Abstract
Background
Soft tissue sarcomas are a heterogeneous group of rare malignant tumors. Advanced soft tissue sarcomas have a poor prognosis, and effective systemic therapies have not been established. Tyrosine kinases are increasingly being used as therapeutic targets for a variety of cancers and soft tissue sarcomas. Although complex karyotype sarcomas typically tend to carry more potentially actionable genetic alterations than do translocation-associated sarcomas (fusion gene sarcomas), based on our database review, we found that leiomyosarcoma and malignant peripheral nerve sheath tumors have lower frequencies of potential targets than other nontranslocation soft tissue sarcomas. We theorized that both leiomyosarcoma and malignant peripheral nerve sheath tumors might be included in any unique translocations. Furthermore, if tyrosine kinase imbalances, especially fusion genes, occur in patients with leiomyosarcomas and malignant peripheral nerve sheath tumors, tyrosine kinase inhibitors might be a drug development target for this sarcoma. In this study, we used a tyrosine kinase screening system that could detect an imbalance in mRNA between 5’- and 3’-sides in tyrosine kinase genes to identify potential novel therapeutic tyrosine kinase targets for soft tissue sarcomas.
Questions/purposes
(1) Are there novel therapeutic tyrosine kinase targets in tumors from patients with soft tissue sarcomas that are detectable using mRNA screening focusing on imbalance expressions between the 5’ and 3’ end of the kinase domain? (2) Can potential targets be verified by RNA sequencing and reverse transcription PCR (RT-PCR)? (3) Will potential fusion gene(s) transform cells in in vitro assays? (4) Will tumors in mice that have an identified fusion gene respond to treatment with a therapeutic drug directed at that target?
Methods
We used mRNA screening to look for novel tyrosine kinase targets that might be of therapeutic potential. Using functional assays, we verified whether the identified fusion genes would be good therapeutic candidates for soft tissue sarcomas. Additionally, using in vivo assays, we assessed whether suppressing the fusion’s kinase activity has therapeutic potential. Study eligibility was based on a patient having high-grade spindle cell and nontranslocation sarcomas, including leiomyosarcoma, malignant peripheral nerve sheath tumor, and high-grade myxofibrosarcoma. Between 2015 and 2019, of the 172 patients with soft tissue sarcomas treated with surgical resection at Juntendo University Hospital, 72 patients had high-grade nontranslocation sarcomas. The analysis was primarily for leiomyosarcoma and malignant peripheral nerve sheath tumors, and there was a limitation of analysis size (reagent limitations) totaling 24 samples at the start of the study. We collected additional samples from a sample bank at the Tokyo Medical and Dental University to increase the number of sarcomas to study. Therefore, in this study, a total of 15 leiomyosarcoma samples, five malignant peripheral nerve sheath tumors samples, and four high-grade myxofibrosarcoma samples were collected to achieve the sample size of 24 patients. To identify tyrosine kinase fusion genes, we designed a NanoString-based assay (NanoString Technologies Inc, Seattle, WA, USA) to query the expression balances regarding transcripts of 90 tyrosine kinases at two points: the 5’ end of the kinase domain and within the kinase domain or 3’ end of the kinase domain. The tumor’s RNA was hybridized to the NanoString probes and analyzed for the expression ratios of outliers from the 3’ to 5’ end of the kinase domain. Presumed novel fusion events in these positive tumors that were defined by NanoString-based assays were confirmed tyrosine kinase fusion genes by RNA sequencing and confirmatory RT-PCR. Functional analyses consisting of in vitro and in vivo assays were also performed to elucidate whether the identified tyrosine kinase gene fusions were associated with oncogenic abilities and drug responses.
Results
We identified aberrant expression ratios regarding the 3’ to 5’ end of the kinase domain ratios in ROS1 transcripts in a leiomyosarcoma in a 90-year-old woman. A novel MAN1A1-ROS1 fusion gene was identified from her thigh tumor through RNA sequencing, which was confirmed with real-time PCR. In functional assays, MAN1A1-ROS1 rearrangement revealed strong transforming potential in 3T3 cells. Moreover, in an in vivo assay, crizotinib, a ROS1 inhibitor, markedly inhibited the growth of MAN1A1-ROS1 rearrangement-induced transformed cells in a dose-dependent manner.
Conclusion
We conducted tyrosine kinase screening to identify new therapeutic targets in soft tissue sarcomas. We found a novel MAN1A1-ROS1 fusion gene that may be a therapeutic target in patients with leiomyosarcoma. This study demonstrates that the mRNA screening system may aid in the development of useful therapeutic options for soft tissue sarcomas.
Clinical Relevance
If novel tyrosine fusions such as MAN1A1-ROS1 fusion can be found in sarcomas from other patients, they could offer avenues for new molecular target therapies for sarcomas that currently do not have effective chemotherapeutic options. Therefore, the establishment of a screening system that includes both genomic and transcript analyses in the clinical setting is needed to verify our discoveries and take the developmental process of treatment to the next step.
Introduction
Soft tissue sarcomas (STSs) are a heterogeneous group of rare malignant tumors. The treatment is primarily surgical resection with or without radiotherapy, but systemic chemotherapy with cytotoxic anticancer agents may improve the prognosis of STS [16, 18]. However, advanced STSs still have a poor prognosis, and effective systemic therapies have not been established. Therefore, identifying novel therapeutic targets that might identify effective therapies is desirable. Tyrosine kinases, especially tyrosine kinase fusion genes including ALK, ROS1, and RET, are increasingly being used as therapeutic targets for a variety of cancers [25, 28]. Recent reports revealed that tyrosine kinase inhibitors specific to NTRK1-3 exhibit antitumor activity in a small population of STSs carrying the NTRK fusion gene [8, 14, 15]. Multitarget assays, mainly DNA-based, with next-generation sequencing have recently become common in clinical diagnostic laboratories in the cancer field [2, 27, 31]. However, DNA-based gene panel assays are sometimes inadequate to detect fusion genes, no matter how well-designed they are [2, 11, 12]. Therefore, multitarget assays with RNA-based systems may be a better alternative for detecting fusion genes.
Genetically, STSs are classified into two types. Translocation-associated sarcomas have been shown to have translocations, activating mutations, and simple gene copy number alterations [26]. These are associated with specific histologic subtypes and are believed to be involved in the oncogenic process in these sarcomas. The second type is complex karyotype (nontranslocation) sarcomas, including leiomyosarcoma and malignant peripheral nerve sheath tumor (MPNST). Unlike translocation-associated sarcomas, for which a potential target could be designed that focuses on these known lesions, these tumors have complex, numerous, and patternless genetic aberrations that make the identification of potential drug targets more difficult. Very few specific molecular events, including receptor tyrosine kinase fusion genes, have been defined in complex karyotype sarcomas [6, 16, 26]. With respect to potentially actionable genetic events, large-scale clinical sequencing revealed that one-third of STSs have actionable genetic events composed of mainly copy number alterations, including MDM2 and CDK4, nonsynonymous mutations such as those for PIK3CA and PTEN, and a small number of receptor tyrosine kinase fusions [31]. Complex karyotype sarcomas tend to carry more potentially actionable genetic alterations than do translocation-associated sarcomas [31]. However, while reviewing the database regarding the frequencies of actionable genetic events in STSs, we found that leiomyosarcoma and MPNST have lower frequencies of potential targets than other nontranslocation STSs, although they have been classified as complex karyotype (nontranslocation) sarcomas [31]. Furthermore, we theorized that these findings might suggest that both leiomyosarcoma and MPNST are included in any unique translocations harboring tyrosine kinase fusion genes. Given this background, we conducted tyrosine kinase screening in complex karyotype sarcomas using NanoString assays (NanoString Technologies Inc, Seattle, WA, USA) to detect an imbalance in the expression of mRNA between the 5’- and 3’-sides in tyrosine kinase genes and to identify novel therapeutic tyrosine kinase targets including tyrosine kinase fusions in STSs, especially leiomyosarcoma and MPNST [28].
We therefore asked: (1) Are there novel therapeutic tyrosine kinase targets in tumors from patients with soft tissue sarcomas that are detectable using mRNA screening focusing on imbalance expressions between the 5’ and 3’ end of the kinase domain? (2) Can potential targets be verified by RNA sequencing and reverse transcription PCR (RT-PCR)? (3) Will potential fusion gene(s) transform cells in in vitro assays? (4) Will tumors in mice that have an identified fusion gene respond to treatment with a therapeutic drug directed at that target?
Materials and Methods
Experimental Overview and Primary/Secondary Study Endpoints
Our primary study goal was the identification of tyrosine kinase fusion genes in STSs (Fig. 1). To achieve this, we conducted NanoString-based transcriptome assay and analyzed 24 tumor samples with STSs, including 15 leiomyosarcomas, five MPNSTs, and four high-grade myxofibrosarcomas (Step 1). This assay screened the expression balances on the transcripts of 90 tyrosine kinases at two points: the 5’ and 3’ endpoints of the kinase domain. RNA sequencing and confirmatory RT-PCR validated the existance of tyrosine kinase fusion genes in presumed novel fusion events, identified by NanoString (Step 2). Our secondary study goals were the confirmation of the identified tyrosine kinase gene fusions that were associated with oncogenic abilities and drug responses (Fig. 1). Then, functional analyses consisting of in vitro (focus formation assay [Step 3]) and in vivo assays (xenograft tumor assays [Step 4]) were conducted.
Fig. 1.
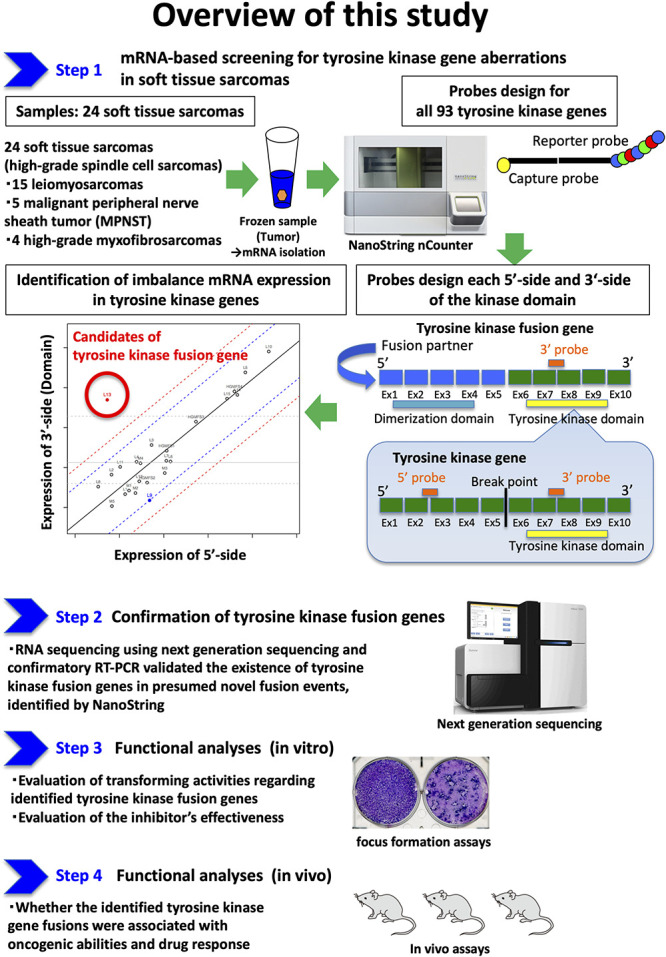
Step 1: To identify tyrosine kinase fusion genes, NanoString-based transcriptome assay analyzed 24 tumor samples with STSs including 15 leiomyosarcomas, five with MPNSTs, and four with high-grade myxofibrosarcomas. This assay screened the expression balances regarding transcripts of 90 tyrosine kinases at two points: the 5’ and 3’ endpoints of the kinase domain. Step 2: RNA sequencing using next generation sequencing and confirmatory reverse transcription-polymerase chain reaction (RT-PCR) validated whether NanoString identified existing tyrosine kinase fusion genes in presumed novel fusion events. Step 3: Functional in vitro analyses were performed to elucidate whether the identified tyrosine kinase gene fusions were associated with oncogenic abilities. Step 4: Functional analyses consisting of in vivo assays were also performed to elucidate whether the identified tyrosine kinase gene fusions were associated with oncogenic abilities and drug responses.
Patient Sample
Data on 24 patients with STSs consisting of 15 patients with leiomyosarcomas, five with MPNSTs, and four with high-grade myxofibrosarcomas were collected and analyzed using a protocol approved by the institutional review boards at Juntendo University (No. 2018135) and Tokyo Medical and Dental University. Informed consent was obtained from the patients in this study (Table 1). Histologic subtypes were determined by two board-certified pathologists (TS, TH) based on histologic features, immunohistochemical staining, and the WHO classification of tumors [16]. Sample collection was conducted between 2015 and 2019, when 172 patients with STSs were treated with surgical resection at Juntendo University Hospital. To be eligible for this study, a patient needed to have high-grade spindle cell and nontranslocation sarcomas, including leiomyosarcoma, MPNST, and high-grade myxofibrosarcoma; 72 patients met these criteria. In planning the analysis of leiomyosarcoma and MPNST, we found analysis size (a total of 24 samples) during the start of the study in 2016 was limited. Therefore, for leiomyosarcoma, we employed 11 leiomyosarcoma samples in the 2015 and 2016 series and excluded those (n = 2) patients who did not have sufficient frozen tumor samples and those who declined to participate, leaving a total of nine patients. We additionally collected six leiomyosarcoma samples from a sample bank at the Tokyo Medical and Dental University Hospital and analyzed a total of 15 leiomyosarcomas in this study. For MPNSTs, all five samples were collected from a sample bank at the Tokyo Medical and Dental University Hospital. To bring the sample size to 24 patients, we added four high-grade myxofibrosarcoma samples in the 2015 series from Juntendo University. These resected samples had not received any pretreatment with radiation or chemotherapy. Imbalances in the expression of mRNA in tyrosine kinase genes were screened in these 24 patients with STSs using NanoString analysis (Table 1).
Table 1.
Characteristics of patients in whom tumors were assessed
| Sample | Age in years | Sex | Location |
| LMS1 | 37 | Male | Thigh |
| LMS2 | 54 | Female | Groin |
| LMS3 | 38 | Female | Groin |
| LMS4 | 73 | Male | Groin |
| LMS5 | 67 | Female | Forearm |
| LMS6 | 47 | Male | Abdominal wall |
| LMS7 | 65 | Female | Forearm |
| LMS8 | 47 | Male | Chest wall |
| LMS9 | 68 | Female | Upper arm |
| LMS10 | 86 | Male | Upper arm |
| LMS11 | 69 | Male | Thigh |
| LMS12 | 83 | Female | Forearm |
| LMS13 | 90 | Female | Thigh |
| LMS14 | 71 | Female | Buttocks |
| LMS15 | 71 | Female | Thigh |
| MPNST1 | 62 | Male | Shoulder |
| MPNST2 | 70 | Male | Knee |
| MPNST3 | 52 | Female | Thigh |
| MPNST4 | ND | ND | ND |
| MPNST5 | ND | ND | ND |
| HGMFS1 | 75 | Female | Chest wall |
| HGMFS2 | 47 | Male | Upper arm |
| HGMFS3 | 78 | Male | Lower leg |
| HGMFS4 | 75 | Male | Upper arm |
LMS = leiomyosarcoma; MPNST = malignant peripheral nerve sheath tumor; HGMFS = high-grade myxofibrosarcoma; ND = not determined.
RNA Extraction and cDNA Synthesis
RNA was extracted from frozen samples using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), and first-strand synthesis was performed using 5 μg of RNA and the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA).
NanoString Analysis
To identify imbalances in the expression of mRNA between the 5’- and 3’-sides of 93 tyrosine kinase genes (Table 2), we used the NanoString system (NanoString Technologies Inc) using our original custom probes based on our previous study [28] (Supplemental Fig. 1; Supplemental Digital Content 1, http://links.lww.com/CORR/A465). In the NanoString system, 400 ng of RNA was hybridized to probes (a reporter probe and a capture probe) at 65°C for 18 to 24 hours using a thermal cycler. Samples were then inserted into the nCounter Prep Station (NanoString Technologies Inc) for removal of excessive probes, purification, and immobilization onto the internal surface of a sample cartridge for 3 hours. Finally, the sample cartridge was transferred to the nCounter Digital Analyzer (NanoString Technologies Inc), where color codes were counted and tabulated for each target molecule. The expression number of the base sequence of the probes was calculated using nSolver Analysis Software (NanoString Technologies Inc) (Supplemental Table 1; Supplemental Digital Content 2, http://links.lww.com/CORR/A466). The raw data were statistically analyzed and plotted on graphs (abscissa: log2 [5’-side expression], ordinate: log2 [3’-side expression]) with 2 and 3 SD lines. If either 3’- or 5’-side expression showed higher than 10-fold SD or plotted on the area higher than 2 SDs of the graph, the abnormality was judged as having imbalanced gene expression.
Table 2.
Tyrosine kinase genes investigated
| Number | Gene name | Number | Gene name | Number | Gene name |
| 1 | AATK | 32 | ERBB4 | 63 | MST1R |
| 2 | ABL1 | 33 | FER | 64 | MUSK |
| 3 | ABL2 | 34 | FES | 65 | NTRK1 |
| 4 | ALK | 35 | FGFR1 | 66 | NTRK2 |
| 5 | ARAF | 36 | FGFR2 | 67 | NTRK3 |
| 6 | AXL | 37 | FGFR3 | 68 | PDGFRA |
| 7 | BLK | 38 | FGFR4 | 69 | PDGFRB |
| 8 | BMX | 39 | FGR | 70 | PTK2 |
| 9 | BRAF | 40 | FLT1 | 71 | PTK2B |
| 10 | BTK | 41 | FLT3 | 72 | PTK6 |
| 11 | CSF1R | 42 | FLT4 | 73 | PTK7 |
| 12 | CSK | 43 | FRK | 74 | RAF1 |
| 13 | DDR1 | 44 | FYN | 75 | RET |
| 14 | DDR2 | 45 | HCK | 76 | ROR1 |
| 15 | EGFR | 46 | IGF1R | 77 | ROR2 |
| 16 | EPHA1 | 47 | INSR | 78 | ROS1 |
| 17 | EPHA2 | 48 | INSRR | 79 | RYK |
| 18 | EPHA3 | 49 | ITK | 80 | SRC |
| 19 | EPHA4 | 50 | JAK1 | 81 | SRMS |
| 20 | EPHA5 | 51 | JAK2 | 82 | STYK1 |
| 21 | EPHA6 | 52 | JAK3 | 83 | SYK |
| 22 | EPHA7 | 53 | KDR | 84 | TEC |
| 23 | EPHA8 | 54 | KIT | 85 | TEK |
| 24 | EPHA10 | 55 | LCK | 86 | TIE1 |
| 25 | EPHB1 | 56 | LMTK2 | 87 | TNK1 |
| 26 | EPHB2 | 57 | LMTK3 | 88 | TNK2 |
| 27 | EPHB3 | 58 | LTK | 89 | TXK |
| 28 | EPHB4 | 59 | LYN | 90 | TYK2 |
| 29 | EPHB6 | 60 | MATK | 91 | TYRO3 |
| 30 | ERBB2 | 61 | MERTK | 92 | YES1 |
| 31 | ERBB3 | 62 | MET | 93 | ZAP70 |
AATK = apoptosis associated tyrosine kinase; ABL1 = ABL proto-oncogene 1, non-receptor tyrosine kinase; ABL2 = ABL proto-oncogene 2, non-receptor tyrosine kinase; ALK = ALK receptor tyrosine kinase; ARAF = a-raf proto-oncogene, serine/threonine kinase; AXL = AXL receptor tyrosine kinase; BLK = BLK proto-oncogene, src family tyrosine kinase; BMX = BMX non-receptor tyrosine kinase; BRAF = b-raf proto-oncogene, serine/threonine kinase; BTK = bruton tyrosine kinase; CSF1R = colony stimulating factor 1 receptor; CSK = c-terminal src kinase; DDR1 = discoidin domain receptor tyrosine kinase 1; DDR2 = discoidin domain receptor tyrosine kinase 1; EGFR = epidermal growth factor receptor; EPHA1 = erythropoietin-producing hepatoma receptor a1; EPHA2 = erythropoietin-producing hepatoma receptor a2; EPHA3 = erythropoietin-producing hepatoma receptor a3; EPHA4 = erythropoietin-producing hepatoma receptor a4; EPHA5 = erythropoietin-producing hepatoma receptor a5; EPHA6 = erythropoietin-producing hepatoma receptor a6; EPHA7 = erythropoietin-producing hepatoma receptor a7; EPHA8 = erythropoietin-producing hepatoma receptor a8; EPHA10 = erythropoietin-producing hepatoma receptor a10; EPHB1 = EPH receptor b1; EPHB2 = EPH receptor b2; EPHB3 = EPH receptor b3; EPHB4 = EPH receptor b4; EPHB6 = EPH receptor b6; ERBB2 = erb-b2 receptor tyrosine kinase 2; ERBB3 = erb-b2 receptor tyrosine kinase 3; ERBB4 = erb-b2 receptor tyrosine kinase 3; FER = proto-oncogene tyrosine-protein kinase FER; FES = FES proto-oncogene, tyrosine kinase; FGFR1 = fibroblast growth factor receptor 1; FGFR2 = fibroblast growth factor receptor 2; FGFR3 = fibroblast growth factor receptor 3; FGFR4 = fibroblast growth factor receptor 4; FGR = FGR proto-oncogene, src family tyrosine kinase; FLT1 = FMS related receptor tyrosine kinase 1; FLT3 = FMS related receptor tyrosine kinase 3; FLT4 = FMS related receptor tyrosine kinase 4; FRK = fyn related src family tyrosine kinase; FYN = FYN proto-oncogene, src family tyrosine kinase; HCK = HCK proto-oncogene, src family tyrosine kinase; IGF1R = insulinlike growth factor 1 receptor; INSR = insulin receptor; INSRR = insulin receptor related receptor; ITK = il2 inducible t cell kinase, JAK1 = janus kinase 1; JAK2 = janus kinase 2; JAK3 = janus kinase 3; KDR = kinase insert domain receptor; KIT = KIT proto-oncogene; receptor tyrosine kinase, LCK = LCK proto-oncogene, src family tyrosine kinase; LMTK2 = lemur tyrosine kinase 2; LMTK3 = lemur tyrosine kinase 3; LTK = leukocyte receptor tyrosine kinase; LYN = LYN proto-oncogene, src family tyrosine kinase; MATK = megakaryocyte-associated tyrosine kinase; MERTK = MER proto-oncogene, tyrosine kinase; MET = MET proto-oncogene, receptor tyrosine kinase; MST1R = macrophage-stimulating 1 receptor; MUSK = muscle-associated receptor tyrosine kinase; NTRK1 = neurotrophic receptor tyrosine kinase 1; NTRK2 = neurotrophic receptor tyrosine kinase 2; NTRK3 = neurotrophic receptor tyrosine kinase 3; PDGFRA = platelet-derived growth factor receptor alpha; PDGFRB = platelet-derived growth factor receptor beta; PTK2 = protein tyrosine kinase 2; PTK2B = protein tyrosine kinase 2 beta; PTK6 = protein tyrosine kinase 6; PTK7 = protein tyrosine kinase 7; RAF1 = raf-1 proto-oncogene, serine/threonine kinase; RET = ret proto-oncogene; ROR1 = receptor tyrosine kinase-like orphan receptor 1; ROR2 = receptor tyrosine kinase-like orphan receptor 2; ROS1 = ROS proto-oncogene 1, receptor tyrosine kinase; RYK = receptor-like tyrosine kinase; SRC = SRC proto-oncogene, non-receptor tyrosine kinase; SRMS = src-related kinase lacking c-terminal regulatory tyrosine and n-terminal myristylation sites; STYK1 = serine/threonine/tyrosine kinase 1; SYK = spleen-associated tyrosine kinase; TEC = tec protein tyrosine kinase; TEK = TEK receptor tyrosine kinase; TIE1 = tyrosine kinase with immunoglobulin-like and egf-like domains 1; TNK1 = tyrosine kinase non-receptor 1; TNK2 = tyrosine kinase non-receptor 2; TXK = TXK tyrosine kinase; TYK2 = tyrosine kinase 2; TYRO3 = TYRO3 protein tyrosine kinase; YES1 = YES proto-oncogene 1, src family tyrosine kinase; ZAP70 = zeta chain of t cell receptor associated protein kinase 70.
Detection of Fusion Genes by RNA Sequencing
Total RNA was extracted from fresh frozen samples using RNA-Bee (Tel-Test Inc, Gainesville, FL, USA), followed by treatment with DNase I (Thermo Fisher Scientific) and poly(A)-RNA selection before cDNA synthesis. The library used for the RNA sequencing was prepared with a NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) in accordance with the manufacturer’s protocol. Sequencing was conducted from both ends of each cluster using the HiSeq 2500 System (Illumina, San Diego, CA, USA). RNA sequencing was aligned to hg19 using TopHat (v2.0.9; https://ccb.jhu.edu/software/tophat/index.shtml). The expression level of each gene was calculated using Cufflinks (v2.1.1; http://cole-trapnell-lab.github.io/cufflinks), and gene fusion was detected using the deFuse pipeline (https://bitbucket.org/dranew/defuse).
Real-time Polymerase Chain Reaction
We performed real-time PCR analyses to confirm the expression of MAN1A1-ROS1 using PCR SuperMix (Thermo Fisher Scientific). MAN1A1-ROS1 was amplified by combining either of the following primers: MAN1A1: 5’- TGGTGGACTACTCTCAGCCT -3’, 5’- ACGCTTTGTTGGTGGACTACT -3’, 5’-GGAGAAGAAGAAGGTGGCCC-3’, and 5’-CGCGAGAAAAGGGCAAAGAT-3’; and ROS1: 5’-TCTTCAGCTTTCTCCCACTGT-3’, 5’-TCAGCTTTCTCCCACTGTATTGA-3’, and 5’-GCAAACACTACTGCAGGATCC-3’. The expression of glyceraldehyde-3-phosphate dehydrogenase was used to control the RNA quality with the following primers: 5’-GAAGGTGAAGGTCGGAGTC-3’ and 5’- GAAGATGGTGATGGGATTT-3’.
Immunohistochemistry
To diagnose leiomyosarcoma accurately, we performed an immunohistochemical analysis with the streptavidin-biotin method using the antibodies desmin (mouse monoclonal, 1:100; Roche, Basel, Switzerland), smooth muscle actin (mouse monoclonal, 1:200; Dako, Santa Clara, CA), M-actin (mouse monoclonal, 1:100; Dako), and h-Caldesmon (mouse monoclonal, 1:1; Dako). Two board-certified pathologists (TS, TH) assessed the immunostaining independently. The assessment was qualitative. When discrepancies arose, the slides were reviewed using a multiheaded microscope to reach a consensus.
Preparation of Retrovirus, Gene Transduction into 3T3 Cells, and Focus Formation Assay
Recombinant plasmids were introduced with packaging plasmids (Takara Bio Inc, Shiga, Japan) into HEK293T cells to obtain recombinant retroviral particles. For the focus formation assay, 3T3 mouse fibroblasts purchased from the American Type Culture Collection were infected with ecotropic recombinant retroviruses using 4 μg/mL of polybrene (Sigma-Aldrich, St Louis, MO, USA) for 24 hours. With respect to focus formation assay, they were further cultured for up to 2 weeks in Dulbecco’s modified eagle medium-nutrient mixture F-12 supplemented with 5% bovine serum albumin. A focus formation assay was performed using 3T3 cells infected with recombinant retroviruses expressing wild-type cDNAs (ROS1), mutant cDNAs (CD74-ROS1: [CD74_exon 6 / ROS1_exon 32] and MAN1A1-ROS1: [MAN1A_exon 5 / ROS1_exon 31]) and green fluorescent protein as a negative control. After 2 weeks, the cells were stained with Giemsa solution and transforming activity was measured. Cell transformation was performed quantitatively in triplicate and assessed by staining with Giemsa solution as quantitative.
Xenograft Tumor Assays
We performed in vivo animal model studies to clarify the antitumor activity of crizotinib in the ROS1 fusion-transfected cells. All animal studies were conducted following the protocols approved by the Animal Ethics Committee of the National Cancer Research Center, Tokyo, Japan. Before injection, 3T3 cells (1.0 × 106) were mixed in phosphate-buffered saline with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) at a 1:1 ratio. The cell suspension was injected subcutaneously (200 µL/mouse) into the back of 6-week-old female BALB/c nude mice (CREA Japan, Tokyo, Japan). In accordance with our previous study, which similarly evaluated the drug efficacy using a 3T3 xenograft model, five age- and sex-matched mice of this strain were randomly assigned to each group (wild-type, ROS1 fusions, and crizotinib treatments) [29]. The mice were inoculated subcutaneously with transfected cells with ROS1 wild-type and ROS1 fusion genes including CD74-ROS1 and MAN1A1-ROS1. These inoculated mice were administered either crizotinib or a vehicle (dimethyl sulfoxide). Additionally, ROS1-transfected cells (control) with no drug administration were also analyzed. When tumors reached approximately 100 to 150 mm3 in size, the mice were randomized into two groups. Either 50 mg/kg of crizotinib or vehicle control was administered by intraperitoneal injection. Both treatments were administered for 5 days per week over a series of weeks. Tumor volume was used as the endpoint to directly evaluate drug efficacy with regard to tumor growth inhibition. The tumor volume (mean ± SD) in each group is expressed in cubic millimeters and was calculated using the formula π/6 × (largest diameter) × (smallest diameter)2. Tumor size assessment was completed by observers (SK, SM, NH) blinded to the study groups of the animals and their treatments. The mice were euthanized when the tumor reached 1.5 cm in diameter, and the collected data were analyzed.
Results
Are There Novel Therapeutic Tyrosine Kinase Targets in Tumors from Patients with STSs That are Detectable Using mRNA Screening?
Using scatter plots constructed with data acquired form the NanoString system, we identified an imbalance in the expression of mRNA in tumors in two patients. These imbalances consisted of ROS1 in a patient with a leiomyosarcoma and NTRK1 in a patient with a high-grade myxofibrosarcoma (Fig. 2).
Fig. 2.
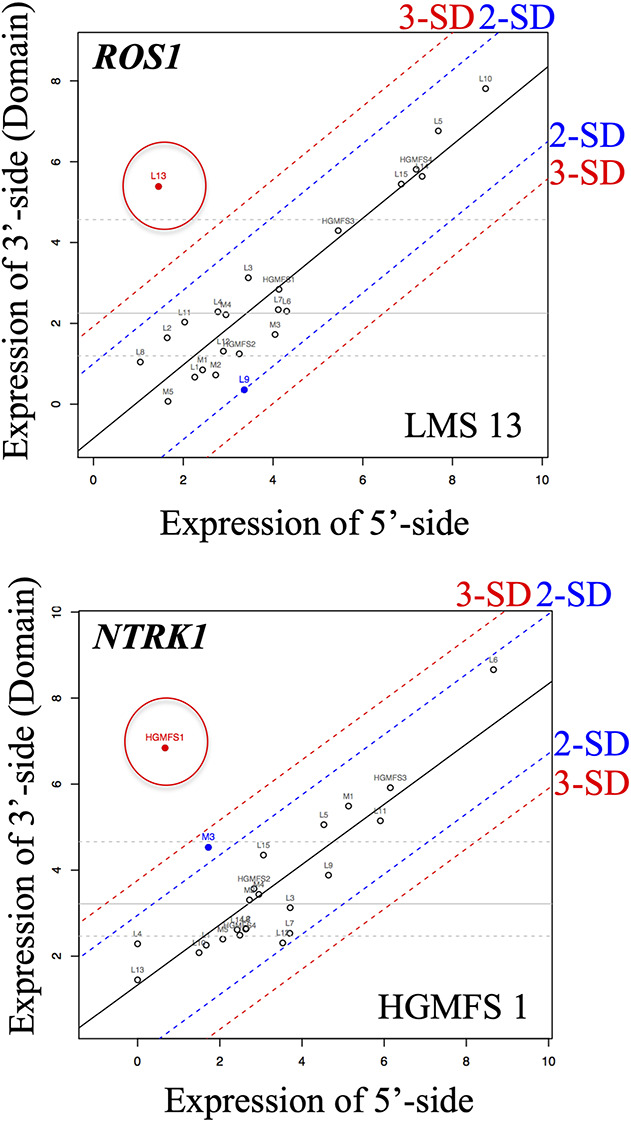
NanoString assay results for ROS1 and NTRK1. The ROS1 gene showed imbalance in the expression of mRNA between the 5’- and 3’- sides in a tumor in a patient with leiomyosarcoma (patient LMS13 in Table 1; 5'-side value: 2.7, 3'-side value: 41.9, the rate of 3'/5'-side value showed over 3 SD). In patients with high-grade myxofibrosarcoma, NTRK1 showed differences in the expression of mRNA between the 5’- and 3’- sides in an HGMFS1 tumor in a patient with high-grade myxofibrosarcoma (patient HGMFS1 in Table 1; 5'-side value: 1.6, 3'-side value: 114.7, the rate of 3'/5'-side value showed over 3 SD). These samples were subjected to RNA sequencing, and MAN1A1-ROS1 was identified in the LMS13 tumor; LMS = leiomyosarcoma, HGMFS = high-grade myxofibrosarcoma, and the dots circled indicate candidates of tyrosine kinase fusion gene.
Can Potential Targets be Verified by RNA Sequencing and RT-PCR?
The results of RNA sequencing in two patients with an imbalance in ROS1 and NTRK1 mRNA expression identified a novel MAN1A1-ROS1 (MAN1A1_exon 5 / ROS1_exon 31) fusion gene in the patient with a leiomyosarcoma (Supplemental Fig. 2; Supplemental Digital Content 3, http://links.lww.com/CORR/A467). Regarding the NTRK1 gene, there was no rearrangement in the tumor in the patient with a high-grade myxofibrosarcoma based on RNA sequencing (Supplemental Fig. 3; Supplemental Digital Content 4, http://links.lww.com/CORR/A468). The in-frame fusion of MAN1A1 exon 5 to ROS1 exon 31 was confirmed with independent RT-PCR, and the RT-PCR product was also sequence-verified (Fig. 3A-B). This tumor was sampled from the thigh of a 90-year-old woman (Fig. 3C-D). Histologically, the tumor was composed of fascicular proliferation of spindle-shaped cells (Fig. 3E). Tumor cells with bizarre giant nuclei were also scattered throughout the lesion (Fig. 3F). Immunohistochemistry revealed that the spindle tumor cells were positive for desmin, smooth muscle actin, M-actin, and h-caldesmon (Fig. 3G-J). Additionally, these tumor cells showed an MIB-1 (Ki-67) index of approximately 60%. Based on the histologic features, immunohistochemistry findings, and imaging studies, this patient had a diagnosis of stage III pleomorphic leiomyosarcoma. The primary lesion in the thigh was resected and used for further analyses. This patient was followed with imaging studies and did not receive adjuvant chemotherapy because of her older age. Two years postoperatively, a metastasis developed in the pelvis. Based on the patient’s genetic profile, the ROS1 inhibitor crizotinib was considered to be beneficial to the patient. However, because the patient was older than 90 years when the metastasis developed, we decided not to use the inhibitor against the ROS1 fusion-positive tumor to avoid any adverse events. At 2.5 years postoperatively, the patient died.
Fig. 3.
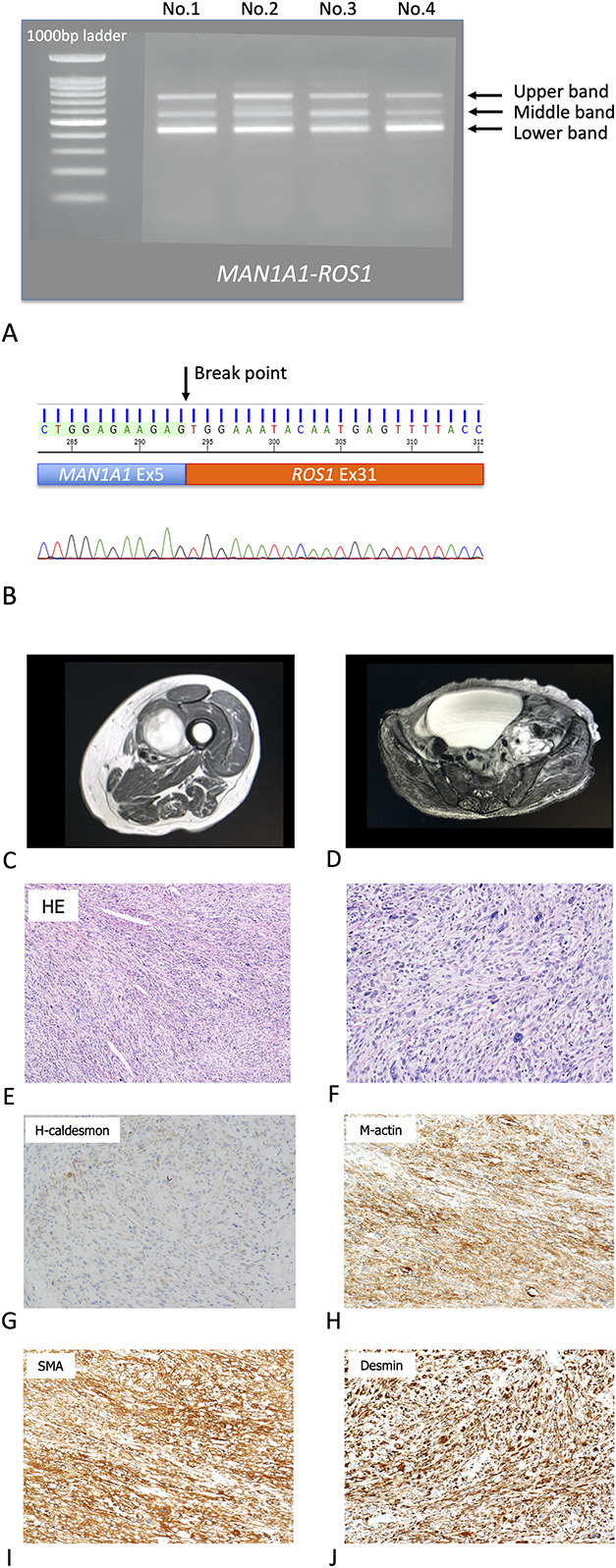
A-J (A) Identification of MAN1A1-ROS1 fusion. Real-time polymerase chain reaction showed three bands in the LMS13 tumor sample. An upper band revealed MAN1A1 exon 5 to ROS1 exon 31 as in-frame. A middle band revealed MAN1A1 exon 5 to ROS1 exon 32 as frameshift. A lower band revealed MAN1A1 exon 5 to ROS1 exon 33 as frameshift. We repeated gels of the same experiment four times (No.1 - 4). (B) Sanger sequencing using the cDNA product showed in-frame fusion of MAN1A1 exon 5 to ROS1 exon 31 in the tumor sample. We repeated gels of the same experiment four times (No.1 - 4). (C) This image shows a soft tissue mass in the left thigh of a 90-year-old woman. T2-weighted MRI showed a 5.0-cm mass in the abductor muscles. (D) After 2 years, a metastasis developed. T2-weighted MRI showed an 8.5-cm mass in the left retroperitoneum. (E) The histologic diagnosis was leiomyosarcoma (the low power microscope of HE). (F) A high-power microscope of HE showed several pleomorphic tumor cells. (G) Immunohistochemical staining was performed using h-Caldesmon, and the immunostaining test result was diffuse positive. (H) Immunohistochemical staining was performed using M-actin, and the immunostaining test showed positive. (I) Immunohistochemical staining was performed using smooth muscle actin, and the result was positive. (J) Immunohistochemical staining was performed using desmin, and tumor cells stained as positive.
Will Potential Fusion Genes Transform Cells in In Vitro Assays?
In the focus formation assay, plate A shows the top row represent GFP (control) and the bottom row represent Empty (control) (Fig. 4A). Plate B shows the top row represent ROS1 wild-type (control) and the bottom row represent EGFR L858R (positive control) (Fig. 4B). Plate C shows the top row represent CD74-ROS1 (positive control) and the bottom row represent MAN1A1-ROS1 (Fig. 4C). Accumulated foci were observed in the CD74-ROS1 and MAN1A1-ROS1 fusion-transfected cells, indicating that a strong transforming potential was associated with these rearrangements (Fig. 4D) compared with the negative control. With respect to the effectivities of the ROS1 inhibitor in vitro, in the ROS1-transfected cells, including MAN1A1-ROS1, CD74-ROS1, and the controls, crizotinib treatment inhibited cell viabilities in ROS1 fusion transfected cells, including MAN1A1-ROS1 and CD74-ROS1 fusions (Supplemental Fig. 4; Supplemental Digital Content 5, http://links.lww.com/CORR/A469). These results demonstrated that crizotinib exerted anticell activities in ROS1 fusion cells, including MAN1A1-ROS1 and CD74-ROS1 fusions in vitro.
Fig. 4.
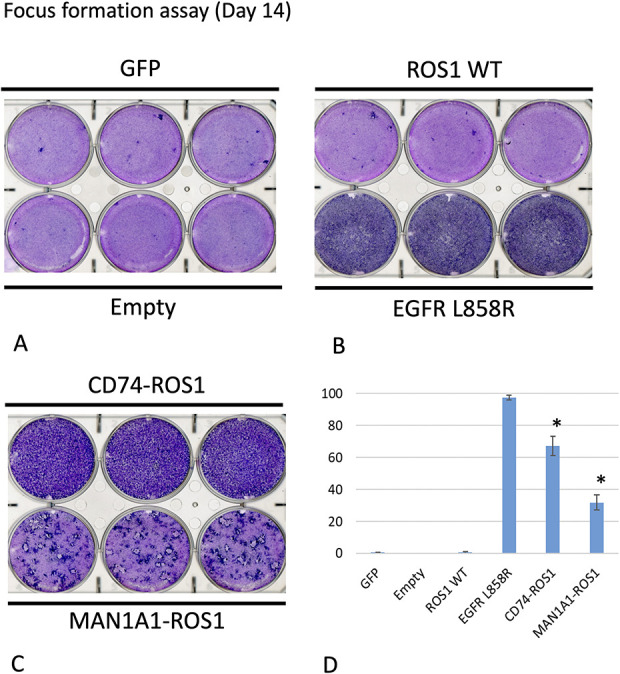
A-D (A) This image shows a functional assay of ROS1 variants. An NIH 3T3 focus formation assay was performed in triplicate. A MAN1A1-ROS1 mutant was transduced into a 3T3 mouse fibroblast cell line to investigate its transforming potential. Plate A shows the top row represent GFP (control) and the bottom row represent Empty (control). (B) Plate B shows the top row represent ROS1 wild-type (control) and the bottom row represent EGFR L858R (positive control). (C) Plate C shows the top row represent CD74-ROS1 (positive control) and the bottom row represent MAN1A1-ROS1. Focus formation after culturing for 14 days in 5% bovine serum albumin was evaluated in 3T3 cells transduced with MAN1A1-ROS1 or control genes (GFP, EGFR L8585R, ROS1 wild-type, or CD74-ROS1). (D) The focus number was quantified using ImageJ software (NIH, Bethesda, MD, USA) and normalized to that of the negative control. *p < 0.01 versus ROS1 WT, t-test. These assays showed the MAN1A1-ROS1 had the highest focus number.
Will Tumors in Mice That Have an Identified Fusion Gene Respond to Treatment with a Therapeutic Drug Directed Against That Target?
To detect whether tumor presence of this fusion gene would respond to a drug treatment with crizotinib, we conducted in vivo assays, which revealed no differences in tumor growth between ROS1 fusion genes (vehicle: CD74-ROS1 and MAN1A1-ROS1) and control (ROS1 wild-type) (Fig. 5). There was a difference in tumor growth between mice with crizotinib treatment and those with the vehicle in ROS1 fusion genes (CD74-ROS1 mean ± SD: 308 ± 178 mm3 versus 1276 ± 533 mm3; 253 mm3 < the mean difference with 95% CI < 1809 mm3; p < 0.01 and MAN1A1-ROS1 mean ± SD: 123 ± 53 mm3 versus 2304 ± 854 mm3; 1156 mm3 < the mean difference with 95% CI < 3206 mm3; p < 0.01) (Fig. 5). These results indicate that the novel MAN1A1-ROS1 fusion gene had oncogenic abilities and crizotinib exhibited anti-tumor activity in the MAN1A1-ROS1 fusion cells.
Fig. 5.
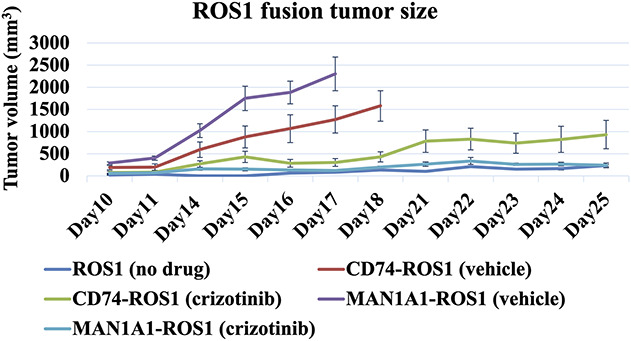
Formation and crizotinib treatment in vivo. 3T3 cells with MAN1A1-ROS1, CD74-ROS1, or ROS1 wild-type were subcutaneously injected into female BALB/c nude mice. The tumor growth with crizotinib or vehicle (dimethyl sulfoxide) treatment was evaluated. This graph shows tumor growth in size/volume versus time for the different groups (no crizotinib treatment: MAN1A1-ROS1, CD74-ROS1, or ROS1 wild-type [control], crizotinib treatment: MAN1A1-ROS1, CD74-ROS1). With respect to comparisons including MAN1A1-ROS1, CD74-ROS1, or ROS1 wild-type (control) with no crizotinib treatment, on day 17, the cells expressing MAN1A1-ROS1 and CD74-ROS1 had bigger tumor volumes than ROS1 wild-type (control). Regarding crizotinib treatments, on day 17, the tumor volume of the cells expressing MAN1A1-ROS1 that were treated with crizotinib were compared with corresponding cells treated with the vehicle. Error bars, SD; *p < 0.01.
Discussion
STSs, including leiomyosarcoma, are malignant tumors that have poor prognoses, and novel systemic therapies are being searched for to improve patient survival. Recently, tyrosine kinase fusion genes, including ALK, ROS1, RET, and NTRK, have been found to be effective therapeutic targets for a variety of cancers [25, 28]. The comprehensive analysis of cancer-related genes that was innovated by Next Generation Sequencing technology made tyrosine kinase targets more accessible, having opened new avenues in the treatment modalities for various tumor types [27, 8, 14, 15]. However, the identified frequencies of tyrosine kinase targets in STSs, including leiomyosarcoma, are still low. DNA-based gene panel assays are the current main tests in clinical settings. However, there are technical issues; DNA-based gene panel assays are often inadequate for detecting tyrosine kinase fusion genes compared with RNA-based panel assays [2, 11, 12]. Therefore, in this study, we conducted a comprehensive transcriptome screening for all tyrosine kinases to identify new therapeutic targets in STSs. We successfully identified a novel MAN1A1-ROS1 fusion gene in a leiomyosarcoma with mRNA screening, and MAN1A1-ROS1 rearrangement revealed a strong transforming potential and was inhibited by crizotinib. These findings potentially provide a novel therapeutic option for this intractable malignancy in STSs.
Limitations
This study has several limitations. We performed tyrosine kinase screening using only 24 nontranslocation-related sarcomas, and in the 24 tumors we investigated, only one had a “targetable” fusion product. To clarify the accuracy of the tyrosine kinase fusion rates in nontranslocation-related sarcomas, we must conduct screening using larger STS cohorts, but we view this study as a first step to encourage more investigations. As a second limitation, with respect to the malignant transformation of the MAN1A1-ROS1 fusion gene, is that we performed a focus formation assay on ROS1 fusions, including CD74-ROS1 and MAN1A1-ROS1, and confirmed that the MAN1A1-ROS1 fusion gene had strong transforming potential. We also confirmed that the strong transforming potential of MAN1A1-ROS1 had the same power as that of the commonly mutated ROS1 fusion gene (CD74-ROS1). Furthermore, in vivo assays successfully verified the malignant potential of MAN1A1-ROS1 as tumor growths. Therefore, since the in vitro and in vivo assays have classified malignant potential with tumor growths, we did not conduct the other cell growth assays, including CCK-8 cell proliferation and transwell migration assays in this study. Additionally, several articles have described these assays using several partner types of ROS1 fusion genes, including CD74-ROS1 fusion, and confirmed their cell proliferation and invasion potentials [10, 21]. Therefore, we assumed that MAN1A1-ROS1 fusion would have their proliferation and invasion potentials. As a third limitation, using in vivo assay, the tumor of MAN1A-ROS1 fusion has not been verified for the differentiation of leiomyosarcomas using immunohistochemistry in this study. However, we confirmed that the tumor cells had MAN1A1-ROS1 fusion (data not shown). As a fourth limitation, with respect to the false-positive detection of NTRK1 fusion in the NanoString assay in this study, the 5’-side probe (NTRK1_Ex1/2 probe) was designed to detect the break point between exon 1 and exon 2 in NTRK1 (Supplemental Table 1; Supplemental Digital Content 2, http://links.lww.com/CORR/A466), and our validation study with RNA sequencing revealed that exon 2 had fewer RNA reads than the other exons in NTRK1 did (Supplemental Fig. 3; Supplemental Digital Content 4, http://links.lww.com/CORR/A468). Therefore, we supposed that the 5’-side probe might have detected a few RNA reads in exon 2 instead of the fusion gene.
Are There Novel Therapeutic Tyrosine Kinase Targets in Tumors from Patients with STSs That Are Detectable Using mRNA Screening?
We wanted to investigate whether the NanoString assay and a confirming RNA panel in a next-generation sequencing assay using RNAs may aid in identifying novel therapeutic gene fusions in STSs, especially nontranslocation-related sarcomas, and detect MAN1A1-ROS1 fusion in a tumor from a patient with leiomyosarcoma. The NanoString nCounter assay is a single assay that can use RNA isolated from frozen and formalin-fixed paraffin-embedded samples [5]. It uses a high-throughput hybridization technique using target-specific probes that can be customized to test for numerous fusion transcripts. A previous study on STSs using a NanoString assay targeting histologic fusion genes, including 174 fusion junctions in 25 sarcoma subtypes, successfully identified histologic fusion genes in 96 of 212 patients with STSs [4]. Furthermore, our previous assay for tyrosine kinase screening identified KIF5B-RET and CAPG-ROS1 fusion genes in non-small cell lung cancers [28]. Based on these previous studies and our current findings, we suggest that mRNA screening might be able to detect novel therapeutic tyrosine kinase targets in STSs.
Can Potential Targets be Verified by RNA Sequencing and RT-PCR?
We confirmed this finding regarding a ROS1 imbalance mRNA expression by an RNA panel in a next-generation sequencing assay. This sequencing identified novel fusion partners of ROS1 rearrangements with MAN1A1 in leiomyosarcoma. Additionally, we confirmed the in-frame fusion of MAN1A1 exon 5 to ROS1 exon 31 with independent RT-PCR. ROS1 is a receptor tyrosine kinase and its fusion products have been observed in several types of cancers including glioblastoma, non-small cell lung cancer, cholangiocarcinoma, ovarian cancer, gastric adenocarcinoma, and colorectal cancer [10, 22, 28]. Fusion partners of ROS1 rearrangements harboring SLC34A2, CD74, TPM3, SDC4, FN1, and GOPC have been described in detail [10, 22, 28]. ROS1 fusion genes have been reported in STSs, including inflammatory myofibroblastic tumors [30], angiosarcomas [17, 23], epithelioid hemangioendotheliomas [10], perivascular epithelioid cell tumors [31], and synovial sarcomas [31]. With respect to sequencing, interestingly, it has been revealed that DNA-based large gene panel Next Generation Sequencing assays could not detect driver gene fusions or oncogenic isoforms perfectly compared with the alterations identified via RNA-based approaches [2, 11, 12]. Even though Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) is FDA cleared and is one of the best examples of DNA-based large gene panel Next Generation Sequencing assays [5, 30], some genes harboring NTRK3 and NRG1 are not included as certain important introns are too large (> 90 Kb each). Furthermore, it is extremely difficult for Next Generation Sequencing to read contained, highly repetitive elements due to their recurring presence across the genome [2, 11]. Based on these data, as current clinical cancer sequencing tests have mainly employed DNA-based large gene panel Next Generation Sequencing assays in the clinical setting, we believe that these (DNA-based) clinical sequencing tests might have been unable to identify this novel ROS1 fusion that we identified.
Will Potential Fusion Genes Transform Cells in In Vitro Assays?
We next demonstrated the overexpression of MAN1A1-ROS1 fusion by focus formation assay, which further substantiated our findings. This study additionally revealed that the novel ROS1 fusion gene had good sensitivity toward the anti-ROS1 inhibitor crizotinib in vitro (Supplemental Fig. 4; Supplemental Digital Content 5, http://links.lww.com/CORR/A469). Therefore, the novel ROS1 fusion gene had oncogenic potential. With respect to the fusion partner as MAN1A1, MAN1A1 encodes Class I mammalian Golgi 1,2-mannosidase, a Type II transmembrane protein. N-glycan maturation involves Golgi mannosidase and plays a major role in cancer progression. In a previous report, ovarian cancers showed poor prognosis in the presence of high MAN1A1 expression [19]. Recent studies showed that MAN1A1 plays a critical role in tumorigenesis in breast and ovarian cancers [1, 20]. Although a low expression of MAN1A1 was associated with poor prognosis in breast cancer, a high expression of MAN1A1 conversely contributed to poor prognosis in ovarian cancer [1, 20]. Therefore, to elucidate its functions in various cancers, further studies are needed.
Will Tumors in Mice That Have an Identified Fusion Gene Respond to Treatment with a Therapeutic Drug Directed Against That Target?
Regarding in vivo analysis, we found that the novel MAN1A1-ROS1 fusion gene had oncogenic abilities, and crizotinib exhibited antitumor activity in the MAN1A1-ROS1 fusion cells. Crizotinib is a multiple tyrosine kinase inhibitor targeting ALK and ROS1 [24]. The clinical response to crizotinib in ROS1-rearranged non-small cell lung cancers has been reported. The FDA recently approved crizotinib for the treatment of a few types of cancers with ROS1 rearrangement, including non-small cell lung cancers [24]. Both RNA- and DNA-based multitarget assays could be critical in developing novel therapeutic strategies for detecting tyrosine kinase fusion genes. Additionally, NanoString, which could detect imbalances in the expression of mRNA between the 5’ and 3’ exons in tyrosine kinase genes, might support these detections. Several reports have demonstrated that tyrosine kinase inhibitors, including crizotinib, exert antitumor activities in STS [8, 13, 14, 15]. Therefore, tyrosine kinase fusion genes would be useful therapeutic targets for STSs.
Clinicopathogenic Futures and Our Patient Who Had a Tumor with MAN1A1-ROS1 Fusion Gene
Leiomyosarcoma originates from the smooth muscles and frequently occurs in the uterus, retroperitoneum, and blood vessels [13, 16]. Leiomyosarcoma is a high-grade sarcoma and may develop into a metastasis with resistance to standard cytotoxic chemotherapy [13, 16]. Recent large-scale studies using a multiplatform genome-wide dataset of patients with leiomyosarcoma revealed several gene alterations, including copy number loss involving PTEN, RB1, CDH1, and TP53; gains involving MYOCD and IGF1R; PTEN mutations; and alternative telomere lengthening in ATRX, RBL2, and SP100 as frequent gene alterations [1, 3, 7]. The genomic landscape of leiomyosarcoma demonstrates that leiomyosarcoma has a genetically complex karyotype; no pathognomonic chromosomal rearrangements have been detected [3, 7]. Furthermore, there is a low incidence of actionable gene alterations and very few oncogenic kinase fusions through clinical sequencing in leiomyosarcoma [9, 13, 31]. To the best of our knowledge, ROS1 rearrangements have not been identified in leiomyosarcoma, and this patient is the first instance of which we are aware of leiomyosarcoma with ROS1 fusion gene. With respect to our patient, these tumors harboring novel MAN1A1-ROS1: fusion genes were diagnosed as pleomorphic leiomyosarcoma based on clinicopathological features. The primary lesion in the thigh was resected. This patient was followed with imaging studies and did not receive adjuvant chemotherapy because of her age. Two years postoperatively, a metastasis developed in the pelvis. Based on the patient’s genetic profile, the ROS1 inhibitor crizotinib was considered to be beneficial to the patient. However, because the patient was older than 90 years when the metastasis developed, we decided not to use the inhibitor against the ROS1 fusion-positive tumor to avoid any adverse events. At 2.5 years postoperatively, the patient died.
Conclusion
We conducted tyrosine kinase screening using NanoString to identify novel therapeutic tyrosine kinase targets including tyrosine kinase fusions in STSs. Additionally, we identified novel MAN1A1-ROS1 fusion in a patient with leiomyosarcoma and elucidated the malignant potential of the fusion gene via functional analyses. This NanoString-based screening system could be useful for detecting tyrosine kinase fusion genes in high-grade STSs and may provide novel therapeutic targets for treating advanced STSs.
Supplementary Material
Acknowledgments
We thank Shunsuke Kato MD, PhD, of the Department of Medical Oncology at Juntendo University School of Medicine, Tokyo, for his clinical support.
Footnotes
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research® editors and board members are on file with the publication and can be viewed on request.
This study was supported by a Grant-in-Aid from the Japan Society for the Promotion of Science Kahenhi (Grant numbers 19H03789, 19K22694, and 15KK0353 to YS; 19K16753 to KA; 18K15329 to TO; and 18K16634 to YK), the Practical Research for Innovative Cancer Control (Grant number JP18ck0106252 to SK), and the Leading Advanced Projects for Medical Innovation (Grant Number JP18am0001001 to HM) from the Japan Agency for Medical Research and Development.
Each author certifies that neither he nor she, nor any member of his or her immediate family, has funding or commercial associations (consultancies, stock ownership, equity interest, patent/licensing arrangements, etc.) that might pose a conflict of interest in connection with the submitted article.
Each author certifies that his or her institution approved the animal protocol for this investigation and that all investigations were conducted in conformity with ethical principles of research.
This work was performed at Juntendo University School of Medicine, Tokyo, Japan.
References
- 1.Agaram NP Zhang L LeLoarer F, et al. Targeted exome sequencing profiles genetic alterations in leiomyosarcoma. Genes Chromosomes Cancer. 2016;55:124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benayed R Offin M Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25:4712-4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950-965.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang KTE Goytain A Tucker T, et al. Development and evaluation of a pan-sarcoma fusion gene detection assay using the nanostring ncounter platform. J Mol Diagn. 2018;20:63-77. [DOI] [PubMed] [Google Scholar]
- 5.Cheng DT Mitchell TN Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibon F Lagarde P Salas S, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16:781-787. [DOI] [PubMed] [Google Scholar]
- 7.Chudasama P Mughal SS Sanders MA, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun. 2018;9:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15:731-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote GM, He J, Choy E. Next-generation sequencing for patients with sarcoma: a single center experience. Oncologist. 2018;23:234-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19:4040-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies KD Lomboy A Lawrence CA, et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol. 2018;13:1474-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies KD Lomboy A Lawrence CA, et al. DNA-based versus RNA-based detection of MET Exon 14 skipping events in lung cancer. J Thorac Oncol. 2019;14:737-741. [DOI] [PubMed] [Google Scholar]
- 13.Davis LE Nusser KD Przybyl J, et al. Discovery and characterization of recurrent, targetable ALK fusions in leiomyosarcoma. Mol Cancer Res. 2019;17:676-685. [DOI] [PubMed] [Google Scholar]
- 14.Doebele RC Drilon A Paz-Ares L, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020;21:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drilon A Laetsch TW Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher CDM, Bridge JA, Hogendoorn P, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone . 4th ed. World Health Organization; 2013. [Google Scholar]
- 17.Giacomini CP Sun S Varma S, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9:e1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gronchi A Stacchiotti S Verderio P, et al. Short, full-dose adjuvant chemotherapy (CT) in high-risk adult soft tissue sarcomas (STS): long-term follow-up of a randomized clinical trial from the Italian Sarcoma Group and the Spanish Sarcoma Group. Ann Oncol . 2016;27:2283-2288. [DOI] [PubMed] [Google Scholar]
- 19.Hamester F Legler K Wichert B, et al. Prognostic relevance of the Golgi mannosidase MAN1A1 in ovarian cancer: impact of N-glycosylation on tumour cell aggregation. Br J Cancer. 2019;121:944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legler K Rosprim R Karius T, et al. Reduced mannosidase MAN1A1 expression leads to aberrant N-glycosylation and impaired survival in breast cancer. Br J Cancer. 2018;118:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama R Kobayashi Y Friboulet L, et al. Cabozantinib overcomes crizotinib resistance in ROS1 fusion-positive cancer. Clin Cancer Res. 2015;21:166-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Nunez O John I Panasiti RN, et al. Infantile inflammatory myofibroblastic tumors: clinicopathological and molecular characterization of 12 cases. Mod Pathol. 2020;33:576-590. [DOI] [PubMed] [Google Scholar]
- 23.Marks EI Pamarthy S Dizon D, et al. ROS1-GOPC/FIG: a novel gene fusion in hepatic angiosarcoma. Oncotarget. 2019;10:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw AT Ou SH Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soda M Choi YL Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561-566. [DOI] [PubMed] [Google Scholar]
- 26.Suehara Y. Proteomic analysis of soft tissue sarcoma. Int J Clin Oncol. 2011;16:92-100. [DOI] [PubMed] [Google Scholar]
- 27.Suehara Y Alex D Bowman A, et al. Clinical genomic sequencing of pediatric and adult osteosarcoma reveals distinct molecular subsets with potentially targetable alterations. Clin Cancer Res. 2019;25:6346-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suehara Y Arcila M Wang L, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suehara Y Kohsaka S Yamaguchi S, et al. Assessment of predictive biomarkers of the response to pazopanib based on an integrative analysis of high-grade soft-tissue sarcomas: analysis of a tumor sample from a responder and patients with other soft-tissue sarcomas. Clin Orthop Relat Res . 2020;478:2461-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto H, Yoshida A, Taguchi K, et al. ALK, ROS1 and NTRK3 gene rearrangements in inflammatory myofibroblastic tumours. Histopathology. 2016;69:72-83. [DOI] [PubMed] [Google Scholar]
- 31.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


