Abstract
In response to replication hindrances, DNA replication forks frequently stall and are remodelled into a four-way junction. In such a structure the annealed nascent strand is thought to resemble a DNA double-strand break and remodelled forks are vulnerable to nuclease attack by MRE11 and DNA2. Proteins that promote the recruitment, loading and stabilisation of RAD51 onto single-stranded DNA for homology search and strand exchange in homologous recombination (HR) repair and inter-strand cross-link repair also act to set up RAD51-mediated protection of nascent DNA at stalled replication forks. However, despite the similarities of these pathways, several lines of evidence indicate that fork protection is not simply analogous to the RAD51 loading step of HR. Protection of stalled forks not only requires separate functions of a number of recombination proteins, but also utilises nucleases important for the resection steps of HR in alternative ways. Here we discuss how fork protection arises and how its differences with HR give insights into the differing contexts of these two pathways.
Keywords: Replication fork protection, BRCA1, BRCA2, RAD51, Fork reversal, Homologous recombination
1. Introduction
Protection of the reversed replication fork from untimely nuclease attack has emerged as a critical process for maintaining genome stability. Recent insights into both the mechanism and fundamental importance of replication fork protection have relied on a few key techniques (see Box 1 and Fig. 1), revealing significant overlap between factors involved in this process and homologous recombination (HR). Much of this work has underscored the relevance of replication fork protection to chemotherapeutic responses, demonstrating the potential clinical impact that a greater understanding of this process could hold. Throughout this review, we examine the key factors involved in the closely linked processes of replication fork reversal and protection, and contrast their distinct functions in fork protection versus HR.
Box 1. Examination of fork reversal, fork protection and post replicative gaps.
Fork reversal can be directly examined by two major techniques: electron microscopy (EM) and neutral-neutral 2D gel electrophoresis. In vivo crosslinking of DNA followed by EM has suggested that fork reversal is a universal response to replication stress in eukaryotic cells [2]. Combining EM with DNA fibre spreading, described below, has suggested replication fork protection follows fork reversal [9]. Neutral-neutral 2D gel electrophoresis separates DNA molecules, allowing X-shaped structures including reversed forks, to be resolved [156]. These are powerful techniques but assigning unique structures to distinct electrophoretic shifts is difficult and whether EM reflects actual proportions of reversed forks is not clear. EM has also been used to study the composition of reversed forks by assessing the relative filament thickness to indicate double-stranded DNA (dsDNA) versus single-stranded DNA (ssDNA), revealing that nucleases contribute to the amount of ssDNA at reversed forks [8].
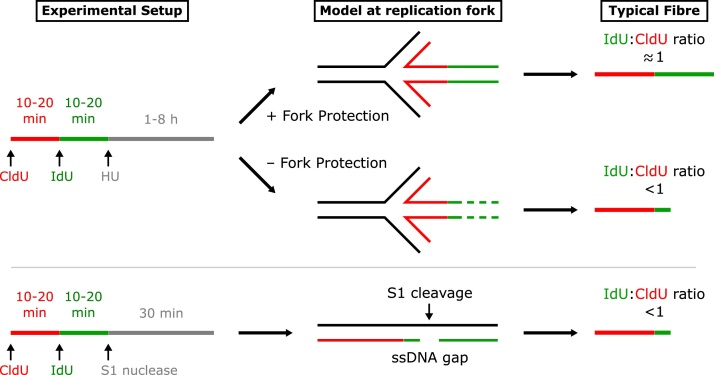
DNA fibre spreading allows replication fork dynamics to be assessed [157,158]. Analysis of fork protection involves introducing the thymidine analogues CldU and IdU sequentially into cells to label adjacent sections of nascent DNA, in combination with incubations with agents that slow or block replication, such as the ribonucleotide reductase inhibitor hydroxyurea (HU), which can be added after the second label [80]. HU reduces dNTP synthesis, leading to depletion of the cellular dNTP pool and global replication fork slowing and stalling [42]. DNA fibres are then spread and stained to measure the lengths of adjacent labels, where a ratio of approximately 1 between the two labels is expected. An alternative approach is to introduce the stalling agent after a single label and compare track lengths with those in cells untreated with the stalling agent [8,80] A shortening of the label on addition of replication stalling agents, combined with an ability to rescue this shortening by nuclease inhibition, is suggestive of a defect in stalled fork protection (Fig. 1). This assay is extensively used to explore the context of factors contributing to fork protection by depleting or inhibiting additional factors to either rescue or exacerbate the defect. Intriguingly, combining the loss of protective factors often appears epistatic even when they defend against different nucleases (e.g. [59,63,71]). Nevertheless, co-depletion of some protective factors results in additive degradation (e.g. RECQL5 + BOD1L [71], CtIP + BRCA1 [63]) therefore revealing evidence for separable pathways using this assay.
EM has also observed occurrence of single-stranded gaps, seen as thin filaments (ssDNA) in replicated duplexes. These gaps are suppressed by loss of nucleases involved in the digestion of stalled replication forks, suggesting gaps may be a consequence of restart of an unprotected fork [8]. To detect these structures in DNA fibre spreading, a modified protocol is used in which the S1 nuclease nicks the ssDNA opposite gaps, converting a ssDNA gap into a double-strand break (DSB). This results in loss of the sequence after the gap, therefore giving a shorter second label in fibre-spreading assays [159].
Alt-text: Box 1
Fig. 1.
Schematic of the DNA fibre-spreading assay. To assess replication fork protection, cells are sequentially incubated with the thymidine analogues CldU and IdU, followed by the addition of a replication stress-inducing agent, most commonly HU. After spreading, fixing and staining, the fibres can be visualised. A second label tract shorter than the first label may be due to nucleolytic degradation of the nascent strand at stalled replication forks, indicating defective fork protection. This protocol can be modified to assay for single-stranded DNA (ssDNA) gaps, by introducing the S1 nuclease which cleaves opposite gaps to result in a shorter second label.
2. Fork reversal and restoration
During DNA replication, fork progression may be hindered by various obstacles, including DNA lesions, torsional stress, secondary structures, nucleotide shortage, transcription complexes and DNA:RNA hybrids [1]. Upon encountering many, if not all, of these obstacles, forks stall and are remodelled into a four-way junction (also called ‘reversed fork’ or ‘chicken foot’) [2]. This is achieved by the re-annealing of parental strands alongside the unwinding and annealing of newly synthesised DNA to form a regressed arm [3]. Fork reversal restrains fork progression under conditions of replication stress [4,5], which may allow time for repair machineries to resolve perturbations [3] and prevent progression of DNA synthesis across lesions, which might otherwise result in DNA double-strand breaks (DSBs) [6]. Fork reversal also places lesions on the parental template back into the context of double-stranded DNA (dsDNA) to facilitate repair [7].
2.1. Fork reversal
Fork reversal is mediated by RAD51 [2,[8], [9], [10]], and the SNF2-family DNA translocases SMARCAL1, ZRANB3 and HLTF (SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily A Like 1, Zinc Finger RANBP2-Type Containing 3 and Helicase Like Transcription Factor, respectively) [[11], [12], [13], [14], [15], [16], [17]]. Inactivation of these SNF2-family remodellers rescues fork protection defects, for example in BRCA1- or BRCA2-deficient cells [9,18], placing remodellers upstream of fork protection.
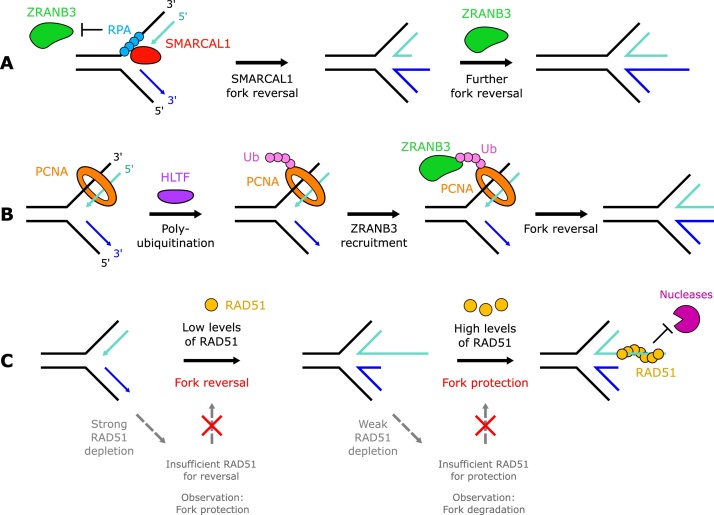
SMARCAL1 localises to stalled replication forks through interaction with the single-stranded binding protein RPA [16,19]. Single-molecule experiments suggest that fork reversal by SMARCAL1 is promoted in bursts with pausing in between [11,20]. This pausing, and the inhibition of fork reversal induced by ATR phosphorylation of SMARCAL1, have been proposed to prevent excessive fork modelling [21,22]. Similarly human RAD52, a single-stranded DNA (ssDNA) binding protein with many roles in HR (reviewed in [23]), acts to restrain SMARCAL1 recruitment to forks [24].
HLTF acts at least in part to oppose the permissive DNA synthesis mediated by FANCJ at stalled forks [5,25]. FANCJ is capable of driving replication through barriers such as secondary DNA structures [26]. Without FANCJ, excessive remodelling by HLTF contributes to fork degradation, whereas in cells lacking HLTF, replication under conditions of replication stress proceeds at a faster rate [25]. In addition to its dsDNA translocase activity, HLTF is an E3 ubiquitin ligase which can polyubiquitinate PCNA [27]. The interaction of ZRANB3 with polyubiquitinated PCNA stimulates ZRANB3 fork reversal activity both in vitro and in vivo [4,12,20,28].
The mechanism of fork regression by SNF2-family translocases is not widely understood, however single-molecule experiments have suggested that Rad5, the budding yeast orthologue of HLTF, unwinds the leading strand at the fork junction upon ATP binding [29]. The four-way junction may then form spontaneously, followed by branch migration to extend the regressed arm [29]. While several SNF2-family translocases are able to catalyse fork reversal, their roles are not redundant [12]. Given their preferences for distinct substrates, it is likely they act upon a variety of replication structures present at stalled forks. HLTF binds to free 3’−OH ends on the nascent leading strand [30], while SMARCAL1 and ZRANB3 bind preferentially to DNA substrates with splayed arms [31,32] but are regulated by RPA in contrasting ways. These remodellers may also act sequentially in common pathways [20,21,33] (Fig. 2A and B).
Fig. 2.
SNF2-family translocases and RAD51 mediate fork reversal. The translocases SMARCAL1, ZRANB3 and HLTF may act in a sequential manner. A: SMARCAL1, recruited and stimulated by RPA, may catalyse initial fork reversal. Once RPA has been evicted, ZRANB3 activity will no longer be suppressed, allowing further fork reversal. B: HLTF polyubiquitinates PCNA, which can recruit ZRANB3 to forks – suggesting ZRANB3 may act downstream of HLTF. C: Differential levels of RAD51 are required for fork reversal versus fork protection. Strong, near complete, RAD51 depletion prevents fork reversal, leading to fork protection. Weaker depletion leaves sufficient RAD51 capable of promoting fork reversal but not enough to support fork protection, leading to degradation by MRE11 and other nucleases.
Other factors have also been shown to catalyse fork reversal in vitro, including the translocases RAD54 [34,35] and FANCM [36,37], and the RecQ helicases BLM, WRN and RECQL5 [[38], [39], [40]], although their relevance in vivo is unclear. The helicase FBH1 is also able to reverse replication forks in vitro and in vivo [10,41], however its role in fork reversal is difficult to define clearly since FBH1 is also involved in modulating RAD51 filament stability (see Section 3.4.4).
RAD51 co-purifies with replication forks and associates with chromatin during S phase in perturbed and unperturbed conditions [[42], [43], [44], [45]]. Depleting RAD51 reduces the proportion of reversed replication structures observed under electron microscopy (EM) and increases the frequency of post-replicative ssDNA gaps at the replication fork, suggesting that ssDNA at forks acts as a precursor for RAD51-mediated fork reversal [2,9]. RAD51 alone is unable to catalyse fork reversal in vitro [35], but may co-operate with remodelling enzymes to mediate reversal. Alternatively, RAD51 binding to reversed forks may capture the DNA ends and shift the equilibrium fork state towards reversal [46]. Intriguingly, the RAD51-T131 P mutant that is unable to form stable filaments can promote fork reversal, suggesting that reversal may not require stable RAD51 filament formation [9]. Furthermore, unlike the function of RAD51 in HR or fork protection, RAD51-mediated fork reversal can occur independently of BRCA2 [9]. In this context, RAD51 may instead be recruited to replication forks by direct interactions with DNA polymerase alpha, RAD54 or RAD51C [15,35,47]. The dual role of RAD51 in mediating fork reversal and then protecting those reversed forks has led to seemingly contradictory observations of protected or degraded forks upon RAD51 depletion [2,9,[48], [49], [50]]. This can be resolved by evidence that suggests low levels of RAD51 are sufficient for fork reversal but are not sufficient for fork protection [51] (Fig. 2C).
2.2. Restoration of forks and suppression of fork reversal
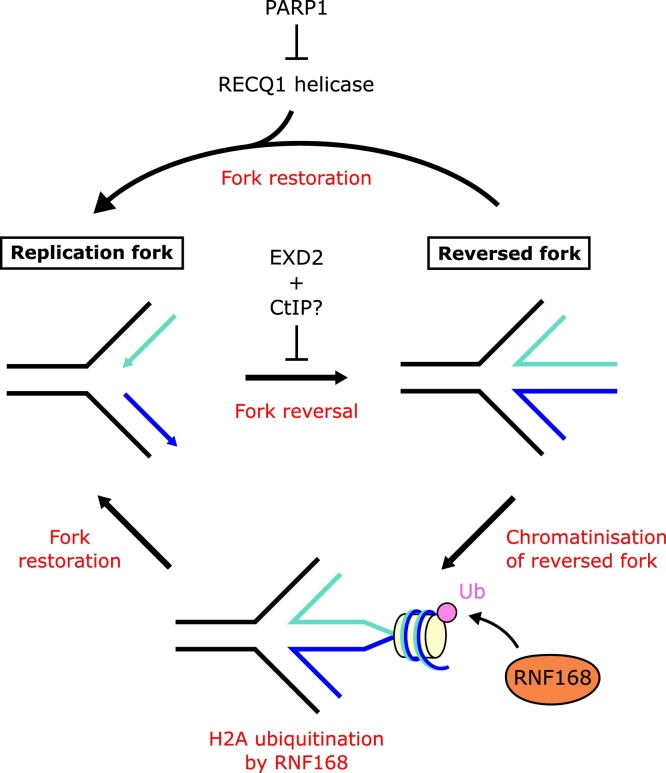
The reversed fork can be restored back into a three-way junction to resume DNA synthesis [52]. Resolution is mediated by the RECQ1 helicase [3,53] and regulated by PARP1, which inhibits RECQ1 activity [53] (Fig. 3).
Fig. 3.
Suppression of replication fork reversal. The nuclease EXD2 may disfavour fork reversal by processing a stalled replication fork into a form that is refractory to remodelling. CtIP may also be involved in suppressing fork reversal. Reversed replication forks can be restored to a canonical fork by RECQ1 helicase. PARP1 negatively regulates RECQ1 activity, controlling the balance between reversal and restoration of replication forks. Chromatinisation of the regressed arm may act as a target for H2A ubiquitination by RNF168, which may also suppress fork reversal and promote restoration of the fork to restart replication.
In yeast and bacteria, the nucleolytic processing of nascent DNA strands at stalled forks inhibits fork reversal and promotes the regeneration of functioning replication forks [54,55]. Human EXD2 is a 3’-5’ exonuclease that functionally interacts with MRE11 during HR, acting to accelerate resection through the 3'-5' exonuclease activity of MRE11 [[56], [57], [58]]. Thus, EXD2 might be expected to also co-operate with MRE11 in the degradation of stalled replication forks. Surprisingly however, recent work suggests EXD2 acts to prevent fork reversal and therefore suppresses subsequent MRE11-mediated degradation [59]. Using PARP1 accumulation as an indication of fork regression [60], Nieminuszczy et al. found that EXD2 loss increases PARP1 accumulation at stalled forks. Moreover, purified EXD2 can degrade substrates with extruded, single-stranded nascent DNA expected to arise at stalled forks [59]. These experiments suggest a model in which degradation of nascent DNA can prevent fork remodelling in mammalian cells.
Similarly, CtIP directs MRE11-mediated short-range DNA resection in HR [61]. As might be expected for a co-factor of MRE11, the degradation of stalled forks in BRCA2-mutant cells was prevented by CtIP loss [62]. However, depletion of CtIP alone has been shown to have conflicting roles, with either no impact on stalled replication structures [8,62] or increasing their degradation [63]. Thus in some contexts CtIP may prevent fork regression or contribute to protection of regressed forks [63]. This activity may be mediated by its specificity in binding Y-shaped DNA structures present at stalled forks [64].
Observations from crosslinked regressed forks suggest that the annealed nascent DNA comprising the ‘toe’ of the regressed fork is chromatinised [65]. These exposed ends, reminiscent of chromatin adjacent to a DSB, may engage the E3 ubiquitin ligase RNF168 for H2A ubiquitination, since RNF168, and factors that it recruits, are able to suppress fork reversal and promote ongoing replication during unperturbed S phase [65].
3. Degradation of reversed forks
Once reversed into a four-way junction, replication forks can be degraded as part of normal cellular physiology. Following prolonged fork stalling in wild-type cells, nascent DNA is progressively degraded by the 5’-3’ exonuclease DNA2, in partnership with WRN ATPase (helicase) function, but is not degraded by MRE11, MUS81, EXO1 or CtIP (although see Section 2.2) [8]. In contrast, in the absence of specific fork protection factors, nascent DNA degradation occurs more rapidly. Degradation of deprotected forks involves several nucleases in addition to DNA2, including structure-specific nucleases (MUS81-EME1, SLX4-XPF-ERCC1) and 3’-5’ exonucleases (MRE11-EXO1). These observations suggest a need to defend the stalled structure at several potential nuclease entry points when protective factors are missing. In reports examining fork-protective factors, a full exploration of which nucleases are not involved is frequently incomplete (Table 1); nevertheless the current data suggests there are different branches of fork protection which defend against particular nucleases. Given the role of DNA2 in degradation of stalled forks in wild-type cells, it is tempting to consider factors that give rise to DNA2 vulnerability as those whose loss further exposes an otherwise ‘normal’ stalled fork state. Meanwhile loss of factors that expose vulnerabilities to other nucleases implies that fork structures in these contexts are abnormal. However, this view is also an oversimplification. For example, the WRN helicase assists DNA2 in degrading regressed forks following HU exposure [8], but WRN exonuclease activity prevents degradation by MRE11 and EXO1 upon exposure to low doses of camptothecin [40]. Thus WRN activities and fork nuclease vulnerabilities differ according to the replicative stress. Our current understanding of the precise substrates and vulnerabilities that define the differences between forks exposed to different agents or lacking different protective factors is poor.
Table 1.
A summary of the known nuclease susceptibilities of fork protection defects triggered by the depletion of specific factors.
| Depletion of nuclease can rescue defect? |
References | ||||
|---|---|---|---|---|---|
| Factor involved in fork protection | MRE11 | DNA2 | MUS81-EME1 | SLX4-ERCC1 | |
| ABRO1 | ✗ | ✓ | ? | ? | [73]. |
| ATRX | ✓ | ? | ? | ? | [83]. |
| BOD1L | ✗ | ✓ | ? | ? | [71]. |
| BRCA1 | ✓ | ✗ | ✗ | ? | [62,76]. |
| BRCA2 | ✓ | ✗ | ✓ | ? | [62,76,80]. |
| Cohesin | ✓ | ? | ? | ? | [77]. |
| CtIP | ✗ | ✓ | ✗ | ✗ | [63]. |
| EXD2 | ✓ | ? | ? | ? | [59]. |
| FANCB | ✓ | ? | ? | ? | [78]. |
| FANCD2 | ✓ | ? | ? | ? | [79]. |
| PALB2 | ? | ? | ? | ? | [103]. |
| PDS5B | ✓ | ? | ? | ? | [77]. |
| RAD51 | ✓ | ? | ? | ? | [48,49]. |
| RAD51C | ✓ | ? | ? | ? | [82]. |
| RECQ1 | ✗ | ✓ | ✗ | ✗ | [8]. |
| RECQL5 | ✓ | ? | ? | ? | [78]. |
| REV1 | ✓ | ? | ? | ? | [84]. |
| RIF1 | ✗ | ✓ | ? | ? | [69]. |
| RNF168 | ✓ | ? | ? | ? | [65]. |
| SETD1A | ✗ | ✓ | ? | ? | [72] |
| WRNIP1 | ✓ (Long) | ✓(short) | ✗ | ✓(short) | [66,67]. |
| XRCC2 | ✓ | ? | ? | ? | [82]. |
| XRCC3 | ✓ | ? | ? | ? | [82]. |
3.1. Protection against SLX4-associated nuclease activity
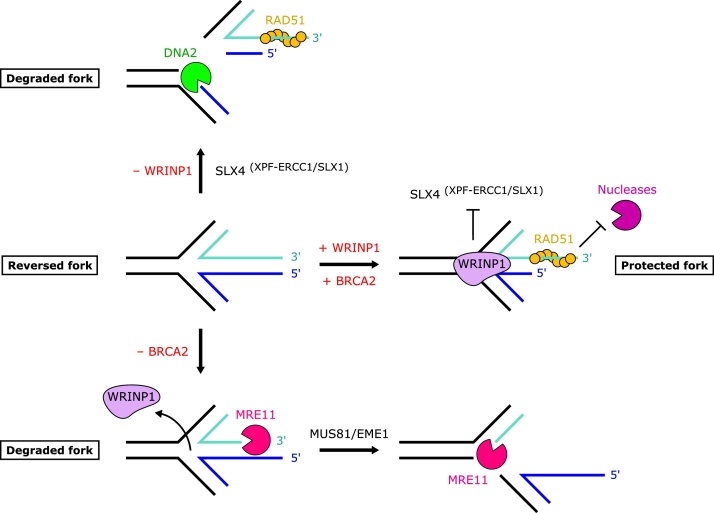
The shorter and less common form of the Werner’s interacting protein, WRNIP1, counteracts degradation mediated by the SLX4-interacting structure-specific nuclease XPF-ERCC1 (and to a lesser extent SLX4-SLX1) [66]. Interestingly, depletion or inhibition of DNA2 also protects forks in WRNIP1-deficient cells [66]. Thus one model is that SLX4 targeting of unprotected replication fork structures generates a substrate for DNA2-mediated digestion. In another context, or perhaps depending on the isoform expressed, WRNIP1 can also protect against MRE11-mediated degradation [67]. Importantly, WRNIP1 could not protect junctions from MUS81-EME1 in vitro and SLX4 is not relevant to fork protection seen in a BRCA2-deficient context [66], indicating WRNIP1 and BRCA2 protect replication forks through different mechanisms (Fig. 4).
Fig. 4.
WRNIP1 protects replication forks in a distinct manner to BRCA2. WRNIP1 is able to bind the four-way junction of a reversed replication fork and protects it from cleavage by SLX4(XPF−ERCC1/SLX1). In the absence of WRNIP1, cleavage of the fork by SLX4(XPF−ERCC1/SLX1) creates a substrate for nascent DNA degradation by DNA2. In contrast, following loss of BRCA2, fork protection is compromised in a distinct manner, whereby initial degradation by MRE11 enables cleavage of the fork by MUS81/EME1, subsequently facilitating extensive MRE11-mediated degradation of DNA.
3.2. Protection against DNA2 activity
A structure-specific role may be relevant to fork protection mediated by Replication Timing Regulatory Factor 1 (RIF1), which binds to cruciform structures in vitro [68]. RIF1 protects structures from DNA2 nuclease activity [69], and this requires its PP1-interaction motif. Loss of PP1 results in fork degradation, possibly through deregulating phosphorylation of DNA2 or its helicase partner WRN [70].
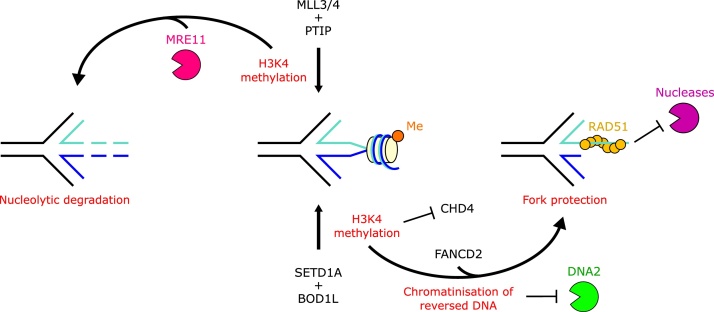
The Biorientation Of Chromosomes In Cell Division 1 Like protein, BOD1L, also protects forks from degradation by DNA2. This is mediated by interaction with the H3K4 histone methyltransferase SETD1A [71,72]. SETD1A has two impacts (Fig. 5); firstly it inhibits recruitment of the chromatin remodeller CHD4 and loss of CHD4, like inhibition of DNA2, rescues fork stability of BOD1L-SETD1A depleted cells; secondly, SETD1A enhances the histone chaperone activity of FANCD2 and the subsequent mobilisation of histones upstream of RAD51 deposition [72].
Fig. 5.
H3K4 methylation can promote distinct outcomes at reversed replication forks depending on the cellular context. In BRCA2-deficient cells, H3K4 methylation by MLL3/4 and PTIP promotes MRE11 recruitment to stalled replication forks and contributes to nascent DNA degradation. In contrast, H3K4 methylation by SETD1A and BOD1L promotes replication fork protection, by preventing the recruitment of CHD4 to the fork, and enhancing the histone chaperone function of FANCD2 to facilitate the chromatinisation of the reversed nascent strand and prevent DNA2-mediated degradation.
ABRO1 is homologous to CCDC98/Abraxas and acts to prevent DNA2/WRN-mediated, but not MRE11-mediated, fork degradation [73]. How this is achieved is unclear, but it is intriguing that the association of ABRO1 with the de-ubiquitinating enzyme USP7 can decrease ubiquitination of MDM2 and p53 [74], and that USP7 in turn is required for ongoing replication [75]. We might speculate ABRO1 acts to prevent inappropriate ubiquitination of vital components at the fork to protect the stalled fork structure.
Thus there are a range of functions in the factors that protect structures from DNA2; some of which bind complex DNA structures (WRNIP1, RIF1) and directly counteract the DNA2-WRN nuclease or decrease the availability of its substrates, while BOD1L-SETD1A regulation of chromatin remodelling suggests correct histone positioning contributes to shielding from DNA2.
3.3. Protection against MUS81-EME1 activity
While SLX4 is not relevant to fork protection in BRCA2-deficient cells, MUS81-EME1 loss can restore fork stability in this context [66,76]. The MUS81-EME1 complex is a structure-specific endonuclease, with a preference for branched DNA structures with a 5'-end at the branch, and plays an important role in rescuing stalled replication forks. Enhancer of zeste homologue 2 (EZH2) methylates histone H3K27, and di- or trimethylated Lys27 (H3K27me2 and me3) interacts with MUS81 and contributes to MUS81 recruitment to stalled forks. EZH2 depletion further improved fork protection conferred by MRE11 depletion in BRCA2-deficient cells, suggesting that EZH2-MUS81 and the MRE11 complex belong in distinct, additive pathways that degrade DNA at stalled replication forks [76].
3.4. Protection against MRE11-EXO1 nucleases
Inhibition of MRE11 3’-5’ exonuclease activity with the small-molecule inhibitor mirin prevents fork degradation in cells lacking cohesin [77], RECQL5 [78], FANCB [78], FANCD2 [79], BRCA1 or BRCA2 [9,15,18,62,[79], [80], [81]], the RAD51 paralogs (RAD51C, XRCC2, XRCC3) [82], WRNIP1 [67], WRN exonuclease [40], RNF168 [65], EXD2 [59], ATRX [83] or REV1 [84]. The 3’-5’ EXO1 nuclease appears to function in the same pathway as MRE11, with EXO1 extending degradation begun by MRE11 in BRCA1- or BRCA2-deficient cells [62].
3.4.1. MRE11 recruitment
In addition to inhibiting fork resolution by RECQ1, PARP1 can be activated by stalled replication forks and interacts with NBS1-MRE11, facilitating MRE11 recruitment to stalled forks [85,86]. Resection by MRE11 in wild-type cells contributes to timely fork restart [86], but if forks are unprotected, MRE11, and PARP1 activity, contribute to fork degradation [81]. Alongside its role in limiting fork reversal, RAD52 interacts with stalled replication-like structures and also contributes to MRE11 recruitment to promote fork degradation [9]. In HR, RAD52 supports RAD51 and alternative HR-mediated repair mechanisms. The findings of entirely separate roles for RAD52 in fork protection is a dramatic illustration of the divergent roles of some proteins between these two contexts.
The PTIP and Mixed-Lineage Leukaemia 3 and 4, MLL3/4 (Lysine Methyltransferases 2C and 2D) complex acts to specifically methylate H3K4 [87] which promotes MRE11 recruitment [81]. Targeting the catalytic SET domain of MLL4 in BRCA1-deficient B cells rescues fork stability, and loss of the MLL3_4/PTIP/MRE11 chromatin modifier pathway protects replication forks from degradation in BRCA1- and BRCA2-deficient cells [81]. As MLL3/4 promotes MRE11-depedent degradation, and SETD1A prevents DNA2-mediated degradation, both apparently through H3K4 methylation [72,81], further exploration is needed of how H3K4me differentially regulates nuclease accessibilities. The nucleosome remodeller CHD4 also mediates MRE11 recruitment in BRCA2-deficient cells, and its depletion reverses fork degradation in addition to rescuing cisplatin sensitivity and chromosome aberrations in these cells [81,88]. Since loss of CHD4 similarly restores fork stability and genome stability in SETD1A-deficient cells (where forks are sensitive to DNA2, not MRE11) [71] there appears to be a toxic influence of remodelling nucleosomes in at least two different contexts of stalled forks.
3.4.2. Protection against MRE11 activity
Cells employ several means to prevent MRE11 nuclease attack of stalled replication forks, including direct inhibition, upregulation of translesion synthesis (TLS) and RAD51-mediated inhibition.
ATRX physically interacts with MRE11 and inhibits excessive MRE11 activity at stalled forks, so that loss of ATRX leads to compromised fork protection [83]. Both the Fanconi core complex and the mono-ubiquitination of FANCD2 contribute to full fork protection from MRE11 [79]. Mono-ubiquitinated FANCD2 binds the TLS polymerase REV1 for bypassing lesions, and depletion of REV1 also results in the poor protection of replication forks from MRE11 [84]. FANCD2 has several further functions (described in Sections 3.2 and 3.4.3), and its positive role is such that its overexpression is found in tumours lacking BRCA1/BRCA2, where it acts to restore the stability of stalled forks [89]. Loss of FANCD2 meanwhile is synthetic lethal with loss of BRCA2 [89,90]. Furthermore, restoration of fork protection by depletion of CHD4 in BRCA2-deficient cells is linked to increased PCNA mono-ubiquitination mediated by the E3 ligase RAD18 [81,88], which drives recruitment of TLS polymerases. These observations imply that less stringent polymerases can act to support exposed forks.
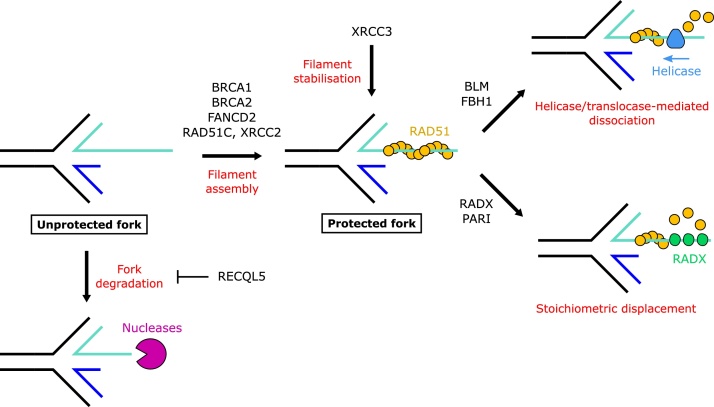
RAD51 stabilisation is a major determinant in protection of forks against nuclease attack. When bound to ATP, RAD51 monomers can associate to form nucleoprotein filaments on ssDNA and dsDNA [91], and these filaments can be disassembled upon ATP hydrolysis [92]. In vitro, RAD51 pre-assembled on model replication forks prevents degradation by MRE11 [15]. Suppressing the chromatin association of RAD51 by expression of the BRCA2-BRC4 peptide also results in MRE11-dependent degradation of nascent DNA in mouse embryonic stem cells [80] and depletion of RAD51 from Xenopus extracts results in MRE11-dependent accumulation of ssDNA gaps at the replication fork [48]. Conversely, overexpression of RAD51, or stabilisation of filaments using an ATPase-defective mutant of RAD51 (RAD51-K133R), rescues fork degradation seen upon HU treatment of BRCA2-and FANCD2-deficient mammalian cells [79,80]. Moreover, RAD51-T131 P and A293 T mutants, which form unstable nucleoprotein filaments [49], were identified as de novo mutations in patients displaying Fanconi anaemia-like features [93,94]. Cells expressing these mutants display extensive fork degradation rescued by inhibition of MRE11, as these are able to mediate fork reversal but are defective in protecting the reversed forks from nucleases [9,49,93,94]. Intriguingly, cells bearing an experimental mutation in RAD51 that disrupts its strand exchange activity (RAD51-II3A) were capable of both fork regression and protection [10]. Therefore while nucleoprotein filaments are needed in HR to invade a homologous duplex DNA (reviewed in [95]), in the context of a stalled fork they appear to confer mechanical protection of DNA from nuclease degradation.
3.4.3. RAD51 stabilisers
During HR, BRCA2 promotes RAD51 loading onto ssDNA for filament formation [[96], [97], [98]], and also stabilises these filaments by limiting ATP hydrolysis by RAD51 [97]. BRCA2 interacts with RAD51 through its eight BRC repeat motifs and a C-terminal site [99,100]. A BRC3-RPA fusion protein linking resected DNA (RPA) to RAD51 loading (BRC3) is sufficient to promote HR [101] but cannot promote replication fork protection, at least in part because the BRCA2 C-terminus is also required for fork protection [9,80].
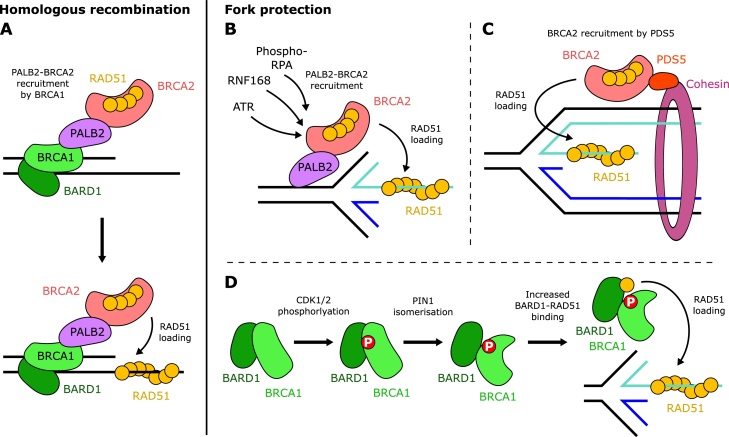
BRCA2 is largely recruited by BRCA1 through a bridging interaction with PALB2, required for HR (reviewed in [102]). However, the BRCA1-PALB2 interaction face is dispensable for replication fork protection [103,104]. Moreover in some lines of PARPi-resistant BRCA1-deficient cells, RAD51 foci formation is supported by PALB2-BRCA2 and ATR, illustrating an ATR-dependent, but BRCA1-independent mechanism for PALB2-BRCA2 recruitment [105]. PALB2 may be recruited to replication forks by phosphorylated RPA [106] and RNF168 [65,107]. In addition, PALB2 can bind chromatin directly [[108], [109], [110]] and the PALB2 WD40 domain also interacts with RAD51 and RAD51C [111] (Fig. 6).
Fig. 6.
BRCA1 and BRCA2 in homologous recombinationversusfork protection. A: In homologous recombination, BRCA1 recruits BRCA2 via PALB2 to double-strand breaks. BRCA2 in turn mediates RAD51 loading and filament formation. B: In fork protection, BRCA2-PALB2 recruitment is not dependent on BRCA1, and may instead be mediated by ATR, RNF168 and phosphorylated RPA. PALB2 may also bind directly to chromatin. C: BRCA2 may also be recruited independently of PALB2, which may involve the cohesin cofactor PDS5. D: BRCA1-BARD1 plays a distinct role in replication fork protection compared to homologous recombination. CDK1/2 phosphorylation of BRCA1 recruits the peptidyl-prolyl isomerase PIN1. Subsequent isomerisation of BRCA1 increases the accessibility of BARD1, resulting in increased BARD1-RAD51 interaction which facilitates RAD51 loading onto reversed forks.
Furthermore, BRCA2 may be recruited to stalled forks independently of PALB2. The cohesin complex entraps sister chromatids after replication and is enriched on nascent DNA following HU treatment [69]. The cohesin cofactor PDS5 directly interacts with BRCA2 [112,113] and may contribute to BRCA2 recruitment for fork protection as loss of PDS5 results in fork degradation, which can be rescued by MRE11 inhibition [77].
We discovered that BRCA1 is regulated in fork protection by the peptidyl-prolyl isomerase PIN1 [103]. PIN1 specifically recognises and isomerizes phosphorylated serine/threonine-proline motifs (reviewed in [114]) and CDK1/2-mediated phosphorylation of BRCA1 regulates PIN1 interaction [103]. The conformational changes resulting from PIN1-mediated isomerisation of BRCA1 increases accessibility to the BRCA1-binding partner BARD1, which in turn enhances direct BARD1-RAD51 interaction and RAD51 recruitment to stalled forks. Moreover, several mutations, including at the BRCA1 phosphorylation site and at the BARD1-RAD51 interaction face, result in a BRCA1-BARD1 complex that is unable to fully protect replication forks, yet is competent in HR [103]. These surprising findings indicate a direct and distinct role for BRCA1 in fork protection compared to HR and add to an emerging theme of unexpected differences between BRCA1 and BRCA2 in the protection of stalled replication forks (see Box 2).
Box 2. Distinct roles of BRCA1 versus BRCA2 in replication fork protection.
Although BRCA1 and BRCA2 protect replication forks from MRE11-mediated degradation through stabilising RAD51 [79], recent data illustrate that fork protection is uncoupled from the canonical BRCA1-PALB2-BRCA2 pathway [103]. Moreover, several other observations highlight the differences between BRCA1 and BRCA2 in replication fork protection. Increased translesion synthesis (TLS) is thought to contribute to the rescue of fork protection in BRCA2-deficient cells when CHD4 is depleted [81,88]. In contrast, CHD4 loss does not rescue BRCA1-deficient cells [88], and instead toxic TLS is thought to explain why USP1 loss is synthetic lethal with BRCA1 depletion [160]. The de-ubiquitinating enzyme USP1 suppresses PCNA ubiquitination, reducing the association of TLS polymerases with forks. Strikingly, USP1 loss or inhibition is not synthetic lethal with BRCA2 loss [160]. Further differences include the finding that CtIP repression worsens fork degradation in BRCA1- but not BRCA2-deficient cells [63]; conversely, depletion of MUS81 confers replication fork protection in BRCA2- but not BRCA1-deficient cells [62,76]. These findings imply different contexts for BRCA1 and BRCA2 functions at stalled replication forks, in addition to their independent roles in supporting RAD51 stability.
Alt-text: Box 2
FANCD2-deficient cells show a greater defect in fork protection than those lacking the ubiquitin ligase responsible for FANCD2 modification [115], suggesting independent as well as ubiquitin-dependent roles of FANCD2 in fork protection. FANCD2 has been implicated in directly stabilising RAD51 nucleoprotein filaments. Upon HU treatment, RAD51 foci formation and association with PCNA is reduced in FANCD2-depleted cells, suggesting that FANCD2 is involved in RAD51 accumulation at stalled replication forks [116]. In vitro assays revealed that the FANCD2-FANCI complex stabilises RAD51 filaments on replication fork substrates, and protects them from degradation by the nuclease FAN1 [116]. Direct stabilisation of RAD51 filaments may therefore represent an additional mechanism by which FANCD2 promotes replication fork protection.
The RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3) share sequence homology with RAD51 [117] and form various subcomplexes with each other [118,119] that interact with RAD51 in vitro and in vivo [120]. RAD51C, XRCC2 and XRCC3 also protect nascent strands from MRE11-dependent degradation in response to HU [82]. These paralogs may function via different mechanisms, as fork protection by RAD51C and XRCC3 requires ATP binding but not hydrolysis, whilst XRCC2 requires neither ATP binding nor hydrolysis [82]. Furthermore, XRCC2-mediated fork protection is epistatic to BRCA2 and FANCD2 pathways, but RAD51C and XRCC3 are not [82].
Nevertheless, RAD51 paralogs may contribute to RAD51 filament stabilisation. U2OS cells individually deficient in each of the RAD51 paralogs show reduced spontaneous RAD51 foci compared to wild-type cells, implying a function in RAD51 filament assembly on chromatin [121], although other studies suggest that loss of XRCC3 has no effect on RAD51 focus formation [82,122].The human RAD51B-RAD51C complex also stabilises RAD51-ssDNA filaments in vitro [123]. The role of RAD51 paralogs in maintaining fork stability through RAD51 stabilisation would correlate with their roles in promoting RAD51 filament formation for HR and inter-strand crosslink (ICL) repair (reviewed in [124]).
3.4.4. Negative regulators of RAD51 stability
RAD51 filaments on the regressed arms of reversed forks may initiate inappropriate, ectopic HR, particularly in repetitive genomes [[125], [126], [127]], necessitating negative regulators for careful control of RAD51. Filament dissociation may also be stimulated to restore replication forks and restart DNA synthesis once replication perturbations have been resolved.
RADX is recruited to sites of replication stress and competitively displaces RAD51 from ssDNA [45,51,128]. Depletion of RADX increases RAD51 accumulation at replication forks and restores fork protection in cells deficient in BRCA1, BRCA2, FANCD2 or BOD1L, indicating that the loss of RAD51 stability in these cells can be rescued by relieving the negative inhibition on RAD51 [45,51]. Interestingly, silencing RADX in BRCA2-deficient cells restores fork protection, but does not restore HR [45].
Additionally, several helicase/translocases associated with regulating HR, through their ability to dissociate RAD51 filaments, have also been implicated in regulating fork protection. The RecQ helicase BLM disrupts inactive, ADP-bound RAD51 filaments in vitro [129,130]. In cells, BLM reduces RAD51 foci formation upon HU treatment [131], consistent with a role of BLM in destabilising RAD51 filaments. Depletion of BLM largely rescues fork degradation in cells with homozygous BRCA1 hypomorphic mutations [132] or those lacking the Fanconi core complex component FANCB [78] or FANCD2 [72], and partially rescues degradation in the absence of BOD1L [71]. Another RecQ helicase, RECQL5, also has RAD51 dissociation activity [133]. Intriguingly, loss of RECQL5 in the absence of FANCB or BOD1L increases fork degradation [71,78], suggesting that RECQL5 plays a role in protecting rather than degrading forks.
FBH1, a member of the UvrD helicase family [134,135], is recruited to ssDNA regions in response to replication stress [136]. FBH1 depletion causes spontaneous RAD51 foci formation [136] and hyperrecombination [137]. In vitro the helicase/translocase activity of FBH1 is required to disrupt RAD51-ssDNA filaments [137]. FBH1 also contains an F-box domain and functions as part of an SCF E3 ubiquitin ligase that ubiquitinates RAD51 [138,139]. Cells expressing ubiquitination-resistant RAD51 (K58/K64R) display increased RAD51 foci in response to HU [139]. K64-RAD51 is involved in binding to DNA [140], suggesting that ubiquitination of this residue may affect RAD51 interaction with DNA for filament assembly [139]. Thus F-box mediated ubiquitination functions alongside the helicase activity of FBH1 to restrain unwarranted RAD51 activity. In the absence of BOD1L, SETD1A or WRNIP1, loss of FBH1 rescues fork degradation [67,71,72]. Similarly a defect in RAD51 foci formation in response to HU is rescued by FBH1 depletion in cells lacking PARP1/2, suggesting a further role for PARP in RAD51 stabilisation [141].
PARI is another UvrD helicase that suppresses RAD51. However, given that it lacks domains required for ATP hydrolysis, and therefore lacks helicase activity, its RAD51 displacement is likely mediated through a non-catalytic, stoichiometric interaction with RAD51 [142]. While PARI deficiency suppresses fork degradation, it also leads to chromosomal instability – demonstrating how dysregulated RAD51 activity, despite appearing to protect replication forks, is detrimental to proliferating cells [143]. PARI recruitment to chromatin is mediated by interaction with PCNA, which may confine PARI activity to periods of cellular replication for regulation of replication stress responses [142].
The abundance of factors involved in fine-tuning RAD51 activity is striking (Fig. 7), and exist presumably to allow adequate fork protection while preventing excessive RAD51 filament formation, thereby safeguarding fork stability and maintaining genome integrity.
Fig. 7.
Factors directly regulating RAD51 filament stability. RAD51 filament formation onto the regressed arm of a reversed replication fork is promoted by BRCA1, BRCA2, FANCD2 and the RAD51 paralogs RAD51C and XRCC2. The RAD51 paralog XRCC3 meanwhile may stabilise filaments downstream of RAD51 recruitment. Multiple factors negatively regulate the stability of RAD51 filaments to promote disassembly. These can act as translocases to strip RAD51 from the DNA (e.g. BLM or FBH1), or may act in a non-enzymatic fashion (e.g. RADX or PARI). In contrast, RECQL5, although able to displace RAD51 filaments, has been implicated in preventing fork degradation.
4. Replication fork protection in cancer and chemoresistance
Several mechanisms contribute to the recovery of stalled forks, however for a fork in which the regressed arms are degraded, fork recovery by RECQ1-mediated resolution is unavailable. If the stalling leads to fork collapse, HR can mediate break-induced replication, involving invasion of the dsDNA end into the sister chromatid. This can be RAD51-dependent, or RAD51-independent using RAD52-POLD3 [62,76]. If forks are not processed to breaks, replication may be restarted through HR-mediated template switching of a blocked replicating strand to the undamaged sister chromatid, PCNA-dependent DNA damage bypass, or PRIMPOL-mediated repriming downstream of the lesion. However, template switching can lead to genetic rearrangements, damage bypass can generate mutation hotspots, and use of repriming leads to ssDNA gaps in the post-replicative genome. Elevated use of these potentially mutagenic fork restart pathways could drive genomic instability in the absence of efficient replication fork protection.
Evidence to support this idea is emerging, with the observation of a delay in the restart of fork progression under conditions of poor fork protection [62,69]. This is often accompanied by the generation of ssDNA gaps behind the fork [8,144] and the appearance of chromosome abnormalities, often breaks and gaps [8,69,103,145]. PRIMPOL-mediated repriming likely contributes to these observations, as its use is associated with a poor quality, gapped, post-replicative genome [146] and in therapy-resistant cells engagement of PRIMPOL may be an adaptive response to suppress fork reversal [147]. That these abnormalities are likely due to poor fork protection is underlined by the fact that aberrations can be prevented by restoring fork protection through inhibition of DNA2 or MRE11 [62,69].
Since replication fork protection acts to suppress genome alterations, a role in cancer predisposition might be anticipated. However current evidence is mixed; some mutations resulting in poor fork protection, such as a BRCA2 C-terminal mutation and ABRO1 loss, are associated with cancer in mouse models [73,148,149] while others are not, such as a fork-protection defective Bard1-allele [150]. Mutation of factors involved in fork reversal have been linked to cancer, with SMARCAL1 mutations associated with a rare, childhood disorder that is sometimes characterized by cancer [11,151]. ZRANB3 mutations have been observed in endometrial cancers [152] and HLTF is commonly found silenced in colorectal cancers [[152], [153], [154]], but whether these mutations are significant in tumorigenesis is unclear.
Given the central role of HR in the recovery of aberrant replication structures, an important question has been whether restoration of fork protection influences the survival of HR-deficient cells. Deletion of factors that promote MRE11 recruitment to stalled forks prior to deletion of BRCA2allows the emergence of BRCA2-null murine cells, when BRCA2 loss is alone is lethal [81,155]. However, in a non-transformed human mammary epithelial cell line, restoration of replication fork protection in BRCA2-mutant cells by similar means was unable to restore normal growth kinetics [50]. Nevertheless, restoration of fork protection may significantly enhance cell survival in a tumour environment. For example, heterozygous PARP1 loss resulted in accelerated tumour growth in mice bearing a conditional deletion of BRCA2, suggesting fork protection may allow survival of cells lacking BRCA2, forming a pool from which tumours emerge [155]. Moreover restoration of fork protection in BRCA-deficient cells reduces chromosome aberrations caused by cisplatin, camptothecin or PARPi, and in platinum-treated patients low expression of PTIP or CHD4, which mediate MRE11 recruitment, correlates with poor patient response to chemotherapy [81,88]. BRCA-deficient but PARPi-resistant mouse tumours [81] and some human cell lines [105] exhibit normal fork protection. Similarly loss of EZH2-MUS81 contributes to both fork protection and resistance to PARPi and cisplatin in BRCA2-deficient cells [76]. A potential explanation is that the protection of forks reduces the need for their HR-mediated recovery, resulting in therapy resistance. Such rewiring in resistant cancer cells could be targeted. For example, repression of ATR can re-sensitize cell lines to HR therapies and drive stalled replication fork degradation [105]. Mechanistically this may occur through inhibition of PALB2-BRCA2 or RAD51-paralog pathways, or by driving hyperactive SMARCAL1-mediated fork regression.
5. Summary
Recent discoveries of the roles that many HR proteins and other factors have in promoting stalled replication fork stability has revealed a pathway critical to genome stability and chemoresistance. Uncovering some of the mechanistic details has surprisingly found that fork protection is more than just the RAD51-loading step of HR (Table 2). In addition to aspects of HR being dispensable, such as the PALB2-BRCA1 interaction or RAD51 strand exchange activity, we are slowly building the view that previously unappreciated functions of known proteins contribute to defending reversed forks. We currently have little knowledge of the precise vulnerabilities of the fork structure resulting from specific stresses or deficiencies, or how different fork protection factors relate to one another. Nevertheless, obtaining greater insights into restored fork protection selected for by exposure to anti-cancer therapies will be important both in understanding the normal process and in identifying strategies to treat tumours that depend on this pathway.
Table 2.
Role of Factors in homologous recombination or inter-strand cross link repair versus replication fork protection.
| Factor | In DNA repair | In fork protection | References |
|---|---|---|---|
| ATR | Promote long range resection | Promote ‘rewired’ fork protection | [105]. |
| BLM | Promote end resection, Holliday junction dissolution and RAD51 dissociation during late HR | Promote RAD51 dissociation | [71,72,78,129,130,132]. |
| BRCA1-BARD1 | Counteract 53BP1-Shieldin and promote PALB2-BRCA2 and RAD51 recruitment | Promote RAD51 recruitment by PIN1 | [79,102,103]. |
| BRCA2 | Stabilise RAD51 | Stabilise RAD51 | [9,79,80,102]. |
| CtIP | Initiate MRE11-mediated resection | Restrict fork reversal/protect reversed forks and aid MRE11-mediated degradation | [61,62,63]. |
| DNA2 | Extend MRE11 resection | Resect ‘wild-type’ and de-protected forks | [8,66,69,70,71,73,161]. |
| EXD2 | Promote MRE11 resection | Suppress fork reversal | [56,57,58,59]. |
| EXO1 | Extend MRE11 resection | Extend MRE11 resection | [62,161]. |
| FANCD2 | Histone mobilisation for ICL repair, recruitment of FAN1 nuclease | Histone mobilisation, increase TLS, direct RAD51 stabilisation | [72,84,116,162]. |
| FBH1 | Dissociate RAD51 filaments | Promote fork reversal, promote RAD51 dissociation | [10,41,67]. |
| MRE11 | Initiate short-range resection | Resect unprotected forks, promote fork restart | [9,15,18,40,59,62,65,66,67,77,78,79,80,81,82,83,84]. |
| PALB2 | Promote BRCA2 recruitment via BRCA1 | BRCA1-independent recruitment of BRCA2 | [103,104,105]. |
| PARP | Break excision repair, Pol-theta mediated end joining | Inhibit fork restoration, recruit MRE11, inhibits fork restoration, RAD51 stabilisation | [53,86,141,163]. |
| PDS5 | Promote BRCA2 recruitment | Promote BRCA2 recruitment | [77,112]. |
| RAD51 | Strand exchange | Promote fork reversal, protect forks from degradation | [2,8,9,10,15,48,49,79,80,93,94,95]. |
| RAD51 paralogs | Stabilise RAD51 | Promote RAD51 recruitment | [82,124]. |
| RAD52 | Promote RAD51 loading, single strand annealing, break-induced-repair | Suppress SMARCAL1 recruitment, promote MRE11 recruitment | [9,23,24]. |
| RECQL5 | Dissociate RAD51 filaments | Protect against degradation | [78,133]. |
| RIF1-PP1 | Part of the Shieldin complex that promotes NHEJ and inhibits DNA resection | Inhibit DNA2/WRN | [69,70]. |
| RNF168 | Recruit 53BP1 | Restrict fork reversal | [65,164,165]. |
| SLX4 | Scaffold for nucleases in Holliday Junction cleavage | Scaffold for nucleases involved in fork cleavage | [66,67]. |
| WRN | Support DNA2 in long-range resection | Support DNA2 (helicase), prevent MRE11-dependent degradation (exonuclease) | [8,40]. |
Funding
This work was supported by the University of Birmingham, Cancer Research UK [C8820/A28283]; and an Imperial College President’s PhD Scholarship.
Declaration of Competing Interest
The authors declare they have no financial or personal relationships with other people or organizations that could inappropriately influence this work.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.semcdb.2020.07.004.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16(1):2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015;208(5):563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neelsen K.J., Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015;16(4):207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 4.Vujanovic M., Krietsch J., Raso M.C., Terraneo N., Zellweger R., Schmid J.A., Taglialatela A., Huang J.W., Holland C.L., Zwicky K., Herrador R., Jacobs H., Cortez D., Ciccia A., Penengo L., Lopes M. Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol. Cell. 2017;67(5):882–890. doi: 10.1016/j.molcel.2017.08.010. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kile A.C., Chavez D.A., Bacal J., Eldirany S., Korzhnev D.M., Bezsonova I., Eichman B.F., Cimprich K.A. HLTF’s ancient HIRAN domain binds 3’ DNA ends to drive replication fork reversal. Mol. Cell. 2015;58(6):1090–1100. doi: 10.1016/j.molcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012;19(4):417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 7.Amunugama R., Willcox S., Wu R.A., Abdullah U.B., El-Sagheer A.H., Brown T., McHugh P.J., Griffith J.D., Walter J.C. Replication fork reversal during DNA interstrand crosslink repair requires CMG unloading. Cell Rep. 2018;23(12):3419–3428. doi: 10.1016/j.celrep.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A., Cejka P., Stewart S., Lopes M., Vindigni A. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015;208(5):545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P., Lopes M. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017;8(1):859. doi: 10.1038/s41467-017-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason J.M., Chan Y.L., Weichselbaum R.W., Bishop D.K. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun. 2019;10(1):4410. doi: 10.1038/s41467-019-12297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betous R., Mason A.C., Rambo R.P., Bansbach C.E., Badu-Nkansah A., Sirbu B.M., Eichman B.F., Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26(2):151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciccia A., Nimonkar A.V., Hu Y., Hajdu I., Achar Y.J., Izhar L., Petit S.A., Adamson B., Yoon J.C., Kowalczykowski S.C., Livingston D.M., Haracska L., Elledge S.J. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell. 2012;47(3):396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan J., Ghosal G., Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell. 2012;47(3):410–421. doi: 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blastyak A., Hajdu I., Unk I., Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 2010;30(3):684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L., Krejci L., Costanzo V. Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell. 2017;67(5):867–881. doi: 10.1016/j.molcel.2017.07.001. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhat K.P., Betous R., Cortez D. High-affinity DNA-binding domains of replication protein a (RPA) direct SMARCAL1-dependent replication fork remodeling. J. Biol. Chem. 2015;290(7):4110–4117. doi: 10.1074/jbc.M114.627083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achar Y.J., Balogh D., Haracska L. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc. Natl. Acad. Sci. U. S. A. 2011;108(34):14073–14078. doi: 10.1073/pnas.1101951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R., Cejka P., Costanzo V., Ciccia A. Restoration of replication fork stability in BRCA1- and BRCA2-Deficient cells by inactivation of SNF2-Family fork remodelers. Mol. Cell. 2017;68(2):414–430. doi: 10.1016/j.molcel.2017.09.036. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bansbach C.E., Betous R., Lovejoy C.A., Glick G.G., Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23(20):2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betous R., Couch F.B., Mason A.C., Eichman B.F., Manosas M., Cortez D. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 2013;3(6):1958–1969. doi: 10.1016/j.celrep.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole L.A., Cortez D. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 2017;52(6):696–714. doi: 10.1080/10409238.2017.1380597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couch F.B., Bansbach C.E., Driscoll R., Luzwick J.W., Glick G.G., Betous R., Carroll C.M., Jung S.Y., Qin J., Cimprich K.A., Cortez D. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27(14):1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalan M., Olsen K.S., Powell S.N. Emerging roles of RAD52 in genome maintenance. Cancers (Basel) 2019;11(7) doi: 10.3390/cancers11071038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malacaria E., Pugliese G.M., Honda M., Marabitti V., Aiello F.A., Spies M., Franchitto A., Pichierri P. Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation. Nat. Commun. 2019;10(1):1412. doi: 10.1038/s41467-019-09196-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng M., Cong K., Panzarino N.J., Nayak S., Calvo J., Deng B., Zhu L.J., Morocz M., Hegedus L., Haracska L., Cantor S.B. Opposing roles of FANCJ and HLTF protect forks and restrain replication during stress. Cell Rep. 2018;24(12):3251–3261. doi: 10.1016/j.celrep.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwab R.A., Nieminuszczy J., Shin-ya K., Niedzwiedz W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 2013;201(1):33–48. doi: 10.1083/jcb.201208009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motegi A., Liaw H.J., Lee K.Y., Roest H.P., Maas A., Wu X., Moinova H., Markowitz S.D., Ding H., Hoeijmakers J.H., Myung K. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. U. S. A. 2008;105(34):12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yusufzai T., Kadonaga J.T. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc. Natl. Acad. Sci. U. S. A. 2010;107(49):20970–20973. doi: 10.1073/pnas.1011196107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin S., Hyun K., Kim J., Hohng S. ATP binding to Rad5 initiates replication fork reversal by inducing the unwinding of the leading arm and the formation of the holliday junction. Cell Rep. 2018;23(6):1831–1839. doi: 10.1016/j.celrep.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Chavez D.A., Greer B.H., Eichman B.F. The HIRAN domain of helicase-like transcription factor positions the DNA translocase motor to drive efficient DNA fork regression. J. Biol. Chem. 2018;293(22):8484–8494. doi: 10.1074/jbc.RA118.002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badu-Nkansah A., Mason A.C., Eichman B.F., Cortez D. Identification of a substrate recognition domain in the replication stress response protein zinc finger ran-binding domain-containing protein 3 (ZRANB3) J. Biol. Chem. 2016;291(15):8251–8257. doi: 10.1074/jbc.M115.709733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weston R., Peeters H., Ahel D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 2012;26(14):1558–1572. doi: 10.1101/gad.193516.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebesta M., Cooper C.D.O., Ariza A., Carnie C.J., Ahel D. Structural insights into the function of ZRANB3 in replication stress response. Nat. Commun. 2017;8:15847. doi: 10.1038/ncomms15847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flaus A., Martin D.M., Barton G.J., Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34(10):2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugreev D.V., Rossi M.J., Mazin A.V. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011;39(6):2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gari K., Decaillet C., Delannoy M., Wu L., Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. U. S. A. 2008;105(42):16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Z., Delannoy M., Ling C., Daee D., Osman F., Muniandy P.A., Shen X., Oostra A.B., Du H., Steltenpool J., Lin T., Schuster B., Decaillet C., Stasiak A., Stasiak A.Z., Stone S., Hoatlin M.E., Schindler D., Woodcock C.L., Joenje H., Sen R., de Winter J.P., Li L., Seidman M.M., Whitby M.C., Myung K., Constantinou A., Wang W. A histone-fold complex and FANCM form a conserved DNA-Remodeling complex to maintain genome stability. Mol. Cell. 2010;37(6):865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machwe A., Xiao L., Groden J., Orren D.K. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45(47):13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 39.Kanagaraj R., Saydam N., Garcia P.L., Zheng L., Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34(18):5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iannascoli C., Palermo V., Murfuni I., Franchitto A., Pichierri P. The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1-dependent degradation. Nucleic Acids Res. 2015;43(20):9788–9803. doi: 10.1093/nar/gkv836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fugger K., Mistrik M., Neelsen K.J., Yao Q., Zellweger R., Kousholt A.N., Haahr P., Chu W.K., Bartek J., Lopes M., Hickson I.D., Sorensen C.S. FBH1 catalyzes regression of stalled replication forks. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 42.Petermann E., Luis Orta M., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-Mediated pathways for restart and repair. Mol. Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014;16(3):281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dungrawala H., Cortez D. Purification of proteins on newly synthesized DNA using iPOND. Methods Mol. Biol. 2015;1228:123–131. doi: 10.1007/978-1-4939-1680-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dungrawala H., Bhat K.P., Le Meur R., Chazin W.J., Ding X., Sharan S.K., Wessel S.R., Sathe A.A., Zhao R., Cortez D. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell. 2017;67(3):374–386. doi: 10.1016/j.molcel.2017.06.023. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhat K.P., Cortez D. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018 doi: 10.1038/s41594-018-0075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gildemeister O.S., Sage J.M., Knight K.L. Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J. Biol. Chem. 2009;284(46):31945–31952. doi: 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17(11):1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zadorozhny K., Sannino V., Belan O., Mlcouskova J., Spirek M., Costanzo V., Krejci L. Fanconi-anemia-Associated mutations destabilize RAD51 filaments and impair replication fork protection. Cell Rep. 2017;21(2):333–340. doi: 10.1016/j.celrep.2017.09.062. [DOI] [PubMed] [Google Scholar]
- 50.Feng W., Jasin M. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 2017;8(1):525. doi: 10.1038/s41467-017-00634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhat K.P., Krishnamoorthy A., Dungrawala H., Garcin E.B., Modesti M., Cortez D. RADX modulates RAD51 activity to control replication fork protection. Cell Rep. 2018;24(3):538–545. doi: 10.1016/j.celrep.2018.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petermann E., Helleday T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010;11(10):683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 53.Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., Marino F., Lucic B., Biasin V., Gstaiger M., Aebersold R., Sidorova J.M., Monnat R.J., Jr., Lopes M., Vindigni A. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 2013;20(3):347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeeles J.T., Poli J., Marians K.J., Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 2013;5(5) doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu J., Sun L., Shen F., Chen Y., Hua Y., Liu Y., Zhang M., Hu Y., Wang Q., Xu W., Sun F., Ji J., Murray J.M., Carr A.M., Kong D. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell. 2012;149(6):1221–1232. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 56.Biehs R., Steinlage M., Barton O., Juhasz S., Kunzel J., Spies J., Shibata A., Jeggo P.A., Lobrich M. DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell. 2017;65(4):671–684. doi: 10.1016/j.molcel.2016.12.016. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broderick R., Nieminuszczy J., Baddock H.T., Deshpande R., Gileadi O., Paull T.T., McHugh P.J., Niedzwiedz W. EXD2 promotes homologous recombination by facilitating DNA end resection. Nat. Cell Biol. 2016;18(3):271–280. doi: 10.1038/ncb3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E., Clark A.B., Kunkel T.A., Harper J.W., Colaiacovo M.P., Elledge S.J. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010;39(1):36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nieminuszczy J., Broderick R., Bellani M.A., Smethurst E., Schwab R.A., Cherdyntseva V., Evmorfopoulou T., Lin Y.L., Minczuk M., Pasero P., Gagos S., Seidman M.M., Niedzwiedz W. EXD2 protects stressed replication forks and is required for cell viability in the absence of BRCA1/2. Mol. Cell. 2019;75(3):605–619. doi: 10.1016/j.molcel.2019.05.026. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margalef P., Kotsantis P., Borel V., Bellelli R., Panier S., Boulton S.J. Stabilization of reversed replication forks by telomerase drives telomere catastrophe. Cell. 2018;172(3):439–453. doi: 10.1016/j.cell.2017.11.047. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anand R., Ranjha L., Cannavo E., Cejka P. Phosphorylated CtIP functions as a Co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016;64(5):940–950. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 62.Lemacon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., Vindigni A. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017;8(1):860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Przetocka S., Porro A., Bolck H.A., Walker C., Lezaja A., Trenner A., von Aesch C., Himmels S.F., D’Andrea A.D., Ceccaldi R., Altmeyer M., Sartori A.A. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol. Cell. 2018;72(3):568–582. doi: 10.1016/j.molcel.2018.09.014. e6. [DOI] [PubMed] [Google Scholar]
- 64.Wilkinson O.J., Martin-Gonzalez A., Kang H., Northall S.J., Wigley D.B., Moreno-Herrero F., Dillingham M.S. CtIP forms a tetrameric dumbbell-shaped particle which bridges complex DNA end structures for double-strand break repair. Elife. 2019;8 doi: 10.7554/eLife.42129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmid J.A., Berti M., Walser F., Raso M.C., Schmid F., Krietsch J., Stoy H., Zwicky K., Ursich S., Freire R., Lopes M., Penengo L. Histone ubiquitination by the DNA damage response is required for efficient DNA replication in unperturbed S phase. Mol. Cell. 2018;71(6):897–910. doi: 10.1016/j.molcel.2018.07.011. e8. [DOI] [PubMed] [Google Scholar]
- 66.Porebski B., Wild S., Kummer S., Scaglione S., Gaillard P.H.L., Gari K. WRNIP1 protects reversed DNA replication forks from SLX4-Dependent nucleolytic cleavage. iScience. 2019;21:31–41. doi: 10.1016/j.isci.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leuzzi G., Marabitti V., Pichierri P., Franchitto A. WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J. 2016;35(13):1437–1451. doi: 10.15252/embj.201593265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sukackaite R., Jensen M.R., Mas P.J., Blackledge M., Buonomo S.B., Hart D.J. Structural and biophysical characterization of murine rif1 C terminus reveals high specificity for DNA cruciform structures. J. Biol. Chem. 2014;289(20):13903–13911. doi: 10.1074/jbc.M114.557843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mukherjee C., Tripathi V., Manolika E.M., Heijink A.M., Ricci G., Merzouk S., de Boer H.R., Demmers J., Van Vugt M., Ray Chaudhuri A. RIF1 promotes replication fork protection and efficient restart to maintain genome stability. Nat. Commun. 2019;10(1):3287. doi: 10.1038/s41467-019-11246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garzon J., Ursich S., Lopes M., Hiraga S.I., Donaldson A.D. Human RIF1-Protein phosphatase 1 prevents degradation and breakage of nascent DNA on replication stalling. Cell Rep. 2019;27(9):2558–2566. doi: 10.1016/j.celrep.2019.05.002. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Higgs M.R., Reynolds J.J., Winczura A., Blackford A.N., Borel V., Miller E.S., Zlatanou A., Nieminuszczy J., Ryan E.L., Davies N.J., Stankovic T., Boulton S.J., Niedzwiedz W., Stewart G.S. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol. Cell. 2015;59(3):462–477. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 72.Higgs M.R., Sato K., Reynolds J.J., Begum S., Bayley R., Goula A., Vernet A., Paquin K.L., Skalnik D.G., Kobayashi W., Takata M., Howlett N.G., Kurumizaka H., Kimura H., Stewart G.S. Histone methylation by SETD1A protects nascent DNA through the nucleosome chaperone activity of FANCD2. Mol. Cell. 2018;71(1):25–41. doi: 10.1016/j.molcel.2018.05.018. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu S., Wu X., Wu L., Castillo A., Liu J., Atkinson E., Paul A., Su D., Schlacher K., Komatsu Y., You M.J., Wang B. Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev. 2017;31(14):1469–1482. doi: 10.1101/gad.299172.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang J., Cao M., Dong J., Li C., Xu W., Zhan Y., Wang X., Yu M., Ge C., Ge Z., Yang X. ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nat. Commun. 2014;5:5059. doi: 10.1038/ncomms6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lecona E., Rodriguez-Acebes S., Specks J., Lopez-Contreras A.J., Ruppen I., Murga M., Munoz J., Mendez J., Fernandez-Capetillo O. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat. Struct. Mol. Biol. 2016;23(4):270–277. doi: 10.1038/nsmb.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rondinelli B., Gogola E., Yucel H., Duarte A.A., van de Ven M., van der Sluijs R. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017;19(11):1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- 77.Morales C., Ruiz-Torres M., Rodriguez-Acebes S., Lafarga V., Rodriguez-Corsino M., Megias D., Cisneros D.A., Peters J.M., Mendez J., Losada A. PDS5 proteins are required for proper cohesin dynamics and participate in replication fork protection. J. Biol. Chem. 2020;295(1):146–157. doi: 10.1074/jbc.RA119.011099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim T.M., Son M.Y., Dodds S., Hu L., Luo G., Hasty P. RECQL5 and BLM exhibit divergent functions in cells defective for the Fanconi anemia pathway. Nucleic Acids Res. 2015;43(2):893–903. doi: 10.1093/nar/gku1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22(1):106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145(4):529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., John S., Day A., Crespo A.V., Shen B., Starnes L.M., de Ruiter J.R., Daniel J.A., Konstantinopoulos P.A., Cortez D., Cantor S.B., Fernandez-Capetillo O., Ge K., Jonkers J., Rottenberg S., Sharan S.K., Nussenzweig A. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535(7612):382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Somyajit K., Saxena S., Babu S., Mishra A., Nagaraju G. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 2015;43(20):9835–9855. doi: 10.1093/nar/gkv880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huh M.S., Ivanochko D., Hashem L.E., Curtin M., Delorme M., Goodall E., Yan K., Picketts D.J. Stalled replication forks within heterochromatin require ATRX for protection. Cell Death Dis. 2016;7:e2220. doi: 10.1038/cddis.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Y., Liu Z., Wang F., Temviriyanukul P., Ma X., Tu Y., Lv L., Lin Y.F., Huang M., Zhang T., Pei H., Chen B.P., Jansen J.G., de Wind N., Fischhaber P.L., Friedberg E.C., Tang T.S., Guo C. FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress. Nucleic Acids Res. 2015;43(17):8325–8339. doi: 10.1093/nar/gkv737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haince J.-F., McDonald D., Rodrigue A., Dery U., Masson J.-Y., Hendzel M.J., Poirier G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283(2):1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 86.Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28(17):2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007;282(28):20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guillemette S., Serra R.W., Peng M., Hayes J.A., Konstantinopoulos P.A., Green M.R., Cantor S.B. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 2015;29(5):489–494. doi: 10.1101/gad.256214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kais Z., Rondinelli B., Holmes A., O’Leary C., Kozono D., D’Andrea A.D., Ceccaldi R. FANCD2 maintains fork stability in BRCA1/2-Deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 2016;15(11):2488–2499. doi: 10.1016/j.celrep.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Michl J., Zimmer J., Buffa F.M., McDermott U., Tarsounas M. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat. Struct. Mol. Biol. 2016;23(8):755–757. doi: 10.1038/nsmb.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van der Heijden T., Seidel R., Modesti M., Kanaar R., Wyman C., Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35(17):5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Mameren J., Modesti M., Kanaar R., Wyman C., Peterman E.J., Wuite G.J. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457(7230):745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K., Abhyankar A., Sougnez C., Gabriel S.B., Auerbach A.D., Kowalczykowski S.C., Smogorzewska A. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell. 2015;59(3):478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ameziane N., May P., Haitjema A., van de Vrugt H.J., van Rossum-Fikkert S.E., Ristic D. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat. Commun. 2015;6:8829. doi: 10.1038/ncomms9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kowalczykowski S.C. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2015;7(11) doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu J., Doty T., Gibson B., Heyer W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17(10):1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467(7316):678–U62. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thorslund T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., West S.C. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17(10):1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wong A.K., Pero R., Ormonde P.A., Tavtigian S.V., Bartel P.L. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997;272(51):31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 100.Esashi F., Galkin V.E., Yu X., Egelman E.H., West S.C. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14(6):468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 101.Saeki H., Siaud N., Christ N., Wiegant W.W., Van Buul P.P., Han M., Zdzienicka M.Z., Stark J.M., Jasin M. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl. Acad. Sci. U. S. A. 2006;103(23):8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park J.Y., Zhang F., Andreassen P.R. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochim. Biophys. Acta. 2014;1846(1):263–275. doi: 10.1016/j.bbcan.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daza-Martin M., Starowicz K., Jamshad M., Tye S., Ronson G.E., MacKay H.L., Chauhan A.S., Walker A.K., Stone H.R., Beesley J.F.J., Coles J.L., Garvin A.J., Stewart G.S., McCorvie T.J., Zhang X., Densham R.M., Morris J.R. Isomerization of BRCA1-BARD1 promotes replication fork protection. Nature. 2019 doi: 10.1038/s41586-019-1363-4. [DOI] [PubMed] [Google Scholar]
- 104.Nacson J., Di Marcantonio D., Wang Y., Bernhardy A.J., Clausen E., Hua X., Cai K.Q., Martinez E., Feng W., Callen E., Wu W., Gupta G.P., Testa J.R., Nussenzweig A., Sykes S.M., Johnson N. BRCA1 mutational complementation induces synthetic viability. Mol. Cell. 2020 doi: 10.1016/j.molcel.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yazinski S.A., Comaills V., Buisson R., Genois M.M., Nguyen H.D., Ho C.K., Todorova Kwan T., Morris R., Lauffer S., Nussenzweig A., Ramaswamy S., Benes C.H., Haber D.A., Maheswaran S., Birrer M.J., Zou L. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31(3):318–332. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murphy A.K., Fitzgerald M., Ro T., Kim J.H., Rabinowitsch A.I., Chowdhury D., Schildkraut C.L., Borowiec J.A. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 2014;206(4):493–507. doi: 10.1083/jcb.201404111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luijsterburg M.S., Typas D., Caron M.C., Wiegant W.W., van den Heuvel D., Boonen R.A. A PALB2-interacting domain in RNF168 couples homologous recombination to DNA break-induced chromatin ubiquitylation. Elife. 2017;6 doi: 10.7554/eLife.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sy S.M., Huen M.S., Zhu Y., Chen J. PALB2 regulates recombinational repair through chromatin association and oligomerization. J. Biol. Chem. 2009;284(27):18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bleuyard J.Y., Fournier M., Nakato R., Couturier A.M., Katou Y., Ralf C., Hester S.S., Dominguez D., Rhodes D., Humphrey T.C., Shirahige K., Esashi F. MRG15-mediated tethering of PALB2 to unperturbed chromatin protects active genes from genotoxic stress. Proc. Natl. Acad. Sci. U. S. A. 2017;114(29):7671–7676. doi: 10.1073/pnas.1620208114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Belotserkovskaya R., Raga Gil E., Lawrence N., Butler R., Clifford G., Wilson M.D., Jackson S.P. PALB2 chromatin recruitment restores homologous recombination in BRCA1-deficient cells depleted of 53BP1. Nat. Commun. 2020;11(1):819. doi: 10.1038/s41467-020-14563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park J.Y., Singh T.R., Nassar N., Zhang F., Freund M., Hanenberg H., Meetei A.R., Andreassen P.R. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene. 2014;33(40):4803–4812. doi: 10.1038/onc.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brough R., Bajrami I., Vatcheva R., Natrajan R., Reis-Filho J.S., Lord C.J., Ashworth A. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012;31(5):1160–1176. doi: 10.1038/emboj.2011.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Couturier A.M., Fleury H., Patenaude A.M., Bentley V.L., Rodrigue A., Coulombe Y., Niraj J., Pauty J., Berman J.N., Dellaire G., Di Noia J.M., Mes-Masson A.M., Masson J.Y. Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic Acids Res. 2016;44(22):10879–10897. doi: 10.1093/nar/gkw921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen Y., Wu Y.R., Yang H.Y., Li X.Z., Jie M.M., Hu C.J., Wu Y.Y., Yang S.M., Yang Y.B. Prolyl isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis. 2018;9(9):883. doi: 10.1038/s41419-018-0844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tian Y., Shen X., Wang R., Klages-Mundt N.L., Lynn E.J., Martin S.K., Ye Y., Gao M., Chen J., Schlacher K., Li L. Constitutive role of the Fanconi anemia D2 gene in the replication stress response. J. Biol. Chem. 2017;292(49):20184–20195. doi: 10.1074/jbc.M117.814780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sato K., Shimomuki M., Katsuki Y., Takahashi D., Kobayashi W., Ishiai M., Miyoshi H., Takata M., Kurumizaka H. FANCI-FANCD2 stabilizes the RAD51-DNA complex by binding RAD51 and protects the 5’-DNA end. Nucleic Acids Res. 2016;44(22):10758–10771. doi: 10.1093/nar/gkw876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin Z., Kong H., Nei M., Ma H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl. Acad. Sci. U. S. A. 2006;103(27):10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu N., Schild D., Thelen M.P., Thompson L.H. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30(4):1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miller K.A., Yoshikawa D.M., McConnell I.R., Clark R., Schild D., Albala J.S. RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J. Biol. Chem. 2002;277(10):8406–8411. doi: 10.1074/jbc.M108306200. [DOI] [PubMed] [Google Scholar]
- 120.Rodrigue A., Lafrance M., Gauthier M.C., McDonald D., Hendzel M., West S.C., Jasin M., Masson J.Y. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006;25(1):222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Garcin E.B., Gon S., Sullivan M.R., Brunette G.J., Cian A., Concordet J.P., Giovannangeli C., Dirks W.G., Eberth S., Bernstein K.A., Prakash R., Jasin M., Modesti M. Differential requirements for the RAD51 paralogs in genome repair and maintenance in human cells. PLoS Genet. 2019;15(10) doi: 10.1371/journal.pgen.1008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chun J., Buechelmaier E.S., Powell S.N. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-Dependent homologous recombination pathway. Mol. Cell. Biol. 2013;33(2):387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Amunugama R., Groden J., Fishel R. The HsRAD51B-HsRAD51C stabilizes the HsRAD51 nucleoprotein filament. DNA Repair (Amst) 2013;12(9):723–732. doi: 10.1016/j.dnarep.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sullivan M.R., Bernstein K.A. RAD-ical new insights into RAD51 regulation. Genes (Basel) 2018;9(12) doi: 10.3390/genes9120629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lambert S., Mizuno K., Blaisonneau J., Martineau S., Chanet R., Freon K., Murray J.M., Carr A.M., Baldacci G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell. 2010;39(3):346–359. doi: 10.1016/j.molcel.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 126.Mizuno K., Lambert S., Baldacci G., Murray J.M., Carr A.M. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009;23(24):2876–2886. doi: 10.1101/gad.1863009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mizuno K., Miyabe I., Schalbetter S.A., Carr A.M., Murray J.M. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature. 2013;493(7431):246–249. doi: 10.1038/nature11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schubert L., Ho T., Hoffmann S., Haahr P., Guerillon C., Mailand N. RADX interacts with single-stranded DNA to promote replication fork stability. EMBO Rep. 2017;18(11):1991–2003. doi: 10.15252/embr.201744877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bugreev D.V., Yu X., Egelman E.H., Mazin A.V. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21(23):3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xue C., Daley J.M., Xue X., Steinfeld J., Kwon Y., Sung P., Greene E.C. Single-molecule visualization of human BLM helicase as it acts upon double- and single-stranded DNA substrates. Nucleic Acids Res. 2019;47(21):11225–11237. doi: 10.1093/nar/gkz810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tripathi V., Nagarjuna T., Sengupta S. BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J. Cell Biol. 2007;178(1):9–14. doi: 10.1083/jcb.200610051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Patel D.S., Misenko S.M., Her J., Bunting S.F. BLM helicase regulates DNA repair by counteracting RAD51 loading at DNA double-strand break sites. J. Cell Biol. 2017;216(11):3521–3534. doi: 10.1083/jcb.201703144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hu Y., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., Jasin M., Vogel H., Sung P., Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21(23):3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim J.H., Kim J., Kim D.H., Ryu G.H., Bae S.H., Seo Y.S. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 2004;32(8):2287–2297. doi: 10.1093/nar/gkh534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chiolo I., Saponaro M., Baryshnikova A., Kim J.H., Seo Y.S., Liberi G. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol. Cell. Biol. 2007;27(21):7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fugger K., Mistrik M., Danielsen J.R., Dinant C., Falck J., Bartek J., Lukas J., Mailand N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 2009;186(5):655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Simandlova J., Zagelbaum J., Payne M.J., Chu W.K., Shevelev I., Hanada K., Chatterjee S., Reid D.A., Liu Y., Janscak P., Rothenberg E., Hickson I.D. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 2013;288(47):34168–34180. doi: 10.1074/jbc.M113.484493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim J., Kim J.H., Lee S.H., Kim D.H., Kang H.Y., Bae S.H., Pan Z.Q., Seo Y.S. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 2002;277(27):24530–24537. doi: 10.1074/jbc.M201612200. [DOI] [PubMed] [Google Scholar]
- 139.Chu W.K., Payne M.J., Beli P., Hanada K., Choudhary C., Hickson I.D. FBH1 influences DNA replication fork stability and homologous recombination through ubiquitylation of RAD51. Nat. Commun. 2015;6:5931. doi: 10.1038/ncomms6931. [DOI] [PubMed] [Google Scholar]
- 140.Aihara H., Ito Y., Kurumizaka H., Yokoyama S., Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999;290(2):495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 141.Ronson G.E., Piberger A.L., Higgs M.R., Olsen A.L., Stewart G.S., McHugh P.J., Petermann E., Lakin N.D. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat. Commun. 2018;9(1):746. doi: 10.1038/s41467-018-03159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Moldovan G.L., D’Andrea A.D. To the rescue: the Fanconi anemia genome stability pathway salvages replication forks. Cancer Cell. 2012;22(1):5–6. doi: 10.1016/j.ccr.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mochizuki A.L., Katanaya A., Hayashi E., Hosokawa M., Moribe E., Motegi A., Ishiai M., Takata M., Kondoh G., Watanabe H., Nakatsuji N., Chuma S. PARI regulates stalled replication fork processing to maintain genome stability upon replication stress in mice. Mol. Cell. Biol. 2017;37(23) doi: 10.1128/MCB.00117-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guillemette S., Branagan A., Peng M., Dhruva A., Scharer O.D., Cantor S.B. FANCJ localization by mismatch repair is vital to maintain genomic integrity after UV irradiation. Cancer Res. 2014;74(3):932–944. doi: 10.1158/0008-5472.CAN-13-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lachaud C., Moreno A., Marchesi F., Toth R., Blow J.J., Rouse J. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science. 2016;351(6275):846–849. doi: 10.1126/science.aad5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guilliam T.A., Doherty A.J. PrimPol-prime time to reprime. Genes (Basel) 2017;8(1) doi: 10.3390/genes8010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Quinet A., Tirman S., Jackson J., Svikovic S., Lemacon D., Carvajal-Maldonado D., Gonzalez-Acosta D., Vessoni A.T., Cybulla E., Wood M., Tavis S., Batista L.F.Z., Mendez J., Sale J.E., Vindigni A. PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-Deficient cells. Mol. Cell. 2020;77(3):461–474. doi: 10.1016/j.molcel.2019.10.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McAllister K.A., Bennett L.M., Houle C.D., Ward T., Malphurs J., Collins N.K., Cachafeiro C., Haseman J., Goulding E.H., Bunch D., Eddy E.M., Davis B.J., Wiseman R.W. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 2002;62(4):990–994. [PubMed] [Google Scholar]
- 149.Donoho G., Brenneman M.A., Cui T.X., Donoviel D., Vogel H., Goodwin E.H., Chen D.J., Hasty P. Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer. 2003;36(4):317–331. doi: 10.1002/gcc.10148. [DOI] [PubMed] [Google Scholar]
- 150.Billing D., Horiguchi M., Wu-Baer F., Taglialatela A., Leuzzi G., Nanez S.A., Jiang W., Zha S., Szabolcs M., Lin C.S., Ciccia A., Baer R. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell. 2018;72(1):127–139. doi: 10.1016/j.molcel.2018.08.016. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Carroll C., Badu-Nkansah A., Hunley T., Baradaran-Heravi A., Cortez D., Frangoul H. Schimke Immunoosseous Dysplasia associated with undifferentiated carcinoma and a novel SMARCAL1 mutation in a child. Pediatr. Blood Cancer. 2013;60(9):E88–90. doi: 10.1002/pbc.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moinova H.R., Chen W.D., Shen L., Smiraglia D., Olechnowicz J., Ravi L., Kasturi L., Myeroff L., Plass C., Parsons R., Minna J., Willson J.K., Green S.B., Issa J.P., Markowitz S.D. HLTF gene silencing in human colon cancer. Proc. Natl. Acad. Sci. U. S. A. 2002;99(7):4562–4567. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sandhu S., Wu X., Nabi Z., Rastegar M., Kung S., Mai S., Ding H. Loss of HLTF function promotes intestinal carcinogenesis. Mol. Cancer. 2012;11:18. doi: 10.1186/1476-4598-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ding X., Ray Chaudhuri A., Callen E., Pang Y., Biswas K., Klarmann K.D., Martin B.K., Burkett S., Cleveland L., Stauffer S., Sullivan T., Dewan A., Marks H., Tubbs A.T., Wong N., Buehler E., Akagi K., Martin S.E., Keller J.R., Nussenzweig A., Sharan S.K. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun. 2016;7:12425. doi: 10.1038/ncomms12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412(6846):557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 157.Petermann E., Maya-Mendoza A., Zachos G., Gillespie D.A., Jackson D.A., Caldecott K.W. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol. Cell. Biol. 2006;26(8):3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Quinet A., Carvajal-Maldonado D., Lemacon D., Vindigni A. DNA Fiber analysis: mind the gap! Methods Enzymol. 2017;591:55–82. doi: 10.1016/bs.mie.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 160.Lim K.S., Li H., Roberts E.A., Gaudiano E.F., Clairmont C., Sambel L.A., Ponnienselvan K., Liu J.C., Yang C., Kozono D., Parmar K., Yusufzai T., Zheng N., D’Andrea A.D. USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol. Cell. 2018;72(6):925–941. doi: 10.1016/j.molcel.2018.10.045. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sato K., Ishiai M., Toda K., Furukoshi S., Osakabe A., Tachiwana H., Takizawa Y., Kagawa W., Kitao H., Dohmae N., Obuse C., Kimura H., Takata M., Kurumizaka H. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 2012;31(17):3524–3536. doi: 10.1038/emboj.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ray Chaudhuri A., Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18(10):610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 165.Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P.J., Stankovic T., Taylor A.M., Durocher D. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.