Abstract
Objectives
The Bio-Rad SARS-CoV-2 ddPCR Kit (Bio-Rad Laboratories) was the first droplet digital polymerase chain reaction (ddPCR) assay to receive Food and Drug Administration (FDA) Emergency Use Authorization approval, but it has not been evaluated clinically. We describe the performance of ddPCR—in particular, its ability to confirm weak-positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) results.
Methods
We clinically validated the Bio-Rad Triplex Probe ddPCR Assay. The limit of detection was determined by using serial dilutions of SARS-CoV-2 RNA in an artificial viral envelope. The ddPCR assay was performed according to the manufacturer’s specifications on specimens confirmed to be positive (n = 48) or negative (n = 30) by an FDA-validated reverse transcription–polymerase chain reaction assay on the m2000 RealTime system (Abbott). Ten borderline positive cases were also evaluated.
Results
The limit of detection was 50 copies/mL (19 of 20 positive). Forty-seven specimens spanning a range of quantification cycles (2.9-25.9 cycle numbers) were positive by this assay (47 of 48; 97.9% positive precent agreement), and 30 negative samples were confirmed as negative (30 of 30; 100% negative percent agreement). Nine of 10 borderline cases were positive when tested in triplicate.
Conclusions
The ddPCR of SARS-CoV-2 is an accurate method, with superior sensitivity for viral RNA detection. It could provide definitive evaluation of borderline positive cases or suspected false-negative cases.
Keywords: COVID-19, SARS-CoV-2, Digital droplet polymerase chain reaction, Real-time polymerase chain reaction
Key Points.
• This study addresses how well a novel platform such as ddPCR works for detecting SARS-CoV-2.
• The ddPCR is one of the most sensitive assays available and can resolve borderline positive cases.
• Interestingly, high-viral-load samples create a smear artifact not typically observed in ddPCR.
The global coronavirus disease 2019 (COVID-19)1 pandemic continues to pose a serious global public health threat. The COVID-19 pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),2 is a single-stranded RNA Betacoronavirus with a 26-kilobase genome. Molecular detection of SARS-CoV-2 targeting the viral genes (eg, Orf1a/b, E, S, N genes) is currently the gold standard for assessing acute infection.3-7
In the United States, the first clinical assay for SARS-CoV-2 was developed by the Centers for Disease Control and Prevention (CDC)3 and approved under a Food and Drug Administration (FDA) Emergency Use Authorization (EUA). As of the first quarter of 2021, multiple testing platforms have obtained EUA and been clinically implemented to diagnose SARS-CoV-2 infection. Although these point-of-care tests are rapid, many are limited by moderate sensitivity and high false-negative rates.8 Real-time polymerase chain reaction (RT-PCR) platforms can detect low levels of virus with high throughput, but weak positives (cycle number [CN] > 35) can be difficult to distinguish from technical artifacts after many PCR cycles. Droplet digital PCR (ddPCR) is an orthogonal method designed to detect and measure precise copy numbers of nucleic acid, but it can be applied to detect extremely low levels of nucleic acid, as well. The Bio-Rad SARS-CoV-2 ddPCR Kit (Bio-Rad Laboratories) was the first assay in this class to receive EUA (Precigenome is the only other company with a ddPCR EUA-approved assay), but data characterizing its clinical performance are limited. One study observed that ddPCR has a sensitivity of 93.33% and a specificity of 100% for both the N1 and N2 gene regions of SARS-CoV-2.9 In this study, we report the limit of detection, artifactual findings, and correlation with other clinically validated methods. Overall, ddPCR is a highly sensitive approach capable of resolving borderline results with reasonable throughput and cost. With its sensitivity, it is well suited to pooled testing as well.
Materials and Methods
RNA Extraction
In Biosafety Level 2 containment, 400 µL of residual nasopharyngeal (NP) swab samples in universal viral transport medium (VTM) (3 mL initial collection, VTM; BD Catalog No 220529 [Becton Dickinson]) or spiked control material in VTM were transferred to conical tubes containing 400 µL lysis buffer, 40 µL proteinase K, and 7.5 µL carrier RNA for lysis/virus inactivation (20 minutes at 56°C) before extraction. Total nucleic acids from pooled or individual residual NP collections and controls were obtained using a Maxwell RSC instrument (Promega), with an offboard lysis protocol and the following volume parameters: 400 µL input, 500 µL magnetic silica, and 35 µL output/elution.
Control Material and Patient Specimens
Control material for endemic SARS-CoV-2 coronavirus strains was obtained from AccuPlex SARS-CoV-2 Reference Material Kit (SeraCare).
Residual deidentified patient samples from a clinical laboratory performing testing with the m2000 RealTime SARS-CoV-2 EUA test (Abbott) included both positive (n = 30) and negative (n = 30) samples. All CN values come from the m2000 system unless specified as Alinity CN, which designates a value coming from the FDA EUA Alinity m SARS-CoV-2 assay (Abbott). The transport medium of these samples was a formulation of VTM. Each patient sample underwent freeze-thaw cycles at least twice before our extraction and PCR. Positive patient specimens, when indicated, were diluted with nuclease-free water after extraction. Borderline positive cases were defined as “Alinity CN > 38” that repeated weak positive by the Alinity SARS-CoV-2 assay (n = 10). Only borderline cases were run in triplicate.
Droplet Digital PCR
All procedures followed the manufacturer’s instructions for the SARS-CoV-2 ddPCR Kit unless otherwise specified.10 A mastermix was prepared from the Bio-Rad 1-Step RT-ddPCR Advanced Kit for Probes and ddPCR Expert Design Assay 2019-nCoV CDC probe. In a 96-well PCR plate, 13 µL of extracted patient sample or control (representing 149 µL of original) were added to 9 µL of mastermix for each reaction. At least 6,000 and 10,000 total droplets were required for positive and negative samples, respectively. After droplet generation, the samples were placed on a Bio-Rad C1000 Touch Thermal Cycler and underwent reverse transcription and PCR amplification. When droplet stabilization had occurred, the plate was immediately read on a Bio-Rad QX200 Droplet Reader and evaluated using the QuantaSoft Analysis Pro 1.0 software.
Primers and probes for the CDC N1 and N2 gene targets were present, along with ribonuclease P protein subunit p30 (RPP30) as an internal control for endogenous human RNA. The N1 probe was labeled with HEX, the N2 probe was labeled with HEX and carboxyfluorescein (FAM), and the RPP30 probes were labeled with higher levels of the FAM fluorophore. Therefore, positive results led to unique clusters of populations. Sometimes, droplets were positive for multiple targets; in this case, the software could anticipate the location and apply gating. Each case was reviewed for automated droplet count and then reviewed manually to determine whether positive droplets fell outside gating parameters. At least 1 of the N1 and N2 droplets (or ≥3 N1 or N2 droplets) was required for a positive result, and at least 10 RPP30 droplets were required for a valid result. A result was considered invalid if (1) fewer than 10 RPP30 droplets were detected; (2) the sample was negative, with fewer than 10,000 negative droplets; or (3) the sample was positive, with fewer than 6,000 droplets.
Results
Limit of Detection
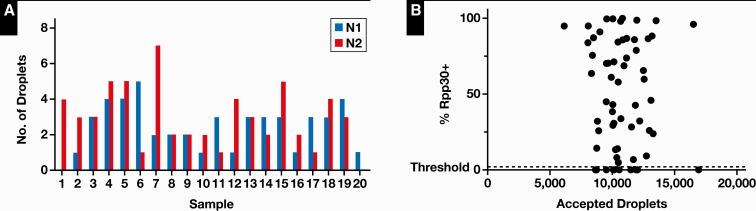
To determine the limit of detection of the assay, triplicate serial dilutions of synthetic SARS-CoV-2 viral particles were prepared (AccuPlex SARS-CoV-2 Reference Material Kit positive reference) ranging from 500 copies/mL to 1 copy/mL Table 1. When all triplicate samples were detected, an extended replicate series of 20 samples was performed Figure 1A. We found a 95% positive rate (19 of 20) at 50 copies/mL, which was 20-fold lower than the kit manufacturer reported. The 1 sample that failed had 1 droplet positive from N1, which was below our cutoff but suggested that even better sensitivity may be possible.
Table 1.
Summary of Limit-of-Detection Analysis Results
| Concentrations, copies/mLa | No. of Copies per Reactionb | N1c | N2c | Totalc |
|---|---|---|---|---|
| 500 | 74.3 | 3/3 | 3/3 | 3/3 |
| 100 | 14.8 | 3/3 | 3/3 | 3/3 |
| 50 | 7.4 | 3/3 | 3/3 | 3/3 |
| 25 | 3.7 | 1/3 | 1/3 | 1/3 |
| 10 | 1.5 | 1/3 | 2/3 | 1/3 |
| 1 | 0.1 | 0/3 | 0/3 | 0/3 |
| VTM | 0 | 0/3 | 0/3 | 0/3 |
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VTM, viral transport medium.
aStarting copy numbers of SARS-CoV-2.
bTheoretical calculated viral copy number in each individual reaction.
cN1, N2, and total positivity rates (positive if at least 3 droplets are detected). The ability to detect SARS-CoV-2 falls at viral loads less than 50 copies/mL.
Figure 1.
A, Limit-of-detection study with a 95% (19 of 20) positivity rate at a concentration of 50 copies/mL. Sample 20 failed, with only 1 positive droplet below our cutoff. B, Rate of positivity of internal control (ribonuclease P protein subunit p30 [RPP30]) compared with the number of accepted droplets. The total accepted droplet count is not related to percent positive internal control. Samples with RPP30 < 5% had high-viral-load artifact present.
Accuracy
Residual SARS-CoV-2–positive patient specimens were tested undiluted or diluted in VTM. Thirty positive and 30 negative clinical samples confirmed by an FDA-approved and clinically validated RT-PCR assay (m2000 RealTime System) were used along with 20 diluted positive specimens. Two positive specimens failed quality metrics (droplet count criteria) and were excluded. All specimens were tested in a blinded manner and showed sufficient expression of the internal control (RPP30). All 30 negative and 47 of 48 positive specimens were concordant with the reference quantitative PCR (qPCR) method (97.9% positive precent agreement [PPA] and 100% negative precent agreement [NPA]). The 1 positive specimen that failed had 2 positive N2 droplets and corresponded to a high CN value (25.23), which was above the reference assay’s cutoff.
High-Viral-Load Artifacts
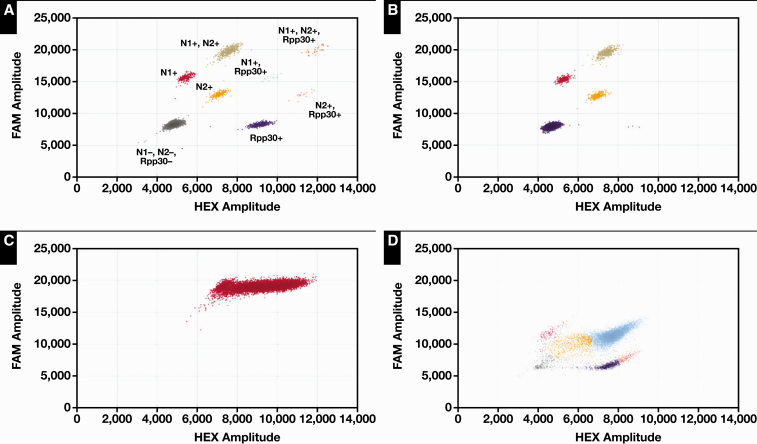
The positive samples for the accuracy study spanned a range of CN values, from 3 to more than 25. Some of the samples exhibited a smear of droplet positions that did not fall into any of the predicted categories for the N1, N2, or RPP30 primers Figure 2B nor into any gates where a combination of primers would be expected Figure 2A. This smear was highly positive for both FAM and HEX and was observed in samples with low CN values (<10), consistent with high viral loads Figure 2C. This phenomenon likely arises from droplets having transcripts labeled with multiple probes for N1 and N2. This unusual finding of a smear population is not commonly reported from ddPCR assays and likely reflects the technology’s deployment for the detection of low-prevalence, inherited genetic variants.
Figure 2.
Examples of severe acute respiratory syndrome coronavirus 2–positive samples. A, All possible clusters displayed and labeled. Each is discrete and easily distinguishable. B, A typical plot of a positive and C, smear artifact observed in cases with high viral load. D, Artifact caused by inhibitors in a positive sample.
Lastly, inhibitors may be present and will present as a smear of droplets from the expected gate position downwards Figure 2D.
Correlation of ddPCR Results to CN Values
A comparison of ddPCR for the detection of SARS-CoV-2 to approved RT-PCR methods has not been described in a clinical laboratory setting. Our current RT-PCR assay has a limit of detection of 100 copies/mL, with a CN value (CN is equivalent to cycle threshold) of 25. Differences in N1 and N2 primer sensitivity have been reported for the RT-PCR method, so we evaluated whether differences existed with the ddPCR testing modality as well.
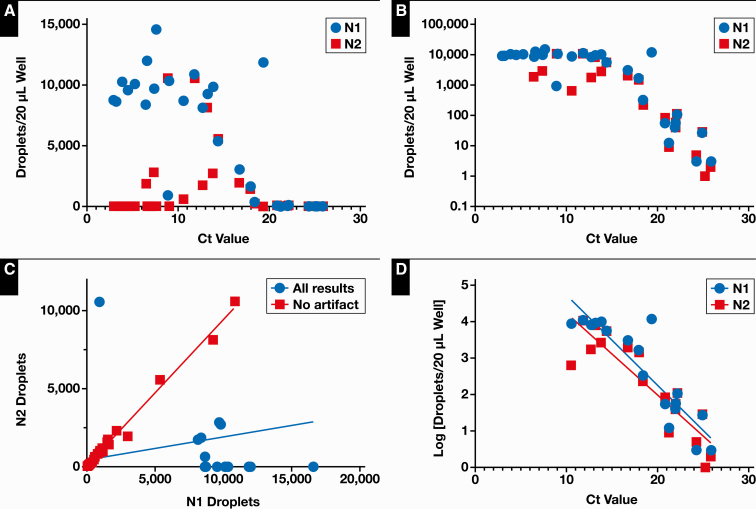
When low CN values were present, the N1 target still worked well, with a high number of positive droplets, but the N2-positive droplets would often be absent Figure 3A and Figure 3B. When the number of N1 and N2 droplets in the same specimen were compared, a bias for high N1 and low N2 was again observed Figure 3C. To determine whether the observed bias for N1-positive droplets is a function of primer or probe compatibility or artifactual, we manually examined the 2-dimensional plots of droplets. For each case of N1 or N2 bias, the result stemmed from smear artifact, which fell into the N1 gate. Thus, there is no apparent bias of N1 to N2 targets.
Figure 3.
A, B, and D compare the concentration of positive droplets vs the cycle threshold (Ct) values for corresponding polymerase chain reaction results. A, Bias toward N1 droplets called positive and N2 called negative at lower Ct values. B, Saturation effect observed at Ct values < 10. C, Comparison of N1-positive droplets with N2-positive droplets in every sample (blue circles) (slope = 0.15; R2 = 0.088). Bias toward N1 or N2 is eliminated when smear artifact data are excluded (red squares) (slope = 0.94; R2 = 0.992). D, Linear relationship for N1 (slope = –0.2475; R2 = 0.81) and N2 (slope = –0.2244; R2 = 0.81) compared with Ct values > 10.
If these samples were excluded, the linear regression of N1-positive to N2-positive droplets (ddPCR) formed a line, with a slope of 0.94 ± 0.01 (R2 = 0.99), indicating strong correlation between probes (Figure 3C). Similarly, when samples with low CN values (<10) were excluded, N1 and N2 had good correlation to CN value (Figure 3D; R2 = 0.815 and 0.806, respectively).
Quality Metrics
Several important quality metrics for ddPCR exist, and they must be considered for a run to be called valid. First, the total number of counted droplets must be over a certain threshold to reach reported sensitivity levels. Internal control RPP30-positive droplets must also be present except when artifact was observed Figure 1B. These cases were considered valid because they were clearly positive for SARS-CoV-2, but they were unquantifiable for RPP30 droplets, which typically require 10 droplets counted.
Variability between runs could emanate from emulsion preparation, which would affect the whole plate. Therefore, we measured variability in droplet formation among each run. There was minimal variability in the number of accepted drops (11,022 ± 1,404) within a run (coefficient of variation [CV], 13%) and between runs (CV, 9%).
Precision Studies
For interrun tests, the mean (SD) SARS-CoV-2 droplet number for the plasmid control (N1 and N2; 4,760,000 copies/mL) and negative control (RPP30) was 1,237 (190) for N1, 1,095 (171) for N2, and 1,096 (120) for RPP30. Mean CV was 15%, 16%, and 11% for N1, N2, and RPP30, respectively. No carryover was observed between positive and negative controls nor between high-viral-load samples and adjacent negative samples.
Resolving Borderline Positive Cases
Specimens with amplification at 38 to 41 CN on the Alinity assay often have poor amplification characteristics and must be repeated to determine whether the sample was a true positive or a false positive. Many times, however, the repeat is positive but still weak, with a high CN value and poor amplification. To resolve this situation, an orthogonal, sensitive assay is required because the implications of a positive result can include quarantine, loss of ability to work, or delay in a medical procedure. Ten specimens with CN greater than 38 repeating weakly positive (CN > 38) were selected and performed on the ddPCR assay. When run as a single specimen, only 50% (5 of 10) of the samples were positive, but repeating the specimens 2 additional times for a total of 3 runs led to detection of positivity in 90% of samples (9 of 10). The “3 positive droplet” criterion was applied additively across all 3 runs. Although the numbers of droplets were at low levels, this finding is consistent with low concentrations of SARS-CoV-2 RNA. Running samples in triplicate improved the sensitivity from 50% to 90%, which resolved our concern regarding repeat weak-positive Alinity samples. Now, we can observe these cases with low viral loads with little concern that a false positive will be present.
Discussion
This clinical validation of ddPCR for COVID-19 molecular testing demonstrates interesting potential artifacts, potentially high throughput, and increased sensitivity (50 copies/mL) compared with an approved and validated qPCR method (100 copies/mL). The sensitivity (97.9% PPA), specificity (100% NPA), precision (9%-13% CV), same-day turnaround time (8 hours for a 96-well plate), and relatively low cost ($29.49 per sample for a full plate; reagent cost and tech time) make the platform appealing for use in pooled sample testing, where high throughput without sacrificing sensitivity is a priority. Overall, ddPCR results for N1 and N2 correlated well with each other and with the qPCR method when viral load was greater than 10 Ct cycles, but cases in which only N1 or N2 have high positive droplet counts warrant manual inspection for potential artifact.
Many of the advantages of ddPCR are derived from its use of end point quantification rather than quantification by real-time amplification used by qPCR.11 This qualitative measurement for each droplet confers increased resistance to inhibitors,12 reduced susceptibility to poor amplification efficiency,13 and compartmentalization of the individual droplets for a higher confidence of low positive results14 because of lower competition for reaction resources with higher-frequency targets.15
To ensure optimal detection, a minimum number of accepted droplets is required for analysis. Our assay requires that a sample have at least 6,000 accepted droplets to be interpreted as SARS-CoV-2 positive and at least 10,000 to be interpreted as SARS-CoV-2 negative. Samples that do not meet the criteria for SARS-CoV-2 detected with fewer than 10,000 droplets were called indeterminate and repeated. Most cases in routine clinical use require a single run in a single well. Detecting human RPP30 served as an internal control for intact RNA collection from a patient. A negative RPP30 can suggest several errors with the sample collection, reagents, or instrument. We noticed that RPP30 could be falsely negative when high-viral-load artifact was present. Positive N1 and/or N2 signals with negative RPP30 suggests low human cell number but still could be resulted as SARS-CoV-2 detected.
Droplet digital PCR technology has been applied widely to circulating tumor DNA detection, but a role in infectious disease and specifically viruses has not been as extensively explored. Examples of viruses that have been detected using ddPCR include hepatitis B virus (HBV), human immunodeficiency virus (HIV), influenza virus, human papillomavirus (HPV), and cytomegalovirus.11,16-21 This technique has been applied to measuring HBV DNA copy number,16 HIV RNA for viral load,19 high-sensitivity detection of influenza A virus, and HPV DNA in cytology and formalin-fixed samples.22 These studies demonstrated increased sensitivity and specificity vs other analytical methods, such as qPCR and serologic testing.16
Multiple studies have shown that ddPCR testing has improved sensitivity and specificity compared with RT-qPCR for the diagnosis of SARS-CoV-2 infection, especially in specimens with low viral load.23-27 In particular, some of the negative RT-PCR samples that were identified as positive using ddPCR assay were confirmed by RT-PCR in follow-up tests.26 It has been reported that ddPCR can also be used to quantitatively monitor a patient’s viral load, evaluate disease progression,27 and directly detect SARS-CoV-2 in crude lysate.9,28 The limits of detection of ddPCR SARS-CoV-2 assays were reported to be between 1.8 and 2.9 copies per reaction,26,29,30 slightly lower than the data reported here.
As ddPCR can detect low levels of SARS-CoV-2 with good sensitivity and specificity, it could potentially be used as a reflex test for equivocally positive RT-PCR results. The clinical significance of low levels of SARS-CoV-2 is still controversial, however. Persistent, asymptomatic SARS-CoV-2 nucleic acid is present in a significant percentage of patients with COVID-19,31 which can delay discharge from the hospital without strong evidence that they are still infectious. Wölfel et al reported that live virus could not be isolated from samples collected after day 8 of symptom onset in patients with COVID-19 despite ongoing high viral RNA load.32 Another study by Bullard et al reported that infectious virus was detected only on respiratory specimens with a high concentration of viral RNA (RT-PCR positive at CT < 24).33 Therefore, it is important to incorporate the ddPCR test results into the clinical contexts of these patients.
Thus, the usefulness of this assay may be best suited to pooled sample testing, where high sensitivity allows for significant pooling. Twenty samples of pooled RNA in this platform would have a theoretical lower limit of detection of ~1,000 copies/mL, sufficient to reliably detect a single positive patient. In SARS-CoV-2 assays, 500 to 1,000 copies/mL is a common benchmark for sensitivity.10,34 False negatives are a concern for pooled specimen testing, but studies35,36 have found that most borderline positive results occur in convalescent patients after 10 to 14 days of symptoms, when infectiousness is thought to be low, because viable virus could not be cultured.37,38 This finding is important for testing large numbers of asymptomatic patients, where a low positivity rate would be expected. Positivity rates would need to be adjusted with pooling numbers to balance the number of retests required.39,40
Disclosure: J.A.S. owns stock in Myriad Genetics.
References
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control Prevention. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. https://www.fda.gov/media/134922/download. Updated December 1, 2020. Accessed March 6, 2021. [Google Scholar]
- 4. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance. https://apps.who.int/iris/bitstream/handle/10665/331501/WHO-COVID-19-laboratory-2020.5-eng.pdf?sequence=1&isAllowed=y.2020. Accessed March 6, 2021.
- 6. Chan JFW, Yip CCY, To KKW, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310-e00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitchell SL, George KS. Evaluation of the COVID19 ID NOW EUA assay. J Clin Virol. 2020;128:104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deiana M, Mori A, Piubelli C, et al. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci Rep. 2020;10:18764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bio-Rad. Bio-Rad SARS-CoV-2 ddPCR test: instructions for use. 2020. [Google Scholar]
- 11. Trypsteen W, Kiselinova M, Vandekerckhove L, et al. Diagnostic utility of droplet digital PCR for HIV reservoir quantification. J Virus Erad. 2016;2:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sedlak RH, Jerome KR. Viral diagnostics in the era of digital polymerase chain reaction. Diagn Microbiol Infect Dis. 2013;75:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pekin D, Skhiri Y, Baret JC, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11:2156-2166. [DOI] [PubMed] [Google Scholar]
- 15. Gutiérrez-Aguirre I, Rački N, Dreo T, et al. Droplet digital PCR for absolute quantification of pathogens. Plant Pathol. 2015;1302:331-347. [DOI] [PubMed] [Google Scholar]
- 16. Huang JT, Liu YJ, Wang J, et al. Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue. Clin Chem. 2015;61:290-296. [DOI] [PubMed] [Google Scholar]
- 17. Larsson GL, Helenius G. Digital droplet PCR (ddPCR) for the detection and quantification of HPV 16, 18, 33 and 45-a short report. Cell Oncol. 2017;40:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mauvisseau Q, Davy-Bowker J, Bulling M, et al. Combining ddPCR and environmental DNA to improve detection capabilities of a critically endangered freshwater invertebrate. Sci Rep. 2019;9:14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strain MC, Lada SM, Luong T, et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8:e55943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rowlands V, Rutkowski AJ, Meuser E, et al. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci Rep. 2019;9:12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruelle J, Yfantis V, Duquenne A, et al. Validation of an ultrasensitive digital droplet PCR assay for HIV-2 plasma RNA quantification. J Int AIDS Soc. 2014;17:19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor SC, Carbonneau J, Shelton DN, et al. Optimization of droplet digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: clinical implications for quantification of oseltamivir-resistant subpopulations. J Virol Methods. 2015;224:58-66. [DOI] [PubMed] [Google Scholar]
- 23. Lu R, Wang J, Li M, et al. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis, treatment monitoring and criteria for discharge [published online March 30, 2020]. medRxiv. doi: 10.1101/2020.03.24.20042689. [DOI] [Google Scholar]
- 24. Liu X, Feng J, Zhang Q, et al. Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg Microbes Infect. 2020;9:1175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Falzone L, Musso N, Gattuso G, et al. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med. 2020;46:957-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suo T, Liu X, Feng J, et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu F, Yan L, Wang N, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71:793-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vasudevan H, Xu P, Servellita V, et al. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification [published online September 3, 2020]. medRxiv. doi: 10.1101/2020.09.02.20186023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alteri C, Cento V, Antonello M, et al. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. 2020;15:e0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dong L, Zhou J, Niu C, et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR [published online March 30, 2020]. medRxiv. doi: 10.1101/2020.03.14.20036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ikegami S, Benirschke R, Flanagan T, et al. Persistence of SARS-CoV-2 nasopharyngeal swab PCR positivity in COVID-19 convalescent plasma donors. Transfusion. 2020;60:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465-469. [DOI] [PubMed] [Google Scholar]
- 33. Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples [published online May 22, 2020]. Clin Infect Dis. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. SoRelle JA, Frame I, Falcon A, et al. Clinical validation of a SARS-CoV-2 real-time reverse transcription PCR assay targeting the nucleocapsid gene. J Appl Lab Med. 2020;5:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lohse S, Pfuhl T, Berkó-Göttel B, et al. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020;20:1231-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. SoRelle JA, Mahimainathan L, McCormick-Baw C, et al. Saliva for use with a point of care assay for the rapid diagnosis of COVID-19. Clin Chim Acta. 2020;510:685-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kujawski SA, Wong KK, Collins JP, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi J, Han D, Zhang R, et al. Molecular and serological assays for SARS-CoV-2: insights from genome and clinical characteristics. Clin Chem. 2020;66:1030-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sahajpal NS, Mondal AK, Njau A, et al. Proposal of RT-PCR-based mass population screening for severe acute respiratory syndrome coronavirus 2 (coronavirus disease 2019). J Mol Diagn. 2020;22:1294-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freire-Paspuel B, Vega-Mariño P, Velez A, et al. Sample pooling of RNA extracts to speed up SARS-CoV-2 diagnosis using CDC FDA EUA RT-qPCR kit. Virus Res. 2020;290:198173. [DOI] [PMC free article] [PubMed] [Google Scholar]