Abstract
Background
Accumulating evidences have suggested that high body fat percentage (BF%) often occurs in parallel with cardiovascular diseases (CVDs), implying a common etiology between them. However, the shared genetic etiology underlying BF% and CVDs remains unclear.
Methods
Using large-scale genome-wide association study (GWAS) data, we investigated shared genetics between BF% (N = 100,716) and 10 CVD-related traits (n = 6968-977,323) with linkage disequilibrium score regression, multi-trait analysis of GWAS, and transcriptome-wide association analysis, and evaluated causal associations using Mendelian randomization.
Results
We found strong positive genetic correlations between BF% and heart failure (HF) (Rg = 0.47, P = 1.27 × 10− 22) and coronary artery disease (CAD) (Rg = 0.22, P = 3.26 × 10− 07). We identified 5 loci and 32 gene-tissue pairs shared between BF% and HF, as well as 16 loci and 28 gene-tissue pairs shared between BF% and CAD. The loci were enriched in blood vessels and brain tissues, while the gene-tissue pairs were enriched in the nervous, cardiovascular, and exo-/endocrine system. In addition, we observed that BF% was causally related with a higher risk of HF (odds ratio 1.63 per 1-SD increase in BF%, P = 4.16 × 10–04) using a MR approach.
Conclusions
Our findings suggest that BF% and CVDs have shared genetic etiology and targeted reduction of BF% may improve cardiovascular outcomes. This work advances our understanding of the genetic basis underlying co-morbid obesity and CVDs and opens up a new way for early prevention of CVDs.
Keywords: Body fat percentage, Cardiovascular diseases, Shared genetics, Genetic correlation, Mendelian randomization
Background
Body fat percentage (BF%) is a proxy for adiposity that is genetically regulated through a leptin or melanocortin pathway in the central nervous system (CNS) [1]. Recent studies found that high BF%, especially with excess visceral adipose tissue, is associated with increased risk of cardiovascular diseases (CVDs) [2, 3], independent of body mass index (BMI) [4, 5]. BF% is biologically important and distinct from other proxies of adiposity such as BMI and waist-to-hip ratio for its capacity to differentiate between fat-free mass (i.e., lean mass, bone mass and fluid mass) and fat mass [6, 7]. One hypothesis to account for the link between high BF% and CVDs is shared genetic etiology. BF% and CVDs may share common genetic variants that influence metabolism or response to environmental risk factors [8]. While the genetic basis for BF% and CVDs is poorly understood, large-scale genome-wide association studies (GWAS) may provide novel insight into specific biological processes underlying their comorbidity.
Genetic correlation analysis estimates the correlation of genetic effects between two clinically related traits and highlights the shared etiologies behind such an association. With methodological advances in molecular genetics and the increased number of available GWAS results, it is now feasible for us to investigate genome-wide genetic correlation and identify significant expression-trait associations for complex traits by using genomics resources (i.e., summary-level statistics from large GWAS [8–14] and the Genotype-Tissue Expression (GTEx) project [15]) and state-of-the-art statistical analysis methods (i.e., linkage disequilibrium score regression (LDSC) [16, 17], multi-trait analysis of GWAS (MTAG) [18], and transcriptome-wide association studies (TWAS) [19]). Furthermore, previous twin and family studies have shown that BF% and CVDs are heritable traits, with heritability estimates ranging from 25 to 40% [10, 20, 21]. Large-scale GWAS have enabled detection of more than 20 susceptibility loci for BF% [8]. In particular, the identified BF%-related loci near FTO predicts long-term CVD risk (OR = 1.895), suggesting that BF% and CVDs might share genetic architecture [22]. Previous post-GWAS analyses conducted in a large European population found that genetically predicted BMI was strongly associated with several CVD outcomes, including heart failure (HF) and coronary artery disease (CAD) [23]. Further, there was suggestive evidence of associations between genetically predicted fat mass index and some CVDs [23]. However, these studies are limited because BMI is an imperfect measure of adiposity which does not directly measure body fat. To our knowledge, no large-scale genome-wide study has systematically reported the shared genetic loci between excess adiposity and CVDs, which is not fully accounted for through BMI evaluation.
Although observational studies have reported associations between BF% and adverse cardiovascular outcomes [24–26], some of the findings have been inconsistent, which may be due to biases such as unmeasured confounding. Mendelian randomization (MR) is a form of instrumental variable analysis that can be used to estimate the causal association under certain assumptions [27, 28], even in the presence of unmeasured confounders. HF is a catabolic state that can lead to weight reduction [29], which indicates that the association between body weight and HF could be bidirectional. Given that inherited genetic variants are unlikely to be influenced by reverse causation or environmental confounders after accounting for population stratification, investigating the directions of these associations is crucially important.
Using genome-wide association study (GWAS) summary-level data from several international consortia (n = 6968-977,323), we investigated the genetic correlation and causality between BF% and 10 CVD-related traits with the overarching goal of characterizing the specific shared genetic loci and biological pathways. Further, we conducted a large-scale, genome-wide cross-trait analysis to explore novel genetic components among these diseases. The biological effects reflected by shared loci may play important roles in the co-occurrence of high BF% and CVDs.
Methods
Study design, data summary and quality control
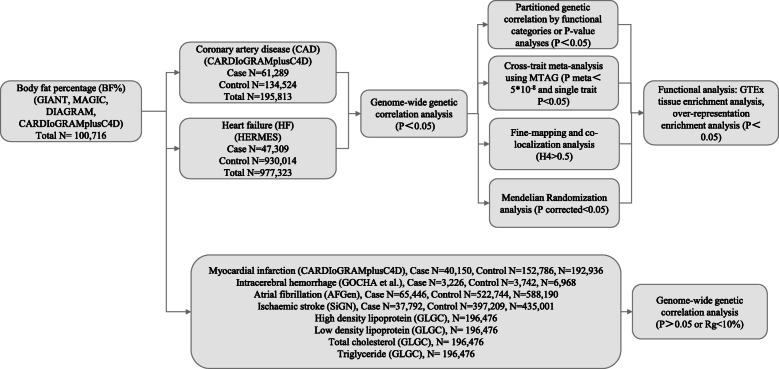
The overall study design is shown in Fig. 1. We retrieved summary statistics from the Genetic Investigation of ANthropometric Traits (GIANT) consortium for BF% (n = 100,716) [8]; the Heart Failure Molecular Epidemiology for Therapeutic Targets (HERMES) for HF (47,309 cases and 930,014 controls) [9]; the Coronary ARtery Disease Genome wide Replication and Meta-analysis (CARDIoGRAM) plus the Coronary Artery Disease (C4D) Genetics (CARDIoGRAMplusC4D) consortium for CAD (60,801 cases and 123,504 controls) [10]; and myocardial infarction (MI) (43,676 cases and 128,197 controls) [10], respectively; from the NINDS Stroke Genetics Network (SiGN) and International Stroke Genetics Consortium (ISGC) for ischemic stroke (IS) (37,792 cases and 397,209 controls) [11]; from the AFGen Consortium for atrial fibrillation (AF) (65,446 cases and 522,744 controls) [12]; from the Genetics of Cerebral Hemorrhage with Anticoagulation (GOCHA) study and the Genetic and Environmental Risk Factors for Hemorrhagic Stroke (GERFHS) studies for intracerebral hemorrhage (ICH) (3226 cases and 3742 controls) [13]; and from the Global Lipids Genetics Consortium (GLGC) consortium (n = 188,578) for lipids such as high-density lipoprotein (HDL) [14], low-density lipoprotein (LDL) [14], total cholesterol (TC) [14], and triglycerides (TG) [14]. Each study has done the study-specific quality control to ensure the criteria of BF% measurement and CVDs similar among the different studies. Baseline characteristics for each study are also provided in previous studies. For example, BF% in each cohort was measured either with bioimpedance analysis or dual energy X-ray absorptiometry as described in detail before [8]. In addition, CAD status was defined by an inclusive CAD diagnosis, including MI, chronic stable angina, acute coronary syndrome, or coronary stenosis > 50% [10]. HF cases included participants with a clinical diagnosis of HF of any etiology with no inclusion criteria based on LV ejection fraction [9]. Details of each of the GWAS studies are present in Table S1.
Fig. 1.
Overall study design. Multiple GWAS data sources were first retrieved. We first conducted genome-wide genetic correlation between BF% and 10 cardiovascular disease (CVD)-related traits. For CVD traits that were shown significant genetic correlation with BF%, we conducted further post-GWAS analyses to investigate genetic overlap between them (variant/region/functional levels, and causal inference). GWAS: genome-wide association study; GIANT: Genetic Investigation of ANthropometric Traits; MAGIC: Meta-Analyses of Glucose and Insulin-related traits Consortium; DIAGRAM: DIAbetes Genetics Replication And Meta-analysis; CARDIoGRAMplusC4D: the Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) plus the Coronary Artery Disease (C4D) Genetics (CARDIoGRAMplusC4D) consortium; HERMES: Heart Failure Molecular Epidemiology for Therapeutic Targets; GOCHA: Genetics of Cerebral Hemorrhage with Anticoagulation; SiGN: the NINDS Stroke Genetics Network; ISGC: International Stroke Genetics Consortium; GLGC: The Global Lipids Genetics; MTAG, multi-trait analysis of genome-wide association studies
The ethical approval and quality control procedures of each consortium have been described in previous studies [8–14]. Informed consent was obtained from all participants of contributing studies. In addition, we restricted the chromosome region to autosomal chromosomes and excluded single-nucleotide polymorphisms (SNPs) in MHC region (chr6:25 Mb–34 Mb).
LDSC analysis
We performed a post-GWAS genome-wide genetic correlation analysis for BF% and CVD-related traits using the LDSC software [30] by assuming that the effect size for each SNP in GWASs represents all SNPs in LD with it [17]. The genetic correlation of two traits (ranging from − 1 to 1) can be computed on LDSC software using the known LD structure of European ancestry reference data from the 1000 Genomes Project. Since low imputation quality may yield lower test statistics, we restricted our analyses to HapMap3 SNPs which seem to be well-imputed in most studies to minimize the bias in our analysis [17]. The mean chi-squared statistics in LDSC is higher in high-LD region compared with low-LD region and cannot be further analyzed when it is below 1.02. The analysis also provides a self-estimated intercept to show the sample overlap between single-trait GWASs [17].
Partitioned genetic correlation analysis
The genetic correlation between BF% and CVDs was partitioned by 13 functional category using partitioned LDSC, including conserved region, DNaseI digital genomic footprinting region (DGF), DNase I hypersensitivity sites (DHSs), fetal DHSs, intron region, repressed region, super enhancers, transcription factor binding site (TFBS), transcribed region, and histone marks H3K4me1, H3K4me3, H3K9ac, and H3K27ac from Roadmap Epigenomics Project [31, 32]. We recalculated the LD scores of SNPs assigned to specific annotation which was used to evaluate the BF%-CVDs genetic correlation for each functional category. Additionally, we conducted another genetic correlation analysis partitioned by SNPs groups at different significant level [33]. The P value of a single SNP was defined a larger one in two GWASs, and then sorted and divided it into five groups in quartile, to further test the proportion of observed genetic correlation explained by each group and the robustness of our findings.
Cross-trait meta-analysis
We applied MTAG [34], a novel approach for conducting meta-analysis of summary statistics from GWAS of multiple traits robust to sample overlap, to identify genome-wide significant loci between BF% and CVD-related traits [18], and detect novel genes by combining GWASs of two correlated diseases [35]. The key assumption of MTAG is that all SNPs share the same variance-covariance matrix of effect sizes among traits. As Turley et al. initially described in 2018, MTAG is a consistent estimator whose effect estimates always have a lower genome-wide mean-squared error than the corresponding single-trait GWAS, even if the assumption is not satisfied [18]. In addition, association statistics from MTAG also yield more statistical power and little inflation of the false discovery rate for each trait analyzed with high correlation, matching theoretical expectations [18].
Fine-mapping credible set analysis
For each of the shared loci between BF% and HF or CAD that meet the cross-trait meta-analysis significance criteria, we extracted variants within 500 kb of the index SNP and then identified a 99% credible set of causal SNPs using the Bayesian likelihood fine-mapping algorithm [36, 37]. This algorithm only maps the primary signal and uses flat prior with steepest descent approximation to identify causal variants, which may reveal molecular mechanisms behind the associations. Details of the method have been described in previous studies [38, 39].
Co-localization analysis
We first extracted summary statistics for variants within 500 kb of the index SNP at each of the shared loci between BF% and HF or CAD and then used R “coloc” package to perform genetic co-localization analysis to calculate the probability that the two traits shared a common genetic causal variant (H4) [40]. We conducted fully Bayesian co-localization analysis using the function “coloc.abf” in the R “coloc” package, which requires regression coefficients for each SNP and variance of these regression coefficients for each trait when only summary data are available [41]. In the present study, we considered loci with probability greater than 0.5 to be co-localized.
GTEx TSEA
To test if shared gene sets were highly enriched or specific expressed in a tissue, we conducted a tissue-specific expression analysis (TSEA) [42, 43]. The analysis was based on the gene lists that were identified from cross-trait meta-analysis with a matching HUGO Gene Nomenclature Committee (HGNC) name. The gene expression data used in TSEA was collected using published RNA-Seq data from the GTEx project [44–46]. The raw GTEx data was derived from 189 post-mortem subjects consisting of 1839 samples from 45 different tissues. Considering the small sample size of GTEx data, we selected suggestive significant loci for this analysis (Pmeta < 1 × 10− 4) to ensure the robustness of TSEA results. In addition, we used Benjamini–Hochberg correction to account for multiple testing [47].
Over-representation enrichment analysis
To obtain biological insights for identified shared genes (Pmeta < 5 × 10− 08) from cross-trait meta-analysis, we used the PANTHER tool to access enrichment of the gene sets in the Gene Ontology (GO) biological process and Reactome pathway [48, 49]. Benjamini–Hochberg procedure was used to account for multiple testing (false discovery rate < 0.05) [47].
Bidirectional MR
We performed a bidirectional MR analysis between BF% and HF and CAD since they are genetically correlated, using inverse-variance weighted (IVW) as the primary method [50–52]. Median-based methods (simple and weighted), MR-Egger, MR-Robust Adjusted Profile Scores (MR-RAPS), and MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) methods were used as sensitivity analyses. The intercept of MR-Egger can be explained as a test of overall unbalanced horizontal pleiotropy [53, 54]. In addition, we also performed single-SNP and leave-one-out analysis to determine whether there was any single SNP that might drive the IVW point estimate. For instrumental variables, we have used the largest and latest GWASs for these traits [8–10]. We only selected independent genetic variants which are not in linkage disequilibrium (LD) (defined as r2 < 0.1) with other genetic variants based on European ancestry reference data from the 1000 Genomes Project. We chose the variant with the lowest P value for association with the exposure when genetic variants were in LD. For SNPs that were not available in outcome GWASs, we used the LD proxy search on the online platform (https://snipa.helmholtz-muenchen.de/snipa3/index.php/) to replace them with the proxy SNPs identified in high-LD (r2 > 0.8) or discard them if the proxies were not available.
TWAS
To identify associations of BF% and CVDs with gene expressions in specific tissues, we conducted a TWAS using FUSION software package based on 44 post-mortal GTEx (version 6) tissue expression weights [19, 55, 56]. We applied Benjamini-Hochberg correction on TWAS P values of all gene-tissue pairs for each trait, and false discovery rate < 0.05 was considered significant [47].
Results
Genome-wide genetic correlation
We found strong positive genetic correlations with BF% for both HF (Rg = 0.47, P = 1.27 × 10− 22) and CAD (Rg = 0.22, P = 3.26 × 10− 07) in large study populations that were predominantly of European ancestry (more than 75%) (Table 1, Table S1). Additionally, we found nominally significant genetic correlation with BF% for ICH (Rg = 0.29; P = 0.021) and HDL (Rg = − 0.326; P = 0.048). However, we did not find evidence of genetic correlation between BF% and IS, MI, AF, TC, TG, and LDL (all P > 0.05) (Table 1). Estimates of SNP-based heritability on the observed scale using GWAS summary statistics are shown in Table S2.
Table 1.
Genetic correlation between BF% and cardiovascular disease-related traits
| Phenotype | Rg | Rg_SE | P |
|---|---|---|---|
| Coronary artery disease | 0.223 | 0.044 | 3.26E−07 |
| Heart failure | 0.473 | 0.048 | 1.27E−22 |
| Myocardial infarction | 0.167 | 0.090 | 0.062 |
| Intracerebral hemorrhage | 0.292 | 0.126 | 0.021 |
| Atrial fibrillation | 0.068 | 0.053 | 0.199 |
| Ischemic stroke | 0.147 | 0.089 | 0.097 |
| High-density lipoproteins | − 0.326 | 0.165 | 0.048 |
| Low-density lipoproteinsa | / | / | / |
| Total cholesterola | / | / | / |
| Triglycerides | 0.127 | 0.099 | 0.200 |
Rg genetic correlation estimate, SE standard error. aOut of bound
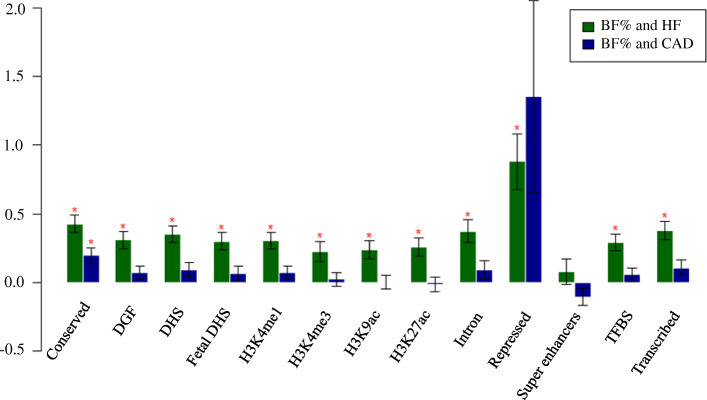
Since high positive genetic correlations between BF% and CVDs was only observed with HF and CAD, we further used 13 functional annotations to evaluate genetic correlations between BF% and HF or CAD in the partitioned genetic correlation analysis by specific functional category. The highest magnitude of significant genetic correlation was in the repressed region (Rg = 0.88; P = 1.51 × 10− 05) for BF% and HF, which could restore the phenotypic effects of a mutant gene. The correlation estimate for BF% and CAD was highest in the conserved region (Rg = 0.20; P = 5.00 × 10− 04), where this region remained almost unchanged during evolution (Fig. 2; Table S3). The shared genetic etiology for BF% and CVDs encourages the exploration of a common pathophysiology, especially in specific functional categories. Subsequently, we conducted a partitioned genetic correlation analysis by SNP groups with different P values and found that the first two groups remained significant (P < 1.00 × 10− 05), suggesting that our findings were robust (Figure S1-S2).
Fig. 2.
Partitioned genetic correlation between BF% and HF or CAD. The vertical axis represents the genetic correlation estimate Rg; the horizontal axis represents 13 functional categories. The asterisk represents significance (P < 0.05), error bars represent the standard error of genetic correlation estimates. DGF: DNaseI digital genomic footprinting; DHS: DNase I hypersensitivity site; TFBS: transcription factor binding sites
MTAG for single traits
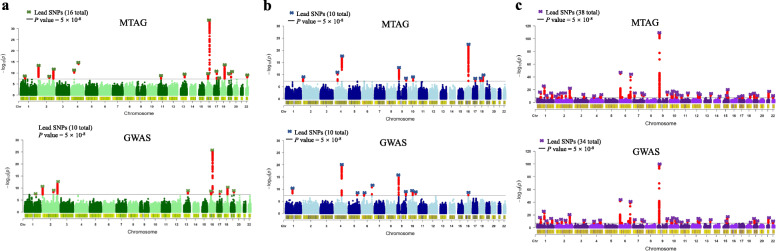
Manhattan plots from the GWAS and MTAG analyses for each trait are shown in Fig. 3. From GWAS to MTAG, the total number of lead SNPs increased from 11 to 16 for BF% and from 42 to 51 for CAD, while no changes were found for HF. We confirmed most of the previously identified loci and found novel associations between BF% and HF and CAD. Out of the 16 independent loci reported for BF% using MTAG, 6 were novel associations and 2 of these fell within protein-coding gene bodies (PLA2G6, RPTOR) (Table S4). Among the 51 loci associated with CAD, 13 were novel loci, 10 of which were mapped to protein-coding genes (i.e., HNRNPUL1, NAA25, FES, TNS1, CYP46A1, ABCG8, IGF2BP1, BCAS3, SMG6, APOE) (Table S4). The proteins encoded by these gene and gene-related pathways are shown in Table S4. Out of the 10 independent loci identified for HF, 8 were novel, 3 of which were mapped to protein-coding genes (i.e., FTO, NPC1, IGF2BP1) (Table S4).
Fig. 3.
The up and down plots display the MTAG and GWAS results for a BF%, b HF, and c CAD, respectively, for a fixed set of SNPs. The x axis shows chromosomal position, and the y axis shows significance on a −log10 scale. The line marks the threshold for genome-wide significance (P = 5 × 10− 8). Each approximately independent genome-wide significant association (lead SNP) is marked by a cross
Cross-trait meta-analysis
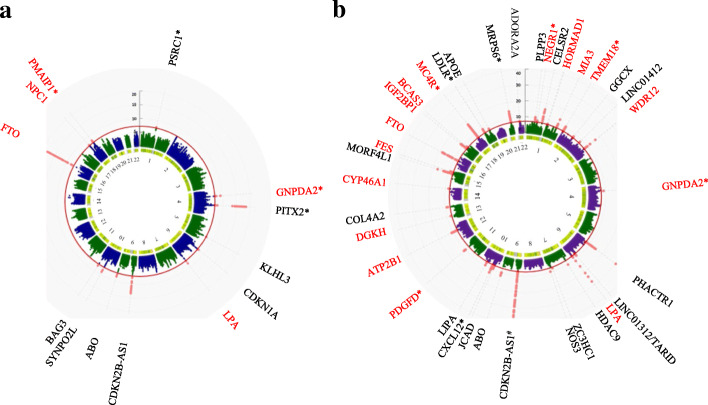
Given the strong genetic correlation between BF% and HF or CAD, we further used MTAG to perform genome-wide meta-analysis to improve our power to identify shared significant genetic loci (meta-analysis P < 5 × 10− 08 and single trait P < 0.05). The genomic control parameter (λ) was 1.16 in the cross-trait meta-analysis for BF% and HF, while λ = 1.20 for BF% and CAD (Figure S3-S4). Illustrative calculations in the two-trait setting were 0.0012 for BF% and CAD and 0.0035 for BF% and HF. The Manhattan plot of these results indicated that shared genetic loci drove the overall significance of the meta-analysis (Fig. 4).
Fig. 4.
Circus Manhattan plot of cross-trait meta-analysis. The first layer of the plot illustrates the chromosome position and the second layer illustrates the representative genes of significant loci. Genes in red are shared genes between two traits [a BF% and HF; b BF% and CAD] with single-trait P value < 0.05. The inside layer illustrates the significance level − log10 (P value) shared markers from cross-trait meta-analysis. The red dots indicate genome-wide significant (P < 5 × 10− 8). Genes at loci in close proximity were assigned one gene label separated by a slash. Asterisks represent the gene closest to index SNP. #: The P value of gene CDKN2B-AS1 (rs4977574) is 9.9 × 10−84 (out of range)
We identified 5 genome-wide significant independent loci for BF% and HF (Table 2). Out of the 5 independent loci, 3 were mapped to protein-coding genes (i.e., FTO, NPC1, LPA). The strongest signal was observed on chromosome 16 at the FTO region (index SNP rs9937053, Pmeta = 1.09 × 10− 18), the first GWAS-identified susceptibility gene for obesity [57, 58]. This locus was not only significant after meta-analysis, but also reached genome-wide significance in both single-trait GWAS of BF% (P = 9.86 × 10− 26) and HF (P = 2.99 × 10− 08). The second strongest signal was in close proximity to an intergenic region closest to the GNPDA2 gene on chromosome 4 (index SNP rs10938397, Pmeta = 4.36 × 10− 10), a critical gene involved in lipid and glucose metabolism. Previous studies found that the expression level of GNPDA2 alters the transcriptome profile of human adipose-derived mesenchymal stem cells [59]. In addition, we found that genetic loci represented by rs10455872 (Pmeta = 3.88 × 10− 09) on LPA are associated with both BF% and HF after meta-analysis. Variation within LPA has shown strong association with Lp(a)-cholesterol levels, which is an independent risk factor for cardiovascular-related events [60].
Table 2.
Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with BF% and HF (Pmeta < 5 × 10− 8; single trait P < 0.05)
| SNP | CHR | Position | Ref. allele | Alt. allele | PBF% | PHF | PMETA | Variant annotation | Genes within clumping region |
|---|---|---|---|---|---|---|---|---|---|
| rs9937053 | 16 | 53799507 | G | A | 9.858E−26 | 2.992E−08 | 1.09E−18 | Intron | AKTIP,CHD9,FTO,FTO-IT1,LOC643802,LOC102723373,RBL2,RPGRIP1L |
| rs1652348 | 18 | 21147509 | C | T | 0.0006127 | 2.477E−06 | 3.35E−08 | Intron | ANKRD29,C18orf8,CABLES1,LAMA3,LOC102724246,NPC1,RIOK3,TMEM241.TTC39C |
| rs7234864 | 18 | 57734857 | C | T | 6.827E−07 | 1.804E−05 | 8.159E−09 | Regulatory region | CCBE1,MC4R,PMAIP1 |
| rs10938397 | 4 | 45182527 | A | G | 1.359E−07 | 2.948E−06 | 4.36E−10 | Intergenic | GNPDA2,GUF1 |
| rs10455872 | 6 | 161010118 | A | G | 0.02079 | 1.892E−11 | 3.879E−09 | Intron | IGF2R,LOC729603,LPA,LPAL2,MAP 3 K4,PLG,SLC22A1,SLC22A2,SLC22A3 |
SNP single-nucleotide polymorphism, CHR chromosome
The cross-trait meta-analysis between BF% and CAD identified 16 genome-wide significant loci (Table 3), 10 of which were in protein-coding gene bodies (i.e., LPA, HORMAD1, MIA3, WDR12, DGKH, CYP46A1, FES, FTO, IGF2BP1, BCAS3). The most significant locus is characterized by the LPA gene (index SNP rs10455872, Pmeta = 3.14 × 10− 28). The second locus (index SNP rs8050136, Pmeta = 4.02 × 10− 19) was mapped to the FTO gene. These loci were also found to be significant in the meta-analysis for BF% and HF, showing genetic overlaps between BF% and CVD-related traits. The third strongest signal was observed closest to the MC4R gene (index SNP rs663129, Pmeta = 3.10 × 10− 18), which is involved in the leptin signaling pathway and its disruption is a causal factor of obesity [61].
Table 3.
Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with BF% and CAD (Pmeta < 5 × 10− 8; single trait P < 0.05)
| SNP | CHR | Position | Ref. allele | Alt. allele | PBF% | PCAD | PMETA | Variant annotation | Genes within clumping region |
|---|---|---|---|---|---|---|---|---|---|
| rs2590942 | 1 | 72885281 | T | G | 4.98E−07 | 1.73E−03 | 6.166E−09 | Intergenic | NEGR1 |
| rs4970926 | 1 | 150673684 | T | C | 1.43E−03 | 3.99E−05 | 4.269E−08 | Intron | ADAMTSL4,ANP32E,ANXA9,APH1A,ARNT,BNIPL,C1orf51,C1orf54,C1orf56,CA14,CDC42SE1,CTSK,CTSS,ECM1,ENSA,FAM63A,GOLPH3L,HORMAD1,KIAA0460,LASS2,LYSMD1,MCL1,MLLT11,MRPS21,PIP5K1A,PRPF3,PRUNE,RP11-68I18.1,SCNM1,SEMA6C,SETDB1,TARS2,TMOD4,TNFAIP8L2,VPS72 |
| rs2133189 | 1 | 222814442 | C | T | 1.99E−02 | 2.42E−12 | 7.714E−09 | Intron | AIDA,C1orf58,DISP1,FAM177B,HHIPL2,MIA3,TAF1A,TLR5 |
| rs13396935 | 2 | 653195 | G | A | 1.42E−09 | 6.43E−04 | 2.493E−11 | Regulatory Region | ACP1,FAM150B,LOC391343,SH3YL1,SNTG2,TMEM18 |
| rs6435169 | 2 | 203753016 | G | A | 4.61E−02 | 7.54E−18 | 7.068E−10 | Intron | ABI2,ALS2CR8,ALS2CR13,BMPR2,CYP20A1,ICA1L,NBEAL1,WDR12 |
| rs10938397 | 4 | 45182527 | A | G | 1.36E−07 | 1.06E−03 | 1.176E−09 | Intergenic | GNPDA2,GUF1 |
| rs10455872 | 6 | 161010118 | A | G | 2.08E−02 | 5.73E−39 | 3.141E−28 | Intron | IGF2R,LPA,LPAL2,MAP 3K4,PLG,SLC22A1,SLC22A2,SLC22A3 |
| rs11226029 | 11 | 103693627 | G | A | 3.54E−02 | 1.14E−09 | 2.967E−11 | Intron | DDI1,DYNC2H1,PDGFD |
| rs4842662 | 12 | 89933446 | T | C | 3.00E−05 | 7.13E−07 | 1.861E−11 | Intron | ATP2B1,DUSP6,GALNT4,WDR51B |
| rs9532984 | 13 | 42634693 | G | A,C,T | 1.93E−02 | 1.62E−06 | 1.802E−08 | Intron | AKAP11,DGKH,KIAA0564 |
| rs3752958 | 14 | 100182687 | C | A T | 2.52E−02 | 7.98E−07 | 1.42E−08 | Intron | BCL11B,CCNK,CYP46A1,DEGS2,EML1,EVL,HHIPL1,SETD3 |
| rs1894400 | 15 | 91428955 | C | T | 1.95E−02 | 1.54E−07 | 1.118E−09 | Intron | BLM,CRTC3,FES,FURIN,HDDC3,IQGAP1,MAN2A2,PRC1,RCCD1,SV2B,UNC45A,VPS33B |
| rs8050136 | 16 | 53816275 | C | A | 1.36E−25 | 4.65E−03 | 4.023E−19 | Intron | AKTIP,CHD9,FTO,RBL2,RPGRIP1L |
| rs9906944 | 17 | 47091420 | C | G,T | 2.86E−08 | 1.17E−05 | 1.935E−12 | Intron | ABI3,ATP5G1,B4GALNT2,C17orf92,CALCOCO2,GIP,GNGT2,HOXB1,HOXB2,HOXB3,HOXB4,HOXB5,HOXB6,HOXB7,HOXB8,HOXB9,HOXB13,IGF2BP1,NGFR,PHB,PHOSPHO1,SNF8,TTLL6,UBE2Z,ZNF652 |
| rs4456565 | 17 | 58921974 | T | C | 4.22E−02 | 6.63E−07 | 3.104E−08 | Intron | APPBP2,BCAS3,C17orf64,LOC729617,PPM1D,USP32 |
| rs663129 | 18 | 57838401 | G | A | 1.47E−10 | 3.20E−08 | 3.097E−18 | Intergenic | CCBE1,MC4R,PMAIP1 |
SNP single-nucleotide polymorphism, CHR chromosome.
GTEx TSEA
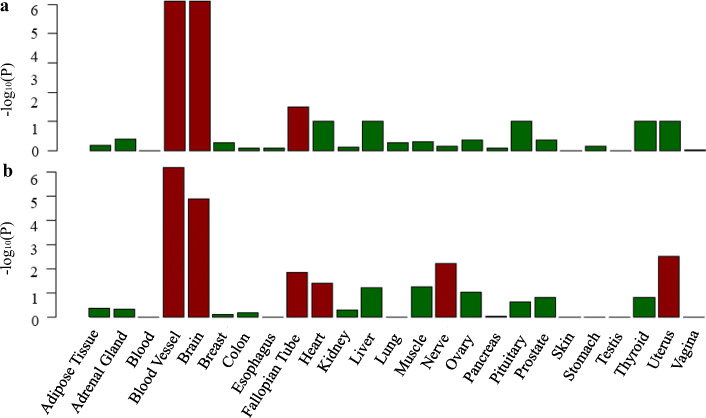
In order to assess whether shared genes between BF% and CVDs are enriched for expression in the disease-relevant tissue, we conducted the TSEA using the GTEx pilot data. We found that shared genes of BF% and HF had three significantly enriched tissues, including blood vessel, brain, and fallopian tube (Fig. 5). The shared genes of BF% and CAD were enriched in six tissues including blood vessel, brain, fallopian tube, heart, nerve, and uterus (Fig. 5). The most strongly enriched tissue for both BF% and HF or CAD was part of the cardiovascular and nervous system (Fig. 5).
Fig. 5.
GTEx tissue enrichment analysis for expression of cross-trait-associated genes (Pmeta < 1 × 10− 4) for BF% and HF (a) and BF% and CAD (b). Red represents significant tissue enrichment after Benjamin-Hochberg correction
Over-representation enrichment analysis
The GO analysis indicated several significant shared biological processes between BF% and HF or CAD (false discovery rate < 0.05) such as glutathione derivative metabolic, glutathione derivative biosynthetic, and nitrobenzene metabolic processes (Table S5-S6). In additional analyses of Reactome pathways, we found that shared association signals for BF% and HF or CAD were significantly enriched in glutathione conjugation and plasma lipoprotein-related pathways (Table S7-S8). In general, the identified shared genes between BF and CVDs showed common significant enrichment in expression for glutathione metabolic-related pathways.
Fine-mapping and co-localization analysis
Lists of credible set SNPs in each shared locus for BF% and HF or CAD from fine mapping are shown in Table S9-S10. The co-localization analysis showed that 4 out of 5 loci and 16 out of 16 share causal variants between BF% and HF or CAD, respectively (Table S11-S12). The results of co-localization analysis were consistent with the cross-trait meta-analysis.
Single-trait TWAS
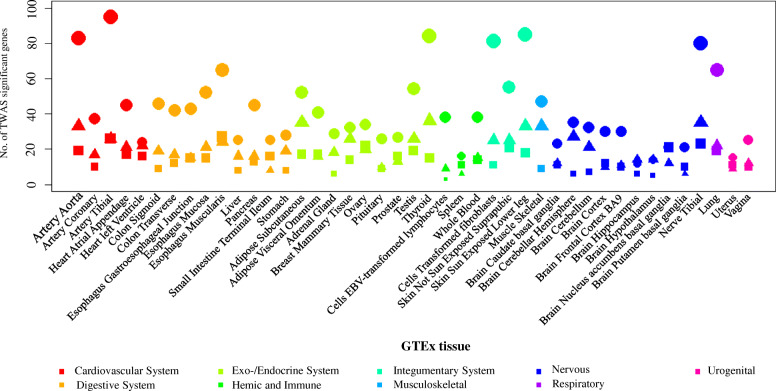
We conducted a TWAS analysis to examine if there are genes whose expression are related with BF%, CAD, and HF, and to determine if these genes are common among these traits. A total of 270 gene-tissue pairs were found across 44 GTEx tissues to be significantly associated with BF% after Benjamini-Hochberg correction (false discovery rate < 0.05) [47], in addition to 786 gene-tissue pairs with CAD, and 270 gene-tissue pairs with HF (Fig. 6). Among these gene-tissue pairs, 32 overlapped between BF% and HF, most of which were observed in nervous and cardiovascular system (Table S17). Notably, C18orf8 and NPC1 are expressed in multiple tissues including brain, nerve, artery, and adipose tissue. Consistent with a previous study [62], NPC1 was validated as a shared genetic component in lipid metabolism and cardiovascular health from our TWAS results. In addition, the methylation levels of C18orf8 were considered as an epigenetic mechanism in maternal obesity and early life origins of CVD and cancers [63]. Furthermore, we identified 28 gene-tissue pairs that were shared by BF% and CAD (Table S17). Most of the associations were found in exo-/endocrine and digestive system. Some genes are expressed among multiple tissues such as ATP5G1, SLC22A3, SNF8, and UBE2Z. ATP5G1 has been shown to encode a subunit of mitochondrial ATP synthase [64] and SLC22A3 have been reported to be expressed at the blood-brain barrier and alters neuronal excitability [65]. By integrating human and mouse results, previous studies have predicted that SNF8 and UBE2Z play a causal role in the development of CAD through a role in the vasculature [66, 67].
Fig. 6.
Number of significant expression-trait associations from transcriptome-wide association study (TWAS). The total number is the significant expression-trait associations after Bonferroni correction (false discovery rate < 0.05). The square represents BF%, the circle represents CAD, and the triangle represents HF. GTEx: genotype-tissue expression project
Bidirectional MR
We conducted a bidirectional MR analysis to evaluate potential causal associations between BF% and HF and CAD. To assess the causal effect of BF% on HF and CAD, we used 10 previously identified genetic variants for BF% (Table S13). To assess the causal effect of HF and CAD on BF%, 5 and 24 previously identified genetic variants were used as instruments for HF and CAD, respectively (Table S14). We found that genetically predicted BF% was causally associated with an increased risk of HF (OR 1.63 per 1-SD increase in BF%, P = 4.16 × 10− 04) (Table 4) after Bonferroni correction. However, HF was not causally associated with BF%. In addition, there were no causal relationships between BF% and the risk of CAD. Sensitivity analysis showed that our results were reliable and were not affected by horizontal pleiotropy or outliers (Table 4, Table S15-S16).
Table 4.
The bidirectional MR analysis of BF% levels and CVD traits
| Outcome | MR methods | Forward | Reserved | ||||
|---|---|---|---|---|---|---|---|
| Causal effect size | SE | P value | Causal effect size | SE | P value | ||
| HF | IVW | 0.488 | 0.138 | 4.16E−04 | − 0.036 | 0.094 | 0.71 |
| SMB | 0.451 | 0.134 | 7.45E−04 | − 0.067 | 0.142 | 0.64 | |
| WMB | 0.456 | 0.132 | 5.46E−04 | 0.027 | 0.118 | 0.82 | |
| MR-RAPS | 0.594 | 0.088 | 1.22E−11 | − 0.036 | 0.099 | 0.72 | |
| MR-PRESSO | 0.488 | 0.138 | 6.42E−03 | −0.036 | 0.077 | 0.67 | |
| MR-Egger | 1.452 | 0.571 | 0.03 | 0.323 | 0.290 | 0.35 | |
| CAD | IVW | 0.264 | 0.330 | 0.42 | 0.014 | 0.051 | 0.78 |
| SMB | 0.437 | 0.175 | 0.01 | − 0.010 | 0.043 | 0.81 | |
| WMB | 0.491 | 0.165 | 2.93E−03 | 0.025 | 0.042 | 0.55 | |
| MR-RAPS | 0.563 | 0.096 | 3.88E−09 | 0.008 | 0.028 | 0.78 | |
| MR-PRESSO | 0.264 | 0.330 | 0.44 | 0.014 | 0.051 | 0.78 | |
| MR-Egger | − 0.152 | 1.617 | 0.93 | − 0.013 | 0.115 | 0.91 | |
SE standard error, IVW inverse-variance weighted, SMB simple median, WMB weighted median, RAPS robust adjusted profile scores, PRESSO pleiotropy residual sum and outlier
Discussion
Using large-scale GWAS data from international consortia, we analyzed shared genetic etiology and causal associations between BF% and CVD-related traits. First, we found strong positive genetic correlations and further identified novel shared genetic loci between BF% and HF or CAD. Second, we found that the shared genetic loci were enriched mainly in blood vessels and brain tissues and glutathione-related metabolic pathway using functional analysis. Third, we found that the shared genes between BF% and HF or CAD are mostly from nervous, cardiovascular and exo-/endocrine system using TWAS analysis. Finally, we found that BF% was causally associated with HF using bidirectional MR analysis results. Taken together, evaluation of the genetic correlation and causality between BF% and CVDs furthers the understanding of the shared loci and biological mechanisms underlying this comorbidity.
BF% is becoming recognized as a more predictive measurement in cardiovascular risk assessment than BMI or waist circumference, and increased BF% was often related to higher CVD risks with the presence of the metabolic abnormalities [25, 68]. Genetic studies have suggested that most of the BF% loci also affect cardiometabolic traits and comorbidities [8]. Compared to GWAS results for single traits, MTAG discovered 6 novel loci for BF%, 8 novel loci for HF, and 13 novel loci for CAD. Importantly, we further conducted genome-wide cross-trait meta-analysis to improve our power to identify specific shared loci, including 3 novel loci between BF% and HF and 8 novel loci between BF% and CAD which did not reach genome-wide significance in previous GWAS. We highlight the potentially interesting functions of the novel associations for NPC1, PMAIP1, and GNPDA2 between BF% and HF, along with NEGR1, HORMAD1, GNPDA2, DGKH, CYP46A1, FES, POC1B, and BCAS3 between BF% and CAD. Most of these loci have significant single-tissue expression quantitative trait loci (eQTL), mainly in the cardiovascular, nervous, and exo-/endocrine system (Table S18-S19).
The only top loci common to the BF%-HF and BF%-CAD meta-analysis was rs10938397 near GNPDA2, which encodes an enzyme that catalyzes the deamination of the glucosamine-6-phosphate involved in the hexosamine signaling pathway [59]. An animal study also suggested that GNPDA2 was involved in the regulation of body weight, fat and energy metabolism in the development of cardiovascular disease [69]. Other top associations for BF%-HF were NPC1 gene variations, which might lead to metabolic diseases by modulating steroid hormone synthesis and lipid homeostasis [70]. NPC1 is known to play a critical role in the atherosclerotic progression, and loss-of-function mutations in NPC1 can cause adiposity in humans [71, 72]. In particular, the NPC1 gene was also confirmed to be shared between BF% and HF using TWAS (Table S17). BCAS3 is a cytoskeletal protein that promotes directional cell migration and angiogenesis in vitro and is implicated in CAD [73]. Additionally, other causal genes are highly expressed or known to act in human brain function (i.e., NEGR1, GNPDA2, CYP46A1, DGKH) [74–76] and various carcinomas (i.e., PMAIP1, HORMAD1, FES, BCAS3) [77–79], indicating the potential role of metabolic dysregulation in the pathogenesis of obesity and cardiac events.
We found that the shared loci for BF% with HF and CAD were highly expressed in blood vessel/heart tissues, indicating that these traits might be caused by dysfunction of the cardiovascular system. We found that the FTO gene on chromosome 16 was associated with both BF%/HF and BF%/CAD. FTO is an essential regulator in the development of obesity-induced metabolic and vascular changes, which is independent of its known function in regulation of obesity [80]. Previous studies found that FTO plays a critical role in cardiac contractile function during homeostasis, remodeling, and regeneration for HF and CAD [81, 82]. Moreover, from the perspective of gene function, overlaps in genetic and molecular pathways advance our understanding of the potential role of brain/nerve tissue diseases in the association between BF% and CVDs. For example, the NEGR1 gene, which is strongly expressed in the brain, has been reported to affect neuronal control of food intake and promote obesity [83]. People who have mutations in the NEGR1 gene may have abnormal fat deposition in various peripheral cells, suggesting a potential molecular target for regulating body fat [84]. In the over-representation enrichment analysis, glutathione-related metabolic pathway was found to be common in shared genes of both BF%/HF and BF%/CAD meta-analyses. Experimental studies have reported that glutathione, which is the most abundant antioxidant in the heart, plays a key role in preventing the damage of redox homeostasis to cause obesity-related cardiovascular complications [85, 86]. More importantly, the protective effect of glutathione on blood vessel and brain tissue against oxidative stress has been previously reported and partially explains our findings [87, 88]. However, the nonsignificant genetic correlations between BF% and the other CVD-related traits suggested heterogeneous genetic architecture among CVD-related traits, which cannot be determined without further study.
In addition to the cardiovascular and nervous system, our TWAS also reported tissues enrichment from the exo-/endocrine and digestive system suggesting that the shared pathway between BF% and CVDs might have significant functions extending beyond brain and blood vessels. Besides a variety of adaptations/alterations in cardiac structure and function, body fat may affect the risk of HF with an altered metabolic profile in exo-/endocrine system [89]. The incidence of CVDs are known to be closely related to endocrine factors such as hormone levels in vivo, which may be altered by body fat distribution and then affect the whole body organs such as ovary, thyroid, colon, and liver. For example, adolescent girls with obesity and polycystic ovary syndrome have increased fasting and postprandial plasma triglycerides and ApoB-lipoprotein remnants, and these indices are highly associated with early subclinical CVD risk [90]. Indeed, an essential cause of CVDs is excessive body fat, which can be distributed in various organs of the body and cause pathological changes there. Thus, TWAS provided additional evidence of the enrichment of BF% and CVD genes expressed are not specific to a certain tissue; rather, it seems to be generalizable across metabolic organs of the whole body.
The bidirectional MR analysis showed a significant association between genetically predicted BF% and increased risk of HF (1.63-fold risk per 1-SD increase in BF%), which was stronger than that observed in a community-based cohort study (n = 100,280) at 1.32-fold per 1-SD increase in BF% [91]. Our findings were also consistent with a large cohort study which was conducted in 5520 community-based, elderly individuals, showing that higher BF% might contribute to the development of HF through increasing ventricular-arterial stiffness [92]. However, clinically, the association between BF% and HF could be bidirectional. In addition, previous studies suggested that BMI or body weight was positively or negatively correlated with HF or clinical outcomes [93, 94]. Although various associations were observed between BF% and CVDs, they were inconsistent possibly due to reverse causation and confounding. Therefore, this MR design is warranted to obtain causality which largely avoided bias such as reverse causation and confounding under MR key assumptions. Our MR findings provided reliable evidence of a causal role of BF% in HF based on large GWAS consortia regardless of unknown confounders existing in observational studies, highlighting the benefit of targeting BF% in the prevention of HF. The potential mechanisms underlying the causal association between BF% and HF require investigation, but the shared loci and related pathway could provide new directions in revealing shared etiologies.
In addition, we observed nominal positive correlation of BF% with intracerebral hemorrhage and negative correlation with HDL from genetic correlation analysis, but not with any other lipid profiles. Previous studies have suggested that low HDL may contribute directly to establishing or maintaining the obese condition due to the role of HDL in cellular lipid transport [95]. HDL contribute to modulating body fat content by controlling the extent of lipolysis in mice model, which appears to be key components of lipid metabolism in adipose tissue and constitute new therapeutic targets in obesity [96]. In cell culture studies, HDL specifically increased catecholamine-induced lipolysis possibly through modulating the adipocyte plasma membrane cholesterol content [96]. In addition, the HERITAGE Family Study has shown that high HDL cholesterol levels are good correlates of the metabolic profile, representing a better index in CVD risk assessment [97].
This study had limitations. First, the study is limited by the quality of data collected including the BF% measurement, CAD diagnosis, medication administration, and so on. However, each previous study has conducted the study-specific quality control to ensure the data quality. In addition, considering the natural and random assortment of genetic variants during meiosis yielding the random distribution of genetic variants in populations, the result and conclusion of this genetic analysis are less likely to be influenced by confounding factors. Second, while BF% accurately reflects the proportion of fat content in the human body, it does not give full insight to distinguish between visceral and subcutaneous fat or fat content in each anatomical part. Further studies are required to explore the genetic association of body fat distribution with CVD risk. Third, we used limited number of SNPs as instrumental variables in the MR analysis, so we cannot exclude the possibility that our findings might have been affected by weak instrument bias, although all genetic instruments were associated with the exposure (F-statistic> 10). Fourth, this study did not assess sex-specific genetic effects using LDSC and MR analysis. Since we conducted the analysis using summary-level GWASs without individual data, it is difficult for us to verify the findings of this study for any variation with sex-specific analysis. Fifth, we could not re-test our hypothesis using additional or alternative large cohort without individual information in the present study. However, the data we used are the largest and latest GWASs for these traits, and the sample sizes of other cohorts are small from which we cannot yield more reliable results. Finally, the current study was limited to assessing shared genetic factors between BF% and CVD-related traits, which could only explain a small proportion of these traits. The effect of gene-environment interaction, in which the genetic variants lead to the occurrence of diseases in high-risk environments, may explain part of the rest variance [98]. Although CAD and MI or stroke vest in similar pathophysiologic mechanisms, the complex roles of interactions between genes and environmental factors may be different [99]. More large-scaled, well-conducted studies on gene-environment interaction in the development of CVDs are warranted.
Conclusions
In summary, our findings provide strong evidence of genetic correlations between BF% and CVDs, including HF and CAD. Additionally, we found a causal association between BF% and HF, which further supports targeted reduction of adiposity for improved cardiovascular outcomes. These results advance our understanding of body fat and provide novel insight into the common genetic basis of BF% and CVDs from molecular and functional levels. Notably, we identified novel genetic loci in both single-trait GWAS and cross-trait meta-analysis for multi-trait GWASs using MTAG. This work reinforces the idea that BF% and CVDs share common biological processes and opens up a new way for early prevention of CVDs.
Supplementary Information
Additional file 1: Table S1. Summary of GWAS data. Table S2. SNP based heritability estimated by LDSC. Table S3. Partitioned genetic correlation between BF% and 2 cardiac traits. Table S4. Novel loci in MTAG for single trait compared with GWAS. Table S5. GO biological process pathway analysis for BF% and HF. Table S6. GO biological process pathway analysis for BF% and CAD. Table S7. Reactome pathways analysis for BF% and HF. Table S8. Reactome pathways analysis for BF% and CAD. Table S9. List of credible set SNPs in each locus from fine mapping. For each of the 5 BF% and HF shared loci, the table lists all SNPs within 500 kb in the 99% credible sets that were calculated. Table S10. List of credible set SNPs in each locus from fine mapping. For each of the 16 BF% and CAD shared loci, the table lists all SNPs within 500 kb in the 99% credible sets that were calculated. Table S11. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with BF% and HF with directional effect (Pmeta < 5×10-8; single trait P < 0.05). Table S12. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with BF% and CAD with directional effect (Pmeta < 5×10-8; single trait P < 0.05). Table S13. Specific characteristics of genetic instruments of MR analysis of BF% levels upon CVD traits. Table S14. Specific characteristics of genetic instruments of MR analysis of CVDs upon BF% levels. Table S15. MR analysis between BF% and cardiac traits in a leave one out approach. Table S16. MR analysis between BF% and cardiac traits for a single snp. Table S17. Significant overlap transcriptome-wide association analysis results between BF% and CVDs. (FDR<0.05). Table S18. Significant single-tissue eQTL from GTEx for 3 representative gene between BF% and HF. Table S19. Significant single-tissue eQTL from GTEx for 8 representative gene between BF% and CAD.
Additional file 2: Figure S1. Partitioned genetic correlation between BF% and HF by genetic variants groups with different P value. Figure S2. Partitioned genetic correlation between BF% and CAD by genetic variants groups with different P value. Figure S3. QQ plot of cross-trait meta-analysis between BF% and HF. Figure S4. QQ plot of cross-trait meta-analysis between BF% and CAD.
Acknowledgements
Not applicable.
Abbreviations
- AF
Atrial fibrillation
- BF%
Body fat percentage
- BMI
Body mass index
- CAD
Coronary artery disease
- CARDIoGRAM
Coronary ARtery Disease Genome wide Replication and Meta-analysis
- CNS
Central nervous system
- CVD
Cardiovascular disease
- DGF
DNaseI digital genomic footprinting region
- DHS
DNase I hypersensitivity sites
- GERFHS
Genetic and Environmental Risk Factors for Hemorrhagic Stroke
- GIANT
Genetic Investigation of ANthropometric Traits
- GLGC
Global Lipids Genetics Consortium
- GOCHA
Genetics of Cerebral Hemorrhage with Anticoagulation
- GTEx
Genotype-Tissue Expression
- GWAS
Genome-wide association study
- HDL
High-density lipoprotein
- HERMES
Heart Failure Molecular Epidemiology for Therapeutic Targets
- HF
Heart failure
- HGNC
HUGO Gene Nomenclature Committee
- ICH
Intracerebral hemorrhage
- IS
Ischemic stroke
- ISGC
International Stroke Genetics Consortium
- IVW
Inverse-variance weighted
- LD
Linkage disequilibrium
- LDL
Low-density lipoprotein
- LDSC
Linkage disequilibrium score regression
- MI
Myocardial infarction
- MR
Mendelian randomization
- MTAG
Multi-trait analysis of GWAS
- PRESSO
Pleiotropy Residual Sum and Outlier
- RAPS
Robust Adjusted Profile Scores
- SiGN
NINDS Stroke Genetics Network
- SNP
Single-nucleotide polymorphism
- TC
Total cholesterol
- TFBS
Transcription factor binding site
- TG
Triglycerides
- TSEA
Tissue-specific expression analysis
- TWAS
Transcriptome-wide association study
Authors’ contributions
ZZ, MY, ZL, and TH designed the research. ZL and TH had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. ZZ and MY performed the data analysis. ZZ, MY, and JW wrote the paper. All authors contributed to the statistical analysis, critically reviewed the manuscript during the writing process, and approved the final version to be published. ZL and TH are the guarantors for the study.
Funding
The study was supported by grants from the Peking University Start-up Grant (BMU2018YJ002), High-performance Computing Platform of Peking University. The funding organization had no role in the preparation of the manuscript.
Availability of data and materials
All data used in the present study were obtained from genome-wide association study summary statistics which were publicly released by genetic consortia.
Declarations
Consent to publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Competing interests
All authors declare: no support from companies for the submitted work; no relationships with companies that might have an interest in the submitted work in the previous 3 years; no spouses, partners, or children have no financial relationships that may be relevant to the submitted work; no non-financial interests that may be relevant to the submitted work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhenhuang Zhuang and Minhao Yao contributed equally to this work.
Contributor Information
Zhonghua Liu, Email: zhhliu@hku.hk.
Tao Huang, Email: huangtaotao@pku.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12916-021-01972-z.
References
- 1.Goodarzi MO. Genetics of obesity: what genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol. 2018;6(3):223–236. doi: 10.1016/S2213-8587(17)30200-0. [DOI] [PubMed] [Google Scholar]
- 2.Oikonomou EK, Antoniades C. The role of adipose tissue in cardiovascular health and disease. Nat Rev Cardiol. 2019;16(2):83–99. doi: 10.1038/s41569-018-0097-6. [DOI] [PubMed] [Google Scholar]
- 3.Piché ME, Poirier P, Lemieux I, Després JP. Overview of epidemiology and contribution of obesity and body fat distribution to cardiovascular disease: an update. Prog Cardiovasc Dis. 2018;61(2):103–113. doi: 10.1016/j.pcad.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Padwal R, Leslie WD, Lix LM, Majumdar SR. Relationship among body fat percentage, body mass index, and all-cause mortality: a cohort study. Ann Intern Med. 2016;164(8):532–541. doi: 10.7326/M15-1181. [DOI] [PubMed] [Google Scholar]
- 5.Romero-Corral A, Somers VK, Sierra-Johnson J, Korenfeld Y, Boarin S, Korinek J, et al. Normal weight obesity: a risk factor for cardiometabolic dysregulation and cardiovascular mortality. Eur Heart J. 2010;31(6):737–746. doi: 10.1093/eurheartj/ehp487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero-Corral A, Somers VK, Sierra-Johnson J, Jensen MD, Thomas RJ, Squires RW, et al. Diagnostic performance of body mass index to detect obesity in patients with coronary artery disease. Eur Heart J. 2007;28(17):2087–2093. doi: 10.1093/eurheartj/ehm243. [DOI] [PubMed] [Google Scholar]
- 7.Szabó T, von Haehling S, Doehner W. Differentiating between body fat and lean mass--how should we measure obesity? Nat Clin Pract Endocrinol Metab. 2008;4(11):E1. doi: 10.1038/ncpendmet0999. [DOI] [PubMed] [Google Scholar]
- 8.Lu Y, Day FR, Gustafsson S, Buchkovich ML, Na J, Bataille V, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020;11(1):1–12. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NINDS Stroke Genetics Network (SiGN) International Stroke Genetics Consortium (ISGC) Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 2016;15(2):174–184. doi: 10.1016/S1474-4422(15)00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50(9):1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94(4):511–521. doi: 10.1016/j.ajhg.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Human genomics The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York) 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia working Group of the Psychiatric Genomics C et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchard C, Pérusse L, Leblanc C, Tremblay A, Thériault G. Inheritance of the amount and distribution of human body fat. Int J Obes. 1988;12(3):205–215. [PubMed] [Google Scholar]
- 21.Lindgren MP, PirouziFard M, Smith JG, Sundquist J, Sundquist K, Zöller B. A Swedish Nationwide adoption study of the heritability of heart failure. JAMA Cardiol. 2018;3(8):703–710. doi: 10.1001/jamacardio.2018.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Äijälä M, Ronkainen J, Huusko T, Malo E, Savolainen ER, Savolainen MJ, et al. The fat mass and obesity-associated (FTO) gene variant rs9939609 predicts long-term incidence of cardiovascular disease and related death independent of the traditional risk factors. Ann Med. 2015;47(8):655–663. doi: 10.3109/07853890.2015.1091088. [DOI] [PubMed] [Google Scholar]
- 23.Larsson SC, Bäck M, Rees JMB, Mason AM, Burgess S. Body mass index and body composition in relation to 14 cardiovascular conditions in UK biobank: a Mendelian randomization study. Eur Heart J. 2020;41(2):221–226. doi: 10.1093/eurheartj/ehz388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina-Inojosa JR, Somers VK, Thomas RJ, Jean N, Jenkins SM, Gomez-Ibarra MA, et al. Association Between Adiposity and Lean Mass With Long-Term Cardiovascular Events in Patients With Coronary Artery Disease: No Paradox. J Am Heart Assoc. 2018;7(10):e007505. doi: 10.1161/JAHA.117.007505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byambasukh O, Eisenga MF, Gansevoort RT, Bakker SJ, Corpeleijn E. Body fat estimates from bioelectrical impedance equations in cardiovascular risk assessment: the PREVEND cohort study. Eur J Prev Cardiol. 2019;26(9):905–916. doi: 10.1177/2047487319833283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pajunen P, Jousilahti P, Borodulin K, Harald K, Tuomilehto J, Salomaa V. Body fat measured by a near-infrared interactance device as a predictor of cardiovascular events: the FINRISK'92 cohort. Obesity (Silver Spring, Md) 2011;19(4):848–852. doi: 10.1038/oby.2010.236. [DOI] [PubMed] [Google Scholar]
- 27.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 28.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey SG. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 29.Lavie CJ, Alpert MA, Arena R, Mehra MR, Milani RV, Ventura HO. Impact of obesity and the obesity paradox on prevalence and prognosis in heart failure. JACC Heart Failure. 2013;1(2):93–102. doi: 10.1016/j.jchf.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 30.LDSC. https://github.com/bulik/ldsc. Accessed 1 Mar 2020.
- 31.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89(5):607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MTAG. https://github.com/JonJala/mtag/. Accessed 4 Mar 2020.
- 35.Chung J, Marini S, Pera J, Norrving B, Jimenez-Conde J, Roquer J, et al. Genome-wide association study of cerebral small vessel disease reveals established and novel loci. Brain. 2019;142(10):3176–3189. doi: 10.1093/brain/awz233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.FM-summary. https://github.com/hailianghuang/FM-summary. Accessed 15 Mar 2020.
- 38.Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547(7662):173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dadaev T, Saunders EJ, Newcombe PJ, Anokian E, Leongamornlert DA, Brook MN, et al. Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat Commun. 2018;9(1):2256. doi: 10.1038/s41467-018-04109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coloc. https://github.com/chr1swallace/coloc. Accessed 15 Mar 2020.
- 41.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.TSEA. http://genetics.wustl.edu/jdlab/tsea/. Accessed 20 Mar 2020.
- 43.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38(13):4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Wells AB, O'Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34(4):1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.GTEx. http://www.gtexportal.org. Accessed 25 Mar 2020.
- 47.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Royal Statist Soc B. 1995;57:289–300. [Google Scholar]
- 48.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, et al. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14(3):703–721. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.PANTHER. http://www.pantherdb.org/. Accessed 25 Mar 2020.
- 50.Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. Jama. 2017;318(19):1925–1926. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 51.TwoSampleMR. https://mrcieu.github.io/TwoSampleMR/. Accessed 28 Mar 2020.
- 52.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-egger method. Eur J Epidemiol. 2017;32(5):377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.FUSION http://gusevlab.org/projects/fusion/. Accessed 1 Apr 2020.
- 57.Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10(1):51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458(7240):894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 59.Wu L, Ma F, Zhao X, Zhang MX, Wu J, Mi J. GNPDA2 gene affects adipogenesis and alters the transcriptome profile of human adipose-derived mesenchymal stem cells. Int J Endocrinol. 2019;2019:9145452. doi: 10.1155/2019/9145452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu W, Cheng YC, Chen K, Wang H, Gerhard GS, Still CD, et al. Evidence for several independent genetic variants affecting lipoprotein (a) cholesterol levels. Hum Mol Genet. 2015;24(8):2390–2400. doi: 10.1093/hmg/ddu731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lotta LA, Mokrosiński J, Mendes de Oliveira E, Li C, Sharp SJ, Luan J, et al. Human gain-of-function MC4R variants show signaling bias and protect against obesity. Cell. 2019;177(3):597–607.e599. doi: 10.1016/j.cell.2019.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welch CL, Sun Y, Arey BJ, Lemaitre V, Sharma N, Ishibashi M, et al. Spontaneous atherothrombosis and medial degradation in Apoe−/−, Npc1−/− mice. Circulation. 2007;116(21):2444–2452. doi: 10.1161/CIRCULATIONAHA.107.701276. [DOI] [PubMed] [Google Scholar]
- 63.Liu X, Chen Q, Tsai HJ, Wang G, Hong X, Zhou Y, et al. Maternal preconception body mass index and offspring cord blood DNA methylation: exploration of early life origins of disease. Environ Mol Mutagen. 2014;55(3):223–230. doi: 10.1002/em.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller TE, Henkels KM, Huddleston M, Salisbury R, Hussain SM, Sasaki AT, et al. Depletion of phosphatidylinositol 4-phosphate at the Golgi translocates K-Ras to mitochondria. J Cell Sci. 2019;132(16):jcs231886. doi: 10.1242/jcs.231886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grube M, Hagen P, Jedlitschky G. Neurosteroid transport in the brain: role of ABC and SLC transporters. Front Pharmacol. 2018;9:354. doi: 10.3389/fphar.2018.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erbilgin A, Civelek M, Romanoski CE, Pan C, Hagopian R, Berliner JA, et al. Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J Lipid Res. 2013;54(7):1894–1905. doi: 10.1194/jlr.M037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y, Wang F, Li L, Gao H, Arckacki S, Wang IZ, et al. Genome-wide linkage analysis of large multiple multigenerational families identifies novel genetic loci for coronary artery disease. Sci Rep. 2017;7(1):5472. doi: 10.1038/s41598-017-05381-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Correa-Rodríguez M, González-Ruíz K, Rincón-Pabón D, Izquierdo M, García-Hermoso A, Agostinis-Sobrinho C, et al. Normal-weight obesity is associated with increased cardiometabolic risk in young adults. Nutrients. 2020;12(4):1106. doi: 10.3390/nu12041106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ouyang H, Zhang H, Li W, Liang S, Jebessa E, Abdalla BA, et al. Identification, expression and variation of the GNPDA2 gene, and its association with body weight and fatness traits in chicken. PeerJ. 2016;4:e2129. doi: 10.7717/peerj.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamri A, Pigeyre M, Garver WS, Meyre D. The extending spectrum of NPC1-related human disorders: from Niemann-Pick C1 disease to obesity. Endocr Rev. 2018;39(2):192–220. doi: 10.1210/er.2017-00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu R, Zou Y, Hong J, Cao M, Cui B, Zhang H, et al. Rare loss-of-function variants in NPC1 predispose to human obesity. Diabetes. 2017;66(4):935–947. doi: 10.2337/db16-0877. [DOI] [PubMed] [Google Scholar]
- 72.Yu XH, Jiang N, Yao PB, Zheng XL, Cayabyab FS, Tang CK. NPC1, intracellular cholesterol trafficking and atherosclerosis. Clin Chim Acta. 2014;429:69–75. doi: 10.1016/j.cca.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 73.Shetty R, Joshi D, Jain M, Vasudevan M, Paul JC, Bhat G, et al. Rudhira/BCAS3 is essential for mouse development and cardiovascular patterning. Sci Rep. 2018;8(1):5632. doi: 10.1038/s41598-018-24014-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrov AM, Lam M, Mast N, Moon J, Li Y, Maxfield E, et al. CYP46A1 activation by Efavirenz leads to behavioral improvement without significant changes in amyloid plaque load in the brain of 5XFAD mice. Neurotherapeutics. 2019;16(3):710–724. doi: 10.1007/s13311-019-00737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matsumoto Y, Suzuki A, Shirata T, Takahashi N, Noto K, Goto K, et al. Implication of the DGKH genotype in openness to experience, a premorbid personality trait of bipolar disorder. J Affect Disord. 2018;238:539–541. doi: 10.1016/j.jad.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 77.Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D'Alessandro A, et al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov. 2019;9(7):910–925. doi: 10.1158/2159-8290.CD-19-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nichols BA, Oswald NW, McMillan EA, McGlynn K, Yan J, Kim MS, et al. HORMAD1 is a negative prognostic indicator in lung adenocarcinoma and specifies resistance to oxidative and genotoxic stress. Cancer Res. 2018;78(21):6196–6208. doi: 10.1158/0008-5472.CAN-18-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang C, Ge S, Wang J, Jing X, Li H, Mei S, et al. Epigenomic profiling of DNA methylation for hepatocellular carcinoma diagnosis and prognosis prediction. J Gastroenterol Hepatol. 2019;34(10):1869–1877. doi: 10.1111/jgh.14694. [DOI] [PubMed] [Google Scholar]
- 80.Krüger N, Biwer LA, Good ME, Ruddiman CA, Wolpe AG, DeLalio LJ, et al. Loss of endothelial FTO antagonizes obesity-induced metabolic and vascular dysfunction. Circ Res. 2020;126(2):232–242. doi: 10.1161/CIRCRESAHA.119.315531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N, et al. FTO-dependent N (6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139(4):518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mo XB, Lei SF, Zhang YH, Zhang H. Detection of m (6) A-associated SNPs as potential functional variants for coronary artery disease. Epigenomics. 2018;10(10):1279–1287. doi: 10.2217/epi-2018-0007. [DOI] [PubMed] [Google Scholar]
- 83.Gamero-Villarroel C, González LM, Gordillo I, Carrillo JA, García-Herráiz A, Flores I, et al. Impact of NEGR1 genetic variability on psychological traits of patients with eating disorders. Pharmacogenomics J. 2015;15(3):278–283. doi: 10.1038/tpj.2014.53. [DOI] [PubMed] [Google Scholar]
- 84.Joo Y, Kim H, Lee S, Lee S. Neuronal growth regulator 1-deficient mice show increased adiposity and decreased muscle mass. Int J Obes. 2019;43(9):1769–1782. doi: 10.1038/s41366-019-0376-2. [DOI] [PubMed] [Google Scholar]
- 85.Bajic VP, Van Neste C, Obradovic M, Zafirovic S, Radak D, Bajic VB, et al. Glutathione “redox homeostasis” and its relation to cardiovascular disease. Oxidative Med Cell Longev. 2019;2019:5028181. doi: 10.1155/2019/5028181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Darroudi S, Fereydouni N, Tayefi M, Ahmadnezhad M, Zamani P, Tayefi B, et al. Oxidative stress and inflammation, two features associated with a high percentage body fat, and that may lead to diabetes mellitus and metabolic syndrome. BioFactors (Oxford) 2019;45(1):35–42. doi: 10.1002/biof.1459. [DOI] [PubMed] [Google Scholar]
- 87.Ruiz-Ramírez A, Ortiz-Balderas E, Cardozo-Saldaña G, Diaz-Diaz E, El-Hafidi M. Glycine restores glutathione and protects against oxidative stress in vascular tissue from sucrose-fed rats. Clin Sci (Lond) 2014;126(1):19–29. doi: 10.1042/CS20130164. [DOI] [PubMed] [Google Scholar]
- 88.González-Fraguela ME, Blanco L, Fernández CI, Lorigados L, Serrano T, Fernández JL. Glutathione depletion: starting point of brain metabolic stress, neuroinflammation and cognitive impairment in rats. Brain Res Bull. 2018;137:120–131. doi: 10.1016/j.brainresbull.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 89.Poirier P, Giles TD, Bray GA, Hong Y, Stern JS, Pi-Sunyer FX, et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss: an update of the 1997 American Heart Association Scientific Statement on obesity and heart disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation. 2006;113(6):898–918. doi: 10.1161/CIRCULATIONAHA.106.171016. [DOI] [PubMed] [Google Scholar]
- 90.Vine DF, Wang Y, Jetha MM, Ball GD, Proctor SD. Impaired ApoB-lipoprotein and triglyceride metabolism in obese adolescents with polycystic ovary syndrome. J Clin Endocrinol Metab. 2017;102(3):970–982. doi: 10.1210/jc.2016-2854. [DOI] [PubMed] [Google Scholar]
- 91.Si S, Tewara MA, Ji X, Wang Y, Liu Y, Dai X, et al. Body surface area, height, and body fat percentage as more sensitive risk factors of cancer and cardiovascular disease. Cancer Med. 2020;9(12):4433–4446. doi: 10.1002/cam4.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fernandes-Silva MM, Shah AM, Claggett B, Cheng S, Tanaka H, Silvestre OM, et al. Adiposity, body composition and ventricular-arterial stiffness in the elderly: the atherosclerosis risk in communities study. Eur J Heart Fail. 2018;20(8):1191–1201. doi: 10.1002/ejhf.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kenchaiah S, Evans JC, Levy D, Wilson PW, Benjamin EJ, Larson MG, et al. Obesity and the risk of heart failure. N Engl J Med. 2002;347(5):305–313. doi: 10.1056/NEJMoa020245. [DOI] [PubMed] [Google Scholar]
- 94.Horwich TB, Fonarow GC, Hamilton MA, MacLellan WR, Woo MA, Tillisch JH. The relationship between obesity and mortality in patients with heart failure. J Am Coll Cardiol. 2001;38(3):789–795. doi: 10.1016/S0735-1097(01)01448-6. [DOI] [PubMed] [Google Scholar]
- 95.Stadler JT, Marsche G. Obesity-related changes in high-density lipoprotein metabolism and function. Int J Mol Sci. 2020;21(23):8985. doi: 10.3390/ijms21238985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wei H, Averill MM, McMillen TS, Dastvan F, Mitra P, Subramanian S, et al. Modulation of adipose tissue lipolysis and body weight by high-density lipoproteins in mice. Nutrition & diabetes. 2014;4(2):e108. doi: 10.1038/nutd.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Couillard C, Bergeron J, Després JP, Gagnon J, Rankinen T, Leon AS, et al. Apolipoprotein AI- and AI:AII-containing lipoproteins in white men and women of the HERITAGE family study: associations with metabolic risk profile variables. Metab Clin Exp. 2003;52(12):1530–1536. doi: 10.1016/j.metabol.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 98.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 99.Heianza Y, Qi L. Impact of genes and environment on obesity and cardiovascular disease. Endocrinology. 2019;160(1):81–100. doi: 10.1210/en.2018-00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Summary of GWAS data. Table S2. SNP based heritability estimated by LDSC. Table S3. Partitioned genetic correlation between BF% and 2 cardiac traits. Table S4. Novel loci in MTAG for single trait compared with GWAS. Table S5. GO biological process pathway analysis for BF% and HF. Table S6. GO biological process pathway analysis for BF% and CAD. Table S7. Reactome pathways analysis for BF% and HF. Table S8. Reactome pathways analysis for BF% and CAD. Table S9. List of credible set SNPs in each locus from fine mapping. For each of the 5 BF% and HF shared loci, the table lists all SNPs within 500 kb in the 99% credible sets that were calculated. Table S10. List of credible set SNPs in each locus from fine mapping. For each of the 16 BF% and CAD shared loci, the table lists all SNPs within 500 kb in the 99% credible sets that were calculated. Table S11. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with BF% and HF with directional effect (Pmeta < 5×10-8; single trait P < 0.05). Table S12. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with BF% and CAD with directional effect (Pmeta < 5×10-8; single trait P < 0.05). Table S13. Specific characteristics of genetic instruments of MR analysis of BF% levels upon CVD traits. Table S14. Specific characteristics of genetic instruments of MR analysis of CVDs upon BF% levels. Table S15. MR analysis between BF% and cardiac traits in a leave one out approach. Table S16. MR analysis between BF% and cardiac traits for a single snp. Table S17. Significant overlap transcriptome-wide association analysis results between BF% and CVDs. (FDR<0.05). Table S18. Significant single-tissue eQTL from GTEx for 3 representative gene between BF% and HF. Table S19. Significant single-tissue eQTL from GTEx for 8 representative gene between BF% and CAD.
Additional file 2: Figure S1. Partitioned genetic correlation between BF% and HF by genetic variants groups with different P value. Figure S2. Partitioned genetic correlation between BF% and CAD by genetic variants groups with different P value. Figure S3. QQ plot of cross-trait meta-analysis between BF% and HF. Figure S4. QQ plot of cross-trait meta-analysis between BF% and CAD.
Data Availability Statement
All data used in the present study were obtained from genome-wide association study summary statistics which were publicly released by genetic consortia.