Highlights
-
•
All tested phytochemicals induce the transcription of at least some P450s.
-
•
Clan 3 (particularly family 6) in insects and clan 2 in mites are most responsive.
-
•
Induction of P450s is mainly studied in polyphagous arthropod herbivores.
-
•
Regulation of P450 induction upon phytochemicals exposure is underexplored.
Abstract
Cytochrome P450 monooxygenases (P450s) play a key role in the detoxification of phytochemicals in arthropod herbivores. We present here an overview of recent progress in understanding the breadth and specificity of gene expression plasticity of P450s in response to phytochemicals. We discuss experimental setups and new findings in mechanisms of P450 regulation. Whole genome transcriptomic analysis of arthropod herbivores, either after direct administration of phytochemicals or after host plant shifts, allowed to integrate various levels of chemical complexity and lead to the unbiased identification of responsive P450 genes. However, despite progress in identification of inducible P450s, the link between induction and metabolism is still largely unexplored, and to what extent the overall response is biologically functional should be further investigated. In the near future, such studies will be more straightforward as forward and reverse genetic tools become more readily available.
Current Opinion in Insect Science 2021, 43:117–127
This review comes from a themed issue on Insect genomics
Edited by May Berenbaum and Bernarda Calla
For a complete overview see the Issue and the Editorial
Available online 26th December 2020
https://doi.org/10.1016/j.cois.2020.12.002
2214-5745/© 2020 The Author(s). Published by Elsevier Inc. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
The interplay between phytophagous arthropods and their host plants is often seen as a text-book example of co-evolution and the driver of species diversification [1], although this concept is sometimes also criticized [2]. Nevertheless, it is clear that an important part of the interactions between arthropods and plants relies on the recognition and response to phytochemicals (allelochemicals) produced by the plant to defend itself against herbivory [1, 2, 3]. Gene expression changes in response to phytochemicals and host shifts have been studied both on the short-term, within a generation (induction), as well as on the long-term (adaptation, genetic accommodation) [4••]. Surprisingly, only few studies characterized gene expression patterns both upon initial exposure and after adaptation to new host plants [5,6,7•,8,9], with the current consensus that, upon adaptation, mainly genes with unknown function or genes involved in core metabolic pathways show constitutive changes in expression, whereas many detoxification-related genes exhibit within-generation environmental plasticity [4••,5]. Therefore, there is little evidence that genes induced upon initial exposure, become constitutively overexpressed after adaption, and thus overall patterns of genetic accommodation in arthropod-plant interactions are rare. The majority of transcriptome studies, however, have only focused on induction and phenotypic plasticity, that is, to what extent gene expression levels change in direct response to phytochemicals, either by a complete host shift, or by a more controlled direct exposure [4••].
The induction of P450 monooxygenases (P450s), a key detoxification enzyme family, has been historically best studied in this context [10], as it is known to metabolize and detoxify a wide range of allelochemicals [11•]. Nevertheless, compared to pesticide resistance, the functional role of induction in adaptation is far less studied. Here, we focus on recent findings and complement earlier work investigating the response of P450s to phytochemicals, in the light of currently available methodologies and with special reference to experimental setup.
P450 transcriptional responses to phytochemicals
The breadth and specificity of arthropod transcriptional responses upon exposure to phytochemicals have been investigated using either, (1) controlled administration (e.g. via fumigation, artificial diet or leaf dip (Table 1), (2) complete host plant shifts or (3) shifts between transgenic or natural mutant plants differing in only a few allelochemicals (overview per species is shown in Table 2, while an overview and visualization of experimental set-ups is given in Figure 1). These studies use different technologies like RT-qPCR, microarray hybridization and next-generation RNA sequencing (RNA-seq). The experimental set-up overview in Table 2 indicates that although RT-qPCR remains a popular tool for expression validation, genome-wide transcriptomics using next-generation RNA-seq is now the method of choice, as it became much more affordable. This allows to study the effect of phytochemicals in the complex context of gene expression and regulation more completely and accurately. In the near future, the newest generation sequencing technologies that offer single molecule sequencing, and thus complete transcripts, will not only allow to drastically improve annotation but will also facilitate the identification of splice variants and measuring allele-specific induction by phytochemicals.
Table 1.
Phytochemical inducers of CYP genes in insects
| Inducer | P450s induced | Reference |
|---|---|---|
| Alkaloids | ||
| Tomatine | CYP6(AB60, AE14, B7)3, CYP4(L4, G75)4, CYP340AB14, CYP339A1M | [13,40,67] |
| Nicotine | CYP4(M1, M3)4 | |
| CYP6-like 53, CYP6CY33 | [14,15,68] | |
| Caffeine | CYP6(A2, A8)3 + 9 Drosophila genes | |
| CYP6-like 43, CYP6(A8, D5)3, CYP12D1M | [68,69] | |
| senita/saguaro cactus alkaloids | CYP28(A1, A2, A3)3, CYP4D104 | |
| Chromenes | ||
| Precocene | CYP15H12, CYP6(FD2, FE1, HL1)3, CYP409A13, CYP4C694 | [28] |
| Derived from phenylpropanoid pathway | ||
| Cinnamic acid | CYP6AE143 | |
| CYP9A403 | [70] | |
| Chlorogenic acid | CYP6(B8, B9, B27, B28)3 | |
| Salicylic acid | CYP6(B8, B9, B27, B28)3, CYP321A13 | |
| Tannic acid | CYP6AE143 | |
| CYP6(CY19, CY22, DA1)3 | [45] | |
| Flavonoids | ||
| Flavone | CYP6(B8, B9, B27, B28)3, CYP321A13 | |
| CYP6-like(2, 5)3, CYP6AB143, EE600001 | [68,71] | |
| Quercetin | CYP6B83 | |
| CYP15H22, CYP6-like(1, 5)3, CYP6(AE10, B6, CY19, CY22)3, CYP9(A11, A40)3, CYP321A83, CYP337B13, CYP4C804, CYP301A1M | [28,38,45,68,70,72] | |
| Rutin | CYP6(B8, B27, B28)3 | |
| CYP6(AB5, K1)3, CYP9E13 | [27,73] | |
| B-naphtoflavone | CYP6B83, CYP321A13 | |
| Coumarins | ||
| Coumarin | CYP6B83, CYP321A13 | |
| CYP6(AB14, AB60)3 | [40,71] | |
| Furanocoumarin | CYP6(AE14, B1, B3, B4, B8, B9, B17, B27, B28)3, CYP9A(2, 4, 5)3, CYP321A13 | |
| CYP15H12, CYP18A12, CYP6(AB14, AB60, AE, AE9, AE14, AE89, B7, B29, B39, B40, FD1, FE1, FG1, HL1, HN1, HQ1)3, CYP9(A, A27, A31, A32, AQ2)3, CYP337(A1, A2)3, CYP321(A7, A8, A9, B1)3, CYP4(AA1, C84)4, CYP301A1M, CYP404D1M, CYP333B4M | [13,28,37,39,40,47,60•,71] | |
| Lectin | ||
| Ricin | CYP6(AE9, B29)3, CYP9A3, CYP321B13, CYP337(A1,A2)3 | [39] |
| Phorbol ester | ||
| Tetradecanoyl-phorbol-13-acetate | CYP6BD63, CYP9(A17, A21)3 | [17] |
| Terpenoids | ||
| α-pinene | CYP6(B2, B7)3 | |
| CYP6(BX1, DJ2)3, CYP345E43 | [19] | |
| β-pinene | CYP6(BX1, DJ2)3, CYP345E43 | [19] |
| 3-carene | CYP6(BX1, DJ2)3, CYP345E43 | [19] |
| Turpentine-oil | CYP6(BX1, DJ2)3, CYP345E43 | [19] |
| Limonene | CYP6A23 | |
| Melaleuca alternifolia essential oil (terpinen-4-ol) | CYP6(BQ36, BW1, BW2, BW3, BW4, BX1, DJ1, DJ2, DJ3, DG1)3, CYP345E23, CYP4(BH1,G56)4, CYP412A14 | [16] |
| Menthol | CYP6B23 | |
| Monoterpenes (peppermint oil) | CYP6B23 | |
| Gossypol | CYP6(A12, A17, AE14, B27)3 | |
| CYP18B12, CYP6(AB9, AE11, AE12 AE14, B7, CY19, CY22, DA1)3, CYP4L114 | [12,13,35,45,60•] | |
| Other phytochemicals | ||
| Ethanol | CYP6(A1, A8)3 | |
| CYP6A83, CYP4E34 | [74] | |
| 2-phenylethanol | CYP6(A2, A8, D5)3, CYP4E34 | [74] |
| Indole | CYP6(B39, B40)3, CYP9A313, CYP321(A7, A8, A9)3, CYP332A13 | [47] |
| Indole-3-carabinol | CYP6(B8, B9, B27, B283)3, CYP9A23, CYP321A13 | |
| CYP6B393, CYP321(A7, A8, A9)3 | [47] | |
| Jasmonic acid | CYP6(B8, B9, B27, B28)3, CYP321A13 | |
| 2-tridecanone | CYP6(A2, B6)3, CYP4M34 | |
| CYP15(H1, H2)2, CYP6(AE, B39, CY19, CY22, DA1, FD1, FE1, FF1, FG1, HL1, HN1, HQ1)3, CYP9(AQ1, AQ2)3, CYP408B13, CYP409A13, CYP4(C69, C73, C79, C80, C84, DH1, FD1, L13)4, CYP3117C14, CYP3118(A1, A2)M, CYP404D1M | [28,45,47,60•] | |
| Glucosinolates | CYP392(A1, A16, D8)2 and several members of Clan 3 (CYP6 and CYP9) | [6,31•,75] |
| Piperamides | 6 drosophila genes | |
| Pyrethrum | CYP9F23, CYP12D1/2M | |
The original data of Feyereisen (2012) is indicated in bold font while newly (since 2012) discovered CYP−inducing phytochemicals or induced CYP genes that were identified via controlled administration (fumigation, artificial diet or leaf dip/spray) are indicated in normal font. Not all CYP gene names follow the official CYP nomenclature (David Nelson, University of Tennesee), but are based on the CYP name of their best BLAST hits. CYP genes are grouped per family and their respective clan is displayed in superscript: (2) Clan2, (3) Clan3, (4) Clan4, (M)Mitochondrial.
Table 2.
P450 transcriptional responses per species, presented by experimental setup, gene identity and level of validation
| Species (order) | Diet breadth | Phytochemicals/hosts/pathway | Differential expression method | P450 genes or families of interest | Validation technique? | Ref |
|---|---|---|---|---|---|---|
| Artificial administration | ||||||
|
Aphis gossypii (Hemiptera) |
P | Gossypol, 2-Tridecanone, Quercetin, Tannic acid | RT-qPCR | CYP6(CY19, CY22, DA1)3 | / | [45] |
|
Depressaria pastinacella (Lepidoptera) |
O | Bergapten, Sphondin, Xanthotoxin | RT-qPCR | CYP6AE893 | Functional expression | [37] |
|
Drosophila melanogaster (Diptera) |
P | Acetic acid, Ethanol, 2-Phenylethanol (Fumigation) |
RNA-seq (Illumina HiSeq 4000) RT-qPCRa |
CYP6(A2, A8, D5)3, CYP4E34 | / | [74] |
|
Helicoverpa armigera (Lepidoptera) |
P | Gossypol | RT-qPCR | CYP18B12, CYP6(AB9, AE14)3, CYP4L114 | RNAi | [35] |
|
Helicoverpa armigera (Lepidoptera) |
P | Xanthotoxin, 2-Tridecanone, Gossypol | RT-qPCR | CYP6AE3 (e.g. CYP6AE(14, 19, 20)) | CRISPR-Cas9 Functional expression |
[60•] |
|
Hyles euphorbiae (Lepidoptera) |
M | 12-Tetradecanoyl-phorbol-13-acetate (TPA) | RNA-seq (Illumina HiSeq 2000)b DeepSuperSAGE |
CYP6BD63, CYP9(A17, A21)3 | / | [17] |
|
Locusta migratoria (Orthoptera) |
P | 12 phytochemicals, model inducers and common insecticides (Leaf-dip) |
RNA-seq (Illumina HiSeq 4000)b RT-qPCR |
43 genes of CYP2, CYP3, CYP4 and mitochondrial clan | / | [28] |
|
Nilaparvata lugens (Hemiptera) |
M | Rice leaf sheath extracts | RT-qPCR | CYP6(AX1, AY1)3, CYP4C614 | RNAi | [76] |
|
Sitophilus zeamais (Coleoptera) |
P | Terpinen-4-ol (Fumigation) |
RNA-seq (BGISEQ-500) RT-qPCRa |
11 members of CYP63, CYP345E23, CYP4(BH1, G56)4, CYP412A14 | / | [16] |
|
Oedaleus asiaticus (Orthoptera) |
O | Rutin (Leaf-dip) |
RNA-seq (Illumina HiSeq 4000) RT-qPCRa |
CYP6K13, CYP9E13 | / | [27] |
|
Spodoptera exigua (Lepidoptera) |
P | Quercetin | RT-qPCR | CYP6AE103 CYP9A113, CYP321A83 | RNAi | [38] |
|
Spodoptera litura (Lepidoptera) |
P | Xanthotoxin, Ricin | RNA-seq (Illumina HiSeq 4000) RT-qPCRa |
CYP6(AE9, B29)3, CYP9A3 CYP321B13, CYP337(A1, A2)3 | RNAi | [39] |
|
Spodoptera litura (Lepidoptera) |
P | Coumarin, Tomatine, Xanthotoxin | RT-qPCR | CYP6AB603 | RNAi | [40] |
|
Spodoptera litura (Lepidoptera) |
P | Tomatine | RNA-seq (Illumina HiSeq 2000) RT-qPCRa |
CYP4(G75, L4)4, CYP340AB14, CYP339A1M | / | [67] |
|
Spodoptera litura (Lepidoptera) |
P | z-ligustilide | RT-PCR (semi-quantitative) | CYP4(M14, S9)4 | / | [18] |
| Complete host plant shift | ||||||
|
Bactrocera oleae (Diptera) |
M | Green and black olive | Microarray RT-qPCRa |
2 B. oleae P450 contigs (contig03604, contig10157) | / | [20] |
|
Bemisia tabaci (Hemiptera) |
P | Eggplant, Pepper, Cassava, Kale | NextSeq 500 RT-qPCRa |
24 P450 s most related to Clan 3 and Clan 4 | / | [26•] |
|
Danaus plexippus (Lepidoptera) |
O | Milkweed (2 species) | RNA-seq (BGIseq-500) | e.g. CYP6AB43 | / | [34] |
|
Helicoverpa armigera (Lepidoptera) |
P | Chili, Cotton, Corn, Soybean | RNA-seq (Illumina HiSeq 2000) RT-qPCRa |
CYP18B12, CYP6(AB9, AE14)3, CYP4L114 | RNAi | [35] |
|
Oedaleus asiaticus (Orthoptera) |
O | Grasses (3 common species), A.frigida | RNA-seq (Illumina HiSeq 2000) RT-qPCRa |
CYP6K13 | / | [23] |
|
Phaedon cochleariae (Coleoptera) |
O | Chinese cabbae, Watercress, White mustard | RNA-seq (Illumina HiSeq 2000)b Microarray |
A lot of not-specified P450 s differentially expressed | / | [7•] |
|
Sitobion avenae (Hemiptera) |
O | Barley, Wheat | RNA-seq (Illumina HiSeq 2500) RT-qPCRa |
CYP6(A13, K1-1, K1-2)3, CYP4(C1, G15)4 | / | [21] |
|
Sitobion avenae (Hemiptera) |
O | Barley, Wheat | RNA-seq (Illumina HiSeq 2500) RT-qPCRa |
e.g. CYP6A133, CYP4C14 | / | [22] |
|
Spodoptera exigua (Lepidoptera) |
P | Cabbage, Maize, Tobacco | RNA-seq (Illumina HiSeq 4000) | e.g. CYP6AE, CYP321 and more Clan 3, Clan 4 P450s | / | [25] |
|
Spodoptera frugiperda (Lepidoptera) |
P | Corn, Rice | RNA-seq (Illumina HiSeq 2500) | e.g. CYP6(AB4, AE9)3, CYP332A13, CYP4(L6, M5)4 | / | [77] |
|
Tetranychus cinnabarinus (Trombidiformes) |
P | Cowpea, Cotton | RNA-seq (Illumina HiSeq 2000) RT-qPCRa |
CYP392A43 | RNAi (transgenic cotton expressing dsRNA) | [9] |
|
Tetranychus urticae (Trombidiformes) |
P | Bean, Tomato | Microarray | CYP392(A1, A3, B1, B2, B3, D3)2, CYP385(C2, C3, C4)3, CYP381A2M | / | [5] |
| Transgenic host-plants | ||||||
|
Heliothis virescens (Lepidoptera) Pieris brassicae (Lepidoptera) |
P O |
Glucosinolates (Arabidopsis) | RNA-seq (Illumina HiSeq 2500) | Several members of Clan 3 (CYP6 and CYP9) | / | [31•] |
|
Tetranychus urticae (Trombidiformes) |
P | Glucosinolates (Arabidopsis) | RT-qPCR | CYP392(A1, A16, D8)2 | RNAi of Tu-CPR | [6] |
|
Tupiocoris notatus (Hemiptera) |
O | Jasmonic acid (Nicotiana attenuata) | RNA-seq (Illumina HiSeq 2000) RT-qPCR |
Several members of Clan 3 and Clan 4 | / | [75] |
The studies reported in this table were published between 2017 and 2020, except for key studies on transgenic host shifts and those with the spider mite T.urticae, representative for chelicerates. This table categorizes recent research (partly) focusing on short-term transcriptional responses to of P450s to phytochemicals based on arbitrarily chosen experimental set-up categories. The P450s of interest in these studies are CYP genes actually responding to the host-shift or phytochemical administered. Not all CYP gene names do follow the official CYP nomenclature (David Nelson, University of Tennesee) but were based on the CYP name of their best BLAST hits. The feeding column indicates the feeding patterns of the respective species (P: polyphagous, O: oligophagous, M: monophagous). CYP genes are grouped per family and their respective clan is displayed in superscript: (2) Clan2, (3) Clan3, (4) Clan4, (M)Mitochondrial. Extra comments on the differential expression methods is indicated as followed: (a) RT-qPCR used for verification of differential gene expression, (b) Used for reference transcriptome assembly.
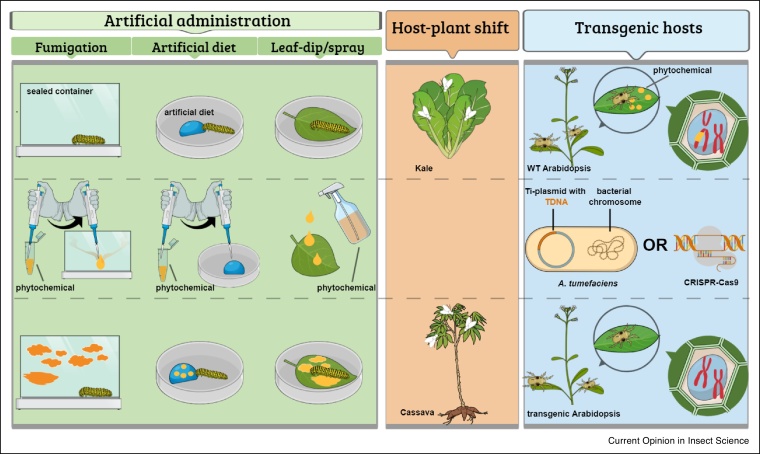
Figure 1.
Visual overview of experimental setups presented in Table 2.
The starting point, experimental procedures and end point are represented from top to bottom. Artificial administration of a phytochemical can be achieved via fumigation, addition to artificial diet or via leaf-dipping/spraying. Upon fumigation the volatile phytochemical is added to a sealed container that contains the insect or mite. For administration via artificial diet, the phytochemical is added to/poured on the (semi)artificial diet. Administration via the leaf is realized by spraying the phytochemical-solution on the leaf or by dipping the complete leaf in a phytochemical-solution. For host-transfer experiments, insect and mites are transferred to a new plant species. For transgenic host experiments, P450 induction by phytochemicals in insects and mites is investigated by interfering with the secondary metabolism pathways of the plant, most often by A. tumefaciens or CRISPR-Cas9 based gene knock-out.
Historically, most studies aimed at understanding P450 induction by specific phytochemicals via controlled administration ([10], Table 1, Table 2). The ease of testing single or selected mixes of phytochemicals and its convenience for dosage control and reproducibility, still makes it one of the most commonly used experimental setups today (Table 2). In 2012, Feyereisen presented an overview of phytochemicals that are known to induce P450 genes [10] and in Table 1 an update of this overview is presented. Recent data underpins the earlier trends towards high diversity at two levels. Not only can P450 genes be induced by a wide variety of chemical classes, but also the induced P450 families and clans they belong to appear to be very diverse. As seen in Table 1, induction of P450s by a certain phytochemical is not even limited to only one clan. Nevertheless, we can clearly notice an enrichment of CYP genes belonging to Clan 3 and 4 in insects and Clan 2 in mites due to numerical dominance in their CYPomes [11•]. Not surprisingly, especially the notorious CYP6 family is strongly represented as every phytochemical present in Table 1 induces expression of at least one CYP6 gene in insects. In particular the list of phytochemicals inducing the CYP6AE subfamily has expanded (Table 1), as next to gossypol, cinnamic acid, tannic acid and furanocoumarins we can now add compounds such as tomatine, quercetin, ricin and 2-tridecanone. The last decade, efforts have been made in more accurately determining dose-responsiveness to phytochemicals whereas this was already common in investigating transcriptional responses to synthetic chemicals like insecticides. In particular, dose-dependent responses of one or more CYP genes to gossypol [12,13], nicotine [14,15], terpinen-4-ol [16], tomatine, xanthotoxin [13], TPA [17], z-ligustilide [18] and several terpenoids [19] were established recently.
Although the above setup of controlled administration is the most precise in terms of direct molecular interactions that lead to induction of a precise number of genes, a potentially important drawback is that the chemical complexity of the whole plant is often not taken into account which complicates and prevents a complete biological interpretation of results. Therefore, it is also crucial to study transcriptional responses after short-term host-plant shifts, and a number of recent studies have quantified genome-wide responses to the biological mix of allelochemicals in planta (Table 2). Transfer of phytophagous arthropods to plants rich in alkaloids or polyphenolic compounds results in induction of several Clan 3 (CYP6) P450 genes [20, 21, 22, 23, 24, 25,26•]. In some cases, the patterns after host-shift were further investigated by controlled administration. For example, rutin is a flavonoid present in Artemisia and RNA-seq expression analysis revealed the upregulation of Clan 3 (CYP6) P450 genes when the grasshopper Oedaleus asiaticus was switched from feeding on grasses to Artemisia [23]. In a follow-up experiment, artificially administering of rutin to O. asiaticus larvae resulted in a similar response of CYP6 genes, suggesting that rutin in Artemisia lies at the basis of the P450 response upon transfer to Artemisia [23,27].
An ideal compromise consists of a set-up where the complexity of the host is preserved while at the same time the specific responses against a certain (set of) phytochemicals can be studied. A convenient method to achieve this for chewing insects like grasshoppers and locusts, relies on direct administration of the chemical on the plant via leaf-dip or spray [27,28] (Table 1, Table 2, Figure 1). However, this method is not suitable to study interactions in species with piercing-sucking mouthparts, as the chemical is only present on the plant-surface and will thus not be ingested by these insects. Transgenic plants that are deficient/enriched in given secondary metabolite pathway circumvent this problem, making it a universal method to achieve the above-mentioned goals. Arabidopsis thaliana allows efficient and high-throughput transformation [29] and is an ideal model, at least for those arthropods than can feed on it. Zhurov et al. used this system to study reciprocal genome-wide transcriptional responses in both A. thaliana and the polyphagous spider mite Tetranychus urticae as a model for host-herbivore interactions. At least 40 genes showed a significant dose-dependent response to glucosinolates, mainly consisting of detoxification enzymes, including P450s and glycosyltransferases [30]. A follow-up study focused on whether these expression changes are indeed associated with host-plant adaptation or whether they are general stress responses [6]. Interestingly, none of three selected P450s (CYP392A1, CYP392A16, CYP392D8) that were initially highly upregulated upon short-term host shift [30] showed a constitutively higher expression after long-term adaptation relative to the non-adapted lines [6], a pattern that was also found by Wybouw et al. for the same mite species and tomato [5]. A. thaliana mutants were also used to compare glucosinolate-induced transcriptomic responses between the generalist Heliothis virescens and Pieris brassicae, a specialist feeding on glucosinolate-rich diets. P450s were significantly enriched in the set of upregulated genes of H. virescens, whereas this was not the case for P. brassicae. [31•]. Unfortunately, the genetic toolkit of A. thaliana is not yet available for other host plants, limiting the use of the transgenic plants for investigating other insect-plant systems. However, as the CRISPR-Cas9 technology is rapidly advancing, the availability of mutant non-reference host plants will most likely improve in the near future [32]. Also, not only transgenic plants but also natural mutants/cultivars enriched/lacking specific compounds could be valuable in this approach [33,34].
Of particular note, many factors in the experimental design of host-shift experiments in the above studies vary, which makes them hard to compare. For example, there is no consensus on what the optimal time-point is for studying short-term transcriptional response after induction/host transfer. Also, some studies focus on whole-insect RNA-sequencing [20,35], others only on responses in certain tissues involved in detoxification [17,34,36, 37, 38, 39, 40]. There is also a significant amount of variability with respect to transcriptome completeness, coverage, differential expression analysis methods and annotation [41]. Hence, these studies only allow us to identify potential candidate P450 genes, providing working hypotheses for further research, and validation of these candidate genes for their role in detoxification remains essential.
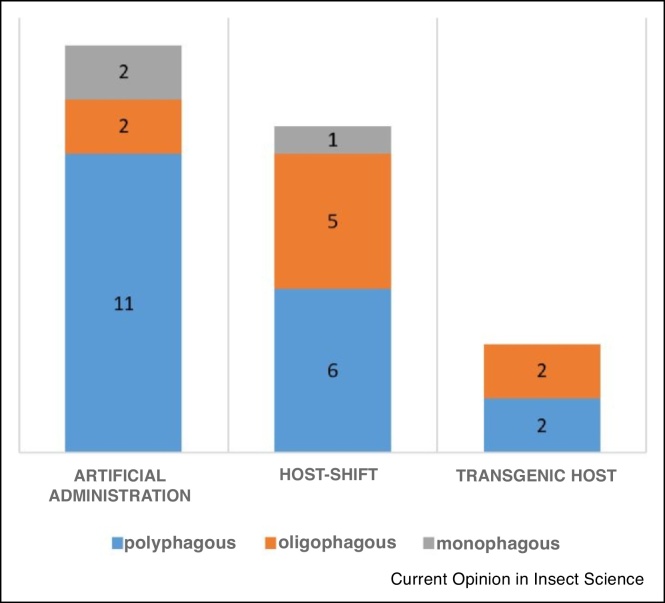
Finally, Figure 2 and Table 2 indicate that most studies cover transcriptional responses of polyphagous species of which the majority belong to the Lepidoptera. However, complete host shifts or transgenic plants are now also being used to investigate transcriptional plasticity in oligo- or monophagous species. When more studies become available, it will be interested to compare more thoroughly the P450 transcriptional induction patterns in relation to diet breadth.
Figure 2.
Graphical representation of the number of studies (represented in Table 2) categorized based on feeding preference of investigated species between different experimental set-ups. The majority of recent studies use administration of phytochemicals via the diet and study polyphagous insects and mites.
Regulation of P450 expression in arthropods
P450 transcriptional responses to phytochemicals have been widely documented, however, the mechanisms of induction, especially the first steps in the signaling cascade, remain a black box. The upregulation of Papilio polyxenes P450 genes in response to xanthotoxin is probably the best case studied so far. In the promoter region of Papilio polyxenes CYP6B1, xenobiotic response elements (XREs, including a XRE to xanthotoxin (XRE-Xan) and XRE to aryl hydrocarbon receptor (XRE-AhR)) were discovered that are responsible for basal and xanthotoxin-inducible expression of CYP6B1. Further, it was shown that binding of a Drosophila heterodimer - consisting of the transcription factor spineless (ortholog of the mammalian AhR) and Tango (ortholog of the mammalian AhR nuclear translocator (ARNT)) - on XRE-AhR, enhanced basal expression of P. polyxenes CYP6B1 [42]. XREs to xanthotoxin and other allelochemicals such as flavones, gossypol and 2-tridecanone, were also identified in the promoter region of aphid and Helicoverpa P450s [43, 44, 45] (and references therein), while recent studies showed that RNAi knockdown of AhR/ARNT significantly reduced the expression of the nicotine metabolizing CYP6C3 in Myzus persicae [46], or dramatically repressed the expression of CYP6DA2, a P450 strongly associated with tolerance of Aphis gossypii to gossypol [44]. Nevertheless, some elements of this P450 regulatory pathway still await elucidation. For example, why there is no correlation between the occurrence of XRE-Xan elements and the xanthotoxin induction profile of Spodoptera frugiperda P450s [47]? What is the identity of the xanthotoxin receptor and how is it connected to the AhR/ARNT complex [42]? In fact, with the exception of phytoecdysteroids [48], direct binding of phytochemicals to insect receptors has not yet been demonstrated while such cases have been reported for vertebrates (e.g. the flavonoid luteolin binding to the nuclear receptor HNF4 [49]).
The Cap “n” collar:Muscle Aponeurosis Fibromatosis (CncC:Maf) is another well-known P450 regulatory pathway. Under normal conditions, the mammalian ortholog of CncC, Nrf2, is present in the cytoplasm and bound to the Kelch-like ECH associated protein 1 (Keap1), while under stress conditions (e.g. oxidative stress caused by exposure to xenobiotic compounds) Nrf2 translocates to the nucleus, forms a complex with Maf, binds to antioxidant responsive elements (AREs) upstream of detoxification genes such as P450s and initiates transcription. However, this pathway has been mainly studied for its role in overexpression of arthropod detoxification genes involved in pesticide resistance (recently reviewed in Refs. [50,51] and, to our knowledge, only two studies have examined its role in upregulation of P450s in response to allelochemicals [52,53••]). Both studies showed that RNAi knockdown of CnCC resulted in decreased expression of P450 gene(s), but while in Kalsi and Palli the investigated Leptinotarsa decemlineata P450s were strongly associated with detoxification of potato leaf allelochemicals [53••], Peng et al. 2016 examined the overexpression of A. gossypii CYP6DA2, implicated in gossypol tolerance [52].
Finally, many alternative P450 regulatory pathways have recently been uncovered, including roles for nuclear hormone receptor 96 (HR96), Hepatocyte Nuclear Factors (HNF-1A and HNF-3/FOXA), bZIP transcription factor CREB, nuclear protein P8 (containing the PFAM10195 domain) and nuclear receptor FTZ-F1 in overexpression of P450s associated with insecticide resistance [54, 55, 56, 57, 58]. Future studies should not only focus on further unraveling existing regulatory pathways (AhR/ARNT, CncC:Maf) but use unbiased approaches to uncover new regulatory mechanisms involved in P450 response to phytochemicals.
Functional validation of induced P450 genes
In contrast to P450s involved in resistance to pesticides, only few recent studies have functionally expressed P450 s to test whether they can metabolize the inducing phytochemical (Table 2) [37,59,60•]. It was shown that (amongst others) gossypol could induce CYP6AE gene expression in H. armigera [60•], but a subsequent study could not show in vitro metabolism by any of the candidate CYP6AEs after being functionally expressed in insect cells [59]. This example clearly indicates that carefulness is necessary when drawing conclusions based solely on the induction of a given P450, as the organism might react in a general stress response upon exposure. Thus, not all or only few of the induced genes might be functionally important and able to metabolize the chemical. On the other hand, in vitro findings of metabolism should also be complemented with in vivo experiments, for example using reverse-genetics based approaches such as RNAi and CRISPR-Cas9 knock-out or knock-down.
However, these genetic tools first need to be tailored for the species under investigation. A variety of dsRNA delivery systems exist, going from direct feeding to dsRNA expressing transgenic plants and microinjection, each with a different efficiency depending on the species [61, 62, 63, 64]. When designing RNAi experiments it is also important to keep in mind that in some species CYP genes can be duplicated and that silencing all paralogs with RNAi is practically unfeasible. In addition, specific silencing of a single P450 always needs to be confirmed, as this might be harder to achieve for members of recent P450 ‘blooms’ by cross-silencing. Silencing the gene encoding cytochrome P450 reductase, an obligatory co-enzyme of P450s, is another strategy that has been explored recently with T. urticae P450s [6,65], although this does not allow to study the effect of a single P450 and may result in pleiotropic effects unrelated to detoxification genes. In vivo validation of the role of P450 induction by RNAi is also complicated by uncertainties in timing of both the induction and the silencing, when quantifying the phenotypic effect. Lastly, RNAi might be straightforward for several coleopteran and orthopteran insects, but is not yet an easy option for all insects and mites. Even if Table 2 indicates many examples of RNAi in Lepidoptera, care should still be taken as other studies indicate difficulties in silencing genes by dsRNA in this insect order [64].
The CRISPR-Cas9 technique might be a valuable alternative for RNAi and can be used for gene knock-out of candidate loci. Although CRISPR-Cas9 is increasingly being used in the field of pesticide mode of action and elucidation of resistance mechanisms [66], only few studies have targeted P450 s involved in insect-plant interactions. In addition, gene knock-out might have more complex consequences compared to lack of induction, which makes this tool possibly too strong to look at subtle effects. Nevertheless, Wang et al. used the CRISPR-Cas9 system to successfully generate a CYP6AE cluster knock-out in H. armigera. Although no effects in viability under rearing conditions could be noted, a clearly increased susceptibility to plant toxins and insecticides was observed. Whether this phenotype is the result of the absence of constitutive expression, and/or lack of induction of this CYP6AE cluster is however hard to determine [60•].
Conclusions
Today, P450s still remain one of the most studied detoxification genes families. The last decade revealed that virtually all tested phytochemicals induced the transcription of some P450s, and the known number of P450s induced by phytochemicals has been drastically expanded, as more and more insect and mite species are studied by the availability of new technologies. Especially Clan 3 P450s and more specifically CYP6 family genes seem to respond to every phytochemical class presented in this review and most likely represent a core P450 family important in adaptation. Whereas studies using controlled administration provide strong insights in the specific responses to certain (sets of) phytochemicals, research that uses complete host-shifts preserves the hosts’ biological complexity. Thus, the use of transgenic plants with altered phytochemical content, might help to study induction in a biological relevant setup. Transcriptomes after short-term exposure to phytochemicals provide a powerful tool for identifying candidate genes and regulatory pathways involved in diet breadth, but unbiased validation of their causal role in detoxification in vivo remains important. As powerful reverse-genetic tools like RNAi and CRISPR-Cas9 are more and more available for non-model organisms, they will soon further dissect the specific role of P450 induction in plant interactions. Finally, most studies focus on P450 induction in polyphagous insects, while comparing both short- and long-term responses after host shifts of arthropods with different diet breath will allow to more completely study the evolutionary mechanisms of arthropod-plant interactions.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We apologize in advance to our many colleagues for the inspiring articles we did not have space to feature. This work was supported by the European Union’s Horizon 2020 research and innovation program [ERC consolidator grant 772026-POLYADAPT and 773902-SuperPests]. We sincerely thank Prof. Dr. René Feyereisen for his remarks and expertise while writing this review.
References
- 1.Ehrlich P.R., Raven R.H. Butterflies and plants: a study in coevolution. Evolution. 1964;18:506–608. [Google Scholar]
- 2.Suchan T., Alvarez N. Fifty years after Ehrlich and Raven, is there support for plant-insect coevolution as a major driver of species diversification? Entomologia Experimentalis et Applicata. 2015;157:98–112. [Google Scholar]
- 3.Janz N. Ehrlich and raven revisited: mechanisms underlying codiversification of plants and enemies. Annu Rev Ecol Evol Syst. 2011;42:71–89. [Google Scholar]
- 4••.Birnbaum S.S.L., Abbot P. Gene expression and diet breadth in plant-feeding insects: summarizing trends. Trends Ecol Evol. 2020;35:259–277. doi: 10.1016/j.tree.2019.10.014. [DOI] [PubMed] [Google Scholar]; Very comprehensive review, summarizing recent research-trends in studying the role of gene expression plasticity in relation to diet breadth in plant-feeding insects. It also provides an experimental framework to study diet breadth in plant-feeding insects.
- 5.Wybouw N., Zhurov V., Martel C., Bruinsma K.A., Hendrickx F., Grbic V., Van Leeuwen T. Adaptation of a polyphagous herbivore to a novel host plant extensively shapes the transcriptome of herbivore and host. Mol Ecol. 2015;24:4647–4663. doi: 10.1111/mec.13330. [DOI] [PubMed] [Google Scholar]
- 6.Salehipourshirazi G., Bruinsma K., Ratlamwala H., Dixit S., Arbona V., Widemann E., Milojevic M., Jin P., Bensoussan N., Gómez-Cadenas A. The generalist herbivore Tetranychus urticae (Koch) adapts to novel plant hosts through rapid evolution of metabolic resistance. bioRxiv. 2020 preprint. [Google Scholar]
- 7•.Muller C., Vogel H., Heckel D.G. Transcriptional responses to short-term and long-term host plant experience and parasite load in an oligophagous beetle. Mol Ecol. 2017;26:6370–6383. doi: 10.1111/mec.14349. [DOI] [PubMed] [Google Scholar]; One of the few studies, together with Wybouw et al. (Ref. [8]), investigating both short-term and long-term transcriptional responses after host transfer.
- 8.Mathers T.C., Chen Y., Kaithakottil G., Legeai F., Mugford S.T., Baa-Puyoulet P., Bretaudeau A., Clavijo B., Colella S., Collin O. Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 2017;18:27. doi: 10.1186/s13059-016-1145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen G.M., Song C.G., Ao Y.Q., Xiao Y.H., Zhang Y.J., Pan Y., He L. Transgenic cotton expressing CYP392A4 double-stranded RNA decreases the reproductive ability of Tetranychus cinnabarinus. Insect Sci. 2017;24:559–568. doi: 10.1111/1744-7917.12346. [DOI] [PubMed] [Google Scholar]
- 10.Feyereisen R. Insect CYP genes and P450 enzymes. In: Gilbert L.I., editor. Insect Molecular Biology and Biochemistry. Elsevier BV; London: 2012. pp. 236–316. [Google Scholar]
- 11•.Dermauw W., Van Leeuwen T., Feyereisen R. Diversity and evolution of the P450 family in arthropods. Insect Biochem Mol Biol. 2020;127 doi: 10.1016/j.ibmb.2020.103490. p. 103490. [DOI] [PubMed] [Google Scholar]; Nearly 3000 arthropod CYPs from 40 arthropod species were carefully annotated and analyzed, allowing to dissect and interpret both quantitative and qualitative variations in arthropod CYPomes over 500 million years of evolution.
- 12.Celorio-Mancera M., Ahn S.J., Vogel H., Heckel D.G. Transcriptional responses underlying the hormetic and detrimental effects of the plant secondary metabolite gossypol on the generalist herbivore Helicoverpa armigera. BMC Genomics. 2011;12:575. doi: 10.1186/1471-2164-12-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharath Chandra G., Asokan R., Manamohan M., Sita T. Cytochrome P450 isoforms transcriptional, larval growth and development responses to host allelochemicals in the generalist herbivore, Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) Curr Sci. 2016;111:901–906. [Google Scholar]
- 14.Ramsey J.S., Elzinga D.A., Sarkar P., Xin Y.R., Ghanim M., Jander G. Adaptation to nicotine feeding in Myzus persicae. J Chem Ecol. 2014;40:869–877. doi: 10.1007/s10886-014-0482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bass C., Zimmer C.T., Riveron J.M., Wilding C.S., Wondji C.S., Kaussmann M., Field L.M., Williamson M.S., Nauen R. Gene amplification and microsatellite polymorphism underlie a recent insect host shift. Proc Natl Acad Sci U S A. 2013;110:19460–19465. doi: 10.1073/pnas.1314122110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y., Liao M., Yang Q., Xiao J., Hu Z., Zhou L., Cao H. Transcriptome profiling reveals differential gene expression of detoxification enzymes in Sitophilus zeamais responding to terpinen-4-ol fumigation. Pest Biochem Physiol. 2018;149:44–53. doi: 10.1016/j.pestbp.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Barth M.B., Buchwalder K., Kawahara A.Y., Zhou X., Liu S., Krezdorn N., Rotter B., Horres R., Hundsdoerfer A.K. Functional characterization of the Hyles euphorbiae hawkmoth transcriptome reveals strong expression of phorbol ester detoxification and seasonal cold hardiness genes. Front Zool. 2018;15:20. doi: 10.1186/s12983-018-0252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi Y., Dou G., Yu Z., He H., Wang C., Li L., Zhou J., Liu D., Shi J., Li G. Z-ligustilide exerted hormetic effect on growth and detoxification enzymes of Spodoptera litura Larvae. Evid-based Complemen Altern Med. 2018;2018 doi: 10.1155/2018/7104513. p. 7104513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai L., Ma M., Gao G., Chen H. Dendroctonus armandi (Curculionidae: Scolytinae) cytochrome P450s display tissue specificity and responses to host terpenoids. Comparative biochemistry and physiology. Part B Biochem Mol Biol. 2016;201:1–11. doi: 10.1016/j.cbpb.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Pavlidi N., Gioti A., Wybouw N., Dermauw W., Ben-Yosef M., Yuval B., Jurkevich E., Kampouraki A., Van Leeuwen T., Vontas J. Transcriptomic responses of the olive fruit fly Bactrocera oleae and its symbiont Candidatus Erwinia dacicola to olive feeding. Sci Rep. 2017;7:42633. doi: 10.1038/srep42633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X., Liu D., Zhang R., Shi X. Transcriptional responses in defense-related genes of Sitobion avenae (Hemiptera: Aphididae) feeding on wheat and barley. J Econ Entomol. 2019;112:382–395. doi: 10.1093/jee/toy329. [DOI] [PubMed] [Google Scholar]
- 22.Wang D., Shi X., Liu D., Yang Y., Shang Z. Transcriptome profiling revealed potentially critical roles for digestion and defense-related genes in insects’ use of resistant host plants: a case study with Sitobion Avenae. Insects. 2020;11 doi: 10.3390/insects11020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X., Whitman D.W., Ma J., McNeill M.R., Zhang Z. Diet alters performance and transcription patterns in Oedaleus asiaticus (Orthoptera: Acrididae) grasshoppers. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186397. e0186397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pym A., Singh K.S., Nordgren A., Davies T.G.E., Zimmer C.T., Elias J., Slater R., Bass C. Host plant adaptation in the polyphagous whitefly, Trialeurodes vaporariorum, is associated with transcriptional plasticity and altered sensitivity to insecticides. BMC Genomics. 2019;20:996. doi: 10.1186/s12864-019-6397-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breeschoten T., Ros V.I.D., Schranz M.E., Simon S. An influential meal: host plant dependent transcriptional variation in the beet armyworm, Spodoptera exigua (Lepidoptera: Noctuidae) BMC Genomics. 2019;20:845. doi: 10.1186/s12864-019-6081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Malka O., Santos-Garcia D., Feldmesser E., Sharon E., Krause-Sakate R., Delatte H., van Brunschot S., Patel M., Visendi P., Mugerwa H. Species-complex diversification and host-plant associations in Bemisia tabaci: aplant-defence, detoxification perspective revealed by RNA-Seq analyses. Mol Ecol. 2018;27:4241–4256. doi: 10.1111/mec.14865. [DOI] [PMC free article] [PubMed] [Google Scholar]; Extensive study providing insights in the evolutionary pattern of a generalist insect herbivore (Bemisia tabaci) and its diversification-promoting hosts. It is hypothesized that a commonly-shared adaptive environmental-response machinery enables survival of species thriving on multiple hosts and thus passively contributing to diversification.
- 27.Huang X., Lv S., Zhang Z., Chang B.H. Phenotypic and transcriptomic response of the grasshopper Oedaleus asiaticus (Orthoptera: Acrididae) to toxic rutin. Front Physiol. 2020;11:52. doi: 10.3389/fphys.2020.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Kang X., Wu H., Silver K., Zhang J., Ma E., Zhu K.Y. Transcriptome-wide survey, gene expression profiling and exogenous chemical-induced transcriptional responses of cytochrome P450 superfamily genes in migratory locust (Locusta migratoria) Insect Biochem Mol Biol. 2018;100:66–77. doi: 10.1016/j.ibmb.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Abdeeva I., Piruzian E., Abdeev R., Bruskin S. INTECH Open Access Publisher; Rijeka: 2012. Transgenic Plants as a Tool for Plant Functional Genomics. [Google Scholar]
- 30.Zhurov V., Navarro M., Bruinsma K.A., Arbona V., Santamaria M.E., Cazaux M., Wybouw N., Osborne E.J., Ens C., Rioja C. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol. 2014;164:384–399. doi: 10.1104/pp.113.231555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Schweizer F., Heidel-Fischer H., Vogel H., Reymond P. Arabidopsis glucosinolates trigger a contrasting transcriptomic response in a generalist and a specialist herbivore. Insect Biochem Mol Biol. 2017;85:21–31. doi: 10.1016/j.ibmb.2017.04.004. [DOI] [PubMed] [Google Scholar]; Comprehensive study in which glucosinolate-induced transcriptional responses were compared between a generalist and a specialist lepidopteran herbivore using transgenic Arabidopsis thaliana.
- 32.Shan S., Soltis P.S., Soltis D.E., Yang B. Considerations in adapting CRISPR/Cas9 in nongenetic model plant systems. Appl Plant Sci. 2020;8 doi: 10.1002/aps3.11314. p. e11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wybouw N., Dermauw W., Tirry L., Stevens C., Grbic M., Feyereisen R., Van Leeuwen T. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. eLife. 2014;3 doi: 10.7554/eLife.02365. p. e02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan W., Acevedo T., Harris E.V., Alcaide T.Y., Walters J.R., Hunter M.D., Gerardo N.M., De Roode J.C. Transcriptomics of monarch butterflies (Danaus plexippus) reveals that toxic host plants alter expression of detoxification genes and down‐regulate a small number of immune genes. Mol Ecol. 2019;28:4845–4863. doi: 10.1111/mec.15219. [DOI] [PubMed] [Google Scholar]
- 35.Jin M., Liao C., Fu X., Holdbrook R., Wu K., Xiao Y. Adaptive regulation of detoxification enzymes in Helicoverpa armigera to different host plants. Insect Mol Biol. 2019;28:628–636. doi: 10.1111/imb.12578. [DOI] [PubMed] [Google Scholar]
- 36.Mittapelly P., Bansal R., Michel A. Differential expression of cytochrome P450 CYP6 genes in the brown marmorated stink bug, Halyomorpha halys (Hemiptera: Pentatomidae) J Econ Entomol. 2019;112:1403–1410. doi: 10.1093/jee/toz007. [DOI] [PubMed] [Google Scholar]
- 37.Calla B., Wu W.Y., Dean C.A.E., Schuler M.A., Berenbaum M.R. Substrate-specificity of cytochrome P450-mediated detoxification as an evolutionary strategy for specialization on furanocoumarin-containing hostplants: CYP6AE89 in parsnip webworms. Insect Mol Biol. 2020;29:112–123. doi: 10.1111/imb.12612. [DOI] [PubMed] [Google Scholar]
- 38.Hafeez M., Qasim M., Ali S., Yousaf H.K., Waqas M., Ali E., Ahmad M.A., Jan S., Bashir M.A., Noman A. Expression and functional analysis of P450 gene induced tolerance/resistance to lambda-cyhalothrin in quercetin fed larvae of beet armyworm Spodoptera exigua (Hubner) Saudi J Biol Sci. 2020;27:77–87. doi: 10.1016/j.sjbs.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng T., Wu J., Wu Y., Chilukuri R.V., Huang L., Yamamoto K., Feng L., Li W., Chen Z., Guo H. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat Ecol Evol. 2017;1:1747–1756. doi: 10.1038/s41559-017-0314-4. [DOI] [PubMed] [Google Scholar]
- 40.Sun Z., Shi Q., Li Q., Wang R., Xu C., Wang H., Ran C., Song Y., Zeng R. Identification of a cytochrome P450 CYP6AB60 gene associated with tolerance to multi-plant allelochemicals from a polyphagous caterpillar tobacco cutworm (Spodoptera litura) Pest Biochem Physiol. 2019;154:60–66. doi: 10.1016/j.pestbp.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Shahjaman M., Manir Hossain Mollah M., Rezanur Rahman M., Islam S.M.S., Nurul Haque Mollah M. Robust identification of differentially expressed genes from RNA-seq data. Genomics. 2020;112:2000–2010. doi: 10.1016/j.ygeno.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Brown R.P., McDonnell C.M., Berenbaum M.R., Schuler M.A. Regulation of an insect cytochrome P450 monooxygenase gene (CYP6B1) by aryl hydrocarbon and xanthotoxin response cascades. Gene. 2005;358:39–52. doi: 10.1016/j.gene.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 43.Xu L., Li D.Z., Luo Y.Y., Qin J.Y., Qiu L.H. Identification of the 2-tridecanone cis-acting element in the promoter of cytochrome P450 CYP6B7 in Helicoverpa armigera. Insect Sci. 2018;25:959–968. doi: 10.1111/1744-7917.12479. [DOI] [PubMed] [Google Scholar]
- 44.Peng T., Chen X., Pan Y., Zheng Z., Wei X., Xi J., Zhang J., Gao X., Shang Q. Transcription factor aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator is involved in regulation of the xenobiotic tolerance-related cytochrome P450 CYP6DA2 in Aphis gossypii Glover. Insect Mol Biol. 2017;26:485–495. doi: 10.1111/imb.12311. [DOI] [PubMed] [Google Scholar]
- 45.Li F., Ma K., Chen X., Zhou J.J., Gao X. The regulation of three new members of the cytochrome P450 CYP6 family and their promoters in the cotton aphid Aphis gossypii by plant allelochemicals. Pest Manage Sci. 2019;75:152–159. doi: 10.1002/ps.5081. [DOI] [PubMed] [Google Scholar]
- 46.Pan Y., Peng T., Xu P., Zeng X., Tian F., Song J., Shang Q. Transcription factors AhR/ARNT regulate the expression of CYP6CY3 and CYP6CY4 switch conferring nicotine adaptation. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20184521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giraudo M., Hilliou F., Fricaux T., Audant P., Feyereisen R., Le Goff G. Cytochrome P450s from the fall armyworm (Spodoptera frugiperda): responses to plant allelochemicals and pesticides. Insect Mol Biol. 2015;24:115–128. doi: 10.1111/imb.12140. [DOI] [PubMed] [Google Scholar]
- 48.Dinan L., Whiting P., Girault J., Lafont R., Dhadialla T.S., Cress D.E., Mugat B., Antoniewski C., Lepesant J.-A. Cucurbitacins are insect steroid hormone antagonists acting at the ecdysteroid receptor. Biochem J. 1997;327:643–650. doi: 10.1042/bj3270643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Inoue J., Choi J.M., Nakamura S., Yan Z., Fushinobu S., Kamada H., Kato H., Hashidume T., Shimizu M. Identification of the Flavonoid luteolin as a repressor of the transcription factor hepatocyte nuclear factor 4alpha. J Biol Chem. 2015;290:24021–24035. doi: 10.1074/jbc.M115.645200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palli S.R. CncC/Maf-mediated xenobiotic response pathway in insects. Arch Insect Biochem Physiol. 2020;104 doi: 10.1002/arch.21674. p. e21674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilding C.S. Regulating resistance: CncC:Maf, antioxidant response elements and the overexpression of detoxification genes in insecticide resistance. Curr Opin Insect Sci. 2018;27:89–96. doi: 10.1016/j.cois.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Peng T., Pan Y., Gao X., Xi J., Zhang L., Yang C., Bi R., Yang S., Xin X., Shang Q. Cytochrome P450 CYP6DA2 regulated by cap’ n’collar isoform C (CncC) is associated with gossypol tolerance in Aphis gossypii Glover. Insect Mol Biol. 2016;25:450–459. doi: 10.1111/imb.12230. [DOI] [PubMed] [Google Scholar]
- 53••.Kalsi M., Palli S.R. Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say) Insect Biochem Mol Biol. 2017;83:1–12. doi: 10.1016/j.ibmb.2017.02.002. [DOI] [PubMed] [Google Scholar]; Pioneering study revealing that potato plant extracts induce Colorado potato beetle P450 expression via the CncC pathway.
- 54.Faucon F., Gaude T., Dusfour I., Navratil V., Corbel V., Juntarajumnong W., Girod R., Poupardin R., Boyer F., Reynaud S. In the hunt for genomic markers of metabolic resistance to pyrethroids in the mosquito Aedes aegypti: An integrated next-generation sequencing approach. PLoS Negl Trop Dis. 2017;11 doi: 10.1371/journal.pntd.0005526. p. e0005526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia H., Peiling L., Yuan H., Wencai L., Zhifeng X., Lin H. P8 nuclear receptor responds to acaricides exposure and regulates transcription of P450 enzyme in the two-spotted spider mite, Tetranychus urticae. Comparative biochemistry and physiology. Toxicol Pharmacol. 2019;224 doi: 10.1016/j.cbpc.2019.108561. 108561. [DOI] [PubMed] [Google Scholar]
- 56.Yang X., Deng S., Wei X., Yang J., Zhao Q., Yin C., Du T., Guo Z., Xia J., Yang Z. MAPK-directed activation of the whitefly transcription factor CREB leads to P450-mediated imidacloprid resistance. Proc Natl Acad Sci U S A. 2020;117:10246–10253. doi: 10.1073/pnas.1913603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X., Shan C., Li F., Liang P., Smagghe G., Gao X. Transcription factor FTZ-F1 and cis-acting elements mediate expression of CYP6BG1 conferring resistance to chlorantraniliprole in Plutella xylostella. Pest Manage Sci. 2019;75:1172–1180. doi: 10.1002/ps.5279. [DOI] [PubMed] [Google Scholar]
- 58.Afschar S., Toivonen J.M., Hoffmann J.M., Tain L.S., Wieser D., Finlayson A.J., Driege Y., Alic N., Emran S., Stinn J. Nuclear hormone receptor DHR96 mediates the resistance to xenobiotics but not the increased lifespan of insulin-mutant Drosophila. Proc Natl Acad Sci U S A. 2016;113:1321–1326. doi: 10.1073/pnas.1515137113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi Y., Wang H., Liu Z., Wu S., Yang Y., Feyereisen R., Heckel D.G., Wu Y. Phylogenetic and functional characterization of ten P450 genes from the CYP6AE subfamily of Helicoverpa armigera involved in xenobiotic metabolism. Insect Biochem Mol Biol. 2018;93:79–91. doi: 10.1016/j.ibmb.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 60•.Wang H., Shi Y., Wang L., Liu S., Wu S., Yang Y., Feyereisen R., Wu Y. CYP6AE gene cluster knockout in Helicoverpa armigera reveals role in detoxification of phytochemicals and insecticides. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07226-6. p. 4820. [DOI] [PMC free article] [PubMed] [Google Scholar]; CRISPR-Cas9 was used to knock-out a cluster of nine P450 genes, significantly reducing the survival rate of the insect when exposed to two classes of host plant chemicals. One of the few studies in which the role of P450s in phytochemical detoxification was functionally validated by reverse genetics.
- 61.Suzuki T., Espana M.U., Nunes M.A., Zhurov V., Dermauw W., Osakabe M., Van Leeuwen T., Grbic M., Grbic V. Protocols for the delivery of small molecules to the two-spotted spider mite, Tetranychus urticae. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180658. p. e0180658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terenius O., Papanicolaou A., Garbutt J.S., Eleftherianos I., Huvenne H., Kanginakudru S., Albrechtsen M., An C., Aymeric J.L., Barthel A. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Vogel E., Santos D., Mingels L., Verdonckt T.W., Broeck J.V. RNA interference in insects: protecting beneficials and controlling pests. Front Physiol. 2018;9 doi: 10.3389/fphys.2018.01912. p. 1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cooper A.M., Silver K., Zhang J., Park Y., Zhu K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manage Sci. 2019;75:18–28. doi: 10.1002/ps.5126. [DOI] [PubMed] [Google Scholar]
- 65.Adesanya A.W., Cardenas A., Lavine M.D., Walsh D.B., Lavine L.C., Zhu F. RNA interference of NADPH-cytochrome P450 reductase increases susceptibilities to multiple acaricides in Tetranychus urticae. Pest Biochem Physiol. 2020;165 doi: 10.1016/j.pestbp.2020.02.016. p. 104550. [DOI] [PubMed] [Google Scholar]
- 66.Douris V., Steinbach D., Panteleri R., Livadaras I., Pickett J.A., Van Leeuwen T., Nauen R., Vontas J. Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis. Proc Natl Acad Sci U S A. 2016;113:14692–14697. doi: 10.1073/pnas.1618258113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Q., Sun Z., Shi Q., Wang R., Xu C., Wang H., Song Y., Zeng R. RNA-Seq analyses of midgut and fat body tissues reveal the molecular mechanism underlying Spodoptera litura resistance to tomatine. Front Physiol. 2019;10:8. doi: 10.3389/fphys.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Halon E., Eakteiman G., Moshitzky P., Elbaz M., Alon M., Pavlidi N., Vontas J., Morin S. Only a minority of broad-range detoxification genes respond to a variety of phytotoxins in generalist Bemisia tabaci species. Sci Rep. 2015;5 doi: 10.1038/srep17975. p. 17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coelho A., Fraichard S., Le Goff G., Faure P., Artur Y., Ferveur J.F., Heydel J.M. Cytochrome P450-dependent metabolism of caffeine in Drosophila melanogaster. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117328. p. e0117328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang R.L., Staehelin C., Xia Q.Q., Su Y.J., Zeng R.S. Identification and characterization of CYP9A40 from the tobacco cutworm moth (Spodoptera litura), a cytochrome P450 gene induced by plant allelochemicals and insecticides. Int J Mol Sci. 2015;16:22606–22620. doi: 10.3390/ijms160922606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang R.L., Xia Q.Q., Baerson S.R., Ren Y., Wang J., Su Y.J., Zheng S.C., Zeng R.S. A novel cytochrome P450 CYP6AB14 gene in Spodoptera litura (Lepidoptera: Noctuidae) and its potential role in plant allelochemical detoxification. J Insect Physiol. 2015;75:54–62. doi: 10.1016/j.jinsphys.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 72.Liu D., Yuan Y., Li M., Qiu X. Effects of dietary quercetin on performance and cytochrome P450 expression of the cotton bollworm, Helicoverpa armigera. Bull Entomol Res. 2015;105:771–777. doi: 10.1017/S0007485315000760. [DOI] [PubMed] [Google Scholar]
- 73.Li B., Zhang H., Ni M., Wang B.B., Li F.C., Xu K.Z., Shen W.D., Xia Q.Y., Zhao P. Identification and characterization of six cytochrome P450 genes belonging to CYP4 and CYP6 gene families in the silkworm, Bombyx mori. Mol Biol Rep. 2014;41:5135–5146. doi: 10.1007/s11033-014-3379-z. [DOI] [PubMed] [Google Scholar]
- 74.Seong K.M., Kim Y., Kim D., Pittendrigh B.R., Kim Y.H. Identification of transcriptional responsive genes to acetic acid, ethanol, and 2-phenylethanol exposure in Drosophila melanogaster. Pest Biochem Physiol. 2020;165 doi: 10.1016/j.pestbp.2020.02.018. p. 104552. [DOI] [PubMed] [Google Scholar]
- 75.Crava C.M., Brutting C., Baldwin I.T. Transcriptome profiling reveals differential gene expression of detoxification enzymes in a hemimetabolous tobacco pest after feeding on jasmonate-silenced Nicotiana attenuata plants. BMC Genomics. 2016;17:1005. doi: 10.1186/s12864-016-3348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng L., Zhao Y., Wang H., Song C., Shangguan X., Ma Y., Zhu L., He G. Functional study of cytochrome P450 enzymes from the brown planthopper (Nilaparvata lugens Stal) to analyze its adaptation to BPH-resistant rice. Front Physiol. 2017;8:972. doi: 10.3389/fphys.2017.00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva-Brandao K.L., Horikoshi R.J., Bernardi D., Omoto C., Figueira A., Brandao M.M. Transcript expression plasticity as a response to alternative larval host plants in the speciation process of corn and rice strains of Spodoptera frugiperda. BMC Genomics. 2017;18:792. doi: 10.1186/s12864-017-4170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]