Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus that has given rise to the devastating global pandemic. In most cases, SARS-CoV-2 infection results in the development of viral pneumonia and acute respiratory distress syndrome, known as ‘coronavirus disease 2019’ or COVID-19. Intriguingly, besides the respiratory tract, COVID-19 affects other organs and systems of the human body. COVID-19 patients with pre-existing cardiovascular disease have a higher risk of death, and SARS-CoV-2 infection itself may cause myocardial inflammation and injury. One possible explanation of such phenomena is the fact that SARS-CoV-2 utilizes angiotensin-converting enzyme 2 (ACE2) as the receptor required for viral entry. ACE2 is expressed in the cells of many organs, including the heart. ACE2 functions as a carboxypeptidase that can cleave several endogenous substrates, including angiotensin II, thus regulating blood pressure and vascular tone. It remains largely unknown if the SARS-CoV-2 infection alters the enzymatic properties of ACE2, thereby contributing to cardiovascular complications in patients with COVID-19. Here, we demonstrate that ACE2 cleavage of des-Arg9-bradykinin substrate analogue is markedly accelerated, while cleavage of angiotensin II analogue is minimally affected by the binding of spike protein. These findings may have implications for a better understanding of COVID-19 pathogenesis.
Keywords: ACE2, COVID-19, SARS-CoV-2, spike protein
Graphical abstract
The recent COVID-19 pandemic caused by the novel coronavirus SARS-CoV-2 represents a significant threat to human health. To date, more than two million people worldwide have died of COVID-19 or COVID-19-related complications. SARS-CoV-2 is both structurally and phenotypically similar to another strain of coronavirus, SARS-CoV-1, that caused an outbreak of severe acute respiratory syndrome in 2003 (1). Similarly to SARS-CoV-1, SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE2) receptor to gain entry to mammalian cells (2, 3). SARS-CoV-2 attaches to ACE2 via glycosylated spike (S) protein trimers on the virion surface (4). Interestingly, SARS-CoV-2 spike protein has a much higher binding affinity to ACE2 than does that of SARS-CoV-1 (KD, app = 3.9 × 10−11 M compared with 1.2 × 10−10 M, respectively) (5). The receptor-binding domain (RBD) of SARS-CoV-2 spike protein is recognized by the extracellular domain of ACE2 mainly through polar residues (6). Interestingly, the virus hijacks the ACE2 proteolytic N-terminal domain, but the binding site itself is located away from the ACE2 active site (7), suggesting that even upon binding of the virus, ACE2 is likely to maintain some proteolytic activity. The binding of spike protein to ACE2 is also conformation-dependent, suggesting that it could affect conformational states of ACE2. X-ray crystallography studies of the unbound or inhibitor-bound forms of ACE2 also reveal a striking difference in the structure of the active site that impacts a number of surrounding residues, including the ACE2 residues implicated in binding to the SARS-CoV-2 spike protein (8).
ACE2 normally functions as a zinc-dependent carboxypeptidase (9), and plays an important cardio-protective role as part of the renin–angiotensin–aldosterone system (RAAS) by catalysing the cleavage of the vasoconstricting peptide angiotensin II into the vasodilating angiotensin 1–7. ACE2 also has other known, and likely unknown, peptide-based substrates, such as apelin 13 (which has known cardioactive functions) (10, 11), des-Arg(9)-bradykinin (an immune modulator) (12, 13) and dynorphin A (13), and thus may contribute to the regulation of blood pressure, vascular tone and various immune pathways. ACE2 is expressed by pulmonary alveolar cells and cells of many other organs and tissues including vascular endothelium, heart and kidneys (9). The lung likely serves as the primary point of viral entry, but it remains unclear to what extent other tissues are directly infected by SARS-CoV-2. Nevertheless, there are numerous clinical reports showing that COVID-19 result in increased mortality in patients with pre-existing conditions, such as hypertension, diabetes, obesity, as well as patients with cancer and a compromised immune system (14–17). A growing number of studies highlight that COVID-19 patients develop acute myocardial injury with potentially chronic effects on the cardiovascular system. In some cases, patients without any pre-existing heart problems had signs of ongoing myocardial inflammation after apparent recovery from COVID-19 (18). However, it remains unknown if the cardiac symptoms are developed as a result of the direct infection of myocardial cells or as a secondary event associated with infection-mediated effects such as cytokine storm (19).
A better understanding of the mechanisms underlying the interaction between ACE2 and SARS-CoV-2 is of high importance. First, it will likely be crucial to developing effective strategies for COVID-19 treatment. Several studies have shown that soluble recombinant ACE2 is able to act as a molecular trap to bind SARS-CoV-2, thus preventing or limiting infection (20, 21). Second, it will help to elucidate how SARS-CoV-2 infection affects the function of ACE2 and downstream signalling. Moreover, SARS-CoV-2 can potentially bind to a catalytically active soluble form of ACE2 (sACE2), which lacks the transmembrane domain, is present in the bloodstream, and is able to catalyse Ang II conversion (9). Altered ACE2 function in the setting of viral infection might contribute to the regulation of proteolytic cascades that could account for some of the many unusual characteristics of COVID-19, including cardiac dysfunction, vascular disease and thrombosis. Hence, we sought to test if SARS-CoV-2 binding to ACE2 affects its enzymatic properties. Here, we show that the binding of SARS-CoV-2 enhances ACE2 peptidase activity in vitro.
Materials and Methods
Recombinant proteins and other reagents
The extracellular catalytic domain of recombinant human ACE2, SARS-CoV-2 spike full-length and spike RBD, SARS-CoV-1 RBD proteins and ACE2/Caspase-1 quenched fluorogenic peptide pseudosubstrate were obtained from R&D systems (cat# 933-ZN, 10549-CV, 10500-CV-100, 10558-CV-100 and ES007, respectively, R&D Systems Inc., MN, USA). The Caspase-1 pseudosubstrate contained an N-terminal 7-methoxycoumarin (MCA) and a C-terminal Lys-linked 2,4-dinitrophenyl quencher [K(Dnp)] in the peptide MCA-YVADAPK(Dnp). Recombinant SARS-CoV-2 RBD spike mutant (Y505C) was obtained from Sino Biological (cat# 40592-V08H72, Sino Biological US Inc., PA, USA). The other quenched fluorogenic substrates were custom synthesized by GenScipt (GenScript Biotech, NJ, USA). SARS-CoV-2 spike full-length and RBD and SARS-CoV-1 spike RBD proteins were used in Fig. 1. Other recombinant RBD spike and RBD (G502D) proteins of SARS-CoV-2 (Arg319-Phe541 with a C-terminal 6-His tag; NCBI Reference Sequence: YP_009724390.1) were created in our laboratory using previously described methods (22) and were used in Fig. 2 and Supplementary Fig. S1. The G502D variant was created by introducing a nucleotide substitution in the RBD-expressing plasmid using the QuikChange II Site-directed mutagenesis kit (cat# 200523, Agilent, CA, USA) with PCR conditions according to manufacturer’s instructions. The plasmid was expressed in 293F cells and the protein was purified as described previously (22). The heat-inactivated SARS-CoV-2 (strain 2019nCoV/USAWA1/2020; inactivated at 65°C for 30 min) was obtained from ATCC (cat# VR1986HK, ATCC, VA, USA). Inactivation at this temperature was intended to leave viral epitopes intact, though partial denaturation may occur (23).
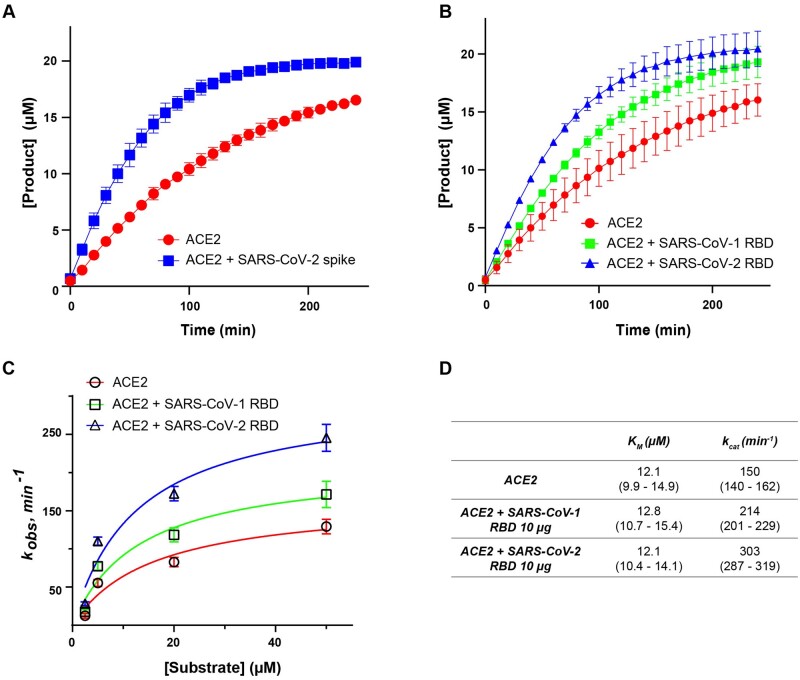
Fig. 1.
Binding of the SARS-CoV-2 spike protein increases the rate of ACE2 activity. (A) Kinetic curves showing the effect of full-length SARS-CoV-2 spike on ACE2 activity. (B) Kinetic curves showing the effect of SARS-CoV-1 and SARS-CoV-2 RBD spike on ACE2 activity. (C) Michaelis–Menten plot showing effect of SARS-CoV-1 and SARS-CoV-2 RBD on catalytic activity of ACE2. kobs, observed rate constant. (D) Catalytic rate (kcat) and KM of ACE2 in the absence or presence of SARS-CoV-1 and SARS-CoV-2 RBD spike protein [mean (95% confidence intervals)]. Pseudosubstrate concentration in (A) and (B) is 20 µM. Assays were conducted in two biological replicates.
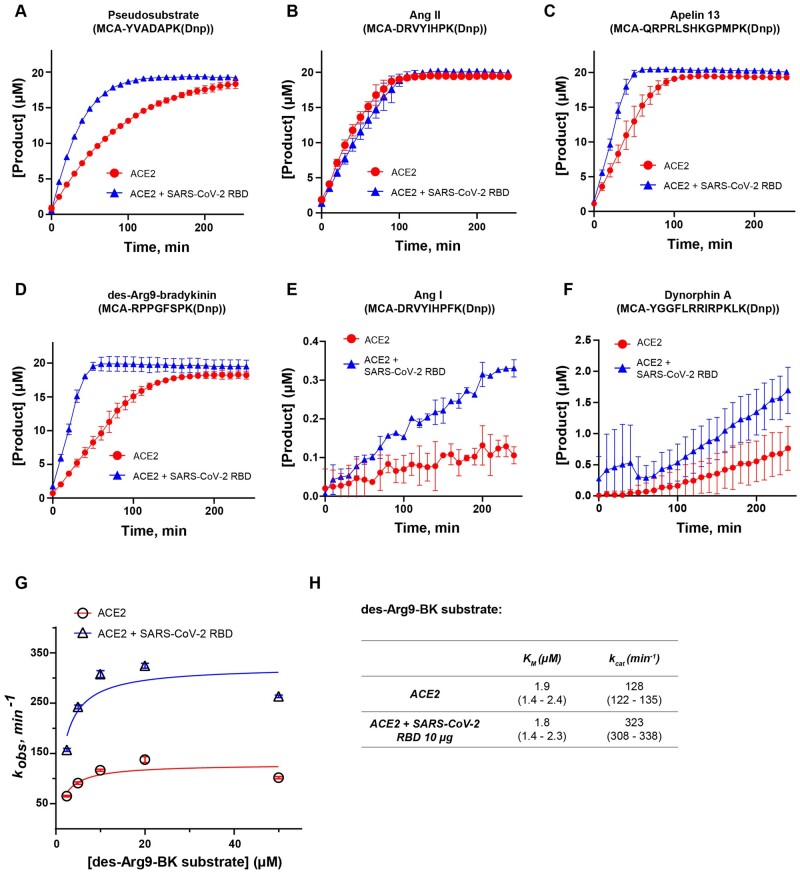
Fig. 2.
SARS-CoV-2 spike protein accelerates the activity of ACE2 in a substrate-dependent manner. Kinetic curves showing the effect of SARS-CoV-2 spike RBD binding on ACE2 activity in the presence of (A) pseudosubstrate MCA-YVADAPK(Dnp); (B) angiotensin II mimic [MCA-DRVYIHPK(Dnp)]; (C) apelin 13 mimic [MCA-QRPRLSHKGPMPK(Dnp)]; (D) des-Arg9-bradykinin mimic [MCA-RPPGFSPK(Dnp)] (E) angiotensin I mimic [MCA-DRVYIHPFK(Dnp)]; (F) dynorphin A mimic [MCA-YGGFLRRIRPKLK(Dnp)] substrates; (G) Michaelis–Menten plot showing effect of SARS-CoV-2 RBD on catalytic activity of ACE2 in the presence of des-Arg9-bradykinin mimic substrate. (H) Catalytic rate (kcat) and KM of ACE2 in the absence or presence of SARS-CoV-2 RBD spike protein and des-Arg9-bradykinin mimic substrate [mean (95% confidence intervals)]. Substrate concentration in (A)–(F) is 20 µM. Assays were conducted in two biological replicates.
Quantification of SARS-CoV-2 viral genome copy (VGC) number
The VGC number was quantified by RT-qPCR using the calculated Ct-value determined from the standard curve. Briefly, 5 µl of the inactivated virus or 5 µl of quantitative synthetic SARS-CoV-2 RNA (cat# VR-3276SD, ATCC, VA, USA) were reverse transcribed using SuperScript III Reverse Transcriptase (cat# 18080044, Invitrogen, CA, USA) following the manufacturer’s instructions. The resulting cDNA from the virus was diluted 1:50; cDNA from the synthetic standards was diluted 1:5, 1:10, 1:50, 1:100, and used to generate a standard curve. The samples were analysed by real-time PCR with SYBR green detection reagent (cat# 4367659, Invitrogen, CA, USA) using StepOne Real-Time PCR System (Applied Biosystems, CA, USA) and the primers targeting N gene (encoding nucleocapsid protein of SARS-CoV-2): F: 5′-ATGCTGCAATCGTGCTACAA-3′; R: 5′-CCTCTGCTCCCTTCTGCGTA-3′.
ACE2 peptidase activity assay
The assay was performed in 96-well black, clear-bottom plates (cat# 3603, Corning, NY, USA). Each reaction included 50 µl (ACE2 + RBD or ACE2 + the inactivated virus) containing 1.5-10 ng (0.17–1.13 nM) of ACE2 diluted in assay buffer [AB, 75 mM Tris, 150 mM NaCl (pH = 7.3)]. ACE2 was preincubated with diluent control, inactivated virus (3.8 × 105 VGC/µl stock), or recombinant spike protein at 28°C for 30 min. To initiate the reaction, 50 µl of the quenched fluorogenic substrate at various concentrations was added. The ACE2 activity was measured in relative fluorescent units (RFUs) with excitation at 320 nm and emission at 405 nm at 28°C in a kinetic mode using Synergy H1 plate reader (BioTek, VT, USA) with readings recorded every 10 min. The RFU values for each reaction were corrected by subtracting the background. Corrected RFUs were then converted into molar concentrations by calculating a conversion factor (CF) established by the generation of a product control. Molar concentrations of the converted product for the less efficient ACE2 substrates, such as Ang I and Dynorphin A, were calculated using the CF for Ang II. The initial rates were calculated based on measurements over the first hour, at which point turnover was linear with respect to time and turnover was limited to <20% substrate conversion. Steady-state kinetic constants were then calculated by fitting to a hyperbolic Michaelis–Menten plot. Assays were conducted for a minimum of two biological replicates.
Statistical analysis
Statistical analysis was performed using linear regression analysis, two-way analysis of variance (Tukey test), nonlinear regression analysis for Michaelis–Menten kinetics, and exponential plateau equation to estimate the maximum of the converted substrate for each kinetic assay using GraphPad PRISM 9.0.0 software (GraphPad Software, Inc.).
Results
Binding of SARS-CoV-2 spike protein to ACE2 accelerates its peptidase activity
As a carboxypeptidase, ACE2 cleaves many biological substrates besides angiotensin II (Ang II). In general, ACE2 hydrolyzes a bond between proline and a C-terminal hydrophobic amino acid in peptides that resembles the following sequence: proline-X(1-3 residues)-proline-hydrophobic amino acid (13). One well-established method to analyse the activity of peptidases in vitro is to measure the concentration of the cleaved product over time in the presence of internally quenched fluorogenic peptides (24, 25). These peptides are usually flanked by a fluorophore and a quencher that inhibits the fluorophore in an intact state. The inhibition is liberated upon cleavage of the peptide by a protease—the peptide fragment with the fluorophore is released from the quencher, and the fluorescent signal is emitted (25). Therefore, to test the effect of the viral spike protein binding on the peptidase activity of ACE2, we first performed experiments to evaluate the ability of ACE2 to cleave a commercially available fluorogenic pseudosubstrate MCA-YVADAPK(Dnp) (commonly used to measure ACE2 activity, and also known to be a substrate for Caspase-1). First, we measured ACE2 activity, with and without preincubation with SARS-CoV-2 trimeric full-length spike protein, in the presence of MCA-YVADAPK(Dnp) substrate. Under these conditions, we observed a significant increase in the ACE2 activity rate in the presence of the spike protein (Fig. 1A). We observed a similar effect by preincubating ACE2 with a shorter version of recombinant spike protein that contains only the RBD (Ala319-Phe541) of SARS-CoV-2 (Fig. 1B), suggesting that RBD spike is sufficient to accelerate ACE2 activity. The RBD domain of SARS-CoV-1 was also able to accelerate ACE2 activity, albeit to a lesser extent than that of SARS-CoV-2, consistent with the known lower binding affinity of SARS-CoV-1 (Fig. 1B). We also quantified ACE2 steady-state kinetics in the absence and presence of SARS-CoV-1 and SARS-CoV-2 spike RBDs (Fig. 1C and D). The addition of SARS-CoV-2 RBD results in an approximate doubling of enzyme activity (kcat) with no change in KM. A smaller activation of kcat (∼42%) was induced by SARS-CoV-1 RBD (Fig. 1C and D). In addition, we demonstrated that SARS-CoV-2 RBD spike affects ACE2 kcat in a dose-dependent manner (Supplementary Fig. S1).
SARS-CoV-2 spike protein accelerates the activity of ACE2 in a substrate-specific fashion
Next, we sought to determine whether the acceleration of ACE2 catalytic activity produced by spike protein binding that is observed when using the commercially available pseudosubstrate is also evident in the presence of endogenous substrates. ACE2 is known to cleave multiple physiological substrates. Besides controlling blood pressure and fluid balance as a part of the RAAS and apelin signalling (26), ACE2 negatively regulates the plasma kallikrein–kinin system (KKS) by cleaving des-Arg9-bradykinin which is released in response to tissue injury or inflammatory stimuli (12). Thus, we synthesized several internally quenched fluorogenic substrates that mimic known biological substrates of ACE2, including angiotensin II (Ang II) [MCA-DRVYIHPK(Dnp)], apelin 13 [MCA-QRPRLSHKGPMPK(Dnp)] and des-Arg9-bradykinin [MCA-RPPGFSPK(Dnp)]. We also included two peptides that are normally less efficiently cleaved by ACE2: dynorphin A [MCA-YGGFLRRIRPKLK(Dnp)] and angiotensin I (Ang I) [MCA-DRVYIHPFK(Dnp)]. We preincubated ACE2 with SARS-CoV-2 RBD spike, added 20 µM of each substrate, and measured the accumulation of product over time (Fig. 2). RBD failed to activate ACE2 catalytic activity when Ang II was used as a substrate (Fig. 2B). However, catalytic activity was increased by RBD when each of the other substrates was utilized (Fig. 2C–F). The greatest increase in the rate of ACE2 activity was observed in the presence of des-Arg9-bradykinin as substrate (Fig. 2D). Taken together, these findings suggest substrate-specific enhancement of ACE2 enzymatic activity produced by spike RBD binding.
Next, we quantified ACE2 steady-state kinetics in the presence or absence of SARS-CoV-2 spike RBD against des-Arg9-bradykinin and found there to be an ∼2.5-fold increase in ACE2 enzymatic activity (kcat) with no significant change in KM produced by addition of spike RBD (Fig. 2G and H). Notably, we observed that ACE2 activity dropped off when using the highest concentration of des-Arg9-bradykinin substrate (50 µM) suggesting a possibility of substrate inhibition (Fig. 2G). When fitting observed rates to an alternative model that includes substrate inhibition, the key conclusions did not change, and we still observed ∼2.7-fold increase in kcat in the presence of spike RBD (data not shown).
In addition, we tested if two spike RBD mutants (G502D and Y505C) with a lower affinity to ACE2 (5) would still affect enzymatic activity. We observed that both mutants accelerated the activity of ACE2 but significantly less than spike RBD wild-type (Supplementary Fig. S2), suggesting that the increase in ACE2 activity also depends on the affinity to spike RBD.
SARS-CoV-2 virus binding to ACE2 enhances catalytic activity and cleavage of des-Arg9-bradykynin
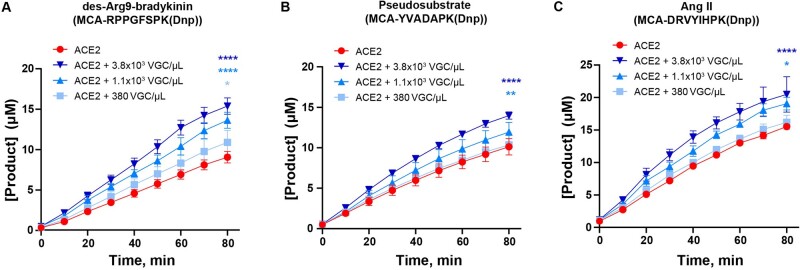
Next, we sought to determine if binding of intact SARS-CoV-2 viral particles to ACE2 would mimic the effects on ACE2 catalytic activity observed when purified spike protein or RBD was used. Therefore, we measured ACE2 activity against three substrates [des-Arg9-bradykinin, the pseudosubstrate MCA-YVADAPK(Dnp), and Ang II] in the presence of varying concentrations of intact virus. A significant effect on ACE2 catalytic activity was observed in the presence of the highest concentration of the virus (3.8 × 103 VGCs/µl) when tested against all three substrates (Fig. 3). At the same time, the addition of the lower concentration of the virus (1.1 × 103 VGC/µl) resulted in a more modest acceleration of ACE2 proteolytic activity towards Ang II and relatively little against the pseudosubstrate MCA-YVADAPK(Dnp) (Fig. 3B and C). In the case of the des-Arg9-bradykinin substrate, ACE2 activity was significantly accelerated even upon the addition of the lowest concentration of the virus (380 VGC/µl) (Fig. 3A).
Fig. 3.
Binding of the heat-inactivated SARS-CoV-2 viral particles accelerates ACE2 catalytic activity. (A–C) Kinetic curves showing the effect of different concentrations of heat-inactivated SARS-CoV-2 on ACE2 activity in the presence of des-Arg9-bradykinin mimic [MCA-RPPGFSPK(Dnp)] substrate (A); MCA-YVADAPK(Dnp) substrate (B); and angiotensin II mimic [MCA-DRVYIHPK(Dnp)] substrate (C). Substrate concentration in (A)–(C) is 20 µM. Assays were conducted in two biological replicates.
Discussion
The COVID-19 pandemic represents a significant threat to public health and there are still numerous unanswered questions regarding the mechanisms of COVID-19 pathogenesis. Our results suggest that SARS-CoV-2 binding to ACE2 can affect its catalytic activity in a substrate-specific manner, suggesting that various signalling cascades normally regulated by ACE2 may be affected in patients suffering from COVID-19. Changes in ACE2 activity could contribute to vascular, thrombotic and cardiac complications of infection by altering local peptide mediators or perturbing the systemic action of peptide hormones. Supporting this hypothesis is one short report showing that plasma ACE2 activity levels are significantly increased and remain elevated up to an average of 114 days post-COVID-19 infection (27).
The results of our study are in general agreement with results recently published by Lu and Sun (28), although we found some significant differences. In that study, the authors also observed enhancement of ACE2 catalytic activity upon SARS-CoV-2 spike RBD binding, with some variation depending on the substrate (Caspase-1 pseudosubstrate versus a bradykinin analogue) (28). However, they reported a significant decrease in KM upon spike binding which we did not reproduce under our assay conditions. Also, Lu and Sun did not directly measure catalytic activity using an Ang II substrate analogue, an important distinction as we did not observe significant enhancement with this substrate. Notably, the authors used a bradykinin analogue substrate [Mca-RPPGFSAFK(Dnp)] that does not fully replicate the sequence of the ‘classic’ bradykinin peptide (RPPGFSPFR, sequence ID: AAH60039.1). It is also known that ACE2 has no catalytic activity against bradykinin (13). Additionally, the rates of catalysis (kcat) that the authors reported with the pseudosubstrate are lower than what we observed, likely attributable to different assay conditions or enzyme and substrate preparations. Importantly, however, our studies support the general observation that spike RBD binding to ACE2 can lead to higher rates of catalysis, and our work extends these findings by emphasizing the significant differences in this effect dependent upon substrate and the affinity to SARS-CoV-2 spike. Furthermore, our results newly demonstrate that the effect is not confined to isolated RBD, but that intact SARS-CoV-2 virus can also enhance ACE2 activity.
The relevance of our in vitro studies to COVID-19 pathogenesis will require further studies including the assessment of both cell surface and soluble ACE2 activity and various cleavage products in infected patients. Prior work has suggested that SARS-CoV-1 infection results in downregulation of cell surface ACE2, likely due to internalization and/or shedding of bound receptor (29, 30). However, similar downregulation upon SARS-CoV-2 binding has not been reported to date in experimental models up to several days after infection (31, 32). Even if viral infection in humans does result in altered cell surface concentrations of ACE2 in some tissues, short-term changes in cell surface ACE2 activity, and that of soluble ACE2, could be clinically relevant. The observation of substrate specificity, with little effect of spike binding when Ang II is the substrate, suggests that the primary effects in vivo may be on pathways other than the renin–angiotensin–aldosterone pathway.
Our findings, together with data published by other groups, point to the importance of additional studies to decipher changes related to ACE2 activity in vivo in the context of COVID-19 infection, such as the overall receptor expression levels or other feedback regulatory mechanisms. Results presented here should spur both diagnostic and therapeutic advances necessary to overcome the COVID-19 pandemic.
Supplementary Data
Supplementary Data are available at JB Online.
Funding
This work was supported by the NIH [R35 HL140018 to J.A.E.], the Cotswold Foundation (to J.A.E.) and the WW Smith endowed chair (to J.A.E.).
Conflict of Interest
None declared.
Supplementary Material
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- CF
conversion factor
- RBD
receptor-binding domain
- RFUs
relative fluorescent units
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2.
References
- 1. LeDuc J.W., Barry M.A. (2004) SARS, the first pandemic of the 21st century. Emerg. Infect. Dis. 10, e26 [Google Scholar]
- 2. Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wan Y., Shang J., Graham R., Baric R.S., Li F. (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. (2020) Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 [DOI] [PubMed] [Google Scholar]
- 8. Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. (2004) ACE2 X-ray structures reveal a large Hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 279, 17996–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. (2020) Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 126, 1456–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang P., Kuc R.E., Brame A.L., Dyson A., Singer M., Glen R.C., Cheriyan J., Wilkinson I.B., Davenport A.P., Maguire J.J. (2017) [Pyr1]apelin-13(1-12) is a biologically active ACE2 metabolite of the endogenous cardiovascular peptide [Pyr1]apelin-13. Front. Neurosci. 11, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu P., Wysocki J., Serfözö P.D., Ye M., Souma T., Batlle D., Jin J. (2017) A fluorometric method of measuring carboxypeptidase activities for angiotensin II and apelin-13. Sci. Rep. 7, 45473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sodhi C.P., Wohlford-Lenane C., Yamaguchi Y., Prindle T., Fulton W.B., Wang S., McCray P.B., Chappell M., Hackam D.J., Jia H. (2018) Attenuation of pulmonary ACE2 activity impairs inactivation of des-arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. Am. J. Physiol. Lung Cell. Mol. Physiol. 314, L17–L31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. (2002) Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 277, 14838–14843 [DOI] [PubMed] [Google Scholar]
- 14. Roshanravan N., Ghaffari S., Hedayati M. (2020) Angiotensin converting enzyme-2 as therapeutic target in COVID-19. Diabetes Metab. Syndr. 14, 637–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Magdy Beshbishy A., Hetta H.F., Hussein D.E., Saati A.A., C. Uba C., Rivero-Perez N., Zaragoza-Bastida A., Shah M.A., Behl T., Batiha G.E.-S. (2020) Factors associated with increased morbidity and mortality of obese and overweight COVID-19 patients. Biology 9, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hussain A., Bhowmik B., do Vale Moreira N.C. (2020) COVID-19 and diabetes: knowledge in progress. Diabetes Res. Clin. Pract. 162, 108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L., Zhu F., Xie L., Wang C., Wang J., Chen R., Jia P., Guan H.Q., Peng L., Chen Y., Peng P., Zhang P., Chu Q., Shen Q., Wang Y., Xu S.Y., Zhao J.P., Zhou M. (2020) Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann. Oncol. 31, 894–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Puntmann V.O., Carerj M.L., Wieters I., Fahim M., Arendt C., Hoffmann J., Shchendrygina A., Escher F., Vasa-Nicotera M., Zeiher A.M., Vehreschild M., Nagel E. (2020) Outcomes of cardiovascular magnetic resonance imaging in patients recently recovered from coronavirus disease 2019 (COVID-19). JAMA Cardiol. 5, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Unudurthi S.D., Luthra P., Bose R.J.C., McCarthy J., Kontaridis M.I. (2020) Cardiac inflammation in COVID-19: lessons from heart failure. Life Sci. 260, 118482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. (2020) Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905–913.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glasgow A., Glasgow J., Limonta D., Solomon P., Lui I., Zhang Y., Nix M.A., Rettko N.J., Zha S., Yamin R., Kao K., Rosenberg O.S., Ravetch J.V., Wiita A.P., Leung K.K., Lim S.A., Zhou X.X., Hobman T.C., Kortemme T., Wells J.A. (2020) Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. USA 117, 28046–28055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flannery D.D., Gouma S., Dhudasia M.B., Mukhopadhyay S., Pfeifer M.R., Woodford E.C., Gerber J.S., Arevalo C.P., Bolton M.J., Weirick M.E., Goodwin E.C., Anderson E.M., Greenplate A.R., Kim J., Han N., Pattekar A., Dougherty J., Kuthuru O., Mathew D., Baxter A.E., Vella L.A., Weaver J., Verma A., Leite R., Morris J.S., Rader D.J., Elovitz M.A., Wherry E.J., Puopolo K.M., Hensley S.E. (2020) SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sci. Immunol. 5, eabd5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loveday E.K., Hain K.S., Kochetkova I., Hedges J.F., Robison A., Snyder D.T., Brumfield S.K., Young M.J., Jutila M.A., Chang C.B., Taylor M.P.. Effect of inactivation methods on SARS-CoV-2 virion protein and structure. bioRxiv 2020.11.14.383026; doi:10.1101/2020.11.14.383026 [DOI] [PMC free article] [PubMed]
- 24. Fields G.B. (2001) Using fluorogenic peptide substrates to assay matrix metalloproteinases. Methods Mol. Biol. 151, 495–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kasperkiewicz P., Poreba M., Groborz K., Drag M. (2017) Emerging challenges in the design of selective substrates, inhibitors and activity-based probes for indistinguishable proteases. FEBS J. 284, 1518–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benjamin E.J., Muntner P., Alonso A., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Das S.R., Delling F.N., Djousse L., Elkind M.S.V., Ferguson J.F., Fornage M., Jordan L.C., Khan S.S., Kissela B.M., Knutson K.L., Kwan T.W., Lackland D.T., Lewis T.T., Lichtman J.H., Longenecker C.T., Loop M.S., Lutsey P.L., Martin S.S., Matsushita K., Moran A.E., Mussolino M.E., O'Flaherty M., Pandey A., Perak A.M., Rosamond W.D., Roth G.A., Sampson U.K.A., Satou G.M., Schroeder E.B., Shah S.H., Spartano N.L., Stokes A., Tirschwell D.L., Tsao C.W., Turakhia M.P., VanWagner L.B., Wilkins J.T., Wong S.S., Virani S.S., and American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee (2019) Heart disease and stroke statistics-2019 update: a report from the American Heart Association. Circulation 139, e56–e528 [DOI] [PubMed] [Google Scholar]
- 27. Patel S.K., et al. (2021) Plasma ACE2 activity is persistently elevated following SARS-CoV-2 infection: implications for COVID-19 pathogenesis and consequences. Eur. Respir. J. 2003730; doi:10.1183/13993003.03730-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu J., Sun P.D. (2020) High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity. J. Biol. Chem. 295, 18579–18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oudit G.Y., Kassiri Z., Jiang C., Liu P.P., Poutanen S.M., Penninger J.M., Butany J. (2009) SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Invest. 39, 618–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. (2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao L., Liu P., Zhao L., Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. (2020) The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583, 830–833 [DOI] [PubMed] [Google Scholar]
- 32. Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R., Gralinski L.E., Schäfer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M.L., Livraghi-Butrico A., Nicely N.I., Cameron M., Cameron C., Kelvin D.J., de Silva A., Margolis D.M., Markmann A., Bartelt L., Zumwalt R., Martinez F.J., Salvatore S.P., Borczuk A., Tata P.R., Sontake V., Kimple A., Jaspers I., O’Neal W.K., Randell S.H., Boucher R.C., Baric R.S. (2020) SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.