Abstract
Exosomes contain cargoes of proteins, lipids, micro-ribonucleic acids, and functional messenger RNAs, and they play a key role in cell-to-cell communication and hold valuable information about biological processes such as disease pathology. To harvest their potentials in disease diagnostics, prognostics, and therapeutics, exosome isolation is a crucial first step in providing pure and intact samples for both research and clinical purposes. Unfortunately, conventional methods for exosome separation suffer from low purity, low capture efficiency, long processing time, large sample volume requirement, the need for dedicated equipment and trained personnel, and high cost. In the last decade, microfluidic devices, especially those that incorporate nanostructures, have emerged as superior alternatives for exosome isolation and detection. In this review, we examine microfluidic platforms, dividing them into six categories based on their capture mechanisms: passive-structure-based affinity, immunomagnetic-based affinity, filtration, acoustofluidics, electrokinetics, and optofluidics. Here, we start out exploring the research and clinical needs that translate into important performance parameters for new exosome isolation designs. Then, we briefly introduce the conventional methods and discuss how their failure to meet those performance standards sparks an intense interest in microfluidic device innovations. The essence of this review is to lead an in-depth discussion on not only the technicality of those microfluidic platforms, but also their strengths and weaknesses with regards to the performance parameters set forth. To close the conversation, we call for the inclusion of exosome confirmation and contamination evaluation as part of future device development and performance assessment process, so that collectively, efforts towards microfluidics and nanotechnology for exosome isolation and analysis may soon see the light of real-world applications.
Keywords: microfluidics, exosomes, lab on a chip, cancer, liquid biopsy, nanotechnology
1. Introduction
Over the last decade, exosomes have received much attention as potential disease biomarkers for diagnostics and prognostics applications. Secreted by all cell types in culture, and found abundantly in biofluids such as blood, urine, and saliva, exosome is a subtype of extracellular vesicles (EVs) released through multivesicular bodies (MVBs) in the endosomal pathway [1–3]. Other EV subtypes include microvesicles (MVs) (100–1000 nm) produced by the imbalances of lipids distributed on the plasma membrane, and apoptotic bodies (>1000 nm) released by apoptotic cells [1, 4]. Exosomes, typically 30–200 nm in size, are membrane-bound carriers with cargoes of proteins, lipids, and nucleic acids such as micro-ribonucleic acids (miRNAs), functional messenger RNAs (mRNAs), and deoxyribonucleic acids (DNAs) as shown in figure 1 [4, 5]. The cargoes of exosomes reflect the cells of origin and their current state [3]. It is believed that exosomes perform key functions in intercellular communication, and therefore, hold valuable information about biological and pathological processes [6–8]. For diseases such as cancer, where early detection and continued disease monitoring are of crucial importance to chance of survival, the information provided by tumor-derived exosomes is an indispensable, molecular-level tumor assessment in a less invasive and more repeatable manner than the gold-standard, tissue biopsy [2].
Figure 1.
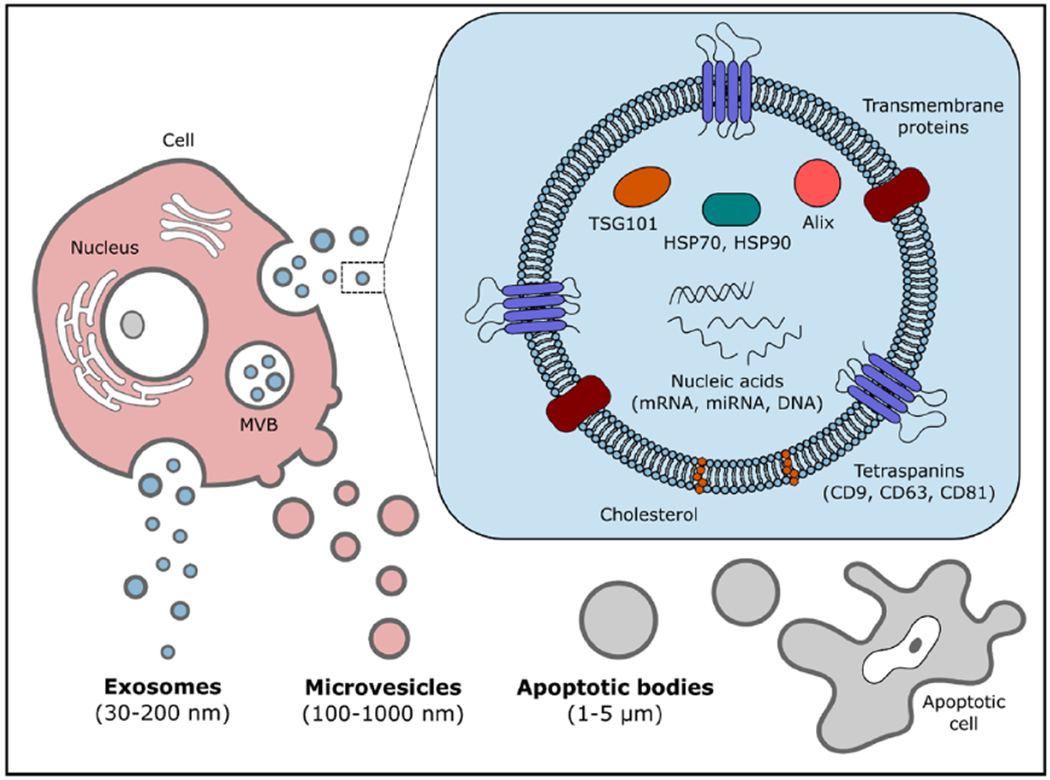
EVs include apoptotic bodies, MVs, and exosomes, which are distinguished from each other based on their origin and size. Exosomes are often categorized as being 30–200 nm in size, containing valuable cargoes of proteins, lipids, and nucleic acids from their parent cells.
Even though the potentials of exosomes are exciting, our current knowledge of the function of this species remain limited. One major reason for this stall in research progress is the immense challenges associated with isolating pure and intact exosomes from biofluids (which are complicated in composition) in a standardized and efficient manner [1, 9, 10]. Even with the limited knowledge we have, the translation of prognostic and diagnostic powers of exosomes to the clinical settings face great difficulties. Along with the need to isolate pure and intact exosomes, clinical technologies also need to provide high capture efficiency at high throughput with high specificity, minimal sample preparation, ease of operation, and low operation time [11, 12].
Currently, conventional methods for exosome isolation include ultracentrifugation, ultrafiltration, polymer-based precipitation, and immunoaffinity-based separation. Unfortunately, as explained in a subsequent section, drawbacks of these conventional methods include the need for expensive instruments such as an ultracentrifuge, prolonged processing time (>4 h), expensive commercial kits and reagents, skilled personnel to handle the equipment and complete laborious protocols, along with hurdles in low isolation efficiency, possible protein contamination, large starting volume and sample loss during the long process [9, 13].
Progresses in microfabrication techniques and nanotechnology have led to the rise of microfluidic devices and the incorporation of nanostructures for isolating the exosomes. From microfluidics, researchers leverage small reagent volumes, reduced analysis cost, feasible integration with accessories/detectors, and large surface-to-volume ratio for surface interactions to design microfluidic devices providing higher isolation efficiency, throughput, and purity, while ensuring a simpler handling, less cumbersome device or equipment [1, 2, 8]. From nanotechnology, researchers create nanoscale multi-functional materials or structures delivering superior sensitivity and specificity while maintaining low cost, portability, and user-friendly operation [14–16]. Together, microfluidics and nanotechnology display advantages that are incredibly important in providing satisfactory exosomes samples for further research applications and clinical use. Thus, while exosome isolation using microfluidic systems is a field still in its infancy, the merging of microfluidics and nanotechnology has catapulted encouraging leaps that deem it a field worthy of recognition and future research efforts.
While there has been a number of reviews already written on the topic of microfluidic platforms for exosome isolation and analysis, most of them have focused more heavily on informing about the technologies, and much less on pointing out how each technology could be improved to take us closer to unlocking the immense potentials of exosomes both in research labs and in clinics [7, 8, 11, 17]. We feel that, with the need for more research in the field, it is important to critically discuss each technology to both commend the novel ideas and point out areas that need improvement. In addition to purely discussing microfluidic systems, we also recognize the emergence of nanostructures being incorporated in these platforms to achieve improved capabilities for nanoscale exosomes.
In this review, we start out with explanations on the performance parameters needed in exosome isolation method evaluations. We then introduce in brief the conventional exosome separation approaches, noting their advantages and drawbacks. The focus of this review will be the in-depth discussion on the various microfluidic platforms, highlighting novel design aspects and accomplishments, and commenting on the integration of subsequent analyses, if applicable. More uniquely, we emphasize areas needing future development with regards to performance parameters critical to enabling exosome research and clinical translation. We end our review with a highlight on the immediate needs for post-isolation confirmation of exosomes and evaluation of contamination using contemporary detection and analysis techniques. Even though the development of microfluidic platforms for exosomes has only started roughly a decade ago, there are numerous platforms already achieving commendable steps forward. With this review, we aim to provide a critical overview of the achievements of the field, and more importantly, reveal to aspiring researchers the possible directions for future device development.
2. Parameters to assess performance of exosome isolation platforms
The design of new exosome isolation platforms should be guided by the end goals of the research. For basic research and clinical translation, the end goals of exosome-based discovery and diagnostics lead to a set of commonly reported parameters used to assess and compare the performance of exosome isolation platforms, including capture efficiency, purity, throughput, and release efficiency. Other criteria that are less often reported include sensitivity, reproducibility, minimal sample preparation, cost of operation, and ease of operation. Realistically, a new design should have a prioritized list of performance requirements to meet the specified research goals. For basic research purposes where the analysis of exosomal RNA cargo, proteomics, lipidomics, and other characteristic dominate, an isolation platform must prioritize high reproducibility, high purity, and high sensitivity [18]. For clinical diagnostic applications where correlations between biomarkers and disease must be clearly established in an efficient manner, an isolation platform must favor high specificity, high sensitivity, high throughput, minimal sample preparation, reduced cost, and ease of operation [9, 10, 19]. In most studies that reported values for these parameters, either exosomes isolated from cell culture supernatant using ultracentrifugation, or other exosome-sized beads, were spiked into healthy bodily fluid or buffered saline for evaluation. In this section, we provide the definition for each performance parameter to lay the common ground for forth-coming conversations on exosome isolation technologies presented in this review.
Capture efficiency, or isolation efficiency, is defined as the number of exosomes captured, or isolated, over the number of exosomes spiked into the sample fluid (equation 1). In other words, capture efficiency is a measure indicating the sensitivity of a method. While high capture efficiency is a parameter extremely crucial for isolating other types of liquid biopsy markers such as circulating tumor cells, it is not of utmost importance for exosome isolation due to the abundance of this species in many bodily fluids [20].
Purity measures the degree of non-contamination of an isolated population. Besides from the non-exosomal EV species in biofluids, there are often other contaminants such as lipoproteins or proteins found in isolated exosome samples [18]. In other words, purity is a measure indicating the specificity of an isolation method. In this review paper, the terms purity and specificity are used interchangeably, unless noted otherwise for a particular isolation platform being introduced. Purity, therefore, is the fraction of exosomes (or a specific targeted subtype of exosomes) captured among the entire heterogeneous ingredients obtained from isolation (equation 2).
Throughput describes how much sample a device can process in a certain amount of time. Although throughput is often reported as sample volume processed per unit time (i.e., flow rate), we believe it is more useful to present both the sample input volume and total processing (or analysis) time as parameters for comparing between different platforms. Presenting these two parameters instead of reporting a single value can highlight the difference between a device designed to handle small volumes and a device for large sample volumes. In addition to the flow rate, the sample throughput should also consider the number of samples processed in a unit time.
Release efficiency depicts the fraction of exosomes released from the device over the total number of exosomes immobilized on the device (equation 3). Though not explicitly stated as a performance requirement for any research end goals, this criterion is of high importance especially for basic research purposes to allow for subsequent downstream analyses of isolated exosomes.
Among the less-often reported parameters, sensitivity and/or limit of detection (LOD) are sometimes reported, while minimal sample preparation is more subtly demonstrated by the type of sample the platform can handle (e.g., whole blood requires almost no preparation, while plasma requires centrifugation steps). It is also important to mention that inconsistencies in the nomenclature for performance parameters do exist. For example, the term recovery refers to the capture or isolation efficiency in some publications [6, 21, 22], but describes the release efficiency in others [12, 23]. These nomenclature inconsistencies may lead to misinterpretation of data or incorrect assessment across different isolation technologies. Therefore, we believe that it is better to adhere to two separate terms (capture efficiency and release efficiency) for their respective definitions. To be even more helpful, each new technology should clearly state the definitions of the performance parameter values in publications.
3. Conventional exosome isolation techniques
In this section, we briefly introduce conventional exosome isolation techniques employed for exosome isolation: ultracentrifugation, ultrafiltration, polymer-based precipitation, and immunoaffinity-based capture. Although achieving some success in the late 1990s and first decade of this century, there are still limitations on the quality of isolated exosomes and the burden of operation that hinder the utilization of these technologies in the clinical or research settings. Here, we briefly discuss basic characteristics of each technique, along with examples of implementation in studies, and commercially available platforms.
Ultracentrifugation, often considered the ‘gold standard’ for exosome isolation, consists of a lengthy series of steps that involves a high-capacity ultracentrifuge rotor operated by a trained personnel. There are two main types of ultracentrifugation often used for exosome isolation: differential and gradient density. Both processes follow the similar protocol of successively eliminating dead cells, cell debris, and contaminating proteins based on size as the centrifugation speed is increased to at least 100 000× g [24, 25]. The main difference between the two processes is the incorporation of a sucrose gradient in gradient density ultracentrifugation which ultimately improves the purity [24]. While the pellet obtained at the end of differential ultracentrifugation will often contain proteins or other non-exosomal aggregates, gradient density ultracentrifugation has been shown by Greening et al to provide isolated exosomes of superior quality [18, 25]. In general, ultracentrifugation often requires a large volume to start with, and a running time of at least 4 h, making it unsuitable for clinical samples [7, 9]. The isolation efficiency of ultracentrifugation, which has been reported to range from 5%–25%, is largely dependent on the starting volume and sample density [24, 26].
A faster isolation method that is often used in combination with ultracentrifugation is ultrafiltration, also a size-based technique. Ultrafiltration often requires the use of physical barriers (e.g. membranes) with a certain pore size to exclude particle larger than the pore size. For example, membranes with 800 nm pore size are often used to eliminate cells and larger EVs, while those with 200 nm pores are used for separating exosomes [18]. After filtration, centrifugation at high speeds is often employed to wash proteins off the isolated exosomes. In a study done by Cheruvanky et al, commercial Vivaspin® polyethersulfone nanomembrane with a pore size of 13 nm was used to isolate exosomes from urine samples [27]. Unfortunately, while being faster and simpler to conduct than ultracentrifugation, ultrafiltration often suffers from clogging of the membrane pores and is suspected to cause significant shear stress on the exosomes [7, 18].
Instead of exploiting size property of exosome, polymer-based precipitation operates on differential solubility by which an added precipitation reagent creates sediments ready to be collected using centrifugation or filtration. Some commercially available examples include Exo-spin™, ExoQuick™ Exosome precipitation, and Invitrogen™ Total Exosome Isolation Reagent Isolation Kit. Though leading to better isolation efficiency compared to ultracentrifugation and ultrafiltration while requiring little hands-on effort from users, one main drawback of precipitation is the risk of compromising the purity and integrity of isolated exosomes due to the presence of the added reagents leading to the formation of polymer matrices [3, 17]. While using precipitation to isolate exosomes from serum, Vlassov et al observed low specificity in the results and difficulty in resuspending the isolated pellet [3]. It is also important to note that the above three methods, ultracentrifugation, ultrafiltration, and precipitation, do more to enrich or concentrate the exosomes in the biofluids, than to isolate them from the biofluids. Therefore, subsequent washing or purifying steps are often recommended for these conventional methods to prevent contaminations.
Of all the conventional isolation methods, perhaps the one that can offer the highest purity is the affinity-based capture method. Taking advantage of the surface proteins present on exosomes, generally accepted exosome biomarkers such as CD9, CD63, CD81, major histocompatibility complex (MHC) class I and II, and epithelial cell adhesion molecule (EpCAM) are often immobilized onto magnetic beads or other matrices to capture exosomes. Clayton et al demonstrated the use of Dynal® paramagnetic beads coated with MHC class II antibodies to isolate exosomes from cell culture supernatant [28]. However, the use of magnetic beads for capturing often leads to the burden of detaching exosomes from them. Furthermore, while the use of surface biomarkers for capturing results in high specificity, there could potentially be problems with a low yield or biased selection due to desired markers not being uniformly present or being ubiquitously present on all exosomes [18, 29].
4. Microfluidics for exosomes isolation
Microfluidic devices for exosome isolation offer many advantages over the conventional methods. Along with the miniaturization of the device comes the benefits of a smaller required sample volume, lower reagent consumption, higher throughput, and higher sensitivity [11, 30]. Additionally, technology development enables the integration of multiple assays, sensors, and other components onto the same chip, eliminating the need for a fully-equipped laboratory as in the case of conventional methods [31].
Microfluidic technologies for exosome isolation can be categorized based on the isolation mechanism using either biological properties or physical properties. The primary biological properties of interest are immunoaffinity and aptamer-enabled specific interactions. The corresponding microfluidic platforms rely on interactions between the capture agents in the device and the surface biomarkers on exosomes, leading to highly specific and pure isolation. The physical properties used for exosome isolation include size, density, and electrical properties. These microfluidic systems rely on filtration, and external acoustic, electrical, and optical forces. Table 1 summarizes the categories of exosome isolation devices and exosome characterization methods introduced throughout this review paper.
Table 1.
Summary of exosome isolation and analysis strategies.
| Isolation methods | Isolation mechanisms | Exosome characterization methods |
|---|---|---|
| Biological properties (e.g., antibodies, lipids) Physical properties (e.g., size) |
• Passive micro- and nanostructures • Immunomagnetic particles • Acoustofluidics • Electrokinetics • Optofluidics |
• Physical properties (e.g., size): ◦Nanoparticle tracking analysis ◦ Electron microscopy • Chemical properties (e.g., proteins): ◦ Bradford assay ◦ Western blot ◦ Enzyme-linked immunosorbent assay ◦ Polymerase chain reaction |
4.1. Biological-property-based microfluidic exosome isolation techniques
Biological-property-based, or affinity-based, exosome separation methods primarily depend on the affinity interactions between immobilized capture agents (e.g. antibodies or aptamers) and exosomes that possess the corresponding surface biomarkers. In microfluidic devices, high surface-area-to-volume ratio is advantageous to their interactions, leading to higher exosome capture. Without many exceptions, most affinity-based exosome isolation devices have utilized surface proteins considered ‘universal’ exosome biomarkers as capture agents. From proteomic studies, it has been reported that although the molecular composition of exosomes depends on the cellular origin, there is a conserved set of exosomal proteins across all cell types which include the tetraspanins CD63, CD81, CD82, CD9, the MVB-related protein TSG101, the heat-shock protein HSP70, and the MHC class I molecules, among others [32–34]. Of these, the surface proteins gaining popular implementation within microfluidic exosome isolation platforms are the CD63, CD81, and CD9, as will be seen in this section.
While affinity-based methods offer high purity, these advantages come at the price of needing to identify appropriate capture agents. These potential capture agents would need to target surface markers that are specific enough to the exosome subpopulation of interest and have an expression level high enough to generate an acceptable binding efficiency [11]. Besides these biomolecular concerns, there are two physics phenomena that put limits on these affinity-based capturing: (a) limit of mass transfer due to the extremely low diffusion/convection ratio in microchannels deterring exosomes from ever coming into contact with the capture agents conjugated on the channel walls, and (b) hydrodynamic resistance as the exosomes in a liquid approach a solid surface [16, 35].
In next two sections, we will see how the affinity-based microfluidic devices have, partially or completely, overcome these physical limits with creative approaches such as fabricating microstructures that induce chaotic mixing, incorporating beads to increase surface area, and utilizing porous materials to reduce near-surface resistance. We divide affinity-based devices into two subcategories based on the location of conjugated capture agents: (a) microfluidic devices with passive structures conjugated with capture agents, and (b) microfluidic devices incorporating magnetic particles conjugated with capture agents.
4.1.1. Exosome isolation with passive micro- and nanostructures
As mentioned previously, the performance of affinity-based isolation is governed by the mass transfer of targets to the surface-conjugated capture agents and the binding efficiency [16]. The miniaturization aspect of microfluidics and nanotechnology offer the high surface-to-volume ratio to produce the large surface area on which these types of devices capitalize. In addition to this inherent advantage, innovative designs of passive micro- and nanostructures in the microchannels are often required to increase mass transfer and reduce near-surface hydrodynamic resistance [16, 36]. Passive-structure-based isolation, along with filtration isolation, boast simple handing of the devices by eliminating the hassles of incorporating magnetic particles for capturing, or actively controlling isolation parameters as will be required for acoustic, electrical, and optical isolation [23, 37].
The earliest known microfluidic work in isolating exosomes was the platform reported by Irimia and his collaborators in 2010 [38]. Expanding on the idea of using herringbone grooves to induce chaotic mixing as introduced in 2002 by Whitesides’ research group [36], this device incorporated herringbone structures on the ceiling of microfluidic channels to increase the surface area, and therefore, enhance the contact of exosomes with anti-CD63-conjugated surfaces. This platform boasted the ability to process 100–400 μl of human glioblastoma or healthy serum in under an hour. Downstream analyses included the use of scanning electron microscopy (SEM) for confirming intact exosome morphology and size, and reverse transcription polymerase chain reaction (RT-PCR) for total RNA count and specific mutation. Even though the purity of isolated population was not investigated and its capture efficiency of 42%–94% prompted further validation, this pioneering work probed the immense advantages of microfluidic platforms over conventional methods in isolating stable disease-specific exosomes and exosomal RNA for diagnosis and monitoring potentials.
The initial demonstration of microfluidics for exosomes paved the way for other innovative immunoaffinity-based platforms such as the ExoChip [39]. Like the previous device, the ExoChip also enhanced the particle-surface contact by extending contact time between sample and surface-immobilized anti-CD63 antibodies. However, instead of adding microstructures to achieve that goal, Nagrath’s research group designed a series of big circular chambers connected together by straight narrow channels—two geometric features that respectively reduced the velocity of incoming exosomes fluid stream to extend the interaction time for immune-capture, and accelerated the exosomes to induce mixing. The ExoChip was able to process 400 μl of undiluted serum samples from pancreatic cancer patients and healthy patients in about 1 h. It is worth noting that the ExoChip design was the first integrated platform to not only successfully provide isolation capability using anti-CD63 antibodies, but also easy on-chip detection using the fluorescence DiO dye, and quantification using a standard plate reader. Subsequent electron microscopy (EM) immune-detection with Rab5 protein, Western blotting, and RT-PCR were also performed on-chip to confirm physical traits and analyze protein and miRNA contents. Although future studies to assess other performance values such as capture efficiency and purity should be done to further increase the credibility of this technology, the convenience of standardized on-chip detection, quantification, and confirmation prove the ExoChip a worthy candidate for a comprehensive clinical diagnostic tool.
Following up with the work on ExoChip, in 2019, Nagrath and her coworkers introduced newExoChip, the lipid-affinity-based microfluidic device for exosomes isolation serving both clinical and research purposes [40]. Being one of the few affinity-based platforms that strayed away from using a tetraspanin (e.g. CD63, CD81, and CD9) as a capture agent, the newExoChip showed a possibility of obtaining greater enrichment of certain lipids, such as phosphatidylserine, in exosomes comparing to cells, and specifically in cancer cell-derived exosomes comparing to normal cell-derived exosomes. Featuring 225 times more circular chambers than the previous ExoChip, and functionalized with annexin V proteins on the device surface, the newExoChip was catered towards cancers where CD63 is down-regulated (e.g. lung cancer or melanoma). Processing 30–100 μl of lung cancer, melanoma, or healthy plasma samples in under 10 min, the device performance was reported with a ~90% capture efficiency, ~75% purity, and ~85% release efficiency. A note of caution is that due to the capture target being exosomal lipids, the isolated population may only be a subset of exosomes representing some unknown biological conditions. Hence, further research and validation must be done on the significance of exosomal lipids before any meaningful clinical conclusions can be made on the exosomes isolated by the newExoChip.
Unlike the newExoChip that explored a different capture agent for affinity capture, the device introduced by Chen et al remained loyal to anti-CD63 antibodies, but explored a more complicated structural design featuring three-dimensional (3D) polydimethylsiloxane (PDMS) scaffolds coated with zinc oxide (ZnO) nanowires [41]. This chip improved capture efficiency by inducing chaotic mixing with the interconnected nanopores of 40–250 nm in size, increasing surface area for affinity capture with the nanowire forest, and providing exclusion-like effect with the nanowires of exosome-size spacing as shown in figure 2(A). To aim towards clinical applications, the chip demonstrated the isolation and detection of exosomes from 100 μl of cancer serum and plasma samples in about 2.5 h using the widely available colorimetric enzyme-linked immunosorbent assay (ELISA). To also cater towards basic research, the release of exosomes was achieved by dissolving the ZnO nanowires with 0.2 M sodium citrate buffer, resulting in ~80% of wires dissolved. Though a novel approach, no concrete values for capture efficiency, purity, release efficiency were reported. Also lacking in quality is the downstream analysis done on captured exosomes, which only consisted of SEM and transmission electron microscopy (TEM) for visualize the exosomes bound to nanowires and confirming the morphology.
Figure 2.
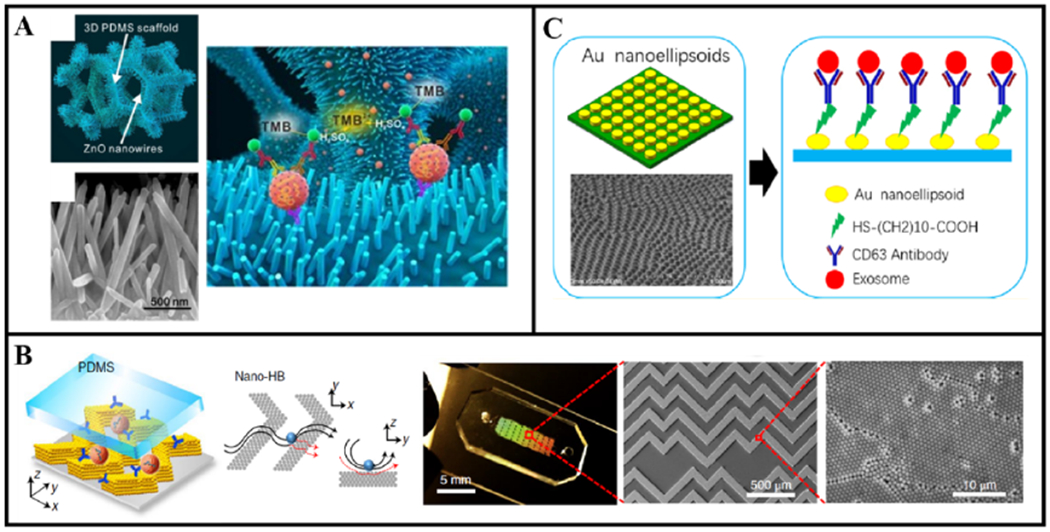
Examples of exosome isolation with passive micro- and nanostructures. (A) The porous 3D PDMS scaffolds coated with ZnO nanowires to provide exclusion-like effect for exosome capture. The two red spheres represent exosomes while TMB stands for 3,3′,5,5′-tetramethylbenzidine, which is for colorimetric detection. Reprinted from [41] with permission from Elsevier. (B) The nano-HB chip with antibody-conjugated nanoporous CNTs posts arranged in the herringbone pattern. Reprinted from [16] with permission from Springer Nature. (C) The low-cost LSPR-based microfluidic biosensor with antibody-conjugated gold nano-ellipsoid arrays. Reprinted from [45] with permission from American Chemical Society.
Also leveraging structural design to enhance affinity capture, Zeng and his colleagues combined the two previous ideas of (a) wires or microposts and (b) well-studied herringbone microstructures into a device featuring 3D nanoporous carbon-nanotube (CNT) posts arranged in the herringbone pattern (nano-HB) (figure 2(B)) [16]. With these physical features, the nano-HB device directly addressed the challenges associated with affinity capture: (a) CNT to increase surface area and enhance target-bioparticle/capture-agent binding efficiency, (b) herringbone structures to create chaotic mixing and enhance mass transfer, and (c) nanopores of CNT posts to drain the fluid and reduce the near-surface hydrodynamic resistance. Using anti-CD81 antibodies as capture agents, the 3D nano-HB technology only required 20 μl of sample to process in 40 min and successfully showed capture efficiencies ranging 80%–85% for various ovarian and breast cancer cell lines. For clinical applications, this device needed a mere 2 μl of ovarian cancer plasma sample diluted 10× in phosphate-buffered saline (PBS). Optimized for a remarkably low LOD of 10 vesicles/μl, the 3D nano-HB chip outcompeted conventional methods in detecting rare but powerful ovarian cancer tumor-related exosomes, such as FRα+ exosomes. Additionally, thorough characterization of the device such as quantifying the capture agent density on the surface of nano-HB, comparing the chaotic mixing performance of nano-HB to a solid-herringbone-patterned device with the nanopores filled, and assessing the permeability of the nanoporous CNT presented a convincing case supporting the achievement of the three goals mentioned above. One last point worth commending was the incorporation of colloidal self-assembly in a microfluidic device fabrication process called multiscale integration by designed self-assembly, removing the needs for expensive and time-consuming standard nanofabrication techniques. As this technology already seemed immensely promising for clinical translation, further studies on the release of exosomes from device and assessment on purity of isolated exosomes for basic research applications may be worth pursuing.
So far, almost all the discussed technologies focused on improving abilities such as on-chip isolation, on-chip detection, and on-chip analysis—abilities critical for clinical translation. In 2018, Hansford and his coworkers saw a critical need for intact and pure exosomes not just for clinical applications, but also for extensive downstream analysis needed in biomarker discovery of early ovarian cancer detection [42]. Inspired by affinity chromatography, their team designed a microfluidic platform that conjugated CD9 or EpCAM antibodies onto the herringbone-patterned surface using covalent bonding, and enabled the release of label-free exosomes using glycine-hydrochloric acid (Glycine-HCl) buffer. This elution technique disrupted the antigen–antibody bond to cleave label-free exosomes, as opposed to other affinity-based approaches which release exosomes-antibody or exosome-magnetic beads pairs with some surface receptors blocked. The microfluidic platform isolated and eluted exosomes from 100 μl of healthy and high-grade serous ovarian cancer (HGSOC) serum samples in under 20 min, achieving a ~60% elution of isolated exosomes. The group also went as far as monitoring the internalization of these eluted exosomes by OVCAR8 cells, proving that those exosomes were indeed intact and biologically active. In a follow-up study in 2019, the same research group demonstrated an in-depth protein analysis of exosomes eluted from this platform to identify HGF, STAT3, and IL6 as being highly elevated in early-stage HGSOC patients, suggesting the potential of exosomes as early detection biomarkers [43]. The initial studies showed the promise of a powerful microfluidic platform serving both clinical and research settings; however, additional characterization regarding capture efficiency and purity, and an improvement upon the elution rate are points to confirm before this device can truly realize its impact.
Turning our attention away from platforms that intensely focused on ways to efficiently isolate exosomes, while relying on popular immunofluorescence or other biochemistry-based techniques for detection, we now look at microfluidic systems that integrate plasmonic sensors optimized for exosomes detection in bodily fluids. In 2014, recognizing the downsides of extensive time consumption by ultracentrifugation and large sample requirement for Western Blotting and ELISA, Lee’s research group diverged from conventions to develop an exosome detection and molecular profiling system called nanoplasmonic exosome assay (nPLEX) catered towards both clinical and research applications [44]. Consisting of arrays of periodic 200 nm diameter nanoholes patterned in 200 nm thick gold film, exosome detection was achieved based on either the shift in the localized surface plasmon resonance (LSPR) wavelength of the sensors (spectral detection) or the change in the intensity of the wavelength (intensity detection) induced by the attachment of the exosomes onto the conjugated antibodies. To prove its advantages over conventional methods, the nPLEX showed the LOD to be ~3000 exosomes, a sensitivity that is 104-fold higher than Western blot and 102-fold higher than ELISA, with a high correlation to protein level quantification by ELISA. The molecular profiling of ovarian cancer cell line-derived exosomes and benign cell-derived exosomes revealed EpCAM and CD24 as potential cancer biomarkers. With the capability of performing parallel detection of 12 protein markers in 150 μl samples in <30 min, the nPLEX offered rapid, highly sensitive, label-free, and real-time exosomes detection, quantification, and profiling. While initial clinical experiments on ovarian cancer ascites were performed to confirm diagnostic potential of EpCAM and CD24, no conclusion was reached due to a small patient cohort. With sensitive detection, high-throughput parallel profiling, and exosome recovery for subsequent quantitative PCR (qPCR) analysis, the innovative nPLEX platform demonstrated its worth for attention and future investigations from the research community. In addition to the proposed plan on improving device fabrication, signal amplification, and patient cohort, the clinical translation of the nPLEX will benefit from optimizations on the capture efficiency, purity, and release of exosomes bound to the nanoholes.
Even though Lee’s research group provided an innovative alternative for exosome detection, the high cost and complexity of plasmonic sensor fabrication deterred the concept from being widely implemented. Lv et al, claiming the lack of cost-effective and sensitive detection methods as reasons to hinder clinical translations, introduced the low-cost LSPR-based microfluidic biosensor that could be cost-effectively and simply constructed using an anodic aluminum oxide (AAO) template [45]. With the AAO method, the group fabricated gold nano-ellipsoid arrays on top of a quartz wafer, and conjugated anti-CD63 antibodies onto those ellipsoids (figure 2(C)). Operating on spectral detection, the low-cost LSPR-based biosensor could detect commercial COLO-1 exosomes in 50 μl samples in <4 h, achieving its translational goals of a low-cost option, rapid detection, low-sample requirement, and high sensitivity. As with the nPLEX, future experiments steps to enhance the potential adaptations of this low-cost technology could include characterizing the capture efficiency, purity, release efficiency, and further downstream analysis using biofluids and clinical samples.
One of the most recent exosome isolation microfluidic efforts involving nano-plasmonics is the templated plasmonics for exosomes (TPEX) device from Wu et al [46]. The TPEX is quite distinctive among all devices in this paper in the way that it targeted multiparametric profiling of exosomes—it detected exosomes based on both physical and biological properties. By tagging fluorescent aptamers of interest (e.g. anti-CD63) onto exosomes, and depositing gold nanoparticles (AuNP) that grew 9 nm thick nanoshells onto exosomes via electrostatic interactions, the TPEX technology could both detect specific aptamer binding via the fluorescence signal changes, and detect particles of diameters 30–150 nm via a unique resultant plasmonic resonance. This microfluidic chip featured serpentine channels for the mixing and incubation of the sample and reagents, along with a smartphone-based optical sensor system. For clinical applications, the TPEX processed 1 μl of colorectal and gastric cancer ascites samples in 15 min. Another unique feature of this nano-plasmonic technology was the selective detection of exosomes, without free-floating proteins, to provide a more meaningful analysis of exosome protein contents in patient samples. Future improvements for this technology could lie in the retrieval (or release) aspect of the detected exosomes for further exosome studies.
4.1.2. Exosome isolation with magnetic particles
Incorporating magnetic beads for affinity capture, also referred to as immunomagnetic isolation, is a popular technique implemented for exosome isolation both by conventional methods and in microfluidic devices. The wide scale of operation of this technique is partly due to its inherent ability to produce high magnetic field gradients even on the microscale [47]. On principle, it relies on permanent or electromagnetic coils to exert forces on magnetically-labeled bioparticles and offer highly efficient separation since all biological materials are diamagnetic or weakly magnetic [47–49]. For exosome isolation, magnetic beads conjugated with antibodies against a surface biomarker of interest are used for exosome capture and manipulation under a magnetic field.
Immunomagnetic-based isolation offers notable advantages over passive-structure-based affinity isolation. The use of beads inherently enhance capture efficiency with the larger surface area, while the flexibility in handling increase the recovery or release efficiency of isolated exosomes from a device to enable robust downstream analysis (e.g. simpler sample preparation for SEM or TEM imaging) [49, 50]. As will be seen in this section, many immunomagnetic-based platforms are somewhat more complicated in design as compared to passive-structure-based platforms. Many of them gravitate towards an integrated design, going beyond simple isolation to include separate chambers for washing, adding reagents, performing on-chip PCR, among other capabilities. However, an alarming downside to using immunomagnetic beads is the additional hassle of detaching intact exosomes from the recovered magnetic beads in the case that exosomes are needed for other purposes.
One of the earliest demonstrations of using magnetic forces for exosome separation from other biofluid components was the integrated platform for clinical application developed by He and her colleagues in 2014 [50]. Recognizing shortcomings of both conventional and microfluidic-based isolation and analysis methods in hindering exosome research and clinical investigation, the research team innovated a circuit-like platform that the promised sensitivity, short processing time, and small sample volume that conventional methods could not offer, while also enabling an on-chip intravesicular content analysis that microfluidic failed to address. In a cascading circuit setup, immunoisolation would first be performed by pre-mixing a plasma sample with antibody-labeled (either EpCAM or CA-125) magnetic beads, followed by exosome lysis and protein capture in a second chamber, and finalized with chemifluorescence detection. Testing its concept on 30 μl of non-small-cell lung cancer and ovarian cancer plasma samples, the device could perform its entire analysis in <1.5 h. The technology showed the ability to quantify five exosome subpopulations, detect proteins with 100-fold increase in sensitivity over ELISA, and isolate exosomes with a lower average size (<150 nm) compared to conventional ultracentrifugation. All these feats established this early work to be quite a revolutionary train of thoughts to be commercialized for the clinics. However, one clear downside to the isolation aspect of this technology was the need for off-chip preparation of the magnetic bead-plasma sample mixture. Along with further quantification of the capture efficiency and purity, improvements should be done on the sample introduction front to allow for the flexible scaling up of sample volume.
A few years after, He and her colleagues attacked this sample introduction problem with the ExoSearch (figure 3(A)), a continuous-flow microfluidic device that isolates exosomes from ovarian cancer blood plasma of volumes ranging 10 μl to 10 ml [51]. Magnetic beads (2.8 μm diameter, 0.1 mg ml−1) conjugated with common exosome markers (CD9, CD81, and CD63) or ovarian cancer-specific markers (EpCAM and CA-125) and a plasma sample were introduced separately into two channels at a Y-shaped entrance. Magnetic beads and the plasma sample uniformly mixed in a long serpentine channel, and beads that captured exosomes were retained at the end of the device by a replaceable magnet. Although this device can accommodate a wide range of sample volume as mentioned previously, this comes at the price of the balancing act between flow rate, hence processing time, and capture efficiency. The ExoSearch gave capture efficiency of 42%–97.3% for flow rates ranging from 50 to 104 nl min−1. At this flow rate of 1 μl min−1, ExoSearch could isolate exosomes from a 20 μl plasma sample in 20 min at a capture efficiency of 72%. The absence of other performance parameter values and downstream molecular analysis in this initial report could serve as starting points for follow-up studies on the ExoSearch.
Figure 3.

Examples of exosome isolation with magnetic particles. (A) The ExoSearch device induces mixing between exosomes and immunomagnetic beads, followed by the capture of bead-exosome with a magnet at the end of the device. Reproduced from [51] under Creative Commons license published by the Royal Society of Chemistry. (B) An exosome isolation device with a microvalve network for easy handling and eliminating the need for external equipment. Reprinted from [52] with the permission of AIP Publishing. (C) Immuno-magnetic exosome RNA (iMER) chip featuring an integrated platform performing operations from exosome isolation to qPCR analysis. Reproduced from [53] under Creative Commons license published by Springer Nature.
Following the similar idea of immunomagnetic bead exosome separation, Chen et al developed an integrated microfluidic device featuring a valve network targeting clinical breast cancer exosome isolation and detection (figure 3(B)) [52]. Magnetic beads (1–1.5 μm, 5 × 107 particles ml−1) conjugate with EpCAM antibodies were incubated with prepared plasma sample before introducing into a well in the microfluidic device. A microvalve network was operated to release the immunomagnetic bead-sample mixture into the reaction chamber, which houses a replaceable magnet, for the capturing and detection of magnetic beads bounded with exosomes. The entire process of isolation and detection for a 2 μl plasma sample was completed in 1.5 h at a capture efficiency of 74.2%. Although the demonstrated capture efficiency was similar to that of the previously mentioned immunomagnetic exosome isolation device, this device features more flexibility over fluid control due to the use of microvalves. With the inherent innovative integrated design, the translational power of this platform will greatly benefit from additional confirmation of exosomes and evaluation of contamination.
Another magnetic-based microfluidic platform that integrated exosome isolation and detection on the same chip is the ExoPCD-chip developed by Xu et al for liver cancer diagnostics [54]. However, rather than using magnetic force to immobilize exosome-bound immunomagnetic beads towards the outlet of the device, the ExoPCD-chip first flowed phosphatidylserine-Tim4 protein-conjugated magnetic beads (0.15 mg) into device and immobilized the beads onto 70 μm high Y-shaped micropillars using a permanent magnet, before introducing serum sample into the device for exosome isolation. The Y-shaped micropillars induced anisotropic flow and mixing to enhance interactions, and thus, binding, between the biofluid sample and protein-conjugated magnetic beads. As with other devices whose immunomagnetic-based capture efficiency is affected by the flow rate, the ExoPCD-chip had to choose an optimal rate of 0.2 μl min−1. The ExoPCD-chip can process a human serum sample of 30 μl within 3.5 h at a 68.5% capture efficiency, a slightly lower efficiency than previously presented magnetic-based microfluidic devices.
Even though integrated microfluidic platforms featuring multiple operations have been discussed, it is worth noting the effort made by Lee and his collaborators in developing an all-inclusive device that combines exosome isolation, exosome RNA isolation, and exosome mRNA qPCR analysis, onto a single microfluidic chip [53]. This device, termed iMER, enriched for and analyzed exosomes from blood samples of glioblastoma multiforme (GBM) patients, serving as a prognosis tool (figure 3(C)). By functionalizing 3 μm microbeads with antibodies against epidermal growth factor receptor (EGFR) and incubating the microbeads with a cell-free serum sample, the device was able to use a magnet to isolate cancer-specific exosomes from a 100 μl sample at a 93% capture efficiency. The iMER device was able to process (from exosome isolation to qPCR analysis) a 100 μl cell-free sample in about 2 h, presenting promising potentials as a prognosis tool in monitoring disease progress and assessing drug resistance in GBM patients.
It should be noted that technologies other than those discussed above have been explored for biological-property-based exosome isolation. For example, Sun and her colleagues used laser-induced thermophoresis for profiling EVs for cancer detection and classification [55]. A serum sample was diluted and then mixed with a panel of seven fluorescent aptamers that are specific for cancer biomarkers such as EpCAM, followed by thermophoretic enrichment and linear discriminant analysis. The team applied the method to a cohort of 102 patients with six cancer types at stages I–IV, and they detected stage I cancers with 95% sensitivity and 100% specificity. For this platform, sensitivity was defined as the true positive rate, and specificity was defined as the true negative rate.
4.2. Physical-property-based microfluidic exosome isolation techniques
While biological-property-based isolation techniques exploit the biochemical properties of exosomes (e.g. surface markers) and use capture agents to isolate target-specific subpopulation, other techniques rely on physical properties (e.g. size, volume, electrical properties, and density) of exosomes and employ filtration and other external forces for isolating exosomes. As reported in literature, exosomes typically range from 30 to 200 nm [4, 5]. It was reported that the buoyant density of exosomes is 1.15–1.19 g ml−1 measured on a continuous sucrose gradient [24]. The zeta potential of exosomes from a plasma sample is about – 11 mV [56], and that of exosomes from MCF-7 breast cancer cell cultures is – 13.4 ± 4.12 mV [57].
Physical-property-based methods free themselves from the hassle of selecting an appropriate surface biomarker as in affinity-based methods. Other advantages to physical-property-based methods include the abilities to achieve a higher yield and non-biased isolation, and preserve the structure and molecular composition of exosomes [8]. However, a less biased, isolated population often comes at the price of a lower purity level due to non-specific isolation of exosomes that possess different identities but resulting in the same physical properties; therefore, further downstream analysis is recommended [49]. Hereafter, we will introduce microfluidic platforms for physical-property-based exosomes isolation in four categories: (a) filtration, (b) acoustofluidics, (c) electrokinetics, and (d) optofluidics. It is worth mentioning that concerns about filter clogging and exosome shear stress-induced deformation as seen in conventional filtration also apply to its microfluidic analog. For the remaining three categories (acoustofluidics, electrokinetics, and optofluidics) that employ external forces for isolation, questions have been raised about their possible internal damages to exosomes. Even though some acoustics platforms have stated to have operated under safe power conditions comparable with medical ultrasound technologies [6, 58], other platforms should also make sure that their operating conditions do not internally harm the bioparticles of interest.
4.2.1. Exosome isolation using filtration and microfluidics
Filtration, or more specifically in this review paper, size exclusion filtration, operates based on the size selection criteria [48]. As noted, a clear advantage of these platforms and surface-based platforms over all other methods is minimal intervention required from the operator, making these easy-to-use devices a clear front-runner in clinical translation. Generally, microfluidic devices incorporating filters for isolating bioparticles can be divided into two subcategories: (a) pillar-based filtration, which uses arrays of micro- or nano-posts, and (b) membrane-based filtration, which relies on nanomembranes [8]. Essentially, these filters allow for particles smaller than the distance between micro- and nano-posts or the pore size of the nanomembranes to pass through, while retaining or displacing those larger than the desired size. While conventional ultrafiltration and many filtration-based microfluidic systems face potential clogging issues and may compromise exosome integrity due to the applied pressure, microfluidic filtration platforms introduced hereafter overcame these setbacks.
In 2013, Zhang and his collaborators published a proof-of-concept on a microfluidic device with ciliated micropillars capable of hierarchical filtration (figure 4(A)) [23]. Determined to overcome the drawbacks of low recovery and protein contamination brought by conventional ultracentrifugation and the need for specific surface proteins for capture required in affinity-based microfluidic devices, the ciliated micropillars ultimately aimed to isolate exosomes from raw, unprocessed, biological fluids. Hierarchical filtration was achieved as micropillars with 900 nm spacings depleted cells, the porous silicon nanowires etched on the micropillars with 30–200 nm spacings excluded the submicron cellular debris while trapping the exosome-like particles, and the entire design let proteins and other small particles pass through. To test this multi-scale filtration concept, the study imitated exosomes with fluorescently labeled 83 nm and 120 nm liposomes, cellular debris with 500 nm fluorescently labeled polystyrene nanoparticles, and proteins with 7 nm fluorescein isothiocyanate labelled bovine serum albumin (FITC-BSA). The device processed 30 μl sample in 10 min, resulting in capture efficiencies of ~60% trapping of 83 nm liposomes, ~45% trapping of 120 nm liposomes, little retention of 500 nm particles, and almost no retention of the FITC-BSA. Being geared towards both clinical and research applications, the device allowed the release of trapped exosome-like particles for further downstream analysis upon dissolving the cilia with PBS. Future confirmation of the device performance using cell line-derived exosomes and clinical samples, as well as the incorporation of capture agents onto the cilia should be done to increase the credibility of this versatile idea and further realize its potential for bridging affinity capture with filtration.
Figure 4.
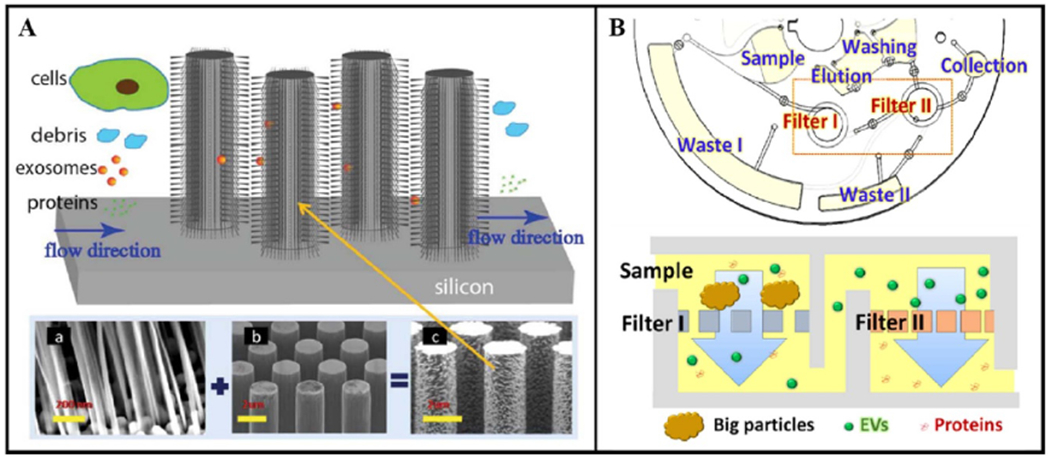
Examples of exosome isolation using filtration and microfluidics. (A) The silicon nanowires on micropillars perform hierarchical filtering to trap exosome-like particles. Republished from [23] with permission of Royal Society of Chemistry. (B) The Exodisc automates the enrichment and protein analysis of EV-size particles using two nanofilters on a disc and subsequent washing and analysis chambers. Adapted from [22] with permission of American Chemical Society.
Rather than employing hierarchical filtering, Wunsch et al applied the deterministic lateral displacement (DLD) to fabricate nanoscale DLD (nano-DLD) for exosome-sized particle sorting [59]. Characterizing nano-DLD arrays with pillars spaced 235 nm apart, they were able to show the complete displacement of 110 nm beads from 50 nm beads. Applying the nano-DLD technology to commercial urine-derived exosomes, these microfluidic devices could separate exosomes with size smaller than 100 nm from the original sample size of ~10 μl in 60 h. The initial characterization of this technology had shown the breakthrough fractioning of particles down to the size of 20 nm. With future incorporation of clinical samples, additional characterization of purity and exosome properties, and improvement on processing time, nano-DLD could be a concept for some nano-bioparticle isolation platforms.
Unlike the above pillar-based microfluidic platforms designed purely for exosome-sized particle isolation, the Exodisc introduced by Woo et al incorporated nanomembranes and demonstrated comprehensive on-disc EV isolation, recovery or release for downstream analysis, and on-chip ELISA for protein detection [22]. This integrated centrifugal microfluidic device was built on a three-layered polystyrene disc, equipped with two nanofilters (600 nm to filter large particles and 20 nm to enrich EVs), washing chambers, and recovery chambers (figure 4(B)). Countering the downsides of laborious process and needing an ultracentrifuge for conventional methods, biased selection in immunoaffinity-based methods, and potential damage in filtration, the Exodisc was designed for table-top centrifuging, label-free EV enrichment, and gentle spin speed (max 500 g). It boasted the ability to process 1 ml of bladder cancer urine samples in <60 min with a >95% isolation efficiency for urinary EVs. Nanoparticle tracking analysis (NTA) and RT-PCR were used for recovery efficiency calculation, while TEM was used to confirm intact EVs, and ELISA was performed to study protein expression of the unbiased enriched population. Purity, herein defined as number of particles per total proteins, was reported as (0.5 ± 16.3) × 107 particles (μg protein)−1. Further improvements to make this device capable of enriching only exosomes would make this technology a useful device not only in the clinics, but also in research lab.
Also following the nanomembrane-based approach, Demirci and his colleagues introduced the ExoTIC device which featured a nanoporous filter membrane housed within a leak-free plastic housing [60]. Using a 50 nm membrane pore size, the device reportedly isolated >90% of EVs present in cell culture media. The performance was also validated by processing 100 μl samples of healthy human plasma and lung cancer plasma, urine, and lavage in about 1 h. A unique advantage of this technology is the flexibility in connecting multiple ExoTIC devices together in series, where subsequent filtering using nanomembranes of pore sizes 30–200 nm can fractionate EV subpopulations. The team also provided an extensive performance comparison between the ExoTIC and ultracentrifugation, and they concluded the superiority of the ExoTIC to provide higher EV isolation efficiency and higher yields for both miRNA and proteins. In addition to the satisfactory performance, simple handling and low-cost manufacturing of ExoTIC made it an attractive technology for both research and especially for point-of-care settings. However, the pre-treatment of clinical samples that included both dilution and filtering may be points worth simplifying to prevent possible sample loss.
4.2.2. Exosome isolation enabled by acoustofluidics
While filtration offered reagent-free isolation, acoustic-based isolation went an extra step to provide a reagent-free and contact-free approach. The integration of acoustics into microfluidics is termed acoustofluidics. Acoustofluidic devices operate based on the principle that particles in an acoustic field experience radiation forces that push them away from the pressure antinodes and toward the pressure nodes, and a counteracting Stokes drag force [6, 21]. The acoustic radiation force is proportional to the volume, while the drag force is proportional to the radius. Therefore, larger particles (e.g. blood cells) under acoustic waves get deflected from their paths more than smaller particles (e.g. exosomes). Although acoustofluidic techniques have been used for micronscale particle separation, a major challenge associated with translating acoustics to exosome separation has been the high radiation force required to manipulate particles as small as exosomes [21, 37]. Nonetheless, there has been commendable progress in applying acoustofluidic principles to exosome isolation.
Lee and his colleagues used acoustofluidics for the separation of MVs. The device, referred to as an acoustic nanofilter, uses a symmetric standing surface acoustic wave to separate cell culture-derived exosomes from larger MVs [21]. Structurally, the device featured one channel for introducing centered sample flow and two side channels for introducing sheath flows. By strategically choosing the acoustic wavelength (60 μm) to place the pressure nodes outside the center flow path, the device allowed for the sheath flows to carry away particles exceeding the desired cutoff size (dc) that had migrated towards the pressure nodes, while retaining the exosome-sized particles with the center flow (figure 5(A)). Aiming to isolate vesicles for both basic research and clinical use, separation of exosomes sized <200 nm was achieved at a capture efficiency of >80% by choosing 300 nm for dc, 1.5 W for acoustic power, and 1.5 mm s−1 for flow speed. This acoustic nanofilter device only required a small sample size of 10 μl, making this an advantage when analyzing biofluid samples of limited quantity such as cerebrospinal fluids. However, no throughput or purity values were reported. Additionally, future post-isolation analysis such as SEM/TEM imaging to assess the successful recovery of intact exosomes from this device may further increase its credibility.
Figure 5.

Examples of exosome isolation enabled by acoustofluidics. (A) The acoustic nanofilter setup and a diagram showing the standing acoustic waves depleting non-exosome particles by directing towards the pressure nodes. Adapted from [21] with permission of American Chemical Society. (B) Two-module acoustofluidic device isolating exosomes from whole blood using taSSAW by successively removing blood cells and then other EVs. Reproduced from [6] with permission of the PNAS Publication. (C) The acoustic trapping of EVs using seeding particles immobilized by acoustic waves. Adapted from [20] with permission of American Chemical Society.
Also taking advantage of particle migration due to standing acoustic waves, Huang’s research group developed an acoustofluidic device that separates exosomes from undiluted whole blood using two connected modules operating on tilted-angle standing surface acoustic waves (taSSAW): (a) cell-removal module, and (b) exosome-isolation module [6]. The taSSAW worked to deflect, or tilt, the path of non-desirable particles away from the desirable particles (figure 5(B)). The cell-removal module relied on 22 Vpp (peak-to-peak) and 19.6 MHz driving frequency to remove particles larger than 5 μm (typical of cells). The exosome-isolation module relied on 45 Vpp and 39.4 MHz to remove particle larger than 140 nm (the chosen cutoff size for exosomes). The two-module setup enabled rapid exosome isolation from 100 μl of sample in 25 min at a capture efficiency of 82% and purity of 98%. It is arguably one of the few microfluidic devices to be able to process undiluted whole blood. In another study, this device successfully isolated exosomes from saliva samples to detect human papilloma viral-associated oropharyngeal cancer-related biomarkers such as HPV16 DNA at an 80% agreement to biopsy results [58]. With its ability to isolate exosomes quickly from a small volume of unprocessed bodily fluid samples, this acoustofluidic device has considerable clinical advantages in serving as a diagnostic or prognostic tool especially when sample pre-preparation must be avoided to prevent loss of sample volume.
Rather than using the primary acoustic force induced by the sinusoidal waves from the transducers similar to the two devices previously described, Ku et al presented another acoustic trapping methods that utilized secondary acoustic forces that allowed for the trapping of EVs (including exosomes and microvesicles) at much weaker transducer settings of 4 MHz, 10 V peak-to-peak sinusoidal wave [20]. This acoustic trapping device generated half-wavelength acoustic standing waves in a microfluidic channel, traps seeding particles (12 μm) using these primary radiation force, then used these immobilized seeding particles to generate a secondary radiation force with EV-sized particles in the biofluids samples that flowed through, inducing a particle aggregation that was removeable when the transducers were turned off (figure 5(C)). The device was characterized with polystyrene beads ranging 0.1–1 μm in size, and additional performance studies were done with cell culture media, urine, and plasma. Interestingly, while this technology gave only a 1% and 5% capture efficiency for 100 nm and 200 nm beads, respectively, Ku et al showed with biofluid samples that even this low of capture efficiency was enough for downstream analysis for protein contents and miRNA due to the abundance of exosomes in biofluids. The device also demonstrated its superior performance compared to conventional ultracentrifugation in isolating more particles of size less than 100 nm from biofluids. With the ability to consume only 300 μl of sample and deliver promising results in 30 min, this acoustic trapping device proves to be a potential alternative to conventional techniques. Future efforts on characterizing the purity levels, and fine tuning the capture efficiency for a narrower range of captured particles may boost the confidence in its performance.
4.2.3. Exosome isolation enabled by electrokinetics
Following the criteria of being label-free and contact-free, some microfluidic devices are designed to perform exosome isolation based on external electrical forces. In particular, the dielectrophoresis (DEP) phenomenon, which involves polarizable particles in a non-uniform electric field, have long been adapted for manipulating micron and submicron scale particles (e.g. cells, other biological components) [49]. The DEP force is a function of the size and dielectric properties of the bioparticles and their surrounding fluid [48]. Thus, DEP is reasoned to be capable of achieving specific isolation [49]. This section introduces microfluidic devices that have creatively incorporated DEP as the main isolation mechanism for exosomes.
Advances in microfabrication has enabled the production of miniaturized DEP electrodes capable of delivering high electric fields for manipulating submicron-scale particles, while consuming low voltage power and producing significantly less heating — qualities well-suited for the biological realm [61]. However, whether the presence of an electric field and the associated heating (however minimal) would interrupt biological processes or cause internal damage to the manipulated bioparticles deserve a more thorough discussion.
One device that took advantage of DEP is the alternating current electrokinetic (ACE) microarray chip developed by members of the Heller Lab for clinical applications [12, 62]. The group designed a chip consisting of over 400 electrodes to isolate exosomes and other nanoparticles of sizes 50–150 nm from 30 to 50 μl of plasma sample within 30 min. When a nonuniform electric field (10 Vpp, 15 kHz) was applied, the differences in the dielectric constant between exosomes, other particles, and the biofluid induced dipoles that force nanosized particles to DEP high-field region around the edges of the electrodes (figure 6(A)). Non-nano-sized particles in low-field region were washed with a 1× tris-ethylenediaminetetraacetic acid (TE) buffer, and isolated exosomes in the high-field region were retrieved using a series of low-frequency electric pulses and a second 1× TE buffer wash. In a follow-up study in 2018, the ACE chip demonstrated its clinical potential by processing undiluted pancreatic ductal adenocarcinoma plasma samples with CD63 and glypican-1 biomarkers within 30 min [62]. Further investigations on ways to decrease unspecific capture of non-exosome nanoparticles would greatly increase the capture efficiency, purity, and reliability of this technology. Overall, the ACE chip proved itself to be a very promising candidate for clinical use by satisfying minimal sample preprocessing, quick isolation, minimal damage of exosomes during isolation, label-free, and viability of nucleic acids and protein biomarkers.
Figure 6.

Examples of exosome isolation enabled by electrokinetics. (A) Cross-sectional view of the ACE microarray chip depicting the electrode with a DEP high-field region where exosomes are captured. Adapted from [12] with permission of American Chemical Society. (B) A diagram explaining the force balance at the pipette tip and the layout of the iDEP device with four micropipettes connected in parallel to the electrodes. Republished from [63] with permission of Royal Society of Chemistry.
Shi et al developed an insulator-based dielectrophoretic (iDEP) device consisted of four glass micropipettes to isolate exosomes from 200 μl of plasma or serum within 20 min (figure 6(B)) [63]. Interestingly, the iDEP used a very low (10 V cm−1) DC field, but was able to create a strong non-uniform electric field needed for DEP owing to the conical shape of the tips of the pipettes (1 μm and 2 μm in diameter). Leveraging the force balance between dielectrophoretic, electroosmosis, and electrophoresis forces, the iDEP device featured a trapping zone near the tip of the pipette where exosomes was trapped when a negative voltage polarity was applied [56]. A notable feat of this device was its simple and low-cost fabrication. With PDMS chambers bonded to a glass slide and glass micropipettes connected in parallel with platinum electrodes, the device forwent the need of dedicated cleanroom equipment as frequently seen for other micro-scale devices [63]. In isolating exosomes from plasma samples, the iDEP demonstrated its ability to enrich exosomes with a concentration higher than that from differential ultracentrifugation by two orders of magnitude, successfully overcoming the disadvantages of conventional methods while maintaining high yield.
Also using an iDEP approach, Ayala-Mar et al developed a two-section direct current-insulator-based dielectrophoretic (DC-iDEP) device that can enrich for exosomes of two different size ranges [57]. The DC-iDEP device used electrically insulating posts to produce a nonuniform electric field when a DC of 2000 V is applied for 20 s across the main channel for exosome concentration and separation. Using two arrays of oval-shaped posts with different gap sizes (15 μm and 10 μm), the device was able to fractionate exosomes of mode sizes 84.34 ± 5.70 nm (first section) and 41.75 ± 9.73 nm (second section). By applying a 200 V DC across the side channels for 1 min separated exosomes could migrate towards the collection sites. Efficiency experiment showed the concentration of collected isolated exosome populations to be 2.4 times (first section) and 2.8 times (second section) the input concentration, demonstrating success in the enrichment of exosomes. Though successful in concentrating exosomes from 100 μl samples in under 2 min and characterizing exosome size, zeta potential, and morphology using dynamic light scattering (DLS) and SEM, Ayala-Mar et al did not show any data for the purity or the molecular characteristics of those collected exosomes. Hence, these aspects should be investigated in the future to confirm the usefulness of the device for real-world exosome isolation and analysis applications.
Featuring a much higher exosome concentration capability, a device developed by Cheung et al used an ion concentration polarization (ICP)-based electrokinetic concentrator aiming to increase the concentration of EVs in biofluids typically small in volume (e.g. cerebrospinal fluid) [64]. As a proof-of-concept, the ICP concentrator was able to increase the concentration of a 30 μl EV sample extracted from MDA-MB-231 breast cancer cell cultures by 100 fold in 30 min using 45 V. With an ion-selective conductive polymer printed onto the PDMS microfluidic channel, this device has the potential to incorporate different mechanism for better capture efficiency and specificity, such as using a SuperAldehyade glass substrate to exploit aldehyde surface binding, printing anti-CD63 antibody on a microarray slide for immunoaffinity-based capturing, or incorporating microtraps for passive mechanical trapping. In 2020, they followed up with this technology by printing anti-CD63 antibody dots onto the device, ultimately combining the idea of using ICP for exosome concentration and immunoaffinity for high-specificity exosome capturing [65]. Unfortunately, even with the newest updates, the technology was not backed by any concrete numbers on the device performance or post-isolation exosome characterization.
Beside from those discussed above for physical-property-based exosome isolation, there is another relatively novel label-free method used for bioparticles separation: optofluidics. Although not much has been published on exosome isolation using this method, we feel that it is important to briefly introduce the 200 nm thick optofluidic plasmonic (OPtIC) microlenses developed by Zhu et al [66]. This technology uses light focused through the lenses to keep larger and higher refractive index particles away from smaller and lower refractive index particles, achieving exosome-sized and exosome-composition bioparticles by negative depletion [66]. Compared to traditional optics-enabled separation technique, the objective-free OPtIC microlenses do not require dedicated instrument or extensive beam focusing effort, ensuring a more accessible and simpler separation technique.
The fundamentals of this device lied in the balancing of the fluid drag force (Fd) and radial drag force (Fd,r) created by the particles moving, and the optical scattering force (Fs), thermo-plasmonic drag force (Ftp), and optical gradient force (Fg) created by the beam focused by the OPtIC microlenses (figure 7). While Fd, Fs, and Ftp competes to keep larger and higher refractive index particles from being collected downstream, Fd,r and Fg balance each other out in the radial direction to achieve alignment along the optical axis. Separation of exosomes, phospholipids, and proteins from larger particles is achieved by tuning the light intensity and flow rate. Although no clinical samples analysis, or other performance parameter data were reported, with its ability to separate exosome-sized bioparticles based on both size and refractive index (related to chemical composition), this proof-of-concept presents many opportunities for further development of the technology and applications in exosome isolation and detection.
Figure 7.
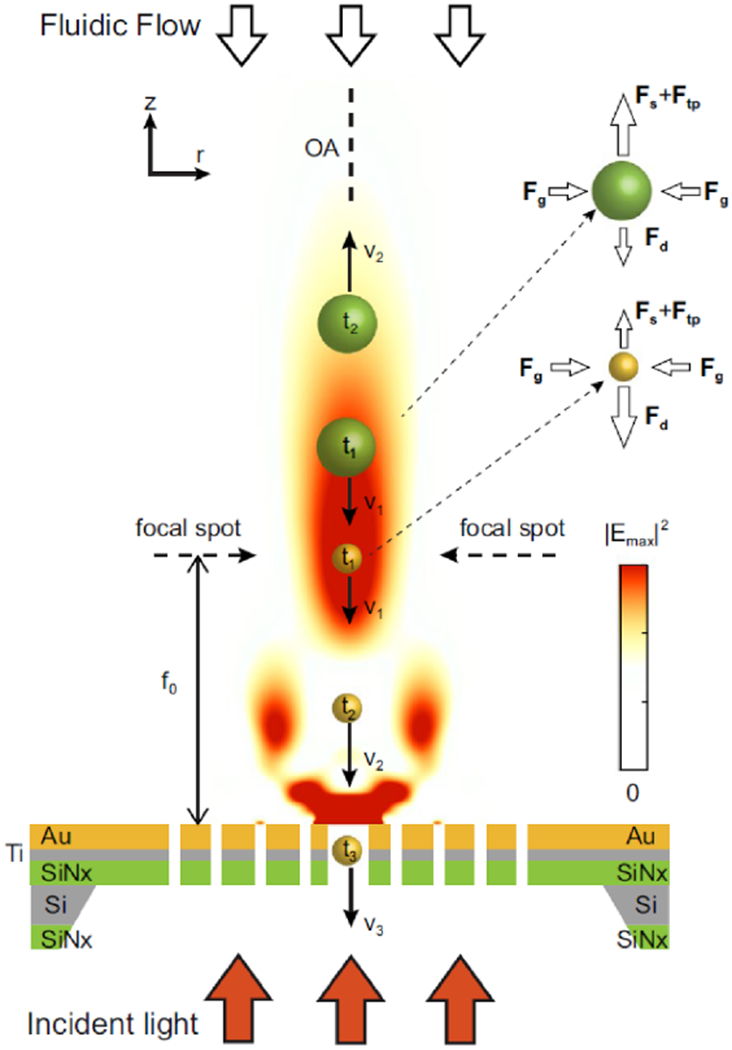
Diagram illustrating the balance between Fd, Fs, and Ftp forces leading to negative depletion of non-exosome particles and the balance between Fd,r and Fg forces leading to the self-alignment property of OPtIC microlenses. Reproduced from [66] under Creative Commons license published by Springer Nature.
5. Confirmation of isolated exosomes and evaluation of contamination
Although the focus of this paper was on the novel exosome isolation mechanism of each microfluidic platform, we briefly review post-isolation exosome characterizations. These exosome characterizations, targeting both physical and molecular characteristics, were typically performed in proving the presence of the exosomes captured on the platform. The brief inclusion of these topics stemmed from two inescapable needs: (a) to confirm the bioparticles isolated by the platform are exosomes, and more often overlooked, (b) to evaluate the level of contamination in the isolated content [18]. Ideally, the device performance should only be assessed using the parameters introduced in section 2 after these two points have been addressed (i.e. one can only truly calculate the capture efficiency or purity of a device after identifying exosome and non-exosome populations in the isolated content).
Over the years, exosome research has continued to unravel the identity of an exosome. Even though researchers have yet to agree on one definition for an exosome, publications have described exosomes to possess (a) a size within range of 30–200 nm [4, 5], (b) a round (and for dehydrated SEM/TEM samples, round and cup-like concavity) morphology [3], (c) a buoyant density of 1.15–1.19 g ml−1 on a continuous sucrose gradient [24], and (d) proteins such as the endocytic Alixand TSG101 [24] and tetraspanins (CD9, CD63, CD81, CD82) [18, 32–34]. It is evident from our review that not all exosome isolation platforms abided to those four criteria when confirming the identity of isolated populations—some only used two or three criteria, while others went beyond that to also look at mRNA expression. Perhaps, a major hindrance to this standardization is the inherent complexity of exosomes that stemmed from the heterogeneity in size, content, function, and origin of exosomes [67]. For example, it has been noted that exosomes derived from different cell types possess quite different proteome profiles [68]. Nonetheless, future isolation platforms should aim to confirm the identity of the isolated populations using as comprehensive of an assessment as possible.
Even when isolation platforms utilize the published identity factors (e.g., size, round morphology, proteins) for exosome isolation, isolated samples are still prone to the contaminations by non-exosomal EVs, non-target exosomes, protein aggregates, lipoprotein particles, and other contaminants. For example, physical-property-based isolation techniques may suffer the contamination of particles overlapping in size or electrical properties with the desired exosomes [29, 33]. For biological-property-based techniques, isolated populations may be plagued by other species also possessing the biomarkers targeted by capture agents [29]. The concerns for biological-property-based devices stemmed from recent studies that have shown the absence of the ‘universal’ exosome biomarkers (e.g. tetraspanins CD63, CD81, CD9, and endocytic proteins TSG101) in some populations of exosomes, and the ubiquity of these markers in bioparticles other than exosomes [29, 33]. Although future exosome research promise to enlighten us on this dilemma, ways to manage contamination include additional washing steps to remove contaminating proteins, the use of a sucrose gradient to further purify the isolated content based on density, and analysis techniques to quantify contamination [18, 24].
Techniques used for the confirmation of desired exosomes and evaluation of contamination can often fall into two categories: (a) physical, and (b) molecular. As listed in table 2, common investigated physical characteristics and the respective characterization methods include (a) morphology with SEM and TEM, (b) size with SEM, TEM, NTA, and DLS, and (c) zeta potential with DLS. Common molecular characteristics and characterization techniques include (a) proteins expression with Western blotting, ELISA, and Bradford assay, and (b) nucleic acids contents with droplet digital PCR (ddPCR), qPCR, and RT-PCR. For example, qPCR has been used to quantify red blood cell (RBC) contamination via known RBC RNA transcripts [6].
Table 2.
Microfluidic platforms for exosome isolation.
| Isolation method | Selection agents | Samples (dilution ratio) | Capture efficiency (%) | Purity (%) | Release efficiency (%) | Sample size (μl) | Analysis time (min) | Particlesize (nm) | Exosome characterization | Characterization methods | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Passive-structure-based affinity isolation | |||||||||||
| Herringbone groove | CD63 antigen | Healthy and glioblastoma serum (undiluted) | 42–94 | N/A | N/A | 100–400 | <60 | N/A | Morphology, size; total RNA count | SEM; RT-PCR, nested PCR | [38] |
| ExoChip | CD63 antigen | Healthy and pancreatic cancer serum (undiluted) | N/A | N/A | N/A | 400 | 60 | 30–300 | Morphology, size, Rab5; other proteins; miRNA | EM; Western blot; RT-PCR | [39] |
| NEWExoChip | Phosphatidylserine lipid | Healthy and lung cancer and melanoma plasma (undiluted) | 90 | 75 | 85 | 30–100 | 30 | N/A | Morphology; size; proteins | SEM; NTA; Western blot | [40] |
| ZnO nanowire-coated 3D scaffold | CD63 antigen | Healthy and cancer serum and plasma (undiluted) | N/A | N/A | N/A | 100 | 150 | 40–250 | Morphology, size; CD63 and CD9 | SEM, TEM, NTA; ELISA | [41] |
| Nano-herringbone | CD81 antigen ovarian cancer plasma (1:9) | Healthy and | 80–85 | N/A | N/A | 20 | 40 | 40–160 | Morphology, size; proteins; mRNA | SEM, confocal microscope; ELISA, Western blot; ddPCR | [16] |
| Label-free exosome release herringbone | EpCAM and CD9 antigen | Healthy and ovarian cancer serum (1:10) | N/A | N/A | 60 | 100 | <20 | <250 | Morphology, size, EpCAM | SEM, NTA; Western blot | [42] |
| nPLEX | EpCAM, CD24, CA-125, CA19-9, HER2, MUC18, EGFR, and CLDN3 | Ovarian cancer ascites (undiluted) | N/A | N/A | N/A | 150 | <30 | <200 | Morphology; mRNA | SEM; qPCR | [44] |
| Low-cost plasmonic bio-sensor array | CD63 antigen | N/A | N/A | N/A | N/A | 50 | <240 | N/A | Morphology, size | SEM | [45] |
| TPEX | CD63, CD24, EpCAM, MUC1, and size | Colorectal and gastric cancer ascites (undiluted) | N/A | N/A | N/A | 1 | 15 | 30–150 | Size; proteins | NTA, TEM; ELISA, Western blot | [46] |
| Immunomagnetic-based affinity isolation | |||||||||||
| Integrated circuit-like platform | EpCAM, IGF-1R, and CA-125 antigen | Healthy, and lung and ovarian cancer plasma (unspecified dilution ratio) | N/A | N/A | >99.9 | 30 | <90 | 40–150 | Morphology; size; total proteins; IGF-1R | TEM; NTA; Bradford assay; ELISA | [50] |
| ExoSearch | CD9, CD81, CD63, EpCAM, and CA-125 antigen | Healthy and ovarian cancer plasma (undiluted) | 72 | N/A | N/A | 20 | 20 | <300 | Morphology, size | TEM | [51] |
| Integrated device with valve network | EpCAM antigen | Healthy and breast cancer plasma (undiluted) | 74 | N/A | N/A | 2 | 90 | ~100 | Morphology; CD9 and CD63 | SEM; Western blot | [52] |
| ExoPCD-chip | Phosphatidylserine-Tim4 | Healthy and liver cancer serum (undiluted) | 68 | N/A | N/A | 30 | 210 | 50–175 | Morphology; size; CD63 | TEM; qNano analysis; Western blot | [54] |
| iMER | EGFR | Healthy and glioblastoma serum | 93 | N/A | N/A | 100 | 120 | N/A | Morphology; proteins; mRNA | SEM; Western blot; qPCR | [53] |
| Filtration | |||||||||||
| Ciliated micropillars | Size | N/A | 45–60 | N/A | N/A | 30 | 10 | 83–120 | N/A | N/A | [23] |
| Nano-DLD | Size | Urine-derived exosomes | 99 | N/A | N/A | 10 | 3600 | <100 | Size | SEM | [59] |
| Exodisc | Size | Healthy and bladder cancer urine (undiluted) | >95 | N/A | N/A | 1000 | <60 | 100–450 | Morphology, size; CD9 and CD81; mRNA | SEM, TEM; ELISA; RT-PCR | [22] |
| ExoTIC | Size | Healthy plasma, and lung cancer plasma, urine, and lavage (1:14) | >90 | N/A | N/A | 10–120 000 | <180 | 30–200 | Morphology, size; proteins; miRNA | SEM, TEM, NTA; Bradford assay; Nanostring eCounter | [60] |
| Acoustofluidics | |||||||||||
| Acoustic nanofilter | Size | RBC units (undiluted) | >80 | N/A | N/A | 10 | N/A | <200 | Size; proteins | NTA, Western blot | [21] |
| taSSAW acoustofluidic | Size | Whole blood (undiluted) | 82 | 98 | N/A | 100 | 25 | <140 | Morphology, size; proteins; mRNA, miRNA | TEM, NTA; Western blot; qPCR | [6] |
| Acoustic trapping | Size | Urine (undiluted) and plasma (1:1) | 1–5 | N/A | N/A | 300 | 30 | <500 | Morphology; size; proteins; miRNA | NTA; TEM; ELISA; qPCR | [20] |
| Electrokinetics | |||||||||||
| ACE microarray chip | Size | Plasma (undiluted) | N/A | N/A | N/A | 30–50 | 30 | 50–150 | Morphology, size; CD63 and TSG101; mRNA | SEM; fluores cence imaging; RT-PCR | [12] |
| iDEP | Size | Plasma, serum, and saliva (undiluted) | N/A | N/A | N/A | 200 | 20 | 50–150 | Morphology; size; proteins | TEM; NTA; ELISA and Western blot | [63] |
| DC-iDEP | Size | N/A | N/A | N/A | N/A | 100 | 2 | <130 | Morphology; size, | SEM; DLS zeta potential | [57] |
| ICP concentrator | Size | N/A | N/A | N/A | N/A | 30 | 30 | <200 | Morphology, size | TEM | [64] |
| Optofluidics | |||||||||||
| OPtIC microlenses | Size | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | [66] |
Lastly, as the field matures and demands for successful clinical and research translation intensifies, one should be extremely precise in establishing the criteria of what is a desired exosome to be isolated even before penning ideas for a novel platform. These criteria, stemming from the end purpose, could be as broad as including both healthy and disease-related exosomes within a specific size range, to as narrow as including only pancreatic cancer-derived exosomes expressing a certain set of proteins or mRNA expression. However broad or narrow the definition for a desired exosome population, laying down clear criteria for both physical and molecular qualities would greatly aid in the design of a novel platform, evaluation using performance parameters, and eventually, successful translation of exosome isolation platforms to clinical and basic research use.
6. Conclusions
Nearly two decades ago, pioneering exosomes research uncovering their promising clinical potentials initiated an interest in isolating these nanoscale bioparticles — a field that has continued to grow immensely today. Over the last decade, the drawbacks experienced by conventional isolation methods inspired efforts in the development of novel microfluidic devices for exosome isolation. As seen throughout this review, there have been significant steps towards designing efficient and user-friendly devices aiming to provide high-quality exosome populations to both exosome basic research and clinical applications. At the beginning, many exosome isolation schemes drew inspirations from devices featuring passive structures, filtration, and magnetic particles that were previously designed for sorting or isolating other bioparticles such as blood cells and circulating tumor cells. As time went on, we noted the development of integrated platforms and witnessed the emergence of novel exosome isolation methods as researchers found creative ways to harness acoustic, electrical, and optical forces. In the last few years, there has been notable efforts in incorporating nanotechnology into exosome isolation platforms and developing low-cost approaches featuring well-established isolation methods. At the present, microfluidic devices for exosome isolation span from biological-property-based platforms that capitalizes on passive micro- and nano-structures, magnetic beads, to physical-property-based methods that incorporate nanofiltration, acoustics, electrokinetics, and optical forces. In the coming years, besides from the imperative quest for high-purity methods, we can expect to see the rise of more integrated and highly translational exosome isolation devices.
From tabulating end clinical and research needs, translating them to specific design and performance requirements, innovating and testing isolation methods, to confirming the identity and integrity of isolated exosomes, evaluating contamination, and incorporation of clinical samples, all steps to development of a successful device involve commitment from both clinicians and researchers. With new information regarding exosomes to be unearthed as time goes by, we can hope that the intense research of today will continue to shape the exosome isolation research of the future. As the field move forwards, it is imperative that the collaborations between clinicians and researchers continue to flourish so that efforts for microfluidic exosome isolation and analysis may soon see the light of real-world applications.
Acknowledgments
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1S42473. We would also like to acknowledge the financial support from US National Institutes of Health (NIH) and the University of Florida.
References
- [1].Contreras-Naranjo JC, Wu HJ and Ugaz VM 2017. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine Lab Chip 17 3558–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].He M and Zeng Y 2016. Microfluidic exosome analysis toward liquid biopsy for cancer J. Lab. Autom 21 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vlassov AV, Magdaleno S, Setterquist R and Conrad R 2012. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials BBA Gen. Subj 1820 940–8 [DOI] [PubMed] [Google Scholar]
- [4].Gurunathan S, Kang MH, Jeyaraj M, Qasim M and Kim JH 2019. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes Cells 8 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shao H, Im H, Castro CM, Breakefield X, Weissleder R and Lee H 2018. New technologies for analysis of extracellular vesicles Chem. Rev 118 1917–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wu M et al. 2017. Isolation of exosomes from whole blood by integrating acoustics and microfluidics Proc. Natl Acad. Set. USA 114 10584–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lin S, Yu Z, Chen D, Wang Z, Miao J, Li Q, Zhang D, Song J and Cui D 2020. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications Small 16 1903916. [DOI] [PubMed] [Google Scholar]
- [8].Su W, Li H, Chen W and Qin J 2019. Microfluidic strategies for label-free exosomes isolation and analysis TRAC Trends Anal. Chem 118 686–98 [Google Scholar]
- [9].Liga A, Vliegenthart ADB, Oosthuyzen W, Dear JW and Kersaudy-Kerhoas M 2015. Exosome isolation: a microfluidic road-map Lab Chip 15 2388–94 [DOI] [PubMed] [Google Scholar]
- [10].Babic A and Wolpin BM 2016. Circulating exosomes in pancreatic cancer: will they succeed on the long, littered road to early detection marker? Clin. Chem 62 307–9 [DOI] [PubMed] [Google Scholar]
- [11].Iliescu FS, Vrtačnik D, Neuzil P and Iliescu C 2019. Microfluidic technology for clinical applications of exosomes Micromachines 10 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ibsen SD et al. 2017. Rapid isolation and detection of exosomes and associated biomarkers from plasma ACS Nano 11 6641–51 [DOI] [PubMed] [Google Scholar]
- [13].Jia Y, Ni Z, Sun H and Wang C 2019. Microfluidic approaches toward the isolation and detection of exosome nanovesicles IEEE Access 7 45080–98 [Google Scholar]
- [14].Zhang P, Wu X, Gardashova G, Yang Y, Zhang Y, Xu L and Zeng Y 2020. Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and metastasis Sci. Transl. Med 12 eaaz2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jurado-Sánchez B. Microscale and nanoscale biosensors. Biosensors. 2018;8:66. doi: 10.3390/bios8030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK and Zeng Y 2019. Ultrasensitive detection of circulating exosomes with a 3d-nanopatterned microfluidic chip Nat. Biomed. Eng 3 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo SC, Tao SC and Dawn H 2018. Microfluidics-based on-a-chip systems for isolating and analysing extracellular vesicles J. Extracell. Vesicles 1 1508271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Witwer KW. et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ferreira MM, Ramani VC and Jeffrey SS 2016. Circulating tumor cell technologies Mol. Oncol 10 374–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ku A, Lim HC, Evander M, Lilja H, Laurell T, Scheding S and Ceder Y 2018. Acoustic enrichment of extracellular vesicles from biological fluids Anal. Chem 90 8011–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee K, Shao H, Weissleder R and Lee H 2015. Acoustic purification of extracellular microvesicles ACS Nano 9 2321–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Woo HK, Sunkara V, Park J, Kim TH, Han JR, Kim CJ, Choi HI, Kim YK and Cho YK 2017. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples ACS Nano 11 1360–70 [DOI] [PubMed] [Google Scholar]
- [23].Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, Zhang JXJ and Liu X 2013. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles Lab Chip 13 2879–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Théry C, Amigorena S, Raposo G and Clayton A 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids Curr. Protoc. Cell Biol 30 3.22.1–9 [DOI] [PubMed] [Google Scholar]
- [25].Greening DW, Xu R, Ji H, Tauro BJ and Simpson RJ 2015. Proteomic Profiling: Methods and Protocols ed Posch A (New York: Springer; ) pp 179–209 [Google Scholar]
- [26].Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C and Le Pecq JB 2002. Production and characterization of clinical grade exosomes derived from dendritic cells J. Immunol. Methods 270 211–26 [DOI] [PubMed] [Google Scholar]
- [27].Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS and Star RA 2007. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator Am. J. Physiol. Renal 292 F1657–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Clayton A, Court J, Navabi H, Adams M, Mason MD, Hobot JA, Newman GR and Jasani B 2001. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry J. Immunol. Methods 247 163–74 [DOI] [PubMed] [Google Scholar]
- [29].Zhang P, Yeo JC and Lim CT 2019. Advances in technologies for purification and enrichment of extracellular vesicles SEAS Technol. 24 477–88 [DOI] [PubMed] [Google Scholar]
- [30].Nguyen NT, Wereley ST and Shaegh SAM 2019. Fundamentals and Applications of Microfluidics (Norwood, MA: Artech House; ) [Google Scholar]
- [31].Manz A, Graber N and Widmer HM 1990. Miniaturized total chemical analysis systems: a novel concept for chemical sensing Sensors Actuators B 1 244–8 [Google Scholar]
- [32].Simpson RJ, Jensen SS and Lim JW 2008. Proteomic profiling of exosomes: current perspectives Proteomics 8 4083–99 [DOI] [PubMed] [Google Scholar]
- [33].Mathivanan S, Ji H and Simpson RJ 2010. Exosomes: extracellular organelles important in intercellular communication J. Proteomics 73 1907–20 [DOI] [PubMed] [Google Scholar]
- [34].Théry C, Zitvogel L and Amigorena S 2002. Exosomes: composition, biogenesis and function Nat. Rev. Immunol 2 569–79 [DOI] [PubMed] [Google Scholar]
- [35].Chen GD, Fachin F, Colombini E, Wardle BL and Toner M 2012. Nanoporous micro-element arrays for particle interception in microfluidic cell separation Lab Chip 12 3159–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Stroock AD, Dertinger SKW, Ajdari A, Mezić I, Stone HA and Whitesides GM 2002. Chaotic mixer for microchannels Science 295 647–51 [DOI] [PubMed] [Google Scholar]
- [37].Wu M, Ozcelik A, Rufo J, Wang Z, Fang R and Jun Huang T 2019. Acoustofluidic separation of cells and particles Microsyst. Nanoeng 5 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M and Irimia D 2010. Microfluidic isolation and transcriptome analysis of serum micro vesicles Lab Chip 10 505–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kanwar SS, Dunlay CJ, Simeone DM and Nagrath S 2014. Microfluidic device (exochip) for on-chip isolation, quantification and characterization of circulating exosomes Lab Chip 14 1891–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kang YT, Purcell E, Palacios-Rolston C, Lo TW, Ramnath N, Jolly S and Nagrath S 2019. Isolation and profiling of circulating tumor-associated exosomes using extracellular vesicular lipid-protein binding affinity based microfluidic device Small 15 e1903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen Z, Cheng SB, Cao P, Qiu QF, Chen Y, Xie M, Xu Y and Huang WH 2018. Detection of exosomes by ZnO nanowires coated three-dimensional Scaffold chip device Biosens. Bioelectron. 122 211–6 [DOI] [PubMed] [Google Scholar]
- [42].Hisey CL, Dorayappan KDP, Cohn DE, Selvendiran K and Hansford DJ 2018. Microfluidic affinity separation chip for selective capture and release of label-free ovarian cancer exosomes Lab Chip 18 3144–53 [DOI] [PubMed] [Google Scholar]
- [43].Dorayappan KDP et al. 2019. A microfluidic chip enables isolation of exosomes and establishment of their protein profiles and associated signaling pathways in ovarian cancer Cancer Res. 79 3503–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R and Lee H 2014. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor Nat. Biotechnol. 32 490–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lv X, Geng Z, Su Y, Fan Z, Wang S, Fang W and Chen H 2019. Label-free exosome detection based on a low-cost plasmonic biosensor array integrated with microfluidics Langmuir 35 9816–24 [DOI] [PubMed] [Google Scholar]
- [46].Wu X, Zhao H, Natalia A, Lim CZJ, Ho NRY, Ong C-AJ, Teo MCC, So JBY and Shao H 2020. Exosome-templated nanoplasmonics for multiparametric molecular profiling Set. Adv 6 eaba2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zborowski M and Chalmers JJ 2011. Rare cell separation and analysis by magnetic sorting Anal. Chem. 83 8050–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shields IVCW, Reyes CD and López GP 2015. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation Lab Chip 15 1230–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Voldman J 2006. Electrical forces for microscale cell manipulation Annu. Rev. Biomed. Eng 8 425–54 [DOI] [PubMed] [Google Scholar]
- [50].He M, Crow J, Roth M, Zeng Y and Godwin AK 2014. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology Lab Chip 14 3773–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhao Z, Yang Y, Zeng Y and He M 2016. A microfluidic exosearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis Lab Chip 16 489–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chen W, Li H, Su W and Qin J 2019. Microfluidic device for on-chip isolation and detection of circulating exosomes in blood of breast cancer patients Biomicrofluidics 13 054113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H and Weissleder R 2015. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma Nat. Commun 6 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Xu H, Liao C, Zuo P, Liu Z and Ye B-C 2018. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes Anal. Chem. 90 13451–8 [DOI] [PubMed] [Google Scholar]
- [55].Liu C et al. 2019. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers Nat. Biomed. Eng 3 183–93 [DOI] [PubMed] [Google Scholar]
- [56].Shi L, Rana A and Esfandiari L 2018. A low voltage nanopipette dielectrophoretic device for rapid entrapment of nanoparticles and exosomes extracted from plasma of healthy donors Set. Rep 8 6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ayala-Mar S, Perez-Gonzalez VH, Mata-Gómez MA, Gallo-Villanueva RC and González-Valdez J 2019. Electrokinetically driven exosome separation and concentration using dielectrophoretic-enhanced pdms-based microfluidics Anal. Chem 91 14975–82 [DOI] [PubMed] [Google Scholar]
- [58].Wang Z et al. 2020. Acoustofluidic salivary exosome isolation: a liquid biopsy compatible approach for human papillomavirus-associated oropharyngeal cancer detection J. Mol. Diagn 22 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G and Astier Y 2016. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 Nm Nat. Nanotechnol 11 936–40 [DOI] [PubMed] [Google Scholar]
- [60].Liu F et al. 2017. The exosome total isolation chip ACS Nano 11 10712–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gonzalez CF and Remcho VT 2005. Harnessing dielectric forces for separations of cells, fine particles and macromolecules J. Chromatogr. A 1079 59–68 [DOI] [PubMed] [Google Scholar]
- [62].Lewis JM, Vyas AD, Qiu Y, Messer KS, White R and Heller MJ 2018. Integrated analysis of exosomal protein biomarkers on alternating current electrokinetic chips enables rapid detection of pancreatic cancer in patient blood ACS Nano 12 3311–20 [DOI] [PubMed] [Google Scholar]
- [63].Shi L, Kuhnell D, Borra VJ, Langevin SM, Nakamura T and Esfandiari L 2019. Rapid and label-free isolation of small extracellular vesicles from biofluids utilizing a novel insulator based dielectrophoretic device Lab Chip 19 3726–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cheung LS, Sahloul S, Orozaliev A and Song Y-A 2018. Rapid detection and trapping of extracellular vesicles by electrokinetic concentration for liquid biopsy on chip Micromachines 9 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kim J, Sahloul S, Orozaliev A, Do VQ, Pham VS, Martins D, Wei X, Levicky R and Song Y-A 2020. Microfluidic electrokinetic preconcentration chips: enhancing the detection of nucleic acids and exosomes IEEE Nanotechnol. Mag. 14 18 [Google Scholar]
- [66].Zhu X, Cicek A, Li Y and Yanik AA 2019. Plasmofluidic microlenses for label-free optical sorting of exosomes Sci. Rep 9 8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kalluri R and LeBleu VS 2020. The biology, function, and biomedical applications of exosomes Science 367 eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Romano E, Netti PA and Torino E 2020. Exosomes in gliomas: biogenesis, isolation, and preliminary applications in nanomedicine Pharmaceuticals 13 319. [DOI] [PMC free article] [PubMed] [Google Scholar]


