Abstract
Small molecule modulators of protein–protein interactions (PPIs) are being pursued as novel anticancer, antiviral and antimicrobial drug candidates. We have utilized a large data set of experimentally validated PPI modulators and developed machine learning classifiers for prediction of new small molecule modulators of PPI. Our analysis reveals that using random forest (RF) classifier, general PPI Modulators independent of PPI family can be predicted with ROC-AUC higher than 0.9, when training and test sets are generated by random split. The performance of the classifier on data sets very different from those used in training has also been estimated by using different state of the art protocols for removing various types of bias in division of data into training and test sets. The family-specific PPIM predictors developed in this work for 11 clinically important PPI families also have prediction accuracies of above 90% in majority of the cases. All these ML-based predictors have been implemented in a freely available software named SMMPPI for prediction of small molecule modulators for clinically relevant PPIs like RBD:hACE2, Bromodomain_Histone, BCL2-Like_BAX/BAK, LEDGF_IN, LFA_ICAM, MDM2-Like_P53, RAS_SOS1, XIAP_Smac, WDR5_MLL1, KEAP1_NRF2 and CD4_gp120. We have identified novel chemical scaffolds as inhibitors for RBD_hACE PPI involved in host cell entry of SARS-CoV-2. Docking studies for some of the compounds reveal that they can inhibit RBD_hACE2 interaction by high affinity binding to interaction hotspots on RBD. Some of these new scaffolds have also been found in SARS-CoV-2 viral growth inhibitors reported recently; however, it is not known if these molecules inhibit the entry phase.
Keywords: drug discovery, protein–protein interaction modulators (PPIMs), machine learning (ML), chemical fingerprints, SARS-CoV-2, docking
INTRODUCTION
Protein–protein interactions (PPI) govern large number of cellular processes such as growth, cell survival, cell adhesion, signal transduction, apoptosis, host–pathogen interactions and immune regulation [1, 2]. Deregulations of PPIs are known to be associated with a number of different physio-pathologies such as cancer development, infectious diseases, neurological disorders, inflammation and oxidative stress disorders. Therefore, modulation of PPIs by small molecule inhibitors is being increasingly recognized as a therapeutic intervention strategy in disease biology and drug discovery research. Choice of PPIs as targets for novel drugs have several additional advantages compared with targeting single proteins or enzymes in terms of better selectivity or lower off target specificity and lower chances of developing resistance [3, 4].
Despite being attractive drug targets, as per a study published in 2011, out of more than 645 000 disease associated PPIs in the human interactome, only about 2% have been targeted for drug development [5]. PPI interfaces are much less conserved compared with active sites of enzymes, lack well defined binding pockets, have larger typically flat interface area (1000–2000 Å2) and high hydrophobicities. Hence, PPI modulators (PPIMs) pose the challenge of low oral bioavailability and low cell permeability associated with large hydrophobic molecules [4]. However, during the last decade, several studies on binding energy landscape of PPIs and emergence of the concept of interaction hot spots (regions on PPI interface with dominant contribution to binding affinity in terms of residues whose mutation results in binding energy change by at least 2 kcal/mol) have opened up the possibilities of targeting PPI hotspots [6, 7] for modulating PPIs using low molecular weight compounds. Experimental studies have revealed that interaction hotspots of PPIs can be successfully targeted by certain class of drug such as low molecular weight chemical compounds [8] for inhibiting several PPI families, thus solving the oral bioavailability and cell permeability problems.
Since design of successful PPI modulators requires special considerations such as correct identification of interaction hotspots for targeting the small molecule, despite use of advanced techniques such as screening of fragment libraries, gene editing-based validation and proteolysis targeting chimera (PROTAC) approach, the success rate for the high throughput screening for PPIs modulators remains low as compared with drugs targeting enzymes or single protein receptors [4]. Another crucial difference between design strategy for PPI modulators and single protein targets is the absence of a native ligand in case of PPI interfaces, which becomes a bottleneck for in silico PPI inhibitor design efforts, as here no structural analogs of natural ligands (small molecules) can be used as starting points.
Even though structure-based methods, such as docking and MD simulations, have been extensively used to screen large libraries of compounds against the target enzyme or PPI partner protein [9], identification of completely novel scaffolds by docking remains a challenge, because of large flat interaction interfaces involving PPIs and poor correlation between docking scores and experimental binding affinities [10]. In recent years, machine learning (ML) is gaining popularity in drug discovery studies [11–14]. It has been used both in combination with docking where it is employed to replace the classical scoring functions for evaluation of docked poses [15, 16] and also as a self-sufficient technique for virtual screening, where ML models based on both protein and ligand features [17, 18] or only ligand features have been trained [19] using a known data set of experimentally identified compound libraries. Interestingly, recent study by Morrone et al. [16] have shown that RF classifiers trained using only ligand features perform as well as models trained on both protein and ligand features using deep learning (DL)-based methods. Sieg et al. [11] have also analyzed the data sets used for benchmarking of the ML methods for structure-based virtual screening and have observed that because of bias in data set construction, ligand features have much more dominant contribution toward the performance of those ML predictors compared with protein structure features. Based on these analysis, Sieg et al. [11] have proposed guidelines for avoiding bias while constructing training and test data sets for developing ML classifiers. All these studies highlight the importance of proper bias control measures for identifying features which govern the performance of ML classifiers. Therefore, ML approaches can be used to identify potential PPI modulators from large compound libraries using ligand features and in the next step to compute intensive docking, and simulation studies can be carried out for lead optimization [20] and subsequent experimental validation of PPI modulators.
However, in contrast to large number of ML-based studies for the identification of drugs targeting single protein enzymes or receptors, there are very few reports on the development of ML methods for prediction of PPI modulators. Hamon et al. [21] have analyzed the properties of 40 orthosteric inhibitors of PPIs and used the DRAGON descriptors with Support Vector Machine (SVM) approach to enrich the chemical libraries for PPIs. Jana et al. [22] have built ML models for classifying modulators for three major classes of PPIs, namely Mdm2/P53, Bcl2/Bak and c-Myc/Max. However, despite the availability of increasing amount of experimental data on PPI modulators in the intervening years, efforts toward the development of ML-based computational methods for identification and design of PPI modulators have not kept pace with increase in experimental data on new scaffolds targeting novel clinically relevant PPI families.
Therefore, in the present work, we have used a large data set of experimentally characterized small molecule PPI modulators to train hierarchical ML classifiers which can first identify potential PPI inhibitors from large compound libraries, and then in the next stage, class-specific predictors can predict PPI modulators for 11 different families of PPIs. The best performing ML classifiers developed in this study for prediction of Small Molecule Modulators of PPI have been implemented in a software package, SMMPPI. Finally SMMPPI software has been used to search large antiviral compound libraries for molecules which can inhibit the interactions between RBD of SARS-CoV-2 and hACE2, thus can be of therapeutic relevance for inhibiting cell entry of SARS-CoV-2.
MATERIAL AND METHODS
Data sets used for development of the ML predictor and its benchmarking
Data sets for building ML classifiers for binary prediction of PPI inhibitors
The data on experimentally characterized small molecule modulators of PPIs were retrieved from PPI modulator databases such as iPP-DB [23] and 2P2Idb v2 [24], Hanson et al. study [25]. This provided a total set of 2578 PPIMs targeting 27 different PPI families, from which redundancy was removed (for details see Supplementary Methods, see Supplementary Data available online at http://bib.oxfordjournals.org/) by clustering the chemical structures of the molecules using Butina clustering module of RDKIT based on chemical structure similarity as measured by Tanimoto score with RDK fingerprints and a clustering radius of 0.9 [26]. The resulting 1324 non-redundant molecules were divided into training (75%) and independent testset (25%). For the negative set, decoys were picked from ChemBL [27] compounds targeting Single proteins (for details see Supplementary Methods, see Supplementary Data available online at http://bib.oxfordjournals.org/).
Data sets for building ML classifiers for prediction of class-specific PPI inhibitors
To build PPI class-specific classifiers, the original data set of 2578 PPIMs was clustered with 0.8 as the Tanimato Score cutoff (for details, see Supplementary Methods, see Supplementary Data available online at http://bib.oxfordjournals.org/) and the non-redundant compounds of each class become the positive set for the respective PPI family. For a given family, all the PPIMs belonging to other remaining classes form the negative set pool from which an equal number of compounds are randomly picked as negative set.
Feature vector for encoding chemical structures of PPI modulators
The two types of feature vectors, namely physico-chemical property descriptors and features representing chemical structure fingerprints (ECFP4) of the small molecules, are calculated with Openbabel [28] and RDKIT [26] using SMILES as input. Supplementary Methods, see Supplementary Data available online at http://bib.oxfordjournals.org/, section provides additional details.
Development of ML models
All ML models development and benchmarking was done using Weka Toolbox [29]. The model’s performance parameters were calculated and plotted using ‘ROCR’ package [30] and the tSNE plot is prepared using ‘tsne’ package [31] in R.
RESULTS AND DISCUSSION
Figure 1 shows a schematic depiction of the workflow of the current study. As described in Materials and methods section, chemical structures for a non-redundant data set of 1324 PPI modulators were compiled from publicly available PPI modulator databases. These 1324 compounds have been shown to modulate PPIs for 27 different PPI families and the number of PPI modulators varied from as high as 361 for Bromodomain and Histone PPI family to as low as 1 for E2 and E1 PPI family (Figure 2). In order to develop a general purpose ML classifier which can distinguish chemical structures of the modulators of various types of PPIs from other types of molecules which target single proteins, all these 1324 known PPI modulators were selected as positive data and negative data sets of different sizes, which were obtained from ChEMBL. While 75% of the positive and negative data were used for training/validation of the ML classifiers, remaining 25% were kept aside for using as independent test set. Effect of different type of feature vectors used to encode chemical structures and types of ML algorithms on the performance of PPIM predictor was analyzed.
Figure 1.
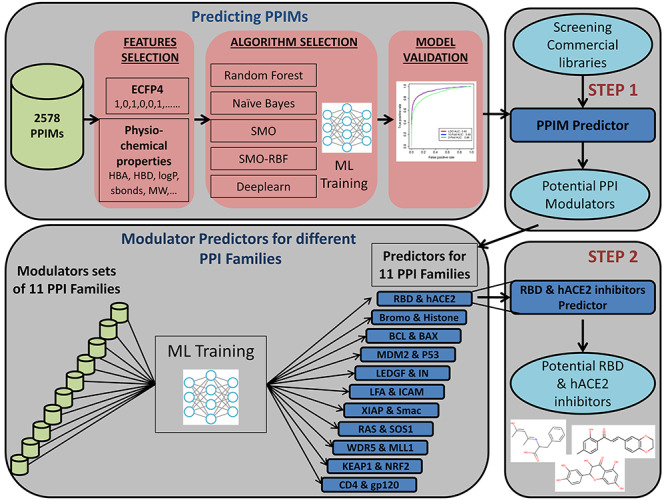
Flowchart depicting hierarchical design of generalized and family specific predictors for identification of modulators of PPI.
Figure 2.
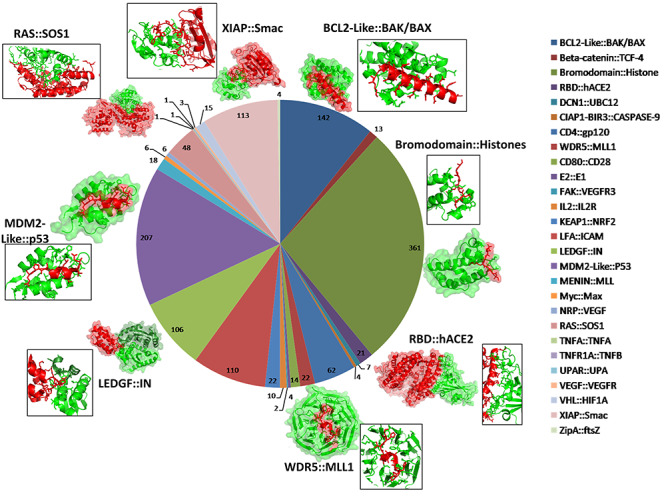
Family wise distribution of non-redundant PPI modulators in the compiled non-redundant data set. Representative three dimensional structures for eight important families with available 3D structures in PDB are shown with insets showing the interface region.
Effect of feature vectors on performance of ML classifiers
In order to identify the types of feature vectors which can best represent chemical structures for ML-based prediction of PPI modulators, RF classifiers were developed using physicochemical descriptor-based feature vectors as well as Morgan fingerprint-based feature vectors. RF classifier was chosen for this task as RF classifier has been known to have superior performance in several chemoinformatics applications. The non-redundant data set of 993 known PPI modulators was used as positive data, while the equal number of single protein targeting inhibitors obtained from ChEMBL database was used as negative data set to train and validate the RF classifier. Supplementary Figure S1, see Supplementary Data available online at http://bib.oxfordjournals.org/, shows Leave One-Out (LOO), 2-fold and 10-fold cross validation (CV) results for RF best prediction of PPI modulators using physicochemical property descriptors (Supplementary Table S1, see Supplementary Data available online at http://bib.oxfordjournals.org/) as feature vectors, while corresponding results using Morgan circular fingerprint feature vectors are shown in Figure 3A. Morgan Fingerprint with a radius size of 2 (radius size of 2 was observed to be performing best after trying other sizes) was used. As can be seen from Supplementary Figure S1, see Supplementary Data available online at http://bib.oxfordjournals.org/, and Figure 3A, the ROC-AUC values for physicochemical property descriptors as feature vectors vary from 0.77 for 2-fold CV to 0.81 for LOO, while ROC-AUC values significantly increases to the range of 0.87–0.93 when Morgan fingerprint feature vector is used. Supplementary Table S2, see Supplementary Data available online at http://bib.oxfordjournals.org/, lists PR-AUC and various other statistical parameters such as SN, SP, FPR, MCC and F1 score at optimum score cutoff for these two different types of fingerprints. As can be seen while Morgan fingerprint feature vector-based predictor shows a sensitivity of 0.77 at FPR of 0.18 for 2-fold CV, the classifier using descriptor-based feature vector shows a sensitivity and an FPR of 0.72 and 0.30, respectively, for 2-fold CV. Other parameters such as MCC, F1 score and PREC at optimum cutoff as well as PR-AUC values also show superior performance of Morgan fingerprint feature vector-based RF classifier. These results indicate that ECFP fingerprints can better capture the patterns in chemical structures, which can distinguish potential PPI modulators from other compounds targeting single proteins. Hence, Morgan circular fingerprints were used in all subsequent studies.
Figure 3.
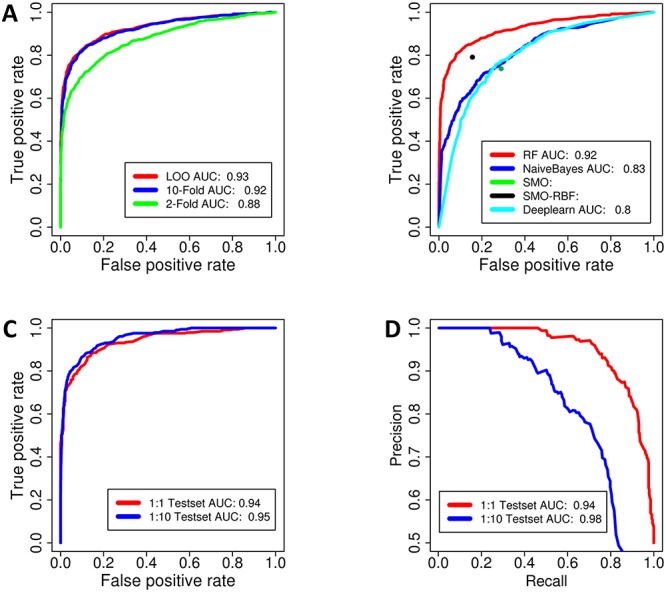
Training and validation of different ML classifiers developed using ECFP fingerprints as feature vectors (A) ROC Plot for n-fold CV of the RF Classifier built with Fingerprints (FP) of chemical structures as feature vectors. (B) ROC curves depicting comparison of the performance of different types of ML algorithms for prediction of PPI modulators-RF and Naïve Bayes, DL. For SMO and SMO-RBF modules of weka, the softwares gives the binary output for two classes, hence no ROC curve could be plotted due lack of continuous scores. Performance of RF Classifier built with Morgan Circular Fingerprints on external test data sets. (C) ROC curve (D) Precision-Recall (PR) curve.
Comparison of the performance of different ML classifiers
It is known that the performance of different ML algorithms often varies depending on the data set which has been used to train and validate the model. In order to identify the best performing ML algorithm on the PPI modulator data set, the performance of the RF classifier for the identification of PPI modulator was compared with three other widely used ML algorithms, namely NaiveBayes, Sequential Minimal Optimization (SMO) and SMO with Radial Basis Function Kernel (SMO-RBF). NaïveBayes is a relatively simplistic algorithm based on Bayes theorem. SMO is optimized training of SVM based on maximum distance calculation between input points. Radial Basis Kernel with SMO provides an additional strength to model by better accounting non-linear dependence of features [32]. All calculations for evaluation of performance of ML classifier were carried out based on training using equal positive and negative data set, and Morgan circular fingerprints were used. In addition to the above-mentioned ML classifiers, performance of a DL-based method for prediction of PPI modulator was also investigated, because in recent years, several studies have reported improved performance of DL-based methods for chemoinformatics problems dealing with large sized and high-dimensional data [33–35]. Figure 3B shows 10-fold CV ROC curve for RF and NaiveBayes predictors, while for SMO and SMO-RBF, only TPR and FPR values at optimum cutoff are shown. It also shows ROC curves for LOO and n-fold CV for the DL-based Dl4jMlp classifier of Weka toolbox with default settings. Table 1 shows ROC-AUC, PR-AUC, SN, SP, FPR, MCC and F1 score values for all five prediction methods. As can be seen, out of the 4 conventional ML algorithms, RF, which is a tree-based algorithm, is found to have best performance. In 10-fold CV, RF classifier has ROC-AUC of 0.93 corresponding to a TPR of 84% at FPR of 88%, while NaiveBayes classifier has ROC-AUC of 0.83 corresponding to a TPR of 70% at FPR of 19% (Figure 3B). The SMO-RBF classifier has a TPR of 77% at FPR of 16% in 10-fold CV test, while SMO has TPR of 76% at FPR of 28%. Thus, RF and SMO-RBF show significantly better performance compared with NaiveBayes and SMO classifiers in terms FPR at optimum cutoff. The same trend is reflected in other statistical measures. The ROC-AUC values for LOO, 2-fold and 10-fold CV for the Dl4jMlp classifier are in the range 0.78–0.80 and corresponding TPR and FPR at optimum cutoff are 77 and 28% respectively. Thus, for the PPI modulator data set, DL-based classifier shows lower prediction accuracy compared with RF and SMO-RBF methods. This counter intuitive observation may be because of the PPI modulator data set being comparatively smaller in size. It is also possible that the performance of DL classifier can be improved by varying the parameters such as the number of epochs, layers, iteration number, etc., which we have not investigated in the current study as default setting of Weka toolbox was used. In order to demonstrate that the observed differences in performances of ML classifiers are statistically significant, we have compared the ROC curves of the respective algorithms and computed the D-score for the difference in AUCs and associated P-value using Bootstrap test which is a standard statistical method to compare any two ROC curves. D-statistic and P-values for AUC differences between RF versus Naïve-Bayes and RF versus DL have been provided in Supplementary Table S3, see Supplementary Data available online at http://bib.oxfordjournals.org/. As can be seen, the P-values are of the order of 10−16 indicating statistically significant differences in AUCs. Thus, our analysis of the role of feature vectors and ML classifiers revealed that RF classifier with Morgan circular fingerprints as feature vectors shows the best performance for ML-based prediction of PPI modulators using chemical structure information alone.
Table 1.
Performance of different types of ML algorithms and DL for prediction of PPI modulators
| ML Algorithm | Positives | Negatives | CV | Sens | Spec | FPR | Prec | F1-Score | MCC | ROC AUC | PRC AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RF | 993 | 993 | 2-Fold | 0.77 | 0.82 | 0.18 | 0.81 | 0.79 | 0.60 | 0.87 | 0.87 |
| 993 | 993 | 10-Fold | 0.84 | 0.88 | 0.12 | 0.88 | 0.86 | 0.72 | 0.93 | 0.92 | |
| 993 | 993 | LOO | 0.84 | 0.88 | 0.12 | 0.88 | 0.86 | 0.72 | 0.93 | 0.92 | |
| NaiveBayes | 993 | 993 | 2-Fold | 0.69 | 0.77 | 0.23 | 0.73 | 0.73 | 0.46 | 0.81 | 0.80 |
| 993 | 993 | 10-Fold | 0.70 | 0.81 | 0.19 | 0.75 | 0.75 | 0.50 | 0.83 | 0.82 | |
| 993 | 993 | LOO | 0.69 | 0.80 | 0.20 | 0.75 | 0.75 | 0.50 | 0.83 | 0.82 | |
| SMO | 993 | 993 | 2-Fold | 0.73 | 0.68 | 0.32 | 0.70 | 0.71 | 0.42 | 0.71 | 0.65 |
| 993 | 993 | 10-Fold | 0.76 | 0.72 | 0.28 | 0.74 | 0.74 | 0.47 | 0.74 | 0.74 | |
| 993 | 993 | LOO | 0.76 | 0.72 | 0.28 | 0.74 | 0.74 | 0.49 | 0.74 | 0.68 | |
| SMO-RBF | 993 | 993 | 2-Fold | 0.71 | 0.83 | 0.17 | 0.77 | 0.77 | 0.54 | 0.77 | 0.71 |
| 993 | 993 | 10-Fold | 0.77 | 0.84 | 0.16 | 0.83 | 0.81 | 0.62 | 0.81 | 0.75 | |
| 993 | 993 | LOO | 0.78 | 0.85 | 0.15 | 0.81 | 0.81 | 0.62 | 0.81 | 0.75 | |
| Deeplearning | 993 | 993 | 2-Fold | 0.75 | 0.68 | 0.32 | 0.71 | 0.72 | 0.43 | 0.78 | 0.76 |
| 993 | 993 | 10-Fold | 0.77 | 0.72 | 0.28 | 0.74 | 0.74 | 0.48 | 0.81 | 0.79 | |
| 993 | 993 | LOO | 0.75 | 0.72 | 0.28 | 0.73 | 0.73 | 0.47 | 0.80 | 0.79 |
Benchmarking on external test set
For further benchmarking of our method, the best performing RF-based predictor of PPI modulators was tested on the external positive and negative data sets, which had not been used in the training of the RF classifier. Figure 3C and D shows ROC and Precision-Recall curves for RF-based prediction of PPI modulators on balanced as well as imbalanced data sets. The performance of the model on data sets having negative data 10 times the positive data was evaluated, because typical chemical libraries in which this classifier will be used to search for PPI modulators are expected to contain negative data several folds higher than the positive data. As can be seen from Figure 3C and D and Supplementary Table S4, see Supplementary Data available online at http://bib.oxfordjournals.org/, for the balanced data sets, the ROC-AUC and PR-AUC values are 0.94 and the TPR and FPR values at optimum cutoff are 84 and 12%, respectively, indicating high prediction accuracy on balanced data sets. On the imbalanced data set with 1:10 ratio of positive and negative data, the ROC-AUC remains almost the same (0.95), but the PR-AUC increases to 0.98 because of increase of precision at high recall values (Figure 3D). This is due to the decrease of FPR to 9% at the same TPR of 84% (Supplementary Table S4, see Supplementary Data available online at http://bib.oxfordjournals.org/). These results indicate that even on external test data sets, the RF classifier for prediction of PPI modulators shows high prediction accuracy both on balanced as well as imbalanced data sets. Comparison of the pair-wise distances between all the compounds in our training, validation and test data sets (Supplementary Figure S2, see Supplementary Data available online at http://bib.oxfordjournals.org/) indicate that vast majority of the pairs in our data set have Tanimoto score below 0.7, and for a given Tanimoto score, the number of positive–positive, negative–negative and positive–negative pairs are almost equal. Thus, distinction between positive and negative points in our data set is not a trivial task.
Assessing bias in model training/testing and evaluation of alternate splitting strategy
Several recent studies on evaluation of performances of chemical structure fingerprint-based ML classifiers have revealed that high prediction accuracy of such classifiers can potentially arise from over-fitting and memorization rather than true learning and ability for generalization across chemical space to deal with unseen data. As our RF classifier for prediction of PPIMs showed very good accuracy, we wanted to analyze the effect of bias in splitting of data into training and test sets and redundancy in training and test data sets arising from the presence of clusters of compounds with similar scaffolds, on ROC-AUC values. Wallach et al. [36] have recently analyzed all ML-based bioactivity predictions carried out using ECFP4 fingerprints and have defined asymmetric validation embedding (AVE) as a measure of bias and have also proposed a debiasing algorithm which can split the data into training and test sets to eliminate AVE bias. Martin et al. [37] have also proposed recently a ‘realistic split’ strategy based on ligand clustering where the training set is built with compounds starting from largest cluster and proceeding successively to smaller cluster until 75% of the compounds are included in training set. The remaining small clusters including singletons constitute the realistic held-out test set. Thus, held-out test sets were constructed using random split as before and debiasing approaches such as AVE split as well as realistic split. RF classifiers were trained and tested using these validation and test sets. ROC-AUC values for the on validation set as well as test sets with three different splitting strategies were compared (Table 2A and B). As can be seen, the RF classifier which has ROC-AUC values above 0.9 both on validation as well as test set has AVE bias value of 0.28, thus indicating the presence of bias in training and test set split. After debiasing using AVE split approach, while validation set ROC-AUC is 0.94, the ROC-AUC on held-out test set reduces to 0.83. Similarly for realistic split, debiasing validation and test set ROC-AUCs are 0.96 and 0.71, respectively. Even though the test set AUC values for random split are higher because of the presence of bias, our RF classifier on debiased test sets have AUCs above 0.7, thus indicating that our RF classifier can predict with reasonable accuracy even on difficult cases involving data sets very different from those used in training and validation. In order to analyze the effect of data redundancy on prediction results, different non-redundant data sets were generated by clustering the compounds (2578 positives and 2578 negatives) at Tanimoto score cutoffs ranging from 0.9 to 0.6, and for each data set, performance of RF classifiers were analyzed for three different splitting strategies and training/test set splits with 1:1 and 3:1 ratios. It is interesting to note that upon removal of redundancy by lowering the Tanimoto score cut off for clustering, the AVE bias with random split also reduces significantly and blind test ROC-AUC for random split also approach closer to AUC values obtained from AVE or realistic splitting approach (Table 2A and B). Supplementary Figure S3, see Supplementary Data available online at http://bib.oxfordjournals.org/, shows ROC curves for the RF classifiers for prediction of general PPIMs on non-redundant data set clustered with Tanimoto score of 0.7 and different types of splitting strategies to remove bias. These results suggest that the removal of redundancy could also be a simplistic debiasing strategy. Both AVE split and realistic split debiasing methods attempt to maximize the distance between training and test sets in chemical space. However, certain types of biological data such as general PPIMs may contain groups/scaffolds which bind and interact with a specific PPI family and positive data set in such cases might be clustered. Since our general PPIM data set contains a number of PPI families with certain families having fewer number of positive data, the entire chemical space representing some particular PPIM families might be completely excluded from the training set in AVE or realistic splitting strategy, which may result in skewed performance like the 3:1 realistic-splitting with clustering at 0.9 Tanimato score giving blind test AUC of 0.58, but it increases to 0.73 upon clustering at 0.7 Tanimoto cutoff (Table 2B). This could be observed more clearly when we repeated this exercise of bias removal for a particular family, namely Bromodomain_Histone PPI. Since distant clusters corresponding to multiple PPI targets will be absent in this case, realistic spitting with clustering at 0.9 Tanimoto score results in a test set AUC of 0.77 compared with 0.58 in case of general PPIM data set (Supplementary Table S5, see Supplementary Data available online at http://bib.oxfordjournals.org/). It may be noted that both AVE and random split give higher AUC values for Bromodomain_Histone PPI data set compared with general PPI. Thus, it is necessary to take the biological context of the problem into consideration while training and testing ML classifiers.
Table 2.
Dependence of ROC-AUC of RF classifier for general PPIM prediction on various types of unbiased splitting of the data into training and test set (A) Training:Test = 1:1 (B) Training:Test = 3:1
| A | |||||||
|---|---|---|---|---|---|---|---|
| Tanimoto cutoff for clustering | AUC-CV (random splitting) | AUC-blind testing (random split) | AVE bias (random split) | AUC-CV (AVE split) | AUC blind testing (AVE split) | AUC-CV realistic-split | AUC blind testing realistic-split |
| 0.90 | 0.91 | 0.90 | 0.28 | 0.94 | 0.83 | 0.96 | 0.71 |
| 0.80 | 0.85 | 0.86 | 0.19 | 0.91 | 0.71 | 0.91 | 0.78 |
| 0.70 | 0.78 | 0.80 | 0.12 | 0.88 | 0.66 | 0.82 | 0.77 |
| 0.60 | 0.74 | 0.76 | 0.07 | 0.81 | 0.68 | 0.78 | 0.75 |
| B | |||||||
| Tanimoto cutoff for clustering | AUC-CV (random splitting) | AUC-blind testing (random split) | AVE bias (random split) | AUC-CV (AVE split) | AUC blind testing (AVE split) | AUC-CV realistic-split | AUC blind testing realistic-split |
| 0.90 | 0.92 | 0.92 | 0.32 | 0.94 | 0.71 | 0.97 | 0.56 |
| 0.80 | 0.88 | 0.87 | 0.22 | 0.93 | 0.68 | 0.92 | 0.67 |
| 0.70 | 0.82 | 0.84 | 0.13 | 0.86 | 0.69 | 0.86 | 0.73 |
| 0.60 | 0.77 | 0.77 | 0.09 | 0.82 | 0.71 | 0.84 | 0.64 |
PPI family-specific classifiers
The generalized ML classifiers for identification of PPI modulators discussed above can identify potential PPI modulators from large libraries of compounds. However, for experimental validation of predictions from such generalized predictors, it will be necessary to predict which PPI complex the predicted modulator is likely to target. Therefore, it is necessary to develop PPI family-specific ML classifiers which can be used in a hierarchical way along with generalized predictor for PPI modulators discussed earlier. Even though information about the PPI family was available for all 1324 PPI modulators in our data set, the number of known modulators for many of the PPI families was not adequate for developing class-specific predictors of PPI modulators. PPI family-specific predictors were developed for 7 PPI families for which at least 40 compounds were available in our non-redundant data set of PPI modulators. In addition, classifiers were also developed for 4 other PPI families for which a number of compounds available were in the range of 18–26. These families are Bromodomain_Histone, BCL2-Like_BAX/BAK, LEDGF_IN, LFA_ICAM, MDM2-Like_P53, RAS_SOS1, XIAP_Smac, RBD_hACE2, WDR5_MLL1, KEAP1_NRF2 and CD4_gp120. As discussed in Materials and methods section, for PPI families with sufficient amount of data (more than 40 non-redundant compounds), total data set for each PPI family was divided into training/validation set and test set in 3:1 ratio. For any given PPI family, the negative data set was chosen from all other PPI families. The Morgan circular fingerprints were used as feature vectors and RF classifiers were developed by LOO and n-fold CV on the training and validation data sets for each of the 11 PPI families. Figure 4A and B and Supplementary Figure S4, see Supplementary Data available online at http://bib.oxfordjournals.org/, show the ROC curves for these 11 family-specific PPI predictors, while ROC-AUC, PR-AUC and all other statistical parameters are listed in Table 3. As can be seen, in CV as well as independent external test data sets, all the 11 classifiers have ROC-AUC and PR-AUC values above 0.90 and at optimum cutoff predictions have sensitivity above 85% with specificity values above 80% except for KEAP_NRF classifier where the optimum sensitivity is 78% and specificity is 89%. The lower performance for this class is primarily due to the very limited number of compounds available for this training this class. These results indicate that our RF classifiers for prediction of class-specific PPI modulators have very high prediction accuracy. In order to increase the chemical space of the negative set, RF classifiers were also trained with unequal sized positive and negative data sets. For each class, the negative data size was restricted to 5 times positive data because of the availability of limited amount of PPI modulator data for different families. It is interesting to note that, with training using unbalanced data sets also, all the classifiers have very good prediction accuracy (Supplementary Figures S5–S8, see Supplementary Data available online at http://bib.oxfordjournals.org/, and Supplementary Table S6). Since relatively less amount of experimental data was available for training of some of the family-specific classifiers, learning curves were analyzed for all the family-specific classifiers and also the general PPIM classifier to validate data set sizes (Supplementary Figure S9, see Supplementary Data available online at http://bib.oxfordjournals.org/). As can be seen, in case of General PPIM predictor as well as majority of the family-specific classifiers except RAS_SOS1, percentage of true positive predictions increases with increase of training data set only upto 40% of the training data, after that there is saturation indicating convergence of learning. However, in case of PPI classes for which less data were available for training, the learning curve has not converged and percentage of correct predictions increase from 60 to 80% as amount of training data increases from 40 to 100%. In case of CD4_gp120, learning curve has converged despite having less data for training. The RF-based ML classifiers developed in the current study can be used hierarchically to first identify potential PPI modulators and then use PPI family-specific classifiers to predict which specific PPI family a given compound is likely to modulate.
Figure 4.
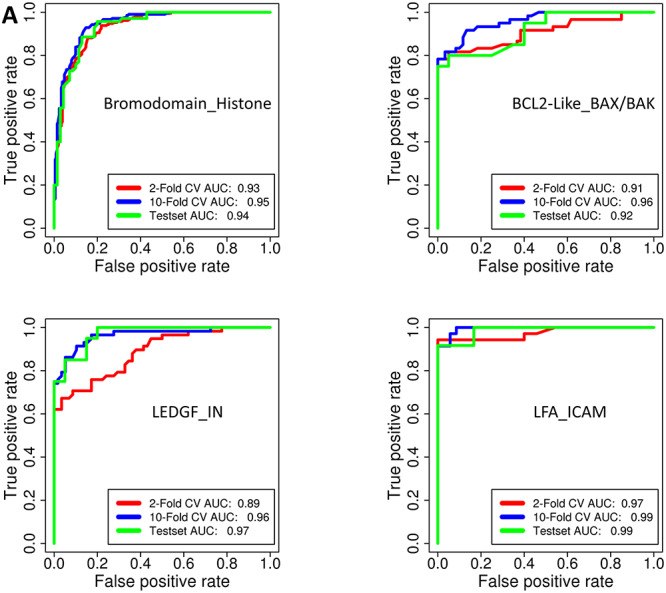
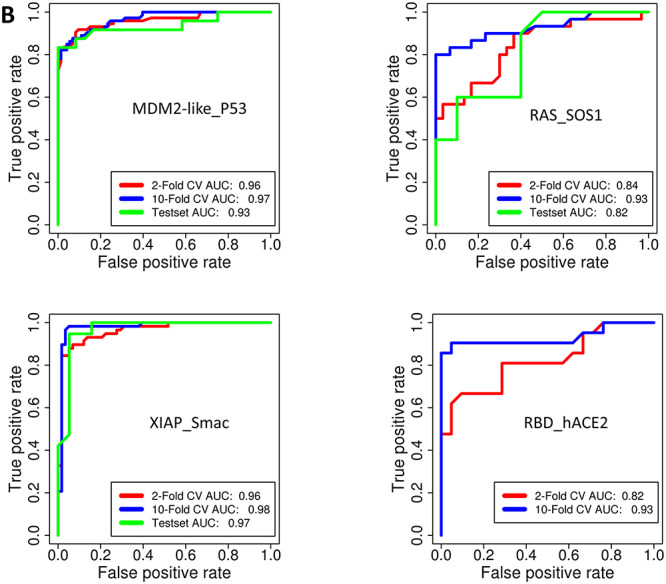
(A) ROC curves of RF classifiers for prediction of family specific PPI modulators for Bromodomain_Histone, BCL2-Like_BAX/BAK, LEDGF_IN and LFA_ICAM PPI families. (B) ROC curves of RF classifiers for prediction of family specific PPI modulators for MDM2-like_P53, RAS_SOS1, XIAP_Smac and RBD_hACE2 PPI families.
Table 3.
Performance of RF model for family specific prediction of PPI modulators
| PPI family model | Testing | Positives | Negatives | Sens | Spec | FPR | Prec | F1-Score | MCC | ROC AUC | PRC AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bromodomain_Histone | 10-Fold CV | 211 | 211 | 0.91 | 0.87 | 0.13 | 0.91 | 0.89 | 0.77 | 0.95 | 0.94 |
| Testset | 70 | 70 | 0.91 | 0.81 | 0.19 | 0.91 | 0.87 | 0.73 | 0.94 | 0.93 | |
| BCL2-Like_BAX/BAK | 10-Fold CV | 60 | 60 | 0.83 | 0.92 | 0.08 | 0.83 | 0.87 | 0.75 | 0.96 | 0.97 |
| Testset | 20 | 20 | 0.8 | 0.9 | 0.1 | 0.8 | 0.84 | 0.7 | 0.92 | 0.94 | |
| LEDGF_IN | 10-Fold CV | 58 | 58 | 0.93 | 0.84 | 0.16 | 0.93 | 0.89 | 0.78 | 0.96 | 0.97 |
| Testset | 20 | 20 | 0.85 | 0.85 | 0.15 | 0.85 | 0.85 | 0.7 | 0.97 | 0.97 | |
| LFA_ICAM | 10-Fold CV | 35 | 35 | 0.94 | 0.94 | 0.06 | 0.94 | 0.94 | 0.89 | 0.99 | 0.99 |
| Testset | 12 | 12 | 0.92 | 1 | 0 | 0.92 | 0.96 | 0.92 | 0.99 | 0.99 | |
| MDM2-Like_P53 | 10-Fold CV | 73 | 73 | 0.88 | 0.93 | 0.07 | 0.88 | 0.9 | 0.81 | 0.97 | 0.97 |
| Testset | 24 | 24 | 0.88 | 0.92 | 0.08 | 0.88 | 0.89 | 0.79 | 0.93 | 0.96 | |
| RAS_SOS1 | 10-Fold CV | 30 | 30 | 0.83 | 0.87 | 0.13 | 0.83 | 0.85 | 0.7 | 0.93 | 0.95 |
| Testset | 10 | 10 | 0.6 | 0.9 | 0.1 | 0.6 | 0.71 | 0.52 | 0.82 | 0.83 | |
| XIAP_Smac | 10-Fold CV | 58 | 58 | 0.98 | 0.95 | 0.05 | 0.98 | 0.97 | 0.93 | 0.98 | 0.97 |
| Testset | 19 | 19 | 0.95 | 0.95 | 0.05 | 0.95 | 0.95 | 0.89 | 0.97 | 0.96 | |
| RBD_hACE2 | 10-Fold CV | 21 | 21 | 0.86 | 0.95 | 0.05 | 0.86 | 0.9 | 0.81 | 0.93 | 0.96 |
| WDR5_MLL1 | 10-Fold CV | 18 | 18 | 0.83 | 0.89 | 0.11 | 0.83 | 0.86 | 0.72 | 0.94 | 0.95 |
| KEAP1_NRF2 | 10-Fold CV | 18 | 18 | 0.78 | 0.89 | 0.11 | 0.78 | 0.82 | 0.67 | 0.93 | 0.94 |
| CD4_gp120 | 10-Fold CV | 26 | 26 | 0.96 | 0.96 | 0.04 | 0.96 | 0.96 | 0.92 | 0.99 | 0.99 |
Even though for class-specific prediction of PPI modulators, the number of PPI families is limited, and most of the PPI families covered have clinical implications, while many are targets for designing cancer therapeutics. Bromodomain and Histone interactions involve binding of Bromo domains to acetylated histones, and thereby, bromodomains mediate the binding of several protein complexes such as histone acetytransferases, chromatin remodeling complexes, specific and general transcription factors, etc., to chromatin and have role in gene activation and regulation [38]. Because of their implications in cancer and inflammation, several groups have been working to design drugs targeting this PPI [39]. Likewise, P53 is a tumor suppressor gene which is inhibited after its binding to MDM2. Hence, inhibitors of p53 and MDM2 interaction help in restoring the p53 function. Some of the compounds such as CGM097 (Novartis) and MK-8242 (SCH 900242) (Merck) have also entered clinical trials [40]. Bcl and Bax interaction is another widely studied target for the development of cancer therapeutics. This interaction decides the fate of cell by regulating the signals for programmed cell death [41]. In addition to the PPIs controlling tumor suppressive genes/proteins, interaction between intracellular adhesion molecule-1 (ICAM-1) and leukocyte function-associated antigen-1 (LFA-1) controls autoimmune diseases because of its role in T-Cell activation and their migration to target tissues. In view of the potential implications in organ transplantation and other autoimmune diseases, some molecules targeting ICAM-1 and LFA-1 interaction are in clinical trials [42]. XIAP_Smac also has role in apoptosis [43], while LEDGF and IN interaction is the target for designing antivirals, specifically anti-HIV compounds as the interaction of human LEDGF with HIV Integrase has role in integration of the viral genome into the host chromatin. Molecules, such as raltegravir, inhibit LEDGF_IN PPI complex and thus help in blocking replication of the virus [44]. In view of the wide application of PPI inhibitors, the family-specific PPI modulator predictors developed in the current study can serve as a valuable resource for quickly screening large compound libraries against these PPI targets to find novel inhibitors in drug discovery research.
Comparison of performance with other similar resources
As mentioned in Introduction section, we developed SMMPPI because no other ML tool for prediction of PPI modulators has been developed incorporating recent experimental data. Since our model has been trained on much larger data set compared with older resources such as PPI-Hunter and PPIMPred, it will not be fair to use the same test data for benchmarking the performance of all three methods. Therefore, we have only compared performance measures reported by those tools to the corresponding measures for SMMPPI on latest test data set. PPI-Hunter, an SVM-based tool trained on a small data set of 40 non-redundant PPI modulators carries out a binary prediction on whether a compound is PPI modulator or not using a feature vector of 11 physiochemical properties [21]. It was reported that PPI-Hunter had a sensitivity of 63% and specificity of 100% in n-fold CV on the small training positive data set of only 40 PPIM compounds with a negative set of 1018 compounds from NCI-Diversity Set II. In contrast to that, because of training using a much larger data set and use of Morgan circular fingerprints as feature vectors, our general PPI modulator predictor shows a sensitivity and specificity of 84 and 91%, respectively, on large independent test data sets with as high as 3641 compounds (Figure 3C and Supplementary Table S4, see Supplementary Data available online at http://bib.oxfordjournals.org/). PPIMPred is the only other ML-based predictor for class-specific prediction of PPI modulators [22]. It can predict small molecule modulators for MDM2_p53, BCL2_Bak and c-Myc_Max using an SVM model and 10 physiochemical properties as feature vector. Jana et al. have reported that the MDM2_p53 predictor of PPIMPred, which was developed using a small data set of 40 non-redundant (clustered with Tanimoto score cutoff of 0.8, same as used in the current study) PPI modulators, had ROC-AUC of 0.62 corresponding to a sensitivity and specificity of 40 and 80%. As discussed earlier, our RF predictor for modulators for MDM2_p53 has ROC-AUC of 0.97 corresponding to a sensitivity and specificity of 88 and 93%, respectively. For Bcl2_Bak, PPIMPred classifier trained with 100 Bcl2_Bak modulators had an ROC-AUC of 0.80 with a sensitivity of less than 80% at specificity of 80%, while our Bcl2_Bak predictor had ROC-AUC of 0.96 corresponding to a sensitivity and specificity of 83 and 92%. Thus, for MDM2_p53 and Bcl2_Bak, the class-specific predictors developed in the current study have superior performance. We have not developed a class-specific predictor for c-Myc_Max because of paucity of data for this PPI family. The other major difference between our class-specific PPI modulator predictors and those from PPIMPred is the choice of negative data set, which covers PPI modulators from 26 PPI families unlike the negative set of PPIMPred which comprised of only three PPI families apart from inhibitors of single proteins from ChEMBL. Thus, because of training using latest large data sets of PPI modulators, the general and class-specific predictors developed in the current study outperform all other available tools for prediction of PPI modulators.
Search for PPI modulators in commercial compound libraries
One of the applications of the ML-based PPI modulator predictors developed in the current work would be to quickly screen large commercially available compound libraries like those from ChemDiv Inc., etc., to select smaller set targeting a specific PPI. The first library selected was the ChemDiv PPI CDI Library 2.0 [45] consisting of a total of 2 22 447 compounds. Search of this library using our general PPI modulator predictor revealed that 33 436 compounds belonging to diverse chemical scaffolds can be ranked as high scoring PPI modulators. Interestingly, despite discarding more than 80% of the compounds off the list, the compounds selected by our predictor retain most of chemical diversity represented in the original library. tSNE plot (Figure 5) depicting coverage of chemical space shows that the selected compounds (green) set covering the almost all the chemical space occupied by the original library with remaining compounds shown in pink color. Similarly, our general PPIM predictor could identify 15 743 compounds as potential antiviral compounds out of 87 043 compounds in Chemdiv Antiviral library.
Figure 5.
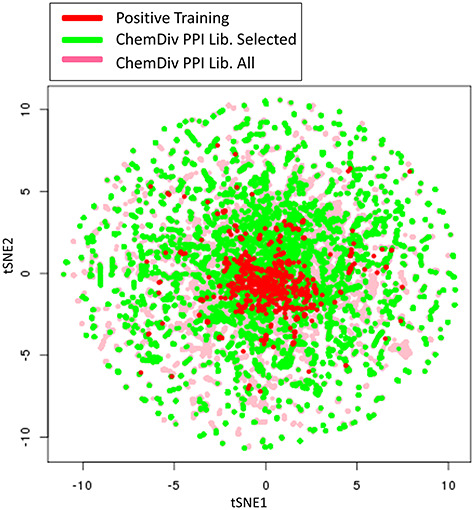
tSNE plot depicting structural diversity of PPI modulators predicted by RF classifier from ChemDiv PPI Library. Clustering has been carried out using Morgan Circular Fingerprints of the compounds. The positive training compounds are shown in green while all ChemDiv Library compounds are shown in pink with selected compounds by classifier shown in green.
We also filtered the specific PPI libraries from ChemDiv with our class-specific PPI predictors. Search in the ChemDiv MDM2-p53 interaction inhibitors library containing 21 903 compounds with our MDM2-p53 PPI predictor, a set of 4697 compounds (21%) were predicted as potential inhibitors of the MDM2_p53 interaction, while 17 206 compounds were rejected by the ML classifier. Similarly for ChemDiv library of small molecule modulators and inhibitors of Bromodomains, a set of 2062 compounds out of 5816 were selected by our ML-based approach for predicting modulators of Bromodomain_Histone PPI. In view of the various known clinical applications of these two PPIs inhibitors, the enriched sets identified by our ML methods can serve as very good starting points for experimental screening studies to identify drugs targeting these PPIs.
Identification of novel inhibitors for RBD:hACE2 interaction
While this work on the development of ML classifiers for prediction of PPI modulators was in progress, the emergence of COVID19 and elucidation of the structure of SARS-CoV-2 spike protein in complex with hACE2 revealed that RBD:hACE2 PPI could be an important target for the development of novel antiviral molecules to block cell entry of this virus. Even though a large number of in silico studies have been carried out using docking and atomistic simulations on RBD:hACE2 complex, we have attempted to develop a ligand-based PPI inhibitor predictor for this newly identified PPI family by training an ML classifier using the available experimental data on RBD:hACE2 small molecule inhibitors. It is encouraging to note that our RBD_ hACE2 classifier can predict the inhibitors of this interaction with a sensitivity of 86% and a specificity of 95% (Figure 4B). We also tested our RBD_hAC2 classifier on an independent data set of 91 inhibitors of SARS-CoV2 viral growth experimentally identified by Huang et al. [46]. Even though targets of these 91 inhibitors are unknown, some of these compounds are very likely to be inhibiting the virus growth by targeting the viral RBD and human ACE2 interaction. Interestingly, 20 out of these 91 compounds were predicted as RBD:hACE2 inhibitors by our RBD_hACE2 classifier. This provided further evidence on prediction accuracy of our ML classifier for an extremely important PPI target. As our ML-based approach is much faster and more easy to use as compared with docking, the classifier can serve as good filtering step to reduce the size of the compound library for or experimental validation or compute intensive structure-based docking and simulation studies as a second layer of in silico screening.
We have here applied our RBD_hACE2 classifier to identify novel inhibitors for this interaction. We screened the SARS-CoV2 ChemDiv compound Library consisting of 21 145 compounds with our general PPI classifier which eliminated large number of compounds from this set and selects 4033 compounds as enriched set for potential PPI modulators. Then, these 4033 compounds are screened with our RBD_hACE2 classifier to predict new scaffold which can inhibit this interaction. Supplementary Figure S10, see Supplementary Data available online at http://bib.oxfordjournals.org/, shows screenshots depicting the usage of SMMPPI for identification of novel inhibitors of RBD:hACE2 interactions. This screening resulted in a set of 674 compounds (SMMPPI_Suppl_Data Sheet) which are most likely to be targeting RBD:hACE2 complex. Our analysis revealed that these compounds belong to 319 structurally diverse clusters (with 0.6 as Tanimato similarity cutoff). Figure 6 shows representative compounds from some of the scaffolds/clusters. Out of these 319 clusters, 5 clusters contain compounds similar to already known inhibitors of RBD:hACE2 or SARS-CoV2 viral growth from the two published experimental studies [25, 46], while all the remaining 314 clusters represent the novel classes of compounds identified with potential to target RBD:hACE interaction. Cluster 120 consisting of 2 compounds shares similarity with Paredrine (4-(2-Aminopropyl)phenol) which has been used as an eye solution for dilation of pupils and controls the release of aqueous humor. Cluster 21 contains 6 compounds with piperidin and indole groups such as the thalidomide drug with immunomodulatory and antineoplastic properties, employed to treat myeloma. Cluster 98 contains 2 compounds sharing similarity with ORAP, an antipsychotics with ability to block dopaminergic recpetors on neurons. These results provide interesting clues on repurposing of known drugs for other indications to treat COVID 19. These sets of 674 compounds are interesting candidates for experimental studies.
Figure 6.
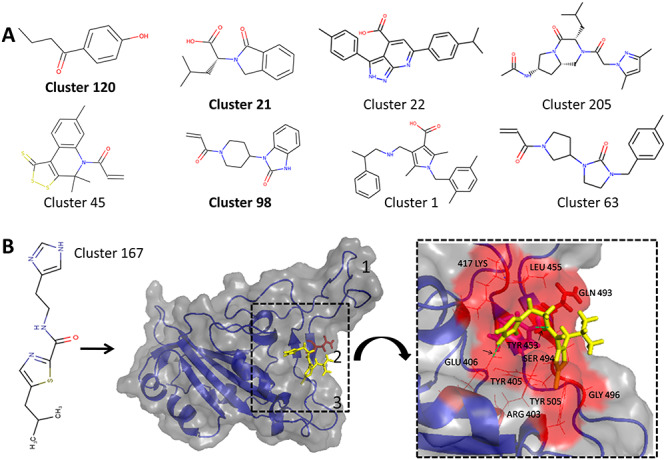
(A) Novel scaffolds which are predicted as inhibitors of RBD:hACE2 interaction by the ML approach. Representatives from some of clusters with similarity to known FDA Approved drugs are shown in bold. (B) Shows the cluster 167 representative compound (yellow sticks) docked onto RBD pocket 2 with residue Q493 shown in red sticks. The inset box shows the interacting residues of docked compound in red sticks with res Y453 shown in pink. The two hydrogen bonds formed are shown with green dashed lines indicated by arrows.
In order to decipher the mechanistic basis of inhibition/modulation of RBD:hACE2 interactions by these 674 molecules identified by SMMPPI, we carried out structural modeling for all these compounds in the binding interface of the crystal structure of RBD:hACE2 complex (PDB ID: 6M0J). It may be noted that structural modeling was carried out based on the assumption of orthosteric mode of action, but some of these compounds might be allosteric modulators as well. Similarly, the orthosteric modulator can bind to either RBD or to hACE2. However, analysis of binding interface of the complex has revealed that solvent exposed helical segments from hACE2 bind to a slightly concave surface of RBD with three sites which interact with residues of hACE2, thus can be interaction hotspots. Thus, it is likely that orthosteric inhibitors can bind to interaction hotspot 2 (Figure 6B) which consists of functionally important residue GLN 493 and TYR 453 involved in hydrogen bonding interactions between SARS-CoV-2 RBD and hACE2. It has been reported that residue GLN 493 makes significant contribution to binding affinity of RBD:hACE interaction and is a crucial determinant of higher binding affinity of RBD:hACE2 interaction in case of SARS-CoV-2 RBD compared with SARS-CoV [47]. Hence, all 674 molecules were docked onto site 2 on RBD using the OpenEye Docking Software [48] (Supplementary Methods, see Supplementary Data available online at http://bib.oxfordjournals.org/). The docked poses were analyzed to check for possible interactions between hotspot residue GLN 493 and the docked ligands. Out of the 314 novel scaffolds or clusters identified by SMMPPI, compounds belonging to 229 clusters had contacts (<3.5 Å distance between any two atoms) with GLN 493, and thus they can potentially block interaction of SARS-CoV-2 RBD with hACE2. Out of these 229 cluster representatives, 5 had OpenEye docking score lower than −6 kcal/mol, thus indicating that they could be high affinity binders to RBD (Supplementary Table S7, see Supplementary Data available online at http://bib.oxfordjournals.org/). The interactions of the top scoring compound (cluster 167) are shown in Figure 6B, which is docked into the RBD pocket 2 with a binding affinity score of −7.3 kcal/mol. Interestingly, the docked compound formed hydrogen bonds with TYR 453 and GLU 406. In addition, the docked compound had interactions with K417, L455, Y495, S494, R403, Q493, G496 and Y505 of which Y495, Q493 and G496 are known to significantly contribute to binding energy of RBD-hACE interaction [47]. The crystal structure of RBD:hACE2 reveals two crucial hydrogen bonds between RBD_GLN493:hACE2_LYS31 and RBD_TYR453:hACE2_HIS34 (Supplementary Figure S11, see Supplementary Data available online at http://bib.oxfordjournals.org/). The top scoring small molecule modulator predicted by SMMPPI and OpenEye docking binds to the same binding hotspot residues of RBD by making hydrogen bonds with TYR453 and GLU406. These results depict mechanistic basis of the inhibition of RBD:hACE2 interactions by small molecules. Similar analysis of other scaffolds with good docking score can identify novel inhibitors of SARS-CoV-2 cell entry, thus providing additional candidates for experimental validation. The current docking study shows how our hierarchical ML-based method SMMPPI can be combined with structure-based analysis to finally shortlist a set of 10–20 for further experimental validation.
Comparison of SMMPPI with docking
In structure-based drug discovery, protein-ligand docking has been the widely used strategy to screen large compound libraries against a target protein. The major advantage of docking is that it can be used based on first principle to search for new inhibitors and predict their binding sites, even in cases where no known inhibitors are there for the given target or binding site on the target is unknown. Even though docking is often used to filter large compound libraries for potential inhibitors, docking score often has poor correlation with experimental binding affinity values and successful prediction of inhibitors by completely automated docking is a difficult task. Therefore, during the last decade, ML-based scoring functions have been developed using known protein-ligand inhibitor data sets. Recent benchmarking studies by Morrone et al. [16] on DUD-E data set revealed that for binder versus non-binder prediction on single proteins/enzymes as targets, ML-based scoring functions (AUC 0.83) performed better compared with scoring functions used in docking (AUC: 0.70). However, as discussed earlier, the apparently superior performance of ML-based methods can often arise from bias in construction of training and test data sets, and such ML methods may not perform well on data sets which are very much different from those used in training. Therefore, comparison of the performance of docking approach with ML methods also requires construction of unbiased data sets [11]. Unlike single protein targets, additional complexity associated with flat interaction interfaces of PPIs [4] and lack of a reference small molecule substrate or native ligand to guide the search strategy, pose major challenges for prediction of modulators of PPIs by docking. Availability of experimentally validated data set of 25 inhibitors for RBD_hACE and a set of 250 compounds which do not inhibit RBD_hACE2 interactions [25] gave us the opportunity to evaluate the performance of a docking approach for this PPI. Docking of these compounds on to the interaction interface of RBD using OpenEye software revealed that out of the 25 inhibitors (binders to RBD) in positive data set, 16 compounds docked to the site with docking score in the range of −2 to −3 kcal/mol, while remaining nine compounds were rejected as potential binders. On the other hand, out of the 250 non-binders, only 84 were rejected by docking, and out of the remaining 166 known non-binders which docked to the RBD, ~82% showed binding score equal to or better than docking scores of the 16 known binders (Supplementary Figure S12B, see Supplementary Data available online at http://bib.oxfordjournals.org/). Supplementary Figure S12A, see Supplementary Data available online at http://bib.oxfordjournals.org/, shows ROC plot based on docking score for 182 compounds which could be docked. Thus, performance of the docking approach is only marginally better than random with ROC-AUC of 0.52. In contrast to docking, ML-based methods trained on known experimental data can identify PPIMs for a given PPI target using conserved patterns in known data. The RF classifier for the prediction of RBD_hACE2 inhibitors was developed using 25 known inhibitors as positive data for training. However, the 250 experimentally validated negative data set, which have not been used in training our ML model, can be used to compare the performance of RBD_hACE2 RF classifier and docking approach for their ability to reject inactive compounds. As can be seen from ROC curve for the RBD_hACE2 RF classifier on this data set, the performance of ML-based approach is superior to docking with AUC value of 0.93. Also, the ML model took just 3 s for screening these 250 molecules, while docking took 1 h 25 min on a desktop workstation. Hence, search of large compound libraries using ML is computationally inexpensive. Even though enrichment by ML over baseline docking is apparent, lack of additional active compounds beyond those used in training is a caveat to our claim. Also, as discussed in the earlier section on assessment bias in training/testing of ML models, superior performance of ML over docking could also arise from similarities between compounds present in the training and test data sets used for performance assessment of ML method. In practice, performance of SMMPPI may degrade when testing sets contain very different molecules from those used in training. In future, similar analysis using unbiased data sets from other PPI families with more numbers of positive and negative data can be carried out for a systematic comparison of docking and ML for prediction PPI modulators.
Even though the ability of docking methods to pick active compounds from large libraries is limited, docking studies have been successfully used in structure-based lead optimization where chemical structures in the neighborhood of a given scaffold are to be explored in the binding site of a target. Therefore, the method utilized in the current work for predicting novel inhibitors of RBD:hACE2, where docking is used in the second stage to filter compounds with favorable interactions in the binding site after first stage screening involving ML methods could be a powerful strategy for identification of small molecule modulators of PPIs.
CONCLUSION
PPIs are increasingly becoming focus for drug discovery due to their extensive involvement in controlling a plethora of disease associated pathways, higher target selectivity of PPI inhibitors and lower probability of drug resistance [5]. Development of newer technologies for discovery of PPI inhibitors have resulted in the experimental characterization of large number of small molecule modulators of various families of PPIs [49]. In the current study, we have utilized these large data sets of PPI modulators and used a data driven approach to develop ML models for in silico prediction of small molecule modulators of PPI. Detailed benchmarking using n-fold CV and completely independent testing on held out data sets have revealed that RF classifier with ECFP4 fingerprints outperforms other ML classifiers including DL-based methods. However, it is possible that high prediction accuracy of the RF classifier arises from possible bias in construction of training and test sets by random split and in case of unseen data performance may be lower. Additional benchmarking using a number of recently proposed methods for construction of unbiased training and test sets revealed that the ML classifier developed in the current study can perform with reasonable accuracy even on data sets very different from those used in training. The general and family-specific predictors of PPI modulators have been implemented in a hierarchical manner to discriminate between modulators of PPI and inhibitors of single protein targets in large compound libraries in the first stage, while PPI family-specific RF classifiers are used in the second stage to predict modulators for 11 different PPI classes with high accuracy. These 11 PPI families not only include several complexes associated with cancer but also RBD:hACE2 associated with cell entry of SARS-CoV-2. To the best of our knowledge, this is the first ML-based method to identify new antivirals for inhibiting cell entry of SARS-CoV-2. Successful identification of 20 out of 91 experimentally validated SARS-CoV-2 viral growth inhibitors from independent data is an encouraging result for the predictions by SMMPPI. However, in absence of any additional experimental data, it is unclear if these molecules would inhibit virus activity in entry phase. Using this RBD: hACE2 predictor, we have identified more than 300 novel small molecule scaffolds, some of which are known drugs, which can be repurposed for SARS-CoV-2. By combining structure-based docking method with results from ML-based screening, we have identified a small set of 5 compounds for experimental validation in collaborative studies. While a potential caveat of the current study is lack of experimental validation of predicted new molecules, the ML classifiers developed in the current study are most efficient tools for high throughput in silico screening of large compound libraries for prioritizing smaller compound lists for compute intensive docking and atomistic simulations to decipher mechanistic details of the modulation of PPIs and subsequent experimental validation. The prediction models developed in this study have been made freely available as downloadable script SMMPPI. Apart from its utility in discovery of new antivirals as demonstrated in this work, SMMPPI will be a valuable resource for ML-based discovery of anticancer molecules.
Availability of software
The SMMPPI program package with documentation and test input/output files is available for download from http://www.nii.ac.in/smmppi.html.
Data Availability
All datasets and scripts used in this study are available for download from http://www.nii.ac.in/smmppi.html.
Key Points
Currently available machine learning (ML)-based computational tools for prediction of PPI modulators cover only three PPI families and have not been updated to keep pace with the increase in the volume of experimental data.
SMMPPI has been developed for an ML-based prediction of small molecule modulators of PPI using a large data set of experimentally validated PPIs. Benchmarking of SMMPPI using different feature vector representation of chemical compounds and different ML algorithms including deep learning revealed that random forest classifier with ECFP4 fingerprints as feature vectors has highest prediction accuracy.
Family-specific PPIM predictors have also been implemented in SMMPPI for 11 clinically important PPI families covering anticancer and antiviral drug targets including SARS-CoV-2 RBD_hACE2 PPI. Currently, no other tool is available for prediction of PPIMs for 9 out of these 11 clinically important classes of PPIs.
Finally, as a test case using SMMPPI, we have identified novel chemical scaffolds as inhibitors for RBD_hACE2 PPI and some of these new scaffolds are in agreement with chemical scaffolds of SARS-CoV-2 viral growth inhibitors reported in independent experimental studies outside our training data.
Supplementary Material
Acknowledgements
We acknowledge the cooperation of Dr. Phillip Roche in providing the updated and curated data sets of PPI modulators.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Priya Gupta is a PhD scholar at NII, New Delhi. Her research work focuses on machine learning-based methods for drug discovery and structural modeling of phosphorylation and protein–protein interaction networks.
Debasisa Mohanty heads the Bioinformatics & Computational Biology research group at NII, New Delhi. His research work focuses on multi-scale modeling of biomolecular systems and applications of machine learning in drug discovery and in silico genome mining.
Contributor Information
Priya Gupta, NII, New Delhi 110067, India.
Debasisa Mohanty, Bioinformatics & Computational Biology research group at NII, New Delhi 110067, India.
Funding
Department of Biotechnology, Government of India grant to National Institute of Immunology, New Delhi; Department of Biotechnology, India under BTIS (BT/BI/03/009/2002 to D.M.) and COE (BT/COE/34/SP15138/2015 to D.M.) projects.
References
- 1. Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 2007;450:1001–9. [DOI] [PubMed] [Google Scholar]
- 2. Fry DC. Small-molecule inhibitors of protein-protein interactions: how to mimic a protein partner. Curr Pharm Des 2012;18(30):4679–84. [DOI] [PubMed] [Google Scholar]
- 3. Voter AF, Keck JL. Development of protein-protein interaction inhibitors for the treatment of infectious diseases. Adv Protein Chem Struct Biol 2018;111:197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ran X, Gestwicki JE. Inhibitors of protein-protein interactions (PPIs): an analysis of scaffold choices and buried surface area. Curr Opin Chem Biol 2018;44:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mabonga L, Kappo AP. Protein-protein interaction modulators: advances, successes and remaining challenges. Biophys Rev 2019;11:559–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol 1998;280:1–9. [DOI] [PubMed] [Google Scholar]
- 7. Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science 1995;267:383–6. [DOI] [PubMed] [Google Scholar]
- 8. Villoutreix BO, Labbe CM, Lagorce D, et al. A leap into the chemical space of protein-protein interaction inhibitors. Curr Pharm Des 2012;18:4648–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sable R, Jois S. Surfing the protein-protein interaction surface using docking methods: application to the design of PPI inhibitors. Molecules 2015;20:11569–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pantsar T, Poso A. Binding affinity via docking: fact and fiction. Molecules 2018;23: Article 1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sieg J, Flachsenberg F, Rarey M. In need of bias control: evaluating chemical data for machine learning in structure-based virtual screening. J Chem Inf Model 2019;59:947–61. [DOI] [PubMed] [Google Scholar]
- 12. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Agrawal P, Bhalla S, Chaudhary K, et al. In silico approach for prediction of antifungal peptides. Front Microbiol 2018;9:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lo YC, Rensi SE, Torng W, et al. Machine learning in chemoinformatics and drug discovery. Drug Discov Today 2018;23:1538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wojcikowski M, Ballester PJ, Siedlecki P. Performance of machine-learning scoring functions in structure-based virtual screening. Sci Rep 2017;7:46710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morrone JA, Weber JK, Huynh T, et al. Combining docking pose rank and structure with deep learning improves protein-ligand binding mode prediction over a baseline docking approach. J Chem Inf Model 2020;60:4170–9. [DOI] [PubMed] [Google Scholar]
- 17. Wen M, Zhang Z, Niu S, et al. Deep-learning-based drug-target interaction prediction. J Proteome Res 2017;16:1401–9. [DOI] [PubMed] [Google Scholar]
- 18. Stepniewska-Dziubinska MM, Zielenkiewicz P, Siedlecki P. Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics 2018;34:3666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu HX, Zhang RS, Yao XJ, et al. QSAR and classification models of a novel series of COX-2 selective inhibitors: 1,5-diarylimidazoles based on support vector machines. J Comput Aided Mol Des 2004;18:389–99. [DOI] [PubMed] [Google Scholar]
- 20. Hoffer L, Muller C, Roche P, et al. Chemistry-driven hit-to-lead optimization guided by structure-based approaches. Mol Inform 2018;37:e1800059. [DOI] [PubMed] [Google Scholar]
- 21. Hamon V, Bourgeas R, Ducrot P, et al. 2P2I HUNTER: a tool for filtering orthosteric protein-protein interaction modulators via a dedicated support vector machine. J R Soc Interface 2014;11:20130860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jana T, Ghosh A, Das Mandal S, et al. PPIMpred: a web server for high-throughput screening of small molecules targeting protein-protein interaction. R Soc Open Sci 2017;4:160501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Labbe CM, Kuenemann MA, Zarzycka B, et al. iPPI-DB: an online database of modulators of protein-protein interactions. Nucleic Acids Res 2016;44:D542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Basse MJ, Betzi S, Morelli X, et al. 2P2Idb v2: update of a structural database dedicated to orthosteric modulation of protein-protein interactions. Database (Oxford) 2016;2016:baw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanson QM, Wilson KM, Shen M, et al. Targeting ACE2-RBD interaction as a platform for COVID19 therapeutics: development and drug repurposing screen of an AlphaLISA proximity assay. ACS Pharmacol Transl Sci 2020;3:1352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. RDKit, Open-Source Cheminformatics .
- 27. Davies M, Nowotka M, Papadatos G, et al. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res 2015;43:W612–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Boyle NM, Banck M, James CA, et al. Open babel: an open chemical toolbox. J Chem 2011;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaufmann M, Frank E, Hall MA, et al. The WEKA Workbench. In: Online Appendix for "Data Mining: Practical Machine Learning Tools and Techniques", 2016.
- 30. Sing T, Sander O, Beerenwinkel N, et al. ROCR: visualizing classifier performance in R. Bioinformatics 2005;21:3940–1. [DOI] [PubMed] [Google Scholar]
- 31. Justin Donaldson JD. Package "tsne", CRAN Repository, 2010.
- 32. Song S, Zhan Z, Long Z, et al. Comparative study of SVM methods combined with voxel selection for object category classification on fMRI data. PLoS One 2011;6:e17191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Korotcov A, Tkachenko V, Russo DP, et al. Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol Pharm 2017;14:4462–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mater AC, Coote ML. Deep learning in chemistry. J Chem Inf Model 2019;59:2545–59. [DOI] [PubMed] [Google Scholar]
- 35. Zhang H, Liao L, Saravanan KM, et al. DeepBindRG: a deep learning based method for estimating effective protein-ligand affinity. PeerJ 2019;7:e7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wallach I, Heifets A. Most ligand-based classification benchmarks reward memorization rather than generalization. J Chem Inf Model 2018;58:916–32. [DOI] [PubMed] [Google Scholar]
- 37. Martin EJ, Polyakov VR, Zhu XW, et al. All-assay-Max2 pQSAR: activity predictions as accurate as four-concentration IC50s for 8558 Novartis assays. J Chem Inf Model 2019;59:4450–9. [DOI] [PubMed] [Google Scholar]
- 38. Josling GA, Selvarajah SA, Petter M, et al. The role of bromodomain proteins in regulating gene expression. Genes (Basel) 2012;3:320–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meslamani J, Smith SG, Sanchez R, et al. Structural features and inhibitors of bromodomains. Drug Discov Today Technol 2016;19:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao Y, Aguilar A, Bernard D, et al. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 inhibitors) in clinical trials for cancer treatment. J Med Chem 2015;58:1038–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ 2018;25:65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anderson ME, Siahaan TJ. Targeting ICAM-1/LFA-1 interaction for controlling autoimmune diseases: designing peptide and small molecule inhibitors. Peptides 2003;24:487–501. [DOI] [PubMed] [Google Scholar]
- 43. Sun H, Stuckey JA, Nikolovska-Coleska Z, et al. Structure-based design, synthesis, evaluation, and crystallographic studies of conformationally constrained Smac mimetics as inhibitors of the X-linked inhibitor of apoptosis protein (XIAP). J Med Chem 2008;51:7169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Christ F, Shaw S, Demeulemeester J, et al. Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob Agents Chemother 2012;56:4365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. https://www.chemdiv.com/complete-list-of-compounds-libraries/. https://www.chemdiv.com/complete-list-of-compounds-libraries/
- 46. Huang R, Xu M, Zhu H, et al. Massive-scale biological activity-based modeling identifies novel antiviral leads against SARS-CoV-2, bioRxiv 2020. doi: 10.1101/2020.07.27.223578, Arxiv biorxiv;2020.07.27.223578v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Othman H, Bouslama Z, Brandenburg JT, et al. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem Biophys Res Commun 2020;527:702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. 'OEDOCKING', OpenEye Scientific Software, Santa Fe, NM .
- 49. Churcher I. Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J Med Chem 2018;61:444–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets and scripts used in this study are available for download from http://www.nii.ac.in/smmppi.html.
Key Points
Currently available machine learning (ML)-based computational tools for prediction of PPI modulators cover only three PPI families and have not been updated to keep pace with the increase in the volume of experimental data.
SMMPPI has been developed for an ML-based prediction of small molecule modulators of PPI using a large data set of experimentally validated PPIs. Benchmarking of SMMPPI using different feature vector representation of chemical compounds and different ML algorithms including deep learning revealed that random forest classifier with ECFP4 fingerprints as feature vectors has highest prediction accuracy.
Family-specific PPIM predictors have also been implemented in SMMPPI for 11 clinically important PPI families covering anticancer and antiviral drug targets including SARS-CoV-2 RBD_hACE2 PPI. Currently, no other tool is available for prediction of PPIMs for 9 out of these 11 clinically important classes of PPIs.
Finally, as a test case using SMMPPI, we have identified novel chemical scaffolds as inhibitors for RBD_hACE2 PPI and some of these new scaffolds are in agreement with chemical scaffolds of SARS-CoV-2 viral growth inhibitors reported in independent experimental studies outside our training data.


