Abstract
Mosaicism, the presence of subpopulations of cells bearing somatic mutations, is associated with disease and aging and has been detected in diverse tissues, including apparently normal cells adjacent to tumors. To analyze mosaicism on a large scale, we surveyed haplotype-specific somatic copy number alterations (sCNAs) in 1,708 normal-appearing adjacent-to-tumor (NAT) tissue samples from 27 cancer sites and in 7,149 blood samples from The Cancer Genome Atlas. We find substantial variation across tissues in the rate, burden and types of sCNAs, including those spanning entire chromosome arms. We document matching sCNAs in the NAT tissue and the adjacent tumor, suggesting a shared clonal origin, as well as instances in which both NAT tissue and tumor tissue harbor a gain of the same oncogene arising in parallel from distinct parental haplotypes. These results shed light on pan-tissue mutations characteristic of field cancerization, the presence of oncogenic processes adjacent to cancer cells.
Somatic mutational events followed by clonal expansion create subpopulations of cells that are genetically distinct from germline (Fig. 1a). This phenomenon, known as clonal mosaicism, plays an important role in aging, infertility and human disease1–4. Genetic profiling of tumors has revealed cancer-site-specific and pan-cancer patterns of somatic mutations5–7. Fewer studies have focused on somatic mutations in pathologically normal tissues8–21. The largest studies of mosaicism in healthy tissue have surveyed existing blood genotype data and have revealed positive associations between blood mosaicism and age, and between blood mosaicism and incidence of hematological cancers22–27. Surveys of the somatic mutational landscape of blood and non-blood tissues have revealed that healthy individuals harbor mutations in cancer-driver genes16–20. The connection between these somatic mutations and disease is an area of ongoing research. Mosaicism has been implicated in other non-cancer chronic conditions, emphasizing the need for comprehensive surveys of mosaicism across human tissues1–3.
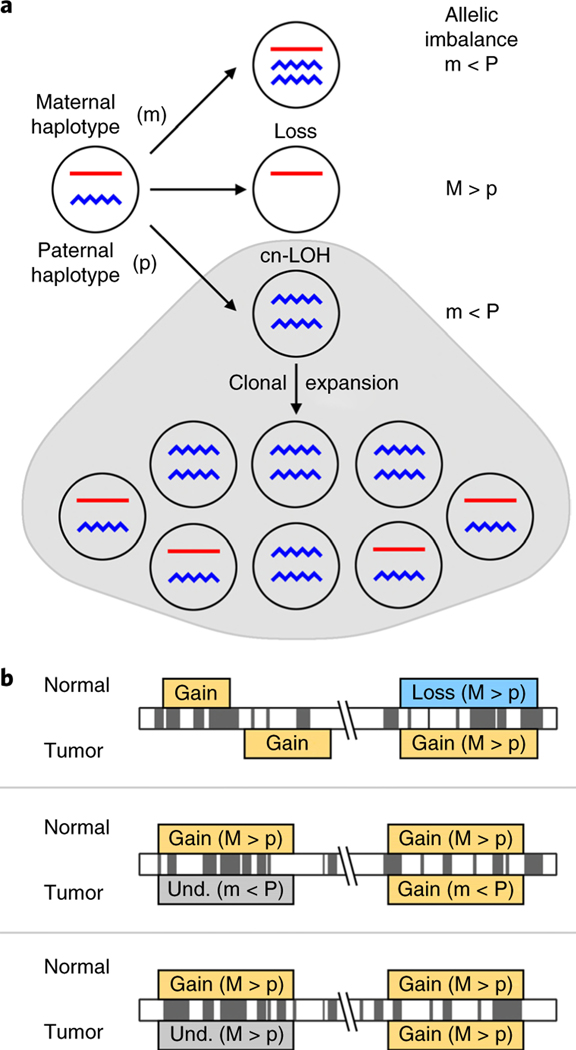
Fig. 1 |. Chromosomal alterations, allelic imbalance and mosaicism.
a, sCnAs lead to deviations from a 1:1 ratio of the maternal to paternal chromosomal segment (haplotype); therefore, the within-sample allele frequencies at heterozygous sites can be used to infer genomic regions with deviations from the 1:1 ratio, which is referred to as allelic imbalance. b, sCnA profiles from non-tumor samples were compared to those from cancers within the same individual. The criteria used for comparison and inference of clonal origin include: event overlap, event type and allelic imbalance direction. The top two panels show examples of events that do not overlap, or that have conflicting event type. The term ‘mirrored allelic imbalance’ has been used to describe instances when samples from the same individual exhibit opposite haplotypes in imbalance, which is shown in the middle panel. As shown in the middle panel, this analysis can help identify conflicting events that are of the same type, or those that are too subtle for classification, which are hereafter referred to as undetermined (und.).
We sought to expand knowledge of mosaicism by analyzing allele-specific megabase-scale sCNAs (gain, loss and copy-neutral loss of heterozygosity (cn-LOH)) in blood and NAT samples from The Cancer Genome Atlas (TCGA). Although NAT tissues appear normal macroscopically, they harbor somatic mutations in tumor-driver genes that are both shared and independent of the adjacent tumor12–14. A previous study examined sCNAs in TCGA non-tumor tissues of ovarian and lung cancer using array comparative genomic hybridization data9. Here we analyze genotype data from Affymetrix 6.0 single-nucleotide polymorphism (SNP) arrays, which are available for more than 10,000 paired control and tumor TCGA samples. By using haplotype-based approaches, we were able to comprehensively study sCNAs in NAT tissues across cancer sites and contrast intraindividual sCNA profiles (Fig. 1b).
Our study uncovered substantial differences in the genomic distribution of sCNAs across tissues (Fig. 2a), with patterns in the blood confirming previous results22,23,25,26. In non-blood tissue, we identified tissue-specific sCNA patterns in NAT tissues from bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), head and neck squamous cell carcinoma (HNSC), stomach adenocarcinoma (STAD) and kidney renal clear cell carcinoma (KIRC). Our comparative analyses of sCNA profiles from matched NAT–tumor samples revealed that these events, detected in NAT tissue,were not always present in the adjacent tumor, including instances of parallel evolution (Fig. 2b). This study lays out a framework for detection, characterization and comparison of sCNAs across human tissues and within tissues from the same donor.
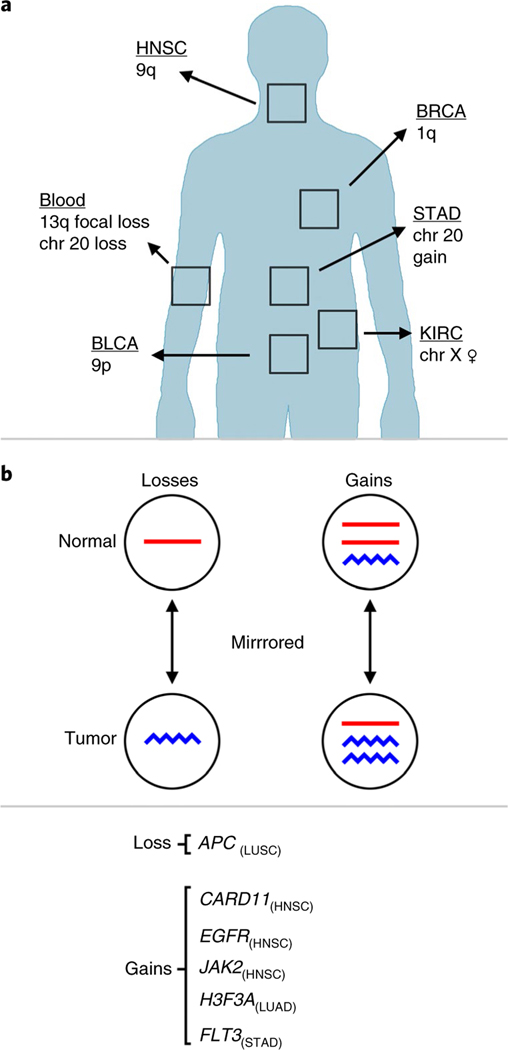
Fig. 2 |. Summary of results.
a, A summary of sCnAs with tissue-specific patterns of enrichment. b, Genes that were gained or lost in nAT tissues and that were also gained or lost in the matched tumor, but show mirrored allelic imbalance and are inferred to have arisen independently in separate clonal lineages.
Results
Detection of somatic copy number alterations.
We profiled the sCNA landscape of blood and NAT samples across TCGA cancer sites (Table 1). We used the hapLOH algorithm for detection of allelic imbalance indicative of sCNA (Fig. 1a). This approach leverages statistically estimated haplotypes for detection of high-confidence megabase-scale sCNAs that are present at low mutant cell fractions (detection limit ~5–10%)28. For each allelic imbalance call, we used the B allele frequency (BAF) and logR ratio (LRR) deviation for classification of sCNAs as gain, loss, cn-LOH or undetermined (where BAF and LRR deviations did not reach the classifications thresholds). We filtered out short high-cell-fraction gains as putative inherited duplications using stringent filtering criteria (Supplementary Fig. 1 and Supplementary Note 1). We validated sCNA calls for a subset of samples with available exome sequencing data29 (Supplementary Fig. 2). Samples with a high rate of missing genotypes and/or potential intra- or interindividual contamination were excluded from downstream analyses. After quality control, we considered samples from 8,437 donors of which 420 had both blood and NAT tissues. These include 1,708 NAT and 7,149 blood samples from 27 TCGA studies (Supplementary Tables 1–3).
Table 1 |.
Mosaicism rates
| Cancer and tCGA study abbreviation | Blood | NAt |
|---|---|---|
| Adrenocortical carcinoma (ACC) | 1/79 (1.3%) | 0/2 (0%) |
| Bladder urothelial carcinoma (BLAC) | 7/343 (2.0%) | 5/28 (18%) |
| Breast invasive carcinoma (BrCA) | 14/885 (1.6%) | 13/114 (11%) |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) | 1/257 (0.4%) | 0/6 (0%) |
| Cholangiocarcinoma (CHOL) | 1/34 (2.9%) | 0/14 (0%) |
| Colon adenocarcinoma (COAD) | 9/346 (2.6%) | 0/83 (0%) |
| Esophageal carcinoma (ESCA) | 2/121 (1.7%) | 1/57 (2%) |
| Glioblastoma multiforme (GBM) | 8/421 (1.9%) | 0/4 (0%) |
| Head and neck squamous cell carcinoma (HNSC) | 7/459 (1.5%) | 18/72 (25%) |
| Kidney chromophobe (KICH) | 0/9 (0.0%) | 0/56 (0%) |
| Kidney renal clear cell carcinoma (KIRC) | 1/94 (1.1%) | 4/332 (1%) |
| Kidney renal papillary cell carcinoma (KIRP) | 7/212 (3.3%) | 2/77 (3%) |
| Liver hepatocellular carcinoma (LIHC) | 2/287 (0.7%) | 3/75 (4%) |
| Lung adenocarcinoma (LUAD) | 14/382 (3.7%) | 4/166 (2%) |
| Lung squamous cell carcinoma (LUSC) | 4/250 (1.6%) | 4/223 (2%) |
| Mesothelioma (MESO) | 4/84 (4.8%) | 0/1 (0%) |
| Ovarian (OV) | 7/390 (1.8%) | 9/100 (9%) |
| Pancreatic adenocarcinoma (PAAD) | 3/149 (2.0%) | 0/33 (0%) |
| Pheochromocytoma and paraganglioma (PCPG) | 4/170 (2.4%) | 1/5 (20%) |
| Rectum adenocarcinoma (READ) | 5/142 (3.5%) | 1/15 (7%) |
| Sarcoma (SARC) | 4/219 (1.8%) | 2/19 (11%) |
| Skin cutaneous melanoma (SKCM) | 5/456 (1.1%) | 1/2 (50%) |
| Stomach adenocarcinoma (STAD) | 9/350 (2.6%) | 8/84 (10%) |
| Thyroid carcinoma (THCA) | 3/401 (0.7%) | 2/93 (2%) |
| Thymoma (THYM) | 0/107 (0.0%) | 0/11 (0%) |
| Uterine corpus endometrial carcinoma (UCEC) | 7/458 (1.5%) | 0/32 (0%) |
| Uterine carcinosarcoma (UCS) | 1/44 (2.3%) | 0/4 (0%) |
| Prostate adenocarcinoma (PRAD) | 7/407 (1.7%) | 0/109 (0%) |
Number of blood and NAT TCGA samples with detectable sCnA/total number of samples tested (percentage of mosaic samples). PrAD was excluded on the basis of TCGA annotation (Methods).
Frequency, burden and size distribution of sCNAs.
A total of 78 NAT and 130 blood samples had at least one autosomal sCNA, with 338 and 178 sCNAs detected in the NAT and blood samples, respectively (Supplementary Tables 2–7). Only one donor (of 420) harbored sCNAs in both blood and NAT tissues; these sCNAs were on different chromosomes. The rate of mosaic NAT samples, those with one or more sCNAs, was more than twice the rate in blood (4.6% versus 1.8%; P = 3 × 10−11, χ2 test); these results held when we used logistic regression (LR) to adjust for age and sex (P = 5.1 × 10−9). NAT mosaic tissues had an average of 4.3 sCNAs (median = 2, minimum = 1, maximum = 44), which was significantly higher than blood with an average of 1.4 sCNAs (median = 1, minimum = 1, maximum = 4; P = 2 × 10−5, Mann–Whitney–Wilcoxon test (MWW)).
The mosaicism rates of NAT tissues were different across cancer sites with BRCA, HNSC and KIRC reaching statistical significance after multiple testing correction (P < 0.05, binomial test; Supplementary Table 3). Among sites with ten or more available NAT samples, HNSC had the highest rate (25%), followed by BLCA (18%), BRCA (11%), sarcoma (11%), STAD (10%) and ovarian (9%) (Supplementary Fig. 3).
Mosaic chromosomal alterations had a median size of 32 Mb and no obvious difference in the distribution of sCNA sizes between blood and NAT samples (P = 0.79, MWW; Supplementary Fig. 4), as well as similar distributions of focal and arm-level sCNAs (Supplementary Fig. 5). In blood, but not NAT tissues, there was an association between sCNA event type and size (P = 2.8 × 10−8 in blood and P = 0.37 in NAT, Kruskal–Wallis rank sum test; Supplementary Figs. 6 and 7).
Chromosome X data in females also allow for the detection of sCNAs leading to allelic imbalance; therefore, we surveyed chromosome X in 4,099 blood and 839 NAT samples from female patients. We observed 67 chromosome X sCNAs in 37 blood samples (~1% of blood samples) and 32 chromosome X sCNAs in 18 NAT samples (~2% of NAT samples) (Supplementary Tables 8–11). Chromosome X sCNAs were present in 7% of NAT samples in KIRC, which was significantly higher than for other sites (adjusted P = 0.004, binomial test, Bonferroni correction for 26 cancer sites; Supplementary Table 9).
Genomic distribution of sCNAs.
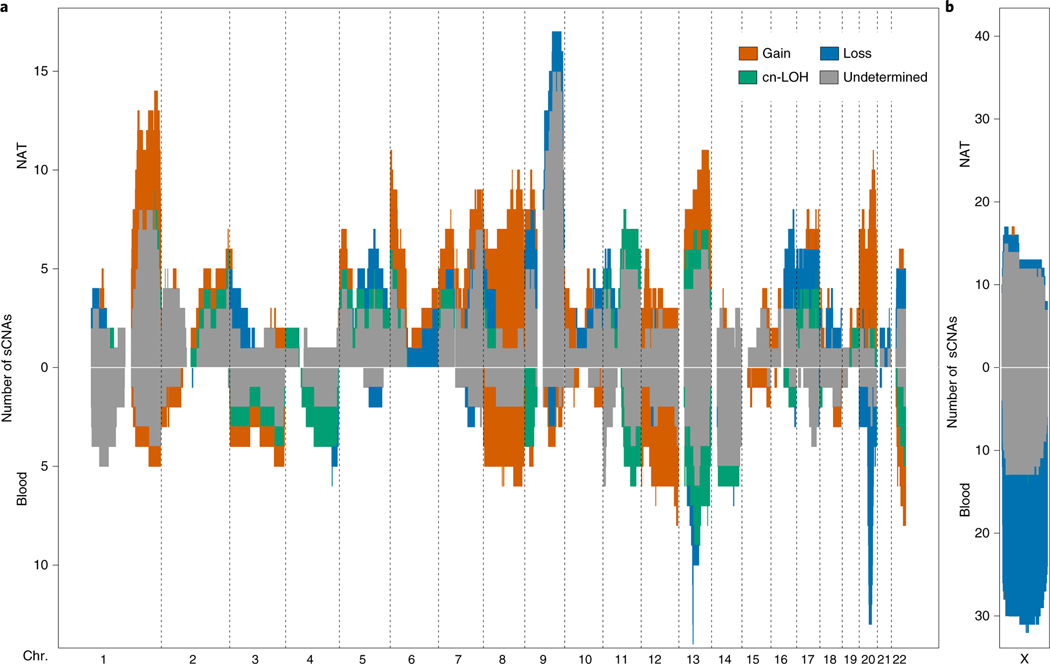
In addition to the differences in frequency and burden of sCNAs between blood and NAT tissues, we observed differences in the genomic distribution of these somatic alterations (Fig. 3a). Alterations on 1q and 9q were the most frequent in NAT tissues, whereas 13q and 20q were the most frequent in blood. To compare these findings, we formally tested for enrichment of alterations at particular genomic loci in blood and in NAT tissues30. In blood, the most significant hits were located in 13q14 (P = 1.7 × 10−4, Q = 0.03) and chromosome 20 (P = 3.2 × 10−4, Q = 0.05; Supplementary Fig. 8 and Supplementary Table 12). These results confirm blood mosaicism profiles previously reported in cohorts of healthy donors and patients with cancer (solid tumors)22–26 (Supplementary Fig. 9). We observe that NAT tissues and blood have different types of sCNAs on chromosomes 13 and 20. Deletions on 13q, which were common in blood, were not observed in NAT tissues; instead, 13q sCNAs in NAT tissues were primarily gains, with no observed losses (Fig. 3a). Alterations of chromosome arm 20q in blood were losses or too subtle for classification (12 losses and 5 undetermined events); by contrast, there were no 20q losses in NAT tissues and the majority of sCNAs were gains (nine gains, one cn-LOH and three undetermined events; Fig. 3a). As for blood, we tested for enrichment of sCNAs at particular genomic loci in NAT tissues; the most significant peaks were located in 6p and 1q (P = 7.7 × 10−4 and P = 6.0 × 10−4, respectively, Q = 0.14 for both; Supplementary Fig. 8 and Supplementary Table 13). The significant genomic interval in 6p overlaps a recurrent pan-cancer deletion peak7. The observed differences between the sCNA landscape in blood and NAT tissues suggests that sCNAs in blood and/or immune cells are rarely detected in NAT samples.
Fig. 3 |. Landscape of sCNAs.
a, The landscape of autosomal sCNAs across blood and NAT tissues. Chromosomes are ordered along the x axis and sCNAs were binned in 1-Mb genomic segments on the basis of their overlap with that region. The number of sCNAs (y axis) is plotted using a different color for each event type. b, The landscape of chromosome X sCNAs in female blood and NAT tissues.
Chromosome X sCNAs were similar in type (predominantly loss or undetermined) in both blood and NAT tissues (Fig. 3b). These alterations were also the most common sCNA in both sets of samples (Fig. 3b). Relative to the most frequent autosomal sCNA in females, the rate of chromosome X sCNAs was 3.7-fold higher (X versus chr. 13q) in blood and 1.3-fold higher in NAT tissues (X versus chr. 1q). This observation of a higher frequency of chromosome X alterations relative to autosomes, has been reported previously by two mosaicism surveys of blood26,27.
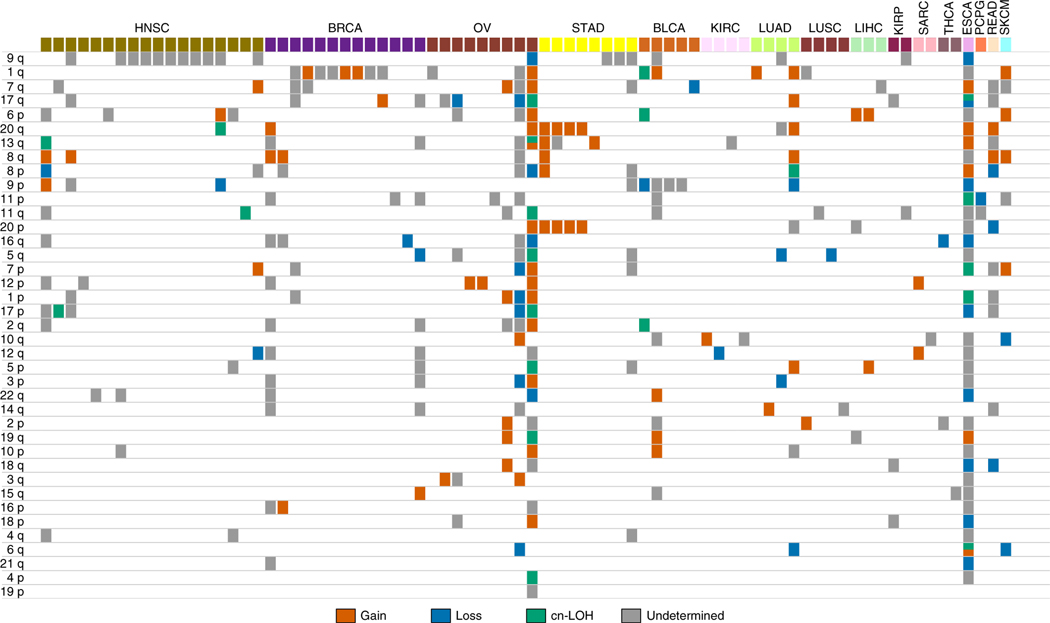
Beyond contrasting the sCNA landscapes of blood and non-blood tissues, we were also interested in cancer-site-specific patterns of sCNA enrichment in NAT tissues (Fig. 4), which was motivated by the well-documented differences in the sCNA profiles among cancers5–7. Owing to limited power at sites with few sCNAs, we formally tested for arm-level sCNA enrichment at cancer sites with five or more mosaic NAT samples. To do so we used an omnibus test to identify chromosome arms with recurrent sCNAs. For each cancer site, we generated a null distribution by permuting the location of sCNAs across chromosome arms of each sample that is within a column in Fig. 4. We then used this null distribution to calculate a P value for the chromosome arm with the highest sCNA rate at that cancer site (Methods). HNSC had a pronounced enrichment for 9q sCNAs in NAT tissues (adjusted P = 1 × 10−7; Bonferroni correction for five cancer sites), BLCA for 9p (adjusted P = 0.01), BRCA for 1q (adjusted P = 3 × 10−5) and STAD had an enrichment for chromosome 20 gains (adjusted = 0.02) (Fig. 4). This set of sCNAs, with NAT-specific enrichment, span chromosome arms that are recurrently altered in tumors from the respective cancer sites (Supplementary Table 14). No statistically significant enrichment was observed for ovarian cancer; however, though not significant (adjusted P = 1), we observed three gains that spanned KRAS, which have been observed in ovarian cancer and in endometriotic epithelium5–7,19.
Fig. 4 |. Arm-level sCNAs in NAT tissues.
Arm-level sCNA summary for each mosaic NAT tissue, arranged by cancer site. Each column represents one NAT sample with detectable sCNAs. Chromosome arms are ordered by sCNA frequency.
We compared the genomic distribution of sCNAs in NAT tissues to patterns of chromosomal alterations reported in pan-cancer surveys5,6. Somatic gains of 8 and 12, which are common across cancers, were present in both blood and NAT tissues (Fig. 3a). Somatic gains of 1q are enriched in epithelial tumors (BRCA, LUAD and LIHC); we observe these gains in BRCA and LUAD NAT tissues (Fig. 4). Co-occurring gains of 8q, 13q and chromosome 20 are enriched in gastrointestinal tumors (ESCA, READ and STAD); NAT samples from those sites show one, two or sometimes all three gains (Fig. 4).
Associations between mosaicism and clinical features.
Studies of clonal mosaicism in blood report an association between sCNAs in autosomes and age22–26. We observe this association in blood (P = 5.5 × 10−13, LR, adjusting for sex; Supplementary Fig. 10). In blood, we also observe the previously reported26,27 association of age with sCNAs in chromosome X (P = 1.5 × 10−5, LR). The TCGA dataset allowed us to examine these associations in NAT tissues, where the presence of sCNAs in autosomes is marginally associated with age (P = 0.10, LR, adjusting for sex and cancer site), and the presence of sCNAs in chromosome X does not show an association (P = 0.30, LR, adjusting for cancer site). Next, we investigated association between mosaicism and cancer stage. When we accounted for differential mosaicism rates among cancer sites, using a permutation-based test, we did not detect an association between the presence of sCNAs in autosomes and cancer stage for blood (P = 0.62) or for NAT tissues (P = 0.31; Supplementary Table 15), which suggests that the observed sCNAs in blood and NAT tissues are not driven by circulating tumor cells or metastases. Additional clinical analyses and a discussion of their limitations are presented in Supplementary Note 2.
Inference of sCNA clonal origins.
As NAT tissue is proximal to tumor tissue, these two tissues may have somatic mutations stemming from a shared clonal lineage. To investigate the similarities between NAT tissue and tumor, we profiled sCNAs in tumor samples of patients with mosaic NAT tissues. We then contrasted intraindividual sCNA profiles by examining whether sCNAs in the NAT tissue overlapped with those in the matched tumor. Owing to inherent difficulties with mapping sCNA boundaries, especially for sCNAs present at a low mutant cell fraction, overlap was defined as an sCNA with greater than 50% overlap with a tumor sCNA (Fig. 1b). Overlapping sCNAs were categorized as conflicting or non-conflicting on the basis of sCNA classification and the direction of allelic imbalance (Fig. 1b). Conflicting sCNAs included those with discordant event type (for example, gain–loss) and those where there was evidence for ‘mirrored’ allelic imbalance, where opposite haplotypes are in imbalance (Fig. 1b and Supplementary Fig. 11), indicating distinct mutations31,32. We define overlapping and non-conflicting pairs of NAT–tumor sCNAs as concordant. We note that concordance is merely suggestive, not conclusive, of a shared clonal lineage.
Of the autosomal sCNAs in blood, 25% were concordant, significantly lower than the 62% of sCNAs in NAT tissues that were concordant (P = 3.7 × 10−15, χ2 test). The majority (59%) of blood samples had zero concordant sCNAs with the tumor, while 38% of NAT samples had zero concordant sCNAs (P = 4.3 × 10−5, χ2 test; Supplementary Table 16). We tested additional, more stringent, overlap criteria, requiring reciprocal overlap between sets of sCNAS with thresholds set to 50%, 75% and 90% (Supplementary Fig. 12). In the blood, 17%, 12% and 8% of sCNAs were concordant at each of the overlap thresholds, respectively. These percentages were more than double (42%, 31% and 25%) those for NAT tissue sCNAs (Supplementary Tables 16 and 17). Overall, autosomal sCNAs in NAT tissues more frequently matched those in the adjacent tumor tissue than did the sCNAs in the blood of patients with cancer (Supplementary Table 16 and Supplementary Fig. 13). For sCNAs in chromosome X, 30% of those detected in blood were concordant (50% overlap) with tumor as compared to 54% for NAT tissues; this difference bordered on significance (P = 0.06, χ2 test).
In contrast to NAT–tumor pairs, we compared sCNA profiles of 80 intraindividual tumor–tumor sample pairs. These included primary tumor samples with a matched recurrence, metastasis or secondary primary tumor sample (Supplementary Table 18). The majority (98%) of tumor samples had one or more concordant sCNAs (50% overlap threshold) with the second tumor sample, significantly higher than for NAT samples (P = 5.8 × 10−8, χ2 test). Extended results for intraindividual sample comparisons are found in Supplementary Notes 3 and 2, and Supplementary Tables 17–21. We did not observe an association between sCNAs in NAT tissues and mutational burden or microsatellite instability in the adjacent tumors (Supplementary Note 5).
Parallel evolution of NAT tissue and tumor clones.
Directional allelic imbalance comparisons between overlapping sCNA calls from paired NAT and tumor tissues revealed examples of independent mutation (Fig. 2b and Supplementary Figs 11 and 13). We identified overlapping sCNAs with matching event type (that is, both loss or gain) that had opposite haplotypes in imbalance (one shows an increase of maternal alleles and the other shows an increase of the paternal alleles), which ruled out the possibility that these events have the same clonal origin; they originated independently of each other within the same organ (Fig. 2b). We observe that 21% (19 of 70) of blood sCNAs and 9% (23 of 247) of NAT tissue sCNAs (50% overlap) exhibit mirrored allelic imbalance (Supplementary Tables 6 and 7). For NAT–tumor samples, these independent mutations include gains of established oncogenes (H3F3A in LUAD; CARD11, EGFR and JAK2 in HNSC; and FLT3 in STAD), as well as two independent losses of APC in the tumor and the NAT tissue from a patient with LUSC (Fig. 2b). Independent mutations targeting APC, an essential driver of colorectal cancer, have been reported in adenomas of the colon33.
Single-nucleotide variants in HNSC NAT tissues and associations with sCNAs.
Owing to the prevalence of sCNAs in HNSC NAT tissues and the previously reported field cancerization in HNSC13, we sought to gain a more comprehensive view of somatic mutations in these tissues. To do so, we used the available exome sequencing data from 59 HNSC NAT samples with paired blood as a reference. As expected, NAT tissues had a lower single-nucleotide variant (SNV; point mutations and indels) burden (median = 18, minimum = 3, maximum = 202) than the tumors (median = 179, minimum = 64, maximum = 754; P = 2.2 × 10−16, MWW). The SNV burden in NAT tissues was not correlated with the SNV burden in the tumor (P = 0.64; Pearson’s R = 0.06) and the presence of one or more sCNAs in NAT tissues did not show an association with sCNA burden in the tumor (arm-level; P = 0.39, LR). SNV and sCNA burden (arm-level) were not correlated in tumors (P = 0.256, Pearson’s R = 0.16). However, in NAT tissues, the SNV burden showed a positive association with sCNAs (presence or absence; P = 0.008, LR; Supplementary Fig. 14).
To gain insights into possible drivers of the positive association between SNV and sCNA burden in NAT tissues, we formally tested for positive selection among all genes, as well as for enrichment of HNSC-driver mutations in NAT tissues with detectable sCNAs. We identified two genes under positive selection in HNSC NAT tissues, PPM1D (Q = 2.3 × 10−4) and FAT1 (Q = 0.02) by applying a maximum-likelihood model of the ratio of synonymous to non-synonymous mutations34. We applied the same model to the adjacent tumor SNV data and detected a significant association for FAT1 (Q = 5.6 × 10−17), but not for PPM1D (Q = 1). PPM1D SNVs (four putatively truncating mutations) were detected in NAT tissues with and without sCNAs, while FAT1 SNVs were detected only in NAT tissues with sCNAs. Three truncating FAT1 mutations in NAT tissues were not detected in the adjacent tumor (Supplementary Fig. 15). Positive selection of FAT1 mutations has been reported in epithelial tissues from healthy individuals16,17. Truncating mutations in PPM1D have been reported at a frequency of 0.7% in blood from patients with cancer (solid tumors)35. We observed that all four NAT tissues with putative HNSC driver mutations (SNVs) that were detected in the adjacent tumor have at least one sCNA (Supplementary Fig. 15 and Supplementary Tables 22 and 23). These shared drivers include a missense PIK3CA SNV recurrent in cancer and truncating mutations in TP53 and CASP8. The CASP8 SNV overlapped an sCNA present in both NAT and tumor tissues (concordant sCNAs), while the TP53 SNV overlapped with an sCNA in the tumor, but not in the NAT tissue (Supplementary Fig. 15). A formal test for enrichment of putative HNSC-driver SNVs (stop–gain, splicing and missense mutations predicted to be deleterious) in NAT tissues with sCNAs revealed a significant association (P = 0.002, LR); this analysis adjusted for SNV burden (SNV count excluding HSNC-driver SNVs). The association between the presence of sCNAs and putative HSNC-driver SNVs complements previous findings of increased genomic alterations in oral premalignant lesions that progress to invasive disease relative to those that do not36.
Discussion
In this study, we detect and characterize the distribution of sCNAs, inferred from allelic imbalance, across non-malignant tissues. We report sCNAs in 1.8% of blood and 4.6% of NAT samples, across 27 TCGA sites. These findings add to a widely accessed resource in the cancer community, highlighting the opportunity to include allelic imbalance in studies of mosaicism, and complementing SNV annotation with larger structural genomic changes across tissues15. This study justifies further investigation of sCNAs in NAT tissues and their potential clinical utility.
The rate of sCNAs and genomic distribution in blood samples generally followed those from two large surveys (30,000 and 150,000 blood samples, respectively), which report autosomal sCNA rates of ~2–5% for individuals 50–70 years of age25,26. NAT tissues had a higher rate of mosaicism and higher sCNA burden than blood. We report tissue-specific patterns of sCNA enrichment (Fig. 2a). Relative to blood, the sCNAs of NAT tissues more closely resembled those of the tumor. The presence of sCNAs in HNSC NAT tissues showed a positive association with mutations (SNVs) in established HNSC-driver genes.
Relative to tumor tissue, characterization of the sCNA landscape of NAT tissues is technically more difficult as mutations are expected to be present in a lower proportion of cells. We used a haplotype-based method to overcome this challenge and identify sCNAs present at cell fractions less than 10%28. This detection limit, which is based on the size and length of sCNAs, mutant cell fraction and mutation type, should be taken into account when comparing rates and profiles across tissues. Our results do not necessarily reflect the rate at which these alterations arise, but rather the rate at which they are present in clonal expansions large enough for detection. Other factors to consider include the anatomical and/or physiological features, cellular structures and the sample collection protocol at each cancer site, particularly for those with a complex structure such as the lung37 (Supplementary Note 6). For example, we did not detect sCNAs in NAT tissues from colon adenocarcinoma (n = 83), which may be due to the structural organization of the colonic mucosa where crypt structures of the colon may constrain clonal expansions4,20. Although challenging, detection and characterization of sCNAs in small clonal expansions may offer a peek into the earliest stages of disease.
This pan-tissue study of sCNAs may support insights into clonal expansions in NAT tissues and how they may be related to the neighboring tumor. Our results suggest that, overall, NAT tissue is more similar to the tumor as compared to blood, which may be partially explained by the closer developmental lineage of NAT tissue and tumor. An alternative explanation, which is not mutually exclusive, is the presence of field cancerization, a field of injury in tissues surrounding tumors at these cancer sites. This explanation is supported by an expression study of a subset of TCGA cancer sites indicating that NAT tissues have unique expression profiles that are distinct from non-tumor-bearing tissues and more closely resemble tumors38. The phenomenon of field cancerization was first described in oral tissues and has since been documented in BLCA, BRCA, HNSC, STAD and other cancers13. Field cancerization may also explain why we observe shared mutations between NAT–tumor pairs. However, these shared mutations are not always clonal in origin as evidenced by our observation of NAT tissue and tumor independently acquiring the same oncogenic gain (‘mirrored’ allelic imbalance). We observe these events less than expected, because on average only half of independent sCNAs of the same mutation type would result in mirrored allelic imbalance. Drivers of such parallel evolution could be extracellular and/or environmental exposures or genetic and epigenetic mutations that are shared among clones at a cancer site. Shared drivers may explain some of the chromosomal alterations with tissue-specific patterns of enrichment.
We present such examples of enrichment in several tissues (Fig. 2a). The challenges of defining how and whether these sCNAs lead to clonal expansions and/or tumors is exemplified by comparing our observations in stomach and head and neck NAT tissues to somatic mutations reported in healthy tissues, premalignant lesions and tumors.
For STAD NAT tissues, we report a marked enrichment of chromosome 20 gains. Gain of chromosome 20 is postulated to target STAD-driver genes and is present in approximately 70% of gastric cancers39. Furthermore, studies of human embryonic stem cells have shown a growth advantage for cells with amplifications in chromosome 20q40. The striking enrichment of HNSC-driver SNVs in NAT tissues with sCNAs suggests that some clones in NAT tissues may be on a path toward malignancy. Although, their path may be in parallel to that of the adjacent tumor as evidenced by NAT tissues with HNSC-driver mutations that are not detected in the tumor (Supplementary Fig. 15).
The potential for these clones to develop into secondary tumors may depend on their specific mutational and epigenetic profiles, as recent studies demonstrate that mutations in cancer-driver genes are common in tissues from healthy individuals, for example, a higher frequency of NOTCH1 mutations in normal esophageal tissues relative to esophageal cancer16–20. NOTCH1 mutations have also been reported in airways that appear to be pathologically normal from patients with lung cancer, as well as premalignant lesions of the lung. In these tissues, NOTCH1 SNVs and sCNAs (9q) are often concurrent14,16,17,41. For HNSC, we observe that two of 11 NAT samples with 9q sCNAs have NOTCH1 mutations. In addition, as reported for ‘healthy’ epithelial tissues, we observe that FAT1 is under positive selection in HNSC NAT tissues16,17. These results suggest that some of the somatic mutations in HNSC NAT tissues are due to clonal expansions that are common in healthy tissues and may not necessarily lead to carcinogenesis. Surveys with larger sample sizes, may help identify sCNAs and SNVs that promote malignant transformation. It is also possible that some of these somatic mutations are found to be protective against progression toward malignancy, serving as markers of cellular age or exposures. Although, not directly relevant to cancer, the latter may provide valuable insights into age-related disease.
Methods
TCGA samples surveyed.
TCGA data comprise tumor and control samples that were derived from blood and NAT tissues (https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/sample-type-codes). We excluded blood samples from five TCGA studies, because these were derived from donors with hematological malignancies and/or did not have NAT samples (brain lower-grade glioma, acute myeloid leukemia, testicular germ cell tumors, uveal melanoma and lymphoid neoplasm diffuse large B cell lymphoma). Our initial survey included 8,459 blood and 2,165 NAT tissues from 28 cancer sites, but one site (PRAD) was excluded from downstream analyses (Supplementary Tables 1, 4 and 5).
Detection of sCNAs.
We used the Birdsuite software to process data from Affymetrix Genome-Wide Human SNP Arrays (one million SNP markers) and generate genotype calls, BAF and LRR at each marker42. The human genome build hg19 (GRCh37) was used as the reference. We phased the genotypes with the MACH software, and used the phased genotypes and BAF data for detection of allelic imbalance with the hapLOH software28,43. We used SyQADA to ensure all samples were processed using the same workflow44. We identified genomic allelic imbalance segments using a threshold of a posterior probability for allelic imbalance ≥0.9 and drew event boundaries at markers where the posterior probability dropped below 0.5. We excluded allelic imbalance events with fewer than ten markers. In comparison to LRR-based methods, hapLOH integrates BAF and haplotype information, thus mitigating the effect of marker-level measurement errors and batch effects, which allowed us to produce a set of high-confidence sCNA calls. This approach has the highest sensitivity for detection of cn-LOH and lowest sensitivity for amplifications (Supplementary Table 24). We estimate a false-positive rate <1%, which is in agreement with previous estimates25. Further details of the specificity and sensitivity analyses are presented in Supplementary Note 7.
Sample-level quality control.
We required that samples had a genotype missing rate <0.05 and α0 <0.52. The α0 parameter is generated by the hapLOH software and is the estimated emission probability of the null state, a background sample-level allelic imbalance rate; thus, an elevated α0 value can indicate cross-individual contamination. In addition, we removed samples that had been redacted from the TCGA dataset as well as blood or NAT samples that did not have an available tumor sample (primary, recurrent or metastasis; https://gdac.broadinstitute.org/, https://portal.gdc.cancer.gov/ and the clinical reference)45. For NAT samples with detectable allelic imbalance we checked the Broad GDAC Firehose annotation files for annotations regarding possible tumor contamination, sample swaps or abnormal pathology (https://gdac.broadinstitute.org/). This led to the exclusion of the following NAT samples from the results: TCGA-V5-AASX-11A-11D-A386–01 (NAT sample is a Barrett’s mucosa with mild dysplasia); TCGA-90–6837-11A-01D-1943–01 (possible tumor–NAT swap); and all NAT samples from prostate adenocarcinoma (PRAD) with detectable sCNAs (contamination with tumor). We removed all PRAD samples (blood and NAT) from downstream analyses. We removed two NAT samples (TCGA-BJ-A28W-11A-11D-A16M-01 and TCGA-BR-6710–11A-01D-1881–01), because tumor samples from these patients had 0 and 1 somatic nucleotide variants, respectively, which is suggestive of possible tumor–NAT sample swaps (https://portal.gdc.cancer.gov/). We report sCNAs for a subset of NAT samples that were excluded in Supplementary Tables 19–21; these include PRAD NAT tissues, a Barrett’s mucosa with mild dysplasia and tissue adjacent to a ductal carcinoma in situ, a premalignant lesion in breast.
It is noted in Broad GDAC Firehose annotation files that a patient with thyroid carcinoma, TCGA-EL-A3H2, with detectable mosaicism in NAT tissue (TCGA-EL-A3H2–11A-11D-A20A-01) had “received radiation in early childhood to the thyroid unrelated to treatment of any malignancy”; this sample was included.
sCNA classification and filtering of putative germline gains.
We used the median BAF deviation and median LRR deviation at allelic imbalance segments for sCNA classification (Supplementary Fig. 1). LRR deviation thresholds were set to ±0.05 for gains and losses. For allelic imbalance segments with an LRR deviation between −0.5 and +0.5, those with BAF deviation >0.10 were classified as a cn-LOH, and the remainder were labeled undetermined, as the BAF and LRR deviations were too subtle for sCNA classification. As previously described, gains detected with the hapLOH method may include small germline gains28. We filtered putative germline gains by removing allelic imbalance segments with LRR deviation >0.08 and size <5 Mb. As an additional filter, we removed gains that had greater than 50% reciprocal overlap with gold-standard gains from the database of genomic variants (Supplementary Fig. 1 and Supplementary Note 1; http://dgv.tcag.ca/dgv/docs/DGV.GS.March2016.50percent.GainLossSep.Final.hg19.gff3). For segments classified as gains we observed an upward shift in segment mean LRR relative to no-call and for losses we observed a downward shift (Supplementary Fig. 16 and Supplementary Note 8). Mosaic NAT tissues had a statistically estimated ploidy of near two (Supplementary Table 25 and Supplementary Note 8).
sCNA mutant cell fraction estimates.
For each sCNA call, we used the BAF deviation of heterozygous markers to estimate the fraction of cells harboring the sCNA. The BAF for a marker is equal to:
As B and A alleles are arbitrary labels we can define B alleles as those present in the over-represented haplotype. Then the theoretical BAF of heterozygous markers in an sCNA segment is:
where μ is equal to the fraction of cells with the sCNA, c is equal to the number of copies of the over-represented haplotype and p is the ploidy at the genomic segment with an sCNA. For cells without an sCNA c = 1 and p = 2. These values are c = 2 and p = 3 for a gain, c = 1 and p = 1 for a loss and c = 2 and p = 2 for a cn-LOH. We relate BAF to BAF deviation as follows:
and then use the median BAF deviation for each sCNA to estimate the mutant cell fraction:
For undetermined sCNAs we report a lower bound estimate (for cn-LOH) and an upper bound estimate (for gain). It is important to note that BAF values are estimated from intensity data and do not represent exact B allele frequencies. In addition, this model assumes that regions of allelic imbalance represent a single sCNA that alters one chromosome segment only, that is, it does not allow for more than two gains or loss of both copies. The latter assumption holds for cn-LOH and deletion events resulting in allelic imbalance. However, it could lead to overestimates of mutant cell fractions for amplifications that generate more than two copies of the same haplotype.
Analyses of chromosome X sCNAs.
We conducted detection and analysis of chromosome X sCNAs using the same approach as for autosomes with the exception that these analyses were restricted to females. Putative germline events were removed using the same criteria used for autosomes. Additionally, we identified two patients with putative germline trisomy X. These gains were detected at high mutant cell fraction in both NAT and blood samples.
HNSC exome sequencing.
BAM files aligned with human reference GRCh37 were downloaded from GDC for patients with HNSC with blood and NAT samples (https://portal.gdc.cancer.gov). We used the MACH software for phasing and hapLOHseq for analyses of allelic imbalance from sequencing data using the blood for germline genotype calls (minimum coverage of ten reads at heterozygous markers)29,43. The hapLOHseq algorithm outputs phase concordance values between adjacent heterozygous markers. A value of 1 indicates that BAF values are shifting toward the same haplotype and a value of 0 indicates they are shifting toward opposite haplotypes. For each genomic segment with a hapLOH call (from array), we tested for the presence of allelic imbalance in the exome sequencing data using a matched sample (with no call from array) as a control. All of the genomic segments with allelic imbalance called from array data had higher phase concordance (indicative of allelic imbalance) as compared to the phase concordance of the matched genomic segment in the control sample, corroborating the allelic imbalance call. In addition, we used a one-sided binomial test (R binom.test() function), with the number of successes equal to the sum of the phase concordance values for the segment with allelic imbalance called from array, and the probability of success set to the sum of the phase concordance values across the same genomic segment (from the matched ‘control’ sample with 0 sCNA calls in the array analysis) divided by the number of marker pairs in the genomic segment being tested. For testing, we selected 43 genomic segments from 16 HNSC NAT samples corresponding to sCNAs detected via SNP array. We performed this test to calculate P values, excluding four segments that had less than 20 informative markers (heterozygous). The average number of heterozygous calls from arrays was tenfold higher than for exome sequences (22,000 heterozygotes). P values for 39 genomic segments are summarized in Supplementary Fig. 2.
Tumor sCNA profiling and intraindividual sCNA profile comparison.
Tumor SNP array data were processed using the same pipeline as for the non-tumor samples with two exceptions. (1) Tumors and non-tumor samples were processed separately with the Birdsuite software42. (2) Genotype calls from the matched non-tumor samples were used for phasing and allelic imbalance detection in the tumor samples. Tumor allelic imbalance segments were classified in the same way as non-tumor samples.
We compared sCNA profiles between samples derived from the same donor using the following three criteria: overlap, event type and direction of allelic imbalance. First, we estimated overlap between sCNAs using the bedtools software intersect function at the following four stringency settings: minimum 50% overlap, minimum 50% reciprocal overlap, 75% minimum reciprocal overlap and 90% minimum reciprocal overlap. Second, we determined whether sCNA calls had conflicting event type or allelic imbalance direction. A set of overlapping sCNAs was deemed non-conflicting if they were both of the same type, for example, both were gains, or if one or both were undetermined. Additionally, we contrasted allelic imbalance profiles with the RECUR software for detection of differences in the allelic shifts of sCNAs called in two samples from the same individual32. This phenomenon has been referred to as ‘mirrored allelic imbalance’, where one sCNA has an increase of the maternal haplotype and the other sCNA has an increase of the paternal haplotype31,32. Input for RECUR included a list of sCNAs from the non-tumor sample to test for mirroring, genotype calls from the non-tumor sample and BAF values from non-tumor and tumor samples. We used the default threshold of P < 0.0001 to identify mirrored genomic segments.
Samples for tumor–tumor sCNA profiling.
We profiled the sCNA landscape of TCGA primary tumors (n = 89) with a matched additional new primary, recurrent or metastasis tumor sample with codes 05, 02 and 06, respectively (https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/sample-type-codes). We examined the overlap of sCNA profiles of 80 tumor–tumor pairs for which there was at least one sCNA call for each tumor sample. The primary tumors were used to generate a list of sCNAs for comparison.
HNSC SNV analyses.
We called somatic mutations with Mutect2 (gatk-4.0.12) following GATK4 best practices for NAT tissue and tumor, these were paired with blood as the germline control sample46. We kept somatic mutation denoted as ‘PASS’ by Mutect2 and used ANNOVAR for annotation47. We removed somatic mutations with population allele frequencies >0.01 in the ExAC database48. We used annotations from Baily et al. to generate a list of pan-cancer (200) and HNSC (33) driver or lineage genes49. These genes are listed in Supplementary Table 22. SNVs in these genes that were deemed putatively deleterious by two or more callers were included in the analysis of driver genes and Supplementary Fig. 15. All but two putative driver SNVs were exonic (one splice and one 5′ UTR). We identify genes under positive selection using a maximum-likelihood model of the ratio of synonymous to non-synonymous mutations and removed GNAQ SNVs from the analyses owing to previously reported false-positive calls34. The sCNA burden in tumor was equal to the number of chromosome arms with one or more sCNAs, as called by hapLOH.
Null model for distribution of sCNAs.
We used GRIN to test for association of sCNAs at particular genomic regions30. This approach creates a null model where the sCNA is represented as occurring with equal probability across the genome.
Statistical analyses.
We defined a mosaic sample as having one or more detectable sCNAs and treated mosaicism as a binary trait. The 95% confidence interval for the rate of mosaic NAT tissues at each cancer site (Supplementary Fig. 3) was estimated using the Wilson score test-based interval and α = 0.05 (probability of type I error; binconf(), Hmisc R package). For logistic regression, we adjusted for cancer site by including the mosaicism rate of NAT tissues of each cancer site as a covariate in the model.
We tested for differences in the mosaicism rates across sites by comparing the observed number of mosaic NAT samples in a site to those expected given the overall mosaicism rate in NAT samples (excluding NAT samples from the site being tested) via binomial distribution, with Bonferroni correction to account for multiple testing (Supplementary Tables 3 and 9). We use adjusted to denote P values that have been adjusted to account for multiple testing.
We obtained sex, age and stage information from a recent pan-cancer TCGA publication with curated clinical data45. In our analyses of the association between mosaicism and stage, we dichotomized stage to early (stage I and II) versus late (stage III or higher). Association of mosaicism with stage was tested by permutation (using χ2 statistic) to account for differences in the late versus early stage cases across cancer sites (Supplementary Note 2). To obtain a null distribution of the χ2 statistic, we permuted mosaicism status within each cancer site (n = 1,000).
We used an omnibus test for chromosome arm enrichment of sCNA calls in NAT tissues from different TCGA studies. These analyses were restricted to cancer sites with five or more mosaic NAT samples, which included BLCA, BRCA, HNSC, STAD and ovarian. We summarized sCNAs as present or absent in each chromosome arm (39 chromosome arms in 22 autosomes). During each permutation, sCNAs were randomly placed in N arms for each sample, where N is the observed number of arms with sCNAs in the sample. For each cancer site, we calculated a P value for the most frequent arm-level sCNA at that site (observed T times), by counting the number of times we observe T or greater number of sCNAs in a chromosome arm in the permutation analysis (n = 1 × 106). This approach allowed us to control for NAT tissue sCNA burden differences among cancer sites and samples.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank D. Swartzlander for help with the graphics and reviewers for their helpful comments. We acknowledge the High Performance Research Computing Center at the University of Texas, MD Anderson Cancer Center. This work was supported by National Institutes of Health grants R25CA057730 (to Y.A.J.), R01HG005855 (to P.S.), R01HG005859 (to P.S.), R01CA181244 (to P.S. and C.D.H.) and P30CA016672 (to MD Anderson) and by the following awards from the Cancer Prevention Research Institute of Texas: RP150079 (to H.K.) and RP160668 (to P.S.).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41587-019-0297-6.
Data availability
The results shown are based on data generated by the TCGA Research Network (http://cancergenome.nih.gov/). All datasets used in this work are available in public repositories (https://portal.gdc.cancer.gov/). A list of TCGA disease sites (Supplementary Table 1) and blood and NAT samples used for the analyses (including case IDs) are included (Supplementary Tables 4 and 5, respectively). Reported sCNAs with case IDs are available in Supplementary Tables (6, 7, 10, 11 and 19–21).
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41587-019-0297-6.
References
- 1.Freed D, Stevens EL & Pevsner J. Somatic mosaicism in the human genome. Genes (Basel) 5, 1064–1094 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forsberg LA, Gisselsson D. & Dumanski JP Mosaicism in health and disease—clones picking up speed. Nat. Rev. Genet 18, 128–142 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Machiela MJ & Chanock SJ The ageing genome, clonal mosaicism and chronic disease. Curr. Opin. Genet. Dev 42, 8–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martincorena I. & Campbell PJ Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Beroukhim R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor AM et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zack TI et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piotrowski A. et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum. Mutat 29, 1118–1124 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Aghili L, Foo J, DeGregori J. & De S. Patterns of somatically acquired amplifications and deletions in apparently normal tissues of ovarian cancer patients. Cell Rep. 7, 1310–1319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genovese G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie M. et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakubek Y. et al. Genomic landscape established by allelic imbalance in the cancerization field of a normal appearing airway. Cancer Res. 76, 3676–3683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtius K, Wright NA & Graham TA An evolutionary perspective on field cancerization. Nat. Rev. Cancer 18, 19–32 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Kadara H. et al. Driver mutations in normal airway epithelium elucidate spatiotemporal resolution of lung cancer. Am. J. Respir. Crit. Care Med 200, 742–750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yadav VK, DeGregori J. & De S. The landscape of somatic mutations in protein coding genes in apparently benign human tissues carries signatures of relaxed purifying selection. Nucleic Acids Res. 44, 2075–2084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martincorena I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martincorena I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore L. et al. The mutational landscape of normal human endometrial epithelium. Preprint at 10.1101/505685v1 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Suda K. et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 24, 1777–1789 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Lee-Six H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Preprint at 10.1101/416800v1 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Lee-Six H. et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurie CC et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet 44, 642–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs KB et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet 44, 651–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machiela MJ et al. Characterization of large structural genetic mosaicism in human autosomes. Am. J. Hum. Genet 96, 487–497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vattathil S. & Scheet P. Extensive hidden genomic mosaicism revealed in normal tissue. Am. J. Hum. Genet 98, 571–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh PR et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 559, 350–355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machiela MJ et al. Female chromosome X mosaicism is age-related and preferentially affects the inactivated X chromosome. Nat. Commun 7, 11843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vattathil S. & Scheet P. Haplotype-based profiling of subtle allelic imbalance with SNP arrays. Genome Res. 23, 152–158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.San Lucas FA et al. Rapid and powerful detection of subtle allelic imbalance from exome sequencing data with hapLOHseq. Bioinformatics 32, 3015–3017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pounds S. et al. A genomic random interval model for statistical analysis of genomic lesion data. Bioinformatics 29, 2088–2095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamal-Hanjani M. et al. Tracking the evolution of non-small-cell lung cancer. New Engl. J. Med 376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Jakubek YA, San Lucas FA & Scheet P. Directional allelic imbalance profiling and visualization from multi-sample data with RECUR. Bioinformatics 35, 2300–2302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gausachs M. et al. Mutational heterogeneity in APC and KRAS arises at the crypt level and leads to polyclonality in early colorectal tumorigenesis. Clin. cancer Res 23, 5936–5947 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martincorena I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machiela MJ et al. Detectible mosaic truncating PPM1D mutations, age and breast cancer risk. J. Hum. Genet 64, 545–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garnis C. et al. Genomic imbalances in precancerous tissues signal oral cancer risk. Mol. Cancer 8, 50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogan BL et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aran D. et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun 8, 1077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura Y. et al. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Mod. Pathol 17, 1328–1337 (2004). [DOI] [PubMed] [Google Scholar]
- 40.The International Stem Cell Initiative Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol 29, 1132–1144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sivakumar S. et al. Genomic landscape of allelic imbalance in premalignant atypical adenomatous hyperplasias of the lung. EBioMedicine 42, 296–303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 42.Korn JM et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet 40, 1253–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Willer CJ, Ding J, Scheet P. & Abecasis GR MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol 34, 816–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fowler J, San Lucas FA & Scheet P. System for quality-assured data analysis: Flexible, reproducible scientific workflows. Genet. Epidemiol 43, 227–237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cibulskis K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lek M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey MH et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.