Abstract
Background:
Oligodendrocytes generate specialized lipid-rich sheaths called myelin that wrap axons and facilitate the rapid, saltatory transmission of action potentials. Extrinsic signals and surface-mediated pathways coordinate oligodendrocyte development to ensure appropriate axonal myelination, but the mechanisms involved are not fully understood. Glycerophosphodiester phosphodiesterase 2 (GDE2 or GDPD5) is a six-transmembrane enzyme that regulates the activity of surface glycosylphosphatidylinositol (GPI)-anchored proteins by cleavage of the GPI-anchor. GDE2 is expressed in neurons where it promotes oligodendrocyte maturation through the release of neuronally-derived soluble factors. GDE2 is also expressed in oligodendrocytes but the function of oligodendroglial GDE2 is not known.
Results:
Using Cre-lox technology, we generated mice that lack GDE2 expression in oligodendrocytes (O-Gde2KO). O-Gde2KOs show normal production and proliferation of oligodendrocyte precursor cells. However, oligodendrocyte maturation is accelerated leading to the robust increase of myelin proteins and increased myelination during development. These in vivo observations are recapitulated in vitro using purified primary oligodendrocytes, supporting cell-autonomous functions for GDE2 in oligodendrocyte maturation.
Conclusions:
These studies reveal that oligodendroglial GDE2 expression is required for controlling the pace of oligodendrocyte maturation. Thus, the cell-type specific expression of GDE2 is important for the coordination of oligodendrocyte maturation and axonal myelination during neural development.
Keywords: glia, terminal differentiation, myelination, brain
Introduction
Oligodendrocytes (OLs) are glial cells that produce myelin, a specialized lipid-rich sheath that wraps axons to facilitate the rapid transmission of electrical signals important for neural circuit function1,2. OLs derive from oligodendrocyte precursor cells (OPCs) that proliferate and migrate to populate the developing nervous system where they differentiate into premyelinating and myelinating OLs3. Because OL production is in part synchronized with neurons and neuronal activity to ensure appropriate axonal myelination, the control of OL maturation is complex4,5. Emergent studies indicate roles for neuronal cues, extrinsic signals and surface membrane proteins in OL differentiation and maturation6–10; however, the mechanisms that regulate the timing of OL maturation and myelination are not fully understood.
OPCs in the brain derive from three waves of OPC production that originate from distinct ventral and dorsal brain areas11. The first wave of OPC generation initiates from the medial ganglionic eminence and the entopeduncular area, and this early population of OPCs is replaced by subsequent waves of OPCs that arise from the caudal ganglionic eminence and from the cortex. Once generated, OPCs proliferate and migrate to populate the nervous system, and on reaching their settling positions, differentiate into premyelinating and myelinating OLs. OPCs exhibit regional diversity in terms of their proliferative and migratory properties, as do OLs with respect to their remyelination capabilities12–14. Interestingly, genetic ablation and transplantation studies indicate that different populations of OPCs are functionally redundant, suggesting that OPCs are plastic and capable of responding to multiple cues that influence their maturation and myelination11,15.
The stages of OPC differentiation and maturation are coordinated with the process of axonal myelination. Accordingly, the mechanisms that control these later aspects of OL development involve communication between neurons and OPCs, as well as cell-autonomous pathways in developing OLs16–18. Both positive and negative regulators of OL differentiation and maturation have been identified in neurons and OLs19–22, suggesting that multiple signaling pathways are integrated to regulate the pace of OL maturation and axonal myelination. In line with this notion, disruptions in OL numbers and myelin production during development are often normalized in juvenile animals suggesting that the tempo of OL differentiation and maturation during development is dynamic and tightly controlled23,24. However, the mechanisms that regulate the pace of OL maturation are not well understood.
Glycerophosphodiester phosphodiesterase 2 (GDE2 or GDPD5) is a six-transmembrane protein that contains an external enzymatic domain that shares homology with bacterial glycerophosphodiester phosphodiesterases (GDPDs)25–27. GDE2 and its two family members, GDE3 and GDE6, are the only known proteins in vertebrates that act on the cell surface to regulate the activity of glycosylphosphatidylinositol (GPI)-anchored proteins by cleavage of the GPI-anchor27–30. GDE2 is primarily expressed in neurons in the developing and adult nervous system, where it plays essential roles in regulating neuronal differentiation and neuronal survival respectively26,27,31–33. Underscoring its role as a regulator of neuronal differentiation, GDE2 can promote the differentiation of neuroblastoma and higher GDE2 expression levels correlates with better outcomes in disease29. A recent study discovered that neuronal GDE2 function contributes to neuron-glial communication pathways that regulate the timing of OL maturation during developmental myelination34. Loss of neuronal GDE2 in the postnatal brain results in delayed OL maturation and myelination. Mechanistically, neuronal GDE2 maintains canonical Wnt signaling in neurons, which is responsible for the release of neuronally-derived soluble factors that promote OL maturation34. Thus, neuronal GDE2 acts non cell-autonomously to positively regulate the pace of OL maturation and myelination during development.
GDE2 is also expressed in a subset of terminally differentiated OLs34, raising the possibility that oligodendroglial GDE2 may play roles in regulating the progress of OL maturation. Here, we examine the requirement of oligodendroglial GDE2 expression in OL development using genetic ablation approaches combined with cell-based models of OL maturation and myelination. In contrast to neuronal GDE2 control of OL development, we find that oligodendroglial GDE2 negatively regulates the pace of OL maturation and myelination. Genetic ablation of GDE2 in oligodendroglia leads to the accelerated production of myelinating OLs and increased myelination during development. This study reveals that the cell-specific expression of GDE2 differentially regulates the timing of OL maturation and myelination and provides new insight into the developmental pathways that ensure appropriate axonal myelination.
Results
Gde2 expression in oligodendroglia is mainly confined to mature OLs.
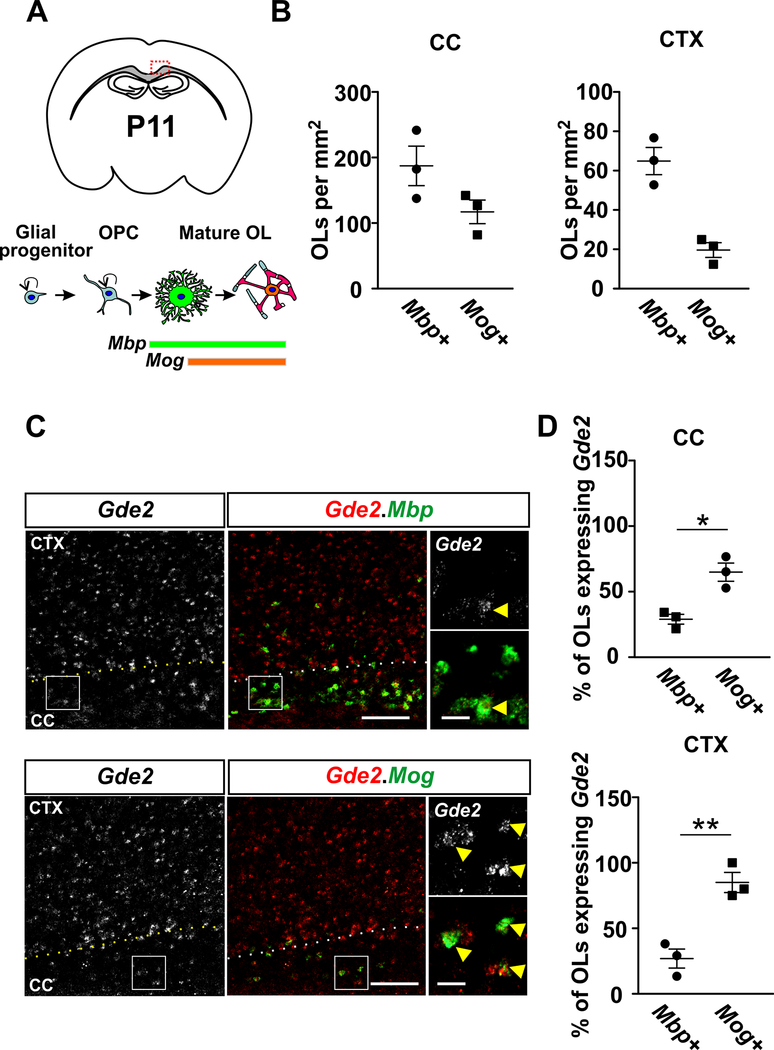
GDE2 protein and transcripts are known to be expressed in neurons and in a subpopulation of Olig2+ oligodendroglia in the postnatal brain34. Gde2 was found to be expressed in OLs with minimal expression in OPCs34, but the precise temporal expression of Gde2 in differentiating OLs in vivo was not addressed. To gain a more precise understanding of the role of GDE2 in OL development, we examined the temporal expression of Gde2 in maturing OLs using fluorescent in situ hybridization (FISH). We focused our analysis on the corpus callosum (CC) and the adjacent motor cortex (CTX) on postnatal day (P) 11 (Fig. 1A). Transcripts for Myelin Basic Protein (Mbp) are expressed by immature OLs35, while mature OLs express transcripts for Myelin Oligodendrocyte Glycoprotein (Mog)36 (Fig. 1A). At this stage in development, the number of Mbp+ immature OLs exceeds the number of mature Mog+ OLs in the CC and CTX (Fig. 1B). Gde2 transcripts are detected in a higher proportion of Mog+ OLs compared to Mbp+ OLs in both CC (55% vs 25%) and CTX (85% vs 25%) (Fig 1C and D). Thus, Gde2 is primarily expressed in mature OLs. This is in line with published transcriptomic analyses, which show the enrichment of Gde2 mRNAs in mature OLs compared with earlier stages of oligodendroglial development37.
Figure 1. Gde2 is expressed in maturing OLs in postnatal brain.
(A) Top: Schematic of a coronal brain section; the red box delineates the defined region of interest (ROI) that includes the corpus callosum (CC) and adjacent motor cortex (CTX). Bottom: schematic showing gene expression profiles associated with the progression of OL development. (B) Graphs quantifying the number of Mbp+ and Mog+ OLs in the CC and CTX. (C) FISH of Gde2 mRNA and Mbp or Mog mRNAs in P11 brain tissue sections. Hatched lines mark the boundary between the CTX and the CC. Boxed areas are shown at higher magnification on the right. Arrows highlight Mbp+ or Mog+ OLs that co-express Gde2 mRNAs. (D) Graphs quantifying the percentage of Mbp+ and Mog+ OLs expressing Gde2 transcripts in CC and CTX. Higher proportions of Mog+ OLs express Gde2 transcripts compared to Mbp+ OLs in CC (*p = 0.0244) and CTX (**p = 0.0052). n = 3. Scale bar: 100 μm, inset: 20 μm.
GDE2 in oligodendroglia does not impact OPC proliferation.
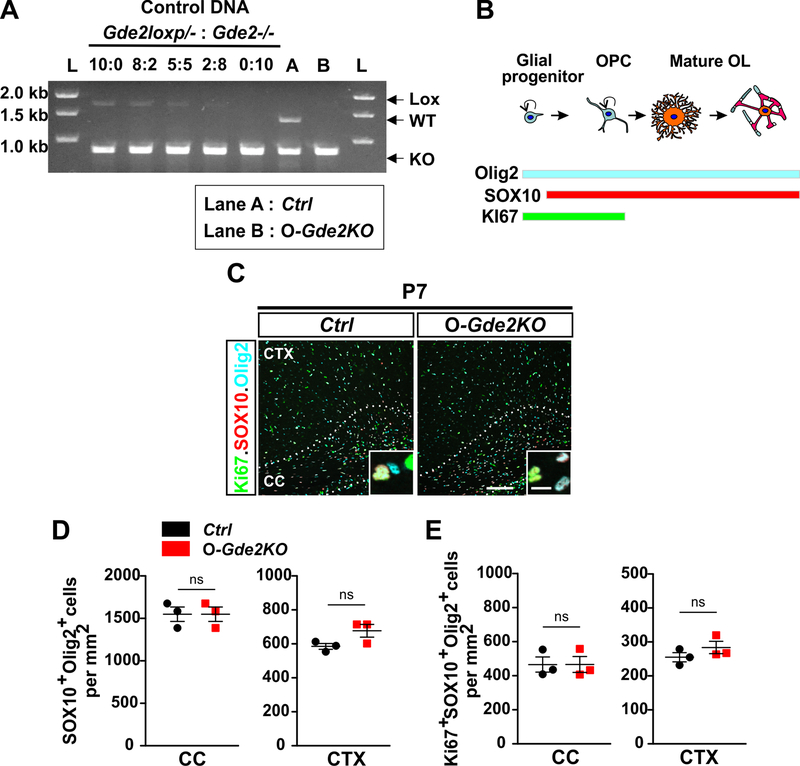
To define the contribution of GDE2 expression in OLs to oligodendroglial development, we took advantage of Cre-lox genetics to ablate GDE2 function in oligodendroglia by utilizing mice harboring a conditional allele of Gde2 (Gde2lox/-)32. Gde2lox/- mice contain lox-P sites flanking exon 11 that encodes the GDE2 GDPD enzymatic domain. Previous studies have shown that Cre-dependent excision of exon 11 results in the complete loss of GDE2 protein expression, validating this strategy to be an effective method to ablate cellular function of GDE2 in vivo32–34. For this study, we utilized an established inducible mouse line (PDGFαR-CreER) where administration of 4 hydroxytamoxifen (4-HT) induces active Cre recombinase expression under the control of Platelet derived growth factor α receptor (PDGFαR) regulatory sequences in OPCs38. Thus, 4-HT treatment of Gde2lox/-; PDGFαR-CreER mice will specifically ablate GDE2 expression in oligodendroglia but will preserve GDE2 function in other cell types (O-Gde2KO). Gde2+/−; PDGFαR-CreER animals that lack the conditional allele and are similarly injected with 4-HT serve as controls (Ctrl). 4-HT was administered once per day intraperitonially into Ctrl and O-Gde2KO pups at P1 and P2. To determine the extent of Cre-mediated recombination of the Gde2lox allele, we first verified the deletion of the Gde2 conditional allele using a competitive PCR strategy32. Genomic DNAs from OPCs isolated from Ctrl and O-Gde2KO P6 cortices were subject to PCR amplification using specific primers that detect the unrecombined Gde2lox allele (Lox), the WT Gde2 allele (WT), and the recombined Gde2lox allele (KO) (Fig. 2A). DNAs prepared from separately generated Gde2lox/- and Gde2−/− pups were used as positive controls. Different serial dilutions of Gde2lox/- DNA with increasing amount of Gde2−/− DNA were prepared so that the total DNA amount was the same across the DNA mixtures (Fig. 2A), and a standard curve of control PCR product that was linear as a function of input DNA was generated32,39. We observed that the amount of Lox PCR product in O-Gde2KO OPCs was similar to the amount amplified when a 2:8 ratio of Gde2lox/- and Gde2−/− control DNAs was utilized, suggesting that approximately 80% of the Gde2lox alleles had undergone recombination in O-Gde2KO OPCs (Fig. 2A, Lane B). This analysis suggests that GDE2 expression is efficiently abrogated in O-Gde2KO OPCs using the 4-HT administration protocol.
Figure 2. Loss of GDE2 in oligodendroglia does not influence OPC proliferation.
(A) Competitive PCR analysis for Gde2 from 4-HT injected Ctrl and O-Gde2KO pups. Control DNA: 5 different ratios of Gde2lox/- and Gde2−/− genomic DNAs. Gde2 PCR primers amplified DNA fragments unique to unrecombined Gde2lox (Lox, 1700bp), WT Gde2 (WT, 1300bp), and recombined Gde2lox (KO, 780bp) alleles. Ctrl (Lane A) shows presence of WT and KO fragments. O-Gde2KO (lane B) shows a similar intensity in the 1700bp fragment to the 2:8 ratio of mixed genomic DNAs. L = DNA ladder. (B) Schematic of marker proteins that distinguish proliferating OPCs. (C) Coronal section of CTX and CC Ctrl and O-Gde2KO P7 animals. Hatched lines mark the CC. Insets show magnified images of OPCs (Ki67+Sox10+Olig2+). (D) Graphs quantifying the number of oligodendroglia (Sox10+Olig2+) between Ctrl and O-Gde2KO CC (ns p =0.0857) and CTX (ns p = 0.1555). (E) Graphs quantifying the number of proliferating OPCs (Ki67+Sox10+Olig2+) between Ctrl and O-Gde2KO CC (ns p =0.5147) and CTX (ns p = 0.2971). n = 3 Ctrl (Gde2+/−; PDGFαR-CreER), 3 O-Gde2KO (Gde2lox/-; PDGFαR-CreER). Graphs: Mean ± sem, two-tailed unpaired Students t-test. Scale bar: 100 μm, inset: 10 μm.
To determine the requirement for GDE2 OL expression in oligodendroglial development, we first characterized the generation and proliferation of OPCs in O-Gde2KO mice compared with Ctrl. Cells that co-express Olig2 and SOX10 mark oligodendroglia while Olig2+SOX10+ cells that co-express the proliferation marker Ki67 constitute proliferating OPCs (Fig. 2B)40,41. At P7, during the peak period of cortical OPC generation and proliferation, the numbers of Olig2+SOX10+ oligodendroglia and Ki67+Olig2+SOX10+ proliferating OPCs in the CC and CTX were equivalent between Ctrl and O-Gde2KO animals (Fig. 2C–E). These observations suggest that GDE2 expression in OLs is not required for the generation of oligodendroglia or OPC proliferation and is consistent with the onset of GDE2 expression in maturing OLs.
Oligodendroglial GDE2 prevents precocious OL maturation.
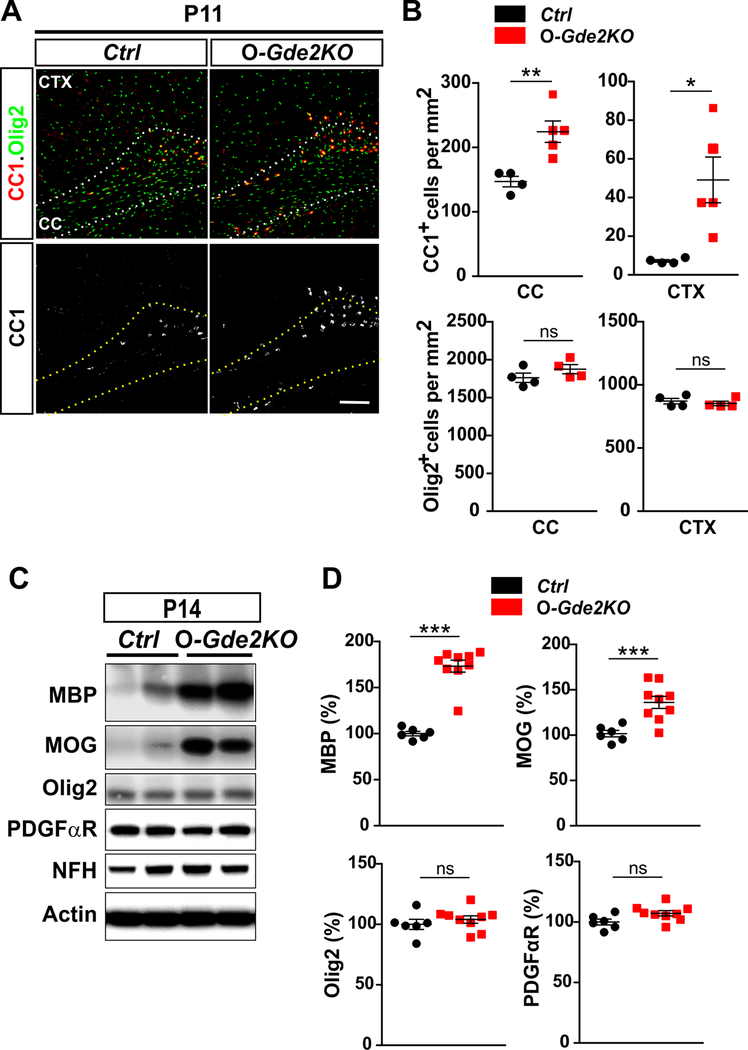
The differentiation of OPCs into mature OLs can be monitored by the temporal expression of specific marker proteins. Premyelinating and myelinating OLs express CC142, while mature OLs express myelin associated proteins that include MBP and MOG35,36. To determine if GDE2 expression in OLs is required for OL maturation, we quantified the number of Olig2+CC1+ cells in Ctrl and O-Gde2KO at P11, during the onset of OL maturation (Fig. 3A and B). Because studies suggest that OLs show region-specific diversity, we analyzed OL maturation in the white matter and grey matter focusing on the CC and CTX respectively. Strikingly, O-Gde2KO animals showed a 50% increase in Olig2+CC1+ OLs in the CC and a marked increase of Olig2+CC1+ OLs in the CTX compared with Ctrl animals with no change in overall numbers of Olig2+ oligodendroglia (Fig. 3A and B). We note that O-Gde2KOs exhibit variability in the numbers of Olig2+CC1+ cells in the CTX at this timepoint. While the basis of this is unclear, it is possible that the pace of OL maturation in absence of GDE2 is subject to tight temporal control such that slight differences in age could contribute to differences in OL maturation rate. Nevertheless, these observations suggest that the loss of GDE2 in OLs enhances OL maturation without altering overall numbers of OLs (Fig. 3A and B). We next measured the amounts of MBP and MOG proteins in Ctrl and O-Gde2KO animals at P14 when myelination is beginning to occur. Western blot analysis of P14 cortical extracts revealed robust increase of MBP and MOG expression in O-Gde2KO mutants compared with Ctrl (Fig. 3C and D). No obvious changes in the levels of the general oligodendroglia marker Olig2 and the OPC marker PDGFαR were detected, suggesting that overall numbers of oligodendroglia are comparable between genotypes (Fig. 3C and D). Amounts of the neuronal protein Neurofilament Heavy Chain (NFH) were equivalent between Ctrl and O-Gde2KO confirming that axons are not disrupted in O-Gde2KO animals (Fig. 3C). Taken together, these observations suggest that the loss of GDE2 expression in OLs leads to the precocious maturation of OPCs into myelinating OLs.
Figure 3. Oligodendroglial GDE2 prevents precocious OL maturation.
(A) Coronal sections of CTX and CC of P11 Ctrl and O-Gde2KO mice. Hatched lines delineate the CC. (B) Graphs quantifying the number of CC1+ and Olig2+ cells in CC and CTX in Ctrl and O-Gde2KO P11 littermates. CC1+ OLs are increased in O-Gde2KOs in CC (**p = 0.0087) and CTX (*p = 0.0242). Olig2+ cell numbers are equivalent between Ctrl and O-Gde2KO in CC (ns p=0.2353) and CTX (ns p=0.5548). n = 4 Ctrl, 5 O-Gde2KO. (C) Western blot of cortical extracts from P14 Ctrl and O-Gde2KO littermates. Actin is used as a loading control. NFH expression shows equivalent axonal content between the groups. (D) Graphs quantifying Western blots show increase of MBP (***p < 0.0001) and MOG (***p = 0.0007) proteins in O-Gde2KO cortical extracts compared with Ctrl. Levels of Olig2 (ns p = 0.4388) and PDGFαR (ns p = 0.0940) are unchanged between genotypes. n = 6 Ctrl, 9 O-Gde2KO. All graphs: Mean ± sem, two-tailed unpaired Students t-test. Scale bar: 100 μm.
GDE2 regulates the tempo of OL maturation in vitro.
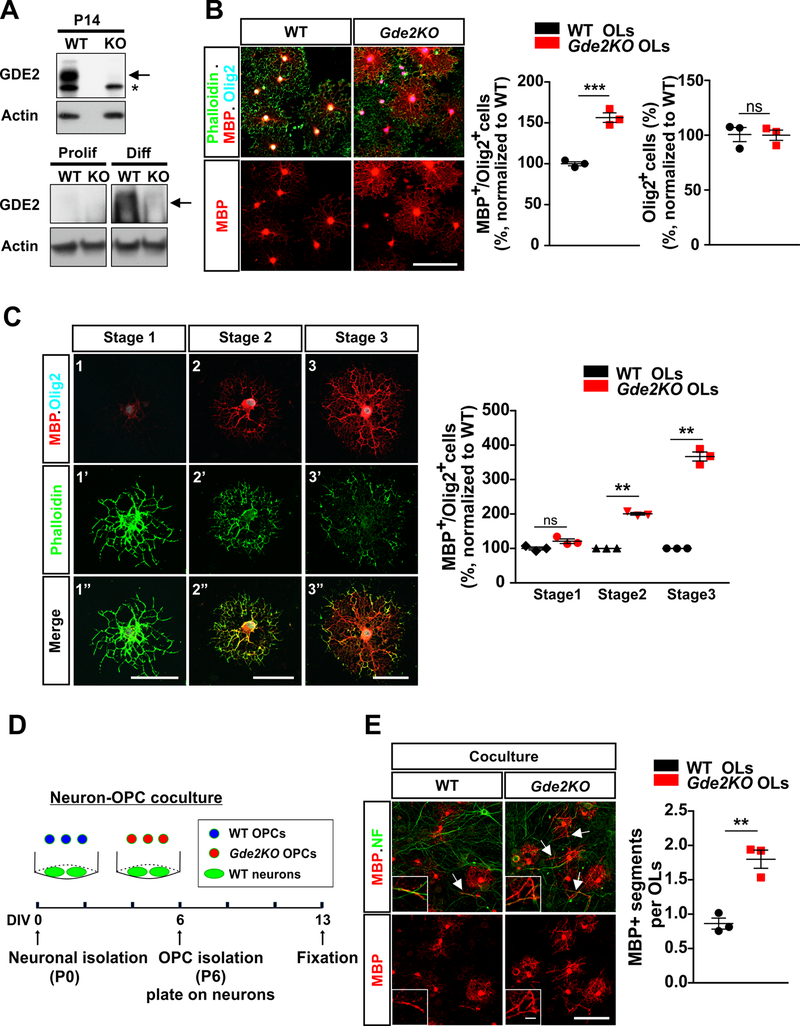
Our observations thus far identify a cell-autonomous function for GDE2 in regulating OL maturation. To better define the requirement for GDE2 in this process, we immunoaffinity-purified OPCs from WT mice and Gde2KO animals that lack GDE2 protein expression (Fig. 4A) and cultured them in vitro using defined culture medium. Western blot analysis confirms that GDE2 is expressed in differentiating OLs and not proliferating OPCs, and that Gde2KO OLs show complete loss of GDE2 protein (Fig. 4A). WT and Gde2KO OPCs were purified from P6 cortices and the same number of OPCs for each condition were cultured in the absence of mitogenic factors such as PDGFα. After 3 days, cultures were fixed and examined for OL maturation based on their morphology and by the expression of MBP (Fig. 4B). We found that the percentage of MBP+Olig2+ OLs was increased by approximately 50% in cultures of Gde2KO OLs compared with WT, with no change in the total number of Olig2+ cells between genotypes (Fig. 4B). These observations recapitulate our in vivo observations that the loss of GDE2 in OLs accelerates OL maturation and supports the cell-autonomy of GDE2 function in this process.
Figure 4. GDE2 regulates the tempo of OL maturation in vitro.
(A) Representative Western blot of P14 cortical lysates (Top) and extracts of proliferating OPCs and differentiated OLs (2 DIV) from WT and Gde2KOs. Arrow highlights GDE2 protein, * marks nonspecific band. (B). Representative images of WT and Gde2KO OLs. Graphs quantifying the percentage of MBP+Olig2+ OLs (***p = 0.0008) and Olig2+ cells (ns p=0.9385) normalized to WT, n = 3 WT, 3 Gde2KO. (C) Representative images of 3 stages of OL maturation in vitro. Graph quantifying the percentage of MBP+Olig2+ OLs. 2-way ANOVA ***p < 0.0001 (Bonferroni correction) ns p >0.05; **p <0.01; n = 3 WT, 3 Gde2KO. (D) Schematic of neuron-OPC co-culture. (E) Representative images of WT and Gde2KO OPCs co-cultured with WT neurons. Insets show higher magnification of myelinated axons. Arrows highlight myelinated axons. Graph quantifying the numbers of MBP+ segments (normalized to WT OLs) **p = 0.0037, n = 3 WT, 3 Gde2KO. All graphs: Mean ± sem. (B, E) two-tailed unpaired Students t-test; Scale bars: (B, E) 100 μm, inset: 10 μm (C) 50 μm.
In addition to increased myelin protein expression, maturing OLs in vitro undergo stereotypic morphological changes manifest by changes in the F-actin network that can be visualized by phalloidin staining. Previous studies have defined three stages of OL maturation based on their morphology, MBP expression and phalloidin staining34,43 (Fig. 4C). Differentiating Olig2+ OLs in vitro initially show arborized morphology with weak MBP expression in and around the cell body with robust phalloidin labeling throughout the cell body and in distal processes (Stage 1, immature). Maturing OLs have increased MBP expression and less strong phalloidin labeling, with some intermittent flattening of the myelin sheath in distal structures (Stage 2, premyelinating). More mature and myelinating OLs show distinct ring-like morphology with strong MBP expression throughout the membrane sheath and disappearance of the actin cytoskeleton from central processes (Stage 3, myelinating). To better define the requirement for GDE2 in OL maturation, we quantified these three stages of OL maturation in WT and Gde2KO OPC cultures. After 3 days in culture we found that Gde2KO OLs showed a 2-fold increase in the percentage of OLs at Stage 2 of maturation and a robust 4-fold increase in the percentage of OLs at Stage 3 (Fig. 4C). However, the percentage of Stage 1 OLs was equivalent between WT and Gde2KO conditions (Fig. 4C). These observations support our findings that the loss of GDE2 leads to an accelerated pace of OL maturation.
To evaluate if Gde2KO OLs are capable of myelinating axons, we utilized an in vitro coculture system. In this paradigm, WT cortical neurons were isolated from P0 animals and were cultured for 6 days in vitro (DIV). O4+ OPCs purified from P6 WT and Gde2KO cortices were then added to the cultured neurons and co-cultured for a further 7 days. Co-cultures were subsequently fixed and stained for myelinated axonal segments (Fig. 4D). Gde2KO OLs showed a significant increase in the number of MBP+ segments that aligned with NFH+ axons compared to WT OLs (Fig. 4E), consistent with enhanced axonal myelination. Thus, these observations suggest that GDE2 functions cell-autonomously to control the progression of OL maturation and myelination.
GDE2 regulates developmental OL myelination.
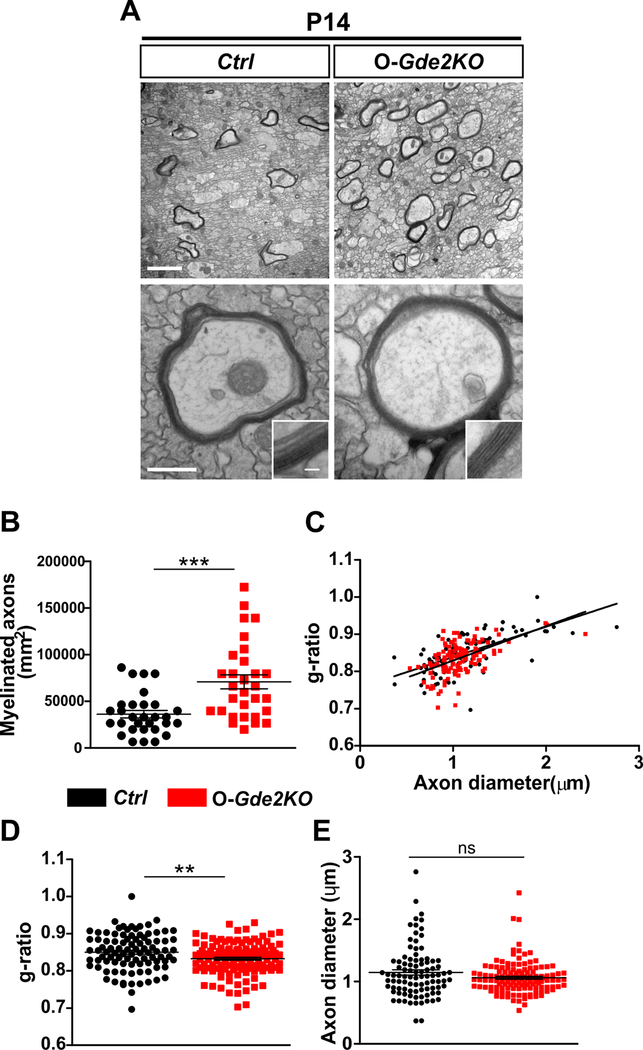
To determine the in vivo consequences of disrupted oligodendroglial GDE2 function on the temporal progression of developmental axonal myelination, we examined myelin ultrastructure in P14 animals using transmission electron microscopy (TEM). Focusing on axons in the CC, we found that animals that lacked GDE2 in OLs showed an increase in the number of axons that are myelinated (Fig. 5A and B). Notably, axons that were myelinated showed an increase in the thickness of the myelin sheath, indicated by a decreased g-ratio, which is the ratio of the axonal diameter to the diameter of the myelinated axon (Fig. 5A, C and D; Ctrl g-ratio= 0.8501 ± 0.005230; Gde2KO g-ratio= 0.8326 ± 0.003728). Axonal caliber is linked to the control of axonal myelination18,44,45. Axonal diameters between O-Gde2KO animals and Ctrl littermates were comparable, suggesting that the increase in myelin thickness and the increased number of myelinated axons in O-Gde2KO animals are not a consequence of alterations in axonal caliber (Fig. 5E). Thus, GDE2 expression in OLs is required for the temporal control of axonal myelination during the peak period of developmental myelination.
Figure 5. GDE2 ablation in oligodendroglia results in increased myelination.
(A) TEM of P14 CC of Ctrl and O-Gde2KOs. Insets show higher magnification images of the myelin sheath. (B-E) Graphs show an increase in myelinated axons (B, ***p <0.0001; points refer to individual regions of interest [ROI] from 3 Ctrl and 3 O-Gde2KOs), and decrease in g-ratios (C, D **p =0.0056; points refer to individual myelinated axons; 3 Ctrl and 3 O-Gde2KOs) in O-Gde2KO compared with Ctrl but no change in axon diameters (E, ns p = 0.0624; points refer to individual axons, 3 Ctrl and 3 O-Gde2KOs). All graphs: Mean ± sem, two-tailed unpaired Students t-test. Scale bars: (A) 2 μm (top), 500 nm (bottom), inset 50 nm.
OL developmental maturation and myelination deficits are restored in the adult.
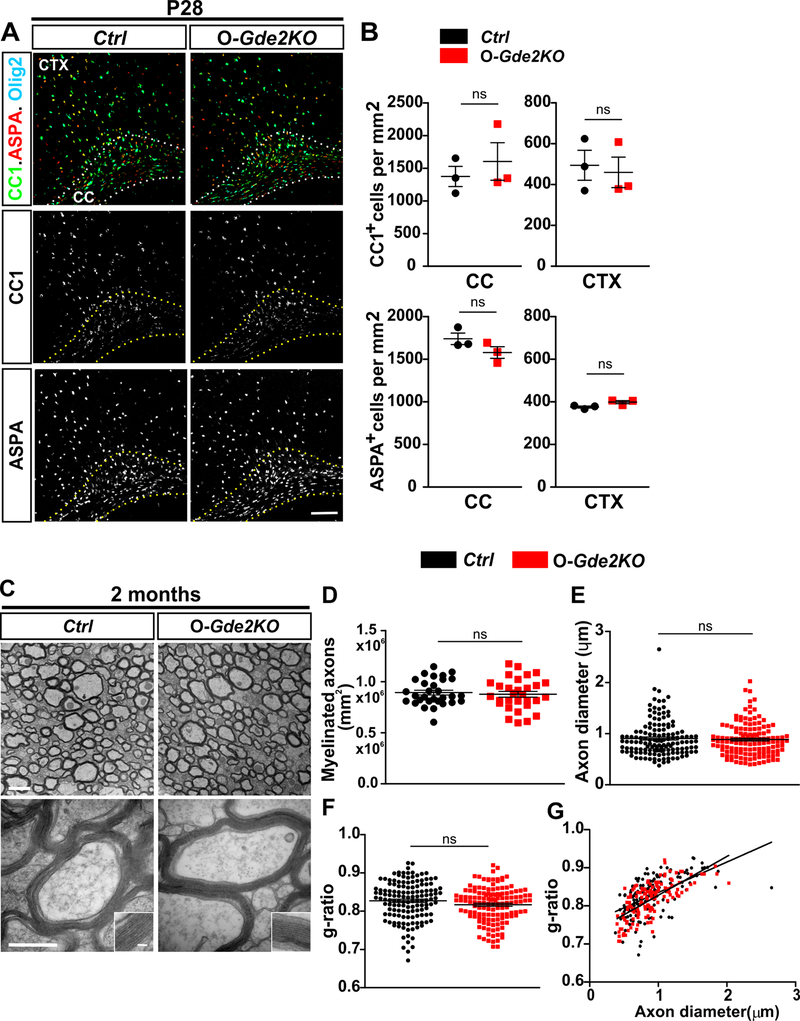
The ablation of GDE2 in oligodendroglia leads to advanced OL maturation and myelination during developmental myelination. Previous studies have established that myelination is a tightly regulated process and that multiple pathways are deployed to compensate for irregularities in OL generation and maturation during development. We thus examined if loss of GDE2 function during development leads to persistent changes in premyelinating and mature OL numbers and axonal myelination in adult animals. CC1 expression identifies premyelinating and myelinating OLs while Aspartocylase (ASPA) marks mature, myelinating OLs46. At P28, the number of CC1+ and ASPA+ OLs was equivalent between Ctrl and O-Gde2KO animals (Fig. 6A and B). Thus, the number of premyelinating and mature OLs in O-Gde2KO mice does not exceed Ctrl levels and has normalized by the time developmental oligodendrogenesis is complete. We next examined axonal myelination at the ultrastructural level in O-Gde2KO animals at 2 months of age when developmental myelination is complete. TEM analysis showed that O-Gde2KO animals had equivalent numbers of myelinated axons to Ctrls, and that myelin thickness was similar between genotypes (Fig. 6C and D, F and G). No difference in axonal diameters between O-Gde2KOs and Ctrls was detected (Fig. 6E). Thus, the level and extent of myelination in animals lacking GDE2 expression in OLs are restored by the time developmental myelination is complete.
Figure 6. O-Gde2KO OL maturation and myelination deficits are restored in the adult.
(A) Coronal sections of mouse CTX and CC from Ctrl and O-Gde2KO P28 animals. Hatched lines delineate the CC. (B) Graphs quantifying the number of mature OLs (CC1+ and ASPA+) show no difference in CC1+ OLs between Ctrl and O-Gde2KO CC (ns = 0.5217) and CTX (ns = 0.7564) and no change in ASPA+ OLs in CC (ns =0.1922) and CTX (ns = 0.0721) at P28. n = 3 Ctrl, 3 O-Gde2KO. (C) TEM of the CC of 2-month Ctrl and O-Gde2KO animals. Insets show higher magnification of the myelin sheath. (D-G) Graphs show comparable numbers of myelinated axons (D, ns P=0.6555; points refer to individual ROI from 3 Ctrl and 3 O-Gde2KOs), g-ratios (F, G, ns p =0.0728; points refer to individual myelinated axons; 3 Ctrl and 3 O-Gde2KOs), and axon diameters (E, ns p = 0.6076; points refer to individual axons, 3 Ctrl and 3 O-Gde2KOs) between genotypes. All graphs: Mean ± sem, two-tailed unpaired Students t-test. Scale bars: (A) 100 μm (C) 1 μm (top), 500 nm (bottom), inset 50 nm.
Discussion
Here we show that GDE2 expression in oligodendroglia is predominantly expressed in mature OLs in the developing nervous system. O-Gde2KO mice that lack GDE2 expression in developing OLs show no differences in the generation of OPCs or OPC proliferation compared with WT counterparts. However, the maturation of OLs into myelinating OLs is accelerated, resulting in the earlier onset of axonal myelination and accelerated myelination in O-Gde2KO mice during the period of developmental myelination. These in vivo studies were recapitulated using primary cultures of purified Gde2KOs OPCs that were differentiated to OLs in vitro, thus confirming the cell-autonomy of GDE2 function. These observations indicate that GDE2 is a physiological pathway that functions in developing OLs to negatively regulate the pace of OL maturation.
Previous studies have shown that GDE2 is highly expressed in neurons and that during the period of developmental myelination, neuronal GDE2 stimulates the release of soluble factors that promote OL maturation without affecting OPC generation or proliferation26,31,34. While GDE2 function in OLs also does not affect OPC generation and proliferation, the differences in the roles of neuronal and oligodendroglial GDE2 in regulating OL maturation are striking. Neuronal and oligodendroglial GDE2 appear to have opposing functions with neuronal GDE2 promoting OL maturation and oligodendroglial GDE2 slowing the development of OLs into myelinating OLs. Analysis of mice that lack global GDE2 expression (Gde2KO) show OL maturation phenotypes that mimic neuronal deletion of GDE2 expression34. This observation suggests that neuronal GDE2 function likely lies upstream of oligodendroglial GDE2 pathways that regulate OL maturation. This is consistent with the temporal profile of GDE2 expression, which initiates in neurons prior to GDE2 expression in premyelinating and myelinating OLs31. These collective observations thus suggest a model where neuronal GDE2 promotes the induction of OL maturation, which consequently results in the onset of GDE2 expression in maturing OLs. Oligodendroglial GDE2 subsequently regulates the pace of OL maturation to ensure the appropriate myelination of axons during the developmental period. We note that the number of mature OLs in O-Gde2KO CC and CTX mice stabilizes by P28 and that the myelin ultrastructure of O-Gde2KO animals at 2 months is equivalent to controls. Because the CC normally myelinates earlier than the CTX, it is possible that the restoration of myelination in O-Gde2KOs occurs earlier in the CC compared with the CTX. These observations suggest that the developmental perturbations in myelination that are elicited by the disruption of GDE2 function in OLs are sensed and ultimately corrected by other regulatory networks. This is in line with other studies that highlight the complex monitoring and dynamic regulation of OL development and maturation to ensure that axons are appropriately myelinated within the timeframe of developmental myelination6,8,47,48.
The mechanism by which GDE2 oligodendroglial expression controls OL maturation remains to be elucidated. Previous RNAseq studies that compare transcriptional changes in spinal cords from WT and Gde2KO animals show that the canonical Wnt signaling pathway is altered in absence of GDE2 function34. Genetic and in vitro experiments have determined that activating canonical Wnt signaling in neurons rescues OL maturation deficits elicited by global or neuronal GDE2 ablation, suggesting that neuronal GDE2 preserves canonical Wnt signaling in neurons to promote OL maturation. In contrast, genetic activation of canonical Wnt signaling in OPCs in Gde2KO mice leads to a more profound slowing of OL maturation, suggesting that elevated Wnt signaling in OLs negatively regulates the pace of terminal OL differentiation34. Although this latter observation is in line with the proposed roles for oligodendroglial GDE2 in controlling OL maturation, it is unlikely that oligodendroglial GDE2 mediates this function by activating Wnt signaling. This is because reporter mice for canonical Wnt pathway activation reveal that Gde2KO animals have dramatically reduced Wnt signaling in neurons and OPCs, but not in OLs during the period of OL maturation. Given that GDE2 is expressed in OLs and not OPCs, these observations suggest that oligodendroglial GDE2 expression likely utilizes mechanisms other than Wnt activation to control the pace of OL maturation. Previous studies have established that GDE2 cleaves the GPI-anchor to release specific GPI-anchored proteins from the cell surface27,29. This suggests that oligodendroglial GDE2 likely regulates OL maturation by regulating the activity of surface GPI-anchored protein substrates and their associated pathways. Several GPI-anchored proteins are expressed in OLs and many of them have known functions in OL development. For example, Contactin (F3) is expressed in migrating, premyelinating and myelinating OLs and is required for OL maturation49, myelination and for the formation of paranodal and nodal structures in myelinated nerves48,50, while TAG1 (Contactin2) is expressed in OLs and is involved in juxtaparanodal organization of K+ channels51. Further investigation to determine the role of GPI-anchored proteins and their relationship to oligodendroglial GDE2 control of OL maturation will be of interest to gain further insight into the mechanisms of OL development and myelination.
In summary, our study identifies GDE2 as a physiological pathway that functions in developing OLs to regulate the pace of OL maturation during developmental myelination. In the adult nervous system, resident OPCs differentiate into mature OLs to facilitate axonal myelination in response to multiple events such as experience, injury and disease; importantly, failure of myelination can severely impact neural circuit viability and function52. Whether GDE2 expression is reinitiated in differentiating OPCs to promote OL maturation and axonal myelination in the adult nervous system warrants further investigation.
Experimental Procedures
Mice
Gde2+/−, Gde2flox mice 32 and PDGFαR-CreER38 mouse lines were used in this study. Mice were housed and treated according to the approved Institutional Animal Care and Use Committee (IACUC) protocol of the Johns Hopkins Medical Institution. Littermates were utilized as controls, and both male and female animals were used for all analyses.
Tissue processing and immunohistochemistry
Brain sections were prepared and analyzed for immunohistochemistry as described34. Briefly, mice were anesthetized with a solution of Avertin and butanol in Phosphate Buffered Saline (PBS) at 0.02ml/g body weight and perfused with 0.1M Phosphate Buffer (PB) transcardially followed by fixation in 4% paraformaldehyde in 0.1M PB. Brains were postfixed in fixation solution overnight at 4°C, transferred to 30% sucrose and stored at 4°C for 48 hrs. Tissues were then embedded in O.C.T. Compound (Tissue-Tek 62550–12), and coronally sectioned (50 μm for P7 brain tissues and 35 μm for the rest) with a Thermo Fisher Scientific HM550 cryostat. Immunofluorescence was performed on free-floating sections. Brain sections were boiled in 10 mM sodium citrate with 0.05% Tween-20 before blocking in 1% normal goat serum, 0.3% Triton X-100 in PBS for 2 hours at room temperature. Sections were incubated in primary antibodies overnight at 4°C, washed and incubated in secondary antibodies for 2 hours at room temperature. Tissue sections were mounted onto slides with mounting reagent (Polysciences 18606) and images were acquired using a Zeiss LSM700 confocal microscope with 20x objective. A total of 4–5 sections per mouse and 3–5 mice were quantified per group, with regions of interest (ROI) chosen based on the typical anatomical structures of CC and CTX using DAPI staining (see section on Quantification and Statistical Analyses).
4-HT preparation and administration
4-hydroxytamoxifen (4-HT, sigma H7904) was prepared as described53. Single dose injections of 0.2 mg of 4HT were administered intraperitoneally on two consecutive days at P0 and P1. Pups were sacrificed at P11, P28, and 2 months of age and relevant tissues were collected.
Fluorescent in situ hybridization (FISH)
Brain tissue sections were processed and subjected to FISH as described34. Briefly, 16 μm cryosections were incubated in 3% hydrogen peroxide to block endogenous peroxidase activity, permeabilized with 0.3% Triton X-100 in PBS and acetylated in 0.3% acetic anhydride. For double FISH to visualize Gde2 transcripts in maturing OLs, tissue sections were hybridized with fluorescein (FITC)-labeled sense and antisense probes that detect Mbp or Mog transcripts and digoxigenin (DIG)-labeled sense and antisense probes for Gde2 transcripts overnight at 70°C. Sections were incubated with anti-FITC-horseradish peroxidase (POD), 1:500 (Roche 11426346910) and FITC-labeled fluorescent signals were developed with TSA Plus FITC system (Akoya NEL741001KT) according to the manufacturer’s instructions. To develop DIG-labeled Gde2 transcripts, sections were first subjected to gradient methanol fixation (30%, 50%, 75%, and 100%) and then incubated in a solution of 3% hydrogen peroxide in methanol for 30 min to inactivate the first POD and then washed in gradient methanol (75%, 50%, 30%) in PBS. To couple FISH with immunohistochemistry, sections were blocked in blocking solution (PerkinElmer FP1020) and incubated with sheep anti-digoxigenin-POD, 1:500 (Roche 11207733910) and rabbit anti-Olig2 (Millipore AB9610) overnight at 4°C. After incubation with secondary antibodies (1 hour, room temperature), fluorescent signals were developed with TSA Plus Cy3 system (Akoya NEL744001KT). Images were acquired on Zeiss LSM800 with 10x and 20x objectives. Oligonucleotide sequences of primers used to generate FISH probes are listed as follows. Gde2: Forward: 5’ CCTCAAGACCGACCCCTT 3’ Reverse: 5’ GGGGCATGATCCAGAGTG 3’; Mbp: Forward: 5’ GAGGCCTGGATGTGATGG 3’ Reverse: 5’ GGGGAACAAGTCAGGGCT 3’; Mog: Forward: 5’ CTCCTTCTCCTCCAGTTGTCAT 3’ Reverse: 5’ CTTCTGCACGAAGTTTTCCTCT 3’.
Transmission Electron Microscopy (TEM)
TEM was performed as described34. Briefly, animals were perfused intracardially with fixative containing 2% paraformaldehyde and 2% glutaraldehyde for 30 min at a rate of 1 ml/min for P14 and 2 ml/min for 2-month old mice. Dissected brains were post-fixed overnight at 4°C, and the CC and adjacent CTX were carefully dissected from 1000μm coronally sectioned brain slices. For TEM imaging of the CC, the sagittal surface near the midline of the CC was further sectioned. All tissues were serially dehydrated, embedded, and sectioned by the JHU SOM Microscope Facility as previously described54. Images were acquired with a Hitachi 7600 TEM. Images (under 15000x and 80000x magnification) were obtained from the CC at random with the operator blinded. ImageJ (National Institutes of Health) software was used to measure the number of myelinated axons and g-ratio per surface area (mm2). For g-ratio analysis, the diameters of axons and outer myelinated axons were calculated from the surface area derived from the circumference of each. Selection of myelinated axons was unbiased. Specifically, a grid was first created, and axons located at grid line intersections were selected for g-ratio analysis. The number of myelinated axons was counted from 10 images (under 15000x magnification) per animal and three animals were used per condition.
Cell culture
Mouse primary OPCs were purified from P6 cortices of WT and Gde2KO pups using Neural Tissue Dissociation and Isolation kits (Miltenyi Biotec 130–090-312) with magnetic beads (Miltenyi Biotec 130–094-543) to positively select O4+ cells according to the manufacturer’s recommendations. Purified OPCs were plated at a density of 3 × 104 cells on glass coverslips coated with poly-D-lysine (0.1 mg/ml in PBS pH 7.4) containing 1% laminin (Sigma-Aldrich L2020). Cells were cultured in DMEM:F12 (Gibco 10565018) with 10 mM HEPES (Gibco 15630080), 2% SM1 supplement, 1% N2B, 0.5% penicillin/streptomycin, 5μg/mL N-Acetyl-Cysteine (Sigma A8199), 5 μM Forskolin (Calbiochem 344270), 10ng/mL CNTF (PeproTech 450–50) for 3 days and then fixed for analysis. For neuron-OL cocultures, cortical neuronal cultures were prepared from P0 WT pups using Neural Tissue Dissociation Kits (Miltenyi Biotec 130–092-628) according to the manufacturer’s recommendations. Cortical preparations were plated at a density of 3 × 105 cells on glass coverslips coated with poly-D-lysine (0.1 mg/ml in 0.1 M Trizma buffer pH 8.5) containing 1% laminin (Sigma-Aldrich L2020) and 1% PureCol Type I Bovine Collagen Solution (Advanced Biomatrix 5005-B). Cells were initially cultured in neurobasal medium (Gibco 21103–049) containing 5% fetal horse serum, 1% penicillin/streptomycin (Gibco 15140122), 1% Glutamax-I Gibco 35050061), 1% sodium pyruvate (Gibco 11360070), 30 mM Glucose, and 2% SM1 supplement (STEMCELL Technologies 5711). The next day on DIV1, the medium was replaced with maintenance medium containing Neurobasal medium with 1% penicillin/streptomycin, 1% Glutamax-I, 1% sodium pyruvate, 30 mM Glucose, 2% SM1 supplement, and 1% N2B (STEMCELL Technologies 7156). Cells were treated on DIV2 with AraC (2.5 μM) for 24 hours to inhibit growth of proliferating cells. Freshly isolated OPCs in coculture media (half DMEM:F12 and half Neural basal media containing 10 mM HEPES (Gibco 15630080), 2% SM1 supplement, 1% N2B, 0.5% penicillin/streptomycin, 5μg/mL N-Acetyl-Cysteine (Sigma A8199), 5 μM Forskolin (Calbiochem 344270), 10ng/mL CNTF (PeproTech 450–50) were added at a density of 30,000 cells on top of DIV6 neurons and cocultured for 7 days.
Immunocytochemistry
Immunocytochemistry was performed as described34. Primary antibodies were used at a dilution of 1:500. Antibodies used in this study are: Rat anti-MBP (Millipore MAB386), rabbit anti-Olig2 (Millipore AB9610), mouse anti-MBP (Covance SMI-99P-100), Rabbit anti-Neurofilament H (Millipore AB1989). After a 1 hour incubation at room temperature with appropriate secondary antibodies, cells were counterstained with DAPI (Invitrogen R37606). Cells were stained with Alexa Fluor 488-phalloidin (Invitrogen A12379) during secondary antibody incubation to visualize F-actin. Cells were mounted on glass slides and imaged on a Zeiss LSM700 confocal microscope with 20x objective or on a Keyence BZ-X710 epifluorescence microscope with 20x objective for tiling (9×9).
Immunoblotting
Tissue samples were prepared, run on SDS-PAGE gels and transferred to PVDF membranes as described34. Transferred membranes were incubated with primary antibodies overnight at 4°C. Primary antibodies used were as follows: Rabbit anti-Olig2 (1:2,000 Millipore AB9610), Mouse anti-MBP (1:2,000 Covance SMI-99P-100), Mouse anti-MOG (1:3,000 Millipore MAB5680), Rabbit anti-Neurofilament H (1:10,000 Millipore AB1989), Rabbit anti-GDE2 (1:1000), Rabbit anti-PDGF receptor α (1:2,000 Cell Signaling Technology 3174), Mouse anti-Actin (1:10,000 Millipore MAB1501). After 1 hour incubation at room temperature with appropriate HRP-conjugated secondary antibodies, membranes were developed by film or by using a digital imaging system (KwikQuant, Kindle Biosciences).
Quantification and statistical analysis
Quantitative analysis was performed as described34. Briefly, quantification was performed using semi-automated Imaris (Bitplane) from 4–5 sections per animal and 3–5 animals per condition. Images were obtained from the CC and adjacent CTX corresponding to the retrosplenial and motor areas. After 3D image reconstruction, the contour function in Imaris was used to create individual ROIs. Spot and surface rendering functions were used to set parameters for size and fluorescent signal intensity threshold to define an object representing a nucleus or a cell body. After manual validation, z-stacks were subjected to automated quantification and manually corrected for false-positive and false negative spots and surfaces. Cell numbers were normalized to ROI surface areas. For quantifying the percent of mature OLs expressing Gde2 transcripts, the numbers of Gde2+ Mbp+ cells and Gde2+Mog+ cells were divided by the numbers of Mbp+Olig2+ cells and Mog+Olig2+ cells respectively after normalization to ROI surface areas. For quantifying the number of MBP+ OLs at Stage1–3 in culture, 9×9 tiled images with 20x objective (6.22mm2 area) were acquired from 2 technical replicates per animal with three biological replicates per experiment. The number of MBP+ cells for each stage was divided by the number of Olig2+ cells in each group and then normalized to control. Plots were generated and statistics were performed using GraphPad Prism 5 software. Mean ± SEM are shown for each plot. Statistical significance was determined by either two-tailed, unpaired Student’s t-test, or by 2-way ANOVA and is shown by * p<0.05, ** p<0.01, *** p<0.001.
Acknowledgements
We thank C. Wladyka and Y. Li for technical assistance; Dr D. Bergles, Dr. P. Calabresi and the Sockanathan lab for discussions; Dr D. Bergles for PDGFαR-CreER mice, and the Multiphoton Imaging Core of the Johns Hopkins P30 Center for Neuroscience Research (NS050274). This work was supported by grants to S.S. (National Institutes of Health, R01NS046336) and to B.C. (Fulbright Graduate Study Award, Science and Engineering).
Grant support: National Institutes of Health RO1NS046336 (S.S); Fulbright Graduate Study Award (Science and Engineering) (B.C).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Hortega PdR Tercera aportaci on al conocimiento morfol ogico e interpretacion funcional de la oligodendroglıa. Mem Real Soc Esp Hist Nat 1928;14:5–122. [Google Scholar]
- 2.Deber CM, Reynolds SJ. Central nervous system myelin: structure, function, and pathology. Clinical biochemistry. April 1991;24(2):113–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. November 11 2010;468(7321):214–22. https://doi.org10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 4.Gibson EM, Purger D, Mount CW, et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science (New York, NY). May 2 2014;344(6183):1252304. https://doi.org10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hines JH, Ravanelli AM, Schwindt R, Scott EK, Appel B. Neuronal activity biases axon selection for myelination in vivo. Nature neuroscience. May 2015;18(5):683–9. https://doi.org10.1038/nn.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Umemori H, Sato S, Yagi T, Aizawa S, Yamamoto T. Initial events of myelination involve Fyn tyrosine kinase signalling. Nature. February 10 1994;367(6463):572–6. https://doi.org10.1038/367572a0. [DOI] [PubMed] [Google Scholar]
- 7.Fraher J, Dockery P. A strong myelin thickness-axon size correlation emerges in developing nerves despite independent growth of both parameters. Journal of anatomy. August 1998;193 (Pt 2):195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens B, Porta S, Haak LL, Gallo V, Fields RD. Adenosine: a neuron-glial transmitter promoting myelination in the CNS in response to action potentials. Neuron. December 5 2002;36(5):855–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harlow DE, Macklin WB. Inhibitors of myelination: ECM changes, CSPGs and PTPs. Experimental neurology. January 2014;251:39–46. https://doi.org10.1016/j.expneurol.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syed YA, Hand E, Mobius W, et al. Inhibition of CNS remyelination by the presence of semaphorin 3A. The Journal of neuroscience : the official journal of the Society for Neuroscience. March 9 2011;31(10):3719–28. https://doi.org10.1523/jneurosci.4930-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nature neuroscience. February 2006;9(2):173–9. https://doi.org10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lentferink DH, Jongsma JM, Werkman I, Baron W. Grey matter OPCs are less mature and less sensitive to IFNgamma than white matter OPCs: consequences for remyelination. Sci Rep. February 1 2018;8(1):2113. https://doi.org10.1038/s41598-018-19934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spitzer SO, Sitnikov S, Kamen Y, et al. Oligodendrocyte Progenitor Cells Become Regionally Diverse and Heterogeneous with Age. Neuron. February 6 2019;101(3):459–471.e5. https://doi.org10.1016/j.neuron.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Power J, Mayer-Proschel M, Smith J, Noble M. Oligodendrocyte precursor cells from different brain regions express divergent properties consistent with the differing time courses of myelination in these regions. Dev Biol. May 15 2002;245(2):362–75. https://doi.org10.1006/dbio.2002.0610. [DOI] [PubMed] [Google Scholar]
- 15.Vigano F, Mobius W, Gotz M, Dimou L. Transplantation reveals regional differences in oligodendrocyte differentiation in the adult brain. Nature neuroscience. October 2013;16(10):1370–2. https://doi.org10.1038/nn.3503. [DOI] [PubMed] [Google Scholar]
- 16.Yellajoshyula D, Liang CC, Pappas SS, et al. The DYT6 Dystonia Protein THAP1 Regulates Myelination within the Oligodendrocyte Lineage. Developmental cell. July 10 2017;42(1):52–67.e4. https://doi.org10.1016/j.devcel.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redmond SA, Mei F, Eshed-Eisenbach Y, et al. Somatodendritic Expression of JAM2 Inhibits Oligodendrocyte Myelination. Neuron. August 17 2016;91(4):824–36. https://doi.org10.1016/j.neuron.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goebbels S, Wieser GL, Pieper A, et al. A neuronal PI(3,4,5)P3-dependent program of oligodendrocyte precursor recruitment and myelination. Nature neuroscience. October 24 2016. https://doi.org10.1038/nn.4425. [DOI] [PubMed] [Google Scholar]
- 19.Wake H, Ortiz FC, Woo DH, Lee PR, Angulo MC. Nonsynaptic junctions on myelinating glia promote preferential myelination of electrically active axons. 2015;6:7844. https://doi.org10.1038/ncomms8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen TJ, Silbereis JC, Griveau A, et al. Oligodendrocyte-encoded HIF function couples postnatal myelination and white matter angiogenesis. Cell. July 17 2014;158(2):383–96. https://doi.org10.1016/j.cell.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebrun-Julien F, Bachmann L, Norrmen C, et al. Balanced mTORC1 activity in oligodendrocytes is required for accurate CNS myelination. The Journal of neuroscience : the official journal of the Society for Neuroscience. June 18 2014;34(25):8432–48. https://doi.org10.1523/jneurosci.1105-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Argaw AT, Gurfein BT, et al. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proceedings of the National Academy of Sciences of the United States of America. November 10 2009;106(45):19162–7. https://doi.org10.1073/pnas.0902834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fancy SP, Baranzini SE, Zhao C, et al. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes & development. July 1 2009;23(13):1571–85. https://doi.org10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodruff RH, Fruttiger M, Richardson WD, Franklin RJ. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol Cell Neurosci. February 2004;25(2):252–62. https://doi.org10.1016/j.mcn.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Yanaka N Mammalian glycerophosphodiester phosphodiesterases. Biosci Biotechnol Biochem. August 2007;71(8):1811–8. https://doi.org10.1271/bbb.70062. [DOI] [PubMed] [Google Scholar]
- 26.Rao M, Sockanathan S. Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science. September 30 2005;309(5744):2212–5. https://doi.org10.1126/science.1117156. [DOI] [PubMed] [Google Scholar]
- 27.Park S, Lee C, Sabharwal P, Zhang M, Meyers CL, Sockanathan S. GDE2 promotes neurogenesis by glycosylphosphatidylinositol-anchor cleavage of RECK. Science. January 18 2013;339(6117):324–8. https://doi.org10.1126/science.1231921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobrowolski M, Cave C, Levy-Myers R, et al. GDE3 regulates oligodendrocyte precursor proliferation via release of soluble CNTFRalpha. Development (Cambridge, England). January 23 2020;147(2). https://doi.org10.1242/dev.180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matas-Rico E, van Veen M, Leyton-Puig D, et al. Glycerophosphodiesterase GDE2 Promotes Neuroblastoma Differentiation through Glypican Release and Is a Marker of Clinical Outcome. Cancer cell. October 10 2016;30(4):548–562. https://doi.org10.1016/j.ccell.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 30.van Veen M, Matas-Rico E, van de Wetering K, et al. Negative regulation of urokinase receptor activity by a GPI-specific phospholipase C in breast cancer cells. Elife. August 29 2017;6. https://doi.org10.7554/eLife.23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez M, Choi J, Park S, Sockanathan S. Gde2 regulates cortical neuronal identity by controlling the timing of cortical progenitor differentiation. Development. October 2012;139(20):3870–9. https://doi.org10.1242/dev.081083. [DOI] [PubMed] [Google Scholar]
- 32.Sabharwal P, Lee C, Park S, Rao M, Sockanathan S. GDE2 regulates subtype-specific motor neuron generation through inhibition of Notch signaling. Neuron. September 22 2011;71(6):1058–70. https://doi.org10.1016/j.neuron.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cave C, Park S, Rodriguez M, et al. GDE2 is essential for neuronal survival in the postnatal mammalian spinal cord. Mol Neurodegener. January 19 2017;12(1):8. https://doi.org10.1186/s13024-017-0148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi BR, Cave C, Na CH, Sockanathan S. GDE2-Dependent Activation of Canonical Wnt Signaling in Neurons Regulates Oligodendrocyte Maturation. Cell reports. May 5 2020;31(5):107540. https://doi.org10.1016/j.celrep.2020.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. The Journal of neuroscience : the official journal of the Society for Neuroscience. October 25 2006;26(43):10967–83. https://doi.org10.1523/jneurosci.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solly SK, Thomas JL, Monge M, et al. Myelin/oligodendrocyte glycoprotein (MOG) expression is associated with myelin deposition. Glia. September 1996;18(1):39–48. . [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. September 3 2014;34(36):11929–47. https://doi.org10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. November 18 2010;68(4):668–81. https://doi.org10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K, Jarry H, Knoke I, Seong JY, Leonhardt S, Wuttke W. Competitive PCR for quantitation of gonadotropin-releasing hormone mRNA level in a single micropunch of the rat preoptic area. Mol Cell Endocrinol. November 1993;97(1–2):153–8. https://doi.org10.1016/0303-7207(93)90222-6. [DOI] [PubMed] [Google Scholar]
- 40.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. January 1 1998;18(1):237–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meijer DH, Kane MF, Mehta S, et al. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nature reviews Neuroscience. December 2012;13(12):819–31. https://doi.org10.1038/nrn3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat RV, Axt KJ, Fosnaugh JS, et al. Expression of the APC tumor suppressor protein in oligodendroglia. Glia. Jun 1996;17(2):169–74. https://doi.org10.1002/(sici)1098-1136(199606)17:2<169::aid-glia8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 43.Zuchero JB, Fu MM, Sloan SA, et al. CNS Myelin Wrapping Is Driven by Actin Disassembly. Developmental cell. July 27 2015;34(2):152–67. https://doi.org10.1016/j.devcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remahl S, Hildebrand C. Changing relation between onset of myelination and axon diameter range in developing feline white matter. Journal of the neurological sciences. April 1982;54(1):33–45. [DOI] [PubMed] [Google Scholar]
- 45.Lee S, Leach MK, Redmond SA, et al. A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nat Methods. September 2012;9(9):917–22. https://doi.org10.1038/nmeth.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madhavarao CN, Moffett JR, Moore RA, Viola RE, Namboodiri MA, Jacobowitz DM. Immunohistochemical localization of aspartoacylase in the rat central nervous system. The Journal of comparative neurology. May 3 2004;472(3):318–29. https://doi.org10.1002/cne.20080. [DOI] [PubMed] [Google Scholar]
- 47.Weng Q, Chen Y, Wang H, et al. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron. February 23 2012;73(4):713–28. https://doi.org10.1016/j.neuron.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colakoglu G, Bergstrom-Tyrberg U, Berglund EO, Ranscht B. Contactin-1 regulates myelination and nodal/paranodal domain organization in the central nervous system. Proceedings of the National Academy of Sciences of the United States of America. January 21 2014;111(3):E394–403. https://doi.org10.1073/pnas.1313769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu QD, Ang BT, Karsak M, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. October 17 2003;115(2):163–75. [DOI] [PubMed] [Google Scholar]
- 50.Boyle ME, Berglund EO, Murai KK, Weber L, Peles E, Ranscht B. Contactin orchestrates assembly of the septate-like junctions at the paranode in myelinated peripheral nerve. Neuron. May 2001;30(2):385–97. [DOI] [PubMed] [Google Scholar]
- 51.Poliak S, Salomon D, Elhanany H, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. September 15 2003;162(6):1149–60. https://doi.org10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergles DE, Richardson WD. Oligodendrocyte Development and Plasticity. Cold Spring Harb Perspect Biol. August 20 2015;8(2):a020453. https://doi.org10.1101/cshperspect.a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience. March 15 2003;23(6):2314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baxi EG, DeBruin J, Tosi DM, et al. Transfer of myelin-reactive th17 cells impairs endogenous remyelination in the central nervous system of cuprizone-fed mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. June 3 2015;35(22):8626–39. https://doi.org10.1523/jneurosci.3817-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]