Abstract
Background
Sustained molecular detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in the upper respiratory tract (URT) in mild to moderate coronavirus disease 2019 (COVID-19) is common. We sought to identify host and immune determinants of prolonged SARS-CoV-2 RNA detection.
Methods
Ninety-five symptomatic outpatients self-collected midturbinate nasal, oropharyngeal (OP), and gingival crevicular fluid (oral fluid) samples at home and in a research clinic a median of 6 times over 1–3 months. Samples were tested for viral RNA, virus culture, and SARS-CoV-2 and other human coronavirus antibodies, and associations were estimated using Cox proportional hazards models.
Results
Viral RNA clearance, as measured by SARS-CoV-2 reverse transcription polymerase chain reaction (RT-PCR), in 507 URT samples occurred a median (interquartile range) 33.5 (17–63.5) days post–symptom onset. Sixteen nasal-OP samples collected 2–11 days post–symptom onset were virus culture positive out of 183 RT-PCR-positive samples tested. All participants but 1 with positive virus culture were negative for concomitant oral fluid anti-SARS-CoV-2 antibodies. The mean time to first antibody detection in oral fluid was 8–13 days post–symptom onset. A longer time to first detection of oral fluid anti-SARS-CoV-2 S antibodies (adjusted hazard ratio [aHR], 0.96; 95% CI, 0.92–0.99; P = .020) and body mass index (BMI) ≥25 kg/m2 (aHR, 0.37; 95% CI, 0.18–0.78; P = .009) were independently associated with a longer time to SARS-CoV-2 viral RNA clearance. Fever as 1 of first 3 COVID-19 symptoms correlated with shorter time to viral RNA clearance (aHR, 2.06; 95% CI, 1.02–4.18; P = .044).
Conclusions
We demonstrate that delayed rise of oral fluid SARS-CoV-2-specific antibodies, elevated BMI, and absence of early fever are independently associated with delayed URT viral RNA clearance.
Keywords: SARS-CoV-2, RT-PCR, viral RNA, COVID-19, antibody
INTRODUCTION
Although severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections chiefly occur in the community, early virus and host response kinetics have been largely described in hospitalized inpatients. The logistics of recruiting and sampling potentially infectious individuals in the outpatient setting have sustained gaps in our knowledge of infectivity and pathogenesis in mild to moderate disease. In particular, it is not known whether antibody kinetics are tightly correlated on an individual level with viral RNA decay in mild to moderate coronavirus disease 2019 (COVID-19). Several case reports of prolonged shedding of infectious virus in immune-compromised people with COVID-19 [1–4] support the hypothesis that adaptive immunity is required for clearance of infectious virus. Two studies report that prolonged viral RNA shedding is correlated with lower peak SARS-CoV-2-specific immunoglobulin G (IgG) titers and lower numbers of B and T cells in blood [5, 6]. However, other studies report no difference in plasma SARS-CoV-2-specific antibody titers in patients testing positive or negative at late time points after symptom onset [7, 8]. Here we leveraged guided home sampling to longitudinally characterize viral and immune kinetics in a prospective cohort with mild to moderate COVID-19 and identified associations with duration of viral RNA shedding from the upper respiratory tract that yield insights into COVID-19 pathogenesis.
METHODS
Patient Consent Statement
Due to the timing of study enrollment and the contagious nature of COVID-19, obtaining a signed informed consent form for participants enrolled in this study was not feasible or safe for study staff. Instead, study staff obtained verbal consent using consent waiver with an alteration of informed consent. All participants provided verbal consent after documentation of understanding, as they were isolating at home due to COVID-19, according to an approved consent script in either English or Spanish. A copy of the informed consent was sent to all participants. This protocol and verbal consent were approved by the Johns Hopkins University School of Medicine Institutional Review Board. All procedures were in accordance with the ethical standards of the Helsinki Declaration of the World Medical Association.
Study Cohort and Sampling
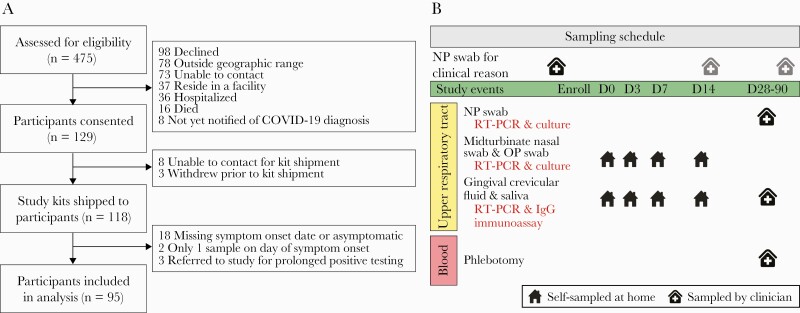
The Johns Hopkins University (JHU) School of Medicine Institutional Review Board approved this study. Participants provided informed consent. The study prospectively enrolled a convenience sample of adults aged ≥30 years within 48 hours of a positive SARS-CoV-2 reverse transcriptase polymerase chain reaction (RT-PCR) test from an outpatient testing site of the Johns Hopkins Health System (JHHS) between April 21, 2020, and July 23, 2020 (Figure 1A) [9, 10]. The 95 symptomatic participants with known symptom onset date and who completed 1 or more RT-PCR tests after symptom onset were included here. Self-sampled midturbinate nasal swabs and oropharyngeal (OP) swabs were obtained with telephone guidance from study staff, and swabs were combined in 3 mL of viral transport medium and frozen. Midturbinate nasal-OP RT-PCR is equivalent to clinician-collected nasopharyngeal swabs [11–15]. Gingival crevicular fluid was obtained using the Oracol device (Malvern Medical Developments Ltd., Worcestershire, UK). Saliva was collected in the tube from June 1, 2020, onwards after validation of the spit saliva sample type [1]. The sampling schedule is shown in Figure 1B. Fifty-three of 95 participants presented to a research clinic visit at 1–4 months post–symptom onset for height, weight, vital signs, blood draw, nasopharyngeal swab, and oral fluid sampling. Symptom reports were collected by telephone at enrollment and on the same days as home and sample collection and are the primary source of symptom data. Symptom reports were cross-checked by reviewing clinical notes for symptom complaints from the electronic medical record.
Figure 1.
CONSORT flow diagram and sampling schedule. A, A convenience sample of 475 adults with recent positive SARS-CoV-2 RT-PCR tests from an outpatient testing site of the Johns Hopkins Health System was assessed for eligibility between April 21, 2020, and July 23, 2020. Preference was given to participants 40 years of age and older. Study kits were sent to 118 participants, and data and samples from 95 participants were included in the analyses presented here. B, Study sampling schedule. Clinical RT-PCR results from the medical record are included in these analyses. Samples on study days 0–14 were self-collected at home with telephone or video guidance by trained study staff. Participants presented to a research clinic for collection of samples a median (range) of 45 (27–88) days after study day 0. Abbreviations: RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Laboratory Procedures
RT-PCR testing of study samples was performed on the Abbott m2000 platform (Abbott Molecular, Des Plaines, IL, USA) in 600-µL volumes [9]. Reported are cycle threshold (Ct) values from 1 of 2 primer/probe regions. Ct values <31.5 were positive. Two hundred–microliter volumes of oral fluid were assayed by RT-PCR. The adequacy of self-collected nasal-OP and oral fluid samples was confirmed via quantitative PCR for GAPDH human gene expression utilizing a TaqMan gene expression assay (ThermoFisher Scientific, Waltham, MA, USA) (data not shown).
Positive nasal-OP samples by RT-PCR were tested for propagation of SARS-CoV-2 in cell culture [16]. Plasma SARS-CoV-2-specific antibody titers were quantified by indirect enzyme-linked immunosorbent assay (ELISA), and neutralizing antibody titers were quantified by microneutralization assay [17, 18]. Quantification of oral fluid SARS-CoV-2-specific IgG and other human coronavirus-specific IgG was obtained from a multiplex SARS-CoV-2 immunoassay based on Luminex technology [19, 20]. The oral fluid SARS-CoV-2-specific IgG readout of this assay is highly correlated with plasma SARS-CoV-2-neutralizing antibodies and plasma S-RBD-specific IgG by ELISA [19, 20].
Statistical Methods
Kaplan-Meier plots and log-rank tests were generated using the “survival” R package [21]. Associations between covariates and clearance of upper respiratory tract SARS-CoV-2 viral RNA were determined via Cox proportional hazards models [21] after multiple imputation and model selection by least absolute shrinkage and selection operator (LASSO). A complete list of covariates for Cox model 1 may be found in Supplementary Table 1. Time to RT-PCR clearance was defined as number of days from symptom onset to the midpoint between the last positive RT-PCR test and the subsequent negative test. Patterns of missing data were investigated, and logistic regression was performed on the most frequent missing patterns to confirm that the data were missing at random (Supplementary Methods, Supplementary Tables 2–3). Twenty data sets, each including all data points for all 95 participants, were imputed using predictive mean matching for univariate imputation and chained equation (MICE) for multivariate imputation using the “mice” R package [22]. Model selection was performed using LASSO on each of the imputed data sets using the “glmnet” R package [23]. Nine variables were included in the model, which used the pooled imputed data set. To determine whether the time from symptom onset to first detection of oral fluid SARS-CoV-2-specific IgG is associated with RT-PCR clearance, we performed model selection using LASSO and ran Cox regression model 2. Further details are provided in the Supplementary Methods.
RESULTS
Study Design
Ninety-five nonhospitalized, symptomatic individuals with a positive nasopharyngeal SARS-CoV-2 RT-PCR test from an emergency room or an ambulatory testing center within the previous 48 hours were enrolled between April 21, 2020, and July 23, 2020 (Figure 1A). Study day 0 was a median (range) of 4 (2–11) days from the collection of the most recent prior positive RT-PCR test and was a median (range) of 9 (2–80) days from symptom onset. Participants presented for a single research clinic visit a median (range) of 45 (27–88) days after study day 0.
Clinical Characteristics
The median age of the participants was 56 years, and 59% were women (Table 1). A total of 39% identified as Black or African American, and 14% as Hispanic/Latinx. The median body mass index (BMI) of participants was 29.3 kg/m2, similar to that of US adults, 29.4 kg/m2 [24]. Eight participants (8.4%) required hospitalization after enrollment in the study [10].
Table 1.
Demographics and Clinical Characteristics of Participants
| Characteristic | Participants (n = 95) |
|---|---|
| Median age at enrollment (IQR), y | 56 (49–64) |
| Age grouping | |
| <65 y | 74 (77.9) |
| ≥65 y | 21 (22.1) |
| Biological sex | |
| Male | 39 (41.1) |
| Female | 56 (58.9) |
| Race | |
| White | 46 (48.4) |
| Black or African American | 37 (38.9) |
| Asian | 5 (5.3) |
| American Indian or Alaska Native | 1 (1.1) |
| Native Hawaiian or other Pacific Islander | 1 (1.1) |
| Other | 5 (5.3) |
| Ethnicity | |
| Hispanic or Latinx | 13 (13.8) |
| Median BMI (IQR), kg/m2 | 29.3 (26.0–35.4) |
| BMI groupa | |
| Normal (<25 kg/m2) | 19 (20.0) |
| Overweight or obese (≥25 kg/m2) | 74 (77.9) |
| Smoking status | |
| Never smoker | 68 (71.6) |
| Ever smoker | 4 (4.2) |
| Missing | 23 (24.2) |
| Hypertension | 39 (41.1) |
| Cardiovascular disease other than hypertension | 15 (15.8) |
| COPD or asthma | 16 (16.8) |
| Chronic kidney disease | 3 (3.2) |
| Diabetes | 15 (15.8) |
| Cancer not in remission | 4 (4.3) |
| Immunocompromisedb | 9 (9.5) |
| Fever as 1 of first 3 COVID-19 symptoms | |
| Yes | 30 (31.6) |
| No | 60 (63.2) |
| Missing | 5 (5.3) |
| Cycle threshold value of first RT-PCRc | |
| Median (IQR) | 18.6 (15.0–23.7) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: IQR, interquartile range; BMI, body mass index; COPD, chronic obstructive pulmonary disease; RT-PCR, reverse transcription polymerase chain reaction.
aTwo participants had missing BMI information.
bImmunocompromised refers to participants who are solid organ or bone marrow transplant recipients, who have primary immunodeficiency or AIDS, or who were taking immune-modulating medications within 3 months of COVID-19 diagnosis such as chemotherapy, including biologics, mycophenolate mofetil, methotrexate, tumor necrosis factor inhibitors, or prednisone ≥20 mg/d.
cThe first SARS-CoV-2 RT-PCR was performed in the clinical laboratory before enrollment. Included here are cycle threshold values from the 59 participants whose first RT-PCR was run on the NeuMoDx platform within 7 days of symptom onset.
Viral Kinetics in COVID-19 Outpatients
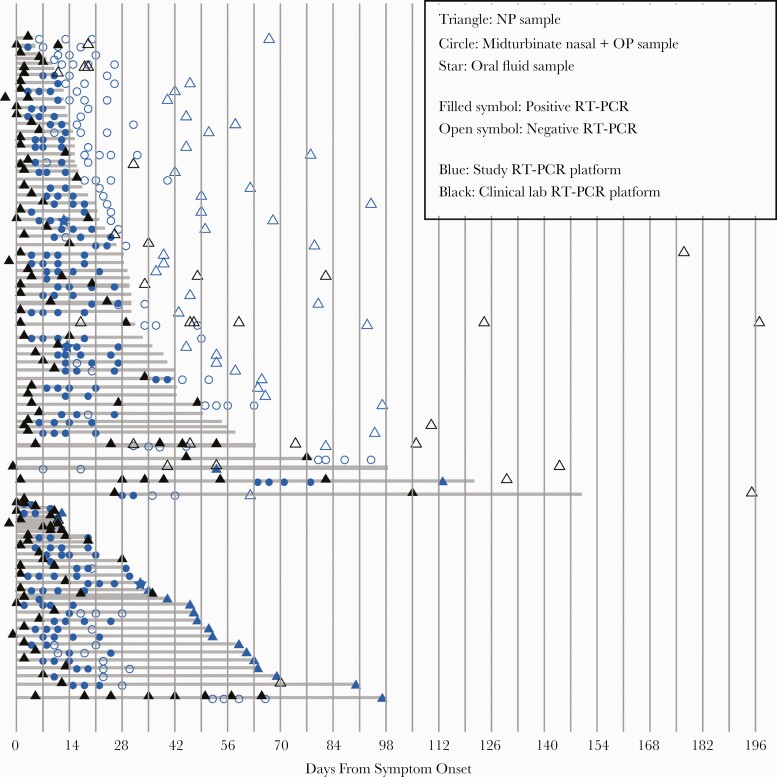
Participants’ SARS-CoV-2 RT-PCR tests from the study and from the medical record were included in these analyses, totaling 507 RT-PCR tests (Figure 2). Midturbinate nasal-OP and nasopharyngeal RT-PCR tests are the primary sample types, while oral fluid RT-PCR tests were included only if nasal samples were missing or in the 3 instances in which the nasal-OP sample was negative while the oral fluid sample was positive. Through December 15, 2020, the 95 participants had a median (interquartile range [IQR]) of 6 (4–6) RT-PCR tests during and after acute COVID-19. Longitudinal Ct values from study samples showed a rapid decay, with several instances of a negative test followed by a positive test (re-positive cases) (Figure 3A, gray lines). Re-positive cases are commonly reported [8, 25, 26] and may be due to poor immune clearance of viral RNA or prolonged low-level replication of SARS-CoV-2. There was no difference in the reference gene GADPDH’s RNA cycle threshold values from samples that tested negative before a re-positive compared with samples from participants who never experienced a re-positive test (Supplementary Figure 1).
Figure 2.
Longitudinal SARS-CoV-2 RT-PCR sampling in outpatients. SARS-CoV-2 RT-PCR test results by day from symptom onset. Each row represents 1 participant. Shown here are nasal samples. Stars indicate a sample time point at which the nasal sample was negative and the oral fluid sample was positive. Included in the top grouping are participants whose last RT-PCR test was negative, and in the bottom group are those whose last RT-PCR test was positive. Shaded gray lines indicate the number of days from symptom onset until the midpoint between the last positive sample and the next negative sample (top grouping) or until the last positive sample (bottom grouping). Nasopharyngeal (NP) swabs collected by a health care worker are indicated by triangles; self-collected midturbinate nasal-oropharyngeal (OP) swab combined in viral transport media are indicated by circles. Abbreviations: RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
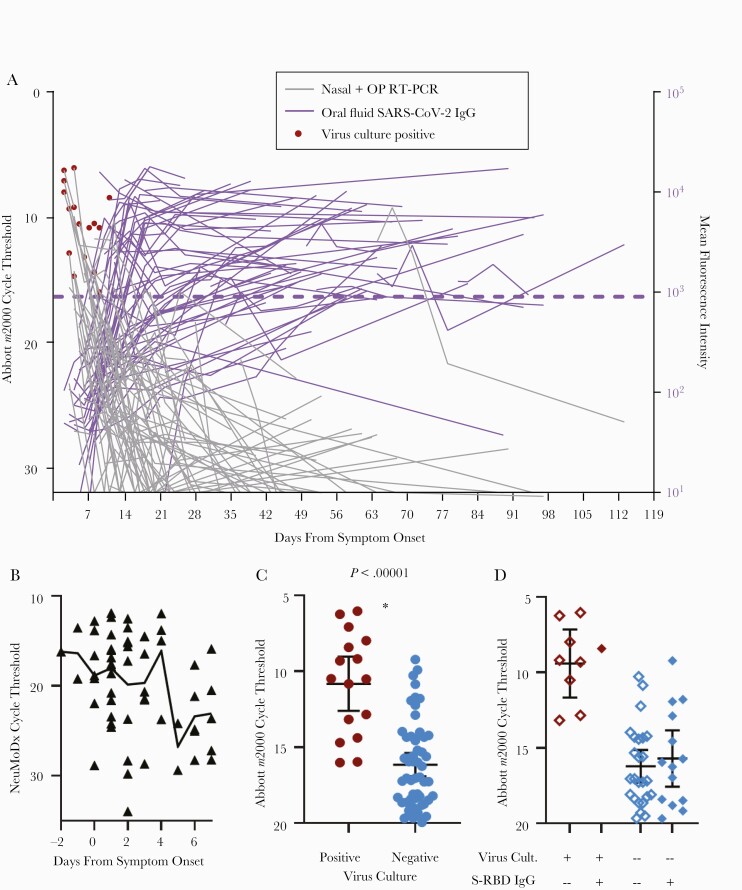
Figure 3.
Kinetics of RT-PCR cycle thresholds, virus culture, and oral fluid SARS-CoV-2 antibodies in mild–moderate COVID-19. A, Longitudinal Abbott m2000 SARS-CoV-2 RT-PCR cycle threshold values from nasal-OP swab study samples (blue circles and triangles in Figure 2) by day from symptom onset are shown in gray. Longitudinal oral fluid antispike (Mt. Sinai) IgG mean fluorescence intensity shown in purple. Lines connect values from the same participant. Each sample with a positive RT-PCR result was assessed for propagation of SARS-CoV-2 on VeroE6-TMPRSS2 cells. Samples with positive virus culture are indicated by red circles. The cutoff for positive anti-S IgG is shown with a purple dotted line (see the “Methods” for cutoff calculation). B, The subset of 59 participants with a nasopharyngeal swab collected within 7 days of symptom onset and run on the NeuMoDx SARS-CoV-2 RT-PCR platform is shown by day from symptom onset. The line connects cycle threshold means. C, The subset of nasal-OP swab samples with a cycle threshold <20 is grouped according to whether virus culture of the sample was positive or negative. Means with 95% CIs are shown. *P < .00001 by 1-tailed Student t test. D, The subset of samples shown in (B) that had a simultaneous adequate oral fluid sample is shown here. Filled diamonds represent study time points at which oral fluid contained detectable IgG antibodies to Mt Sinai S-RBD, and open diamonds represent study time points at which oral fluid was negative for anti-S-RBD IgG. Abbreviations: Ab, antibody; COVID-19, coronavirus disease 2019; Cult, culture; IgG, immunoglobulin G; OP, oropharyngeal; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; S-RBD, spike receptor binding domain.
To examine Ct values as an indicator of viral load in the URT from very early time points, RT-PCR Ct values were obtained from the participant’s first positive clinical RT-PCR test, the majority of which were run on the NeuMoDx platform. The mean Ct value was lowest—meaning the viral burden was highest—before and just after symptom onset (Figure 3B). The first 8–10 cycles on the Abbott RT-PCR platform are not read, and so the Ct values of the Abbott (study samples) and NeuMoDx (clinical samples) platforms are not comparable.
Virus Culture Positivity Is Detected Through 11 Days Post–Symptom Onset
Sixteen (of 183 RT-PCR-positive) study samples cultured for SARS-CoV-2 on VeroTMPRSS2 cells [16, 27] from 14 participants were virus culture positive (Figure 3A, red circles). In these 14 participants, the median time from symptom onset to last positive virus culture (range) was 5 (2–11) days. No sample tested positive for virus culture beyond 11 days from symptom onset, even in the case of an immunocompromised participant who had positive RT-PCRs with low Ct values 2 months post–symptom onset (Figure 3A). None of the re-positive RT-PCR samples were positive for virus culture. Virus culture was positive only in samples with Ct values <17 on the Abbott m2000 platform. The mean Ct value of samples positive for virus culture was significantly lower than that of samples negative for viral culture (Figure 3C).
Positive Oral Fluid SARS-CoV-2-Specific IgG Is Associated With Negative Virus Culture
To determine whether oral fluid anti-SARS-CoV-2 IgG could be used to predict which samples with low Ct values were negative for virus culture, we examined sample time points with (1) nasal Abbott RT-PCR Ct values <20, (2) nasal virus culture results, and (3) oral fluid samples with adequate total IgG for assessment (total IgG >10 µg/mL or detectable oral fluid anti-S-RBD IgG). Fourteen of 15 samples positive for oral fluid anti-S-RBD IgG were negative for virus culture (Figure 3D). The 1 culture-positive sample was collected on day 11 after symptom onset, which is at or shortly after the expected first detection of this antibody (Supplementary Table 4).
Oral Fluid SARS-CoV-2-Specific IgG Is First Detected Between 8 and 13 Days From Symptom Onset
Oral fluid SARS-CoV-2-specific IgG to multiple viral antigens was assessed (Supplementary Figures 2–4). The longitudinal kinetics of oral fluid anti-SARS-CoV-2 S IgG by day from symptom onset are shown in purple in Figure 3A. Supplementary Table 4 shows the means and 95% CIs of number of days from symptom onset to rise above cutoff for each of the 8 SARS-CoV-2 antigens (8–13 days depending on the antigen), similar to that previously described [19].
Kinetics of SARS-CoV-2 Viral RNA Clearance in Outpatients
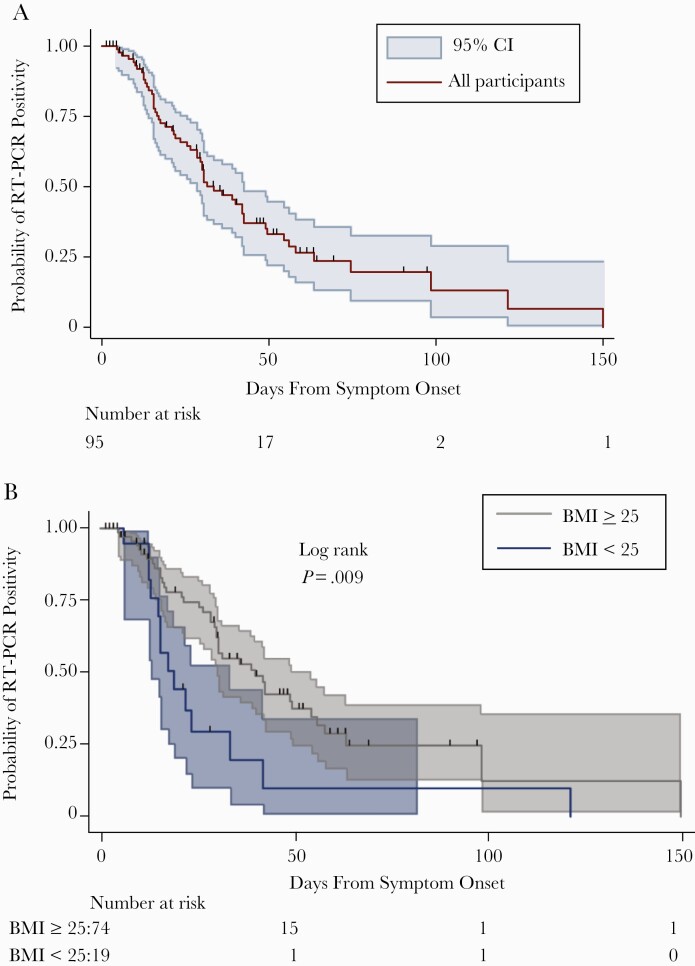
The median time to clearance (range, IQR) was 33.5 (4.5–150, 17–63.5) days (Figure 4A), similar to hospitalized COVID-19 patients with a similar mean age [28].
Figure 4.
Kaplan-Meier plots SARS-CoV-2 RT-PCR positivity by day from symptom onset in mild–moderate COVID-19. A, Kaplan-Meier survival curve with 95% CIs for positive upper respiratory tract SARS-CoV-2 RT-PCR by day from symptom onset. B, Kaplan-Meier survival curve by BMI category. Abbreviations: BMI, body mass index; COVID-19, coronavirus disease 2019; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Elevated BMI Is Associated With Longer Time to Viral RNA Clearance
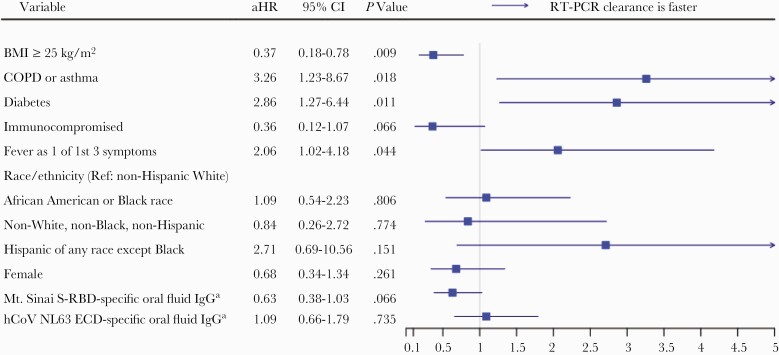
To determine host and immune factors associated with longer time to RT-PCR clearance, we used a Cox proportional hazards model after multiple imputation and model selection by LASSO. Nine covariates were selected for inclusion in the Cox model. Shown in Figure 5 are the univariable and adjusted hazard ratios of clearance and the 95% confidence intervals. BMI ≥25 kg/m2 was independently associated with longer time to viral RNA clearance in outpatients with COVID-19 in the main analysis (adjusted hazard ratio [aHR], 0.37; 95% CI, 0.18–0.78; P = .009) (Figures 4B and 5) and in several sensitivity analyses using different methods to estimate clearance (Supplementary Tables 5–9). This association did not appear to be explained by comorbidities such as hypertension and diabetes as inclusion of hypertension and diabetes in the model did not attenuate the BMI association. Further, hypertension was not significantly associated with time to viral RNA clearance, and diabetes was significantly positively associated with clearance, which is in the opposite direction of elevated BMI.
Figure 5.
Cox proportional hazards model for viral RNA clearance. An aHR <1 denotes longer time to SARS-CoV-2 viral RNA clearance in the upper respiratory tract as measured by RT-PCR. To account for missing data points, 20 data sets were imputed using PPM MICE. Model selection using LASSO was performed on each of the 20 data sets. The top 9 variables of 35—selected 15 or more times out of 20 (Supplementary Table 2)—were chosen for inclusion in the Cox proportional hazards model, which was performed using the pooled imputed data set. aStandardized log mean fluorescence intensity and treated as a time-varying variable. Abbreviations: aHR, adjusted hazard ratio; ECD, spike ectodomain; hCoV, human coronavirus; LASSO, least absolute shrinkage and selection operator; MICE, multiple imputation by chained equation; PPM, predictive mean matching; RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; S-RBD, spike receptor binding domain.
There was a trend toward immunocompromised hosts (n = 9) having a longer time to viral RNA clearance (Figure 5). There was additionally a trend toward greater magnitude of oral fluid anti-S-RBD IgG (treated as a time-varying variable) being associated with longer time to RT-PCR clearance (Figure 5). Larger studies are needed to assess this association. The magnitude of oral fluid antihuman coronavirus IgG for the 4 circulating non-SARS human coronaviruses was not associated with time to viral RNA clearance (Figure 5; Supplementary Table 1).
COPD/Asthma, Early Fever, and Diabetes Are Associated With Faster Viral RNA Clearance
COPD/asthma, diabetes, and fever reported as 1 of the first 3 COVID-19 symptoms were each associated with faster viral RNA clearance (Figure 5). In 4 of 5 sensitivity analyses using different methods to estimate clearance, fever as 1 of the first 3 reported COVID-19 symptoms remained associated with viral RNA clearance, whereas the association of diabetes and COPD/asthma was significant in fewer such analyses (Supplementary Tables 5–9).
Longer Time to First Detection of Oral Fluid SARS-CoV-2-Specific IgG Is Associated With Longer Time to Viral RNA Clearance
In a time-dependent covariate Cox model (Cox model 2) for viral RNA clearance, a longer time to first detection of oral fluid spike-specific IgG is associated with longer time to viral RNA clearance (aHR, 0.96; 95% CI, 0.92–0.99; P = .020) (Table 2; Supplementary Tables 8–9).
Table 2.
Time-Dependent Covariate Cox Proportional Hazards Model for Viral RNA Clearance (Cox Model 2)
| Variable | HR | aHR | 95% CI | P Value |
|---|---|---|---|---|
| Change from undetectable to detectable Mt. Sinai S-specific oral fluid IgG | 0.96 | 0.96 | 0.92–0.99 | .020 |
| BMI ≥25 kg/m2 (referent: BMI <25 kg/m2) | 0.44 | 0.29 | 0.14–0.61 | .001 |
| Diabetes | 2.11 | 2.49 | 1.09–5.67 | .030 |
| Immunocompromised | 0.68 | 0.35 | 0.12–1.02 | .055 |
| Fever as 1 of first 3 COVID-19 symptoms | 1.72 | 2.23 | 1.05–4.71 | .036 |
| Race/ethnicity (referent: non-Hispanic White) | ||||
| African American or Black race, any ethnicity | 0.73 | 0.85 | 0.42–1.73 | .659 |
| Non-White, non-Black, non-Hispanic | 1.20 | 0.56 | 0.14–2.23 | .408 |
| Hispanic of any race except Black | 0.79 | 1.48 | 0.41–5.39 | .549 |
Abbreviations: aHR, adjusted hazard ratio; BMI, body mass index; COVID-19, coronavirus disease 2019; HR, hazard ratio; IgG, immunoglobulin G; S, SARS-CoV-2 spike protein.
Plasma SARS-CoV-2-Specific Antibody Titer in Convalescence Is Not Associated With Time to Viral RNA Clearance
Titers of plasma anti-S-RBD IgG, anti-S IgG, or neutralizing antibody in convalescence (measured once at 1–4 months post–symptom onset and treated as a baseline variable) were not significantly associated with viral RNA clearance in either direction. Additionally, the presence or absence of viral RNA by RT-PCR in the nasopharynx at 1–4 months post–symptom onset was not associated with plasma antibody titer (data not shown).
DISCUSSION
In a racially and ethnically diverse cohort of 95 adult outpatients with symptomatic COVID-19, we report that a longer time to first detection of oral fluid SARS-CoV-2-specific antibodies is independently associated with a longer time to viral RNA clearance. We find that the presence of SARS-CoV-2 antibodies in oral fluid may be a predictor of negative virus culture from nasal samples, even in samples with low RT-PCR Ct values. We demonstrate that the mean time from symptom onset to first detection of oral fluid SARS-CoV-2 S-RBD-IgG is 9–11 days, and virus culture–positive samples may be detected through day 11 post–symptom onset in adults with mild to moderate COVID-19, 1 day longer than the current recommended isolation period [29].
The strengths of this cohort are its size, its prospective and intensive longitudinal sampling design beginning in the acute phase of illness, its long duration of follow-up, and its simultaneous testing of samples for SARS-CoV-2 RT-PCR, virus culture, and oral fluid antibodies. Two studies of mostly outpatients included more cultured specimens but were not longitudinal [30, 31]. Our findings extend to the outpatient domain those described in 129 patients with severe COVID-19 by van Kampen and colleagues [32], who report a mean time from symptom onset to last positive virus culture of 8 days (maximum 20); in this cohort, virus cultures were more likely to be negative when plasma-neutralizing antibodies were elevated [32]. Our results suggest that outpatients testing positive for SARS-CoV-2 who have detectable oral fluid SARS-CoV-2-specfic antibodies are not likely to be transmissible, but larger studies are needed for confirmation.
We demonstrate that elevated BMI is also independently associated with longer time to viral RNA clearance. This association is not likely explained by increased prevalence of comorbidities such as hypertension or diabetes. In fact, diabetes, along with early fever and COPD/asthma, is associated with faster time to viral RNA clearance. Numerous studies report strong associations of COVID-19 severity and older age with viral RNA shedding duration [17, 33, 34]. Neither severity nor age was significantly associated with duration of viral RNA shedding in our study, likely because the vast majority of our cohort participants were in the same severity category and age range (49–64 years). Importantly, we found no effect of race or ethnicity with time to viral RNA clearance after adjustment for other demographic and comorbidities. Obesity is associated with testing positive for SARS-CoV-2, with severity of COVID-19, and with COVID-19 mortality [35–38]. Dysregulated immunity is observed in severe COVID-19, aging, and obesity, and it may be the unmeasured key correlate of viral RNA shedding duration. Aging drastically increases the chronic state of inflammation present in obesity [39], perhaps explaining why our study of middle-aged and older participants uncovered the strong association of elevated BMI and viral RNA shedding duration. The other risk factors that correlated with longer time to viral RNA clearance in our study—absence of early fever and longer time to detection of oral fluid antibodies—are consistent with impaired early innate and adaptive immune responses. Silva et al. report that a delay in antibody production is linked to high salivary viral loads in hospitalized patients [40]. We hypothesize that the chronic inflammatory state of aging and obesity hinders early innate and adaptive immune responses to SARS-CoV-2, leading to higher viral loads, which require higher antibody titers and more time to clear, while also presenting a cytokine milieu in which it is stochastically more likely for a patient to tip into the positive feedback loops of hyperinflammation that portend disease severity. Mechanistic studies are needed to determine how obesity prolongs viral RNA shedding.
Our study has limitations, including that it is undersampled at the time of symptom onset and at >1 month from symptom onset. We address this by including clinician-ordered samples outside of the study, but the time to viral RNA clearance may be less precisely described in participants whose study samples at 1–3 months remained positive than in others. Our analytical approaches—Kaplan-Meier estimate, log-rank test, and Cox regression analyses—were designed to properly handle and correct bias caused by censored data. We minimize bias in estimated clearance by defining clearance as the midpoint between the last positive and subsequent negative test. Several sensitivity analyses using different clearance definitions demonstrate that our conclusions are supported by the evidence. There was a nonsignificant trend toward immunocompromised host status being associated with viral RNA shedding duration, which may appear to contrast with reports of prolonged viral carriage in immunocompromised people [1–4]. This likely occurred because we grouped all immunocompromised people together, whereas there is probably a specific immunocompromised phenotype that leads to prolonged viral RNA detection. The associations of diabetes and COPD/asthma with faster time to viral RNA clearance were not found to be independent of other covariates in all sensitivity analyses (diabetes) or our second Cox model (COPD/asthma), and they conflict with 1 report of asthma being associated with longer viral RNA shedding [41]. Larger studies are needed to determine whether diabetes, COPD, and asthma are associated with time to viral RNA clearance. Finally, our study excluded people with asymptomatic SARS-CoV-2 infection, so it is not yet clear whether these results are generalizable to asymptomatic infection.
In sum, we report viral and immune kinetics in an intensively characterized cohort of 95 adult outpatients with mild to moderate COVID-19. We demonstrate that elevated BMI, longer time to detection of oral SARS-CoV-2 IgG, and the absence of early fever are independently associated with longer time to viral RNA clearance, suggesting that functional early innate and adaptive immunity is critical for timely viral RNA clearance.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We acknowledge Jeffrey Holden, Tyrone Howard, Andrew Karaba, Anastasia Lambrou, Lucy Li, Manuela Plazas Montana, Caroline Popper, and Amanda Tuchler for their assistance.
Financial support. This work was supported by the JHU COVID-19 Research and Response Program Fund and the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases Discovery Program. This research was facilitated by infrastructure and resources provided by the JHU Center for AIDS Research. This work was supported by the National Institutes of Health (grant numbers 1P30AI094189 to C.P., K08AI143391 to A.A., U54EB007958-12 to Y.M., U5411090366 to Y.M., U54HL143541-02S2 to Y.M., UM1AI068613 to Y.M., K23AI135102 to J.T., contract HHS N272201400007C to A.P., U54CA260492 to A.C. and S.K., R21AI139784 to C.H. and N.P., R01ES026973 to C.H., N.P., and K.K., R01AI130066 to C.H., U24OD023382 to C.H., K08HS025782 to S.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the National Institute of Infectious Diseases, Japan, for providing VeroE6TMPRSS2 cells and acknowledge the Centers for Disease Control and Prevention, BEI Resources, NIAID, NIH, for SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-5228. Funding was provided by the FIA Foundation to C.H. and N.P. and by the GRACE Communications Foundation to C.D.H., N.P., and K.K. H.M. reports research collaboration and contribution of equipment and reagents from Bio-Rad Laboratories and DiaSorin Molecular.
Potential conflicts of interest. S.H.M. received speaker fees from Gilead Sciences. All other authors report no conflicts relevant to this article. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 2020; 383:1283–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baang JH, Smith C, Mirabelli C, et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021; 223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aydillo T, Gonzalez-Reiche AS, Aslam S, et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med 2020; 383: 2586–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakajima Y, Ogai A, Furukawa K, et al. Prolonged viral shedding of SARS-CoV-2 in an immunocompromised patient. J Infect Chemother 2021; 27:387–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li K, Huang B, Wu M, et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun 2020; 11:6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu B, Han J, Cheng X, et al. Reduced numbers of T cells and B cells correlates with persistent SARS-CoV-2 presence in non-severe COVID-19 patients. Sci Rep 2020; 10:17718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang C, Jiang M, Wang X, et al. Viral RNA level, serum antibody responses, and transmission risk in recovered COVID-19 patients with recurrent positive SARS-CoV-2 RNA test results: a population-based observational cohort study. Emerg Microbes Infect 2020; 9:2368–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu J, Peng J, Xiong Q, et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine 2020; 59:102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manabe YC, Reuland C, Yu T, et al. Self-collected oral fluid saliva is insensitive compared to nasal-oropharyngeal swabs in the detection of SARS-CoV-2 in outpatients. Open Forum Infect Dis 2020; XXX:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blair PW, Brown DM, Jang M, et al. The clinical course of COVID-19 in the outpatient setting: a prospective cohort study. Open Forum Infect Dis 2020; XXX:XXX–XX. [DOI] [PMC free article] [PubMed]
- 11. Lee RA, Herigon JC, Benedetti A, et al. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J Clin Microbiol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. LeBlanc JJ, Heinstein C, MacDonald J, et al. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS-CoV-2. J Clin Virol 2020; 128:104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altamirano J, Govindarajan P, Blomkalns AL, et al. Assessment of sensitivity and specificity of patient-collected lower nasal specimens for severe acute respiratory syndrome coronavirus 2 testing. JAMA Netw Open 2020; 3: e2012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kojima N, Turner F, Slepnev V, et al. Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tu YP, Jennings R, Hart B, et al. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med 2020; 383:494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pekosz A, Parvu V, Li M, et al. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klein SL, Pekosz A, Park HS, et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J Clin Invest 2020; 130:6141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schaecher SR, Stabenow J, Oberle C, et al. An immunosuppressed Syrian golden hamster model for SARS-CoV infection. Virology 2008; 380:312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pisanic N, Randad PR, Kruczynski K, et al. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. J Clin Microbiol 2020; 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heaney CD, Pisanic N, Randad PR, et al. Comparative performance of multiplex salivary and commercially available serologic assays to detect SARS-CoV-2 IgG and neutralization titers. medRxiv 2021.01.28.21250717 [Preprint]. 2 February 2021. Available at: 10.1101/2021.01.28.21250717. Accessed 6 April 2021. [DOI] [PMC free article] [PubMed]
- 21. Therneau TM, Grambsch PM.. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 22. van Buuren S, Groothuis-Oudshoorn K. mice: multivariate imputation by chained equations in R. J Stat Softw 2011; 45:1–67. [Google Scholar]
- 23. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 24. Fryar CD, Kruszon-Moran D, Gu Q, Ogden CL. Mean body weight, height, waist circumference, and body mass index among adults: United States, 1999–2000 through 2015–2016. Natl Health Stat Report 2018; 1–16. [PubMed] [Google Scholar]
- 25. Gniazdowski V, Morris CP, Wohl S, et al. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Centers for Disease Control and Prevention. Findings from investigation and analysis of re-positive cases. Available at: https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030&act=view&list_no=367267&nPage=1#. Accessed 2 April 2021.
- 27. Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 2020; 117:7001–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou C, Zhang T, Ren H, et al. Impact of age on duration of viral RNA shedding in patients with COVID-19. Aging (Albany NY) 2020; 12:22399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention. COVID-19: isolate if you are sick. Available at: https://www.cdc.gov/coronavirus/2019-ncov/if-you-are-sick/isolation.html. Accessed 19 February 2021.
- 30. Basile K, McPhie K, Carter I, et al. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Kampen JJA, van de Vijver D, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 2021; 12:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fontana LM, Villamagna AH, Sikka MK, McGregor JC. Understanding viral shedding of severe acute respiratory coronavirus virus 2 (SARS-CoV-2): review of current literature. Infect Control Hosp Epidemiol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cevik M, Tate M, Lloyd O, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021; 2:e13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bennett TD, Moffitt RA, Hajagos JG, et al. The National COVID Cohort Collaborative: clinical characterization and early severity prediction. medRxiv 2021.01.12.21249511 [Preprint]. 23 January 2021. Available at: 10.1101/2021.01.12.21249511. Accessed 6 April 2021. [DOI]
- 36. Ho JSY, Fernando DI, Chan MY, Sia CH. Obesity in COVID-19: a systematic review and meta-analysis. Ann Acad Med Singap 2020; 49:996–1008. [PubMed] [Google Scholar]
- 37. Eastment MC, Berry K, Locke E, et al. Body mass index (BMI) and outcomes of SARS-CoV-2 among US veterans. Obesity (Silver Spring). In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peres KC, Riera R, Martimbianco ALC, et al. Body mass index and prognosis of COVID-19 infection. A systematic review. Front Endocrinol (Lausanne) 2020; 11:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frasca D, Blomberg BB, Paganelli R. Aging, obesity, and inflammatory age-related diseases. Front Immunol 2017; 8:1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Silva J, Lucas C, Sundaram M, et al. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv 2021.01.04.21249236 [Preprint]. 10 January 2021. Available at: 10.1101/2021.01.04.21249236. Accessed 6 April 2021. [DOI]
- 41. Knight D, Downes K, Munipalli B, et al. Symptoms and clinical outcomes of coronavirus disease 2019 in the outpatient setting. SN Compr Clin Med. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.