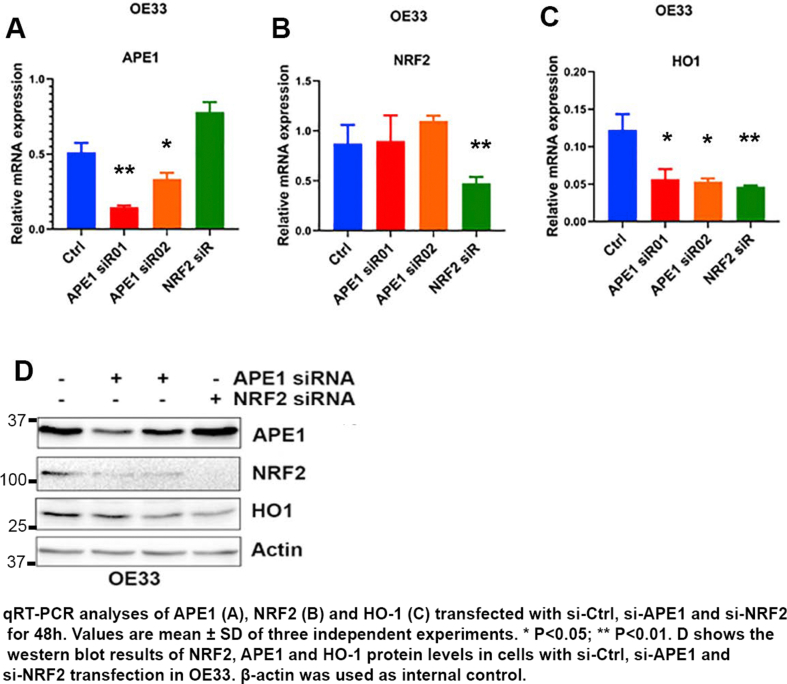
3.5. APE1 is required for maintenance of NRF2 stability
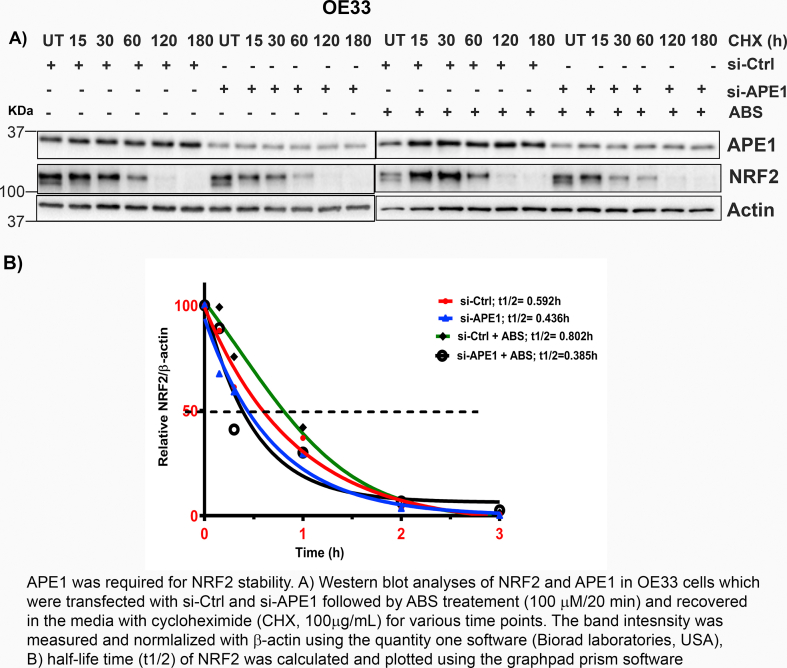
APE1 knockdown downregulated NRF2 protein and its target gene HO-1 levels. At the same time, we did not detect significant changes in NRF2 mRNA expression levels compared to control cells (Supplementary Fig. S7). We, therefore, hypothesized that APE1 could play a role in promoting NRF2 protein stability. To validate this assumption, we first determined NRF2 protein stability by using cycloheximide (CHX) assay in OE33 and FLO1 cells, treated with or without ABS. CHX treatment inhibits protein synthesis [36]. Treatment with ABS led to a significant increase in NRF2 half-life time, as compared with control cells (Fig. 5A and B for OE33, C and D for FLO1). Next, we investigated if APE1 played a role in regulating NRF2 protein stability using APE1 knockdown. Indeed, NRF2 protein degraded much faster in APE1 knockdown cells than control cells (Fig. 5E and F for OE33; G and H for FLO1). To validate whether APE1 played a role in NRF2 stability under ABS conditions, we transfected OE33 cells with si-Ctrl or si-APE1 and treated cells with ABS (100μM/20 min) followed by recovery in regular media in the presence of CHX. Cells treated with ABS showed an increase in NRF2 stability in control cells (si-Ctrl), whereas APE1 knockdown cells (si-APE1) showed a reduction in NRF2 stability (Supplementary Fig S8). These results demonstrated that APE1 is required for the maintenance of NRF2 protein stability in reflux conditions.
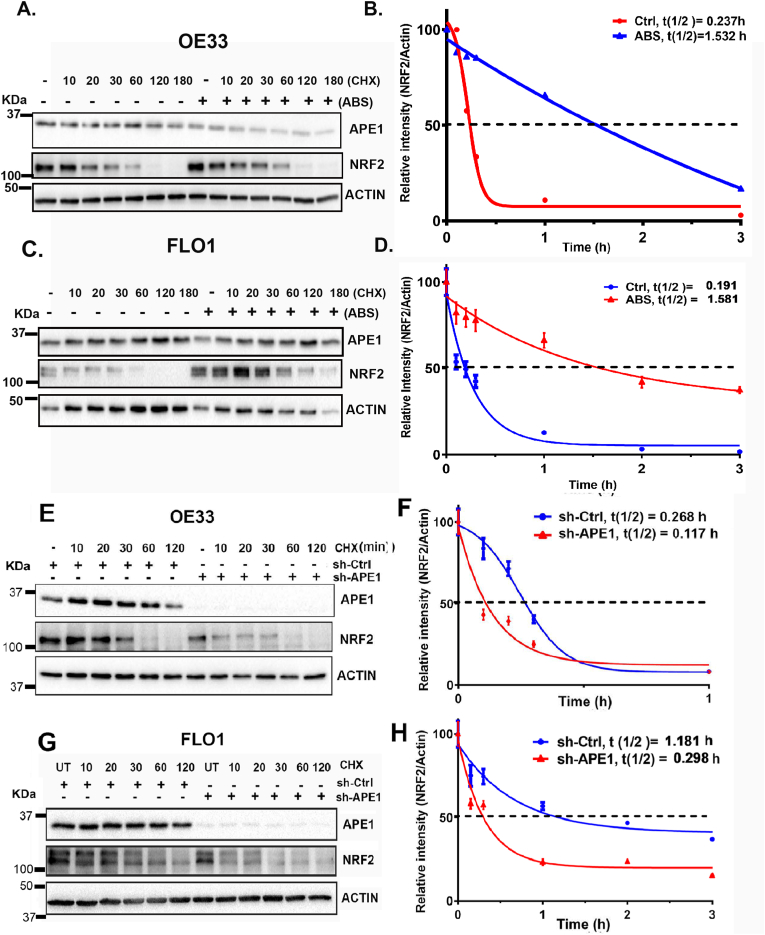
Fig. 5.
APE1 was required for NRF2 stability. (A, C) OE33 and FLO1 cells were treated with or without ABS (100 μM) for 20 min, followed by recovery in complete media with cycloheximide (CHX, 100 μg/mL) at the indicated time points. (E, G) OE33 (E) and FLO1(G) cells with stable knockdown of APE1 using sh-APE1 or sh-Ctrl were treated with cycloheximide (CHX, 100 μg/mL) at the indicated time points. The levels of APE1 and NRF2 were determined by western blotting. The band intensity of NRF2 was measured and normalized with the actin using the Quantity One software (BioRad Laboratories, USA) and normalized to β-actin of the same samples. (B, D, F and H) Half-life time (t1/2) of NRF2 was calculated and plotted using GraphPad Prism software, corresponding to A, C, E and G, respectively.
3.6. APE1-mediated activation of NRF2 in response to ABS was through inhibition of GSK-3β
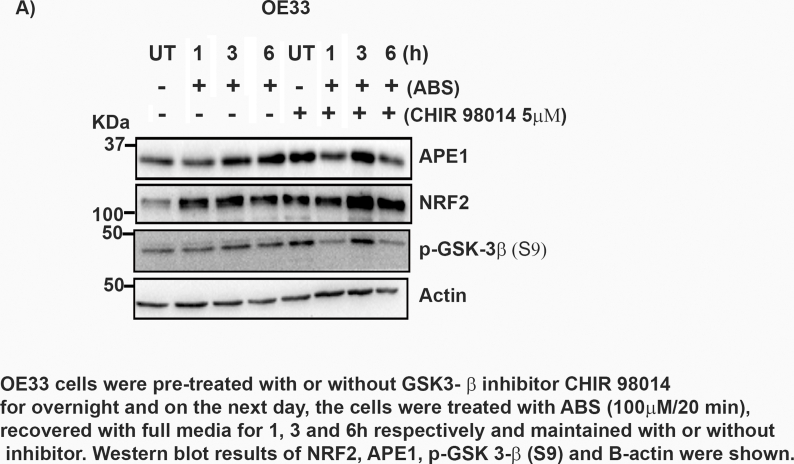
It is known that NRF2 protein stability is mainly regulated by KEAP1, a negative regulator of NRF2 [37]. However, we did not observe notable changes in KEAP1 levels following ABS treatment (Fig. 2C andD, Supplementary Fig. S3A) and in APE1 knockdown cells (Fig. 4 B, D and F). The fact that KEAP1-dependent regulation of NRF2 is a transient rapid process [38], whereas high levels of NRF2 persisted for several hours following ABS, suggested that NRF2 prolonged increase in levels may be independent of KEAP1. On the other hand, active GSK-3β has been reported to regulate NRF2 degradation in a KEAP1-independent manner [39,40]. Therefore, we examined the role of GSK-3β in our cell models. We observed an increase in inactive p-GSK-3β (S9) level at 1 h and 3 h time points in control (si-Ctrl) cells, a similar pattern to NRF2 protein levels after exposure to ABS (Fig. 6 A and B). At the same time, the knockdown of APE1 blocked ABS-induced phosphorylation of GSK-3β (S9) with a decrease in NRF2 levels in these cells (Fig. 6 A and B). These results suggested that the inactivation of GSK-3β by phosphorylation might mediate the increase in NRF2 protein stability. To confirm this result, we used a GSK-3β inhibitor, lithium chloride (LiCl; 10 mM), in our cell models. CPB and OE33 cells were pretreated with or without LiCl (10 mM) overnight and then exposed to ABS (100 μM/20 min) followed by recovery time points. Cells with LiCl treatment followed by ABS exposure showed more NRF2 induction than LiCl untreated cells (Fig. 6C and D). Next, we wanted to address that the induction of NRF2 by APE1 is through inactivating GSK-3β. For this, we have transfected the OE33 cells with or without FLAG-APE1. On the next day, cells were pretreated with or without LiCl overnight and then treated with ABS. Cells treated with LiCl showed more induction of NRF2 when compared to untreated cells (Fig. 6E). To validate the above findings, we used another GSK-3β inhibitor, CHIR-98014, a potent GSK-3 α/β inhibitor [41]. OE33 cells were pretreated with or without CHIR-98014 overnight. On the next day, cells were treated with ABS (100 μM), followed by various recovery time points. Cells treated with CHIR-98014 displayed higher levels of NRF2 (Supplementary Fig S9). Collectively, our findings indicated that inactivation of GSK-3β played a role in APE1-mediated accumulation of NRF2.
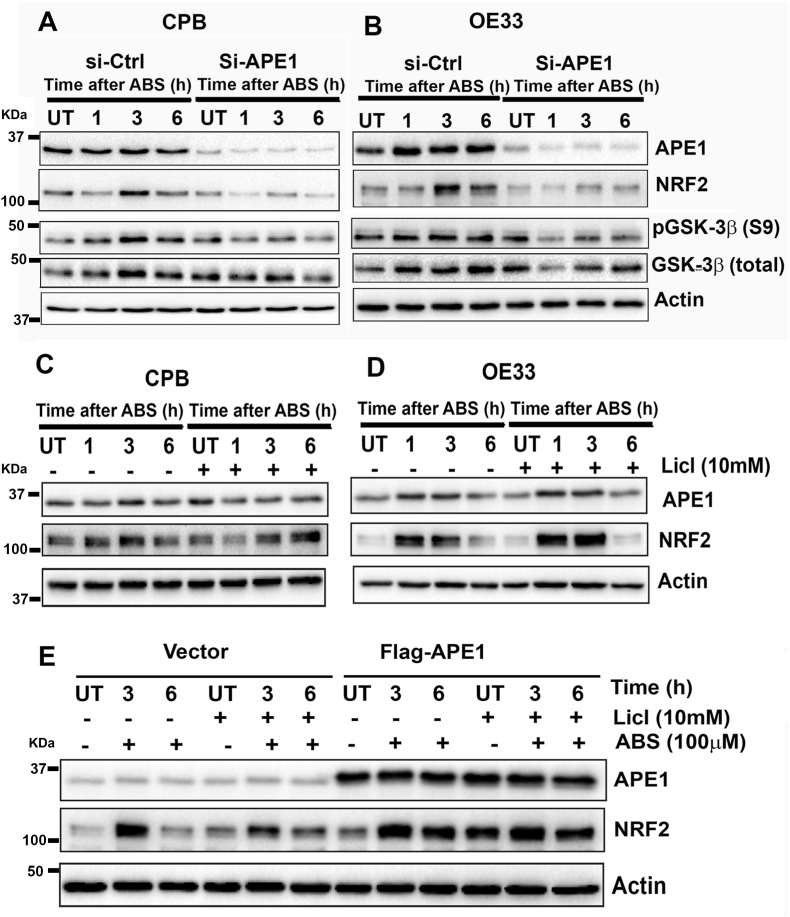
Fig. 6.
APE1-dependent inhibition of GSK-3β promoted the increase in NRF2. (A and B) Knockdown of APE1 blocked ABS-induced NRF2 upregulation. CPB (A) and OE33 (B) cells were transfected with si-Ctrl and si-APE1 followed by exposure to 100 μM ABS for 20 min, then recovery for 1, 3, 6 h in complete media. Immunoblot results of APE1, NRF2, p-GSK-3β (S9), total GSK-3β and β-actin levels were shown. (C and D) CPB and OE33 cells were pretreated with or without 10 mM LiCl (GSK-3β inhibitor) overnight and on the next day cells were treated with ABS (100μM/20min) followed by recovery time points. Immunoblots of APE1, NRF2, and β-actin levels were shown. (E) OE33 cells were transfected with 1 μg of pcDNA flag-APE1, whereas control cells received an empty vector. After 24 h transfection, the cells were split and pretreated with 10 mM LiCl overnight. On the next day, the cells were treated with ABS, followed by different recovery time points in the presence and absence of LiCl. Immunoblots of APE1, NRF2 and β-actin levels were shown.
3.7. APE1 regulation of GSK-3β is dependent on APE1 redox function
To determine if the APE1 redox function is required for the proper regulation of GSK-3β, we first applied (E)-3-[2-(5,6-dimethoxy-3-methyl-1,4-benzoquinonyl)]-2-nonyl propenoic acid (E3330), a known APE1 redox function inhibitor [42]. Treatment of CPB, FLO1, and OE33 cells with E3330 led to a significant decrease in the transcriptional activity of NRF2, as measured by the ARE-luciferase reporter assay (P < 0.05) (Fig. 7A–C). Western blot analyses demonstrated that NRF2 and p-GSK-3β (S9) induction by ABS exposure was diminished in cells treated with E3330 (Fig. 7D and E), strongly supporting the notion that ABS-induced NRF2 activation is dependent on APE1 redox function. To validate this result, we transfected OE33 cells (with relatively low APE1) with wild type APE1 (Flag-APE1) or redox-defective APE1 mutant (C65A). As shown in Fig. 7F, overexpression of wild type APE1 increased NRF2 and p-GSK-3β (S9), while overexpression of mutant APE1 (C65A) did not. To further confirm this finding, we did similar experiments in stable APE1 knockdown cells (shRNA). The reconstitution of wild-type APE1 (Flag-APE1) restored NRF2 and p-GSK-3β (S9) expression in APE1 knockdown cells, whereas reconstitution of mutant APE1 (C65A) failed to rescue the p-GSK-3β (S9) level (Fig. 7G). These data demonstrated that APE1 redox function was required for dysfunction of GSK-3β and NRF2 protein stability (see Fig. 8).
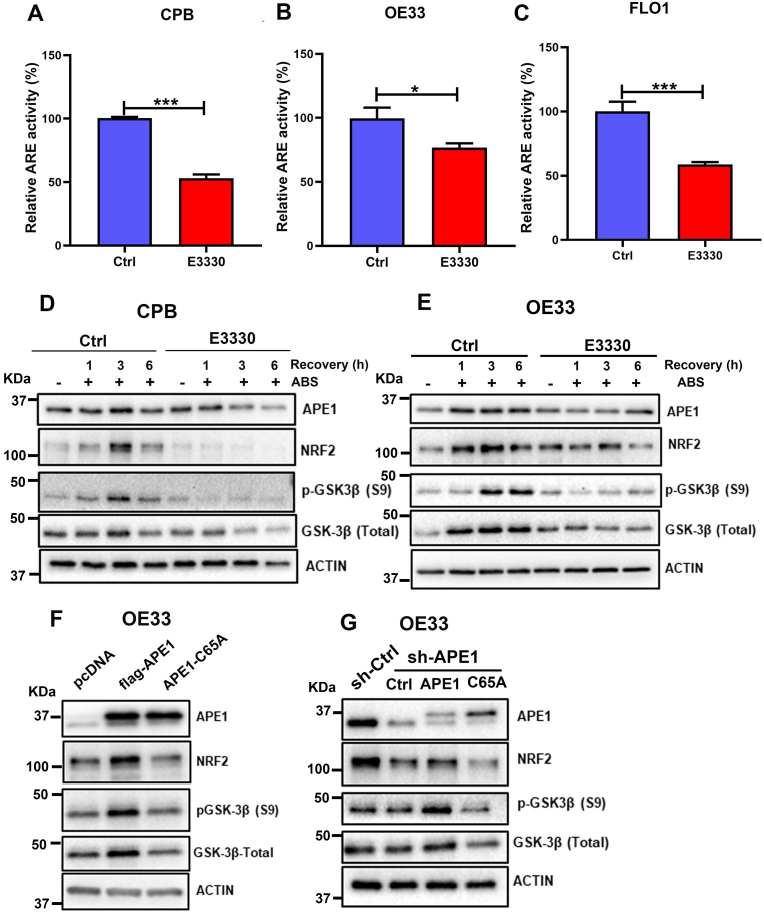
Fig. 7.
APE1 redox function was required for GSK-3β-mediated APE1 regulation of NRF2. (A–C) CPB, OE33 and FLO1 cells were pre-treated with or without E3330, an APE1 redox inhibitor for 24 h. Cells were then transfected with PGL3-NRF2-ARE-Luc and β-gal with or without E3330 for another 24 h. The relative ARE luciferase reporter activity was measured and is shown as a percentage relative to controls (set as 100%). Values are mean ± SD of three independent experiments. *P < 0.05; ***P < 0.001. (D and E) CPB and OE33 cells were pre-treated with or without E3330 (100 μM) for overnight followed by treatment with ABS (100 μM) for 20 min and recovery in full media for 1, 3 and 6 h post-treatment with or without E3330. Western blot analyses were used to determine the levels of APE1, NRF2, GSK-3β (Total) and p-GSK-3β (S9). β-actin was used as a loading control. (F) OE33 cells were transfected with wild type APE1 with the flag (flag-APE1) or mutant APE1 C65A (APE1-C65A, an APE1 redox mutant). Western blot was applied for the levels of NRF2, APE1, GSK-3β (Total) and pGSK-3β (S9). (G) OE33 cells with stable knockdown of APE1 using sh-APE1 were reconstituted with wild type APE1 (APE1) or mutant APE1 C65A (C65A). Western blot was used to analyze the protein levels of NRF2, APE1, GSK-3β (Total) and pGSK-3β (S9).
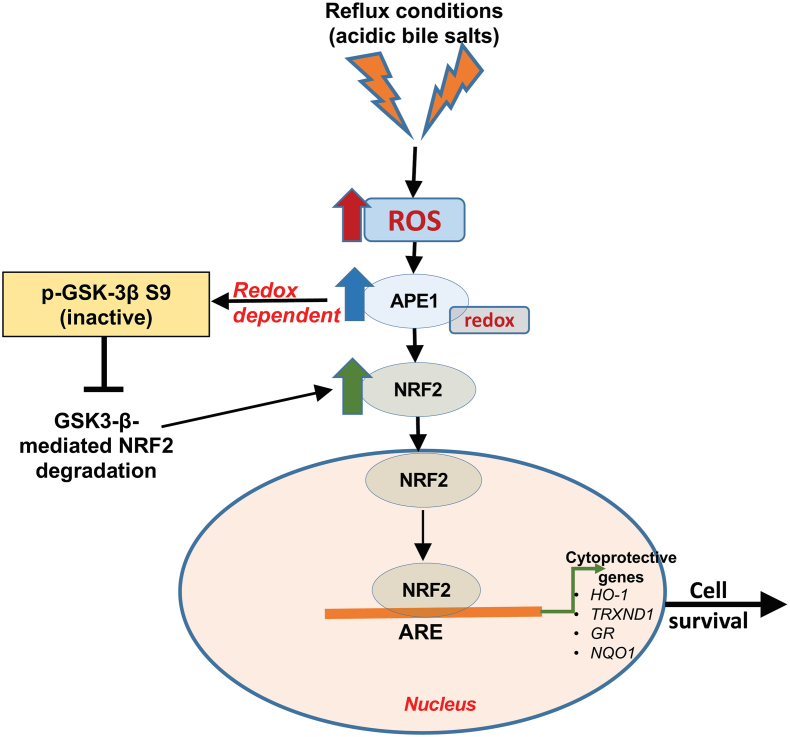
Fig. 8.
A schematic diagram illustrating the role of APE1 in activating NRF2 under reflux conditions. Exposure of cells to reflux conditions (acidic bile salts, ABS) generates high levels of ROS. High ROS and oxidative stress levels induce APE1-dependent increase in NRF2, leading to its release and translocation into the nucleus. In the nucleus, NRF2 is stabilized and regulated by APE1 redox function that inactivates GSK-3β dependent degradation of NRF2. Loss of redox function of APE1 mediates inactivation of GSK-3-β to promote accumulation and activation of NRF2.
4. Discussion
Chronic GERD is a major risk factor for developing Barrett's esophagus and esophageal adenocarcinoma [43]. During the BE-dysplasia-EAC cascade, exposure to reflux conditions mediates an increase in ROS that causes oxidative stress and DNA damage [33,44,45]. In addition, cancer cells contain a higher ROS level than non-neoplastic cells [46]. However, with oxidative effects of chronic reflux conditions, uncontrolled ROS levels can reach lethal levels and induce cell death. Therefore, neoplastic cells must develop mechanisms to counteract persistent higher ROS levels to survive the harsh oxidative reflux environment. NRF2 is one of the most important transcription factors involved in cellular reactions to oxidative stress.
On the other hand, APE1 has also been shown to be involved in the cellular reaction to oxidative stress. Its redox function maintains the activity of several redox-sensitive transcription factors [20]. In the present study, we discovered that the exposure of esophageal cells to reflux conditions led to the induction of APE1, which in turn activated the master antioxidant transcription factor, NRF2. Activation of the APE1-NRF2 axis was essential in counteracting the oxidative stress imposed by exposure to reflux conditions in EAC. The results also suggested that inhibition of GSK-3β by APE1 was a plausible cause for the increased stability and activity of NRF2 in EAC (Fig. 8).
APE1 is a multifunctional protein with DNA base excision repair in its C-terminal domain and redox activity in the N-terminal domain [47,48]. We have previously reported that APE1 base excision repair function is involved in ABS-induced DNA damage repair in EAC cells [21]. However, it is not clear if APE1 can play a role in regulating ROS levels. Herein, we demonstrated that knockdown of APE1 significantly increased intracellular ROS levels in esophageal cells, particularly in neoplastic esophageal cells, exposed to reflux conditions. Consequently, the knockdown of APE1 led to a significant increase in oxidative DNA damage and double-strand breaks and promoted more cell death upon exposure to reflux conditions. We also found that NRF2, a master transcriptional factor involving cellular anti-oxidant and redox homeostasis regulation, was similarly upregulated in neoplastic cells. We hypothesized that regulation of cellular ROS levels by APE1 is through NRF2. To this point, we confirmed that APE1 is required for ABS-induced NRF2 induction and activation in a ROS-dependent manner. We further demonstrated that APE1 regulation of NRF2 is through promoting NRF2 protein stability. It is known that KEAP1, the negative physiological inhibitor of NRF2, plays a crucial role in regulating NRF2 protein stability through the cullin3 ubiquitination pathway [37]. However, this regulation is rapid and transient [49] and cannot explain the sustained increase in NRF2 protein levels for several hours post-exposure to reflux conditions. A number of KEAP1-independent NRF2 degradation pathways have been reported, among which the GSK-3β-TrCP pathway has drawn much attention [50]. We observed a significant upregulation of inactive p-GSK-3β (S9) in cells exposed to ABS. It has been reported that p-GSK-3β (S9) frees NRF2 from β-TrCP-mediated degradation complex [51]. We found that GSk-3β phosphorylation was abrogated in APE1 knockdown cells, suggesting that APE1 was required for proper phosphorylation of GSK-3β. Further studies using APE1 specific redox function inhibitor, E3330 and reconstitution of wild-type or redox-defective APE1 mutant (C65A) demonstrated that APE1 redox function was required for proper phosphorylation of GSK-3β and activation of NRF2. It is known that APE1 acts as a reducing donor and its cysteine 65 (C65)-redox activity is required to maintain a reduced status on specific cysteine residues of APE1-associated transcriptional factors and other signaling proteins [22,52]. Whether APE1 redox function is needed for GSK-3β itself for proper phosphorylation or its upstream regulators, such as AKT, require further investigation.
We detected high constitutive levels of APE1 and NRF2 proteins in EAC cells and primary EAC samples. Based on these findings, approaches that modulate APE1-NRF2 using specific inhibitors of APE1 and/or NRF2 in EAC may have clinical therapeutic benefit, which warrants further investigation. Our studies were conducted mainly using in vitro cell models. Our findings call for additional validation using in vivo models of EAC, which are currently limited.
In summary, the present study highlighted the consequences of reflux-induced oxidative stress in EAC. APE1 emerged as a key player in regulating NRF2 antioxidant activity to maintain oxidative stress below lethal levels and protect cellular homeostasis in reflux conditions.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by grants from the U.S. National Institutes of Health (R01CA206563 and R01CA224366) and the U.S. Department of Veterans Affairs (1IK6BX003787 and I01BX001179). The use of the Flow Cytometry and Biostatistics Shared Resources was supported by the NCI-funded Sylvester Comprehensive Cancer Center (P30CA240139). This work's content is solely the responsibility of the authors. It does not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health, or the University of Miami.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101970.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
figs8.
figs9.
References
- 1.Hongo M., Nagasaki Y., Shoji T. Epidemiology of esophageal cancer: orient to Occident. Effects of chronology, geography and ethnicity. J. Gastroenterol. Hepatol. 2009;24(5):729–735. doi: 10.1111/j.1440-1746.2009.05824.x. [DOI] [PubMed] [Google Scholar]
- 2.Parkin D.M. Estimating the world cancer burden: globocan 2000. Int. J. Canc. 2001;94(2):153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M. Investigating cervical, oesophageal and colon cancer risk and survival among migrants in The Netherlands. Eur. J. Publ. Health. 2013;23(5):867–873. doi: 10.1093/eurpub/cks146. [DOI] [PubMed] [Google Scholar]
- 4.Thrift A.P. The epidemic of oesophageal carcinoma: where are we now? Cancer Epidemiol. 2016;41:88–95. doi: 10.1016/j.canep.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 5.He H. Trends in the incidence and survival of patients with esophageal cancer: a SEER database analysis. Thorac Cancer. 2020;11(5):1121–1128. doi: 10.1111/1759-7714.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souza R.F. From reflux esophagitis to esophageal adenocarcinoma. Dig. Dis. 2016;34(5):483–490. doi: 10.1159/000445225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins G.J. Deoxycholic acid at neutral and acid pH, is genotoxic to oesophageal cells through the induction of ROS: the potential role of anti-oxidants in Barrett's oesophagus. Carcinogenesis. 2007;28(1):136–142. doi: 10.1093/carcin/bgl147. [DOI] [PubMed] [Google Scholar]
- 8.Titi M. Development of subsquamous high-grade dysplasia and adenocarcinoma after successful radiofrequency ablation of Barrett's esophagus. Gastroenterology. 2012;143(3):564–566 e1. doi: 10.1053/j.gastro.2012.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sajadimajd S., Khazaei M. Oxidative stress and cancer: the role of Nrf2. Curr. Cancer Drug Targets. 2018;18(6):538–557. doi: 10.2174/1568009617666171002144228. [DOI] [PubMed] [Google Scholar]
- 10.Baird L., Dinkova-Kostova A.T. The cytoprotective role of the Keap1-Nrf2 pathway. Arch. Toxicol. 2011;85(4):241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 11.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Canc. Cell. 2018;34(1):21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta T. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Canc. Res. 2008;68(5):1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 14.Shibata T. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135(4):1358–1368. doi: 10.1053/j.gastro.2008.06.082. 1368 e1-4. [DOI] [PubMed] [Google Scholar]
- 15.Konstantinopoulos P.A. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Canc. Res. 2011;71(15):5081–5089. doi: 10.1158/0008-5472.CAN-10-4668. [DOI] [PubMed] [Google Scholar]
- 16.Chu X.Y. KEAP1/NRF2 signaling pathway mutations in cervical cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22(14):4458–4466. doi: 10.26355/eurrev_201807_15497. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y.R. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 2010;220(4):446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 18.Wang H. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci. Transl. Med. 2016;8(334):334ra51. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- 19.Padmanabhan B. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21(5):689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Bhakat K.K., Mantha A.K., Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxidants Redox Signal. 2009;11(3):621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong J. APE1-mediated DNA damage repair provides survival advantage for esophageal adenocarcinoma cells in response to acidic bile salts. Oncotarget. 2016;7(13):16688–16702. doi: 10.18632/oncotarget.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhat A.A. Exposure of Barrett's and esophageal adenocarcinoma cells to bile acids activates EGFR-STAT3 signaling axis via induction of APE1. Oncogene. 2018;37(46):6011–6024. doi: 10.1038/s41388-018-0388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu H. APE1 upregulates MMP-14 via redox-sensitive ARF6-mediated recycling to promote cell invasion of esophageal adenocarcinoma. Canc. Res. 2019;79(17):4426–4438. doi: 10.1158/0008-5472.CAN-19-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fishel M.L. Apurinic/apyrimidinic endonuclease/redox factor-1 (APE1/Ref-1) redox function negatively regulates NRF2. J. Biol. Chem. 2015;290(5):3057–3068. doi: 10.1074/jbc.M114.621995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan J.L. APE1 promotes antioxidant capacity by regulating Nrf-2 function through a redox-dependent mechanism. Free Radic. Biol. Med. 2015;78:11–22. doi: 10.1016/j.freeradbiomed.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Peng D.F. Glutathione peroxidase 7 suppresses bile salt-induced expression of pro-inflammatory cytokines in barrett's carcinogenesis. J. Canc. 2014;5(7):510–517. doi: 10.7150/jca.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng D.F. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett's adenocarcinoma. Gut. 2009;58(1):5–15. doi: 10.1136/gut.2007.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bras G.F. Activin A balance regulates epithelial invasiveness and tumorigenesis. Lab. Invest. 2014;94(10):1134–1146. doi: 10.1038/labinvest.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu D., Yotnda P. Production and detection of reactive oxygen species (ROS) in cancers. JoVE. 2011;(57) doi: 10.3791/3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z. Activation of EGFR-DNA-PKcs pathway by IGFBP2 protects esophageal adenocarcinoma cells from acidic bile salts-induced DNA damage. J. Exp. Clin. Canc. Res. 2019;38(1):13. doi: 10.1186/s13046-018-1021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng D. NRF2 antioxidant response protects against acidic bile salts-induced oxidative stress and DNA damage in esophageal cells. Canc. Lett. 2019;458:46–55. doi: 10.1016/j.canlet.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhardwaj V. Activation of NADPH oxidases leads to DNA damage in esophageal cells. Sci. Rep. 2017;7(1):9956. doi: 10.1038/s41598-017-09620-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huo X. In Barrett's epithelial cells, weakly acidic bile salt solutions cause oxidative DNA damage with response and repair mediated by p38. Am. J. Physiol. Gastrointest. Liver Physiol. 2020;318(3):G464–G478. doi: 10.1152/ajpgi.00329.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasai H. Analysis of a form of oxidative DNA damage, 8-hydroxy-2'-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat. Res. 1997;387(3):147–163. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98(3):1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider-Poetsch T. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010;6(3):209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li R., Jia Z., Zhu H. Regulation of Nrf2 signaling. React Oxyg Species (Apex) 2019;8(24):312–322. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D.D. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell Biol. 2004;24(24):10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar M. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 2006;281(21):14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 40.Rada P. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol. Cell Biol. 2012;32(17):3486–3499. doi: 10.1128/MCB.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naujok O. Cytotoxicity and activation of the Wnt/beta-catenin pathway in mouse embryonic stem cells treated with four GSK3 inhibitors. BMC Res. Notes. 2014;7:273. doi: 10.1186/1756-0500-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley M.R. Functional analysis of novel analogues of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref-1. Antioxidants Redox Signal. 2011;14(8):1387–1401. doi: 10.1089/ars.2010.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang R.H. From reflux esophagitis to Barrett's esophagus and esophageal adenocarcinoma. World J. Gastroenterol. 2015;21(17):5210–5219. doi: 10.3748/wjg.v21.i17.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lechner S. Bile acids mimic oxidative stress induced upregulation of thioredoxin reductase in colon cancer cell lines. Carcinogenesis. 2002;23(8):1281–1288. doi: 10.1093/carcin/23.8.1281. [DOI] [PubMed] [Google Scholar]
- 45.Peng D. Glutathione peroxidase 7 protects against oxidative DNA damage in oesophageal cells. Gut. 2012;61(9):1250–1260. doi: 10.1136/gutjnl-2011-301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacker P.T. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Canc. Cell. 2006;10(3):175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Xanthoudakis S., Miao G.G., Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc. Natl. Acad. Sci. U. S. A. 1994;91(1):23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barzilay G., Hickson I.D. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays. 1995;17(8):713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- 49.Kansanen E. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/beta-TrCP. Free Radic. Biol. Med. 2015;88(Pt B):147–157. doi: 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 51.Hayes J.D. Dual regulation of transcription factor Nrf2 by Keap1 and by the combined actions of beta-TrCP and GSK-3. Biochem. Soc. Trans. 2015;43(4):611–620. doi: 10.1042/BST20150011. [DOI] [PubMed] [Google Scholar]
- 52.Vascotto C. Knock-in reconstitution studies reveal an unexpected role of Cys-65 in regulating APE1/Ref-1 subcellular trafficking and function. Mol. Biol. Cell. 2011;22(20):3887–3901. doi: 10.1091/mbc.E11-05-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]