Abstract
Demonstrated improvements in patient outcomes will facilitate the clinical implementation of pharmacogenetic testing. Using the association between solute carrier organic anion transporter family member 1B1 (SLCO1B1) and statin-associated muscle symptoms (SAMS) as a model, we conducted a systematic review of patient outcomes after delivery of SLCO1B1 results. Using PubMed and Embase searches through December 19, 2017, we identified 37 eligible records reporting preliminary or final outcomes, including 6 studies delivering only SLCO1B1 results and 5 large healthcare system-based implementation projects of multi-pharmacogene panels. Two small trials have demonstrated at least short-term improvements in low-density lipoprotein cholesterol after SLCO1B1 testing among previously statin intolerant patients. Evidence from large implementation projects suggests that SLCO1B1 results may change prescribing patterns for some high-risk patients. No study has reported improvements in SAMS or cardiovascular events or tracked the economic outcomes of SLCO1B1 testing. Ongoing studies should collect and report outcomes relevant to pharmacogenetics stakeholders.
Keywords: Pharmacogenetics, hydroxymethylglutaryl-CoA reductase inhibitors, solute carrier organic anion transporter family member 1b1 (SLCO1B1), comparative effectiveness research
Introduction
The field of pharmacogenetics is one beneficiary of the last decade’s accelerated pace of genomic discovery(1, 2). Hundreds of drug-gene associations have now been identified that have the potential to help prescribers and patients optimize the risk-benefit ratio of pharmacotherapy(3). These pharmacogenetic associations could be ideal candidates for the translation of genomic discovery into patient care, since test results might have ready actionability to inform drug selection and dose(4, 5) and might lend themselves to clinical decision support (CDS) interventions in the electronic health record (EHR)(6-9). Some healthcare systems in the United States and worldwide are making large investments to implement clinical pharmacogenetics programs into their healthcare delivery systems(10-14), but most health care is still delivered in settings without the routine use of pharmacogenetic testing. Innovation generally diffuses slowly throughout medical practice(15), and for pharmacogenetics, this lag is exacerbated in part by healthcare providers and insurers who remain unconvinced of its value in improving the health care and outcomes of patients(16-19). Evidence that pharmacogenetic testing improves patient outcomes is needed to break this impasse(16-19).
One well validated drug-gene association is the interaction between simvastatin and the solute carrier organic anion transporter family member 1B1 (SLCO1B1) gene for the risk of statin-associated muscle symptoms (SAMS). Statins, or 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors, are cholesterol-lowering medications used by over 30 million Americans that have dramatically reduced the risk of cardiovascular disease (CVD) and death in the U.S.(20). Statins are generally well tolerated, but up to 20% of patients describe muscle aches or weakness(21, 22) and up to 1 in 10,000 experience life-threatening myopathy(23-25). In 2008, a high-profile genome-wide association study using data from the Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH) trial identified a robust association between a common genetic variant in SLCO1B1 and simvastatin-related myopathy(26). Each copy of the C minor allele at rs4149056 (contained within the SLCO1B1*5, *15, and *17 haplotypes) increased myopathy risk by a factor of 4.5, such that CC homozygotes had a 16.9-fold increased risk compared to TT homozygotes(26). Numerous other studies have gone on to replicate the association between SLCO1B1 and SAMS, particularly with simvastatin(27-30). In 2012, the Clinical Pharmacogenetics Implementation Consortium (CPIC) issued its first recommendations for simvastatin prescribing and dosing when a patient’s SLCO1B1 genotype is known(31), and some early-adopter healthcare systems are now incorporating SLCO1B1 genotyping into their clinical and research pharmacogenetics programs(10-14).
Although it makes intuitive sense that the use of SLCO1B1 testing in clinical care would help prescribers and patients avoid statin-related adverse effects, intuition might not be sufficiently persuasive to promote its widespread adoption among providers and payers. Several reviews have examined the validity of the SLCO1B1-SAMS association(30, 32, 33), but none has reviewed the prospective outcomes of integrating SLCO1B1 genotype information into clinical care. Given the primacy of outcomes data in determining the clinical utility of pharmacogenetic testing, we performed a systematic review of studies reporting outcomes after the delivery of SLCO1B1 results.
Results
Search results
The database searches described in the Methods section below yielded 5,374 unique records (Figure 1). Manual author and reference searches identified another 57 potentially eligible records, including one personal author communication providing unpublished results. After full-text review, we identified 37 records describing 16 eligible studies with completed or preliminary outcomes, including 2 nonrandomized trials and 4 randomized controlled trials (RCT). Table 1 presents the characteristics of these studies. We identified 17 records describing another 10 studies with potentially eligible study designs that have not yet reported outcomes.
Figure 1:
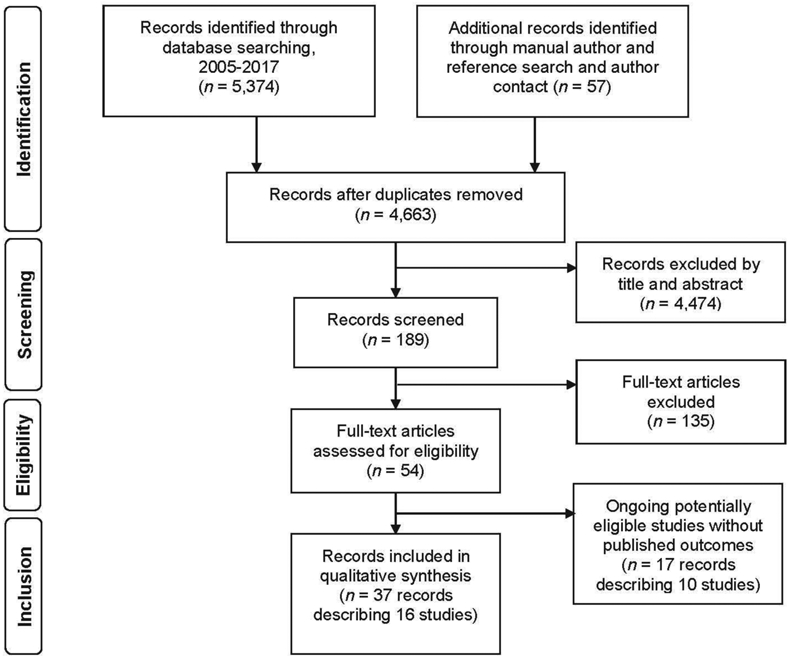
PRISMA flow diagram of search results
Table 1.
Characteristics of eligible studies
| Study/institution | Design (country) |
Population (n) | Genotype(s) reported | Experimental intervention | Decision support | Control group (n) |
|---|---|---|---|---|---|---|
| 1200 Patients Project*(11, 44-49) | Intervention study (U.S.A.) | Patients aged ≥65 years receiving care from one of 17 providers at 8 primary or specialty care clinics (1,108) | ABCB1, ADD1, ADRB1, AGT, CACNA1C, CYP3A4, CYP2C9, CYP2C19, CYP2D6, GNB3, GRK4, KIF6, LDLR, LTC4S, REN, SLCO1B1, VKORC1 | • Genotyping of prospectively enrolled cohort • PGx results available to participating providers in institutional PGx CDS system. • At participants’ office visits, providers alerted verbally or by chart flagging to PGx results |
• CDS system outside EHR including green/yellow/red alerts with clinical summaries and interpretation • Providers could query CDS system for information for other drugs by name or by disease |
--- |
| AltheaDx(61, 63) | Intervention study (U.S.A.) | Patients at several long-term care facilities taking ≥5 medications (132) | COMT, CYP1A2, CYP2C9,CYP2C19, CYP2D6, CYP3A4/CYP3A5, F2, F5, HTR2A, HTR2C, MTHFR, OPRM1, SLC6A2, SLC6A4, SLCO1B1, VKORC1 | • Potential drug-drug drug-environment, and drug-gene interactions reported to physicians • Internal medication management assessment by pharmacists |
Reports listed commonly used medications categorized as “Use as directed” or “Use with caution and/or increased monitoring" | --- |
| Duke University 1 (39) | Non-randomized trial (U.S.A.) | Primary care patients with a history of statin nonadherence (58) | SLCO1B1 | PGx results available to providers through EHR and to patients through patient portal | • Genotype-specific information about patient’s myopathy risk • Recommendations for statin prescribing |
Concurrent controls: patients with prior statin prescription but without statin use in prior 3 months (59) |
| Duke University 2(40) | Intervention study (U.S.A.) | Patients receiving new or recurrent simvastatin prescriptions from pharmacists at 5 community pharmacies (19) | SLCO1B1 (+/− CYP2C19) | • PGx results accessible through lab database and faxed to pharmacist and prescriber • Pharmacists reviewed results with patients and prescribers |
Test interpretation included CPIC guidelines | --- |
| Duke University 3(37, 38) | Intervention study (U.S.A.) | Patients at 2 cardiology clinics (30) • Taking simvastatin and/or clopidogrel • No prior PGx testing or MTM in prior 3 years |
CYP2C9, CYP2C19, CYP2D6, SLCO1B1, VKORC1 | • Pre- and post-test MTM sessions with pharmacist for patients • MTM also included recommendations for lifestyle modification and OTC medications • Patients and referring cardiologists received PGx results |
Pharmacist discussed recommendations based on FDA and/or CPIC guidelines with cardiologist before sharing action plan with patients | --- |
| Duke University 4(34-36) | Non-randomized trial (U.S.A.) | Primary care patients at 2 internal medicine practices (63) | CYP2C9, CYP2C19, CYP2D6, HLA-B*1502, SLCO1B1, VKORC1 | • Site 1: Pharmacist on call: physician consulted pharmacist about PGx testing; pharmacists screened patients and notified physicians about eligibility • Site 2: Pharmacist in-house: pharmacist screened patients and alerted physicians to availability of PGx testing for relevant medications |
All participating physicians first attended a 1-hour CME session about PGx. | --- |
| Duke University 5(42, 43) | Randomized control trial (U.S.A.) | Patients not currently taking statins due to history of adverse effect, ineligible if prior rhabdomyolysis or CK >10xULN (167) | SLCO1B1 | • Genotyping at a research visit. • Patients and their physicians received PGx results by email |
Physician reports: • SLCO1B1 results • Risk of rhabdomyolysis • General expectations of LDL-C reduction from different statin types & doses Patient reports: • SLCO1B1 results • Reassurance about which statins should be tolerable and lower CVD risk |
Physician reports: • General information about LDL-C reductions from different statins Patient reports: • General information about statins and CVD risk |
| First Moscow State Medical University(41) | Intervention study (Russia) | Patients with hyperlipidemia already on statin therapy(35) | SLCO1B1 | Patients received results | Not specified | --- |
| INGENI0US**(13, 50, 51) | Randomized control trial (U.S.A.) | Safety-net hospital system of 10 clinics with common EHR (2,000 planned) • Age ≥18 years • New prescription for one of 28 index medications |
CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, CYP4F2, DPYD, G6PD, IFNL3, ITPA, SLCO1B1, TPMT, VK0RC1 | • Enrolling prescriber receives PGx reports • PGx results uploaded to EHR and viewable by all providers |
• Reports include alternative prescribing recommendations for index medication based on CPIC guidelines • Subsequent prescription prompts an alert notifying provider a genotype report is available and gives dosing recommendations • Providers may consult PGx service that documents recommendations in EHR |
Patient does not undergo genotyping (4000 planned) |
| La Paz University Hospital(10) | Intervention study (Spain) | Patients receiving specialty care at a tertiary care teaching hospital (600) | ABCB1, ABCC2, ABCG2, APOE, COMT, CFTR, CYP2C19, CYP2C8, CYP2C9, CYP2D6, CYP3A4, CYP3A5, CYP4F2, DPYD, ERCC1, EPHX1, FCGR2A, HTR2A, IL10, IL23R, KCNJ6, MTHFR, POR, SLC15A2, SLC22A1, SLC22A2, SLC22A6, SLCO1B1, TLR2, TLR9, TNF, TP53, TPMT, UGT1A1, UGT2B7, VKORC1, XPC, XRCC1 | Providers request testing from PGx unit, which generates report according to prespecified drug-gene protocol or after PGx consultation | Recommendation from PGx unit, based on CPIC and DPWG reviews | --- |
| Marshfield Clinic***(12, 52) | Intervention study (U.S.A.) | Adults aged ≥50 years with healthcare system primary care physician and no prior use of simvastatin, warfarin, or clopidogrel (750 planned) | CYP2C9, CYP2C19, SLCO1B1, VKORC1 | • Providers receive PGx results • Patients have access to website with information about their PGx results |
Active CDS alerts with CPIC dosing recommendation triggered by prescription in EHR | --- |
| MedSeq Project(66, 67) | Randomized control trial (U.S.A.) | Generally healthy adult primary care patients (100) | Genome sequencing including monogenic disease variants, carrier status, 8 polygenic risks, and 5 PGx results: ABCB1, C11orf65, CYP2C9/VKORC1, CYP2C19, SLCO1B1 | Patients discussed interpreted genome report and family history pedigree with physician | Report included statement about simvastatin-associated myopathy risk | Patients discussed family history pedigree alone with physician |
| OSU-Coriell Personalized Medicine Collaborative*(11, 49, 58, 59, 62, 64, 65) | Intervention study (U.S.A.) | Participants with heart failure and hypertension enrolled in RCT of genomic counseling for polygenic risk estimates and CYP2C19 (208). | Polygenic risk estimates for 8 diseases plus PGx results for CYP2C9, VK0RC1, CYP4F2, CYP2C19, SLCO1B1 | • Patients received genetic reports by mail and by patient web portal • Reports also uploaded to EHR • Half of patients were randomly allocated to in-person genomic counseling; half could access a genetic counselor by phone if requested |
Reports to patients and in EHR included CPIC recommendations | --- |
| PRIMER(60) | Intervention study (U.S.A.) | Patients of 27 providers with likelihood of exposure to a relevant medication (705) | COMT, CYP2D6, CYP2C19, CYP2C9, CYP1A2, CYP3A4, CYP3A5, F2, F5, MTHFR, OPRM1, SLCO1B1, SLC6A4, VKORC1 | Providers ordered PGx panel testing and received report | Report with drug-drug and drug-gene interactions categorized as contraindicated, major, moderate, or minor, some with explanatory annotations. | --- |
| RIGHT Protocol***(8, 11, 12, 53-56) | Intervention study (U.S.A.) | Health system patients, including biobank participants, likely to initiate statin treatment within 3 years (3,788) | CYP2C9, CYP2C19, CYP2D6, HLA-B*1502, HLA-B*5701, SLCO1B1, TPMT, VKORC1 | Preemptive genotyping, with results available to provider in EHR and to patients through patient portal | Active CDS • Alerts triggered when simvastatin ordered on high-risk patients • Alerts sent to provider and added to problem list Passive CDS • Internal online medical info system, AskMayoExpert |
--- |
| Yale University(57) | Intervention study (U.S.A.) | Series of consecutive high-risk cardiovascular patients (32) | CYP2C19, CYP2C9, CYP2D6, CYP3A4, CYP3A5, F2, F5, MTHFR, SLCO1B1, VKORC1 | Results reported to clinicians | Not specified | --- |
Part of the Pharmacogenomics Research Network Translational Pharmacogenetics Program(11)
Part of the Implementing Genomics in Practice (IGNITE) Consortium(13)
Part of the eMERGE-PGx Consortium(12)
Abbreviations: CDS (clinical decision support), CK (creatine kinase), CME (continuing medical education), CPIC (Clinical Pharmacogenetics Implementation Consortium), CVD (cardiovascular disease), DPWG (Dutch Pharmacogenetics Working Group), EHR (electronic health record), FDA (Food & Drug Administration), LDL-C (low-density lipoprotein cholesterol), MTM (medication therapy management), OSU (The Ohio State University), OTC (over-the-counter), PGx (pharmacogenetics), ULN (upper limit of normal)
Five pilot studies and one RCT have studied the delivery of SLCO1B1 results specifically(34-43), all but one of which were conducted by investigators at Duke University(34-40, 42, 43). Three of the Duke pilot studies involved pharmacists in the identification of patients for SLCO1B1 genotyping and/or the formulation of medication recommendations based on SLCO1B1 results(34-38, 40). A fourth Duke study, a pilot nonrandomized trial, enrolled 58 patients with prior statin nonadherence and studied the impact of SLCO1B1 results delivery on statin prescriptions and medication adherence, compared to concurrent controls(39). This pilot informed the design of a larger trial, the only published RCT designed specifically to examine the clinical impact of SLCO1B1 testing. In this RCT by Voora and colleagues(42, 43), 167 patients with prior statin intolerance were randomly allocated to SLCO1B1 results delivery to patients and providers at baseline versus at study end. The study was powered to detect a 1-point difference in the primary outcome of a medication adherence scale; secondary outcomes included low-density lipoprotein cholesterol (LDL-C) at 3 and 8 months, the Brief Pain Inventory, and the Short Form Health Survey (SF-12) quality-of-life measure(42, 43).
In addition, five large healthcare system-based pharmacogenetics implementation projects of multigene panels including SLCO1B1 have reported preliminary or final results: four in the U.S. (1,200 Patients Project(11, 44-49); INGENIOUS(13, 50, 51); the Marshfield Clinic(12, 52); and the RIGHT protocol(8, 11, 12, 53-56)) and one from La Paz University Hospital in Spain(10)(Table 1). Most of these are using passive or active CDS in the EHR to support prescriber utilization of pharmacogenetic results in several different clinically actionable pharmacogenes. The remaining studies identified by our search included smaller single-arm intervention studies of multigene panels(11, 49, 57-65) and one pilot RCT of genome sequencing(66, 67)(Table 1).
Study quality and risk of bias
The included studies generally had poor to moderate quality and risk of bias (Table S1). Most intervention studies suffered from poor comparability on the Newcastle-Ottawa Scale due to the absence of a comparator group.
Reported outcomes
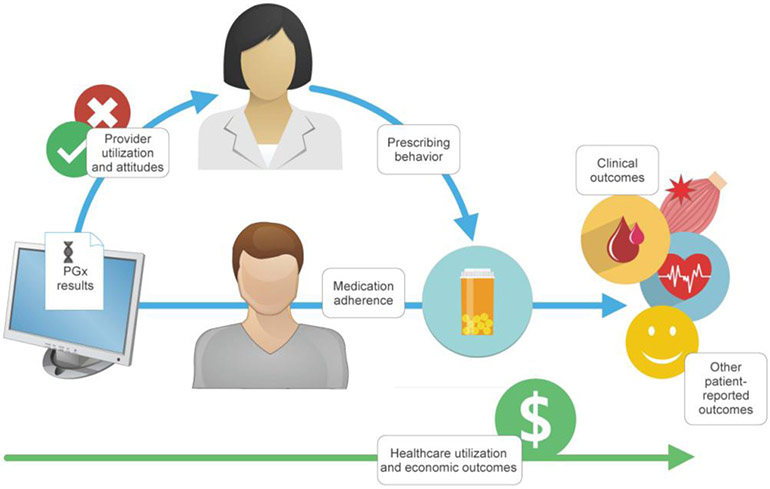
Review of the outcomes reported in the eligible studies led to the conceptual model of patient and healthcare outcomes shown in Figure 2, which guided the creation of the 6 outcome categories in Table 2 and our presentation of the results below. In this conceptual model, SLCO1B1 results might act on provider and patient attitudes and behaviors to effect a change in clinical, economic, and other outcomes (Figure 2). As presented below, we categorized all reported outcomes as either utility outcomes (including clinical outcomes and healthcare utilization and economic outcomes) or process outcomes that might mediate the relationship between SLCO1B1 testing (including provider utilization and attitudes, prescribing behavior and prescriptions, medication adherence, and other patient-reported outcomes.
Figure 2:
Conceptual model of patient and healthcare outcomes after delivery of SLCO1B1 pharmacogenetic test results
Table 2.
Utility and process outcomes after SLCO1B1 genotyping in eligible studies
| Utility outcomes | Process outcomes | ||||||
|---|---|---|---|---|---|---|---|
| Study/Institution | Clinical outcomes | Healthcare utilization and economic outcomes |
Provider utilizatio n and attitudes |
Prescribing behavior and prescriptions |
Medication adherence |
Other patient- reported outcomes |
Key findings |
| 1200 Patients Project*(11, 44-49) | X | X | • Among 868 patients over 3 years, CDS influenced simvastatin discontinuation in 8 cases | ||||
| AltheaDx(61, 63) | X | • 54/112 (48%) of patients had ≥1 medication change | |||||
| Duke University 1 (39) | X | X | X | • 47% vs. 15% of intervention vs. control patients taking statin after 4 months (p< 0.001) • 1-year LDL-C reduction −12 ± 45 mg/dL in invention group vs. +6 ± 38 mg/dL in controls (p=0.06) |
|||
| Duke University 2(40) | X | X | X | X | • No prescription changes observed | ||
| Duke University 3(37, 38) | X | X | X | X | • Pharmacist medication management and PGx visit lasted mean 16 (range 8-29) minutes • 3 PGx-based medication changes were made among 28 patients |
||
| Duke University 4(34-36) | X | X | X | X | X | • Providers consulted pharmacist for 15 cases, averaging 5.7 minutes per consult. • 1/63 patients tested had simvastatin dose halved due to SLCO1B1*5 result. |
|
| Duke University 5(42, 43) | X | X | X | • LDL-C lower in the intervention group vs. controls at 3 months (132 ± 42 mg/dL vs.144 ± 43 mg/dL, p=0.04) but not 8 months (129 ± 38 mg/dL vs. 141 ± 44 mg/dL, p= 0.07) • No between-group differences in pain or quality of life |
|||
| First Moscow State Medical University(41) | X | X | • No medication side effects reported | ||||
| INGENIOUS**(13, 50, 51) | X | • Genotyping prompted PGx consults 10/106 (9%) patients | |||||
| La Paz University Hospital(10) | X | X | X | • Clinical PGx service cost €202,140 over 3 years, with each consult averaging €216 | |||
| Marshfield Clinic***(12, 52) | X | X | • Simvastatin prescriptions have triggered 5 CDS alerts over 3 years, prompting 1 medication change | ||||
| MedSeq Project(66, 67) | X | X | X | X | X | • No between-group differences in 6-month healthcare costs (mean $1490 vs. %1142). • No medication side effects reported |
|
| OSU-Coriell Personalized Medicine Collaborative*(11, 49, 58, 59, 62, 64, 65) | X | X | • Mean patient willingness-to-pay for clinical PGx service was $56 ($81) | ||||
| PRIMER(60) | X | X | X | • 42% of patients not willing to pay out-of-pocket costs for PGx tests; 58% of remainder reported willingness-to-pay of $100 | |||
| RIGHT Protocol***(8, 11, 12, 53-56) | X | X | • No medication side effects reported | ||||
| Yale University(57) | X | X | • No medication side effects reported | ||||
Outcomes are categorized as utility outcomes from SLCO1B1 testing or the process outcomes that might mediate the relationship between SLCO1B1 testing and utlity outcomes. Clinical outcomes include biomarker changes, morbidity, and mortality, while healthcare utilization and economic outcomes include willingness-to-pay and the healthcare costs or other resources required to implement the intervention or resulting from its implementation. Provider utilization and attitudes include frequency of test ordering by providers, their use of the information, and their attitudes about its value. Prescribing behavior includes medication prescriptions, while medication adherence measures patient use of prescribed medications. Other patient-reported outcomes include patient recall of test results, concern or distress about results, and patient perceived utility of the information. Bolded text in the Key Findings column refer to utility outcomes (either clinical or healthcare utilization/economic outcomes). Abbreviations: CDS, clinical decision support; LDL-C, low-density lipoprotein cholesterol; PGx, pharmacogenetic
Part of the Pharmacogenomics Research Network Translational Pharmacogenetics Program(11)
Part of the Implementing Genomics in Practice (IGNITE) Consortium(13)
Part of the eMERGE-PGx Consortium(12)
Utility outcomes
Clinical outcomes
Only two studies to date have quantitatively reported clinical outcomes after SLCO1B1 testing. The Duke nonrandomized pilot trial among previously statin-intolerant patients reported a non-significantly greater reduction in LDL-C in the intervention group (−12 ± 45 mg/dL) compared to concurrent controls (+6 ± 38 mg/dL, p=0.06) after one year(39). In the subsequent RCT, LDL-C values were significantly lower in the intervention group compared to the control group at 3 months (132 ± 42 mg/dL vs. 144 ± 43 mg/dL, p=0.04) but not at 8 months (129 ± 38 mg/dL vs. 141 ± 44 mg/dL, p=0.07)(43). Improvements observed in total cholesterol, but not high-density lipoprotein cholesterol or triglycerides, were consistent with these LDL-C changes. In a follow-up analysis, when patients in the usual care arm received their SLCO1B1 results at the end of the study, they had a greater decrease in LDL-C values compared to intervention patients during the same post-study period, such that the two arms ultimately achieved similar LDL-C reductions from baseline. This RCT found no significant differences in medication side effects between the intervention arms, as measured by pain and quality-of-life instruments(43). Other studies of pharmacogenetic testing have made general qualitative statements that no participants experienced medication side effects during the observation periods(41, 57, 60, 66). No study has reported creatinine kinase values, SAMS, or cardiovascular events after SLCO1B1 testing.
Healthcare utilization and economic outcomes
Some studies have tracked the costs and resources required to conduct their pharmacogenetics projects or those incurred as a result of that implementation(10, 38, 50, 66). The clinical pharmacogenetic service in Spain cost the national health system €202,140 over 3 years, with the cost of each consultation averaging€216(10). Preliminary results from the INGENIOUS RCT of pharmacogenetic panel testing show that genotyping the first 106 participants generated 25 actionable genotypes and prompted 10 (9%) consult requests by physicians(50). Two studies have reported patient willingness-to-pay for multigene pharmacogenetic testing that included SLCO1B1. In one, participants in the OSU-Coriell Personalized Medicine Collaborative RCT of genomic counseling, 28% of whom had an actionable SLCO1B1 result, reported a mean (SD) willingness to pay of $56 ($81) for a clinical pharmacogenetics service(59). In the second, the RIGHT Protocol at the Mayo Clinic, a survey of 869 participants who had undergone panel pharmacogenotyping found that 42% were not willing to incur out-of-pocket costs for pharmacogenetic tests; 58% of the rest reported a maximum willingness-to-pay of $100(54). No study has reported downstream healthcare costs after receipt of SLCO1B1 results specifically, although the MedSeq Project pilot RCT found no differences in 6-month healthcare costs between participants receiving genome sequencing including SLCO1B1 genotyping versus no genome sequencing (mean $1490 vs. $1142, excluding the costs of sequencing and interpretation)(66).
Process outcomes
Provider utilization and attitudes
Studies in which results were delivered to prescribers enabled an examination of how frequently they interfaced with the information and their attitudes about its value. Studies in which providers initiated SLCO1B1 testing generally reported low testing uptake(10, 34-36). Studies delivering SLCO1B1 results to providers through the EHR without provider initiation have reported providers’ EHR transactional data. For example, the 1,200 Patients Project of more than a dozen drug-gene pairs reported that 69% of 2,279 patient visits over 3 years were associated with a provider log-in to the pharmacogenetics CDS system within 72 hours(48). About one-third of patients’ active medications had associated pharmacogenetic alerts (0.5% red, 13% yellow, and 21% green)(48); a 10-month analysis in the first 608 patients reported that providers clicked on 100%, 72%, and 20% of red, yellow, and green alerts, respectively(47). However, SLCO1B1 transactions were not specifically reported. During the first 14 months that SLCO1B1 CDS was in production in the RIGHT Protocol, there were 0.7 interruptive alerts per month for simvastatin orders attempted for rs4149056 TC or CC patients among 3,788 patients seen by 1,247 unique providers(55). Studies surveying providers about their experiences have reported overall positive attitudes about pharmacogenetics, including its clinical relevance and impact on management(10, 47, 60), although no study reported provider attitudes about SLCO1B1 testing specifically.
Prescribing behavior and prescriptions
Most studies have reported or are actively collecting data on the impact of SLCO1B1 results on medication prescriptions, measured either from the EHR or provider or patient report Some studies of pharmacogenetic panel testing have reported only composite medication changes(10, 60, 61, 63), while small pilot studies have reported specific cases where SLCO1B1 results guided therapy(34, 35, 38, 40, 57, 66). Large studies with SLCO1B1 CDS alerts have reported counts of medication changes attributed to pharmacogenetic results. The 1,200 Patients Project reported that 25% of 2,279 visits over 3 years had medication changes; simvastatin was the drug with the highest percentage of changes influenced by CDS (69%), although this represented only 8 simvastatin discontinuations in SLCO1B1 C carriers among 868 patients(48). In the first 3 years of the Marshfield Clinic project, there have been 5 CDS alert recommendations triggered by simvastatin prescriptions, only one of which was followed, prompting the provider to prescribe atorvastatin instead (personal communication, Terrie Kitchner, December 6, 2017). Similarly, pharmacists in a Duke pilot study did not recommend any simvastatin prescription changes to the providers caring for 6 patients with carrier or homozygous SLCO1B1*5 results(40). Two controlled studies have examined prescribing behavior. In the Duke nonrandomized pilot trial, 55% of patients with a history of statin non-adherence had statin prescriptions 4 months after receiving SLCO1B1 results, compared to 20% of concurrent controls with statin prescriptions after one year (p<0.001)(39). In the subsequent RCT, more participants receiving SLCO1B1 results were on statin therapy at 3 months compared to usual care (55% vs. 38%, p=0.04), but this difference was not statistically significant after 8 months (54% vs. 37%, p=0.07)(43).
Medication adherence
While prescriptions largely reflect provider behavior, medication adherence is a patient behavior. Small pilot studies have either found no impact of SLCO1B1 testing on statin adherence or did not collect data to enable pre-post or between-group comparisons(34, 35, 38, 41). The Duke nonrandomized pilot study among patients with prior statin discontinuation found that 47% of intervention patients reported taking a statin after 4 months compared to 15% of concurrent controls after one year (p<0.001)(39). The subsequent RCT, however, found no differences in adherence or the Medication Possession Ratio after 3 or 8 months between the subsets of patients in both arms reinitiated on statin therapy(43). The authors reported that intervention patients perceived higher necessity of their medications than control patients at 3 months, but not at 8 months(43), consistent with observations from the pilot study(39).
Other patient-reported outcomes
Uncontrolled studies have reported that some patients had difficulty recalling their specific SLCO1B1 results(35, 38) and had variable understanding of them(10, 40, 59). Nonetheless, patients generally perceived the information as useful to their providers(35, 38, 54, 59). Pilot studies have also reported that patients generally had no concerns or distress after receiving pharmacogenetic results(35, 40, 66).
Discussion
Ten years after the publication of the association between SLCO1B1 and SAMS(26), we found few high-quality studies reporting patient outcomes after the delivery of SLCO1B1 results. Most notably, a pilot trial and subsequent small RCT among previously statin intolerant patients observed at least short-term improvements in LDL-C after SLCO1B1 testing. Although these findings require replication, the 10-mg/dL reduction in LDL-C the investigators observed, if sustained, would result in a 5% lower 5-year risk of major CVD event(68). Apart from this, while the proposed benefit of SLCO1B1 testing is the avoidance of SAMS, it is worth noting that no study has empirically demonstrated this outcome or the impact of SLCO1B1 testing on CVD events. Evidence from small pilot studies and large healthcare system implementation projects does suggest that SLCO1B1 results may change providers’ prescribing patterns for some, but not all, high-risk patients receiving simvastatin, but to date, the number of potential opportunities to observe prescription changes in large healthcare systems with SLCO1B1 CDS in the EHR has been small. Receipt of SLCO1B1 test results seems generally well tolerated by providers and patients. No study has specifically tracked the economic impact of SLCO1B1 testing and its downstream outcomes.
The clinical validity of the SLCO1B1-SAMS association has been well established; that is, the observed association between rs4149056 in SLCO1B1 and statin-associated myotoxicity of varying severity has been replicated in numerous studies, particularly for simvastatin(30). The PharmGKB knowledge resource rates the genotype-phenotype association between SLCO1B1 and simvastatin myopathy as having the highest level of evidence (Level 1A)(69). On the other hand, the clinical utility of SLCO1B1 testing, or its ability to inform a change in clinical management that demonstrably improves patient outcomes, is less certain. A 2013 review by Stewart found no studies comparing clinical outcomes between patients whose statin prescriptions were guided or not guided by SLCO1B1 results(32), and Sorich found no studies of the cost-effectiveness of SLCO1B1 genotyping(70). A more recent review examined 89 studies purporting to address either the clinical validity or clinical utility of pharmacogenetic testing for statin use and found almost all claims of clinical utility to be lacking when examined against benchmarks such as number needed to genotype, the effect and risks of the intervention, and costs per quality adjusted life year(33). Many of the studies we identified in the present review are collecting data on prescription changes, a measure of the actionability of pharmacogenetic results. Still, the absence of prospective outcomes data for pharmacogenetic testing will continue to make health insurers reluctant to cover the costs of testing(71) and many clinicians reluctant to incorporate pharmacogenetics into their practices(72).
Our review of patient outcomes after SLCO1B1 testing prompts the following recommendations for future research. First, although RCT evidence may not be necessary to justify every clinical application of pharmacogenetic testing(73-75), prospectively collected outcomes data might be, ideally from studies with suitable control groups. Investigators are encouraged to identify and collect data from concurrent matched controls for the participants in ongoing and planned pharmacogenetic projects, to enable a less biased determination of the impact of testing. Second, these outcomes should include those of interest to patients, providers, and payers, including clinical outcomes, quality of life, and costs. Third, even with the multigene pharmacogenetic panels that some implementation projects are using, it is important to report outcomes specific to individual pharmacogenetic tests, such as SLCO1B1 for statins and cytochrome P450 family 2 subfamily C member 19 (CYP2C19) for clopidogrel(76). Very few studies using panels in our review reported outcomes pertaining to SLCO1B1 results specifically. Although panels enable efficiencies of scale in genotyping, additional costs such as the development and implementation of CDS for each drug-gene pair are not trivial(77). Locus-specific outcomes data will enable a determination of the returns on those investments. Fourth, as more outcomes data accrue, the effect of context on those outcomes should be examined, including the degree of patient and provider engagement in the process, the type of CDS used in results delivery, and the characteristics of patients most likely to benefit. To date, the strongest evidence supporting the use of SLCO1B1 testing derives from an RCT among previously statin-intolerant patients(43). This finding is consistent with the French National Network of Pharmacogenetics recommendation that SLCO1B1 testing is potentially useful for patients experiencing SAMS after statin initiation or with at least one SAMS risk factor; it does not recommend routine preemptive SLCO1B1 testing before general simvastatin initiation(78). Further research should examine the clinical utility of SLCO1B1 testing among statin-naïve patients and among patients already tolerating statin therapy.
With the recommendations above, ongoing projects using SLCO1B1 genotyping in research or clinical care in the U.S. and internationally have a tremendous opportunity to contribute to the lack of evidence for its clinical utility. We identified 10 institutions with ongoing studies whose designs and planned outcomes would have been eligible for this review(11, 12, 14, 79-82). Most of these represent multi-institutional efforts such as the Electronic Medical Records and Genomics (eMERGE)-PGx Consortium(12), the Pharmacogenomics Research Network Translational Pharmacogenetics Program (TPP)(11), the Implementing Genomics in Practice (IGNITE) Consortium(13), and the seven-country Ubiquitous Pharmacogenomics (U-PGx) Consortium(14). These projects are collecting a range of prescription, clinical, and economic outcomes, and their large scale will enable more precise estimates of the clinical utility of pharmacogenetic testing. For example, while preliminary reports suggest that individual healthcare systems may observe few instances where SLCO1B1 results change medication prescriptions, a recent update from the TPP reported that 14,508 SLCO1B1 results have been reported in the EHR of five participating institutions, of which 3,513 (24%) were actionable(11). In addition to these large projects of pharmacogenetics panels, we are conducting the Integrating Pharmacogenetics in Clinical Care Study, an RCT specifically examining the impact of SLCO1B1 genotyping on LDL-C and concordance with statin therapy guidelines among statin-naïve patients (ClinicalTrials.gov Identifier: NCT02871934).
This review has a few limitations to note. The paucity of published clinical utility outcomes and the heterogeneity in other outcomes reported after SLCO1B1 testing precluded meta-analysis or between-study comparisons. We examined outcomes data from only a single specific gene-drug pair as an in-depth case study, paradigmatic of the state of the evidence for most other pharmacogenetic tests. It is unknown how the use of multigene pharmacogenetic panels would change the impact of SLCO1B1 information alone, since multiple genetic test results can interact in unpredictable ways on patient outcomes(83). While SLCO1B1 testing might be increasingly common in medical practice outside academic centers, we were only able to examine outcomes published in the biomedical literature.
In conclusion, despite advances bringing pharmacogenetic testing to clinical care, we found few patient outcomes reported after the delivery of SLCO1B1 results, a well validated pharmacogenetic locus. Outcomes data are needed to accelerate the pace of this clinical translation.
Methods
Protocol and registration
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Guidelines were followed for this systematic review (see PRISMA checklist, Table S2, in the Supplemental Information). We performed initial scoping searches of PubMed and Embase on June 22, 2017 before registering the review protocol on the PROSPERO register of systematic reviews on August 28, 2017 (CRD42017074795). After our initial searches and record review, we updated our searches on December 19, 2017 to identify new records.
Search strategy
We searched the PubMed and Embase databases for published reviews, meta-analyses, and primary studies published in or after 2005 using combinations of the following search strategy concepts: pharmacogenetics/pharmacogenomics, precision medicine, SLCO1B1, statins, cardiovascular disease. The full search strategies are included in the Supplemental Information.
Scope and eligibility criteria
The primary aim of the systematic review was to review the evidence for the clinical utility of SLCO1B1 testing, as determined by patient outcomes observed after SLCO1B1 genotyping. We included intervention studies in which 1) participants were directly or indirectly (e.g. via their providers) given their SLCO1B1 genotype results as a part of a research study or clinical care and 2) subsequent outcomes were prospectively collected and reported. Our scoping search enabled us to determine the appropriate breadth of eligibility criteria for both study designs and outcomes. Identifying few eligible randomized trials, we chose to additionally include pilot studies, implementation projects, and nonrandomized trials. We excluded case reports. We excluded studies reporting the association between SLCO1B1 genotype and statin effects (i.e. the clinical validity of the SLCO1B1 genotyping), as this has been reviewed in detail elsewhere, particularly by CPIC(30, 32, 33). We excluded retrospective observations among cohorts who had undergone direct-to-consumer pharmacogenetic testing (reviewed in (84)). We excluded records reporting only the frequency of SLCO1B1 genotypes among participants or in which SLCO1B1 genotype results were used to estimate hypothetical recommendations for medication changes (e.g. (5)). We included records in any language. After our scoping search identified few studies with eligible designs that reported clinical outcomes such as biomarker changes, morbidity, or mortality, we defined an eligible outcome broadly as any provider- or patient-reported outcome, EHR-derived outcome, or other study outcome measured after an intervention that involved the reporting of SLCO1B1 results to providers and/or patients. Our approach to categorizing these clinical utility and process outcomes is described above in the Results. Any record describing an ongoing study whose design and planned outcomes would be eligible for inclusion was noted so that authors could be contacted for more information.
Review process
The titles and abstracts of all search records were screened for potential eligibility by two independent reviewers; discrepancies were resolved by discussion and consensus among the study team. Potentially eligible records progressed to full record review for determination of eligibility. The references of review papers and eligible studies were manually searched for additional eligible studies.
Data abstraction
A Microsoft Excel database was used to abstract the following from each eligible study: country; study design; patient population and number; the genotyping intervention, including any genotypes other than SLCO1B1 reported and any associated decision support; any control group; any quantitative or qualitative outcome reported, including the method of collection and results; and the current status of the study. We categorized any intervention study with a control group as a nonrandomized trial if historical or concurrent controls were used or as a randomized controlled trial if participants were randomly allocated to the study arms(85). All other eligible studies were categorized as intervention studies, which included pharmacogenetics interventions delivered through pilot studies or operational clinical innovation programs.
Author communication
For each ongoing study with a potentially eligible study design (typically identified through a manuscript describing the study design and rationale), we emailed the corresponding author(s) a link to a brief survey requesting any published or unpublished results from the study referenced in the records we identified (Supplemental Information). Each author was sent up to three requests, each separated by at least 7 days. We also performed targeted author searches to identify any additional records from these ongoing studies.
Assessing study quality and risk of bias
Bias and study quality were systemically assessed using the Newcastle-Ottawa Scale (NOS) for the quality of intervention studies and nonrandomized studies, with greater scores indicative of higher study quality(86). For the randomized controlled trials, the Jadad scale(87) was used for the study quality assessment.
Supplementary Material
Acknowledgements
JLV is supported by Career Development Award IK2 CX001262 from the United States Department of Veterans Affairs (VA) Clinical Sciences Research and Development Service. This work does not reflect the views of the VA or the United States government, which had no role in the research.
Footnotes
Conflict of interest
The authors declared no competing interests for this work.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1002/cpt.1223
References
- (1).Manolio TA et al. Implementing genomic medicine in the clinic: the future is here. Genetics in Medicine 15, 258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Relling MV & Evans WE Pharmacogenomics in the clinic. Nature 526, 343–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).United States Food and Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labels. <https://www.fda.gov/Drugs/ScienceResearch/ucm572698.htm> (2018). Accessed March 1, 2018 2018.
- (4).Snyder SR, Mitropoulou C, Patrinos GP & Williams MS Economic evaluation of pharmacogenomics: a value-based approach to pragmatic decision making in the face of complexity. Public Health Genomics 17, 256–64 (2014). [DOI] [PubMed] [Google Scholar]
- (5).Van Driest SLS, Y.; Bowton E; Schildcrout J; Peterson J; Pulley J; Denny J; Roden D Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clinical Pharmacology and Therapeutics 95, 423–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bell GC et al. Development and use of active clinical decision support for preemptive pharmacogenomics. Journal of the American Medical Informatics Association 21, e93–e9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Goldspiel BR et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J Am Med Inform Assoc 21, 522–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dunnenberger HM et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annual Review of Pharmacology and Toxicology 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hinderer M et al. Integrating clinical decision support systems for pharmacogenomic testing into clinical routine-a scoping review of designs of user-system interactions in recent system development. BMC Medical Informatics and Decision Making 17, 81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Borobia AM. et al. Clinical Implementation of Pharmacogenetic Testing in a Hospital of the Spanish National Health System: Strategy and Experience Over 3 Years. Clin Transl Sci, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Luzum JA et al. The pharmacogenomics research network translational pharmacogenetics program: outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clinical Pharmacology & Therapeutics, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Rasmussen-Torvik LJ et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clinical Pharmacology & Therapeutics 96, 482–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Weitzel KW et al. The IGNITE network: a model for genomic medicine implementation and research. BMC medical genomics 9, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).van der Wouden CHC-T, et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clinical Pharmacology and Therapeutics 101, 341–58 (2017). [DOI] [PubMed] [Google Scholar]
- (15).Rogers EM Diffusion of Innovations. 5th edition edn. (The Free Press: New York, 2003). [Google Scholar]
- (16).Mrazek DA & Lerman C Facilitating clinical implementation of pharmacogenomics. JAMA 306, 304–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Feero WG, Manolio TA & Khoury MJ Translational research is a key to nongeneticist physicians' genomics education. Genet Med 16, 871–3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Patel HN, Ursan ID, Zueger PM, Cavallari LH & Pickard AS Stakeholder views on pharmacogenomic testing. Pharmacotherapy 34, 151–65 (2014). [DOI] [PubMed] [Google Scholar]
- (19).Hess GP, Fonseca E, Scott R & Fagerness J Pharmacogenomic and pharmacogenetic-guided therapy as a tool in precision medicine: current state and factors impacting acceptance by stakeholders. Genet Res (Camb) 97, e13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).National Center for Health Statistics. Health, United States; 2013: With Special Feature on Prescription Drugs. <http://www.cdc.gov/nchs/data/hus/hus13.pdf - listfigures> (2014). Accessed February 2, 2018. [PubMed]
- (21).Zhang H et al. Discontinuation of statins in routine care settings: a cohort study. Ann Intern Med 158, 526–34(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Buettner C, Davis RB, Leveille SG, Mittleman MA & Mukamal KJ Prevalence of musculoskeletal pain and statin use. Journal of General Internal Medicine 23, 1182–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Silva MA, Swanson AC, Gandhi PJ & Tataronis GR Statin-related adverse events: a meta-analysis. Clin Ther 28, 26–35 (2006). [DOI] [PubMed] [Google Scholar]
- (24).Law M & Rudnicka AR Statin safety: a systematic review. Am J Cardiol 97, 52C–60C (2006). [DOI] [PubMed] [Google Scholar]
- (25).Armitage J The safety of statins in clinical practice. Lancet 370, 1781–90 (2007). [DOI] [PubMed] [Google Scholar]
- (26).Link EP, S.; Armitage J; Bowman L; Heath S; Matsuda F; Gut I; Lathrop M; Collins R SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med 359, 789–99 (2008). [DOI] [PubMed] [Google Scholar]
- (27).Donnelly LA et al. Common nonsynonymous substitutions in SLCO1B1 predispose to statin intolerance in routinely treated individuals with type 2 diabetes: a go-DARTS study. Clin Pharmacol Ther 89, 210–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Puccetti L, Ciani F & Auteri A Genetic involvement in statins induced myopathy. Preliminary data from an observational case-control study. Atherosclerosis 211, 28–9 (2010). [DOI] [PubMed] [Google Scholar]
- (29).Brunham LR et al. Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. The Pharmacogenomics Journal 12, 233–7 (2012). [DOI] [PubMed] [Google Scholar]
- (30).Ramsey LBJ, S. G; Caudle KE; Haidar CE; Voora D; Wilke RA; Maxwell WD; McLeod HL; Krauss RM; Roden DM; Feng Q; Cooper-DeHoff RM; Gong L; Klein TE; Wadelius M; Niemi M The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clinical Pharmacology and Therapeutics 96, 423–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wilke RA et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clinical Pharmacology and Therapeutics 92, 112–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Stewart A SLCO1B1 Polymorphisms and Statin-Induced Myopathy. PLoS Curr 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Jansen ME et al. Review of the Reported Measures of Clinical Validity and Clinical Utility as Arguments for the Implementation of Pharmacogenetic Testing: A Case Study of Statin-Induced Muscle Toxicity. Frontiers in Pharmacology 8, 555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Haga SB, Mills R, Moaddeb J, Allen LaPointe N, Cho A & Ginsburg GS Primary care providers' use of pharmacist support for delivery of pharmacogenetic testing. Pharmacogenomics 18, 359–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Haga SB, Mills R, Moaddeb J, Lapointe NA, Cho A & Ginsburg GS Patient experiences with pharmacogenetic testing in a primary care setting. Pharmacogenomics (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Haga SB, et al. Pilot study of pharmacist-assisted delivery of pharmacogenetic testing in a primary care setting. Pharmacogenomics 15, 1677–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Haga SB, et al. Pilot study: incorporation of pharmacogenetic testing in medication therapy management services. Pharmacogenomics 15, 1729–37 (2014). [DOI] [PubMed] [Google Scholar]
- (38).Haga SB, et al. Incorporation of pharmacogenetic testing into medication therapy management. Pharmacogenomics 16, 1931–41 (2015). [DOI] [PubMed] [Google Scholar]
- (39).Li JHJ, S. V; Haga SB; Orlando LA; Kraus WE; Ginsburg GS; Voora D Genetically guided statin therapy on statin perceptions, adherence, and cholesterol lowering: a pilot implementation study in primary care patients. Journal of Personalized Medicine 4, 147–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Moaddeb JM, R.; Haga SB Community pharmacists' experience with pharmacogenetic testing. J Am Pharm Assoc (2003) 55, 587–94 (2015). [DOI] [PubMed] [Google Scholar]
- (41).Rumyantsev NAK, V. G; Kazakov RE; Rumyantsev AA; Sychev DA [Use of pharmacogenetic testing to prevent adverse drug reactions during statin therapy]. Ter Arkh 89, 82–7 (2017). [DOI] [PubMed] [Google Scholar]
- (42).Singh KP, B.; Trujillo G; Milazzo N; Savard D; Haga SB; Musty M; Voora D Rationale and design of the SLCO1B1 genotype guided statin therapy trial. Pharmacogenomics, (2016). [DOI] [PubMed] [Google Scholar]
- (43).Voora D (2017). Genetically Guided Statin Therapy. (Report no. AFRL-SA-WP-TR-2017-0005) (Wright-Patterson Air Force Base, OH, 2017). [Google Scholar]
- (44).Hussain SK, B. B; Danahey K; Lee YM; Galecki PM; Ratain MJ; O'Donnell PH Disease–drug database for pharmacogenomic-based prescribing. Clinical Pharmacology and Therapeutics (179–190). (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lee YMM, P. R; Borden BA; Klammer CE; Ratain MJ; O'Donnell PH Assessment of patient perceptions of genomic testing to inform pharmacogenomic implementation. Pharmacogenetics and Genomics 27, 179–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).O'Donnell PH et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clinical Pharmacology & Therapeutics 92, 446–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).(2014). Adoption of a clinical pharmacogenomics implementation program during outpatient care-initial results of the University of Chicago “1,200 Patients Project”. American Journal of Medical Genetics Part C: Seminars in Medical Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).O'Donnell PH et al. Pharmacogenomics-Based Point-of-Care Clinical Decision Support Significantly Alters Drug Prescribing. Clinical Pharmacology & Therapeutics, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Shuldiner A et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Overcoming Challenges of Real-World Implementation. Clinical Pharmacology & Therapeutics 94, 207–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Eadon M et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clinical Pharmacology & Therapeutics 100, 63–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sperber NR et al. Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC medical genomics 10, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Vitek CRR et al. Healthcare provider education to support integration of pharmacogenomics in practice: the eMERGE Network experience. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Bielinski SJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 89, 25–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Bielinski SJ, et al. Are patients willing to incur out-of-pocket costs for pharmacogenomic testing? Pharmacogenomics Journal 17, 1–3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Caraballo PJ et al. Multidisciplinary model to implement pharmacogenomics at the point of care. Genetics in Medicine (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Ji Y et al. Preemptive Pharmacogenomic Testing for Precision Medicine: A Comprehensive Analysis of Five Actionable Pharmacogenomic Genes Using Next-Generation DNA Sequencing and a Customized CYP2D6 Genotyping Cascade. Journal of Molecular Diagnostics 18, 438–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Allen AJ; Wetmore JM; Maturi Allen MF; Talreja DR Evaluation of genetic profiles in medication adjustments. Cardiology (2016) 134 Supplement 1, 247 (2016). [Google Scholar]
- (58).Gharani NK, M. A; Stack CB; Hodges LM; Schmidlen TJ; Lynch DE; Gordon ES; Christman MF The Coriell personalized medicine collaborative pharmacogenomics appraisal, evidence scoring and interpretation system. Genome Medicine 5:10 Article Number: 93. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Mukherjee C, Sweet KM, Luzum JA, Abdel-Rasoul M, Christman MF & Kitzmiller JP Clinical pharmacogenomics: patient perspectives of pharmacogenomic testing and the incidence of actionable test results in a chronic disease cohort. Personalized Medicine 14, 383–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Reynolds KK, Pierce DL, Weitendorf F & Linder MW Avoidable drug-gene conflicts and polypharmacy interactions in patients participating in a personalized medicine program. Personalized Medicine 14, 221–33 (2017). [DOI] [PubMed] [Google Scholar]
- (61).Saldivar JST, D.; Sugarman EA; Cullors A; Garces JA; Oades K; Centeno J Initial assessment of the benefits of implementing pharmacogenetics into the medical management of patients in a long-term care facility. Pharmacogenomics and Personalized Medicine 9, 1–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Sturm AC, Sweet K & Manickam K Implementation of a clinical research pharmacogenomics program at an academic medical center: Role of the genetics healthcare professional. Pharmacogenomics 14, 703–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Sugarman EAC, A.; Centeno J; Taylor D Contribution of Pharmacogenetic Testing to Modeled Medication Change Recommendations in a Long-Term Care Population with Polypharmacy. Drugs and Aging 33, 929–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Sweet K et al. Design and implementation of a randomized controlled trial of genomic counseling for patients with chronic disease. Journal of Personalized Medicine 4, 1–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Sweet K et al. Outcomes of a Randomized Controlled Trial of Genomic Counseling for Patients Receiving Personalized and Actionable Complex Disease Reports. Journal of Genetic Counseling, 1–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Vassy JL et al. The impact of whole-genome sequencing on the primary care and outcomes of healthy adult patients: a pilot randomized trial. Ann Intern Med 167, 159–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Vassy JL et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials 15, 85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Baigent C et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 366, 1267–78 (2005). [DOI] [PubMed] [Google Scholar]
- (69).Whirl-Carrillo M et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Sorich MJC, M.; Pekarsky BA Indirect estimation of the comparative treatment effect in pharmacogenomic subgroups. PLoS One 8, e72256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Pezalla EJ Payer view of personalized medicine. Am J Health Syst Pharm 73, 2007–12 (2016). [DOI] [PubMed] [Google Scholar]
- (72).Feero WG Clinical application of whole-genome sequencing: proceed with care. JAMA 311, 1017–9 (2014). [DOI] [PubMed] [Google Scholar]
- (73).Luzum JA et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clinical Pharmacology and Therapeutics 102, 502–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Khoury MJ Dealing with the evidence dilemma in genomics and personalized medicine. Clin Pharmacol Ther 87, 635–8 (2010). [DOI] [PubMed] [Google Scholar]
- (75).Pirmohamed M et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 369, 2294–303 (2013). [DOI] [PubMed] [Google Scholar]
- (76).Scott SA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94, 317–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Mathias PC, Tarczy-Hornoch P & Shirts BH Modeling the costs of clinical decision support for genomic precision medicine. AMIA Jt Summits Transl Sci Proc 2016, 60–4 (2016). [PMC free article] [PubMed] [Google Scholar]
- (78).Lamoureux FD, T. Pharmacogenetics in cardiovascular diseases: State of the art and implementation-recommendations of the French National Network of Pharmacogenetics (RNPGx). Therapie 72, 257–67 (2017). [DOI] [PubMed] [Google Scholar]
- (79).Borobia AM et al. Ipharmgx project: National collaborative study to evaluate effectiveness and efficiency of pharmacogenetics implementation in the clinical practice. Basic and Clinical Pharmacology and Toxicology 119 (Supplement 1), 32 (2016).26663750 [Google Scholar]
- (80).Gottesman OS, S. A; Ellis SB; Overby CL; Ludtke A; Hulot JS; Hall J; Chatani K; Myers K; Kannry JL; Bottinger EP The CLIPMERGE PGx program: Clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clinical Pharmacology and Therapeutics 94, 214–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Sissung TM et al. Pharmacogenomics Implementation at the National Institutes of Health Clinical Center. J Clin Pharmacol 57, S67–S77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Weitzel KW et al. Clinical Pharmacogenetics Implementation: Approaches, Successes, and Challenges. American Journal of Medical Genetics Part C Seminars in Medical Genetics 0, 56–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Christensen KD et al. Disclosing Pleiotropic Effects During Genetic Risk Assessment for Alzheimer Disease: A Randomized Trial. Ann Intern Med 164, 155–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Chua EW & Kennedy MA Current state and future prospects of direct-to-consumer pharmacogenetics. Frontiers in Pharmacology 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Friedman LM, Furberg CD & DeMets DL Basic Study Design. In: Fundametnals of Clinical Trials 4th edn. 67–96 (Springer, New York, 2010). [Google Scholar]
- (86).Wells G et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. <http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp> (2014). Accessed April 18, 2018. [Google Scholar]
- (87).Jadad AR et al. Assessing the quality of reports of randomized clinical trials: is blinding necessary? Controlled Clinical Trials 17, 1–12 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.