Abstract
While some biomolecules have been explored to identify potential biomarkers for the prognosis of COVID-19 patients, there is no reliable prognostic indicator of the disease progression and severity. We aimed to evaluate the ability of the C-reactive protein (CRP) to predict COVID-19 infection outcome. This retrospective study was conducted on 429 patients diagnosed with COVID-19 between March 30, 2020, and April 30, 2020. The study population was divided into severe (n = 175) and nonsevere cases (n = 254). Data on demographic characteristics, clinical features, and laboratory findings on admission were collected. The proportion of patients with increased CRP levels was significantly higher in severe cases than in nonsevere patients. Analysis of the receiver operating characteristic (ROC) curve found that CRP could be used as an independent factor in predicting the severity of COVID-19. Also, patients with CRP >64.75 mg/L were more likely to have severe complications. In conclusion, CRP serum levels can predict the severity and progression of illness in patients with COVID-19.
1. Introduction
Since December 2019, a new type of coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing coronavirus disease 2019 (COVID-19), has been identified in China [1]. The COVID-19 pandemic then spread quickly around the world [2, 3]. By March 30, 2021, 127,349,248 confirmed cases of COVID-19, including 2,787,593 deaths, were reported to the World Health Organization (WHO) [4]. The rapid spread of the SARS-CoV-2, rapid changes in clinical features, and increased mortality have become the world's biggest concern. Furthermore, there are no reliable prognostic indicators for predicting disease severity and progression. Recognizing markers of disease severity may thus profoundly help to detect at-risk patients. Recently, some studies have reported that C-reactive protein (CRP) levels can be used in the early diagnosis of pneumonia and that higher CRP levels were associated with severe pneumonia [5].
Hence, the current study aims to evaluate the correlation between CRP levels and disease progression to provide a reference for the clinical management of COVID-19 patients.
2. Materials and Methods
2.1. Study Design, Participants, and Definition
The present retrospective study was approved by the Ethics Committee of Babol University of Medical Sciences (code: IR.MUBABOL.REC.1399.041). The three affiliated hospitals of Babol University of Medical Sciences have been designated to treat patients with COVID-19. A total of 429 adult cases were confirmed at these centers from March 30 to April 30, 2020. All patients with COVID-19 who enrolled in the recent study were diagnosed according to the WHO interim guidance for COVID-19 (6th edition) [6]. In other words, all patients with the physician- and laboratory-confirmed (positive nasopharyngeal/throat swab specimens by reverse transcription-polymerase chain reaction (RT-PCR)) COVID-19 infection were included, while suspected cases with similar clinical symptoms were excluded. All cases were monitored using the clinical data collected until March 30, 2020. One of the following criteria was used to determine severe COVID-19 illness: respiratory rate ≥30 bpm, oxygen saturation ≤93%, arterial oxygen partial pressure (PaO2)/oxygen concentration (FiO2) ≤300 mm Hg, and intensive care unit (ICU) admission.
2.2. Data Collection
Patient medical records were reviewed by an experienced team of clinicians of the Infectious Diseases and Tropical Medicine Research Center of Babol University of Medical Sciences. The data on epidemiological, clinical, laboratory, radiological findings, and outcomes were collected using a data collection checklist from electronic medical records. Moreover, recorded patient data, such as demographic characteristics, past medical history (PMH), underlying medical conditions, symptoms, and signs, were collected.
2.3. Statistical Analysis
The statistical data were analyzed using SPSS version 16.0 (IBM, Chicago, IL, USA). Continuous and categorical variables were presented as median (IQR) and n (%), respectively. Mann–Whitney U-test and Student's t-test were used to compare continuous and categorical variables. The predictive value of the CRP was evaluated by measuring the area under the receiver operating characteristic curve (AUC). The optimal threshold value was obtained by calculating the Youden index. A multivariate Cox proportional risk model was used to determine predictive factors for disease risk.
3. Results
There were 429 patients with COVID-19 in our study. Of these, 175 patients (40.8%) were assigned to the severe group, while 254 patients (59.2%) were allocated to the nonsevere group. The demographic and clinical characteristics of the patients are summarized in Table 1. The mean age was 57.21 ± 16.18 years, with a range of 16–99 years. The average age was higher in the severe group than in the nonsevere group (P=0.111). One hundred and eighty-six patients (43.4%) were female. The severity ratio for males was higher than for females, but this difference was not significant (P=0.122). The median duration from illness onset to discharge was seven days. Overall, dyspnea (72.5%) was the most common initial symptom, followed by fever (61.3%) and dry cough (57.6%). However, there was no significant difference in the symptoms ratio between the two groups.
Table 1.
Demographic and baseline characteristics of patients with COVID-19.
| All patients (n = 429) | Severe (n = 175) | Nonsevere (n = 254) | P value | ||
|---|---|---|---|---|---|
| Age, mean ± SD (range) | 57.21 ± 16.18 (16–99) | 58.75 ± 15.88 (22–97) | 56.18 ± 16.32 (16–99) | 0.111 | |
|
| |||||
| Sex (%) | Male | 241 (56.4) | 106 (60.9) | 135 (53.4) | 0.122 |
| Female | 186 (43.6) | 68 (39.1) | 118 (46.6) | ||
|
| |||||
| Hospitalization (days) (range) | 7 (1–37) | 10 (1–37) | 6 (1–31) | <0.001 | |
|
| |||||
| Clinical symptoms (%) | Fever | 263 (61.3) | 105 (60) | 158 (62.2) | 0.645 |
| Fatigue | 161 (37.5) | 70 (40) | 91 (35.8) | 0.380 | |
| Headache | 76 (17.7) | 31 (17.7) | 45 (17.7) | 1.000 | |
| Dry cough | 247 (57.6) | 98 (56) | 149 (58.7) | 0.584 | |
| Sore throat | 36 (8.4) | 19 (10.9) | 17 (6.7) | 0.126 | |
| Expectoration | 83 (19.3) | 34 (19.4) | 49 (19.3) | 0.972 | |
| Hemoptysis | 7 (1.6) | 3 (1.7) | 4 (1.6) | 0.911 | |
| Chest pain | 61 (14.2) | 22 (12.6) | 39 (15.4) | 0.417 | |
| Dyspnea | 311 (72.5) | 126 (72) | 69 (72.8) | 0.849 | |
| Nausea | 81 (18.9) | 37 (21.1) | 44 (17.3) | 0.320 | |
| Diarrhea | 25 (5.8) | 12 (6.9) | 13 (5.1) | 0.450 | |
| Constipation | 24 (5.6) | 9 (5.1) | 15 (5.9) | 0.736 | |
| Anorexia | 118 (27.5) | 53 (30.3) | 65 (25.6) | 0.284 | |
| Arthralgia | 48 (11.2) | 20 (11.4) | 28 (11) | 0.896 | |
| Stomach ache | 39 (9.1) | 16 (9.1) | 23 (9.1) | 0.975 | |
| Dizziness | 54 (12.6) | 21 (12) | 33 (13) | 0.761 | |
| Loss of smell and taste | 18 (4.2) | 9 (5.1) | 9 (3.5) | 0.417 | |
|
| |||||
| Comorbidities (%) | Hypertension | 98 (22.8) | 32 (18.3) | 66 (26) | 0.062 |
| Cardiovascular disease | 107 (24.9) | 38 (21.7) | 185 (27.2) | 0.200 | |
| Diabetes | 119 (27.7) | 49 (28) | 70 (27.6) | 0.920 | |
| Cancer | 10 (2.3) | 3 (1.7) | 7 (2.8) | 0.482 | |
| Chronic liver disease | 5 (1.2) | 3 (1.7) | 2 (0.8) | 0.379 | |
| Chronic kidney disease | 17 (4) | 2 (1.1) | 15 (5.9) | 0.013 | |
| Brain disease | 9 (2.1) | 6 (3.4) | 3 (1.2) | 0.110 | |
| COPD | 14 (3.3) | 7 (4) | 7 (2.8) | 0.476 | |
|
| |||||
| Complications (%) | Nosocomial pneumonia | 8 (1.9) | 2 (1.1) | 6 (2.4) | 0.359 |
| Urinary tract infection | 6 (1.4) | 2 (1.1) | 4 (1.6) | 0.708 | |
| Shock | 5 (1.2) | 5 (2.8) | 0 (0) | 0.073 | |
| Acute heart failure | 28 (6.5) | 9 (5.1) | 19 (7.5) | 0.335 | |
| Arrhythmia | 19 (4.4) | 8 (4.6) | 11 (4.3) | 0.905 | |
| ARDS | 18 (4.2) | 18 (10.3) | 0 (0) | 0.101 | |
| Acute kidney failure | 1 (0.4) | 0 (0) | 1 (0.4) | 0.406 | |
Abbreviations: ARDS, acute respiratory distress syndrome; COPD, chronic obstructive pulmonary disease.
Moreover, nearly half of the patients (213, 49.7%) had comorbidities, such as diabetes (27.7%), cardiovascular disease (24.9%), and hypertension (22.8%). Sixty-two patients (14.5%) had a complication, including the occurrence of acute respiratory distress syndrome (ARDS) (18 (10.3%) vs. 0 (0), P=0.101), acute heart failure (9 (5.1%) vs. 19 (7.5%), P=0.335), and arrhythmia (8 (4.6%) vs. 11 (4.3%), P=0.335) (Table 1). Laboratory findings of the patients are presented in Table 2. Median levels of lymphocyte count, erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), and lactate dehydrogenase (LDH) were not in the normal range for the severe group. These patients had a significantly lower lymphocyte count (MD: 12 vs. MD: 22.7, P < 0.001) and a significantly higher ESR (MD: 57.5 vs. MD: 40, P=0.005), CRP (MD: 97 vs. MD: 50, P < 0.001), and LDH (MD: 783.5 vs. MD: 459, P < 0.001) levels compared to the nonsevere group.
Table 2.
Laboratory findings of severe and nonsevere COVID-19 patients.
| Normal range | Severe (n = 175) | Nonsevere (n = 254) | P value | |
|---|---|---|---|---|
| White blood cell count, median (range) (×103/μL) | 4,500–11,000 | 9,100 (1,700–42,300) | 6,500 (2,100–157,000) | <0.001 |
| Lymphocytes, %, median (range) | 26–46 | 12 (2–36) | 22.7 (3–75) | <0.001 |
| Hemoglobin, median (range) (g/dL) | 13.5–18 | 11.95 (6–21) | 12.5 (5.3–22.3) | 0.032 |
| Erythrocyte sedimentation rate, median (range) (mm/h) | 0–15 | 57.5 (3–128) | 40 (2–140) | 0.005 |
| C-reactive protein, median (range) (mg/L) | 0–10 | 97 (1–440) | 50 (4–392) | <0.001 |
| Platelet count, median (range) (count/µL) | 140,000–450,000 | 197,000 (42,000–568,000) | 197,000 (8,000–108,1000) | 0.839 |
| Blood urea nitrogen, median (range) (mg/dL) | 10–20 | 19 (5–158.2) | 15 (4–150) | <0.001 |
| Creatinine, median (range) (mg/dL) | 0.7–1.4 | 1 (0.5–7.4) | 1 (0.5–7.3) | 0.360 |
| Lactate dehydrogenase, median (range) (U/L) | 140–280 | 783.5 (146–2,436) | 459 (30–2,500) | <0.001 |
| Sodium, median (range) (mEq/L) | 135–145 | 135 (117–146) | 136 (120–152) | 0.144 |
| Potassium, median (range) (mEq/L) | 3.7–5.2 | 4.1 (3–8.2) | 4.1 (2.9–5.5) | 0.961 |
Furthermore, analysis of the ROC curve illustrated an 0.706 area under the curve (AUC) for CRP levels as a predictor of disease severity (95% CI: 0.649–0.764; P < 0.001). The AUC of this biomarker indicated a high diagnostic value for clinical severity, with the optimal threshold value being 64.75 mg/L with a sensitivity of 71.32% and a specificity of 60% (Table 3 and Figure 1). We reclassified patients into two groups according to the optimal CRP threshold (cutoff: 64.75 mg/L). The proportion of severe patients with a CRP level higher than the optimal threshold was significantly different from that of a lower CRP level (P < 0.001).
Table 3.
The area under the ROC curve (AUC) and the optimal cutoff value of CRP.
| AUC | Optimal cutoff value (mg/L) | Sensitivity (%) | Specificity (%) | Predictive value | Likelihood ratio | ||
|---|---|---|---|---|---|---|---|
| Positive (%) | Negative (%) | Positive | Negative | ||||
| 0.706 | 64.75 | 71.32 | 60 | 78.11 | 50.55 | 0.48 | 1.76 |
Abbreviations: AUC, area under the curve.
Figure 1.
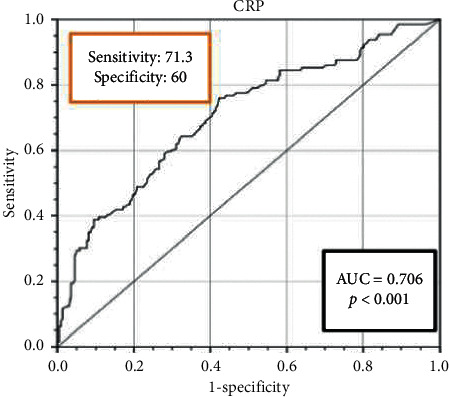
Receiver operating characteristic (ROC) curves of C-reactive protein for predicting the disease severity in COVID-19 patients.
The univariate analysis used in the logistic model indicated the severity was associated with hospital admission (OR: 1.166; 95% CI: 1.119–1.216; P < 0.001), BUN (OR: 1.027; 95% CI: 1.014–1.040; P < 0.001), lymphocyte count (OR: 0.917; 95% CI: 0.895–0.939; P < 0.001), and CRP level (OR: 3.647 95% CI: 2.288–5.813; P < 0.001). As determined by multivariate analysis, hospital admission (OR: 1.185; 95% CI: 1.086–1.293; P < 0.001) and CRP level (OR: 3.826; 95% CI: 1.166–12.560; P=0.027) were significantly associated with severity, so that the patients with a CRP level >64.75 mg/L were more likely to develop the severe form of the disease (Table 4).
Table 4.
Univariate and multivariate analyses of the logistic regression model.
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value | |
| Age | 1.010 (0.998–1.022) | 0.112 | 1.003 (0.963–1.030) | 0.762 |
| Hospital admission (days) | 1.166 (1.119–1.216) | <0.001 | 1.185 (1.086–1.293) | <0.001 |
| Lymphocytes | 0.917 (0.895–0.939) | <0.001 | 0.990 (0.934–1.050) | 0.741 |
| BUN | 1.027 (1.014–1.040) | <0.001 | 1.045 (0.994–1.097) | 0.082 |
| Diabetes | 1.022 (0.665–1.571) | 0.920 | 2.543 (0.809–7.998) | 0.110 |
| CRP | 3.647 (2.288–5.813) | <0.001 | 3.826 (1.166–12.560) | 0.027 |
ARDS, acute respiratory distress syndrome; COPD, chronic obstructive pulmonary disease; BUN, blood urea nitrogen; CRP, C-reactive protein; CI, confidence interval.
4. Discussion
In the present retrospective study, the clinical characteristics of severe COVID-19 patients were compared with those of nonsevere patients and analyzed the possible factors associated with disease progression and severity. Furthermore, the prognostic value of the CRP in the progression of COVID-19 cases has been revealed. The current study evaluated the association between CRP and COVID-19 infection, and the findings indicated that a patient with a CRP level >64.75 mg/L was more likely to develop the severe form of the disease. In other words, ROC analysis confirmed CRP as a valuable predictor of COVID-19 progression and severity.
In response to infections, the liver synthesizes significant quantities of acute-phase proteins (APPs), such as CRP [7, 8]. This acute inflammatory protein is a highly sensitive biomarker for inflammation, tissue damage, and infection [9]. It has been shown that CRP levels are correlated with levels of inflammation [10]. CRP levels can promote phagocytosis and activate the complement system [11]. In other words, CRP binds to microorganisms and promotes their removal through phagocytosis [12].
Moreover, the serum CRP levels increase during inflammatory responses. As shown previously, this biomarker may be raised by viral or bacterial infections. It is important to note that CRP levels were significantly increased in bacterial infections than in viral infections [13]. The current study revealed significantly higher CRP levels in severe cases than in nonsevere patients suggesting that the CRP level may be a biomarker of disease severity and progression in patients with COVID-19. Liu et al. reported that more severe cases infected with COVID-19 expressed significantly higher CRP levels than nonsevere patients [14]. Qin et al. observed higher CRP levels in severe COVID-19 patients than in nonsevere cases, suggesting that this biomarker can be monitored to evaluate disease progression [15]. Sahu et al. performed a meta-analysis to assess CRP levels as a potential biomarker of the COVID-19 prognosis. Their results indicated that CRP concentrations remain high in expired patients and could be a promising biomarker for assessing mortality [16]. Also, some studies showed that some frequent complications in severe and expired COVID-19 patients, such as shock, ARDS, acute kidney injury, and acute cardiac injury, were correlated with higher CRP levels [17].
5. Limitations and Future Suggestions
The current study encompasses a short sample size. Thus, it may lack generalizability. Furthermore, the retrospective nature of this study and the consequent missing clinical data was another limitation. Therefore, subsequent clinical studies with larger sample sizes and multiple CRP level measurements, especially at different treatment times, should be performed to confirm our findings.
6. Conclusions
Our findings suggest that serum CRP levels could be used as an essential indicator of the progression and the severity of COVID-19. Also, the present study suggests that patients with higher CRP levels should be carefully monitored throughout their disease course.
Acknowledgments
The authors wish to thank the Department of Infectious Diseases of Babol University of Medical Sciences.
Data Availability
All data are available upon reasonable request to the corresponding author.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Javanian M., Bayani M., Shokri M., et al. Clinical and laboratory findings from patients with COVID-19 pneumonia in Babol North of Iran: a retrospective cohort study. Romanian Journal of Internal Medicine. 2020;58(3):161–167. doi: 10.2478/rjim-2020-0013. [DOI] [PubMed] [Google Scholar]
- 2.Hsiang S., Allen D., Annan-Phan S., et al. The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature. 2020;584(7820):262–267. doi: 10.1038/s41586-020-2404-8. [DOI] [PubMed] [Google Scholar]
- 3.Mas-Coma S., Jones M. K., Marty A. M. COVID-19 and globalization. One Health. 2020;9 doi: 10.1016/j.onehlt.2020.100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Coronavirus Disease ( COVID-19) Geneva, Switzerland: WHO; 2020. Situation Report. [Google Scholar]
- 5.Ling W. C-reactive protein levels in the early stage of COVID-19. Médecine et Maladies Infectieuses. 2020;50:332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X.-W., Wu X.-X., Jiang X.-G., et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020:p. 368. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil R. H., Al-Humadi N. Types of acute phase reactants and their importance in vaccination. Biomedical Reports. 2020;12(4):143–152. doi: 10.3892/br.2020.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamal E. M., Abd El-Hakeem M. A., El Sayed A. M., Ahmed M. M. Validity of C-reactive protein and procalcitonin in prediction of bacterial infection in patients with liver cirrhosis. Minia Journal of Medical Research. 30(3):124–132. [Google Scholar]
- 9.Sproston N. R., Ashworth J. J. Role of C-reactive protein at sites of inflammation and infection. Frontiers in Immunology. 2018;9:p. 754. doi: 10.3389/fimmu.2018.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rainer T. H., Chan C. P. Y., Leung M. F., et al. Diagnostic utility of CRP to neopterin ratio in patients with acute respiratory tract infections. Journal of Infection. 2009;58(2):123–130. doi: 10.1016/j.jinf.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Gershov D., Kim S., Brot N., Elkon K. B. C-reactive protein binds to apoptotic cells, protects the cells from assembly of the terminal complement components, and sustains an antiinflammatory innate immune response. Journal of Experimental Medicine. 2000;192(9):1353–1364. doi: 10.1084/jem.192.9.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Povoa P., Pereira J., Coelho L. C-Reactive Protein: New Research. 2009. C-reactive protein: structure, synthesis and function; pp. 307–324. [DOI] [Google Scholar]
- 13.Coster D., Wasserman A., Fisher E., et al. Using the kinetics of C-reactive protein response to improve the differential diagnosis between acute bacterial and viral infections. Infection. 2020;48(2):241–248. doi: 10.1007/s15010-019-01383-6. [DOI] [PubMed] [Google Scholar]
- 14.Liu F., Li L., Xu M., et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. Journal of Clinical Virology. 2020;127 doi: 10.1016/j.jcv.2020.104370.104370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin C., Zhou L., Hu Z., et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China. Clinical Infectious Diseases. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahu B. R., Kampa R. K., Padhi A., Panda A. K. C-reactive protein: a promising biomarker for poor prognosis in COVID-19 infection. Clinica Chimica Acta. 2020;509:91–94. doi: 10.1016/j.cca.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Y., Liu W., Liu k., et al. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 in Wuhan, China: a retrospective study. Chinese Medical Journal. 2020;133(11):1261–1267. doi: 10.1097/cm9.0000000000000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available upon reasonable request to the corresponding author.


