Abstract
Background
Previous studies demonstrated that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA can be detected for weeks after infection. The significance of this finding is unclear and, in most patients, does not represent active infection. Detection of subgenomic RNA has been proposed to represent productive infection and may be a useful marker for monitoring infectivity.
Methods
We used quantitative reverse-transcription polymerase chain reaction (RT-qPCR) to quantify total and subgenomic nucleocapsid (sgN) and envelope (sgE) transcripts in 185 SARS-CoV-2–positive nasopharyngeal swab samples collected on hospital admission and to relate to symptom duration.
Results
We find that all transcripts decline at the same rate; however, sgE becomes undetectable before other transcripts. The median duration of symptoms to a negative test is 14 days for sgE and 25 days for sgN. There is a linear decline in subgenomic compared to total RNA, suggesting that subgenomic transcript copy number is dependent on copy number of total transcripts. The mean difference between total and sgN is 16-fold and the mean difference between total and sgE is 137-fold. This relationship is constant over duration of symptoms, allowing prediction of subgenomic copy number from total copy number.
Conclusions
Subgenomic RNA may be no more useful in determining infectivity than a copy number threshold determined for total RNA.
Keywords: SARS-CoV-2, RT-qPCR, subgenomic RNA
Severe acute respiratory syndrome coronavirus 2 subgenomic RNA has been proposed as a marker of infectivity in patients with COVID-19. Our analysis of subgenomic RNA transcripts in 185 samples suggests that it is no more useful in determining infectivity than a total RNA copy number threshold.
The emergence of the severe acute respiratory syndrome virus 2 (SARS-CoV-2) led to the rapid development of diagnostic tests for infection. Many of these tests rely on quantitative reverse-transcription polymerase chain reaction (RT-qPCR) to detect total viral RNA. These assays are highly sensitive and specific, with a limit of detection of approximately 5–500 copies of viral RNA per reaction [1–3]. Unfortunately, the relationship between a positive RT-qPCR and viral infectivity is unclear, particularly as infection progresses over time. While the median duration of detection of SARS-CoV-2 viral RNA in the upper respiratory tract is roughly 14.5 days from symptom onset, studies have shown that SARS-CoV-2 RNA can be detected for many weeks, long past the time when most people are infectious [4].
Because of persistent test positivity, in most cases the Centers for Disease Control and Prevention (CDC) no longer recommends a test-based strategy for discontinuing transmission-based precautions. Current guidelines recommend that patients with mild to moderate disease remain isolated for 10 days from symptom onset, while those with either severe disease or an immunocompromising condition remain isolated for 10–20 days from symptom onset. These guidelines are largely supported by studies of viral infectivity in clinical samples over time [5–7]. However, recent studies have demonstrated that some immunosuppressed patients may shed infectious virus for weeks, regardless of symptoms [8–10]. Symptom-based strategies for isolation may also be problematic in patients incidentally found to be positive for SARS-CoV-2. In cases of severe immunosuppression and in asymptomatic patients, a test-based strategy may still be useful for discontinuation of transmission-based precautions [11].
There is no high-throughput, rapid test that distinguishes those who are infectious from those who are not. While viral culture is perhaps the most reliable way to determine infectivity, it is neither timely nor practical in most clinical laboratories. There has been intense interest in whether other molecular markers can be used as correlates of infectivity or active replication, including the detection of subgenomic RNA [12–14]. SARS-CoV-2 has a positive-sense, single-stranded RNA genome of nearly 30 kb. Negative-sense RNA intermediates serve as templates for the synthesis of positive-sense genomic RNA. The viral polymerase also makes subgenomic messages that have a common 5′ leader fused to downstream open reading frames, which are then translated into 9 proteins, including the nucleocapsid (N), spike (S), envelope (E), and membrane (M), and accessory proteins (3a, 6, 7a, 7b, and 8) [15]. Given this genomic arrangement, subgenomic RNA can be distinguished from total RNA by placement of alternate PCR primers. Because subgenomic transcripts are not packaged into virions, their presence is thought to indicate productively infected cells.
A few studies have evaluated the kinetics of subgenomic RNA during the course of infection. A small study of 9 patients found that subgenomic RNA could be detected in the throat up to 5 days after symptom onset [6]. A larger study of 35 patients showed that subgenomic RNA and culturable virus were not detectable after 8 days from symptom onset in mild COVD-19 disease, but total SARS-CoV-2 transcripts persisted for weeks [16]. Less is known about subgenomic RNA kinetics in hospitalized patients with severe disease and in immunosuppressed patients.
To determine the utility of subgenomic RNA as a correlate of SARS-CoV-2 infectivity, we used RT-qPCR to amplify the total and subgenomic nucleoprotein gene (N) and envelope gene (E) from 185 individual patient samples collected on admission to the hospital. We show that subgenomic transcripts become undetectable before total transcripts when evaluated in relation to duration of symptoms. Because expression levels of total and subgenomic transcripts are highly correlated, the added benefit of measuring subgenomic RNA is limited.
MATERIALS AND METHODS
Samples and Participants
Nasopharyngeal (NP) samples were obtained from patients admitted to the University of Michigan Hospital between 13 March 2020 and 10 June 2020. Dacron swabs were placed in 3 mL viral transport media and transported to the clinical microbiology laboratory for testing. Residual samples were stored in a central biorepository at –80°C. In total, 185 patients meeting a syndromic case definition for acute respiratory illness (a new or worsening cough or sputum production with onset within previous 10 days) and with RT-qPCR–confirmed SARS-CoV-2 were included in our study. Clinical data, including date of symptom onset, basic demographic data, and outcome were abstracted from clinician notes. This study protocol was reviewed and approved by the University of Michigan Institutional Review Board (IRB). The IRB determined the study to be exempt from requirement for informed consent given the retrospective nature of this study and this use of stored biospecimens and deidentified data.
Quantitative RT-PCR
Residual samples from NP swabs were centrifuged at 1200g and 200 µL of sample was used for RNA extraction. RNA was extracted with the Invitrogen PureLink Pro 96 Viral RNA/DNA Purification Kit (Invitrogen, Carlsbad, California). Samples were eluted in 100 µL and stored at –80°C.
Amplification of total and subgenomic transcripts for nucleocapsid (N) and envelope (E) genes was performed using conditions outlined in the CDC 2019-Novel Coronavirus Emergency Use Authorization protocol [17]. We diluted our clinical samples 1:5 in nuclease-free H2O and used 5 µL of this dilution for 1 µL equivalent total RNA per reaction. This was done to preserve clinical sample, and the dilution was accounted for in copy number calculations. Reactions were preformed using Taqpath 1-step RT-qPCR master mix (Thermo Fisher, Waltham, Massachusetts), 500 nM of each primer, and 250 nM of each probe in a total reaction volume of 20 µL. Cycling conditions were as follows: activation of uracil-N-glycosylase for 2 minutes at 25°C, reverse transcription for 15 minutes at 50°C, denaturation for 2 minutes at 95°C, and 45 cycles of 3 seconds at 95°C and 30 seconds at 55°C on an Applied Biosystems 7500 FAST real-time PCR system. Cycle threshold (Ct) was determined uniformly across PCR runs.
Subgenomic transcripts were amplified by substituting subgenomic leader sequence sgLeadSARSCoV2-F: 5′-CGATCTCTTGTAGATCTGTTCTC-3′ [6] as the forward primer for E or N together with the reverse primers and probes for N and E genes. For total E, the primers and probes were from the Charite/Berlin protocol [2]. The N gene was amplified using the CDC N1 primer-probe set [17]. Probe sequences were FAM labeled with Iowa Black quencher (Integrated DNA Technologies, Coralville, Iowa). Primer-probe sets used here for E and N were previously shown to have a limit of detection of approximately 5–500 copies of SARS-CoV-2 RNA per reaction [1–3]. We observed a similar analytic sensitivity of approximately 10–100 copies per reaction based on our standard curves for N, E, subgenomic E (sgE), and subgenomic N (sgN) (data not shown).
Droplet Digital PCR
Absolute copy numbers of N, E, sgN, and sgE were determined by droplet digital PCR (ddPCR) using SARS-CoV-2 RNA as template from HuH-7–infected cells. HuH-7 cells were infected with SARS-CoV-2 strain WA-1. RNA was harvested at 48 hours postinfection using Trizol and Zymo columns. ddPCR was performed using the QX200 ddPCR System (Bio-Rad, Hercules, California). Sample reactions contained HuH-7 RNA/sgRNA template (5.5 μL), the respective forward and reverse primers (final concentration 900 nM each), and the respective FAM-labeled probe (final concentration 250 nM). The other reaction components common to all reactions were added at the final concentration as per the manufacturer’s recommendation (Bio-Rad #1864021; 1X Supermix, 20 units/μL reverse transcriptase, and 15 mM dithiothreitol). Each 20 μL of reaction mix was partitioned into droplets using the QX200 droplet generator (Bio-Rad), transferred into a 96-well plate, sealed, and cycled in a C1000 Thermal Cycler (Bio-Rad). The cycling conditions used were as per the manufacturer’s recommendation (Bio-Rad’s expert design assay for SARS-CoV-2, #dEXD28563542; hold at 25°C for 3 minutes, reverse transcription at 50°C for 60 minutes, enzyme activation for 10 minutes at 95°C for 1 cycle, denaturation at 94°C for 30 seconds, and annealing/extension at 60°C for 60 seconds for 40 cycles, enzyme deactivation for 10 minutes at 98°C for 1 cycle, followed by a hold at 4°C). All steps were performed with a 2°C/second ramp rate and the lid temperature was set at 105°C. Droplets were read using QuantaSoft Software in the QX200 reader (Bio-Rad). Once copy number was determined, a 10-fold dilution series (from 1 × 107 copies to 10 copies/20 μL reaction) of this sample was performed to generate standard curves and used to determine copy number of clinical samples. Copy number was corrected for dilution of sample to reflect copies per milliliter of viral transport media from the original NP swab sample.
Statistical Analyses
Continuous variables were expressed as median and interquartile range (IQR) or mean and standard deviation (SD). Simple linear regression, Kaplan–Meier analysis, and log-rank test were performed using SPSS software version 27 (IBM Corporation) or GraphPad Prism. Statistical significance was set as P < .05.
RESULTS
Patient Characteristics
We obtained NP swabs from 185 SARS-CoV-2–positive patients admitted to the University of Michigan Hospital between 13 March 2020 and 10 June 2020. We were able to obtain clinical information from all 185 patients. In our sample, 56.8% were men (105/185). The mean age was 62.11 (SD, 15.63) years. The median days from symptom onset to SARS-CoV-2 testing was 5 days (IQR, 3–8 days). Close to half of patients required intensive care unit–level care (91/185). See Supplementary Table 1 for additional patient characteristics.
Symptom Duration Compared to Total and Subgenomic Transcripts
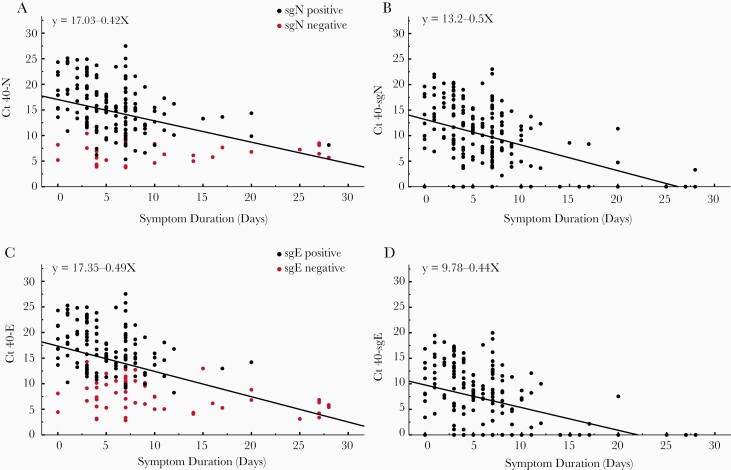
We found that there was no difference in the rate of decline of SARS-CoV-2 transcripts when compared to symptom duration with all slopes equal (P > .05) (Figure 1). We did, however, find a significant difference in the X-intercepts for the regression line of viral RNA transcript decline over time. There was no difference in intercepts between total E and N (P > .72). When total N was compared to sgN, the X-intercepts were significantly different (P < .0001), as was the comparison of E to sgE (P < .0001). Finally, there was a significant difference between intercepts for sgN and sgE (P < .001). The differences in X-intercepts reflect relative differences in days until RNA is no longer detectable based on a threshold of 40 cycles.
Figure 1.
Comparison of cycle threshold (Ct) vs day from symptom onset for clinical samples obtained from 185 inpatients. Vertical axis represents 40 minus the Ct. Total N (A), subgenomic N (sgN; B), total E (C), and subgenomic E (sgE; D). Red dots in A and C represent subgenomic-negative samples and black dots represent subgenomic-positive samples. Of the 185 patients, 57 were negative for sgE and 28 were negative for sgN (shown on y-axis). Pearson correlation coefficients: N: –0.420, P < .0001; sgN: –0.457, P < .0001; E: –0.468, P < .0001; sgE: –0.416, P < .0001. Linear regression equations are indicated in each panel.
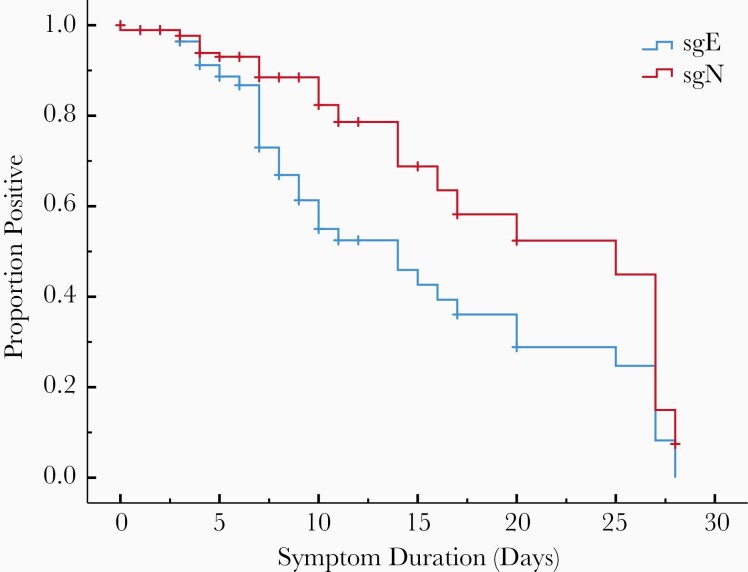
We used a Kaplan–Meier analysis to further investigate the relationship between symptom duration and a negative subgenomic RNA test. The median duration from symptom onset to a negative sgN RT-qPCR was 25 days (95% confidence interval [CI], 11.6–38.4 days) and 14 days for sgE (95% CI, 9.6–18.4 days). The difference in curves was statistically significant (P = .001) (Figure 2). All 185 patients were positive for total RNA regardless of symptom duration. By ≥14 days from symptom onset, only 12.5% (2/16) of patients were positive for sgE compared to 31.3% (5/16) for sgN.
Figure 2.
Kaplan–Meier analysis showing proportion of patients with positive quantitative reverse-transcription polymerase chain reaction (RT-qPCR) for subgenomic transcripts vs day from symptom onset. The median duration from symptom onset to a negative subgenomic N (sgN) RT-qPCR was 25 days and 14 days for subgenomic E (sgE). This difference between the curves by log-rank test was significant (P = .001). Vertical hash marks indicate censored cases.
Recent studies have demonstrated that the total copy number threshold that correlates with a loss of culture infectivity is between 5.5 and 6.5 log10 copies/mL [6, 7, 18]. We determined copy number in clinical samples using standard curves generated with known copy number from ddPCR experiments for total N, total E, sgE, and sgN. At 6.5 log10 total E copies, 47% (57/119) of samples were negative for sgE. At a total N copy number of 6.5 log10, 24% (28/119) were negative for sgN. There we no samples negative for subgenomic RNA at a copy number >6.5 log10 total copies/mL (Supplementary Figure 1).
Relationship of Total to Subgenomic Transcripts
To further understand the relationship between total and subgenomic transcripts for the N and E genes, we compared Ct values from 185 clinical samples. We found that 157 of 185 (85%) patients had both a detectable N and sgN, and 128 of 185 (69%) patients had both a detectable E and sgE. Subgenomic transcripts declined linearly along with total RNA transcripts (Supplementary Figure 2). There was no difference in the slope of subgenomic to total RNA regression (linear regression, slope of N vs sgN –0.99 and slope of E vs sgE –1.02; P > .05). We were unable to detect sgE RNA transcripts once total E reached 32 cycles, and we were unable to detect sgN transcripts once total N reached 35 cycles.
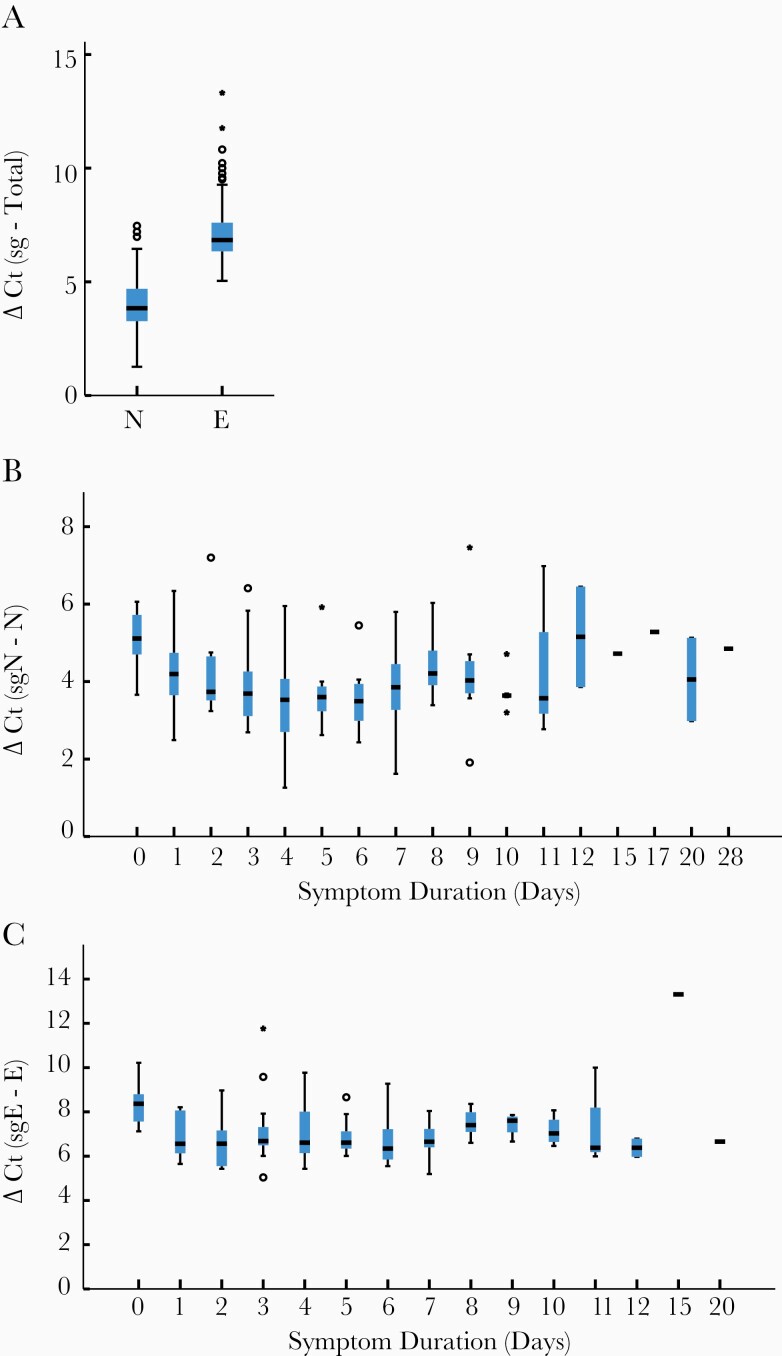
We found there was very little difference between expression levels of total N and E. The mean Ct value of E was 25.76 (SD, 5.69 cycles) and N was 25.6 (SD, 5.6 cycles). In clinical samples, we found that total RNA levels were higher than subgenomic RNA levels. Total N was expressed at a 16-fold (4.0 [SD, 1.1] cycles) higher level than sgN transcripts and E was expressed at a 137.2-fold (7.1 [SD, 1.3] cycles) higher level than sgE when all clinical samples were compared together (Figure 3). This relationship remained unchanged throughout the course of infection (Figure 3). Furthermore, the large fold difference in subgenomic compared to total transcripts indicates that subgenomic RNA contributes little to the overall signal of total transcripts.
Figure 3.
Box plots comparing difference in cycle threshold (Ct) values, delta Ct (subgenomic Ct – total Ct) for clinical samples. Total N was expressed at 16-fold (4.0 [standard deviation {SD}, 1.1] cycles) greater than subgenomic N (sgN) and total E was expressed at 137.2-fold (7.1 [SD, 1.3] cycles) greater than subgenomic E (sgE) expression (A). This relationship was found at all times after symptom onset for N (B) and E (C). Horizontal hash represents median value; if no interquartile range (IQR) is shown, then n = 1 for that sample. Moderate (1.5 × IQR) and extreme outliers (3 × IQR) are noted by circles and stars, respectively.
Subgenomic RNA in a Persistently Infected Patient
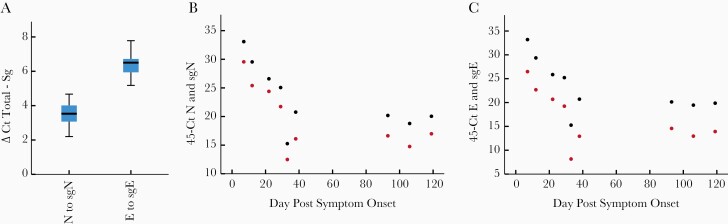
Emerging data indicate that some immunosuppressed patients can be persistently positive for SARS-CoV-2 by RT-qPCR and shed infectious virus for weeks. We evaluated total and subgenomic RNA from a patient with mantle cell lymphoma on B-cell–directed monoclonal antibodies who was persistently infected with SARS-CoV-2 for at least 119 days [9]. This patient showed continued expression of total and subgenomic transcripts throughout this time. The mean fold difference of total N to sgN was 11.3-fold (3.5 cycles) and the mean fold difference of total E to sgE was 84-fold (6.4 cycles) (Figure 4). This ratio was maintained over the 119-day course.
Figure 4.
Relationship between total and subgenomic RNA in a persistently infectious patient. Total N was expressed at a 11-fold (3.5 standard deviation {SD}, 0.75] cycles) higher level than subgenomic N (sgN), and total E was expressed at an 84-fold (6.4 [SD, 0.8] cycles) higher level than subgenomic E (sgE) when all time points were combined (A). Time course showing 45 minus cycle thresholds (Ct) for total (black) and subgenomic RNA (red) over time. This patient showed persistently positive total and subgenomic RNA for 119 days. The ratio of total N and sgN of 3 to 4 cycles was maintained over the duration of infection (B). This was also observed for E and sgE with the ratio of 6 to 7 cycles maintained thorough the course of infection (C).
Discussion
In this study, we found that subgenomic transcripts become undetectable at an earlier timepoint after infection than total transcripts. The median duration of symptoms until a negative RT-qPCR test was 14 days for sgE and 25 days for sgN. Total RNA was positive at all time points in our study up to 28 days. We found a strong correlation between total and subgenomic transcript levels. They decline at the same rate and have a fixed ratio at all time points after symptom onset. The predictability of subgenomic copy number from total copy number indicates that detection of subgenomic RNA does not add additional information about infectivity that cannot be gained from total copy number alone. Our findings support other studies that question the utility of subgenomic RNA in determining infectivity [12, 18].
A recent study in hospitalized patients showed that <5% of patients are infectious 15 days after the onset of symptoms [18]. We find that sgE becomes undetectable at a median of 14 days and only 12% (2/16) of patients were positive for sgE after this time point. Our results suggest that the disappearance of sgE RNA may correlate with a time when patients are no longer infectious.
Despite our finding that a negative sgE RT-qPCR result seems to correlate with a time when patients are not infectious, the supposition that any negative subgenomic RNA transcript can be used to predict infectivity is problematic. In our analysis, the median number of days from symptom onset to a negative sgN RT-qPCR was 25 days. Using sgN as a marker for infectivity would significantly prolong isolation and not predict infectivity. This difference is likely due to the higher levels of expression of sgN relative to sgE.
A negative subgenomic RNA test likely indicates that patients are not infectious simply because a negative subgenomic RNA test correlates with a low total RNA copy number. Previous studies have shown a copy number <6.5 log10 copies/mL is a threshold for culturable virus. In fact, we found that all negative subgenomic transcripts were in samples with total copy number below this threshold. However, we also find many samples that are below 6.5 log10 copies/mL but still express subgenomic E and N. This is in line with a recent study that showed subgenomic RNA could be detected from samples where infectivity in cell culture was negative [18]. It is likely that subgenomic RNA also has a copy number threshold below which infectious virus cannot be isolated. Because of the strong correlation in copy number of total and subgenomic transcripts throughout the course of infection and in a persistently infected patient, we do not believe that subgenomic RNA provides additional information not gained by total copy number.
Previous research suggested that subgenomic RNA is a suitable marker for active infection because it degrades more rapidly than total RNA. One study inoculated animals with infectious and inactivated SARS-CoV-2, which had high levels of total and subgenomic RNA. Total RNA copy number on day 1 postinfection from inactivated virus ranged from roughly 4 to 6 log10 copies/mL, but no sgE RNA was detected. This was interpreted to mean that subgenomic RNA rapidly degrades without replication but total RNA does not [13]. In our study, at a total copy number of 4 log10, no sgE could be detected, and at <6 log10 per mL of total RNA, 48% (57/119) of samples were negative for sgE. We suspect this finding is not due to differential degradation, but rather to differential detection of transcripts. Furthermore, if there was a difference in rates of degradation, we would expect to see the ratio of total RNA to subgenomic RNA increase over time, which we did not see.
Although there are emerging data supporting isolation guidelines in mild/moderate and severe COVID-19 infection, little is known about infectivity in immunosuppressed patients. In this study we showed persistent total and subgenomic RNA expression up to 119 days with a previous study demonstrating that infectious virus could be isolated through this course [9]. Because low levels of subgenomic RNA can be detected in the absence of infectiousness, we do not endorse using subgenomic RNA to determine infectivity in these patients. As with nonimmunosuppressed patients, a copy number threshold of total RNA would likely provide the same information that can be gained from copy number on subgenomic RNA.
Our study is limited by the fact that we did not attempt to isolate virus in culture from clinical samples and correlate to total or subgenomic copy number. We did, however, investigate correlation of transcripts to symptom duration, which has been used as a benchmark for infectivity. We are aware that recall bias may affect reported symptom duration, particularly in ill hospitalized patients. Recall bias may explain the large range in viral load at any given day postinfection, with some patients showing low viral load even when they report that they are early in the disease course. Alternatively, low viral load early in the reported course of infection may reflect RNA degradation in some samples. This is mitigated to some degree by our large sample size. Differences in copy number observed could be the result of differences in efficiency of RT-qPCR reactions between total and subgenomic transcripts. We confirmed differences in copy number using absolute copy number determined using standard curves generated with a known copy number using ddPCR for N, E, sgE, and sgN. Furthermore, our findings are in agreement with direct RNA sequencing studies showing the subgenomic N is expressed at higher levels than subgenomic E [15]. It is possible that our results might not be entirely generalizable if newer SARS-CoV-2 variants or strains express different levels of subgenomic messages.
In this study of inpatients infected with SARS-CoV-2, we find that sgE transcripts become undetectable before sgN, largely because they are transcribed at far lower levels. There is a fixed relationship between copy number of total and subgenomic transcripts over the course of infection. Because of this consistent relationship between total and subgenomic RNA, we do not believe subgenomic RNA detection adds additional information regarding infectivity that cannot be gained from copy number threshold validated to infectivity determined by cell culture.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the University of Michigan Clinical Microbiology Laboratory and the University of Michigan Central Biorepository for their assistance in providing samples.
Financial support. This work was supported by a University of Michigan COVID-19 Response Innovation Grant (to A. S. L.), the National Institutes of Health (grant number K01AI141579 to J. G. P.), and the Centers for Disease Control and Prevention (grant number U01 IP000974 to E. T. M.).
Potential conflicts of interest. A. S. L. reports receiving consulting fees from Sanofi on antiviral drugs and is a paid member of a steering committee for a clinical trial of baloxavir (Roche). All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Vogels CBF, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol 2020; 5:1299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. SoRelle JA, Frame I, Falcon A, et al. Clinical validation of a SARS-CoV-2 real-time reverse transcription PCR assay targeting the nucleocapsid gene. J Appl Lab Med 2020; 5:889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walsh KA, Jordan K, Clyne B, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect 2020; 81:357–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arons MM, Hatfield KM, Reddy SC, et al. ; Public Health–Seattle and King County and CDC COVID-19 Investigation Team . Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 2020; 382:2081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 7. Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 2020; 71:2663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Avanzato VA, Matson MJ, Seifert SN, et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–12.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baang JH, Smith C, Mirabelli C, et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021; 223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. Discontinuation of transmission-based precautions and disposition of patients with SARS-CoV-2 infection in healthcare settings. Atlanta, GA: CDC, 2021.
- 12. Alexandersen S, Chamings A, Bhatta TR. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat Commun 2020; 11:6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Speranza E, Williamson BN, Feldmann F, et al. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Transl Med 2021; 13:eabe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dagotto G, Mercado NB, Martinez DR, et al. Comparison of subgenomic and total RNA in SARS-CoV-2 challenged rhesus macaques [manuscript published online ahead of print 20 January 2021]. J Virol 2021. doi:10.1128/JVI.02370-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell 2020; 181:914–21.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perera RAPM, Tso E, Tsang OTY, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 2020; 26:2701–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Centers for Disease Control and Prevention. CDC 2019-novel coronavirus (2019-nCov) real-time RT-PCR diagnostic panel. Atlanta, GA: CDC, 2020.
- 18. van Kampen JJA, van de Vijver DAMC, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 2021; 12:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.