Abstract
The novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2 or COVID-19) has led to a world-wild pandemic. The replication of SARS-CoV-2 RNA genome involves the core replication-transcription complex (RTC, nsp12-nsp7-nsp8) and the proofreading complex (nsp14-nsp10) that can correct mismatched base pairs during replication. Structures and functions of SARS-CoV-2 RTC have been actively studied, yet little is known about SARS-CoV-2 nsp14-nsp10. Here, we purified, reconstituted, and characterized the SARS-CoV-2 nsp14-nsp10 proofreading nuclease in vitro. We show that SARS-CoV-2 nsp14 is activated by nsp10, functioning as a potent RNase that can hydrolyze RNAs in the context of single- and double-stranded RNA and RNA/DNA hybrid duplex. SARS-CoV-2 nsp14-nsp10 shows a metal-dependent nuclease activity but has different metal selectivity from RTC. While RTC is activated by Ca2+, nsp14-nsp10 is completely inhibited. Importantly, the reconstituted SARS-CoV-2 nsp14-nsp10 efficiently removed the A:A mismatch at the 3′-end of the primer, enabling the stalled RTC to restart RNA replication. Our collective results confirm that SARS-CoV-2 nsp14-nsp10 functions as the RNA proofreading complex in SARS-CoV-2 replication and provide a useful foundation to understand the structure and function of SARS-CoV-2 RNA metabolism.
Keywords: SARS-CoV-2, COVID-19, nsp14, nsp10, RNA replication, Proofreading
1. Introduction
The novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2 or COVID-19) has led to a worldwide pandemic and caused over two million deaths and over one hundred million infections (as of January 28th, 2021). SARS-CoV-2 and other related coronaviruses, including SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV), belong to the Coronaviridae family and are enveloped, single-stranded positive-sense RNA viruses [1].
In coronaviruses (CoVs), nonstructural protein 12 (nsp12) has an RNA-dependent RNA polymerase (RdRp) activity and forms a “core” replication-transcription complex (RTC) together with nsp7 and nsp8 [[2], [3], [4]]. This low-fidelity RNA polymerase complex (nsp12:nsp7:nsp8 of 1:2:1) allows CoVs to effectively and dynamically adapt to changes in environments and selective pressures by providing high genomic diversity. To compensates for the high error rate of the nsp12-nsp7-nsp8 complex, CoVs have a unique proofreading function provided by the nsp14-nsp10 complex [5]. Notably, nsp14 is highly conserved across CoVs and contains 3′-to-5′ exonuclease (ExoN) and guanine-N7-methyltransferase (N7-MTase) domains (Fig. S1). It was previously shown that the RNase activity of SARS-CoV-1 and SARS-CoV-2 nsp14 is strongly stimulated by interaction with nsp10 [[6], [7], [8], [9]].
In CoVs, the balance between the low-fidelity RNA replication by RTC and the proofreading function by nsp14 is critical for viral viability and genome maintenance [10,11]. Genetic deletion or inactivating mutations in nsp14-ExoN is lethal to SARS-CoV-2 [10,11]. This suggests that intervention of the nsp14-ExoN activity can be an effective strategy to combat SARS-CoV-2. Furthermore, the exonuclease activity of nsp14-ExoN has been proposed as a barrier to the development of nucleoside analogs, such as Remdesivir, as antiviral drugs [11]. So far, structural and biochemical analyses of the SARS-CoV-2 RTC-RNA complexes have been active research area [[2], [3], [4],12,13]. However, despite the biomedical importance, the substrate specificity, metal selectivity, and proofreading function of the SARS-CoV-2 nsp14–nsp10 nuclease remain poorly understood.
Here, as a first step to understand the RNA proofreading function of SARS-CoV-2, we purified and reconstituted the functional SARS-CoV-2 nsp14–nsp10 proofreading complex in vitro. In the presence of nsp10, nsp14 efficiently cleaves RNAs, but not DNA, either in a single- or double-stranded form. Importantly, the reconstituted nsp14–nsp10 proofreading complex removed the A:A mismatch at the 3′ end of the primer, a site of extension by RTC, enabling the stalled RTC complex to restart the RNA replication. Our collective results prove SARS-CoV-2 nsp14-nsp10 as a functional RNA proofreading complex and provide a useful foundation to study the structure and function of SARS-CoV-2 replication in the future.
2. Results
2.1. Purification and reconstitution of SARS-CoV-2 nsp14-nsp10 exonuclease
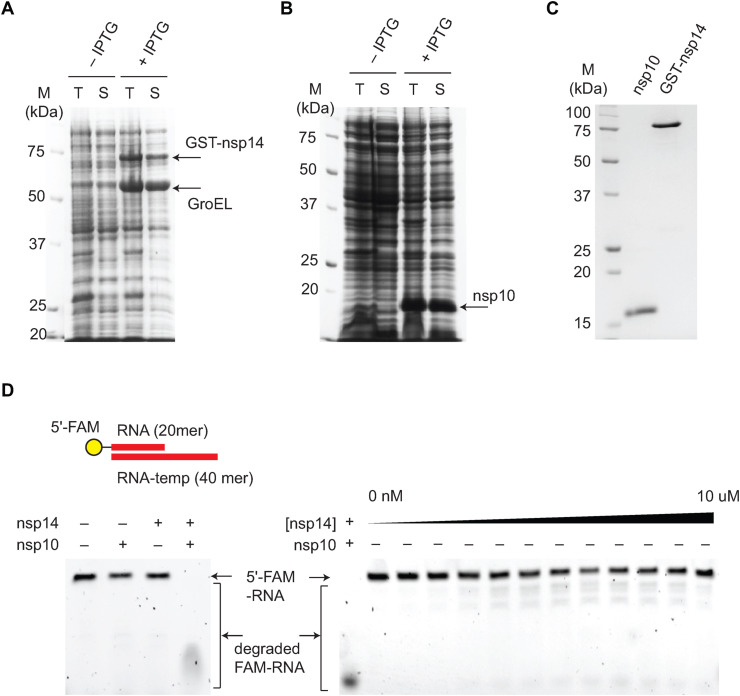
SARS-CoV-2 nsp14 and nsp10 are highly conserved in CoVs and show 95% and 97% sequence identity to SARS-CoV-1 nsp14 and nsp10, respectively (Fig. S1). Consistent with this high degree of sequence conservation, substitution of two catalytic residues Asp90 and Glu92 (Fig. S1, Asp113 and Glu115 in reference 9) with alanine completely abolishes the nuclease activity of nsp14 [9]. To purify recombinant SARS-CoV-2 nsp14 and nsp10 proteins, codon-optimized genes encoding nsp14 and nsp10 were cloned with N-terminal GST- and his-tag, respectively. We found that the expression level, solubility, and stability during purification of nsp14 were substantially improved with the N-terminal GST-tag when co-expressed with GroESL chaperon (Fig. 1 A), in comparison to his- or strep-tagged nsp14 (Fig. S2). Unlike the previous report [9], SARS-CoV-2 nsp10 alone was successfully purified.
Fig. 1.
Purification of recombinant SARS-CoV-2 nsp14 and nsp10. (A) GST-tagged SARS-CoV-2 nsp14 was overexpressed in E. coli BL21 (DE3) expressing GroESL. (B) SARS-CoV-2 nsp10 was overexpressed in E. coli BL21 (DE3). (C) Purified SARS-CoV-2 nsp10 and nsp14.1 μg of nsp10 and nsp14 are loaded. (D) SARS-CoV-2 nsp10 is required for the nuclease activity of nsp14. The nuclease activity of nsp14 is limited when titrated up to 10 μM (right). However, the addition of nsp10 dramatically stimulates the nuclease activity of nsp14 (left). The annealed double-stranded primter/template RNA is shown. This substrate is also used for experiments in Fig. 2, Fig. 3.
Both SARS-CoV-2 nsp14 and nsp10 proteins were purified to near homogeneity (Fig. 1C and S3). The SARS-CoV-2 nsp14 was purified using a three-step purification protocol (Glutathione-agarose, anion-exchange, and size exclusion chromatography), which yields 0.5 mg of purified nsp14 per liter of culture. The SARS-CoV-2 nsp10 was purified using a three-step purification protocol (Ni-NTA, anion-exchange, and size exclusion chromatography), which yields 0.7 mg of purified nsp10 per liter of culture. While nsp10 exists as a monomer, GST-nsp14 appears to be a dimer, presumably due to the dimerization of GST (Fig. S4).
Using purified SARS-CoV-2 nsp14 and nsp10, we tested the nuclease activity of nsp14 against a double-stranded RNA (dsRNA) substrate that consists of a 40-mer RNA template and 5′-fluorescein (FAM)-labeled 20-mer RNA primer (Fig. 1D). SARS-CoV-2 nsp14 alone shows a limited RNase activity when titrated up to 10 μM (Fig. 1D). However, consistent with SARS-CoV-1 nsp14 [8], the addition of nsp10 dramatically stimulates the nuclease activity of nsp14, forming a functional nuclease (Fig. 1D).
2.2. Different metal selectivity between SARS-CoV-2 nsp14-nsp10 nuclease and RTC
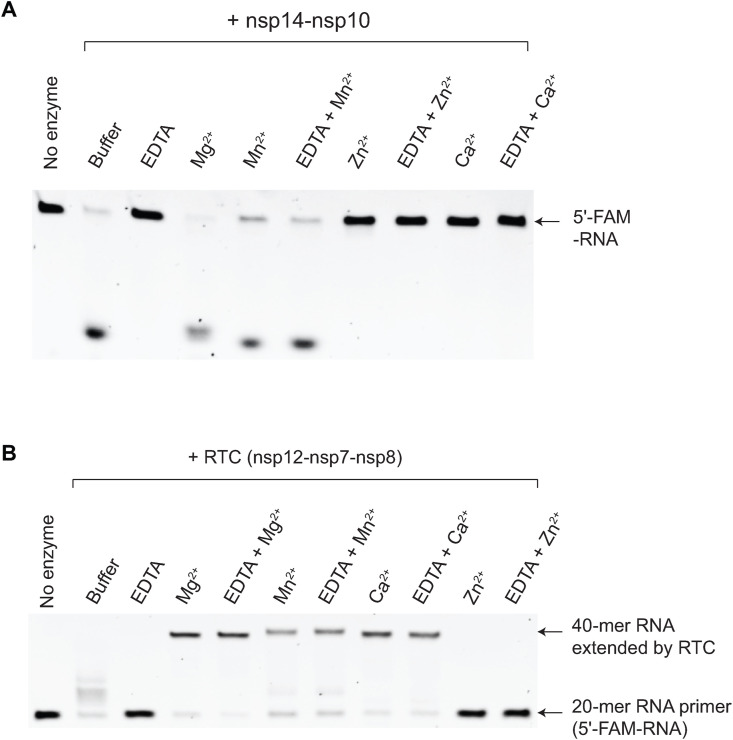
The nuclease activity of SARS-CoV-1 nsp14 is metal-dependent [7,8,14,15]. Therefore, we tested the metal selectivity of the SARS-CoV-2 nsp14-nsp10 nuclease complex. As shown in Fig. 2 A, the SARS-CoV-2 nsp14-nsp10 complex shows a robust RNase activity even in the absence of any divalent metals, whereas treatment of EDTA completely abolishes this activity. This suggests that the recombinant SARS-CoV-2 nsp14 was purified in a metal-bound form. We compared the nuclease activity of the nsp14-nsp10 complex in the presence of different divalent metals. As expected, Mg2+ promotes the RNase activity of nsp14-nsp10. While Mn2+ supports the nsp14-nsp10 nuclease activity, the addition of Zn2+ and Ca2+ completely inhibits. To further confirm these results, we pre-incubated nsp14-nsp10 with EDTA (1 mM) to keep this enzyme in a metal-free state. We obtained the same results when an excess amount (2 mM) of Mn2+, Zn2+, and Ca2+ are added to the reactions (Fig. 2A).
Fig. 2.
Different metal selectivity of SARS-CoV-2 nsp14-nsp10 exonuclease and RTC. (A) Metal selectivity of SARS-CoV-2 nsp14-nsp10. The nuclease activity of SARS-CoV-2 nsp14-nsp10 was assayed in the presence of EDTA or divalent metals. (B) Metal selectivity of SARS-CoV-2 RTC. SARS-CoV-2 RTC extends 20-mer primer to the full-length 40-mer product. In both nsp14-nsp10 and RTC, to make a “metal-free” enzyme state, EDTA (1 mM) was preincubated with nsp14-nsp10 and then excess divalent metals (2 mM) were added. The substrate in Fig. 1D was used for these experiments. Experiments were repeated twice and the representative assay is shown here.
We wondered whether SARS-CoV-2 RTC shares the same metal selectivity with the nsp14-nsp10 nuclease. To this end, we purified SARS-CoV-2 nsp12, nsp7, and nsp8 proteins to near homogeneity and reconsituted SARS-CoV-2 RTC in vitro. Like the nsp14-nsp10 nuclease, Mg2+ and Mn2+ support the RNA-dependent RNA polymerase (RdRp) activity of SARS-CoV-2 RTC, whereas Zn2+ inhibits (Fig. 2B). However, in contrast to the inhibitory effect in the nsp14-nsp10 nuclease, Ca2+ fully supports the RdRp activity of RTC (Fig. 2B), suggesting a potential regulatory role of Ca2+ in SARS-CoV-2 RNA metabolism.
2.3. Substrate specificity of the SARS-CoV-2 nsp14–nsp10 complex
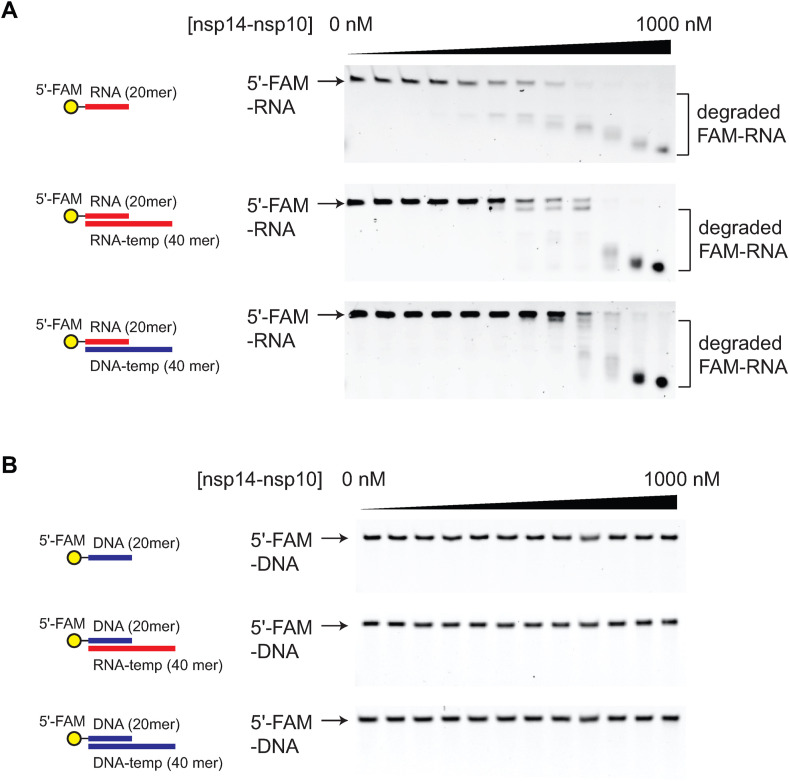
The substrate specificity of SARS-CoV-2 nsp14-nsp10 has not been extensively determined. We compared the nuclease activity of SARS-CoV-2 nsp14-nsp10 against a series of RNA and DNA oligonucleotides (Fig. 3 and Table S1). The observed RNA laddering in the 5′-end-labeled RNA is consistent with the 3′-to-5′ exonuclease activity (Fig. 3, Fig. 4 B) [7,8]. SARS-CoV-2 nsp14-nsp10 efficiently hydrolyzes single- and double-stranded RNAs. Interestingly, SARS-CoV-2 nsp14-nsp10 can also effectively cleave RNA in the context of the RNA/DNA hybrid duplex (Fig. 3A). In contrast, SARS-CoV-2 nsp14-nsp10 cannot catalyze the nucleotide excision from any types of DNAs (Fig. 3B), suggesting that SARS-CoV-2 nsp14-nsp10 is an obligate RNase.
Fig. 3.
Substrate specificity of SARS-CoV-2 nsp14-nsp10 exonuclease. (A) SARS-CoV-2 nsp14-nsp10 (0–1000 nM, 2-fold dilution) was treated to either single-stranded RNA (top) or double-stranded RNA (middle) or RNA/DNA hybrid duplex (bottom). (B) SARS-CoV-2 nsp14-nsp10 (0–1000 nM, 2-fold dilution) was treated to either single-stranded DNA (top) or DNA/RNA hybrid duplex (middle) or double-stranded DNA (bottom). SARS-CoV-2 nsp14-nsp10 efficiently cleaves RNA substrates in different forms, but not DNA.
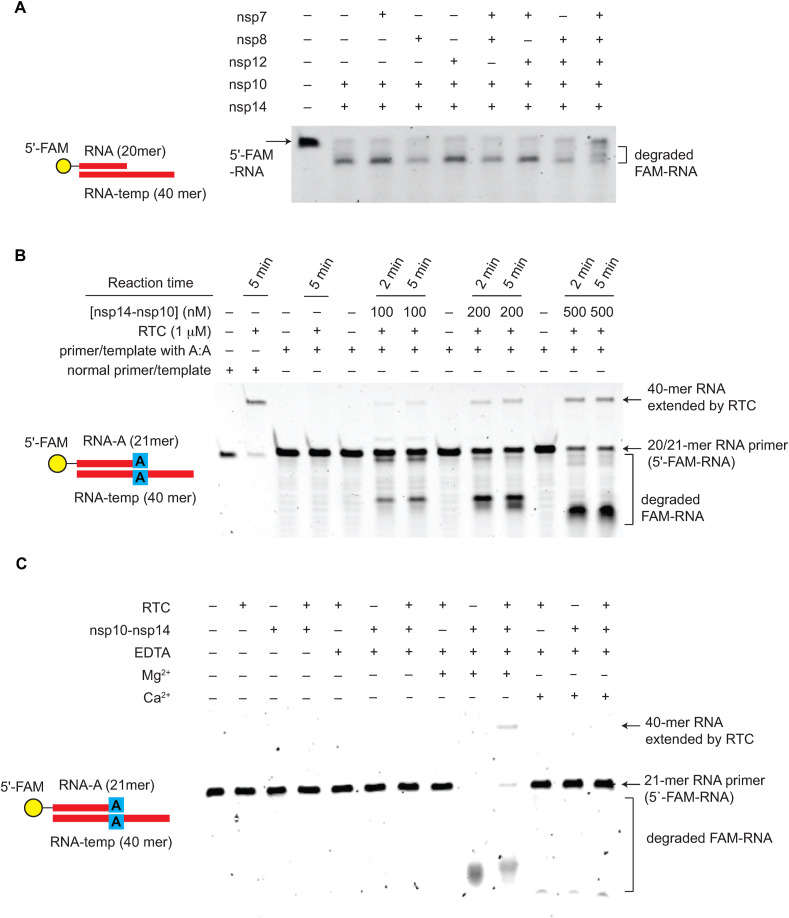
Fig. 4.
SARS-CoV-2 proofreading complex is required to restart the stalled replication-transcription complex. (A) The exonuclease activity of nsp14-nsp10 in the presence and absence of RTC complex. The SARS-CoV-2 RTC complex (nsp12-nsp7-nsp8) partially protects the RNA primer (RNA under extension) from degradation by nsp14-nsp10. The experiment was done in the absence of NTPs using a normal primer/template RNA. (B) SARS-CoV-2 nsp14-nsp10 functions as a proofreading complex during replication. The primer/template RNA with a A:A mismatch at the 3′ end of the primer is extended only in the presence of SARS-CoV-2 nsp14-nsp10. The extended 40-mer product was detected in a nsp14-nsp10 dose-dependent manner. (C) Ca2+ inhibits the restart of the mismatch-induced stalled RNA replication by RTC. The addition of Ca2+ suppresses the nsp14-nsp10 nuclease function and inhibits the primer extension of the mismatch-containing dsRNA by RTC.
2.4. RNA proofreading function of the SARS-CoV-2 nsp14–nsp10 complex is required to restart the stalled RNA replication
Finally, we tested the proofreading function of SARS-CoV-2 nsp14-nsp10 in the context of the stalled RNA replication complex. We used an RNA primer/template that contains an A:A mismatch at the 3′ end of the primer [6]. In the absence of NTPs, the addition of RTC partially protects the dsRNA from degradation by nsp14-nsp10 (Fig. 4A). To minimize the degradation of extended dsRNA by a highly potent nsp14-nsp10 nuclease, we used a sub-stoichiometric amount of nsp14-nsp10 (Fig. 4B).
SARS-CoV-2 RTC cannot extend this primer due to the A:A mismatch at the 3′ end of the primer (Fig. 4B). However, when SARS-CoV-2 nsp14-nsp10 is added to reactions, the full-length extension product (40-mer) starts to be detected in a nsp14-nsp10 dose-dependent manner (Fig. 4B). This further confirms that SARS-CoV-2 nsp14-nsp10 has a 3′-to-5′ exonuclease activity. In contrast to Mg2+, the addition of Ca2+ inhibits the primer extension by RTC, presumably due to the Ca2+-dependent inhibition of nsp14-nsp10 (Fig. 2), further supporting the different metal selectivity between SARS-CoV-2 RTC and the proofreading complex. Collectively, these results indicate that the recombinant SARS-CoV-2 nsp14-nsp10 functions as a proofreading complex that can remove the mismatched base pairs during replication. Furthermore, SARS-CoV-2 nsp14-nsp10 is required to restart the stalled RNA replication machinery during the SARS-CoV-2 genome replication.
3. Discussion
CoVs have a highly conserved nsp14-nsp10 proofreading complex that can correct the mismatched base pairs during replication and thereby compensates for the high error rate of RTC. Here, we purified in a milligram quantity, reconstituted in vitro, and characterized the nuclease activity and proofreading function of SARS-CoV-2 nsp14-nsp10. The overall nuclease activity of the SARS-CoV-2 nsp14-nsp10 complex is consistent with the previous report (reference 9, a preprint in BioRxiv) (Fig. 2, Fig. 3). However, we successfully purified both nsp14 and nsp10 individual proteins for characterization. Importantly, our data visualized the metal selectivity and proofreading functions in the context of SARS-CoV-2 RNA replication with RTC and nsp14-nsp10 nuclease (Fig. 4).
Like SARS-CoV-1 nsp14 [[6], [7], [8]], SARS-CoV-2 nsp14 requires the nsp10 cofactor to become an active nuclease (Fig. 1D). We confirmed that SARS-CoV-2 nsp14-nsp10 is an efficient 3′-to-5′ exonuclease and is able to hydrolyze RNAs, but not DNAs, in the context of single- or double-stranded RNA and RNA/DNA hybrid duplex (Fig. 3).
In addition, we found that divalent metals, such as Mg2+ or Mn2+, fully promotes both the nuclease activity of nsp14-nsp10 and the RdRp activity of RTC (Fig. 2). However, interestingly, Ca2+ shows differential effects: Ca2+ inhibits SARS-CoV-2 nsp14-nsp10 but promotes RTC (Fig. 2). Consistent with this observation, the addition of Ca2+ suppresses the restart of mismatch-induced stalled RNA replication by RTC by inhibiting the proofreading function of nsp14-nsp10 (Fig. 4C). Since Ca2+ is the most abundant cation in human body [16], it will be interesting to see in the future whether Ca2+ is involved in regulation of SARS-CoV-2 replication.
Importantly, we proved that SARS-CoV-2 nsp14-nsp10 functions as a proofreading complex for the SARS-CoV-2 replication machinery. The RNA primer/template that contains an A:A mismatch at the 3′ end of the primer is unable to be extended by SARS-CoV-2 RTC. However, SARS-CoV-2 nsp14-nsp10 is able to correct this mismatch using its 3′-to-5′ exonuclease activity, and thereby enables RTC to resume the stalled replication (Fig. 4B).
In summary, we provide an important foundation to understand the function of the SARS-CoV-2 nsp14-nsp10 proofreading complex, whose structure has not yet been determined. The reconstituted SARS-CoV-2 nsp14-nsp10 can also be used as a tool to identify new small-molecule probes targeting the proofreading function of SARS-CoV-2. Indeed, the 3′-to-5′ exonuclease activity of nsp14-nsp10 is a key player in several crucial processes in the life cycle of CoVs. ExoN of nsp14-nsp10 is structurally and functionally conserved across CoVs, which makes this protein a vulnerable target for the development of broad-spectrum anti-CoV agents. Therefore, it is likely that inhibiting ExoN activity will synergize with current nucleoside analog drugs, such as ATP-analog drug Remdesivir [4], by reducing the viral replication fidelity, by enhancing the effectiveness of Remdesivir incorporation into the viral genome, and by increasing the barrier to drug resistance.
4. Experimental procedures
Plasmids and protein purification: The gene for SARS-Cov-2 nsp14 was synthesized (geneuniversal. Inc) and cloned into a modified pET21b vector with an N-terminal tandem his-tag (pET21b-tHis-nsp14) and GST-tag (pET21b-GST-nsp14). Although his-nsp14 was solubly expressed in E. coli (Fig. S2), this his-nsp14 seems to be unstable alone and we experienced a significant loss of protein during purification. However, the stability of nsp14 during purification was remarkably improved when the GST-nsp14 was used and co-expressed with GroESL. The pET21b-GST-nsp14 plasmid was introduced into E. coli BL21 (DE3) expressing GroESL. Cells were grown in LB medium to an OD600 of 0.6–0.8 at 37 °C and then the temperature was reduced to 16 °C followed by the addition of 0.5 mM IPTG for expression. After overnight induction, the harvested cell pellets were resuspended in buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM PMCF, 1 mM Benzamdine, and 5 mM BME. Cells were disrupted by sonication and the lysate was centrifuged at 13000 rpm for 70 min to obtain the supernatant containing the recombinant protein. SARS-Cov-2 nsp14 was purified by affinity capture on a GSH-Sepharose column (Gold biotechnology. Inc). After elution with a buffer containing 10 mM glutathione, SARS-CoV-2 nsp14 was purified on a Heparin column (GE Lifesciences) using a NaCl gradient (0.1–1 M NaCl). SARS-Cov-2 nsp14 was further purified with a Superdex 200 (GE Lifesciences) size-exclusion column using a buffer containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mm DTT, and 5% glycerol. Purified SARS-Cov-2 nsp14 was concentrated and stored at − 80 °C.
The gene for SARS-Cov-2 nsp10 was synthesized (geneuniversal. Inc) and cloned into a modified pET21b vector with an N-terminal his tag [17] (pET21b-His-nsp10). The pET21b-His-nsp10 plasmid was introduced into E. coli BL21 (DE3). Cells were grown in LB medium to an OD600 of 0.6–0.8 at 37 °C and then induced at 16 °C by the addition of 0.1 mM IPTG. After overnight induction, the harvested cell pellets were resuspended in buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM PMCF, 1 mM Benzamdine, and 5 mM BME. Cells were disrupted by sonication and the lysate was centrifuged at 13000 rpm for 70 min to obtain the supernatant containing the recombinant protein. SARS-Cov-2 nsp10 was purified by affinity capture on a Ni-NTA column (GE Lifesciences). After elution with a buffer containing 250 mM imidazole, SARS-Cov-2 nsp10 was purified with a Heparin column (GE Lifesciences) using a NaCl gradient (0.1–1 M NaCl). SARS-Cov-2 nsp10 was further purified on Superdex 200 (GE Lifesciences) size-exclusion column using a buffer containing 25 mm Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT, and 5% glycerol. Purified SARS-Cov-2 nsp10 was concentrated and stored at − 80 °C.
Bacterial expression constructs for nsp12, nsp7, and nsp8 (pET22b-nsp12, pET28a-nsp7, and pET28a-nsp8 plasmids) are kindly provided by Dr. Zihe Rao [2]. SARS-CoV-2 nsp12, nsp7, and nsp8 proteins were purified as described previously [2]. The purified SARS-CoV-2 nsp12, nsp7, and nsp8 proteins were concentrated and then stored at − 80 °C.
Nuclease assay: Synthetic RNAs (Table S1, 5′-FAM-20mer-RNA primer, 40mer-RNA template, 5′-FAM-21mer-RNA-A primer [for A:A mismatch], 5′-FAM-20mer-DNA-primer, 40mer-DNA template) were purchased from Integrated DNA Technologies, Inc. The 5′-FAM-20mer-RNA primer and 5′-FAM-20mer-DNA-primer were annealed to either 40mer-RNA template or 40mer-DNA template. The annealing reaction is started by heating the oligos in a buffer containing 5 mM MES (pH 6.5) and 20 mM NaCl at 100 °C for 5 s and then slowly cooled down to room temperature. The concentrations of primer and template are 1 μM and 2 μM, respectively.
For the exonuclease assay, 20 μl reactions containing up to 1 μM SARS-CoV-2 nsp10 and nsp14, 100 nM annealed RNA oligos were incubated in a buffer containing 10 mM Tris pH 7.5, 10 mM KCl, 1 mM β-mercaptoethanol, and 2 mM MgCl2 for 30 min in 30 °C. Reactions were quenched by the addition of an 80 μl loading buffer (formamide with 50 mM EDTA) followed by boiling at 95 °C for 5 min. After cooling down to 4 °C, reaction products were analyzed in 13% polyacrylamide gels containing 8 M urea, 89 mM Tris-borate (pH 8.0), and 2.0 mM EDTA. RNA products were visualized by fluorescence using ThermoFisher iBrightFL1000.
For the metal-dependent exonuclease assay, to make enzymes in a metal-free state, SARS-CoV-2 nsp14-nsp10 was preincubated with 1 mM EDTA for 2 min in a buffer containing 10 mM Tris (pH 7.5), 10 mM KCl, 1 mM β-mercaptoethanol for 2 min in 30 °C. Then metals (2 mM MgCl2, 2 mM MnCl2, 2 mM CaCl2 and 2 mM ZnCl2) were added to each reaction at a final concentration of 2 mM. The reaction was started by adding 100 nM annealed dsRNA for 30 min in 30 °C. All reactions were quenched by adding an 80 μl loading buffer (formamide with 50 mM EDTA) followed by boiling at 95 °C for 5 min. After cooling down to 4 °C, reaction products were analyzed in 13% polyacrylamide gels containing 8 M urea, 89 mM Tris-borate (pH 8.0), and 2.0 mM EDTA. RNA products were visualized by fluorescence using ThermoFisher iBrightFL1000.
RNA polymerase and proofreading assays: For the RNA polymerase assay, 20 μl reaction mixtures containing 1 μM SARS-CoV-2 nsp7, 2 μM SARS-CoV-2 nsp8, and 1 μM SARS-CoV-2 nsp12, and 100 nM annealed RNA oligos were preincubated in a buffer containing 10 mM Tris (pH 7.5), 10 mM KCl, 1 mM β-mercaptoethanol, 2 mM MgCl2 for 2 min in 30 °C. The reaction was initiated by adding 2 mM each NTP. The reactions were incubated at 30 °C for 30 min and then quenched by adding an 80 μl loading buffer (formamide with 50 mM EDTA) followed by boiling at 95 °C for 5 min. After cooling down to 4 °C, the RNA polymerization products were analyzed in 13% polyacrylamide gels containing 8 M urea, 89 mM Tris-borate (pH 8.0), and 2.0 mM EDTA. RNA products were visualized by fluorescence using ThermoFisher iBrightFL1000.
For the RNA proofreading assay, 5′-FAM-21mer-RNA-A primer (for A:A mismatch) was annealed to 40mer-RNA template. Reactions were mixtures (20 μl) containing 1 μM SARS-CoV-2 nsp7, 2 μM SARS-CoV-2 nsp8, 1 μM SARS-CoV-2 nsp12, and 100 nM annealed RNA oligos were preincubated in a buffer containing 10 mM Tris (pH 7.5), 10 mM KCl, 1 mM β-mercaptoethanol, 2 mM MgCl2 for 2 min in 30 °C. Then up to 500 nM of SARS-CoV-2 nsp14-nsp10 was added to reactions. The reaction was initiated by adding 2 mM each NTP. Reactions were quenched and RNA products were visualized as described above.
For the metal-dependent RNA polymerase activity assay, Reaction mixtures (20 μl) containing 1 μM SARS-CoV-2 nsp7, 2 μM SARS-CoV-2 nsp8, 1 μM SARS-CoV-2 nsp12, and 100 nM annealed RNA oligos were preincubated in a buffer containing 10 mM Tris (pH 7.5), 10 mM KCl, 1 mM β-mercaptoethanol for 2 min in 30 °C. Then 2 mM CaCl2, 2 mM MgCl2, 2 mM ZnCl2, 2 mM MnCl2, and 1 mM EDTA were added to the reaction. To make enzymes in a metal-free state, RTC was preincubated with 1 mM EDTA for 2 min in 30 °C, and then metals were added to each reaction at a final concentration of 2 mM. The reaction was started by adding 2 mM each NTP and incubated for 30 min at 30 °C. Reactions were quenched and RNA products were visualized as described above.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
We thank Dr. Zihe Rao for providing pET22b-nsp12, pET28a-nsp7, and pET28a-nsp8 plasmids for protein expression. I.K.K. was supported by American Cancer Society RSG Grant (133405-RSG-19-200-01-DMC to I.K.K.), V foundation V scholar Grant (V2018-25 to I.K.K), and Marlene Harris Ride Cincinnati Breast Cancer Pilot Grant program (to I.K.K).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pep.2021.105894.
Author statements
Zhijun Ma: Methodology, Data curation, Formal analysis, Writing – original draft. Yasin Pourfarjam: Methodology, Data curation. In-Kwon Kim: Conceptualization, Supervision, Writing – review & editing.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Hobartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Sullivan J., Tedim Ferreira M., Gagne J.P., Sharma A.K., Hendzel M.J., Masson J.Y., Poirier G.G. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 2019;10:1182. doi: 10.1038/s41467-019-08859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baddock H.T., Brolih S., Yosaatmadja Y., Ratnaweera M., Bielinski M., Swift L.P., Cruz-Migoni A., Morris G.M., Schofield C.J., Gileadi O., McHugh P.J. Characterisation of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: implications for its role in viral genome stability and inhibitor identification. bioRxiv. 2020 doi: 10.1101/2020.08.13.248211. 2020.2008.2013.248211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogando N.S., Zevenhoven-Dobbe J.C., van der Meer Y., Bredenbeek P.J., Posthuma C.C., Snijder E.J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W., Wang Q., Rao Z., Lou Z. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193. doi: 10.1016/j.cell.2020.11.016. e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3'->5' exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P., Jiang M., Hu T., Liu Q., Chen X.S., Guo D. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J. Biochem. Mol. Biol. 2007;40:649–655. doi: 10.5483/bmbrep.2007.40.5.649. [DOI] [PubMed] [Google Scholar]
- 16.Pozzan T., Rizzuto R., Volpe P., Meldolesi J. Molecular and cellular physiology of intracellular calcium stores. Physiol. Rev. 1994;74:595–636. doi: 10.1152/physrev.1994.74.3.595. [DOI] [PubMed] [Google Scholar]
- 17.Pourfarjam Y., Ventura J., Kurinov I., Cho A., Moss J., Kim I.K. Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition. J. Biol. Chem. 2018;293:12350–12359. doi: 10.1074/jbc.RA118.003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.