Chen et al. show how kinetochores integrate mechanics and biochemistry using optogenetic manipulation of Aurora B kinase. Aurora B promotes microtubule depolymerization at low tension or microtubule release at high tension. Thus, tension is a signal inducing distinct error-correction pathways.
Abstract
To ensure accurate chromosome segregation, interactions between kinetochores and microtubules are regulated by a combination of mechanics and biochemistry. Tension provides a signal to discriminate attachment errors from bi-oriented kinetochores with sisters correctly attached to opposite spindle poles. Biochemically, Aurora B kinase phosphorylates kinetochores to destabilize interactions with microtubules. To link mechanics and biochemistry, current models regard tension as an input signal to locally regulate Aurora B activity. Here, we show that the outcome of kinetochore phosphorylation depends on tension. Using optogenetics to manipulate Aurora B at individual kinetochores, we find that kinase activity promotes microtubule release when tension is high. Conversely, when tension is low, Aurora B activity promotes depolymerization of kinetochore–microtubules while maintaining attachment. Thus, phosphorylation converts a catch-bond, in which tension stabilizes attachments, to a slip-bond, which releases microtubules under tension. We propose that tension is a signal inducing distinct error-correction pathways, with release or depolymerization being advantageous for typical errors characterized by high or low tension, respectively.
Introduction
To maintain genome integrity during cell division, kinetochores of sister chromatids attach to opposite spindle poles—bi-orientation—to ensure accurate segregation. This process relies on robust mechanisms to identify errors by distinguishing correct from incorrect attachments and to correct errors by changing the connections between kinetochores and microtubules. To identify errors, tension from opposite spindle poles is widely accepted as a signal indicating correctly bi-oriented sister kinetochores, whereas lack of tension signals an error, based on findings in several model systems (Nicklas, 1997; Sarangapani and Asbury, 2014; Lampson and Grishchuk, 2017; Mukherjee et al., 2019). Correcting errors depends on destabilizing kinetochore–microtubule interactions so that new attachments can form. Aurora B kinase is a key regulator that destabilizes these interactions by phosphorylating kinetochore substrates that bind microtubules (Lampson and Cheeseman, 2011; Carmena et al., 2012; Krenn and Musacchio, 2015). To couple error identification and correction, current models propose that phosphorylation of Aurora B substrates depends on tension through several possible mechanisms: separation of kinetochores from Aurora B at the inner centromere (Liu et al., 2009; Yoo et al., 2018; García-Rodríguez et al., 2019; Zaytsev et al., 2016), regulation of Aurora B activity (Asai et al., 2019), or Aurora B localization to a kinetochore binding site (Campbell and Desai, 2013; Broad et al., 2020; Broad and DeLuca, 2020).
An alternative model that has not been tested is that tension could regulate the downstream response to Aurora B substrate phosphorylation. Two models by which Aurora B destabilizes chromosome attachments have been proposed: release or depolymerization (Fig. S1 A; Lampson and Grishchuk, 2017). The release model, in which kinetochores detach from microtubules, is supported by abundant in vitro evidence of Aurora B substrate phosphorylation lowering the kinetochore–microtubule affinity (Carmena et al., 2012; Krenn and Musacchio, 2015; Sarangapani and Asbury, 2014). Alternatively, cellular studies suggest that Aurora B kinase induces depolymerization of kinetochore-attached microtubules without full detachment. When sister kinetochores are attached to a single pole—syntelic attachment—depolymerization pulls both toward the pole (Lampson et al., 2004), where they subsequently detach due to pole-localized activities (Ye et al., 2015; Chmátal et al., 2015). Furthermore, in the presence of mutations that mimic phosphorylation of the outer kinetochore protein Hec1 by Aurora B, kinetochores track the plus-ends of depolymerizing microtubules with minimal detachment (Long et al., 2017).
Figure S1.
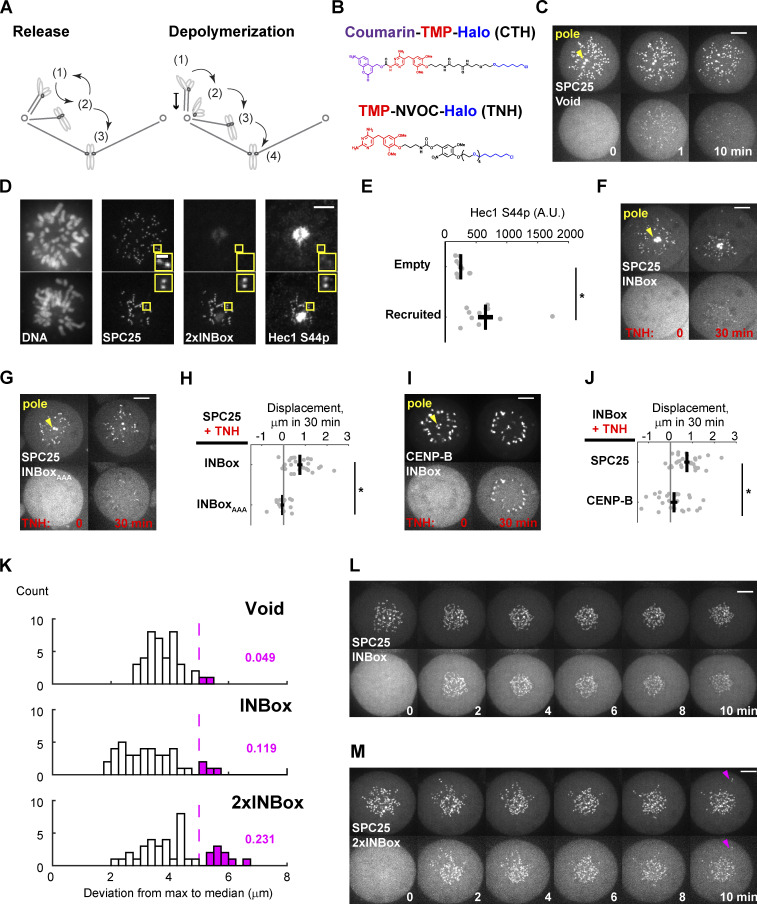
Related to Fig. 1. (A) Two proposed models of error correction mediated by Aurora B kinase. State 1 is the initial incorrect state. In the release model, microtubules detach from kinetochores (state 2) to allow new attachments to form. The process is iterative until bi-oriented attachments (state 3) are stabilized. In the depolymerization model, microtubules depolymerize while maintaining kinetochore attachment, generating poleward chromosome movement. Microtubules detach near the pole (state 2), followed by translocation along another K-fiber to the spindle equator (state 3), and binding of a microtubule from the opposite pole to achieve bi-orientation (state 4). (B) The photocaged and uncaged chemical dimerizers used in this study. CTH consists of the following components: A coumarin photocage; trimethoprim (TMP), which binds eDHFR; and a Halo ligand, which binds covalently to the HaloTag protein. TNH, consisting of TMP linked to NVOC (6-nitroveratryl oxycarbonyl) and a Halo ligand, was used for dimerization without requiring uncaging. (C) Representative images of the negative control (Video 2; yellow triangles label spindle poles). (D and E) Hec1-Ser44 phosphorylation after 2×INBox recruitment with TNH. Images (D) show immunostaining with a phospho-specific antibody in the presence or absence of 2×INBox recruitment to kinetochores. Quantification of Hec1-S44p signals shows higher phosphorylation with 2×INBox recruitment (n = 10 and 11 cells for nonrecruited and recruited kinetochores, respectively). Black lines: Mean ± SEM. (F–J) Depolymerization depends on Aurora B kinase activity and the location of the recruitment site. Representative images show INBox (F; n = 27) or INBoxAAA (G; n = 15) recruitment to the outer kinetochore anchor SPC25, or INBox recruitment to the inner centromere anchor CENP-B (I; n = 24), using TNH. INBoxAAA denotes alanine substitutions at the INBox TSS motif to prevent kinase activation. Images are maximum-intensity projection across z-slices that cover spindle poles for analyses. Displacements (H and J) quantified as in Fig. 1 G. Black lines: Mean ± SEM. (K–M) Aurora B kinases recruitment to kinetochores on monopolar spindles, using CTH, partially triggers microtubule release (same data as Fig. 1, E and F, Fig. S1 C, and Videos 2, 3, and 4). (K) Radial distance from the outermost kinetochores to the population median in the 10-min time window (same dataset as Fig. 1 G). Most kinetochores move toward the pole after activation, but kinetochores that release microtubules should be pushed away from the pole, leading to a large deviation (M, magenta triangles). Magenta bars indicate two of 41 control (Void-recruited) cells (4.9%), with at least one kinetochore pushed away. Cells with INBox recruitment contain fewer such kinetochores (L; 11.9%) than with 2×INBox recruitment (M; 23.1%). *, P < 0.05. Scale bars: 5 µm or 1 µm in insets.
Results and discussion
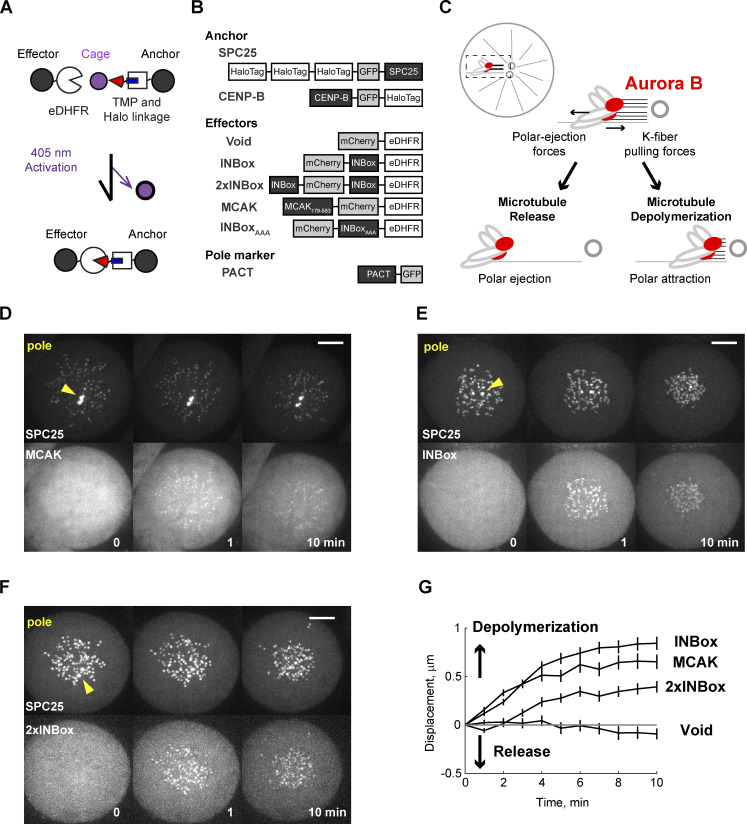
To distinguish between the release and depolymerization models, we acutely recruited Aurora B kinase to kinetochores using a photocaged small molecule that heterodimerizes HaloTag and Escherichia coli dihydrofolate reductase (eDHFR) fusion proteins (coumarin-TMP-Halo [CTH]; Figs. 1 A and S1 B; Ballister et al., 2014; Zhang et al., 2017). We fused HaloTag to the kinetochore protein SPC25 (the anchor) and eDHFR to the INBox subdomain of INCENP (the effector). INBox recruits and activates Aurora B kinase as part of the chromosome passenger complex (Carmena et al., 2012) and thus can be used to control localized Aurora B activity (Fig. 1 B; Zaytsev et al., 2016; Banigan et al., 2015). The release and depolymerization models predict different outcomes of Aurora B kinase activation. Microtubule release reduces or eliminates forces exerted by kinetochore fibers (K-fibers), whereas depolymerization increases the force pulling the kinetochore toward the attached pole. Using kinetochore movement as a readout, we tested these predictions with both monopolar and bipolar spindles, representing low- and high-tension conditions, respectively.
Figure 1.
Recruitment of Aurora B to syntelic kinetochores triggers microtubule depolymerization. (A) Light-induced dimerization schematic. The dimerizer has a Halo ligand (blue) linked to the eDHFR ligand trimethoprim (TMP; red), which is protected by a photoactivatable cage (purple). The kinetochore protein SPC25 anchors HaloTag at the outer kinetochore, and effectors are fused to eDHFR. Effectors are recruited to kinetochores by uncaging the HaloTag-bound dimerizer with light. (B) Constructs for this study. Dimerizer uncaging recruits eDHFR-tagged effector proteins (or Void as a negative control). The PACT domain targets to centrosomes to label spindle poles (yellow triangles; Gillingham and Munro, 2000). (C) Schematic of monopolar spindle assay, with chromosomes under a tug of war between K-fiber depolymerization and polar ejection forces. Red circles indicate Aurora B–activated kinetochores. After full or partial release of kinetochore microtubules, polar ejection forces dominate and chromosomes move away from spindle poles. In contrast, depolymerization increases poleward forces. (D–F) Representative images before and after uncaging at t = 0 (Videos 1, 2, 3, and 4). (G) Kinetochore displacement over time. For each cell, kinetochore-pole distances are measured and averaged at every time point. Displacements are defined relative to t = 0, with poleward movement defined as the positive direction (Void: n = 41 cells; MCAK: n = 46; INBox: n = 42; 2×INBox: n = 39; mean ± SEM). Scale bars, 5 µm.
Inhibition of kinesin-5 (Eg5) with S-trityl-L-cysteine (STLC) generates monopolar spindles so that kinetochores are under low tension with frequent syntelic attachments (Kapoor et al., 2000; Skoufias et al., 2006). Normally, the combination of K-fiber dynamics and polar ejection forces dictates chromosome positions relative to the poles (Fig. 1 C). After Aurora B kinase activation, the release model predicts chromosome movement away from the pole due to reduced forces from K-fibers, whereas the depolymerization model predicts movement toward the pole due to increased pulling forces from K-fibers. High-amplitude oscillations on monopolar spindles make it difficult to distinguish activation-induced movement from noise. To improve the signal-to-noise ratio in our analysis, we activated the entire cell to average movements over all kinetochores. As a positive control, we recruited the microtubule depolymerase MCAK (kinesin-13; Walczak et al., 2013) to kinetochores and observed net poleward movement, as expected for depolymerization (Fig. 1, D and G; and Video 1). Recruitment of eDHFR alone (Void) as a negative control had no effect (Figs. 1 G and S1 C; and Video 2). INBox recruitment to kinetochores increased phosphorylation of a known Aurora B substrate, Hec1-Ser44 (DeLuca et al., 2011), as expected for increased Aurora B activity (Fig. S1, D and E). INBox recruitment also triggered poleward chromosome movement (Fig. 1, E and G; and Video 3), consistent with Aurora B inducing microtubule depolymerization, as shown previously for syntelic attachments (Lampson et al., 2004). This poleward movement depends on Aurora B kinase activity, as recruitment of a mutant INBox in which the TSS motif is mutated to AAA to prevent Aurora B activation (Honda et al., 2003; Sessa et al., 2005; Bishop and Schumacher, 2002), had no effect (Fig. S1, F–H). INBox recruitment to inner centromeres rather than kinetochores, using CENP-B as an anchor (Fig. S1, F, I, and J), did not induce detectable kinetochore movement, indicating that phosphorylation of outer kinetochore substrates drives poleward movement. Finally, increasing Aurora B recruitment using a construct with two copies of INBox (2×INBox; Fig. 1 B) induced net poleward movement, indicating depolymerization (Fig. 1, F and G; and Video 4). The average displacement with 2×INBox is less than that with single INBox because a few kinetochores moved away from the pole (Fig. S1, K–M), suggesting that high Aurora B kinase activity can occasionally promote kinetochore–microtubule release under low tension.
Video 1.
Related to Fig. 1 D. MCAK recruitment triggers poleward chromosome movement on a monopolar spindle. Left: 3×Halo-GFP-SPC25. Right: MCAK-mCherry-eDHFR.
Video 2.
Related to Fig. S1 C. Void recruitment as a negative control for optogenetic recruitment on a monopolar spindle. Left: 3×Halo-GFP-SPC25. Right: mCherry-eDHFR.
Video 3.
Related to Fig. 1 E. INBox recruitment triggers poleward chromosome movement on a monopolar spindle. Left: 3×Halo-GFP-SPC25. Right: mCherry-INBox-eDHFR.
Video 4.
Related to Fig. 1 F. 2×INBox recruitment triggers poleward chromosome movement on a monopolar spindle. Left: 3×Halo-GFP-SPC25. Right: INBox-mCherry-INBox-eDHFR.
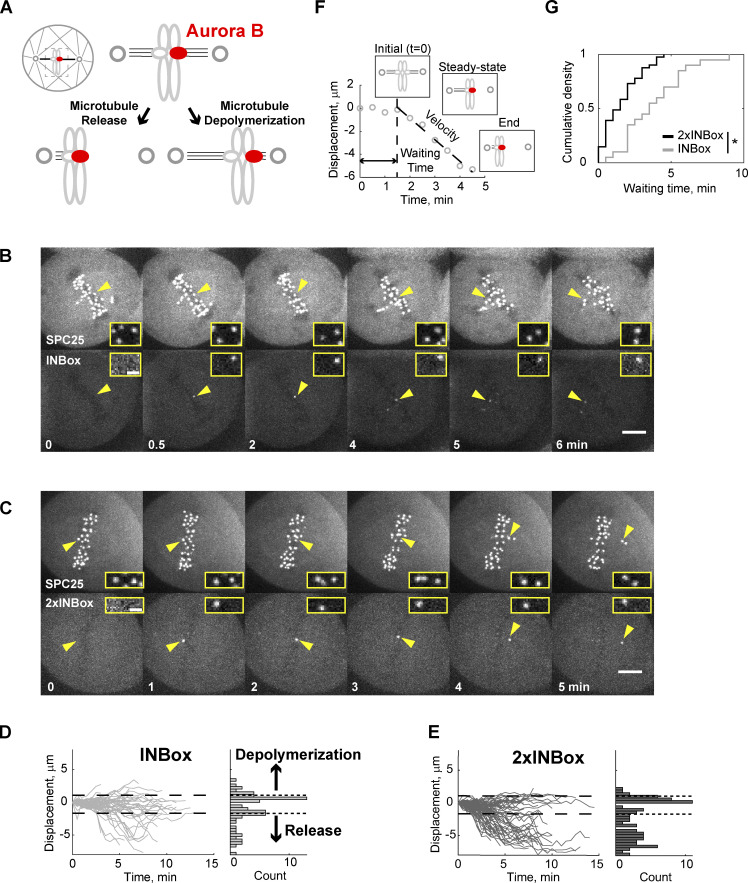
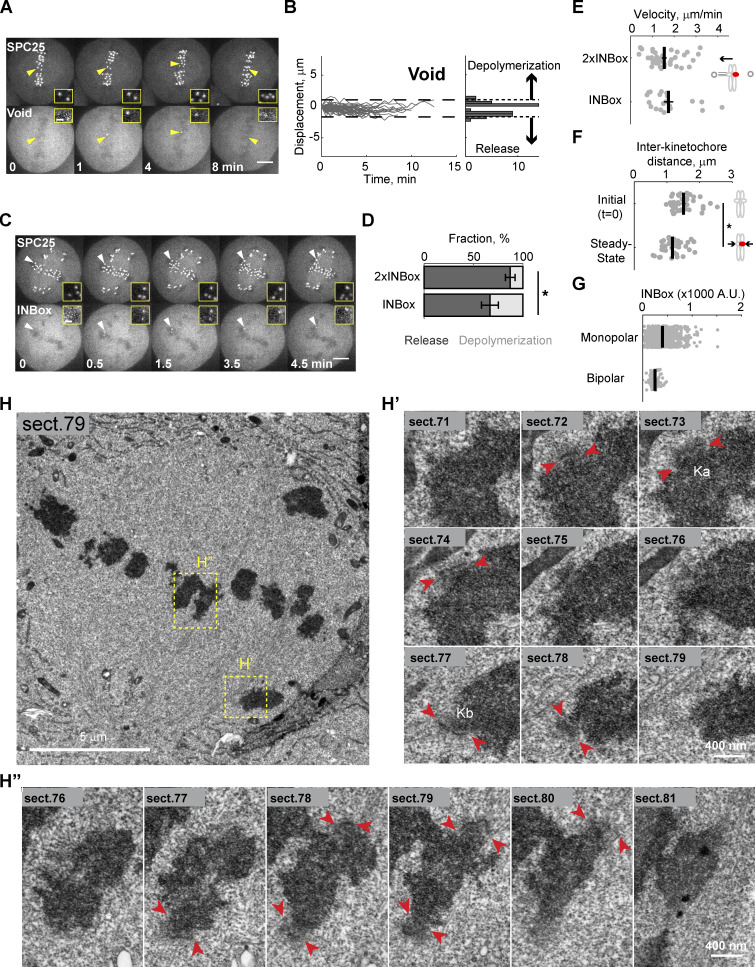
Next, we examined the effects of Aurora B kinase recruitment when sister kinetochores are bi-oriented on bipolar spindles and under high tension. We targeted individual kinetochores in this configuration because the release and depolymerization models make distinct predictions when a single kinetochore of a pair is activated (Fig. 2 A). Microtubule release should lead to reduced pulling forces from the activated kinetochore and movement away from the pole to which it was initially attached, whereas depolymerization has the opposite effect. We found that ∼50% of activated kinetochores moved beyond the bounds defined by the range of naturally occurring chromosome oscillations (Fig. S2, A and B; and Video 5). We defined each of these events as depolymerization or release, denoted as positive or negative direction displacement, respectively. Upon INBox recruitment, release events were more frequent than depolymerization events (67% vs. 33%; Fig. 2, B and D; and Fig. S2, C and D; and Videos 6 and 7). Increasing Aurora B recruitment with 2×INBox further increased the proportion of release events to 87% (Fig. 2, C and E, Fig. S2 D, and Video 8), consistent with our finding in the monopolar spindle assay that higher activity promotes more microtubule release. After an initial lag phase, the released kinetochores moved with a steady-state velocity (∼1.5 µm/min) consistent with kinetochore ablation assays mimicking release (Figs. 2 F and S2 E; Sikirzhytski et al., 2014; Long et al., 2017). Furthermore, interkinetochore distances decreased after Aurora B kinase recruitment (Fig. S2 F), consistent with the relaxed tension predicted by microtubule release from one kinetochore. We found lower INBox recruitment in the bipolar spindle assay compared with the monopolar spindle assay (Fig. S2 G), indicating that the high frequency of release events on bipolar spindles was not due to higher Aurora B activity. Together, these results indicate that bi-oriented kinetochores under tension primarily release their attached microtubules upon phosphorylation.
Figure 2.
Recruitment of Aurora B to a single kinetochore of a bi-oriented pair triggers microtubule release. (A) Schematic of bipolar spindle assay. The bi-oriented sisters are under a tug of war between the two attached K-fibers. Microtubule release or depolymerization at the activated kinetochore (red circles) leads to movement in opposite directions. (B and C) Images from representative experiments showing INBox (B; Video 6) or 2×INBox (C; Video 8) recruitment after activation of a single kinetochore (yellow triangles) at t = 0. Insets show the targeted kinetochore pair at higher magnification. (D and E) Displacement of the activated kinetochore from the metaphase plate over time: Each trace represents a single kinetochore after INBox (D; n = 64) or 2×INBox (E; n = 79) recruitment, with the starting location defined as zero. Dashed lines show the range of chromosome dynamics covering 96% of control (Void recruited) kinetochores (Fig. S2, A and B; and Video 5). Histograms show maximal displacement for each trace. (F and G) Analyses of released kinetochores after INBox or 2×INBox recruitment. Example trace of 2×INBox recruitment (F) shows waiting time after activation, followed by movement at steady-state velocity (Fig. S2 E). (G) Waiting time distribution (2×INBox: median = 1.5 min, n = 41; INBox: median = 3.5 min, n = 20). *, P < 0.005. Scale bars, 5 µm or 1 µm in insets.
Figure S2.
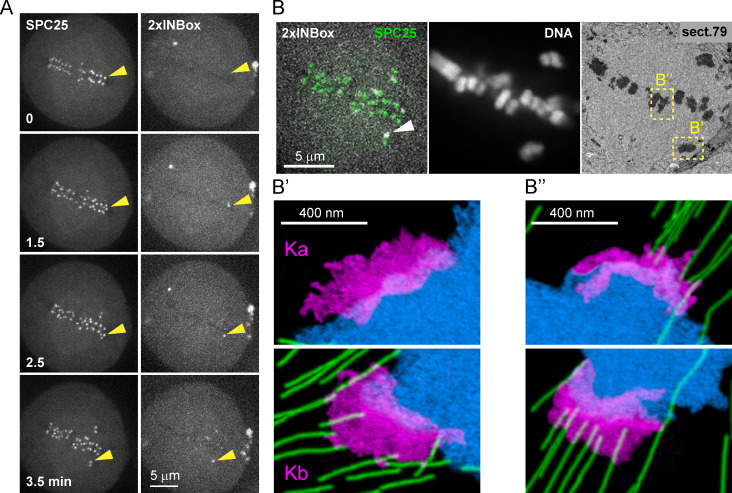
Related to Figs. 2 and 3. (A and B) Representative images (A) and displacements (B) for Void recruitment to a single kinetochore (yellow triangles) of a bi-oriented pair, as a negative control. Displacements, plotted as in Fig. 2, D and E, were used to define the range of naturally occurring chromosome oscillations (dashed lines cover 45 of 47 traces, 96%; Video 5). (C) Representative images showing kinetochore movement indicating microtubule depolymerization after recruiting INBox to a single kinetochore (white triangles; Video 7). (A and C) Inset: The targeted kinetochore pair at higher magnification. Scale bars in A and C: 5 µm or 1 µm in insets. (D) The fraction of release vs. depolymerization events from Fig. 2, D and E (2×INBox: 87 ± 5% mean ± SEM, n = 47; INBox: 67 ± 9% mean ± SEM, n = 30; *, P < 0.05, one tailed). (E and F) Steady-state velocities (E) and interkinetochore distances (F) for released kinetochores. Distances are plotted before activation and at steady-state after 2×INBox recruitment. Each data point represents a single kinetochore or pair of sisters. Black lines: Mean ± SEM (n = 41 for 2×INBox, n = 20 for INBox; *, P < 0.05). (G) INBox recruitment on monopolar and bipolar spindles, measured as mCherry intensity (mean ± SEM). (H) Higher magnification of the image shown in Fig. 3 B. Full series of sections through the centromeres of chromosomes within yellow boxes are shown in H′ and H′′. (H') The activated kinetochore Ka and and its non-activated sister kinetochore Kb. Microtubules have been released from Ka but are present at Kb. Arrowheads denote kinetochore plates. (H′′) Non-irradiated sister kinetochores of a properly aligned chromosome within the metaphase plate. Similar number of microtubules are attached to both kinetochores.
Video 5.
Related to Fig. S2 A. Void-recruitment defines naturally occurring chromosome oscillations on a bipolar spindle. Left: 3×Halo-GFP-SPC25. Right: mCherry-eDHFR.
Video 6.
Related to Fig. 2 B. INBox recruitment triggers microtubule release on a bipolar spindle. Left: 3×Halo-GFP-SPC25. Right: mCherry-INBox-eDHFR.
Video 7.
Related to Fig. S2 C. Example of INBox recruitment triggering microtubule depolymerization on a bipolar spindle. Left: 3×Halo-GFP-SPC25. Right: mCherry-INBox-eDHFR.
Video 8.
Related to Fig. 2 C. 2×INBox recruitment triggers microtubule release on a bipolar spindle. Left: 3×Halo-GFP-SPC25. Right: INBox-mCherry-INBox-eDHFR.
To directly visualize microtubules after single kinetochore activation, we performed correlative serial-section EM. Cells were fixed after the activated kinetochore moved out of the metaphase plate, indicating release (Fig. 3 A). We defined the activated kinetochore as lagging and the sister kinetochore as leading because it moves toward its attached pole (Figs. 3 B and S2 H). We found microtubules bound to the leading kinetochore (Kb), but not to the activated lagging kinetochore (Ka; Figs. 3 B′ and S2 H′). In contrast, kinetochore pairs that were not activated aligned properly on the metaphase plate, with K-fibers attached on both sides (Figs. 3 B′′ and S2, H and H′′). Thus, the activated kinetochore releases its attached microtubules, losing the tug of war to the sister.
Figure 3.
Activated kinetochores lack end-on microtubule attachments. (A) Live imaging showing 2×INBox recruitment and chromosome movement after activation of a single kinetochore (yellow triangles) at t = 0, as in Fig. 2 C. (B) The same cell was fixed at 3.5 min and examined by fluorescence and serial-section EM. Top: A single focal plane shows 2×INBox overlaying SPC25 at the activated kinetochore (white arrow) and chromosomes by fluorescence, and the corresponding section from the 3D EM dataset. Kinetochores of the activated chromosome pair (B’) and a nonactivated aligned chromosome (B′′) are shown at higher magnification below (blue: DNA; green: microtubules; magenta: kinetochores). Microtubules are bound to the leading kinetochore (Kb), but not the activated lagging kinetochore (Ka). Full EM series through these two kinetochores are shown in Fig. S2 H.
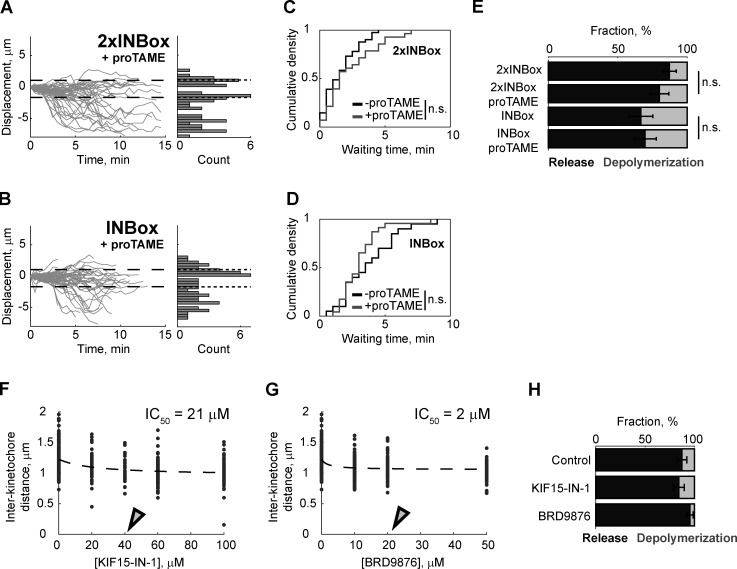
The coexistence of release and depolymerization events after Aurora B kinase activation (Fig. 2, B–E) suggests an underlying kinetic race, with the release rate dominant on bipolar spindles. In addition, we found a shorter waiting time during the lag phase after recruitment of 2×INBox (median, 1.5 min) vs. INBox (median, 3.5 min), indicating that higher Aurora B activity further increases the microtubule release rate and the fraction of release events (Figs. 2 G and S2 D). To explain the differences between monopolar and bipolar spindles, one possibility is that time in mitosis affects the outcome, as Eg5-inhibited cells are arrested in mitosis for up to 2–6 h in our experiments. To test this possibility, we activated individual kinetochores in the presence of the anaphase-promoting complex/cyclosome (APC/C) inhibitor proTAME to delay mitotic exit in cells with bipolar spindles (Fig. S3, A and B). The fraction of release vs. depolymerization events and the release kinetics remained unchanged under these conditions (Fig. S3, C–E), indicating that time in mitosis is not a key variable.
Figure S3.
Related to Fig. 4. (A and B) Displacement of the activated kinetochore, plotted as in Fig. 2, D and E, with 20 µM proTAME to delay mitotic exit (2×INBox: n = 54; INBox: n = 56). (C and D) Waiting time of the released kinetochores after recruiting 2×INBox (control: median = 1.5 min, n = 41; proTAME: median = 1.5 min, n = 28) or INBox (control: median = 3.5 min, n = 20; proTAME: median = 3.0 min, n = 23). (E) The fraction of release vs. depolymerization events after recruiting 2×INBox or INBox. Differences in the presence or absence of proTAME (C–E) are not statistically significant (P > 0.05; mean ± SEM). (F and G) Interkinetochore distances at varying concentrations of rigor inhibitors for KIF15 (KIF15-IN-1: IC50 = 21 µM) or Eg5 (BRD9876: IC50 = 2 µM). Open triangles represent the concentrations used for 2×INBox recruitment assays in Fig. 4, B and C (KIF15-IN-1: 40 µM; KIF15-IN-1: 20 µM). (H) The fraction of release vs. depolymerization events from Fig. 4, B and C (control: 87 ± 5% mean ± SEM, n = 47; KIF15-IN-1: 84 ± 6% mean ± SEM, n = 44; BRD9876: 95 ± 3% mean ± SEM, n = 43).
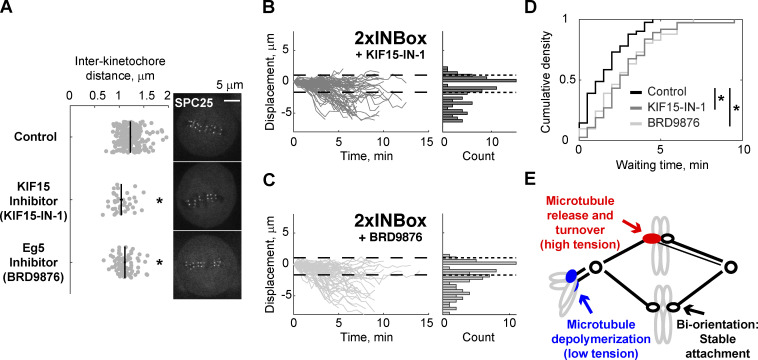
We next tested whether differences in tension, which is high on bipolar spindles but low on monopolar spindles, might explain the different outcomes in these two contexts. If tension promotes microtubule release upon Aurora B kinase activation, then we predict that experimentally reducing tension on bipolar spindles would inhibit microtubule release. Reducing microtubule cross-linkers leads to an ∼10% decrease in interkinetochore distance (Elting et al., 2017; Polak et al., 2017), suggesting that interpolar microtubule arrays are mechanically coupled to K-fibers. We therefore tested whether the microtubule–cross-linking motors Eg5 or KIF15 (kinesin-12) mediate interkinetochore tension. Eg5 motors cross-link and slide microtubules apart to maintain proper pole-to-pole distance (Kapitein et al., 2005; Shimamoto et al., 2015; Uteng et al., 2008), while KIF15 motors additionally cross-link K-fibers to control chromosome movement (Sturgill and Ohi, 2013; Drechsler et al., 2014). Blocking nucleotide binding of these motors entraps them in a rigor state, which maintains cross-linking activity but diminishes the powerstroke (Elting et al., 2017; Needleman et al., 2010; Dumas et al., 2019). We found that both the Eg5 rigor inhibitor, BRD9876, and the KIF15 rigor inhibitor, KIF15-IN-1, reduce interkinetochore distances by ∼10%, indicating reduced tension (Figs. 4 A and S3, F and G). These inhibitors can stabilize microtubules against depolymerization (Chen et al., 2017); therefore, we did not use the fraction of release vs. depolymerization events after single kinetochore activation as our readout (Fig. S3 H). Instead, we measured the waiting time from activation to release. Addition of either inhibitor increased the median waiting time from 1.5 to 2.5 min (Fig. 4, B–D). These findings indicate that reduced tension slows the Aurora B–induced microtubule release rate, although microtubule stabilization by the inhibitors could also contribute.
Figure 4.
Reducing interkinetochore tension slows release of kinetochore microtubules. (A) Representative images of cells treated with a KIF15 inhibitor (40 µM KIF15-IN-1), Eg5 inhibitor (20 µM BRD9876), or control (no inhibitor). In the plot, each data point represents a single kinetochore pair. Black lines: Mean ± SEM (n = 220 kinetochores for control; n = 40 for KIF15-IN-1; n = 81 for BRD9876; ∼8 pairs measured per cell). Entire titration curves are plotted in Fig. S3, F and G. (B and C) Displacement of the activated kinetochore after 2×INBox recruitment with KIF15 (B; n = 93) or Eg5 (C; n = 74) inhibition, plotted as in Fig. 2, D and E. (D) Waiting time distribution of the released kinetochores (control: median = 1.5 min, n = 41; KIF15-IN-1: median = 2.5 min, n = 37; BRD9876: median = 2.5 min, n = 41). (E) Model for distinct error-correction pathways at merotelic (red) and syntelic (blue) attachments in response to Aurora B activation. *, P < 0.05.
Overall, our results demonstrate contrasting responses to Aurora B activity: depolymerization of kinetochore microtubules under low tension and release under high tension. This increase of the release rate with tension is opposite to the catch-bond behavior of unphosphorylated kinetochores, in which force stabilizes attachments (Akiyoshi et al., 2010). These seemingly contradictory effects of tension can be explained by a two-state model, in which the detachment rate is lower for polymerizing compared with depolymerizing microtubule plus-ends, as shown in vitro (Sarangapani and Asbury, 2014; Akiyoshi et al., 2010). At unphosphorylated kinetochores, the catch-bond is due to tension promoting the polymerizing state, which is more strongly attached (Akiyoshi et al., 2010). Kinetochore substrate phosphorylation promotes microtubule depolymerization (Umbreit et al., 2012), preventing the transition to the more strongly attached state, which converts the catch-bond into a more conventional slip-bond that breaks under increasing force. Thus, phosphorylated kinetochores maintain attachment while depolymerizing under low tension, but release under higher tension (Huis in’t Veld et al., 2019). Phosphorylation also increases the detachment rate for polymerizing microtubules (Sarangapani et al., 2013), and there may be additional complexity in the cell if phosphorylation of distinct Aurora B kinase substrates (e.g., the Ndc80 or Ska complexes or MCAK) depends on tension, promoting either microtubule depolymerization or release (Carmena et al., 2012; Krenn and Musacchio, 2015). For example, partial release may be an intermediate state leading to depolymerization while maintaining attachment of the remaining kinetochore microtubules under low tension. Furthermore, centromeric microtubule depolymerases might confer distinct functions under the control of Aurora B (Cimini et al., 2006; Bakhoum et al., 2009)—driving poleward chromosome movement or generating tension at syntelic or bi-oriented attachments, respectively.
By integrating spindle mechanics and kinetochore biochemistry, we propose that the tension-dependent response facilitates correction of distinct attachment errors (Fig. 4 E). Lower tension is associated with increased Aurora B activity at syntelic attachments (Welburn et al., 2010; Liu et al., 2009), which typically scatter around spindle poles. Microtubule release would leave chromosomes to be pushed away from the spindle by polar ejection forces, whereas microtubule depolymerization pulls chromosomes toward the spindle, where they subsequently detach near the pole and congress by gliding along K-fibers of other chromosomes (Ye et al., 2015; Chmátal et al., 2015; Kapoor et al., 2006). Aurora B kinase is also activated at merotelic attachments, in which one kinetochore attaches to both spindle poles, either by recruitment or by interactions with microtubules (Knowlton et al., 2006; Salimian et al., 2011; Trivedi et al., 2019). Because tension is higher with microtubules pulling in opposite directions, phosphorylation would promote microtubule release and turnover to achieve bi-orientation (Zaytsev and Grishchuk, 2015; Cimini et al., 2006; Bakhoum et al., 2009). This mechanism can explain the failure to correct merotelic errors associated with defective spindle mechanics, such as disruptions of poleward microtubule flux, tissue architecture, or tubulin homeostasis (Ganem and Compton, 2006; Knouse et al., 2018; Lin et al., 2020).
Materials and methods
Dimerizers
Synthesis and characterization of the photo-caged (CTH) and uncaged (TNH) dimerizers followed published protocols (Zhang et al., 2017, 2018). Both stocks were dissolved in DMSO at 20 mM and distributed in amber-colored 1.5-ml microcentrifuge tubes at −80°C for long-term storage. For CTH, a stock aliquot was diluted in growth medium to a 10-µM working concentration with 1 ml vol and then kept at −80°C until use. For TNH, a stock aliquot was diluted to 10 µM in the L-15 medium without phenol red (Invitrogen) containing 10% FBS and 1% penicillin/streptomycin.
Plasmids
All constructs for anchors and effectors were integrated into the pEM705 backbone containing a CAG promoter for constitutive expression (Khandelia et al., 2011; Zhang et al., 2017). The kinetochore protein, SPC25, was chosen as an anchor because of its outer kinetochore localization and slow cytosolic exchange. The N terminus of SPC25 was fused to three tandem copies of HaloTag and to GFP to make 3×Halo-GFP-SPC25. The full-length inner centromere protein, CENP-B, was fused to GFP and a HaloTag at its C terminus to make CENP-B-GFP-Halo. To recruit and activate Aurora B kinase, the INBox domain (human INCENP819-918) was fused to mCherry at its N terminus and to eDHFR at its C terminus to make mCherry-INBox-eDHFR. To prevent Aurora B activation upon recruitment, the TSS motif of INBox—corresponding to residues 892–894 of INCENP—was replaced with AAA to make mCherry-INBoxAAA-eDHFR. To maximize Aurora B kinase activity, a second INBox domain was fused to the N terminus of mCherry-INBox-eDHFR to make INBox-mCherry-INBox-eDHFR (2×INBox). As a negative control, mCherry was fused to the N terminus of eDHFR to make mCherry-eDHFR (Void). As a positive control, human MCAK179-583 (Maney et al., 1998; Talapatra et al., 2015) was fused to the N terminus of mCherry-eDHFR to make MCAK-mCherry-eDHFR. To mark the spindle poles, the PACT (Pericentrin and AKAP Centrosome Targeting) domain was fused to GFP (PACT-GFP) in the pcDNA3.1 backbone containing cytomegalovirus and T7 promoters (Gillingham and Munro, 2000).
Cell cultures and transfection
For kinetochore recruitment, assays were performed with HeLa RMCE (recombination-mediated cassette exchange) acceptor cells (with LoxP and Lox2272 recombination sites; obtained from E.V. Makayev, Nanyang Technological University, Singapore) stably expressing 3×Halo-GFP-SPC25 (Khandelia et al., 2011; Ballister et al., 2014; Zhang et al., 2017). In brief, acceptor cells were cotransfected with a plasmid expressing Cre recombinase (10 ng) and another donor plasmid containing 3×Halo-GFP-SPC25 and puromycin selection marker inserted in a transgenic cassette flanked by LoxP and Lox2272 sites (1 µg). Cells were cultured at 37°C in growth medium containing DMEM plus 10% FBS (Clontech) and 1% penicillin/streptomycin (Life Technologies), with 5% CO2 and a humidified atmosphere. For selection purposes, 1 µg/ml puromycin was further added into this growth medium. For bipolar spindle assays, cells were grown on 22 × 22-mm coverslips (Thermo Fisher Scientific) coated by poly-D-lysine (Sigma-Aldrich) for at least 16 h and then transfected with Lipofectamine 2000 (Invitrogen) plus 1–2 µg eDHFR-tagged constructs—Void, INBox, or 2×INBox—for another 24 h. For monopolar spindle assays, 250 ng of PACT-GFP and/or CENP-B-GFP-Halo plasmid was cotransfected with 1–2 µg of the eDHFR-tagged constructs—Void, INBox, 2×INBox, MCAK, or INBoxAAA—for 24–36 h.
Small-molecule inhibitors
Stocks were prepared in DMSO and stored at −20°C. Stock concentrations were 10 mM STLC (Eg5 inhibitor; Sigma-Aldrich), 50 mM BRD9876 (Eg5 rigor inhibitor; Tocris Bioscience), 20 mM proTAME (APC/C inhibitor; Boston Biochem), and 20 mM KIF15-IN-1 (KIF15 rigor inhibitor; APExBIO). Fresh aliquots were used for all experiments.
Immunostaining
Cells stably expressing 3×Halo-GFP-SPC25 with transiently expressed INBox-mCherry-INBox-eDHFR were incubated with 20 µM STLC plus 500 nM TNH for 2 h, followed by 1 h TNH washout to remove unbound dimerizers. After quickly rinsing with PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 4 mM Mg2SO4 [pH 6.9]), cells were permeabilized for 1 min in freshly prepared lysis buffer (PHEM + 0.5% Triton X-100) containing 100 nM microcystin (Sigma-Aldrich), fixed by 4% formaldehyde (Thermo Fisher Scientific) in PHEM, washed 5 × 5 min in PHEM-T (0.1% Triton X-100 in PHEM), and then blocked in 20% boiled donkey serum (BDS; Sigma-Aldrich) with 2% BSA (Fisher BioReagent) in PHEM for 1 h. The primary rabbit antibody against phosphorylated Hec1-Ser44 (DeLuca et al., 2011) was incubated for 1 h after 2,000× dilution with 10% BDS plus 1% BSA in PHEM. Next, cells were washed out with 5 × 5 min in PHEM-T and then incubated with the secondary donkey anti-rabbit antibody conjugated to Alexa Fluor 647 (1:1,000 dilution in 10% BDS + 1% BSA; A-31573; Life Technologies) for another hour. Before infiltrating the coverslips with DAPI-included mounting media (Vectashield; Vector Labs), cells were washed 5 × 5 min in PHEM-T and three times in PHEM to reduce nonspecific binding. All processes before the end of fixation were performed in the dark with care at 37°C and at RT for the rest of the procedure.
Photoactivation and dimerization
For photoactivation experiments, cells were incubated with CTH for 1 h, followed by a 30-min washout with growth medium to remove unbound molecules, as previously described (Zhang et al., 2017). For dimerization experiments without photoactivation (Fig. S1, D–J), 500 nM TNH was added after t = 0. For imaging, coverslips were mounted in a magnetic chamber (Chamlide CM-S22-1; LCI) with L-15 medium without phenol red (Invitrogen) containing 10% FBS and 1% penicillin/streptomycin, and then placed on a heated stage in a 37°C environmental chamber (Incubator BL; PeCon). Targeted uncaging was performed using a 405-nm laser (model #DL405-050-O, 27-mW output after fiber coupling; CrystaLaser LC) under the control of an iLas2 software module (Roper Scientific) within MetaMorph (Molecular Devices). To uncage CTH globally over the entire cell, 8% laser power and 20 repetitions were used for monopolar spindle assays. For single kinetochore targeting, 7% laser power with 50 repetitions was used in a 590-nm diameter region. The region size was chosen to minimize off-target activation based on the diffraction limit, the motion of kinetochores, and the time lag between imaging and manual activation.
For monopolar spindle assays or metaphase-arrested bipolar spindle assays, 20 µM STLC (Skoufias et al., 2006) or 20 µM proTAME (Zeng et al., 2010) was added during CTH incubation and imaging. Kinesin rigor inhibitors were added by media exchange directly on the stage, followed by a 30-min incubation to equilibrate the system. To avoid unwanted CTH uncaging during preparation stage, care was taken to minimize light and heat exposure, using low-luminescence or red light in the room and a long-pass filter to find cells under differential interference contrast microscopy.
Image acquisition
Live imaging at 37°C was performed with a confocal microscope (DM4000; Leica), equipped with a 100× 1.4 NA oil immersion objective (Leica), an XY Piezo-Z stage (Applied Scientific Instrumentation), a CSU10 spinning disk (Yokogawa), an electron multiplier charge-coupled device camera (ImageEM; Hamamatsu Photonics), and a laser merge module (LMM5; Spectral Applied Research) equipped with 488- and 593-nm lasers, as previously described (Zhang et al., 2017). To minimize photobleaching in monopolar spindle assays, images were acquired with a 1-min time interval, with 1-µm spacing for GFP and mCherry z-stacks covering 15 µm total. To improve precision for the bipolar spindle assays, the time interval was 30 s with 0.5-µm z-spacing covering 3 µm total, so not all kinetochore pairs are visualized. For fixed cell imaging, the same microscope was used with a four-line laser module (405 nm, 488 nm, 561 nm, and 639 nm; Vortran Stradus VersaLase 4).
Image processing and data analyses
Images are shown as maximum-intensity z-projections. The Fiji plug-in, TrackMate, was used to define kinetochore coordinates globally. For monopolar spindle assays, the identified objects in the targeted cell were separated as kinetochores and poles based on their intensity and quality. Kinetochore and pole coordinates were imported into MATLAB (MathWorks). The distance from each kinetochore to the center of the monopolar spindle, defined by averaging the pole coordinates, was calculated to determine the average distance at each time point. The reported displacements are average distances at each time point relative to the average distance at t = 0, with poleward movement defined as the positive direction.
For bipolar spindle assays, the position of the metaphase plate was determined by fitting the interior kinetochore ensemble using linear regression and minimizing the mean squares of x- and y-deviation, and the kinetochores of unaligned chromosomes were omitted. The Fiji plug-in, MtrackJ, was used to manually track the activated single kinetochores and identify the sister kinetochores. The relative distances between the activated kinetochore and the fitted metaphase plate were calculated in MATLAB. Movement toward the pole attached to the sister kinetochore was defined as the negative direction. Hence, the depolymerization or release models predict positive or negative directional movements, respectively, for both monopolar and bipolar spindle assays. Classification of each event in the bipolar spindle assay is based on the earliest time point distinguishable from the negative control so that the two models are mutually exclusive and in a kinetic race. For example, if an event shows depolymerization followed by release, then it is classified as a depolymerization event. The steady-state period of a released kinetochore was manually defined by the best linear fit of each trajectory moving beyond the negative threshold (see example in Fig. 2 F). Experiments were repeated at least three times on different days, and data points across repeats were pooled for statistical analysis. To compare INBox recruitment in the monopolar and bipolar spindle assays (Fig. S2 G), mCherry intensity at kinetochores at 2 min after activation was quantified after background subtraction.
Correlative EM
Cells were fixed for 30 min in PBS containing 2.5% glutaraldehyde (Sigma-Aldrich) immediately after the last frame of live imaging. Complete Z-series at 0.2-µm steps were then recorded to map positions of chromosomes in differential interference contrast and Hoechst 33342 fluorescence (0.1 µg/ml; Life Technologies). The images were acquired on a Nikon eclipse Ti2E microscope with a Plan Apochromat 100× 1.45 NA objective lens and Photometrics 95B Prime camera at 43-nm XY pixels. EM embedding, relocation of cells, and serial sectioning were done as previously described (Rieder and Cassels, 1999). 80-nm sections were imaged on a JEM 1400 microscope (JEOL) operated at 80 kV using a side-mounted 4.0 megapixel XR401 sCMOS AMT camera (AMT). Complete image series recorded at 10K magnification were used to reconstruct partial volumes containing activated chromosomes. These volumes were aligned with the light microscopy images by matching positions of prominent landmarks, such as chromosome arms. Serial higher-magnification images (40K) were then collected to detail the distribution of microtubules in the vicinity of activated kinetochores. In two cells, we successfully activated a single kinetochore, identified the activated kinetochore based on 2×INBox fluorescence signal in fixed cells, and analyzed it by correlative EM. In both cases, the activated kinetochore lacked end-on microtubule attachments, as shown in the representative sample in Fig. 3.
Statistical tests
A nonparametric Kolmogorov-Smirnov test was used to compare the cumulative waiting time distributions, and a two-sample Z-test was used to compare the fractions of release vs. depolymerization events. Titration curves were fit by a rectangular hyperbola to measure the IC50 of interkinetochore distance for KIF15-IN-1 and BRD9876. The two-sample comparisons of velocity, interkinetochore distance, kinetochore displacement, kinetochore phosphorylation level, or INBox recruitment level were performed using Student’s t test. Unless otherwise specified in figure legends, two-tailed tests were used to compare samples.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
All MATLAB codes for plotting and data analyses are available as Data S1 and Data S2.
Online supplemental material
Fig. S1 relates to Fig. 1 and shows detailed properties of kinetochores in monopolar spindle assays. Fig. S2 relates to Figs. 2 and 3 and shows biophysical properties and EM images of activated kinetochores in bipolar spindle assays. Fig. S3 relates to Fig. 4 and shows bipolar spindle assays under inhibitor treatments. Video 1 relates to Fig. 1 D and shows that MCAK recruitment triggers poleward chromosome movement on a monopolar spindle. Video 2 relates to Fig. S1 C and shows Void recruitment as a negative control for a monopolar spindle. Video 3 relates to Fig. 1 E and shows that INBox recruitment triggers poleward chromosome movement on a monopolar spindle. Video 4 relates to Fig. 1 F and shows that 2×INBox recruitment triggers poleward chromosome movement on a monopolar spindle. Video 5 relates to Fig. S2 A and shows Void recruitment as a negative control on a bipolar spindle. Video 6 relates to Fig. 2 B and shows that INBox recruitment triggers microtubule release on a bipolar spindle. Video 7 relates to Fig. S2 C and shows an example of INBox recruitment triggering microtubule depolymerization on a bipolar spindle. Video 8 relates to Fig. 2 C and shows that 2×INBox recruitment triggers microtubule release on a bipolar spindle. Data S1 shows the MATLAB code to calculate the average distance of all kinetochores to monopolar spindle poles in a single cell. Data S2 shows the MATLAB code to calculate the kinetochore displacement relative to the metaphase plate of a bipolar spindle.
Supplementary Material
shows the MATLAB code to calculate the average distance of all kinetochores to monopolar spindle poles in a single cell.
shows the MATLAB code to calculate the kinetochore displacement relative to the metaphase plate of a bipolar spindle.
Acknowledgments
We thank Dr. Jennifer DeLuca (Colorado State University, Fort Collins, CO) for the Hec1 S44p antibody, Dr. Ekaterina Grishchuk for commenting on the manuscript, and the Philly Chromo Club, Physical Sciences Oncology Center, and Penn Center for Genome Integrity for discussions.
This work was supported by National Institutes of Health grants GM130298 (A. Khodjakov), GM118510 (D.M. Chenoweth), and GM122475 (M.A. Lampson); National Cancer Institute grant U54-CA193417; and the Penn Center for Genome Integrity.
The authors declare no competing financial interests.
Author contributions: G.-Y. Chen designed and conducted cell biology experiments and wrote the manuscript. H. Zhang and A. Gokden tested the feasibility of this project in the initial stage. F. Renda and A. Khodjakov acquired and analyzed EM images. D.Z. Wu and D.M. Chenoweth synthesized and characterized the CTH dimerizer. M.A. Lampson designed experiments and edited the manuscript.
Footnotes
A preprint of this paper was posted in bioRxiv on June 2, 2020.
References
- Akiyoshi, B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., and Biggins S.. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579. 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, Y., Fukuchi K., Tanno Y., Koitabashi-Kiyozuka S., Kiyozuka T., Noda Y., Matsumura R., Koizumi T., Watanabe A., Nagata K., et al. 2019. Aurora B kinase activity is regulated by SET/TAF1 on Sgo2 at the inner centromere. J. Cell Biol. 218:3223–3236. 10.1083/jcb.201811060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum, S.F., Thompson S.L., Manning A.L., and Compton D.A.. 2009. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 11:27–35. 10.1038/ncb1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister, E.R., Aonbangkhen C., Mayo A.M., Lampson M.A., and Chenoweth D.M.. 2014. Localized light-induced protein dimerization in living cells using a photocaged dimerizer. Nat. Commun. 5:5475. 10.1038/ncomms6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan, E.J., Chiou K.K., Ballister E.R., Mayo A.M., Lampson M.A., and Liu A.J.. 2015. Minimal model for collective kinetochore-microtubule dynamics. Proc. Natl. Acad. Sci. USA. 112:12699–12704. 10.1073/pnas.1513512112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, J.D., and Schumacher J.M.. 2002. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277:27577–27580. 10.1074/jbc.C200307200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad, A.J., and DeLuca J.G.. 2020. The right place at the right time: Aurora B kinase localization to centromeres and kinetochores. Essays Biochem. 64:299–311. 10.1042/EBC20190081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad, A.J., DeLuca K.F., and DeLuca J.G.. 2020. Aurora B kinase is recruited to multiple discrete kinetochore and centromere regions in human cells. J. Cell Biol. 219:e201905144. 10.1083/jcb.201905144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, C.S., and Desai A.. 2013. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 497:118–121. 10.1038/nature12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena, M., Wheelock M., Funabiki H., and Earnshaw W.C.. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13:789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.-Y., Kang Y.J., Gayek A.S., Youyen W., Tüzel E., Ohi R., and Hancock W.O.. 2017. Eg5 Inhibitors Have Contrasting Effects on Microtubule Stability and Metaphase Spindle Integrity. ACS Chem. Biol. 12:1038–1046. 10.1021/acschembio.6b01040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal, L., Yang K., Schultz R.M., and Lampson M.A.. 2015. Spatial Regulation of Kinetochore Microtubule Attachments by Destabilization at Spindle Poles in Meiosis I. Curr. Biol. 25:1835–1841. 10.1016/j.cub.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini, D., Wan X., Hirel C.B., and Salmon E.D.. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16:1711–1718. 10.1016/j.cub.2006.07.022 [DOI] [PubMed] [Google Scholar]
- DeLuca, K.F., Lens S.M.A., and DeLuca J.G.. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634. 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler, H., McHugh T., Singleton M.R., Carter N.J., and McAinsh A.D.. 2014. The Kinesin-12 Kif15 is a processive track-switching tetramer. eLife. 3:e01724. 10.7554/eLife.01724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, M.E., Chen G.Y., Kendrick N.D., Xu G., Larsen S.D., Jana S., Waterson A.G., Bauer J.A., Hancock W., Sulikowski G.A., and Ohi R.. 2019. Dual inhibition of Kif15 by oxindole and quinazolinedione chemical probes. Bioorg. Med. Chem. Lett. 29:148–154. 10.1016/j.bmcl.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting, M.W., Prakash M., Udy D.B., and Dumont S.. 2017. Mapping Load-Bearing in the Mammalian Spindle Reveals Local Kinetochore Fiber Anchorage that Provides Mechanical Isolation and Redundancy. Curr. Biol. 27:2112–2122.e5. 10.1016/j.cub.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., and Compton D.A.. 2006. Functional roles of poleward microtubule flux during mitosis. Cell Cycle. 5:481–485. 10.4161/cc.5.5.2519 [DOI] [PubMed] [Google Scholar]
- García-Rodríguez, L.J., Kasciukovic T., Denninger V., and Tanaka T.U.. 2019. Aurora B-INCENP Localization at Centromeres/Inner Kinetochores Is Required for Chromosome Bi-orientation in Budding Yeast. Curr. Biol. 29:1536–1544.e4. 10.1016/j.cub.2019.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and Munro S.. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529. 10.1093/embo-reports/kvd105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, R., Körner R., and Nigg E.A.. 2003. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 14:3325–3341. 10.1091/mbc.e02-11-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in’t Veld, P.J., Volkov V.A., Stender I.D., Musacchio A., and Dogterom M.. 2019. Molecular determinants of the Ska-Ndc80 interaction and their influence on microtubule tracking and force-coupling. eLife. 8:e49539. 10.7554/eLife.49539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein, L.C., Peterman E.J.G., Kwok B.H., Kim J.H., Kapoor T.M., and Schmidt C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kapoor, T.M., Mayer T.U., Coughlin M.L., and Mitchison T.J.. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988. 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelia, P., Yap K., and Makeyev E.V.. 2011. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl. Acad. Sci. USA. 108:12799–12804. 10.1073/pnas.1103532108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse, K.A., Lopez K.E., Bachofner M., and Amon A.. 2018. Chromosome Segregation Fidelity in Epithelia Requires Tissue Architecture. Cell. 175:200–211.e13. 10.1016/j.cell.2018.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton, A.L., Lan W., and Stukenberg P.T.. 2006. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 16:1705–1710. 10.1016/j.cub.2006.07.057 [DOI] [PubMed] [Google Scholar]
- Krenn, V., and Musacchio A.. 2015. The Aurora B kinase in chromosome bi-orientation and spindle checkpoint signaling. Front. Oncol. 5:225. 10.3389/fonc.2015.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson, M.A., and Cheeseman I.M.. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140. 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson, M.A., and Grishchuk E.L.. 2017. Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology (Basel). 6:1. 10.3390/biology6010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson, M.A., Renduchitala K., Khodjakov A., and Kapoor T.M.. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237. 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lin, Z., Gasic I., Chandrasekaran V., Peters N., Shao S., Mitchison T.J., and Hegde R.S.. 2020. TTC5 mediates autoregulation of tubulin via mRNA degradation. Science. 367:100–104. 10.1126/science.aaz4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Vader G., Vromans M.J., Lampson M.A., and Lens S.M.. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353. 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A.F., Udy D.B., and Dumont S.. 2017. Hec1 Tail Phosphorylation Differentially Regulates Mammalian Kinetochore Coupling to Polymerizing and Depolymerizing Microtubules. Curr. Biol. 27:1692–1699.e3. 10.1016/j.cub.2017.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney, T., Hunter A.W., Wagenbach M., and Wordeman L.. 1998. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142:787–801. 10.1083/jcb.142.3.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., Sandri B.J., Tank D., McClellan M., Harasymiw L.A., Yang Q., Parker L.L., and Gardner M.K.. 2019. A Gradient in Metaphase Tension Leads to a Scaled Cellular Response in Mitosis. Dev. Cell. 49:63–76.e10. 10.1016/j.devcel.2019.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman, D.J., Groen A., Ohi R., Maresca T., Mirny L., and Mitchison T.. 2010. Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol. Biol. Cell. 21:323–333. 10.1091/mbc.e09-09-0816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas, R.B. 1997. How cells get the right chromosomes. Science. 275:632–637. 10.1126/science.275.5300.632 [DOI] [PubMed] [Google Scholar]
- Polak, B., Risteski P., Lesjak S., and Tolić I.M.. 2017. PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep. 18:217–230. 10.15252/embr.201642650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., and Cassels G.. 1999. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods Cell Biol. 61:297–315. 10.1016/S0091-679X(08)61987-1 [DOI] [PubMed] [Google Scholar]
- Salimian, K.J., Ballister E.R., Smoak E.M., Wood S., Panchenko T., Lampson M.A., and Black B.E.. 2011. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr. Biol. 21:1158–1165. 10.1016/j.cub.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani, K.K., and Asbury C.L.. 2014. Catch and release: how do kinetochores hook the right microtubules during mitosis? Trends Genet. 30:150–159. 10.1016/j.tig.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani, K.K., Akiyoshi B., Duggan N.M., Biggins S., and Asbury C.L.. 2013. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc. Natl. Acad. Sci. USA. 110:7282–7287. 10.1073/pnas.1220700110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa, F., Mapelli M., Ciferri C., Tarricone C., Areces L.B., Schneider T.R., Stukenberg P.T., and Musacchio A.. 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 18:379–391. 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Shimamoto, Y., Forth S., and Kapoor T.M.. 2015. Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Microtubules. Dev. Cell. 34:669–681. 10.1016/j.devcel.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikirzhytski, V., Magidson V., Steinman J.B., He J., Le Berre M., Tikhonenko I., Ault J.G., McEwen B.F., Chen J.K., Sui H., et al. 2014. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J. Cell Biol. 206:231–243. 10.1083/jcb.201401090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias, D.A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R.H., Hackney D., and Kozielski F.. 2006. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 281:17559–17569. 10.1074/jbc.M511735200 [DOI] [PubMed] [Google Scholar]
- Sturgill, E.G., and Ohi R.. 2013. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr. Biol. 23:1280–1290. 10.1016/j.cub.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talapatra, S.K., Harker B., and Welburn J.P.I.. 2015. The C-terminal region of the motor protein MCAK controls its structure and activity through a conformational switch. eLife. 4:e06421. 10.7554/eLife.06421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, P., Zaytsev A.V., Godzi M., Ataullakhanov F.I., Grishchuk E.L., and Stukenberg P.T.. 2019. The binding of Borealin to microtubules underlies a tension independent kinetochore-microtubule error correction pathway. Nat. Commun. 10:682. 10.1038/s41467-019-08418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit, N.T., Gestaut D.R., Tien J.F., Vollmar B.S., Gonen T., Asbury C.L., and Davis T.N.. 2012. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. USA. 109:16113–16118. 10.1073/pnas.1209615109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uteng, M., Hentrich C., Miura K., Bieling P., and Surrey T.. 2008. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J. Cell Biol. 182:715–726. 10.1083/jcb.200801125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., Gayek S., and Ohi R.. 2013. Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 29:417–441. 10.1146/annurev-cellbio-101512-122345 [DOI] [PubMed] [Google Scholar]
- Welburn, J.P.I., Vleugel M., Liu D., Yates J.R. III, Lampson M.A., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, A.A., Deretic J., Hoel C.M., Hinman A.W., Cimini D., Welburn J.P., and Maresca T.J.. 2015. Aurora A Kinase Contributes to a Pole-Based Error Correction Pathway. Curr. Biol. 25:1842–1851. 10.1016/j.cub.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, T.Y., Choi J.M., Conway W., Yu C.H., Pappu R.V., and Needleman D.J.. 2018. Measuring NDC80 binding reveals the molecular basis of tension-dependent kinetochore-microtubule attachments. eLife. 7:e36392. 10.7554/eLife.36392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev, A.V., and Grishchuk E.L.. 2015. Basic mechanism for biorientation of mitotic chromosomes is provided by the kinetochore geometry and indiscriminate turnover of kinetochore microtubules. Mol. Biol. Cell. 26:3985–3998. 10.1091/mbc.E15-06-0384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev, A.V., Segura-Peña D., Godzi M., Calderon A., Ballister E.R., Stamatov R., Mayo A.M., Peterson L., Black B.E., Ataullakhanov F.I., et al. 2016. Bistability of a coupled Aurora B kinase-phosphatase system in cell division. eLife. 5:e10644. 10.7554/eLife.10644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., Sigoillot F., Gaur S., Choi S., Pfaff K.L., Oh D.C., Hathaway N., Dimova N., Cuny G.D., and King R.W.. 2010. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 18:382–395. 10.1016/j.ccr.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Aonbangkhen C., Tarasovetc E.V., Ballister E.R., Chenoweth D.M., and Lampson M.A.. 2017. Optogenetic control of kinetochore function. Nat. Chem. Biol. 13:1096–1101. 10.1038/nchembio.2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Chenoweth D.M., and Lampson M.A.. 2018. Optogenetic control of mitosis with photocaged chemical dimerizers. Methods Cell Biol. 144:157–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the MATLAB code to calculate the average distance of all kinetochores to monopolar spindle poles in a single cell.
shows the MATLAB code to calculate the kinetochore displacement relative to the metaphase plate of a bipolar spindle.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.