Abstract
Background
We investigated frequency of reinfection with seasonal human coronaviruses (HCoVs) and serum antibody response following infection over 8 years in the Household Influenza Vaccine Evaluation (HIVE) cohort.
Methods
Households were followed annually for identification of acute respiratory illness with reverse-transcription polymerase chain reaction–confirmed HCoV infection. Serum collected before and at 2 time points postinfection were tested using a multiplex binding assay to quantify antibody to seasonal, severe acute respiratory syndrome coronavirus (SARS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike proteins and SARS-CoV-2 spike subdomains and N protein.
Results
Of 3418 participants, 40% were followed for ≥3 years. A total of 1004 HCoV infections were documented; 303 (30%) were reinfections of any HCoV type. The number of HCoV infections ranged from 1 to 13 per individual. The mean time to reinfection with the same type was estimated at 983 days for 229E, 578 days for HKU1, 615 days for OC43, and 711 days for NL63. Binding antibody levels to seasonal HCoVs were high, with little increase postinfection, and were maintained over time. Homologous, preinfection antibody levels did not significantly correlate with odds of infection, and there was little cross-response to SARS-CoV-2 proteins.
Conclusions
Reinfection with seasonal HCoVs is frequent. Binding anti-spike protein antibodies do not correlate with protection from seasonal HCoV infection.
Keywords: seasonal coronavirus, SARS-CoV-2, COVID-19, reinfection, antibody, waning, household cohort, serology, correlates of protection, immunity
Reinfection with seasonal coronaviruses was frequent over 8 years. Anti-spike protein binding antibody levels to seasonal coronaviruses were high, with little increase postinfection, and were maintained over time. These antibodies did not correlate with protection from infection.
Prior to the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), it was recognized that coronaviruses that infect humans (HCoVs) could be separated into the 4 seasonal, or common, coronaviruses (229E, OC43, NL53, and HKU1), which regularly cause mainly mild respiratory illnesses, and severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), which have caused epidemics of severe lower respiratory disease [1–4]. The current coronavirus disease 2019 (COVID-19) pandemic, the first recognized to be caused by an HCoV, has focused attention on the seasonal coronaviruses in comparison to SARS-CoV-2. Of particular importance are questions around possible cross-protection or enhancement of COVID-19 disease from prior seasonal virus infection and duration of infection following SARS-CoV-2 infection [5–7]. Few have examined antibody response to seasonal infection, antibody waning, or cross-response between the 4 seasonal HCoVs or with SARS-CoV-2 in large prospective cohort studies [8–10].
We have previously reported on 8 years of seasonal HCoV infection among persons in the continuing Household Influenza Vaccine Evaluation (HIVE) study being conducted in Michigan [11]. Over that period, 2010–2018, 1004 infections were detected by reverse-transcription polymerase chain reaction (RT-PCR). The infections were most frequent in children, but substantial numbers of infections were identified in adults. This, and past studies of HCoVs suggested that these agents, like most respiratory viruses, reinfect through life [12, 13]. In this report, we characterize RT-PCR–documented, symptomatic reinfections with these viruses and investigate antibody response to infection, including cross-reactivity and persistence.
MATERIALS AND METHODS
Study Population
The complete methods of the HIVE cohort have been published previously [14]. Households with children receiving primary care from Michigan Medicine were recruited from Ann Arbor, Michigan and surrounding communities beginning in the summer of 2010. Households were retained as long as possible with replacement households enrolled and returning households reengaged in the spring or summer of each year; participant and household characteristics were recorded at this time. Adult participants provided informed consent for themselves and their children, and children ≥7 years of age provided verbal assent prior to participating. This study was reviewed and approved by the University of Michigan Medical School Institutional Review Board.
Seasonal Coronavirus Surveillance
Each study year, participants were asked to report all acute respiratory illnesses (ARIs) defined by ≥2 symptoms as soon as they occurred [14]. Participants were also actively questioned regarding their illness status via weekly calls or emails. Although year-round surveillance did not begin until the fall of 2014, complete coronavirus epidemics were likely captured in each study year because of their sharp seasonality [11]. Participants with ARI attended an illness visit within 7 days of symptom onset where study staff collected nasal and throat swabs (nasal only in children <3 years of age) combined in a single vial of viral transport media; asymptomatic infections were not assessed. Specimens were assayed for detection of respiratory viruses, including the 4 seasonal coronavirus types (229E, OC43, HKU1, and NL63). Specimens collected prior to the 2016–2017 study year were tested by singleplex RT-PCR using primers and probes developed by the CDC Division of Viral Diseases, Gastroenteritis, and Respiratory Viruses [15]. Specimens collected in the 2016–2017 and 2017–2018 years were tested using the FTD Respiratory Pathogen 33 Multiplex PCR Kit (Fast Track Diagnostics). Coinfections were defined as ARI associated with the detection of 2 or more HCoVs in the same specimen. Reinfection was defined as detection of the same or different HCoV type during an ARI with new onset of symptoms 14 or more days from the onset of a previously reported illness.
Serologic Studies
Beginning in the fall of 2011, participants aged ≥13 years were invited to provide blood specimens for serologic studies in the fall of each year prior to the respiratory virus season, and in the spring or summer following each respiratory virus season. Eligibility for serologic studies was expanded to children ≥6 months of age in the fall of 2016. We selected all individuals with PCR-confirmed common coronavirus infection between 2011 and 2018 who had paired serum collected in the fall or summer prior to their infection and in the spring or summer following their infection for serologic studies. In addition to the pair pre- and postinfection serum specimens, a subsequent postinfection specimen was selected from a later study year for each individual when available.
These serum specimens were tested in a 10-Plex electrochemiluminescence immunoassay (ECLIA) (Meso Scale Discovery, Rockville, Maryland) to measure antibody binding to the following antigens: the 4 seasonal coronavirus spike proteins (229E, OC43, HKU1, and NL63), SARS-CoV spike protein, SARS-CoV-2 spike protein, SARS-CoV-2 spike protein receptor binding domain, SARS-CoV-2 spike protein N-terminal domain, SARS-CoV-2 N protein, and a bovine serum albumin (BSA) negative control [16–19]. On the day of the assay, the plate was blocked for 60 minutes with MSD Blocker A (5% BSA). The blocking solution was washed off and test samples were applied to the wells at 4 dilutions (1:100, 1:800, 1:3200, and 1:12 800) and incubated with shaking for 2 hours. Plates were washed, and Sulfo-tag–labeled anti–immunoglobulin G antibody was applied to the wells and allowed to associate with complexed coated antigen-sample antibody. Plates were washed to remove unbound detection antibody, and a read solution containing ECL substrate was applied. In an MSD Sector instrument, a current was applied to the plate and areas of well surface where sample antibody has complexed with coated antigen and labeled reporter will emit light in the presence of the ECL substrate. An MSD Sector instrument quantitated the amount of light emitted and reported this ECL unit response, which is directly proportional to binding antibody. The area under the curve (AUC) was calculated using Prism software (GraphPad Prism, San Diego, California).
Antibody binding to the 4 common coronavirus spike proteins was also measured in singleplex ECLIA assays. The correlation between the singleplex and multiplex assays was generally high and patterns of antibody response were similar for the common coronaviruses comparing the mulitplex and singleplex assays (Supplementary Tables 1 and 2). Therefore, the results of only the multiplex analysis are presented here.
Statistical Analysis
For ease of calculation in time-to-event analyses, individuals were considered to contribute time at risk from 1 July through 30 June for each study year they were enrolled even though ARI surveillance was not carried out during the summer months in all years. Mean, median, minimum, and maximum times from 1 July of the first study year of enrollment to first infection and reinfection were estimated overall and for specific HCoV types and genera. Kaplan–Meier curves summarizing time to reinfection following each previous infection were also generated. Individuals reentered the data after each infection with time at risk of reinfection beginning at the day of onset of symptoms of their prior infection (time = 0). Individuals who were lost to follow-up and reenrolled in a later season were censored during the period during which they were lost to follow-up.
Hazard ratios (HRs) and 95% confidence intervals (CIs) of the effect of infection in the prior year on infection in the following year were estimated in Cox proportional hazards models, stratified by year, with respective overall, type-specific, and genus-specific covariates specified as 1 if the individual was infected in the previous study year and 0 otherwise. Results of unadjusted models and models adjusted for age group (0–8, 9–17, ≥18 years) and sex are reported. Individuals contributed to a year of data if they participated in that and the prior study year. For each study year, only an individual’s first HCoV infection was considered in respective any type, type-specific, and genus-specific analyses; subsequent HCoV infections during the same study year were ignored. Because individuals could contribute to multiple years and be infected multiple times, robust variance was calculated for Cox proportional hazards models.
AUC values from the ECLIA were adjusted by subtracting the BSA AUC value, and if any negative values resulted, adding a constant such that the minimum value for each target antigen was equal to 1. AUC values for each antigen were log2 transformed and the geometric mean was calculated at each time point: preinfection, postinfection, and subsequent postinfection. Changes in antibody binding from pre- to postinfection were calculated as the difference in geometric mean AUC. The number and proportion of individuals with 2-fold and 4-fold increases in AUC from pre- to postinfection were also calculated. Analyses of geometric means and infection response were only calculated among individuals with single infections; participants who had coinfections or >1 infection per year were excluded. The rate of antibody waning and half-life was estimated in linear generalized estimating equation models predicting log2 AUC by time. Pearson correlation coefficients were calculated comparing multiplex and singleplex AUC at each time point (Supplementary Tables 1 and 2).
The association between antibody binding levels and subsequent infection risk was estimated using a case test-negative design analysis. Log2 AUC was compared between cases of single infection with a specific HCoV type and controls that were singly infected with the other 3 HCoV types in logistic generalized estimating equation regression models with exchangeable correlation structure clustered on the individual and adjusted for age, influenza vaccination, and high-risk health status. Inclusion of study year in adjusted models did not substantially change point estimates. Odds ratios (ORs) estimated from these models were interpreted as the reduction in odds of infection associated with a 2-fold increase in AUC.
RESULTS
RT-PCR–Confirmed HCoV Coinfections and Reinfections Over Time
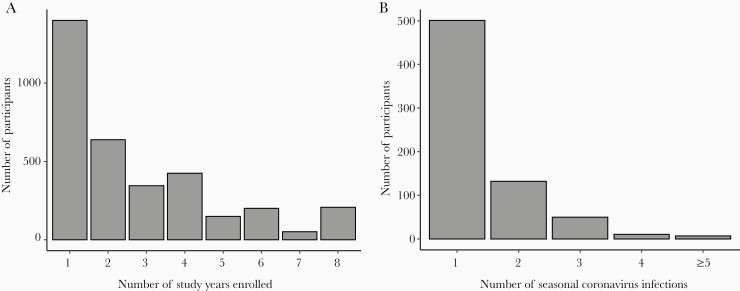
In total, 3418 individuals participated for 1–8 study years (median, 2 [interquartile range, 1–4] years) contributing a total of 9378 person-years of observation (Figure 1). There were 1378 (40%) individuals who were under study for ≥3 years. Between 8.3% and 16.3% of the cohort had an ARI associated with seasonal HCoV infection each year. In total, 1004 ARIs associated with coronavirus infection were identified. The beta genus OC43 was most common (n = 390), followed by the alpha genus NL63 (n = 328). Less common were the beta HKU1 (n = 194), and alpha 229E (n = 152). The mean time from enrollment to first HCoV infection was 542 days. Times from enrollment to first infection for each seasonal HCoV were consistent with their relative incidence—that is, shorter times for the more common OC43 and NL63, and longer for the less common HKU1 and 229E (Supplementary Table 3).
Figure 1.
Distributions of follow-up time (A) and observed acute respiratory illnesses (B) associated with reverse-transcription polymerase chain reaction (RT-PCR)–confirmed seasonal coronavirus infection, Household Influenza Vaccine Evaluation (HIVE) cohort, 2010–2018. The number of observed acute respiratory illnesses associated with RT-PCR–confirmed seasonal coronavirus infection per individual ranged from 1 to 13.
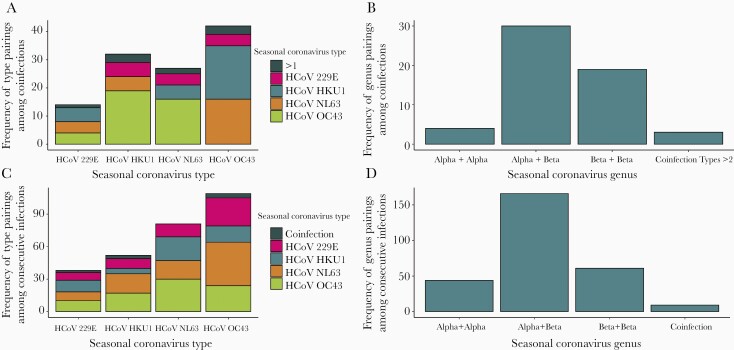
Of the 1004 HCoV-associated ARIs, there were 53 (5.3%) instances of coinfection with 2 HCoV types detected from the same specimen and 3 (0.3%) in which 3 different types were detected. Combinations of all 4 HCoV types were observed in these coinfections, but alpha and beta genus coinfections were most common (Figure 2A and 2B).
Figure 2.
Frequency of seasonal human coronavirus (HCoV) pairings in coinfections by type (A) and genus (B), and frequency of seasonal HCoV pairings in consecutive reinfections by type (C) and genus (D).
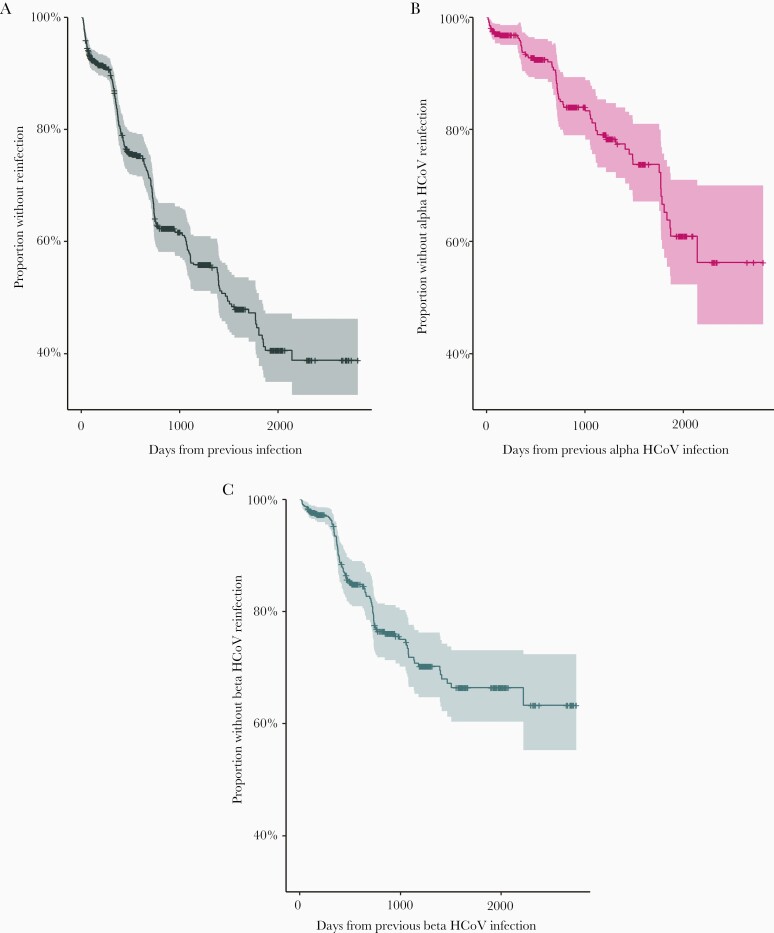
HCoV reinfections were common in the cohort (Figure 3; Supplementary Figure 1). Overall, the 1004 ARI with HCoV occurred among 701 individuals; 303 (30%) of these represented documented reinfection. The number of ARI episodes with HCoV infection ranged from 1 to 13 per individual (Figure 1). Of the overall 303, 81 any type reinfections were identified in the same study year (1 July–30 June), a relatively short period given their seasonality. Of the 81 reinfections during the same study year, 12 were of the same HCoV type potentially representing prolonged shedding (range of days between illnesses, 14–152). Considering any type reinfections, the mean time to reinfection was 505 days. The mean time to same type reinfection was estimated at 983 days for 229E, 578 days for HKU1, 615 days for OC43, and 711 days for NL63 (Supplementary Table 3). Distributions of HCoV type pairings in consecutive any type reinfections were similar to those for coinfections, with consecutive alpha and beta genus infections most common (Figure 2C and 2D).
Figure 3.
Kaplan–Meier curves of time to reinfection following a prior seasonal human coronavirus (HCoV) infection for any seasonal HCoV reinfection (A), and for same-genus alpha (B) and beta (C) coronavirus reinfections.
Overall, the hazard of infection with any coronavirus was more than twice as high among subjects with documented infection of any type in the immediately prior study year relative to those who were not (HR, 2.2 [95% CI, 1.7–2.7]). Similarly, the hazard of reinfection with a beta genus HCoV was more than twice as high among subjects infected with a beta genus HCoV in the previous study year; this effect was consistent for both beta genus viruses, HKU1 and OC43 (Table 1). There was no statistically significant effect of prior season alpha genus HCoV infection on hazard of alpha genus reinfection in the following season. To investigate the possibility that the increased risk of reinfection might be due to confounding by unmeasured risk factors for infection, we performed a sensitivity analysis restricting to only those individuals with any seasonal HCoV infection in the prior year. HRs consistently shifted lower in this analysis (Table 1); the increased hazard of beta coronavirus reinfection was attenuated and no longer statistically significant, while the reduced hazard of alpha coronavirus reinfection was strengthened and statistically significant.
Table 1.
Hazard Ratios of Coronavirus Infection by Prior Year Infection Status, Overall and by Type and Genus
| Cohort and Infection | No. of Infections/Person-Years Not Infected in Previous Year (%) | No. of Infections/Person-Years Infected in Previous Year (%) | HRa (95% CI) | Adjusteda,b HR (95% CI) |
|---|---|---|---|---|
| Full cohort | ||||
| All HCoV infections | 429/5170 (8.3) | 113/639 (17.7) | 2.3 (1.8–2.9) | 2.2 (1.7–2.7) |
| 229E infections | 118/5695 (2.1) | 1/114 (0.9) | 0.7 (.1–5.3) | 0.7 (.1–5.0) |
| HKU1 infections | 118/5690 (2.1) | 9/119 (7.6) | 5.6 (2.5–12.3) | 4.8 (2.2–10.4) |
| NL63 infections | 170/5605 (3.0) | 4/204 (2.0) | 0.8 (.3–2.2) | 0.7 (.3–1.8) |
| OC43 infections | 179/5517 (3.2) | 18/292 (6.2) | 2.6 (1.6–4.1) | 2.3 (1.4–3.6) |
| Alpha genus infections | 275/5494 (5.0) | 14/315 (4.4) | 0.9 (.5–1.7) | 0.8 (.5–1.5) |
| Beta genus infections | 256/5421 (4.7) | 50/388 (12.9) | 2.9 (2.1–4.1) | 2.7 (1.9–3.7) |
| Subset with infection in previous year | ||||
| 229E infections | 18/525 (3.4) | 1/114 (0.8) | 0.5 (.1–3.7) | 0.5 (.1–3.4) |
| HKU1 infections | 23/520 (4.4) | 9/119 (7.6) | 2.7 (1.2–6.1) | 2.4 (1.1–5.2) |
| NL63 infections | 29/435 (6.7) | 4/204 (2.0) | 0.3 (.1–.8) | 0.2 (.1–.6) |
| OC43 infections | 25/347 (7.2) | 18/292 (6.2) | 1.1 (.6–2.0) | 1.0 (.6–1.8) |
| Alpha genus infections | 38/324 (11.7) | 14/315 (4.4) | 0.4 (.2–.7) | 0.3 (.2–.6) |
| Beta genus infections | 22/251 (8.8) | 50/388 (12.9) | 1.5 (.8–2.5) | 1.4 (.8–2.4) |
Abbreviations: CI, confidence interval; HCoV, human coronavirus; HR, hazard ratio.
aEstimated in study year–stratified Cox proportional hazards model predicting time to infection by prior year infection status.
bModel adjusted for age group (0–8, 9–17, ≥18 years) and sex.
Serologic Studies
We selected all individuals with common coronavirus infection between 2011 and 2018 who had paired serum collected in the in the fall or summer prior to their infection and in the spring or summer following their infection for serologic studies (201 of the 1004 total infections). Of these 201, 42 were from individuals infected with 229E, 41 with HKU1, 40 with NL63, and 78 with OC43. Of these, 167 had a subsequent postinfection specimen collected in a later study year. The ages of individuals included in serologic studies ranged from 3 to 67, mainly from younger adults (median age, 38 years). Preinfection specimens were collected between 2 and 216 days prior to infection (median, 60 days); postinfection specimens were collected between 19 and 244 days after infection (median, 110 days); and subsequent specimens were collected between 79 and 517 days after infection (median, 198 days).
Antibody Against Seasonal Coronaviruses
Individuals included in the serologic portion of this analysis were infected despite high binding antibody levels against the coronavirus with which they were infected. In general, preinfection antibody levels were high against all 4 seasonal HCoVs, with geometric mean AUC ranging from 155 751 against NL63 to 2 225 663 against OC43 (Table 2; Supplementary Figure 2). These geometric mean AUCs were approximately 25- to 360-fold higher than those binding the SARS-CoV-2 spike protein. Geometric mean AUCs were remarkably similar overall at preinfection, postinfection, and subsequent time points (Supplementary Figure 2).
Table 2.
Geometric Mean Area Under the Curve Levels of Antibodies Binding Seasonal Coronavirus Spike Protein at Preinfection and at 2 Postinfection Time Points for Individuals Infected With 229E, HKU1, NL63, or OC43; and the Proportion of Individuals Who Demonstrated a ≥2-Fold or ≥4-Fold Rise in Binding Antibody Following Infection
| Antigen | ||||
|---|---|---|---|---|
| Infection | 229E | HKU1 | NL63 | OC43 |
| 229E (n = 42) | ||||
| Preinfection GM-AUC | 1 131 162.57 | 784 535.98 | 15 5751.62 | 2 225 663.81 |
| Postinfection GM-AUC | 2 581 588.76 | 882 948.38 | 177 840.92 | 2 532 165.45 |
| Subsequent GM-AUC | 2 091 271.52 | 821 827.33 | 168 956.51 | 2 179 466.83 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 22 (52.4) | 8 (19.0) | 5 (11.9) | 5 (11.9) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 10 (23.8) | 3 (7.1) | 2 (4.8) | 3 (7.1) |
| HKU1 (n = 41) | ||||
| Preinfection GM-AUC | 1 175 392.03 | 837 183.13 | 211 015.36 | 2 572 590.25 |
| Postinfection GM-AUC | 1 315 880.69 | 1 115 981.78 | 202 850.69 | 2 474 157.99 |
| Subsequent GM-AUC | 1 459 890.11 | 104 3611.88 | 230 423.33 | 2 513 233.77 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 5 (12.2) | 9 (22.0) | 1 (2.4) | 2 (4.9) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 3 (7.3) | 1 (2.4) | 0 (0.0) | 1 (2.4) |
| NL63 (n = 40) | ||||
| Preinfection GM-AUC | 1 449 510.47 | 764 070.12 | 143 224.93 | 1 964 950.38 |
| Postinfection GM-AUC | 1 139 895.79 | 750 090.91 | 187 930.95 | 2 035 442.24 |
| Subsequent GM-AUC | 1 209 333.46 | 685 540.48 | 182 482.86 | 2 251 171.93 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 0 (0.0) | 4 (10.0) | 7 (17.5) | 6 (15.0) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 0 (0.0) | 1 (2.5) | 0 (0.0) | 0 (0.0) |
| OC43 (n = 78) | ||||
| Preinfection GM-AUC | 123 6257.52 | 774 334.37 | 162 126.04 | 2 183 705.59 |
| Postinfection GM-AUC | 1 344 293.21 | 811 925.45 | 173 449.60 | 2 802 698.81 |
| Subsequent GM-AUC | 1 339 786.71 | 830 427.15 | 177 626.98 | 2 740 582.39 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 9 (11.5) | 9 (11.5) | 6 (7.7) | 14 (17.9) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 3 (3.8) | 3 (3.8) | 2 (2.6) | 0 (0.0) |
Statistically significant differences (paired t test P < .05) between preinfection and postinfection GM-AUC are shown in bold.
Abbreviation: GM-AUC, geometric mean area under the curve.
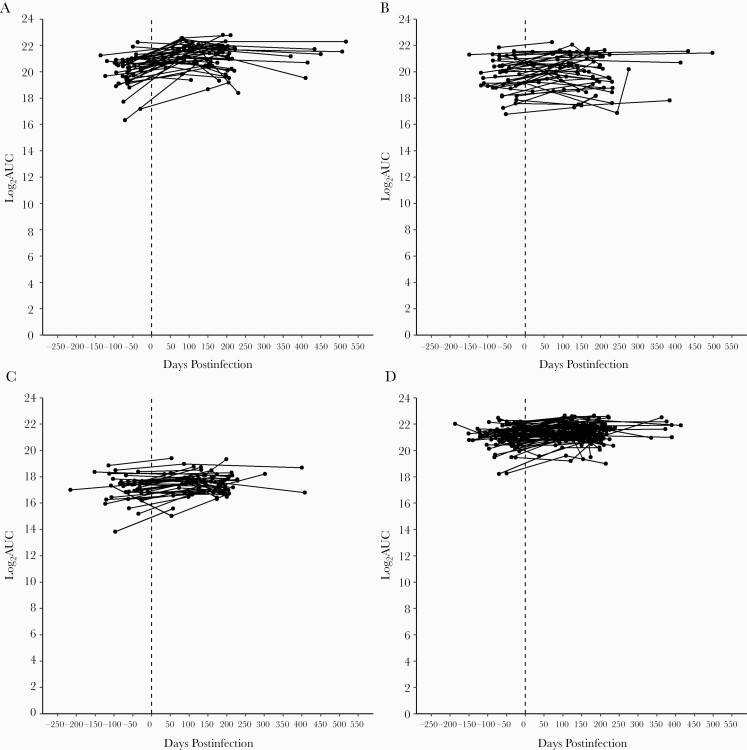
Only modest increases in antibody levels to homologous HCoV types were observed following single infection (Table 2; Figure 4). Homologous geometric mean AUC values increased from pre- to postinfection by 1 450 426 for individuals infected with 229E, by 278 799 with HKU1, by 44 706 with NL63, and by 618 993 with OC43. Antibody levels did not substantially change from pre- to postinfection for other nonhomologous common coronaviruses, although sporadic individuals had cross-reactive responses following infection. Antibody levels postinfection were durable; statistically significant waning of homologous antibody was not observed (Figure 4; Supplementary Table 4).
Figure 4.
Infection-homologous log2 area under the curve (AUC) values preinfection and at 2 postinfection time points for individuals infected with human coronaviruses 229E (A), HKU1 (B), NL63 (C), and OC43 (D).
There was no significant effect of antibody level on odds of infection detected in test-negative analyses comparing those testing positive for 1 coronavirus type to those testing positive for other types. A doubling in homologous AUC was estimated to reduce odds of infection by 7% (OR, 0.93 [95% CI, .76–1.14]) for 229E, 18% (OR, 0.82 [95% CI, .61–1.08]) for NL63, and by 6% (OR, 0.94 [95% CI, .68–1.31]) for OC43; a doubling in AUC was estimated to increase odds of infection by 9% (OR, 1.09 [95% CI, .83–1.45]) for HKU1.
Cross-Reactive Antibody to Epidemic Coronaviruses
Antibody levels were also measured against SARS-CoV spike protein, SARS-CoV-2 spike protein, SARS-CoV-2 spike protein receptor binding domain, SARS-CoV-2 spike protein N-terminal domain, and SARS-CoV-2 N protein (Table 3). With the exception of the SARS-CoV-2 N protein, antibody levels were low against these targets and did not substantially change from pre- to postinfection with each of the 4 seasonal coronaviruses. While much lower than antibody levels to the seasonal coronaviruses, the preinfection geometric mean AUC for the SARS-CoV-2 N protein was higher than for other SARS-CoV targets. Modest, but not statistically significant, increases in geometric mean AUC for SARS-CoV-2 N protein were observed for those infected with 229E where nearly 20% had ≥4-fold rise in log2 AUC.
Table 3.
Geometric Mean Area Under the Curve Levels of Antibodies Binding Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and SARS-CoV-2 Antigens at Preinfection and at 2 Postinfection Time Points for Individuals Infected With 229E, HKU1, NL63, or OC43; and the Proportion of Individuals Who Demonstrated a ≥2-Fold or ≥4-Fold Rise in Binding Antibody Following Infection
| Antigen | |||||
|---|---|---|---|---|---|
| Infection | SARS-CoV Spike | SARS-CoV-2 Spike | SARS-CoV-2 Spike-NTD | SARS-CoV-2 Spike-RBD | SARS-CoV-2 N Protein |
| 229E (n = 42) | |||||
| Preinfection GM-AUC | 5562.88 | 6134.71 | 2552.83 | 7301.94 | 23 461.49 |
| Postinfection GM-AUC | 4331.25 | 7226.25 | 1880.80 | 6143.84 | 42 016.96 |
| Subsequent GM-AUC | 5657.59 | 6236.92 | 2413.06 | 8303.43 | 29 981.52 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 3 (7.1) | 7 (16.7) | 6 (14.3) | 4 (9.5) | 13 (31.0) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 3 (7.1) | 3 (7.1) | 2 (4.8) | 1 (2.4) | 8 (19.0) |
| HKU1 (n = 41) | |||||
| Preinfection GM-AUC | 5609.93 | 4303.93 | 2212.49 | 5489.75 | 23 466.27 |
| Postinfection GM-AUC | 4868.87 | 4447.64 | 2347.31 | 7045.39 | 19 975.46 |
| Subsequent GM-AUC | 6530.69 | 4668.69 | 2979.96 | 6795.63 | 22 969.35 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 2 (4.9) | 4 (9.8) | 9 (22.0) | 11 (26.8) | 7 (17.1) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 0 (0.0) | 1 (2.4) | 4 (9.8) | 7 (17.1) | 1 (2.4) |
| NL63 (n = 40) | |||||
| Preinfection GM-AUC | 4270.94 | 6733.11 | 1975.73 | 5610.12 | 18 421.41 |
| Postinfection GM-AUC | 3959.94 | 6056.06 | 2665.88 | 5926.12 | 20 615.65 |
| Subsequent GM-AUC | 4088.78 | 6005.97 | 2036.92 | 7221.81 | 24 803.20 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 3 (7.5) | 2 (5.0) | 10 (25.0) | 7 (17.5) | 9 (22.5) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 1 (2.5) | 1 (2.5) | 4 (10.0) | 2 (5.0) | 1 (2.5) |
| OC43 (n = 78) | |||||
| Preinfection GM-AUC | 6374.95 | 6479.42 | 3260.21 | 6444.89 | 25 972.48 |
| Postinfection GM-AUC | 5052.53 | 5831.20 | 2119.52 | 6190.94 | 19 376.76 |
| Subsequent GM-AUC | 4602.15 | 6287.42 | 1928.70 | 5624.17 | 18 284.01 |
| ≥2-fold rise from pre- to postinfection, No. (%) | 3 (3.8) | 8 (10.3) | 5 (6.4) | 11 (14.1) | 10 (12.8) |
| ≥4-fold rise from pre- to postinfection, No. (%) | 1 (1.3) | 1 (1.3) | 2 (2.6) | 7 (9.0) | 4 (5.1) |
Statistically significant differences (paired t test, P < .05) between preinfection and postinfection GM-AUC are shown in bold.
Abbreviations: GM-AUC, geometric mean area under the curve; NTD, N-terminal domain; RBD, receptor binding domain; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
During an influenza pandemic, past experience is useful in many areas of planning, but the novel COVID-19 pandemic has left us with few precedents for a number of critical subjects, particularly duration of immunity postinfection or vaccination [20]. Prior experience with epidemic HCoVs offers little help; SARS-CoV was eliminated and MERS-CoV has not spread widely [21, 22]. As a result, we have looked to the 4 seasonal HCoV viruses causing typically mild disease to gain insights on how SARS-CoV-2 might behave going forward. We have previously demonstrated that these HCoVs are truly seasonal, transmitting mainly in the months between November and May and peaking in December–March, and only time will tell if the SARS-CoV-2 occurrence will begin to follow the same pattern as immunity increases in the population [11].
While it was clear from studies conducted years ago that reinfection with seasonal HCoVs, like other common respiratory viruses, occurred throughout life, their frequency had not been a matter of great interest [12, 13]. This has become more urgent currently given the importance of knowing how long immunity might last after SARS-CoV-2 infection and vaccination. Given the age structure of our cohort, and high levels of preexisting antibody, nearly all infections observed in this study are likely reinfections. However, nearly a third of all identified infections were confirmed reinfections during enrollment with an average duration of 505 days between infections. In primary analyses, we observed that the risk of infection with beta genus coronaviruses was higher if an individual had a beta coronavirus infection in the immediately prior study year. This effect was attenuated in sensitivity analyses conditioning only on those with a prior infection, suggesting that this result is likely due to confounding by unmeasured shared risk factors for infection rather than a specific biological effect. Regardless, this finding underscores the frequency of reinfection in this cohort.
While there have been several recent studies on antibody to the seasonal HCoVs in those infected with SARS-CoV-2, there have been few looking at the antibody response of RT-PCR–confirmed seasonal infection [23, 24]. We used a multiplex binding assay that included SARS-CoV-2, SARS-CoV, and seasonal coronavirus prefusion-stabilized spike protein constructs made using techniques developed for studying vaccine response [19, 25–27]. We found high levels of antibody binding the spike protein of the seasonal viruses and very low levels binding SARS-CoV-2 antigens in this pre–COVID-19 population. Individuals in this study were infected with seasonal HCoV despite these high levels of binding antibody, and had very modest increases in antibody following infection, suggesting a ceiling effect. Consistent with this observed infection despite high antibody levels, there was no significant evidence that homologous binding antibody correlated with protection. This analysis was limited by lack of antibody measurements in a completely uninfected control group and by a relatively narrow distribution of preinfection antibody binding levels in this population. We also did not measure neutralizing antibody, which is likely to be a better correlate of protection. While binding assays for SARS-CoV-2 have correlated well with neutralizing antibodies in recent studies of response to infection or vaccination with the novel virus [26, 28–31], it is possible that is not the case with the repeat infection that occurs with the seasonal viruses.
We also found that antibody was remarkably persistent over time with little indication of waning. This stability of antibody is in contrast to several studies that have observed rapidly waning immunity following SARS-CoV-2 infection [8, 28, 32, 33], although other studies do suggest a more persistent antibody response [29, 30, 34]. As with correlates of protection, it may be the case that persistence of neutralizing antibody differs from that of binding antibodies.
There were few responses in SARS-CoV-2 antibodies following seasonal HCoV infection, indicating that the cross-response is infrequent. An exception was higher preinfection antibody to the SARS-CoV-2 N protein, known to be more conserved among the HCoVs [35]. Measuring antibody directed to the SARS-CoV-2 N protein has been suggested as a way to distinguish individuals who are infected vs vaccinated, as both will exhibit spike-directed antibody. The antigen we used was the full N protein; using epitopes restricted to SARS-CoV-2 may be more specific for this strategy.
This study has demonstrated that seasonal HCoV reinfection frequently occurs over a relatively short time period and is possibly affected by the prior infecting virus type. However, we have not determined precisely how frequently this occurs in a broad population nor is it evident whether this will apply to SARS-CoV-2 reinfection or infection after vaccination. Still, it does suggest that duration of immunity will probably be limited by either waning immunity or viral antigenic drift, at least in some of the population, and supports careful determination of the need for booster vaccinations as time from original immunization increases. Careful monitoring of the ways in which repeated vaccination and reinfection shape the development of immunity is also warranted. This has implications for the use of immune assays as it is possible that the close relation between binding and neutralizing antibodies for SARS-CoV-2 relates to the novelty of the virus. The need for periodic revaccination should not be viewed with alarm, since it has been the practice for many years with influenza. Such booster immunizations, through updated reformulation, could also address possible antigenic changes, now becoming a major concern as novel variants continue to be identified globally.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Jennifer Mehalko, Matthew Drew, and Kelly Snead for their technical assistance.
Disclaimer. The content of this publication does not necessarily reflect the official views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant numbers R01 AI097150 and R56 AI097150 to A. S. M.; K01 AI141579 to J. G. P.) and the National Cancer Institute (contract number 75N910D00024, task order number 75N91019F00130).
Potential conflicts of interest. E. T. M. has received grant support from Merck and consultancy fees from Pfizer for work unrelated to this report. A. S. M. has received consultancy fees from Sanofi and Seqirus for work unrelated to this report. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci U S A 1967; 57:933–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monto AS. Medical reviews. Coronaviruses. Yale J Biol Med 1974; 47:234–51. [PMC free article] [PubMed] [Google Scholar]
- 3. Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348:1967–76. [DOI] [PubMed] [Google Scholar]
- 4. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367:1814–20. [DOI] [PubMed] [Google Scholar]
- 5. Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun 2020; 11:4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beretta A, Cranage M, Zipeto D. Is cross-reactive immunity triggering COVID-19 immunopathogenesis? Front Immunol 2020; 11:567710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aran D, Beachler DC, Lanes S, Overhage JM. Prior presumed coronavirus infection reduces COVID-19 risk: a cohort study. J Infect 2020; 81:923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med 2020; 26:1691–3. [DOI] [PubMed] [Google Scholar]
- 9. Ringlander J, Nilsson S, Westin J, Lindh M, Martner A, Hellstrand K. Low incidence of reinfection with endemic coronaviruses diagnosed by real-time PCR [manuscript published online 10 October 2020]. J Infect Dis 2021;223(11):2013–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galanti M, Shaman J. Direct observation of repeated infections with endemic coronaviruses. J Infect Dis 2020; 223:409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Monto AS, DeJonge PM, Callear AP, et al. Coronavirus occurrence and transmission over 8 years in the HIVE cohort of households in Michigan. J Infect Dis 2020; 222:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monto AS, Lim SK. The Tecumseh study of respiratory illness. VI. Frequency of and relationship between outbreaks of coronavirus infection. J Infect Dis 1974; 129: 271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamre D, Beem M. Virologic studies of acute respiratory disease in young adults. V. Coronavirus 229E infections during six years of surveillance. Am J Epidemiol 1972; 96:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monto AS, Malosh RE, Evans R, et al. ; HIVE Study Research Staff . Data resource profile: Household Influenza Vaccine Evaluation (HIVE) Study. Int J Epidemiol 2019; 48:1040–1040g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakthivel SK, Whitaker B, Lu X, et al. Comparison of Fast-Track Diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. J Virol Methods 2012; 185:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson M, Wagstaffe HR, Gilmour KC, et al. Evaluation of a novel multiplexed assay for determining IgG levels and functional activity to SARS-CoV-2. J Clin Virol 2020; 130:104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Majdoubi A, Michalski C, O’Connell SE, et al. Antibody reactivity to SARS-CoV-2 is common in unexposed adults and infants under 6 months. medRxiv [Preprint]. Posted online 6 November 2020. doi: 10.1101/2020.10.05.20206664. [DOI] [Google Scholar]
- 18. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020; 367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsieh CL, Goldsmith JA, Schaub JM, et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020; 369:1501–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Monto AS, Fukuda K. Lessons from influenza pandemics of the last 100 years. Clin Infect Dis 2020; 70:951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lipsitch M, Cohen T, Cooper B, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science 2003; 300:1966–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hui DS, Azhar EI, Kim YJ, Memish ZA, Oh MD, Zumla A. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis 2018; 18:e217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anderson EM, Goodwin EC, Verma A, et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. medRxiv [Preprint]. Posted online 10 November 2020. doi:10.1101/2020.11.06.20227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poston D, Weisblum Y, Wise H, et al. Absence of SARS-CoV-2 neutralizing activity in pre-pandemic sera from individuals with recent seasonal coronavirus infection [manuscript published online ahead of print 3 December 2020]. Clin Infect Dis 2020. doi:10.1093/cid/ciaa1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jackson LA, Anderson EJ, Rouphael NG, et al. ; mRNA-1273 Study Group . An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med 2020; 383:1920–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson EJ, Rouphael NG, Widge AT, et al. ; mRNA-1273 Study Group . Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med 2020; 383:2427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Widge AT, Rouphael NG, Jackson LA, et al. ; mRNA-1273 Study Group . Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N Engl J Med 2021; 384:80–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seow J, Graham C, Merrick B, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol 2020; 5:1598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020; 370:1227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iyer AS, Jones FK, Nodoushani A, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol 2020; 5:eabe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol 2020; 101:791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vabret N. Antibody responses to SARS-CoV-2 short-lived. Nat Rev Immunol 2020; 20:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lumley SF, Wei J, O’Donnell D, et al. The duration, dynamics and determinants of SARS-CoV-2 antibody responses in individual healthcare workers [manuscript published online 6 January 2021]. Clin Infect Dis 2021. doi: 10.1093/cid/ciab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J Virol 2020; 94:e00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.