Abstract
The Coronavirus Disease 2019 (COVID-19) pandemic has fueled unprecedented development of animal models to understand disease pathogenesis, test therapeutics, and support vaccine development. Models previously developed to study severe acute respiratory syndrome coronavirus (SARS-CoV) have been rapidly deployed to study SARS-CoV-2. However, it has become clear that despite the common use of ACE2 as a receptor for both viruses, the host range of the 2 viruses does not entirely overlap. Distinct ACE2-interacting residues within the receptor binding domain of SARS-CoV and SARS-CoV-2, as well as species differences in additional proteases needed for activation and internalization of the virus, are likely sources of host differences between the 2 viruses. Spontaneous models include rhesus and cynomolgus macaques, African Green monkeys, hamsters, and ferrets. Viral shedding and transmission studies are more frequently reported in spontaneous models. Mice can be infected with SARS-CoV; however, mouse and rat ACE2 does not support SARS-CoV-2 infection. Murine models for COVID-19 are induced through genetic adaptation of SARS-CoV-2, creation of chimeric SARS-CoV and SARS-CoV-2 viruses, use of human ACE2 knock-in and transgenic mice, and viral transfection of wild-type mice with human ACE2. Core aspects of COVID-19 are faithfully reproduced across species and model. These include the acute nature and predominantly respiratory source of viral shedding, acute transient and nonfatal disease with a largely pulmonary phenotype, similar short-term immune responses, and age-enhanced disease. Severity of disease and tissue involvement (particularly brain) in transgenic mice varies by promoter. To date, these models have provided a remarkably consistent template on which to test therapeutics, understand immune responses, and test vaccine approaches. The role of comorbidity in disease severity and the range of severe organ-specific pathology in humans remains to be accurately modeled.
INTRODUCTION
The majority of human emerging infectious diseases originate in animals, particularly wild populations. Cross-species transmission of viruses with pandemic potential is likely to parallel increasing animal-human ecological overlap. The last 2 decades have witnessed the emergence of 3 coronaviral diseases with severe regional to global impact on human populations, suggesting that these viruses may be on an accelerating trajectory to cause future novel disease outbreaks in humans. Severe acute respiratory syndrome coronavirus (SARS-Co-V) emerged in 2002 in the Guangdong province of China,1 followed by the appearance of SARS-CoV-2 in China’s Hubei province in late 2019.2 Since then, SARS-CoV-2 has rapidly superseded SARS-CoV in dissemination and mortality to become the worst pandemic in living memory. Animal models of COVID-19 are essential to understand its pathogenesis and support the urgent need to develop vaccines. This review presents a comprehensive overview of comparative aspects of viral biology and species susceptibility to SARS-CoV-2 and its predecessor, SARS-CoV. Key clinical, radiologic, and pathologic features of human COVID-19 are summarized, followed by a comparison with spontaneous and genetically altered animal models of SARS-CoV-2 infection.
BIOLOGY OF CORONAVIRUSES
Coronaviruses infect a rich diversity of mammalian and avian hosts. This characteristic, together with a large genome size and relatively high rate of mutation, particularly in the spike (S) protein region responsible for viral-receptor binding, provide an ongoing source of potential emerging zoonotic disease.
Taxonomy and Host Range
The 4 genera of Coronaviridae are Alphacoronavus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus (Table 1). Human coronaviruses (HCoV) belong to the genera Alphacoronavirus and Betacoronavirus, along with other viruses that cause disease in a wide range of mammalian species, such as alpaca, bats, camels, cats, cows, dogs, ferrets, mice, pigs, and rats. Avian species are the main hosts for Deltacoronaviruses and Gammacoronaviruses.3,4 SARS-CoV-2 emerged in China in 20192 and causes a respiratory disease with systemic manifestations named Coronavirus Disease 2019 (COVID-19). SARS-CoV-2 is a Betacoronavirus and is closely related to 2 other highly pathogenic respiratory human betacoronaviruses that have emerged in the last 20 years: SARS-CoV-1 in 20021 and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012.5 SARS-CoV-1 and MERS-CoV caused 778 and 858 deaths, respectively,6 whereas SARS-CoV-2 has caused more than 2.7 million deaths and over 100 million cases worldwide at the time this review was written (https://coronavirus.jhu.edu/).
Table 1.
Coronaviruses, Their Natural Hosts, and Diseasesa
| Virus Name | Genus | Host | Disease |
|---|---|---|---|
| Alpaca coronavirus | Betacoronavirus | Alpaca | Gastroenteritis |
| Alpaca respiratory coronavirus | Alphacoronavirus | Alpaca | Mild respiratory |
| Bat coronaviruses (many) | Alphacoronavirus | Bat | Subclinical |
| Bat coronaviruses (many) | Betacoronavirus | Bat | Subclinical |
| Dromedary camel alphacoronavirus | Alphacoronavirus | Camel | Mild respiratory |
| Dromedary camel coronavirus HKU23 | Betacoronavirus | Camel | Gastroenteritis |
| Feline coronavirus | Alphacoronavirus | Cat | Enteritis |
| Feline infectious peritonitis virus | Alphacoronavirus | Cat | Peritonitis |
| Bovine coronavirus | Betacoronavirus | Cow | Severe enteritis, respiratory (shipping fever) |
| Infectious bronchitis virus | Gammacoronavirus | Chicken | Bronchitis, nephritis, reproductive failure |
| Canine coronavirus | Alphacoronavirus | Dog | Enteritis |
| Canine respiratory coronavirus | Betacoronavirus | Dog | Mild respiratory |
| Ferret coronavirus | Alphacoronavirus | Ferret | Enteritis |
| Ferret systemic coronavirus | Alphacoronavirus | Ferret | Peritonitis, perivasculitis |
| Equine coronavirus | Betacoronavirus | Horse | Gastroenteritis |
| Human coronaviruses 229E and NL63 | Alphacoronavirus | Human | Mild respiratory |
| Human coronaviruses OC43 and HKU1 | Betacoronavirus | Human | Mild respiratory |
| Middle East respiratory syndrome-related coronavirus | Betacoronavirus | Human | Mild to severe pneumonia |
| Severe acute respiratory syndrome-related coronavirus | Betacoronavirus | Human | Severe pneumonia |
| Severe acute respiratory syndrome coronavirus 2 | Betacoronavirus | Human | Severe pneumonia with systemic complications |
| Mink coronavirus | Alphacoronavirus | Mink | Enteritis |
| Mouse hepatitis virus | Betacoronavirus | Mouse | Enteritis, hepatitis, encephalitis |
| Porcine deltacoronavirus | Deltacoronavirus | Pig | Acute gastroenteritis |
| Porcine epidemic diarrhea virus | Alphacoronavirus | Pig | Acute gastroenteritis |
| Porcine hemagglutinating encephalomyelitis virus | Betacoronavirus | Pig | Vomiting, wasting, encephalomyelitis |
| Porcine respiratory virus | Alphacoronavirus | Pig | Mild respiratory |
| Swine acute diarrhea syndrome virus | Alphacoronavirus | Pig | Acute gastroenteritis |
| Transmissible gastroenteritis virus | Alphacoronavirus | Pig | Acute gastroenteritis |
| Rat coronavirus | Betacoronavirus | Rat | Sialodacryoadenitis, mild respiratory |
| Turkey coronavirus | Gammacoronavirus | Turkey | Enteritis |
aAdapted from Decaro and Lorusso.3
The emergence of SARS-CoV-1, MERS-CoV, and SARS-CoV-2 is believed to occur when a betacoronavirus jumps from an animal host into humans.3,4 Animals, such as bats, can serve as reservoirs for coronaviruses because they are persistently infected without any signs of illness. In these reservoirs, populations of variant viruses are constantly evolving. As illustrated with SARS-CoV-2, new variants that successfully infect humans may generate novel HCoV with pandemic potential. Of the 20 species of alphacoronaviruses and betacoronaviruses recognized by the International Committee of Viral Taxonomy, 11 are found only in bats, and these bat coronaviruses are believed to be the source of many pathogenic coronaviruses.7 SARS-CoV-1 is believed to have jumped from Himalayan palm civets, or raccoon dogs, to humans. However, there is no evidence that civets in the wild had SAR-CoV-1 infection and bat coronaviruses with high sequence homology to SARS-CoV-1 have been found. This makes it likely that the origin of SARS-CoV-1 was in bats and civets served as an intermediate host.8 MERS-CoV is transmitted from dromedary camels, because they are a known reservoir for the virus.9 Bat coronaviruses with high sequence homology to MERS-CoV have also been found, suggesting that bats may also be a reservoir for MERS-CoV.10 Based on sequence homology, it is believed that SARS-CoV-2 is a bat coronavirus that may use Malayan pangolins as an intermediate host prior to entry into humans. Human alphacoronaviruses, HCoV-229E and HCoV-NL63, are also believed to have jumped from bats and become endemic in the human population.11,12 In contrast, endemic HCoV-OC43, which like bovine coronavirus, equine coronavirus, porcine hemagglutinating encephalomyelitis virus, dromedary camel coronavirus HKU23, and HCoV-HKU, is a Betacoronavirus 1 believed to have evolved from a rodent coronavirus.12 Bats are also believed to be the source of swine acute diarrhea syndrome virus and porcine epidemic diarrhea virus.13,14 As human populations more frequently come in contact with bats or other wild animals that are harboring coronaviruses due to loss of natural habitats and urbanization, other coronaviral pandemics may occur.3,4,14 Clinically relevant coronaviruses are given in Table 1.
Genomic Structure and Replication
Coronaviruses have the largest nonsegmented, positive-sense, single-stranded RNA genome (27–32 000 nucleotides).6,15 The genome contains a 5′ cap structure and a 3′ poly A tail. Coronavirus genomes are arranged similarly with the nonstructural replicase gene encoded in the 5′ two-thirds of the genome and the genes for the structural proteins and several small accessory proteins encoded in the 3′ third of the genome. On entry and uncoating of the RNA genome in the cytoplasm, the 2 large open reading frames (ORF1a and ORF1b) of the replicase gene are translated into 2 large polyproteins via a translational frameshift. These polyproteins are co-translationally cleaved to produce proteins with RNA-dependent RNA polymerase, proteinase, single-stranded RNA binding, exonuclease, endoRNase, and ribose methyltransferase activities. These enzymes initiate the transcription of negative-strand copies of the genome. From these new negative-strand copies, new full-length genomes and a nested set of 3′ coterminal mRNAs are transcribed. In general, only the 5′ end of each mRNA in the nested set is translated. The large genome size as well as an RNA-dependent polymerase, which does not have a proofreading function, means that during coronavirus infection a population of mutant viruses is produced each with 1 or more changes to the genome. Additionally, coronaviruses undergo both homologous and nonhomologous recombination, which is linked to the strand-switching ability of the polymerase. Polymerase errors and recombination play a role in the evolution of coronaviruses.
All coronaviruses encode the following 4 structural proteins: spike/surface (S), envelope/small membrane (E), membrane (M), and nucleocapsid (N) proteins.6,15 Some betacoronaviruses also encode a hemagglutinin-esterase (HE) protein. The S glycoprotein forms the characteristic spikes protruding from the envelope of the virus. It binds to cell surface receptors to mediate viral entry into the cell. The specificity of the receptor-binding domain of the S protein is the primary determinant of tissue tropism and host range. S protein–induced fusion of the viral membrane with the cell membrane releases the viral genome into the cytoplasm of the cell. S protein that is not assembled into virions can be transferred to cellular plasma membranes, where it mediates cell–cell fusion leading to the production of giant multinucleated syncytia. The S protein induces neutralizing and cell-mediated immunity. Although the E protein is a minor component of the virion, it plays an important role in the assembly of the virion. The M protein is an integral membrane glycoprotein and directs the protein–protein interactions required for assembly of coronavirus particles. The M protein also has a role in the induction of the interferon response to coronavirus infection. The N phosphoprotein is a basic RNA-binding protein that coats the RNA genome to form the helical capsid. The N protein interacts with the M protein during assembly of virions. The N protein as well as several small accessory proteins have been reported to be type I interferon agonists. The HE protein forms a shorter spike on the envelope of the virus and has sialic acid binding and acetyl esterase activities, which may enhance S protein–mediated cell entry.
SARS-CoV-2 Receptor Tropism Across Species
Use of the ACE2 receptor by SARS-CoV-2 was suspected on the basis of sequence similarity of its spike receptor binding domain (RBD) to that of SARS-CoV.16,17 Phylogenetic analysis of ACE2 residues interacting with SARS-CoV-2 RBD18 and functional in vitro assays19 confirm that ACE2 from multiple species can support viral entry in vitro, however with vastly different efficiency. Identical sequence similarity between human, ape, and Old World monkey ACE220 is consistent with susceptibility of humans and macaques to SARS-CoV-2 infection and predicts high risk of disease in threatened great apes. Species susceptibility appears to be determined by sequence similarity of particular viral binding residues in ACE2 rather than the total number of similar residues. For example, the ACE2 viral binding domain of the common marmoset differs from its human counterpart by only 3 residues, of which 2 (Y41H, Q42E) are predicted to be critical for viral binding.18,20 The marmoset exhibits limited shedding and no clinical disease or pathology after SARS-CoV-2 infection.21 In contrast, the viral binding region of ACE2 in the highly susceptible Syrian hamster differs by 4 residues; however, the critical residues for entry are similar to human ACE2 sequence.18 The pig, differing across only 7/29 residues, is not susceptible to SARS-CoV-2 infection22 despite replication of SARS-CoV-2 in porcine cells in vitro.19 However, the ferret, which differs from human ACE2 in 12/29 residues within the virus-binding region, is susceptible.22 Mice and rats have relatively low similarity to human ACE2 in this region and are also resistant to significant SARS-CoV-2 infection. These functional differences are proposed to reside in a small number of key residues important for ACE2-SARS-CoV-2 RBD binding, particularly K353H in rodents.19
SARS-CoV-2 Exhibits a Narrower Host Range Than SARS-CoV
Animals susceptible to SARS-CoV infection are not necessarily susceptible to infection by SARS-CoV-2 despite both viruses binding to the same host receptor, ACE-2. Again, differences in critical ACE2 binding domains of the viral S protein are implicated.23 SARS-CoV–infected marmosets develop interstitial pneumonia but may also develop hepatic, gastrointestinal, and renal pathology.24 Wild-type mice are susceptible to SARS-CoV infection and develop interstitial pneumonia that can be ameliorated by murine Ace2 ablation.25 Pigs infected with SARS-CoV will shed virus and develop antibodies but not develop clinical disease.26 Apart from differences between SARS-CoV-2 and SARS-CoV RBD binding to ACE2 across species, species differences exist in tissue distribution of additional cell entry factors, such as TMPRSS2 and furin. Both SARS-CoV-2 and SARS-CoV employ the cellular serine protease TMPRSS227,28 for S protein priming to gain cell entry. Unlike SARS-CoV, SARS-CoV-2 also utilizes proprotein convertase furin29 for spike preactivation. These cofactors present addition potential drug targets30 and may contribute to species differences in SARS-CoV-2 susceptibility. Whereas ACE2 is broadly expressed, co-expression of ACE2 and TMRPSS2 is more restricted and highest in ciliated and club cells of the respiratory tract, nasal goblet cells, type II pneumocytes, pulmonary macrophages, ileal enterocytes, and the proximal tubule cells of the kidney.31 This distribution is consistent with respiratory, enteric,32 and renal33 manifestations of COVID-19. Lastly, the crystal structure of SARS-CoV-2 and SARS-CoV RBD in complex with ACE229 reveals differences that allow relatively greater ACE2-binding affinity of SARS-CoV-2 RBD; however, its RBD is structurally hidden compared with SARS-CoV.34 This combination is thought to support its relatively greater efficiency for cell entry and its potential to elude immune surveillance.29
COVID-19 IN HUMANS
Viral Shedding
Viral shedding of SARS-CoV-2 begins 2–3 days before symptoms appear and declines by 7 days post symptom onset. This pattern is unlike that of SARS-CoV,35 in which peak infectiousness occurs after symptom onset. Asymptomatic carriers can potentially shed virus for approximately 3 weeks after exposure.36 In COVID-19 patients, virus is detected most consistently in bronchoalveolar lavage samples, followed by nasopharyngeal swabs, saliva, rectal swabs, blood, and less commonly in urine.37 The highest viral loads are detected at the time of symptom onset and decline gradually over the next 3 weeks, coincident with appearance of antibodies in most individuals by 14 days after onset.38 Viral shedding can persist for several days longer in stool than in respiratory samples39 and allows accurate community tracking of test results and hospitalization using wastewater.40 In the majority of reports, only viral RNA is detected; however, there are rare reports of extraction of live virus from patient stool.41 This raises the possibility of fecal-oral transmission, of particular concern in nursing home settings.
Spectrum of Clinical Presentation
The majority of people infected with SARS-CoV-2 experience asymptomatic or mild disease, characterized by fever, cough and chest tightness, myalgia, or gastrointestinal signs. Hospitalized patients are typically diagnosed with pneumonia and are discharged after approximately 2 weeks.42 Common clinicopathologic characteristics on admission include lymphopenia and, less commonly, thrombocytopenia and leukopenia, elevation of hepatic enzymes, elevated inflammatory markers (C-reactive protein, IL-6, procalcitonin) and elevated D-dimer. Severe clinical COVID-19 most commonly manifests as acute respiratory distress syndrome driven by a positive feedback cytokine loop43 that may precipitate multi-organ failure.44 Additional variants include coagulopathy45 and attendant thromboses.46 Cardiovascular, enteric,32,47 renal,48 and neurologic49 complications are reported in humans. Histologic evidence of SARS-CoV-2 infection in olfactory neurons,50,51 brain,51–54 and gastrointestinal tract18 has been noted in animal models; however, the associated clinical disease spectrum noted in humans is not accurately modeled. Similarly, no animal model for multisystem inflammatory disease in children55 exists.
Pulmonary Imaging and Pathology
Computed tomography of COVID -19 patients56 identified well-demarcated ground glass, reticulated, or solid opacities affecting central or peripheral aspects of lower lung regions. Severity of CT abnormalities correlates with mortality,56 which is high (ranging from 60–88%) for COVID-19 patients requiring mechanical ventilation.57 Diffuse alveolar damage is the most common histologic pattern of acute respiratory distress syndrome58 and manifests in 3 stages: acute (typified by hyaline membrane formation), proliferative, and fibrotic. In SARS-CoV-2 infection, diffuse alveolar damage corresponding to the phase of disease prevails.59 Histologic variations include lymphocytic viral pneumonia in patients who die early (within a week), or acute fibrinous and organizing pneumonia, characterized by lack of hyaline membranes and extensive intra-alveolar “fibrin balls.”60
Factors Altering Disease Severity
Hospitalization rates are highest among older adults and men. Duration of viral shedding is greater with severe disease, patients older than 60 years, and male patients.39 High viral loads may be correlated with increasing age or severity of dyspnea but are not influenced by co-morbidities.38 Of those patients requiring hospitalization, the majority have underlying conditions, including hypertension, obesity, chronic lung disease, diabetes mellitus, and cardiovascular disease.42 The mechanisms underlying the capacity for these comorbidities to influence SARS-CoV-2 pathogenesis are not clear but may center on the role of ACE2. In animals, older age is associated with more severe disease;23,61–64 however, the effect of comorbidities has not yet been described.
Strains of SARS-CoV-2
The mutation rate of SARS-CoV-2 is relatively low (approximately one-half of that for influenza viruses65). To date, several strains have evolved, of which the D614G variant has garnered the most attention through its reported potential for greater infectivity.66 However, the impact of various mutations on transmission, disease phenotype, and vaccine and therapeutic development are as yet unclear.65
MODELING COVID-19 IN SPONTANEOUS ANIMAL MODELS
The core aspects of SARS-CoV-2 infection in humans are modeled faithfully in spontaneous animal models (Syrian hamster, ferret, and macaque, and, to a lesser degree, cat). These include the acute nature and predominantly respiratory source of viral shedding, acute transient and nonfatal disease with a predominantly pulmonary phenotype, and similar short-term immune responses. Systemic inflammation and hematologic abnormalities characteristic of severe COVID-19 in humans are less readily demonstrated in animals. Viral shedding and transmission studies are more frequently reported in spontaneous models than in mice. Spontaneous animal models for COVID-19 are summarized in Table 2. It should be noted that studies in animal models have been performed using differently sourced animals (eg, Indian and Chinese origin macaques); different strains of virus; different infectious doses; and variable combinations of intratracheal, intranasal, and conjunctival infection. Nevertheless, a relatively consistent disease phenotype is evident across studies.
Table 2.
Clinical Signs, Pulmonary and Extrapulmonary Pathology, and Viral Distribution in Spontaneous Animal Models of COVID-19
| Viral Replication and Tissue Tropism | Clinical Signs | Pulmonary Pathology | Extrapulmonary Pathology | Immunohistochemical Localization of SARS-CoV-2 |
|---|---|---|---|---|
| Rhesus macaque21,54,60,61 | ||||
| Viral RNA: Nasal wash, throat swabs, bronchoalveolar lavage, respiratory tissues, lymphoid tissue, and intestine Infectious virus: Bronchoalveolar lavage, shedding peaks in first few days, declines thereafter | Asymptomatic or fever, weight loss, occasional cough, reduced activity and appetite; peak within first week, resolution at 9-14 dpi; age enhanced disease | Gross pulmonary hemorrhage, radiographically evident interstitial pneumonia with diffuse alveolar damage | In some animals, lymphoid hyperplasia, pericardial effusion | Respiratory: Type I and II pneumocytes, alveolar macrophages; nasal and airway epithelium Intestine: lymphocytes and macrophages in lamina propria |
| Cynomologus macaque21,55,62 | ||||
| Viral RNA: Abundant nasal shedding by 2 dpi (and up to 21 dpi), intermittent throat shedding, rare fecal shedding; no virus detectable in blood | Asymptomatic or fever, weight loss in first week dpi; no age enhanced disease | Acute or proliferative advanced diffuse alveolar damage with epithelial syncitia; serial CT reveals ground-glass opacities, reticulation, paving, or alveolar consolidation | No | Respiratory: Type I and II pneumocytes, ciliated epithelial cells of airways and nasal mucosa |
| African Green monkeys66 | ||||
| Viral RNA: Nasal, bronchoalveolar lavage, oral and rectal swabs from 2–3 dpi, absent from blood; shedding persisted for 15 (nasal) to 21 (rectal) d | No overt clinical signs, reduced appetite | Diffuse alveolar damage and interstitial pneumonia with hyaline membrane formation, type II pneumocyte hyperplasia, pulmonary edema, and hemorrhage | No | Respiratory: Type I and II pneumocytes, bronchial epithelial cells |
| Common marmoset21 | ||||
| Viral RNA: Nasal, low levels, 2 wk dpi | None | Very mild interstitial mononuclear infiltration | No | Not done |
| Ferret74,75 | ||||
| Viral RNA: Nasal, low copy numbers in rectal swabs; nasal turbinate, soft palate, tonsils; absent in lung Infectious virus: Nasal washes only | Rare clinical signs of fever, weight loss, and inappetance 10 and 12 dpi | Interstitial pneumonia with hyaline membrane formation, type II pneumocyte hyperplasia, lymphoplasmacytic perivasculitis and vasculitis | No | Respiratory: epithelial cells nasal turbinate, trachea, pulmonary interstitum Intestinal: intestinal lamina propria |
| Mink63,76,77 | ||||
| Viral RNA: Conchae, lung, throat, and rectal swabs, rare liver and intestine | Respiratory signs, anorexia, death | Diffuse alveolar damage and interstitial pneumonia with hyaline membrane formation | No | Respiratory: epithelial cells nasal turbinate, trachea, pulmonary epithelium, macrophages |
| Syrian hamster18,67–69,71 | ||||
| Viral RNA: Nasal turbinates, trachea, lungs (high), intestine (mid), salivary glands, heart, liver, spleen, lymph nodes, kidney, brain, blood (low) up to 7 dpi Infectious virus: Lung up to 4 dpi | Hunched, lethargic, rapid breathing, piloerection, weight loss in first week; age enhanced disease | Rhinitis, tracheitis, diffuse alveolar destruction, hyaline membrane formation, marked mononuclear cell infiltration; consolidation and hemorrhage, syncitia, peak at 4 dpi | Intestinal: necrosis, damaged and lamina propria mononuclear cell infiltration at 4 dpi Spleen: white pulp atrophy and apoptosis (2–4 dpi) followed by lymphoid hyperplasia (7 dpi) Heart: myofiber degeneration and interstitial edema | Respiratory: bronchiolar epithelial cells, alveolar macrophages, and type I and II pneumocyte, Intestinal: enterocytes |
| Deer mouse73 | ||||
| Viral RNA: Lungs, up to 14 dpi Infectious virus: Lungs, 3 and 6 dpi |
No clinical signs | 3 dpi: broncho-interstitial pneumonia and leucocytoclastic vasculitis, multinucleate epithelial cells, fibrinopurulent rhinitis and sinusitis | CNS: neutrophilic infiltration of afferent ethmoidal, olfactory, maxillary, and other sensory nerves; frontal cortical encephalitis, gliosis, and immunoreactivity for SARS-CoV-2 in brainstem nuclei is evident | Nasal: sustentacular, olfactory, basal cells Tongue: mucosal epithelium CNS: sensory ganglia, olfactory bulb, brain, microglia Eye: retinal ganglion cells, inner nuclear layer Intestinal: crypt epithelium, lamina propria, macrophages, ileum and duodenum |
| Cat22 | ||||
| Viral RNA: nasal turbinate, soft palate, tonsil, trachea, lungs, small intestine, rarely rectal, up to 12 dpi Infectious virus: nasal turbinates, soft palates, tonsils, tracheas, lungs up to 6 dpi |
No clinical signs | 3 dpi: massive lesions in nasal and tracheal mucosa, and lungs | No | Not done |
| Dog22 | ||||
| Viral RNA: intermittent rare rectal, up to 6 dpi Infectious virus: none |
No studies to date | No studies to date | No studies to date | No studies to date |
Abbreviations, CT: Computed tomography, Dpi: days post-infection, RNA: ribonucleic acid
Macaques
In rhesus macaques,21,67,68 viral shedding peaks within the first 3–5 days post infection (dpi) and declines by 10–14 dpi, coincident with the appearance of neutralizing antibodies. Similar to humans, viral load is highest in nasal and bronchoalveolar lavage samples, followed by throat and fecal/rectal samples. Shedding, although quite rare, can be prolonged in fecal samples.68 Infectious virus is recovered at a much lower rate than detection of viral RNA. Viral RNA is inconsistently detected in blood, urine, and urogenital swabs.21,68 Similar dynamics are evident in cynomolgus macaques.21,62
Rhesus macaques exhibit clinical signs of fever, weight loss, irregular breathing patterns, occasional cough, and reduced activity and appetite.21,68 These appear as soon as 1 dpi and peak within 1 week, with resolution 2 to 3 weeks after infection. Clinical signs are similar but less severe21 or absent62,69 in cynomolgus macaques. One study in rhesus macaques68 reported a transient stress leukogram by 1 dpi, transient neutropenia in some animals, and early-onset (1 dpi) repression of red cell indices that did not return to original baseline by 21 dpi. On CT, the ground glass, reticulated, or solid opacities affecting central or peripheral lung regions in COVID-19 patients are very similar in cynomolgus macaques69 starting 2 dpi and resolving by 6–12 days dpi. Radiographs of rhesus macaques61 reveal comparably mild foci of interstitial pneumonia.
Histologic evidence of mild to moderate interstitial pneumonia centered on bronchioles is evident in rhesus and cynomolgus macaques.62,68 Lesions are characterized by interstitial and alveolar edema, hyaline membranes, fibrin, macrophages and neutrophils, and minimal type II pneumocyte hyperplasia. Multinucleate syncitial cells of epithelial origin are evident in airways and parenchyma. Multifocal lymphocytic perivascular cuffs are present.
Although no gender bias in SARS-CoV-2 susceptibility is noted in spontaneous animal models, age susceptibility is evident in nonhuman primates and hamsters.70 In middle-aged (15 years) rhesus macaques,61 SARS-CoV-2 causes more severe interstitial pneumonia, more prolonged lymphopenia, and more prolonged (by 14 days) nasopharyngeal, anal, and pulmonary viral replication. In similarly middle-aged cynomolgus macaques,62 increased age does not affect disease outcome; however, there was prolonged shedding of greater amounts of virus in the upper respiratory tract of aged animals. This apparent correlation of viral load, duration of viral shedding, and disease severity and/or age is evident in human patients.
Rhesus macaques treated with remdesivir remain asymptomatic and have reduced pulmonary infiltrates on radiographs. Virus shedding from the upper respiratory tract is not reduced despite reduced virus titers in bronchoalveolar lavages.71 Hydroxychloroquine fails to protect SARS-CoV-2–infected cynomolgus and rhesus macaques.72,73
African Green Monkeys
SARS-CoV-2–infected animals74 do not develop overt clinical signs apart from reduced appetite. Transient lymphopenia and thrombocytopenia are evident in some animals. Viral RNA and infectious virus are present in nasal, bronchoalveolar lavage (BAL), oral, and rectal swabs from 2–3 dpi; however, virus is absent from blood. Shedding persisted for 15 (nasal) to 21 (rectal) days. Multifocal interstitial pneumonia with diffuse alveolar damage, hyaline membranes, and multinucleate epithelial cells is evident by 5 dpi.
Hamsters
Syrian hamster ACE2 receptor binds to the SARS-CoV-2 spike protein, rendering this species highly susceptible to infection with doses as low as 5 TCID50.75 Hamsters experience moderate but recoverable respiratory disease following intranasal infection with SARS-CoV-2 and have become a popular and well-characterized spontaneous model.18,63,75–77 Following infection, animals experience a 2-week disease course characterized by transient weight loss, tachypnea, and depression. Viral RNA is shed at high levels from nasal turbinates, trachea, and lungs and low levels from a wide range of other tissues, including the brain,63 in which destruction of olfactory neurons has been demonstrated.50 Infectious viral shedding peaks within the first 3–5 dpi75 and is limited by appearance of neutralizing antibodies by approximately 1 week post infection. Bilateral lung consolidation is evident on micro-computed tomography. In this study, severity of pulmonary disease was proportional to infective dose.63 In another, disease course was faster with higher infective dose; however, disease severity appeared dose independent.75 Histopathology is characterized by progressive broncho-interstitial pneumonia centered on bronchioles and adjacent parenchyma and corresponding to distribution of viral antigen on immunohistochemistry (Figure 1).75 Necrosis of bronchiolar and tracheal epithelium is accompanied by epithelial syncitia and accumulation of macrophages and neutrophils within bronchiolar lumen. Interstitial pneumonia is characterized by interstitial and alveolar influx of macrophages and neutrophils, alveolar edema, fibrinous effusion, and perivascular lymphocytic cuffing. Migration of lymphocytes through tunica media and elevation of endothelium is noted in pulmonary vessels.75 The most severe pathology is evident at approximately day 518,75 followed by elimination of viral antigen, reduced inflammation, and marked type II pneumocyte hyperplasia by 10 dpi.18,75 In contrast to other models, enteric necrosis and proprial inflammation are present as well as cardiac myofiber degeneration.18 Clinical signs of enteritis are not seen; however, antigen is evident in the duodenum and viral RNA is shed in feces.76 Lack of differential sex or age susceptibility is reported in 1 study,75 with delayed recovery of pulmonary disease and greater weight loss in older (32–34 weeks) hamsters reported in another.70 Transmission between animals occurs efficiently via direct contact or aerosol and less so via fomite.76 A surgical mask partition reduces transmission between hamsters.78 Viral replication can be suppressed by passive transfer of convalescent serum.63 Hydroxychloroquine does not improve clinical outcome or reduce viral shedding in SARS-CoV-2–infected hamsters.73
Figure 1.
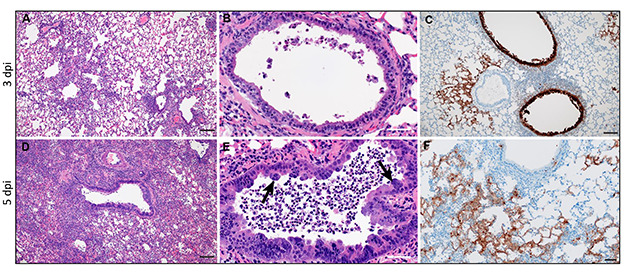
Pulmonary histopathology at 3 and 5 days post infection (dpi) with SARS-CoV-2 (103 TCID50). A–C: 3 dpi. Histopathology is characterized by broncho-interstitial pneumonia centered on bronchioles and adjacent parenchyma (A), corresponding to distribution of viral antigen (C). Bronchiolar epithelial necrosis is accompanied by efflux of neutrophils and macrophages into bronchiolar lumen (B). D–F: 5 dpi. Worsened broncho-interstitial pneumonia results in regional parenchymal consolidation (D), and progressive bronchiolitis is accompanied by syncytial epithelial cells (arrows, E).Viral immunoreactivity is most prominent in alveolar epithelial cells (F). Hematoxylin and eosin (A, B, D, E); immunohistochemistry using GenScript U864YFA140–4/CB2093 NP-1 at a 1:1000 dilution (C, F); Bar = 50 μm (A, C, D), 20 μm (B, E, F). Panel reconstructed from Rosenke K, Meade-White K, Letko M, Clancy C, Hansen F, Liu Y, Okumura A, Tang-Huau TL, Li R, Saturday G, Feldmann F, Scott D, Wang Z, Munster V, Jarvis MA, Feldmann H. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg Microbes Infect. 2020 Nov 29:1–36. doi: 10.1080/22221751.2020.1858177. PMID: 33251966. Permissions obtained under Creative Commons CC BY license (https://creativecommons.org/licenses/by-nc-nd/4.0/).
Deer Mice
Intranasal inoculation of deer mice results in subclinical pulmonary disease characterized by live virus recovery, broncho-interstitial pneumonia, and leucocytoclastic vasculitis within the first week, and resolution by 14 dpi.51 Fibrinopurulent rhinitis and sinusitis and neutrophilic infiltration of afferent ethmoidal, olfactory, maxillary, and other sensory nerves are evident. Suppurative glossitis with necrosis of gustatory buds is evident, consistent with reported loss of smell and taste in human patients. Neuroinvasion characterized frontal cortical encephalitis, gliosis, and immunoreactivity for SARS-CoV-2 in brainstem nuclei is evident. SARS-CoV-2 antigen is evident in nasal and oropharyngeal mucosa, sensory ganglia, brain, microglia, retina, and intestinal mucosa and propria.
Ferrets and Mink
Ferrets develop no or mild (sneezing, lethargy) clinical signs following infection. Virus is evident in nasal turbinate, soft palate, tonsils, lung, and intestine.79 Nevertheless, pulmonary pathology includes lymphoplasmacytic perivasculitis and vasculitis, type II pneumocyte hyperplasia, macrophages, neutrophils in alveolar septa and lumen, and mild peribronchitis. Viral transmission via direct and indirect contact occurs.79 Azathioprine-immunosuppressed ferrets exhibit a longer period of clinical illness, higher virus titers in nasal turbinate, delayed virus clearance, and lower serum neutralization antibody titers.80 Outbreaks of SARS-CoV-2 infection have occurred in mink farms in the Netherlands81–83 and Denmark84 associated with COVID-19–like symptoms in workers. Outbreaks have also been reported in mink farms in Utah and Spain, with high mortality, particularly in older animals occurring in Utah (https://www.avma.org/javma-news/2020-11-15/sars-cov-2-kills-thousands-minks-utah). Animals experience respiratory signs, anorexia, and increased mortality with an acute disease course (2–3 days). Lesions include severe diffuse interstitial pneumonia with diffuse alveolar damage and hyaline membranes,83 with no pathology noted in extra-respiratory tissues. Recently, all 17 million mink in Denmark were culled following mink-human transmission of SARS-CoV-2 after concern that spike protein mutations acquired during passage through mink would imperil efficacy of newly developed vaccines.85
Felines and Canines
Companion animals and zoo felids are the recipients of COVID- 19 from humans (https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/sa_ one_health/sars-cov-2-animals-us) and develop no to mild upper respiratory disease. Cats are permissive to infection and airborne transmission.22
MODELING COVID-19 IN GENETICALLY ALTERED ANIMAL MODELS
Several approaches have been employed to increase susceptibility to SARS-CoV-2 infection in laboratory mice. These include genetic alteration of the virus so that it is able to bind mouse Ace2, expression of human ACE2 under a variety of promoters, and transfection of mice with human ACE2 cDNA using viral vectors. Murine models for SARS-CoV-2 are summarized in Table 3.
Table 3.
Clinical Signs, Pulmonary and Extrapulmonary Pathology, and Viral Distribution in Mouse Models of COVID-19
| Viral Replication and Tissue Tropism | Clinical Signs | Pulmonary Pathology | Extrapulmonary Pathology | Immunohistochemical Localization of SARS-CoV-2 |
|---|---|---|---|---|
| Mouse-adapted SARS-CoV-223 | ||||
| Viral RNA: lung, nasal turbinate, trachea, feces, heart, and liver up to 7 DPI Infectious virus: nasal turbinate, lung up to 2 DPI (young), 4 DPI (aged) |
Age enhanced disease; decrease in body weight by 3 dpi in aged mice only | Not reported | Focal exudation and hemorrhage, interstitial pneumonia | Lung: CC10 (club cell) bronchus and bronchioles, alveolar type 1 cells |
| Chimeric SARS-Co-V/SARS-CoV-282 | ||||
| Viral RNA: Lung | Remdesivir ameliorates loss of pulmonary function (whole-body plethysmography) | Lung hemorrhage at 5 dpi reduced by remdesivir treatment | No | Not examined |
| Murine mAce2 exon 2-hACE2 knockin mouse81 | ||||
| Viral RNA: Predominantly in trachea, lung, brain | Marked weight loss in older animals | Interstitial pneumonia | Vascular system injury | Lung: CC10+ Clara cells in airways, surfactant protein C positive (SPC+) alveolar type II cells |
| Murine mAce2 promoter-hACE2 transgenic mouse82 | ||||
| Viral RNA: Virus is shed from lung for 1 wk post infection and briefly from intestine 1–7 dpi | Transient weight loss and recovery by 7 dpi | Modest interstitial pneumonia, interstitial and intra-alveolar mononuclear and granulocytic inflammation | Vasculitis in extrapulmonary organs | Lung: macrophages and T lymphocytes |
| HFH4/FOXJ1 promoter-hACE2 transgenic mouse83,84 | ||||
| Viral RNA: Lung, eyes, heart, brain | Binary clinical phenotype: weight loss, respiratory distress, and neurological symptoms, die by 6 dpi; others asymptomatic and survive | Mild to severe interstitial inflammation with hyaline membrane formation | Cardiomyocyte edema and sporadic neuroinvasion in brains of deceased mice | Lung: bronchial epithelial cells and alveolar cells |
| K18 promoter-hACE2 transgenic mouse85–91 | ||||
| Viral RNA: expressed at highest levels in lung and brain, evident in colon and serum in a subset of animals by 7 dpi | Significant weight loss by 4 dpi, some animals becoming moribund by 7–8 dpi | Edema, alveolar, interstitial, and perivascular infiltration of neutrophils and mononuclear cells, and consolidation | Encephalitis, vasculitis, and meningitis after 5 dpi | Lung: alveolar epithelial cells, macrophages, nasal epithelium Eye: inner nuclear layer Brain: olfactory bulb, extensive in neurons |
| Adenovirus 5- hACE2 transfected mouse57,78 | ||||
| Viral RNA: lung, low levels in spleen, heart, brain | Transient weight loss in older BALB/c mice | Perivascular to interstitial inflammation, necrotic debris, alveolar edema and vascular congestion and hemorrhage, most severe at 5 dpi | Not described | Lung: alveolar and bronchiolar epithelial cells |
| Adeno-associated virus (AAV9)-hACE2 mouse93 | ||||
| Viral RNA: limited to respiratory tract | No weight loss or clinical illness | Interstitial pneumonia | Not described | Lung: alveolar epithelial cells |
Wild-Type and Immune-Compromised Mice
Wild-type mice do not support significant SARS-CoV-2 infection due to insufficient interaction between viral S protein and murine ACE2.86 This finding in consistent across several inbred strains, including C57BL/6 J,64,87,88 BALB/c, DBA/2J,87 and ICR88 mice. Viral binding to murine ACE2 is not entirely absent, however, and very limited viral replication is evident in lung in Stat1−/− mice,87 Rag1 −/− mice lacking mature B and T cells,87 SCID mice, Il28r−/− mice, Ifnar1−/− mice,77 and AG129 (type I and II interferon receptor-deficient mice87).
Mouse-Adapted SARS-CoV-2
Dinnon et al23 engineered Q498T/P499Y into the SARS-CoV-2 S gene to generate a recombinant virus (SARS-CoV-2 MA) that could utilize mouse ACE2 for entry. SARS-CoV-2 MA replicates in upper and, to a lesser extent, lower airways of both young adult and aged BALB/c mice. Compared with young mice, SARS-CoV-2 MA–infected old mice develop more severe interstitial pneumonia with greater impairment of pulmonary function in life.
Chimeric SARS-CoV/SARS-CoV-2 Viruses
A chimeric virus composed of mouse-adapted SARS-CoV (permitting infection via mACE2 binding) and SARS-CoV-2 RNA-dependent RNA polymerase allows testing of antiviral efficacy of remdesivir.89 Incorporation of active metabolite remdesivir triphosphate occurs preferentially to natural substrate ATP and results in chain termination 3 nucleotides downstream of incorporation. Mice produce a serum esterase, carboxyl esterase 1c (Ces1c), that is absent in humans and reduces half-life of remdesivir. Therefore mouse studies with remdesivir must be performed in transgenic C57Bl/6 Ces1c−/− mice. In this model,89 histopathology is not shown; however, significant reduction of grossly observable lung hemorrhage at 5 dpi with remdesivir treatment is reported despite similar weight loss in vehicle and remdesivir-treated groups.
Murine mAce2 Exon 2-hACE2 Knockin Mouse
Using CRISPR-Cas9 technology, Sun et al52 inserted hACE2 cDNA into Exon 2 of the mAce2 gene to disrupt mAce2 gene expression and drive expression of hACE2 under control of the mAce2 promoter. hACE2 expression occurs in lung, small intestine, spleen, and kidney. In lung, hACE2 is expressed in the CC10+ Clara cells in airways as well as a subpopulation of surfactant protein C positive alveolar type II cells. High viral loads of SARS-CoV-2 are evident in lung, trachea, and brain (despite low hACE2 expression) on intranasal infection. Young inoculated animals do not display obvious clinical symptoms but develop interstitial pneumonia and vascular system injury. More severe disease is seen in aged hACE2. These exhibit more marked weight loss, more prolonged viral shedding (including from feces), and more severe pneumonia accompanied by stronger cytokine responses. Intragastric inoculation of SARS-CoV-2 induces productive infection and pulmonary disease.
hACE2 Transgenic Mice
Mice transgenic for hACE2 were developed over a decade ago to study SARS-CoV and to allow creation of significantly greater disease than that induced in wild-type mice. The same models also permit infection with SARS-CoV-2, and their effects differ by the promoter driving expression of hACE2.
Murine mAce2 Promoter-hACE2 Transgenic Mouse
Originally described by Yang et al,90 SARS-CoV–infected mice expressing hACE2 in lung, heart, kidney, and gut permit greater viral replication and develop more severe pulmonary lesions than wild-type mice. Vasculitis in extrapulmonary organs and viral antigen in brain is noted, but encephalitis does not occur. ICR-hACE2 transgenic mice inoculated with SARS-CoV-288 experience transient weight loss and recovery by 7 dpi. Virus is shed from lung for 1 week post infection and briefly from intestine 1–7 dpi. Relatively mild interstitial pneumonia develops and is characterized by interstitial and intra-alveolar mononuclear and granulocytic inflammation. SARS-CoV-2 is evident immunohistochemically in macrophages and T lymphocytes.
HFH4/FOXJ1 Promoter-hACE2 Transgenic Mice
Originally developed in 2016,91 lung ciliated epithelial cell-specific HFH4/FOXJ1 promoter drives hACE2 expression at high levels of hACE2 in the lung and to varying degrees in brain, liver, kidney, and gastrointestinal tract. SARS-CoV-2–inoculated mice92 experience binary clinical phenotypes, in which some lose weight, display respiratory distress and neurological symptoms, and die by 6 dpi, whereas others survive without evidence of clinical distress. Interestingly, the former are male, possibly recapitulating male susceptibility to COVID-19 mortality. A spectrum of pulmonary pathology ranging from mild to severe interstitial inflammation with hyaline membrane formation is evident. Cardiomyocyte edema and sporadic neuroinvasion in brains of deceased mice is evident. Dying mice experience lymphopenia, 1 of the key clinical hallmarks of severe COVID-19 disease.
K18 Promoter-hACE2 Transgenic Mice
Mice expressing hACE2 under control of the K18 promoter were developed to study SARS-CoV.93,94 When infected with SARS-CoV-2,95,96 pulmonary pathology is among the most severe of that described in murine models96,97 and is accompanied by profound perivascular mononuclear inflammation with minimal viral endothelial infection.97 Significant lethality and male bias in disease severity are noted.97 In hemizygous K18-hACE2 mice,96 expression is highest in lung, followed by heart, brain, colon, and kidney with lower levels in duodenum, ileum, and spleen. Pulmonary levels of hACE2 decline over the disease course.96 K18-hACE2 mice given intranasal SARS-CoV-2 exhibit significant weight loss by 4 dpi,95,96 with many animals becoming moribund by 7 dpi.96,97 Viral RNA is expressed at highest levels in lung and is evident in colon and serum only in a subset of animals by 7 dpi. Pulmonary histopathology is severe and characterized by consolidation, edema, alveolar, and interstitial and perivascular infiltration of neutrophils and mononuclear cells. Viral RNA is evident by in situ hybridization in pulmonary alveolar epithelial cells and macrophages97 as well as nasal epithelia. K18-hACE2 TG mice experience significant central nervous system infection following infection with both SARS-CoV93,94 and SARS-Cov-2.53,54,97 Similar cytokine (lung) and chemokine (brain) elevations are described in SARS-CoV-2–infected K18 mice.54 Individual mice express viral RNA in the retina97 and olfactory bulb and throughout the brain,53,97accompanied by encephalitis, vasculitis, and meningitis by 5 dpi.96–98 By in situ hybridization, viral RNA is present in NeuN + cells, implicating neuronal infection in mice97 and human COVID-19 patients.53 Rare thromboses are noted in murine brain.97 Viral presence in murine endothelial cells within brain is lacking;53 however, vascular remodeling is noted in regions of neuronal viral infection. In contrast, endothelial invasion by virus is evident in COVID-19 patients experiencing ischemic and hemorrhagic infarcts.53 Neurologic lesions in SARS-Cov-2–infected mice are consistent with original studies using SARS-Co-V in this model in which extrapulmonary virus spreads to the brain via the olfactory tract and killed all mice by 7 dpi.93,94 Medullary infection is associated with aspiration pneumonia that complicates interpretation of pulmonary pathology. Both lung and brain express increased inflammatory cytokines (chemokine (C-X-C motif) ligand 1 (CXCL-1), chemokine (C-X-C motif) ligand 10 (CXCL-10), Interleukin-6 (IL-6), and Interleukin-61 beta (IL-1beta)). This is accompanied by significant inflammation in the lung but neuronal death without inflammation in the brain.94 Interestingly, SARS-CoV-2–infected regions of human brain similarly lack inflammation.53
Viral Transduction of hACE2 Using Adenovirus 5
Adenovirally transduced hACE2 BALB/c and C57BL/6 mice infected with SARS-CoV-264 experience acute but transient disease duration of 7–10 days. Pulmonary lesions include perivascular to interstitial inflammation, necrotic debris, alveolar edema, and vascular congestion and hemorrhage, most severe at 5 dpi. Upregulation of genes encoding inflammatory mediators and components of adaptive and innate responses occurs by 2 dpi and is followed by appearance of virus-neutralizing antibodies and CD4 and CD8 responses to N and S viral proteins, respectively, by 10 dpi. Viral clearance is impaired and lung phenotype worsened by CD4/CD8 depletion or by impaired interferon signaling conferred by Stat1−/− genotype. Conversely, viral clearance is accelerated, and phenotype improved by vaccination, type I IFN induction using poly I:C, passive transfer of antibodies from recovered mice, or transfer of serum from COVID-19 patients. In a similar model, lung pathology in AdV-hACE2–transduced, SARS-CoV-2–infected mice can be worsened given anti-Ifnar1 mAb or improved by pretreatment with an anti-SARS-CoV-2 mAb.87
Adeno-associated Virus-hACE2 Mice
Mice transduced with adeno-associated virus-hACE2 and subsequently infected by SARS-CoV-2 experience no weight loss or clinical illness.99 Interstitial pneumonia is accompanied by upregulation of cytokines and interferon stimulated genes and infiltration by monocyte-derived macrophages and activated lymphoid cells. Neutralizing antibodies appear by 7 dpi.
CONCLUSIONS
As infection continues to spread through the United States and globally, the development of several effective vaccines appears likely to stem the COVID-19 pandemic. Animal models have already proved indispensable in our understanding of pulmonary and immune phenotype central to preclinical testing of vaccines and therapies for SARS coronaviruses. Continued evolution of these models is likely to continue to play a critical role in the upcoming challenges regarding vaccine efficacy, potential side effects, long-term consequences of SARS-CoV-2 infection in recovered patients, and organ-specific pathology.
Potential conflicts of interest
All authors: No reported conflicts.
Contributor Information
Caroline J Zeiss, Department of Comparative Medicine, Yale University School of Medicine, New Haven, Connecticut, USA.
Susan Compton, Department of Comparative Medicine, Yale University School of Medicine, New Haven, Connecticut, USA.
Rebecca Terilli Veenhuis, Department of Comparative Medicine, John Hopkins School of Medicine, Baltimore, Maryland, USA.
References
- 1. Ksiazek TG, Erdman D, Goldsmith CS et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348(20):1953–1966. [DOI] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W et al. A novel coronavirus from patients with pneumonia in China. N Eng J Med 2020; 382(8):727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Decaro N, Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet Microbiol 2020; 244:108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019; 17(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zaki AM, van Boheemen S, Bestebroer TM et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367(19):1814–1820. [DOI] [PubMed] [Google Scholar]
- 6. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Meth Mol Biol (Clifton, NJ) 2015; 1282:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong ACP, Li X, Lau SKP et al. Global epidemiology of bat coronaviruses. Viruses 2019; 11(2):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lau SK, Woo PC, Li KS et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A 2005; 102(39):14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reusken CB, Raj VS, Koopmans MP et al. Cross host transmission in the emergence of MERS coronavirus. Cur Op Virol 2016; 16:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lelli D, Papetti A, Sabelli C et al. Detection of coronaviruses in bats of various species in Italy. Viruses 2013; 5(11):2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tao Y, Shi M, Chommanard C et al. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol 2017; 91(5):e01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corman VM, Muth D, Niemeyer D et al. Hosts and sources of endemic human coronaviruses. Adv Virus Res 2018; 100:163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang XC, Zhang JX, Zhang SY et al. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol 2006; 80(15):7481–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woo PC, Lau SK, Li KS et al. Molecular diversity of coronaviruses in bats. Virology 2006; 351(1):180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv Virus Res 2011; 81:85–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lan J, Ge J, Yu et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020; 581(7807):215–220. [DOI] [PubMed] [Google Scholar]
- 17. Wan Y, Shang J, Graham R et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 2020; 94(7):e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan JF, Zhang AJ, Yuan S et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin Infect Dis 2020; 71(9):2428–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao X, Chen D, Szabla R et al. Broad and differential animal ACE2 receptor usage by SARS-CoV-2. J Virol 2020; 94(18):e00940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Melin AD, Janiak MC, Marrone F 3rd et al. Comparative ACE2 variation and primate COVID-19 risk. Commun Biol 2020; 3(1):641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu, S, Zhao, Y, Yu, W, et al. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv 2020:2020.2004.2008.031807. [Google Scholar]
- 22. Shi J, Wen Z, Zhong G et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science (New York, NY) 2020; 368(6494):1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinnon KH, Leist SR, Schäfer A et al. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. Nature 2020 Oct; 586(7830):560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greenough TC, Carville A, Coderre J et al. Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am J Pathol 2005; 167(2):455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuba K, Imai Y, Rao S et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 2005; 11(8):875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weingartl H, Czub M, Czub S et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol 2004; 78(22):12672–12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heurich A, Hofmann-Winkler H, Gierer S et al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 2014; 88(2):1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoffmann M, Kleine-Weber H, Schroeder S et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181(2):271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shang J, Wan Y, Luo C et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 2020; 117(21):11727–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bestle D, Heindl MR, Limburg H et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 2020; 3(9):e202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ziegler CGK, Allon SJ, Nyquist SK et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020; 181(5):1016–1035, e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mönkemüller K, Fry L, Rickes S. COVID-19, coronavirus, SARS-CoV-2 and the small bowel. Rev Esp Enferm Dig 2020; 112(5):383–388. [DOI] [PubMed] [Google Scholar]
- 33. Cheng Y, Luo R, Wang K et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int 2020; 97(5):829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wrapp D, Wang N, Corbett KS et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, NY) 2020; 367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riley S, Fraser C, Donnelly CA et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science (New York, NY) 2003; 300(5627):1961–1966. [DOI] [PubMed] [Google Scholar]
- 36. Yan X, Han X, Fan Y et al. Duration of SARS-CoV-2 viral RNA in asymptomatic carriers. Crit Care 2020; 24(1):245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang W, Xu, Gao R et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020; 323(18):1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. To KK, Tsang OT, Leung WS et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20(5):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng S, Fan J, Yu F et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ (Clinical Research Ed) 2020; m1443:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peccia J, Zulli A, Brackney DE et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol 2020; 38(10):1164–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amirian ES. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int J Infect Dis 2020; 95:363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guan WJ, Ni ZY, Hu et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382(18):1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tay MZ, Poh CM, Renia L et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 2020; 20(6):363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Du, Tu, Zhu P et al. Clinical features of 85 fatal cases of COVID-19 from Wuhan. A retrospective observational study. Am J Respir Crit Care Med 2020; 201(11):1372–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mucha SR, Dugar S, McCrae K et al. Coagulopathy in COVID-19. Cleve Clin J Med 2020; 87(8):461–468. [DOI] [PubMed] [Google Scholar]
- 46. Klok FA, Kruip M, van der Meer NJM et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res 2020; 191:145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol 2020; 35(5):744–748. [DOI] [PubMed] [Google Scholar]
- 48. Pei G, Zhang Z, Peng J et al. Renal involvement and early prognosis in patients with COVID-19 pneumonia. J Am Soc Nephrol 2020; 31(6):1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mao L, Jin H, Wang M et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA neurology 2020; 77(6):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang AJ, Lee AC, Chu H et al. SARS-CoV-2 infects and damages the mature and immature olfactory sensory neurons of hamsters. Clin Infect Dis 2020; Jul 15:ciaa995. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fagre A, Lewis J, Eckley M et al. SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: Implications for reverse zoonosis to new world rodents. bioRxiv 2020; 2020.2008.2007.241810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun SH, Chen Q, Gu et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 2020; 28(1):124–133.e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chakravarty D, Das Sarma J. Murine-β-coronavirus-induced neuropathogenesis sheds light on CNS pathobiology of SARS-CoV2. J Neurovirol 2021;5: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oladunni FS, Park J-G, Tamayo PP et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin converting enzyme 2 transgenic mice. Nat Commun 2020; 11(1):6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheung EW, Zachariah P, Gorelik M et al. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in new York City. JAMA. 2020; 324(3):294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yuan M, Yin W, Tao Z et al. Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan, China. PLoS One 2020; 15(3):e0230548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Richardson S, Hirsch JS, Narasimhan M et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the new York City area. JAMA. 2020; 323(20):2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beasley MB. The pathologist's approach to acute lung injury. Arch Path Lab Med. 2010; 134(5):719–727. [DOI] [PubMed] [Google Scholar]
- 59. Xu, Shi L, Wang Y et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020; 8(4):420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Copin MC, Parmentier E, Duburcq T et al. Time to consider histologic pattern of lung injury to treat critically ill patients with COVID-19 infection. Intensive Care Med 2020; 46(6):1124–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu P, Qi F, Xu Y et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med 2020; 3(1):93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rockx B, Kuiken T, Herfst S et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science (New York, NY) 2020; 368(6494):1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Imai M, Iwatsuki-Horimoto K, Hatta M et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A 2020; 117(28):16587–16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun J, Zhuang Z, Zheng J et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 2020; 182(3):734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Callaway E. The coronavirus is mutating - does it matter? Nature 2020; 585(7824):174–177. [DOI] [PubMed] [Google Scholar]
- 66. Korber B, Fischer WM, Gnanakaran S et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182(4):812–827 e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. New drug and biological drug products; evidence needed to demonstrate effectiveness of new drugs when human efficacy studies are not ethical or feasible. Final rule Federal Register 2002; 67(105):37988–37998. [PubMed] [Google Scholar]
- 68. Munster VJ, Feldmann F, Williamson BN et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020; 585(7824):268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Finch CL, Crozier I, Lee JH et al. Characteristic and quantifiable COVID-19-like abnormalities in CT- and PET/CT-imaged lungs of SARS-CoV-2-infected crab-eating macaques (Macaca fascicularis). bioRxiv 2020. [Google Scholar]
- 70. Osterrieder N, Bertzbach LD, Dietert K et al. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses 2020; 12(7):779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Williamson BN, Feldmann F, Schwarz B et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 2020; 585(7824):273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maisonnasse P, Guedj J, Contreras V et al. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 2020; 585(7826):584–587. [DOI] [PubMed] [Google Scholar]
- 73. Rosenke K, Jarvis MA, Feldmann F et al. Hydroxychloroquine proves ineffective in hamsters and macaques infected with SARS-CoV-2. JCI Insight 2020; 5(23):e143174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Woolsey C, Borisevich V, Prasad AN et al. Establishment of an African green monkey model for COVID-19. Nat Immunol 2020; 22(1):86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rosenke K, Meade-White K, Letko M et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg Microbes Infect 2020; 9(1):2673–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sia SF, Yan LM, Chin AWH et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020; 583(7818):834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Boudewijns R, Thibaut HJ, Kaptein SJF et al. STAT2 signaling as double-edged sword restricting viral dissemination but driving severe pneumonia in SARS-CoV-2 infected hamsters. Nat Commun 2020 Nov 17; 11(1):5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chan JF, Yuan S, Zhang AJ et al. Surgical mask partition reduces the risk of non-contact transmission in a golden Syrian hamster model for coronavirus disease 2019 (COVID-19). Clin Infect Dis 2020; 71(16):2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim YI, Kim SG, Kim SM et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020; 27(5):704–709.e702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Park SJ, Yu, Kim YI et al. Antiviral efficacies of FDA-approved drugs against SARS-CoV-2 infection in ferrets. MBio 2020; 11(3):e01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Enserink M. Coronavirus rips through Dutch mink farms, triggering culls. Science (New York, NY) 2020; 368(6496):1169. [DOI] [PubMed] [Google Scholar]
- 82. Oreshkova N, Molenaar RJ, Vreman S et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill 2020; 25(23):2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Molenaar RJ, Vreman S, Hakze-van der Honing RW et al. Clinical and pathological findings in SARS-CoV-2 disease outbreaks in farmed mink (Neovison vison). Vet Pathol 2020; 57(5):653–657. [DOI] [PubMed] [Google Scholar]
- 84. Hammer AS, Quaade ML, Rasmussen TB et al. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg Infect Dis 2020; 27(2):547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect Dis 2020; 21(1):18–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020; 5(4):562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hassan AO, Case JB, Winkler ES et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 2020; 182(3):744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bao L, Gao H, Deng W et al. Transmission of SARS-CoV-2 via close contact and respiratory droplets among hACE2 mice. J Infect Dis 2020; 222(4):551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pruijssers AJ, George AS, Schäfer A et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep 2020; 32(3):107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yang XH, Deng W, Tong Z et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med 2007; 57(5):450–459. [PubMed] [Google Scholar]
- 91. Menachery VD, Yount BL Jr, Sims AC et al. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 2016; 113(11):3048–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jiang RD, Liu MQ, Chen Y et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 2020; 182(1):50–58.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. McCray PB Jr, Pewe L, Wohlford-Lenane C et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 2007; 81(2):813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Netland J, Meyerholz DK, Moore S et al. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol 2008; 82(15):7264–7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Moreau GB, Burgess SL, Sturek JM et al. Evaluation of K18-hACE2 mice as a model of SARS-CoV-2 infection. Am J Trop Med Hyg 2020; 103(3):1215–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Winkler ES, Bailey AL, Kafai NM et al. SARS-CoV-2 infection in the lungs of human ACE2 transgenic mice causes severe inflammation, immune cell infiltration, and compromised respiratory function. bioRxiv 2020; 2020.07.09.196188. [Google Scholar]
- 97. Golden JW, Cline CR, Zeng X et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight 2020; 5(19):e142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rathnasinghe R, Strohmeier S, Amanat F et al. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg Microbes Infect 2020; 9(1):2433–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Israelow B, Song E, Mao T et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med 217(12):e20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]


