Abstract
Hepatocellular carcinoma (HCC) is one of the most aggressive human cancers. The rising incidence of HCC worldwide and its resistance to pharmacotherapy indicate that the prevention of HCC development may be the most impactful strategy to improve HCC-related morbidity and mortality. Among the broad range of chemopreventive agents, the use of dietary and nutritional agents is an attractive and promising approach; however, a better understanding of the mechanisms of their potential cancer suppressive action is needed to justify their use. In the present study, we investigated the underlying molecular pathways associated with the previously observed suppressive effect of butyrate-containing structured lipids (STLs) against liver carcinogenesis using a rat “resistant hepatocyte” model of hepatocarcinogenesis that resembles the development of HCC in humans. Using whole transcriptome analysis, we demonstrate that the HCC suppressive effect of butyrate-containing STLs is associated with the inhibition of the cell migration, cytoskeleton organization, and epithelial-to-mesenchymal transition (EMT), mediated by the reduced levels of RACGAP1 and RAC1 proteins. Mechanistically, the inhibition of the Racgap1 and Rac1 oncogenes is associated with cytosine DNA and histone H3K27 promoter methylation. Inhibition of the RACGAP1/RAC1 oncogenic signaling pathways and EMT may be a valuable approach for liver cancer prevention.
Keywords: Butyrate-containing structured lipids, chemoprevention, epigenetics, epithelial-to-mesenchymal transition, hepatocarcinogenesis
1. Introduction
Hepatocellular carcinoma (HCC) is the primary histological type of liver cancer, which accounts for 70 - 90% of primary liver tumors and is the second most lethal cancer worldwide [1, 2]. The global incidence of HCC is rising and projected to continue in the near future, surpassing 1 million cases per year [2]. Etiological factors associated with HCC include chronic viral hepatitis B and C infections, nonalcoholic fatty liver disease, alcohol drinking, and exposure to chemicals [3]. HCC is a multistep and long-term disease process characterized by the progressive evolution of preneoplastic lesions (chronic liver injury, necro-inflammation and regeneration, small cell dysplasia, and dysplastic nodules) into full-fledged HCC [4]. The molecular pathogenesis of HCC is a complex and involves genetic, genomic, and epigenetic alterations, including mutation-driven activation of the telomerase reverse transcriptase (TERT) gene, viral integration, focal amplifications, mutations in β-catenin (CTNB1) and tumor suppressor TP53 genes, and numerous transcriptomic aberrations [2, 5, 6]. Unfortunately, most of these abnormalities are not yet druggable targets [2], which substantially limits the pharmacological options for treating this devastating disease. Only 20 - 30% of patients are diagnosed at early stages of HCC, where the 5-year survival rate is approximately 70%; however, patients with advanced HCC have less than a 10% 5-year survival [2, 7]. Early detection and prevention of HCC are the most impactful strategies in reducing the incidence and mortality of HCC [8, 9].
Two strategies have been suggested for the prevention of HCC, both of which aim to halt or impede the development of HCC. Primary prevention focuses on the elimination of the etiological factors associated with the HCC; such approaches include vaccination against viral HBV and HCV infections, modification of the lifestyle and diet (e.g., reduction in the consumption of alcohol and aflatoxin-contaminated grains, and environmental interventions) [8, 9]. Secondary prevention aims to suppress the development of preneoplastic liver lesions or attenuate their progression to low-grade dysplastic nodules and carcinomas [7]. For this latter approach, identification of the molecular targets relevant to liver tumor progression that can be targeted with chemopreventive pharmacotherapy is of great importance.
The use of dietary and nutritional agents as secondary chemopreventive agents is an attractive and promising approach for liver cancer prevention? [8]; however, a better understanding of the mechanisms of their potential chemopreventive action is needed to justify their use. In a previous study, using a rat “resistant hepatocyte” model of hepatocarcinogenesis [10] that resembles the development of HCC in humans [11], we demonstrated that butyrate-containing structured lipids (STLs), synthesized by enzymatic interesterification of flaxseed oil using tributyrin, inhibited the formation of hepatic preneoplastic foci [12]. In this study, we have focused on the identification and investigation of the underlying molecular pathways associated with the previously observed suppressive effect of butyrate-containing STLs against experimental liver carcinogenesis. We now show that the chemopreventive action of butyrate-containing STLs is mediated by the inhibition of Rac1 and epithelial-to-mesenchymal transition (EMT) markers.
2. Materials and Methods
2.1. Experimental design
To investigate the molecular mechanisms associated with the chemopreventive activity of butyrate-containing STLs, we performed transcriptomic, proteomic, and epigenetic analysis in the liver samples of male Wistar rats submitted to a model of hepatocarcinogenesis and treated with butyrate-containing STLs [12]. Briefly, Heidor et al., [12] used the resistant hepatocyte model of hepatocarcinogenesis to demonstrate the chemopreventive activity of butyrate-containing STLs. This model consists of a short-term in vivo assay that allows studying the mechanism of development of liver cancer in vivo, because it reproduces the multistage carcinogenic process observed in humans, i.e. from hepatic preneoplastic lesions to development of HCC [10, 11]. Thus, male Wistar rats that were submitted to the “resistant hepatocyte” cancer model and treated for 8 weeks with 1.65 g/kg bw butyrate-containing STLs daily by gavage (butyrate-containing STLs group) displayed an inhibition in the development of hepatic preneoplastic lesions when compared to rats submitted to the “resistant hepatocyte” cancer model and treated with 3.0 g/kg body weight (bw) maltodextrin (control group). The dose of maltodextrin was isocaloric to a dose of butyrate-containing STLs [12]. Additional details of this study can be found elsewhere [12].
To confirm the relevance of our findings, we analyzed liver tumor samples of mice submitted to a model of fibrosis- and inflammation-associated HCC described by Uehara et al. [13]. Briefly, male C57BL/6J mice were subjected to fibrosis- and inflammation-associated hepatocarcinogenesis induced by N,N-diethylnitrosamine (DEN) and carbon tetrachloride (CCl4) [13].
2.2. RNA extraction and gene expression analysis using microarray technology
Total RNA was extracted from rat liver tissue samples (n = 4) using miRNeasy Mini kits (Qiagen, Valencia, CA). The whole-genome gene expression in the livers was determined by using a SurePrint G3 Unrestricted Gene Expression 8x60K Microarray (Agilent Technologies, Santa Clara, CA). Sample labeling, microarray processing, and data analysis were performed as described in de Conti et al. [14]. The raw data were uploaded into ArrayTrack [15]. To identify genes that were differentially expressed between control and butyrate-containing STLs-treated rats, Benjamini-Hochberg adjusted p-values were calculated [16]. Genes that changed by more than 1.5-fold and had Benjamini-Hochberg adjusted P-value < 0.05 were considered statistically significant. The microarray gene expression data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) database (accession number GSE109685).
2.3. Pathway analysis of differentially expressed genes
The list of differentially expressed genes in the livers of rats treated with butyrate-containing STLs was uploaded into Ingenuity Pathway Analysis (IPA, version 28820210; Ingenuity, Qiagen) and g:Profiler (Functional Profiling of Gene List from large-scale experiments, version 2016; [17]). The IPA “core analysis” function was used to interpret the differentially expressed genes and categorize them into molecular pathways and gene networks. For each molecular pathway, a P-value was calculated based on a right-tailed Fisher’s exact test that indicated whether a pathway was overrepresented by calculating overlap of the predicted genes in a given pathway with experimental significantly differentially expressed genes. Only those pathways with a P-value < 0.05 and with > 10 genes in the data set were considered further. The g:Profiler was used to generate a network of enriched pathways, with the following criteria being applied: (i) a minimum and maximum size of a functional category of 3 and 500 genes, respectively, and (ii) a minimum size of an intersection of 2 genes. For each molecular pathway, a P-value was calculated based on a cumulative hypergeometric probability that determined whether a pathway was over-represented by calculating overlap of the predicted genes in a given pathway with experimental significantly differentially expressed genes and using a Benjamini-Hochberg false discovery rate (FDR) as a multiple testing correction algorithm. The networks of enriched pathways were visualized using Cytoscape [18] and Enrichment Map [19]. Clusters of nodes functionally related to the same biological process were identified and annotated using AutoAnnotate [20].
2.4. Methylated DNA immunoprecipitation assay
Methylated DNA immunoprecipitation (MeDIP) was performed using MethylMiner Methylated DNA Enrichment kits (Invitrogen, Carlsbad, CA). The methylation status of the CpG islands located within the promoter region of the Rac GTPase-activating protein 1 (Racgap1) and Rac family small GTPase 1 (Rac1) genes was determined by quantitative PCR (qPCR) of DNA from immunoprecipitated and unbound DNA. The results were normalized to the amount of unbound DNA and presented as fold change for each DNA in the liver of rats from experimental groups relative to that in control rats.
2.5. Chromatin immunoprecipitation assay
Formaldehyde cross-linking and chromatin immunoprecipitation (ChIP) assays, with primary antibodies against histone H3K9me3 and H3K27me3 (Abcam, Cambridge, MA), were performed by using a Chromatin Immunoprecipitation Assay Kit (Millipore Corporation, Billerica, MA). Purified DNA from immunoprecipitated and input DNA was analyzed by qPCR with the same primer sets for Racgap1 and Rac1 that was used in the MeDIP assay. The results were normalized to the amount of input DNA and presented as fold change for each DNA in the liver of rats from experimental groups relative to that in control rats.
2.6. Western blot analysis
Whole liver tissue lysates containing equal quantities of proteins from control and butyrate-containing STLs-treated rats were separated by 7 - 15% SDS-PAGE and transferred to PVDF membranes. The levels of RACGAP1, RAC1, SRY (sex-determining region Y)-box 9 (SOX-9), SRY-box 2 (SOX2), Yes-associated protein 1 (YAP1), matrix metallopeptidase 2 (MMP2), snail family transcriptional repressor 1 (SNAIL1), snail family transcriptional repressor 2 (SLUG; SNAI2), vimentin (VIM), β-catenin (CTNNB1), NOTCH, E-cadherin (CDH1), N-cadherin (CDH2), extracellular regulated MAP kinase (ERK1/2), phosphorylated extracellular regulated MAP kinase (p-ERK1/2), p38 kinase (p38), mitogen-activated protein kinase 9, stress-activated protein kinases (SAPK)/Jun amino-terminal kinases (JNK) and β-actin (ACTB) proteins were determined by Western blot analysis as described previously [14]. Primary antibodies and their dilutions used for Western blotting are listed in Supplementary Table 1. IRDye 800CW-labeled anti-rabbit or IRDye 680RD-labeled anti-mouse secondary antibodies (LI-COR Biosciences, Lincoln, NE) were used for visualization. Fluorescence was measured using the Odyssey CLx Infrared Imager (LI-COR Biosciences). The images were quantified using ImageStudio 4.0 Software (LI-COR Biosciences). For equal loading control, the relative amount of the protein of interest was normalized against β-actin.
2.7. Analysis of human RACGAP1, RAC1, and SOX9 gene expression data from the online database
RACGAP1, RAC1, and SOX9 gene expression and clinical human HCC data were downloaded from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov) database. All samples were median-centered, and Benjamini-Hochberg adjusted P-values [16] were calculated to control the FDR.
2.8. Transfection of Hep3B cells with RAC1 siRNA
SK-Hep-1, Hep3B, SNU448, PLC/PRF/5, and HepG2 human liver cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured according to the ATCC’s recommendations. Hep3B cells were transfected with 50 nM of either Silencer® Select RAC1 siRNA (n = 3; Life Technologies) or Silencer® Select negative control siRNA (n = 3) using Lipofectamin™ 3000 transfection reagent (Life Technologies). Seventy-two hours post-transfection, adherent cells were harvested by mild trypsinization, re-seeded in 100 mm dishes, and the transfection was repeated. Adherent cells were harvested 48 hours after the second transfection and the RNA and proteins were extracted. The experiments were repeated two times in triplicate.
2.9. Statistical analyses
Results are presented as mean ± SD, n = 3 - 4. Significant differences between groups for non-genomic data were evaluated using an unpaired 2-tailed Student’s t-test (p < 0.05 was considered significant). When necessary, data were natural log-transformed before conducting the analyses to maintain equal variance or normal data distribution. RACGAP1, RAC1, and SOX9 expression data in human HCC were analyzed by a non-parametric Wilcoxon-signed rank test.
3. Results
3.1. Global gene expression alterations in the livers of rats treated with butyrate-containing STLs
In our previous study [12], we demonstrated that the administration of butyrate-containing STLs during the initiation and promotion stages of liver carcinogenesis inhibited the hepatocarcinogenic process in male Wistar rats subjected to a “resistant hepatocyte” cancer model [12]. This HCC-suppressing activity of butyrate-containing STLs was associated with the inhibition of major hepatocarcinogenesis-related oncogenes, including Bcl2, Ccnd1, Fos, Kras, Src, Stat3, Tnf2, and Myc [12]. To investigate further the underlying molecular mechanisms of the tumor-suppressing activity of butyrate-containing STLs, we analyzed the transcriptomic profiles in the livers of control and butyrate-containing STLs treated animals.
Compared to control animals, 1583 differentially expressed genes (920 up-regulated and 663 down-regulated) were found in the livers of rats treated with butyrate-containing STLs (Supplementary Table 2). Pathway enrichment analysis of these genes demonstrated a strong effect of butyrate-containing STLs, based on the significance and numbers of differentially expressed genes, on canonical molecular pathways associated with the deregulation of nuclear receptor signaling, activation of hepatic stellate cells, gap junction signaling, adhesion and diapedesis, and production of reactive oxygen species (Figure 1A). Further analysis of the molecular functions of the differentially expressed genes showed their involvement in the regulation of several biological functions, among which cell migration was the most prominent (Figure 1B). These findings were confirmed by a parallel analysis of the differentially expressed genes using g:Profiler that demonstrated alterations in pathways related to cell migration, such as cytoskeleton organization, cell motility, and calcium signaling (Figure 1C). Detailed analysis of the differentially expressed genes in selected pathway clusters showed that several cancer-related genes were over-represented, among which the oncogenes Racgap1 and Sox9 were among the most down-regulated (Table 1). A gene network interaction analysis identified Racgap1 and Sox9 as key down-regulated genes associated with the inhibition of cell migration and cytoskeleton organization in the livers of butyrate-containing STLs-treated rats (Figure 1D).
Figure 1. Pathway analysis of differentially expressed genes in the livers of rats treated with butyrate-containing STLs.
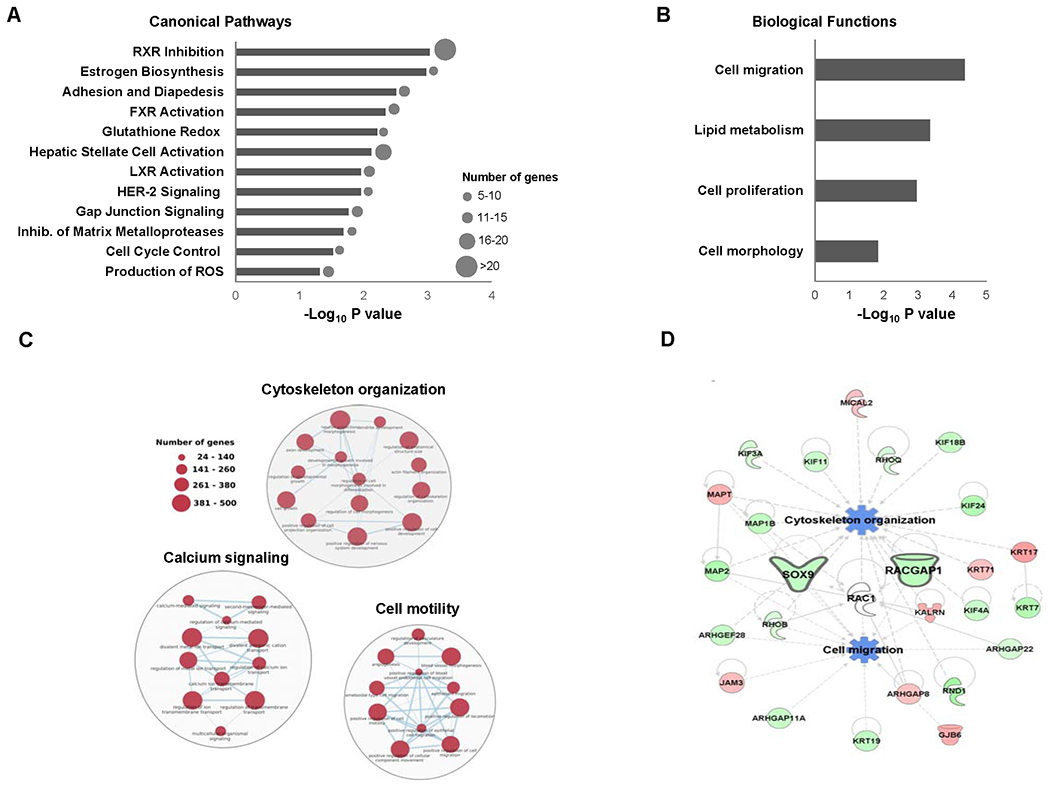
(A) Canonical pathway analysis of differentially expressed genes. (B) Biological functions enriched by differentially expressed genes. (C) Pathway networks of differentially expressed genes related to cell migration. (D) The molecular network of differentially expressed genes related to down-regulated Racgap1 and Sox9. Z-scores were calculated using IPA to predict activation or inhibition of biological functions. Genes in red color are up-regulated, genes in green color are down-regulated, and pathways in blue are inhibited.
3.2. Butyrate-containing STLs down-regulate RACGAP1 and RAC1
Next, we sought to confirm whether changes in mRNA transcripts of the identified genes corresponded to changes in the levels of proteins. Protein levels of RACGAP1, SOX9, and RAC1, a downstream target of RACGAP1 and a well-known oncogene [21], were investigated. Figure 2A shows that the treatment of rats undergoing liver carcinogenesis with butyrate-containing STLs resulted in a decrease of protein levels for RACGAP1 and RAC1 by 65% and 54%, respectively, whereas no effect was observed on the level of SOX9.
Figure 2. Expression of the RACGAP1, RAC1, and SOX9 oncogenes in HCC.
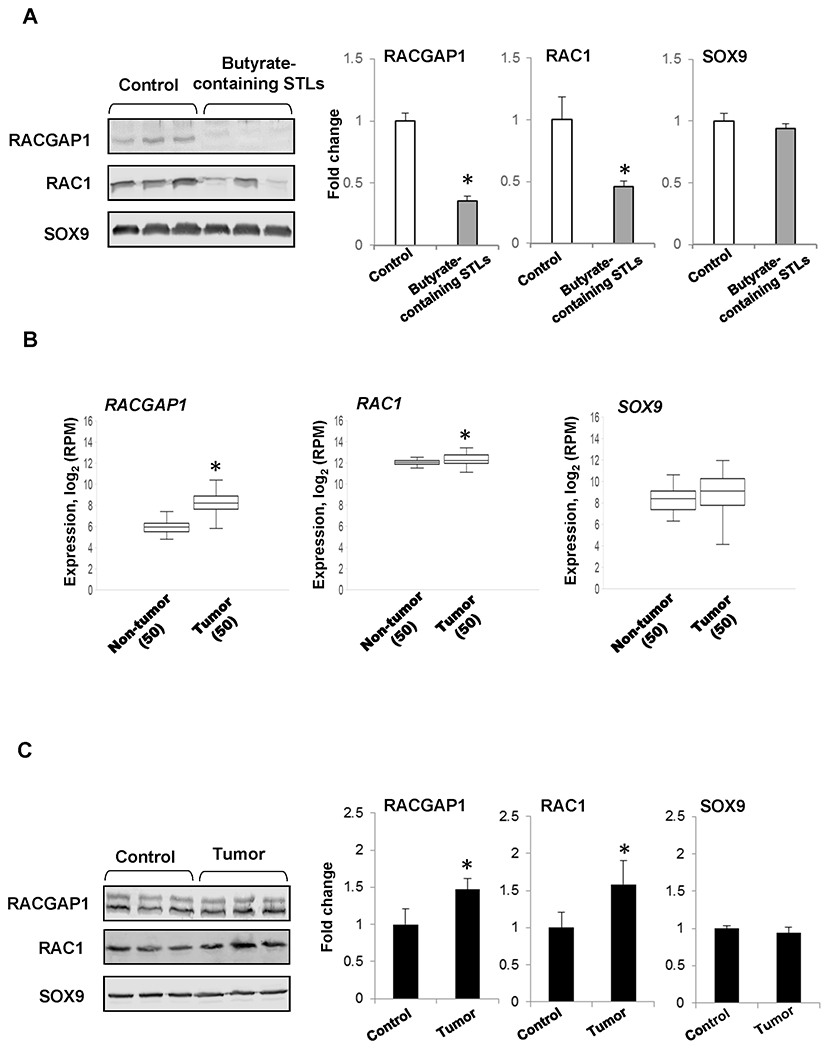
(A) Level of RACGAP1, RAC1, and SOX9 proteins in the livers of rats treated with butyrate-containing STLs. The Western blot results are presented as an average fold change in the level of each protein in the livers of rats treated with butyrate-containing STLs relative to that in the livers of rats submitted to a “resistant hepatocyte” model of hepatocarcinogenesis, which were assigned a value 1. Representative Western blot images of three different samples are shown. Values are mean ± SD, n=3. Asterisks (*) denotes a statistically significant difference between the groups. (B) Expression of RACGAP1, RAC1, and SOX9 in paired HCC tissue samples and non-tumor liver tissue samples obtained from the same HCC patient. Asterisks (*) denote a statistically significant difference between groups as determined by a Wilcoxon-signed rank test, n = 50. (C) Level of RACGAP1, RAC1, and SOX9 proteins in HCC from mice. The Western blot results are presented as an average fold change in the level of each protein in HCC relative to that in the livers of control mice, which were assigned a value 1. Representative Western blot images of three different samples are shown. Values are mean ± SD, n = 3. Asterisks (*) denote a statistically significant difference between groups.
Because RACGAP1, RAC1, and SOX9 are oncogenes involved in the regulation of cytoskeleton function and cytokinesis, cell differentiation, and migration [21–23], we used the TCGA Research Network database (http://cancergenome.nih.gov) to analyze their expression in paired HCC tissue samples and non-tumor liver tissue samples obtained from the same patients. We found a higher level of the RACGAP1 and RAC1 transcripts in HCC, whereas no changes were detected in SOX9 expression (Figure 2B). Additionally, we found that RACGAP1 and RAC1 proteins were also elevated in a mouse model of fibrosis- and inflammation-associated HCC (Figure 2C), another human-relevant model of liver carcinogenesis [13].
3.3. Mechanism of butyrate-containing STLs-mediated down-regulation of RACGAP1 and RAC1
To determine the mechanism of Racgap1 and Rac1 down-regulation by butyrate-containing STLs, we investigated the status of cytosine DNA methylation and histone H3K9 and H3K27 trimethylation in the promoter regions of the Racgap1 and Rac1 genes. Figure 3 shows that in the livers of rats treated with butyrate-containing STLs, the levels of cytosine DNA methylation and histone H3K27me3 were increased by 3.2- and 1.8 -fold, respectively, in the promoter region of Racgap1. Likewise, histone H3K27me3 was increased by 4.7-fold in Rac1.
Figure 3. DNA methylation and histone H3K9 and H3K27 trimethylation in the promoter region of Racgap1 and Rac1 genes in the livers of rats treated with butyrate-containing STLs.
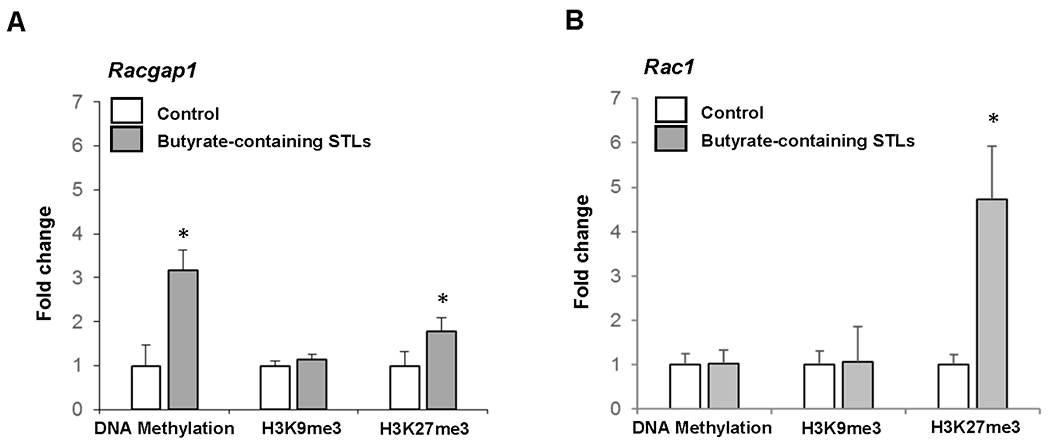
Values are mean ± SD, n = 3. Asterisks (*) denote a statistically significant difference between groups.
3.4. Effect of butyrate-containing STLs on EMT
Recent studies have shown a fundamental role of RACGAP1 and RAC1 in the regulation of YAP1 [24,25], a key effector of the Hippo signaling pathway [25, 26]. The Hippo pathway is essential for tumor initiation and progression of HCC [27, 28], due to its critical role in cancer stem biology, especially in EMT. Hence, the levels of YAP1 and several other key EMT-related proteins were analyzed by Western blotting in the livers of control rats and rats treated with butyrate-containing STLs. Figure 4A shows the concurrent expression of protein-markers of epithelial (e.g., E-cadherin) and mesenchymal (e.g., SNAIL1, MMP2, and VIM) phenotypes in the livers of rats submitted to the “resistant hepatocyte” liver carcinogenesis model (control group), indicating the presence of hybrid epithelial/mesenchymal phenotypes in liver cells. It has been suggested that these hybrid epithelial/mesenchymal cells may possess a high tumor-initiating, cell migrating, and metastatic potential [29]. Treatment of rats undergoing liver carcinogenesis with butyrate-containing STLs resulted in a decrease in YAP1 protein levels by 70% and mesenchymal phenotype markers SNAIL1, MMP2, and VIM by 40%, 64%, and 56%, respectively, while no changes were observed in the epithelial marker E-cadherin (Figure 4A). Additionally, butyrate-containing STLs significantly decreased the levels of proteins related to EMT-inducing signaling pathways [30–32], especially CTNNB1 and NOTCH by 63% and 67%, respectively.
Figure 4. Level of proteins related to (A) EMT and (B) MAP kinase pathways in the livers of rats treated with butyrate-containing STLs.
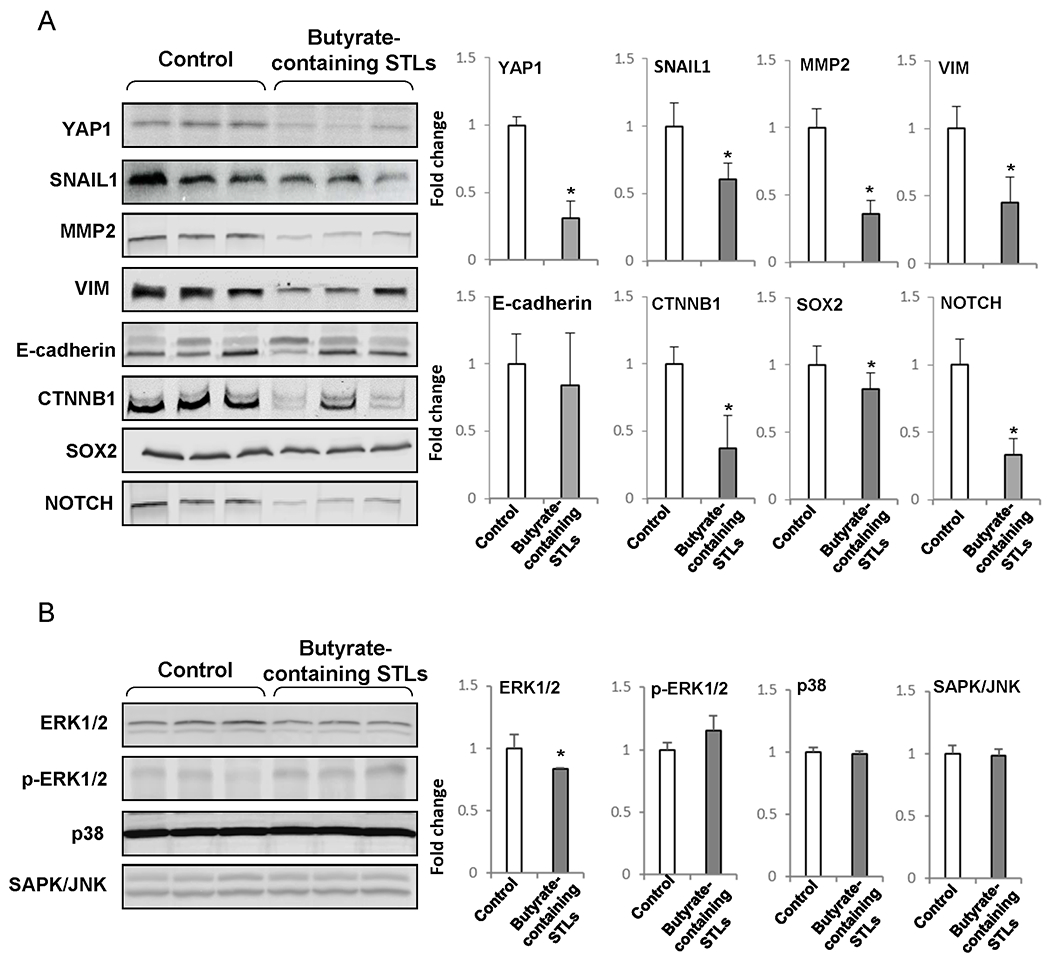
The Western blot results are presented as an average fold change in the level of each protein in the livers of rats treated with butyrate-containing STLs relative to that in the livers of rats submitted to a “resistant hepatocyte” model of hepatocarcinogenesis, which were assigned a value 1. Representative Western blot images of three different samples are shown. Values are mean ± SD, n = 3. Asterisks (*) denote a statistically significant difference between groups.
RACGAP1 and RAC1 are also known to control the mitogen-activated protein kinase pathway [33, 34]. Figure 4B shows that contrary to the pronounced effect of butyrate-containing STLs on EMT, there was no effect on key players (ERK1/2, p-ERK1/2, p38, and SAPK/JNK) in the ERK/MAPK pathway.
3.5. Inhibition of RAC1 decreases EMT in human Hep3B HCC cells
Next, we sought independent confirmation of the hypothesis that inhibition of RAC1 gene decreases EMT in liver cancer. First, we evaluated the status of RAC1 in five human liver cancer cell lines, SK-Hep-1, SNU448, PLC/PRF/5, Hep3B, and HepG2. Figure 5A shows that the Hep3B cell line exhibited the highest level of RAC1. We then knocked down expression and protein for RAC1 by transfection of Hep3B cells with siRNA (Figures 5B and 5C). Silencing of RAC1 resulted in the reduction of mesenchymal phenotype proteins YAP1, SNAIL1, and N-cadherin (CDH2), by 49%, 34%, and 44%, respectively (Figure 5D).
Figure 5. In vitro analysis of RAC1 down-regulation in human HCC cells.
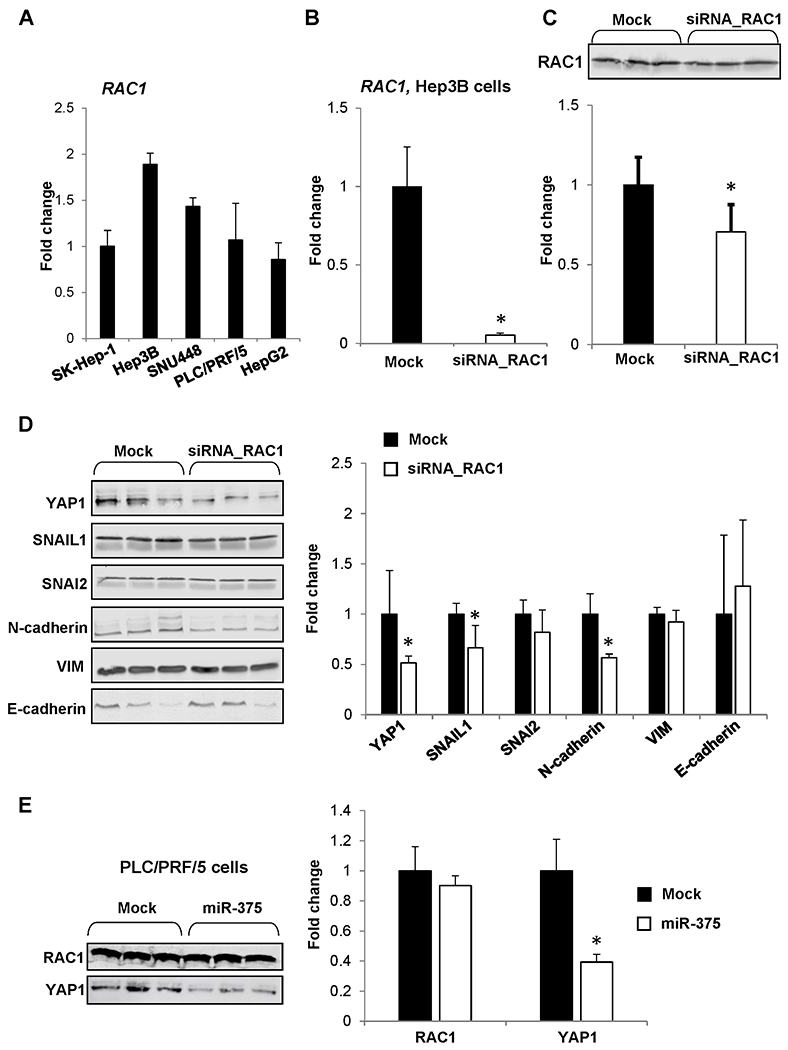
(A) RAC1 expression in SK-Hep-1, Hep3B, SNU448, PLC/PRF/5, and HepG2 human liver cancer lines. (B) Level of RAC1 mRNA and (C) RAC1 protein in Hep3B cells transfected with RAC1 siRNA. Values are mean ± SD, n = 3. Asterisks (*) denote a statistically significant difference between groups. (D) Level of YAP1, SNAIL1, SNAI2, N-cadherin, VIM, and E-cadherin proteins in Hep3B human liver cancer cells transfected with RAC1 siRNA. Values are mean ± SD, n = 3. Asterisks (*) denote a statistically significant difference between groups. (E) Level of RAC1 and YAP1 proteins in PLC/PRF5 human liver cancer cells after transfection of microRNA miR-375. The results are presented as an average fold change in the level of each protein in miR-375-transfected PLC/PRF/5 cells relative to that in the mock group, which were assigned a value 1. Values are mean ± SD, n = 3. Asterisks (*) denote a statistically significant difference between groups.
Recent studies have demonstrated a tight connection between RACGAP1/RAC1-YAP1 pathways and suggested the existence of a functional regulatory feedback loop [24,35]. To confirm further that inhibition of YAP1 is mediated through RAC1 signaling, we utilized PLC/PRF5 cells, a cell line that highly expresses YAP1. The PLC/PRF5 cells were transfected with microRNA miR-375, which directly targets YAP1 [36]. Figure 5E shows that the level of RAC1 protein in miR-375-transfected cells did not differ from that in mock-transfected cells, while the level of YAP1 was markedly reduced. This further supports a causative role for RAC1 in butyrate-containing STLs-associated downregulation of YAP1.
4. Discussion
The rising incidence of HCC globally and its resistance to pharmacotherapy indicate that primary and secondary prevention of HCC development may be the most impactful strategies to improve HCC-related morbidity and mortality [2, 7]. Among several secondary prevention factors, the importance of nutrition, including bioactive food components, is widely recognized [37]. One of the key aspects in developing effective cancer prevention is the identification of molecular drivers that can be used as targets to inhibit the carcinogenic process. Recent advances in large-scale omics-related technologies, combined with bioinformatics tools, have revolutionized our ability to interrogate cancer-related pathways to identify chemopreventive targets [38]. In the present study, using whole transcriptome analysis, we demonstrate that the chemopreventive effect of butyrate-containing STLs against liver carcinogenesis is associated with the inhibition of the cell migration and cytoskeleton organization pathways mediated by the down-regulation of RACGAP1 and RAC1 proteins.
Up-regulation of RACGAP1 [39, 40] and RAC1 [41] has been reported in human HCC. Moreover, the over-expression of RACGAP1 and RAC1 is associated with the poor prognosis in HCC patients [39, 41, 42] and the early recurrence of human HCC [43]. Similarly, our results showed the over-expression of RACGAP1 and RAC1 genes in human HCC. Furthermore, our data, showing that similar molecular pathways are affected in human-relevant rat [12] and mouse [13] models of liver cancer, indicate that findings in the rodent models of liver cancer are informative for human disease and chemoprevention strategies.
One of the pathophysiological consequences of the up-regulation of RACGAP1 and RAC1, which promotes oncogenic progression, is the activation of the EMT program. In the present study, we showed that treatment of rats undergoing liver carcinogenesis with butyrate-containing STLs reduced levels of RACGAP1 and RAC1 proteins, and inhibited EMT markers, as evidenced by marked down-regulation the mesenchymal phenotypic markers VIM, SNAIL1, MMP2, and especially YAP1.
YAP1 has emerged as a key regulator of normal liver development and normal liver physiology [27]; however, its overexpression has been documented in HCC in humans [44]. Several studies have shown that YAP1 is a key driver in the development of HCC, as evidenced by the fact that induction of YAP1 leads to the rapid development of liver tumors in experimental animals [45–48]. In the present study, we demonstrated that butyrate-containing STLs down-regulated YAP1 and inhibited EMT markers in the livers of rats undergoing hepatocarcinogenesis and that this is mediated through the RACGAP1/RAC1 signaling pathway. This was evidenced down-regulation of YAP1, SNAIL1, and N-cadherin in Hep3B HCC cells transfected with RAC1 siRNA. In contrast, transfection with microRNA miR-375, which targets directly YAP1 [36], did not affect the levels of RAC1. This indicates that the inhibitory activity of butyrate-containing STLs on EMT markers is associated with down-regulation RAC1. These findings are in agreement with the results of previous studies showing that inhibition of RACGAP1 and RAC1 reduced cell migration and invasion, induced cytokinesis failure, and increased cell death [35, 42, 49, 50]. Mechanistically, down-regulation of YAP1 by a low level of RACGAP1 may be attributed to RACGAP1-mediated inhibition of the Hippo pathway [35], or in case of RAC1, to impaired nuclear localization of YAP1 [24].
Recently, two small-molecule inhibitors of RAC1, MDQ-167 and EHop-016, have been tested as potential anticancer drugs. Humphries-Bickley et al. [51] reported that treatment of metastatic MDA-MB-231 breast cancer cells with MDQ-167 inhibited RAC1 activity. This resulted in the reduction of the aggressive cancer cell phenotype, evidenced by lower viability of cancer cells and spheroid formation in vitro. More importantly, MDQ-167 inhibited the progression of GFP-HER2-BM xenograph tumors by 90% in immunocompromised mice [51]. Similar findings of the reduction of spheroid formation and cancer stem cell phenotype in vitro were demonstrated in HCCLM3 and SMMC7721 HCC cell lines treated with EHop-016 [50]. In light of this, the results of our study showing the ability of butyrate-containing STLs to inhibit the RAC1 pathway and prevent the progression of liver carcinogenesis indicate that targeting RACGAP1/RAC1 signaling may have utility in liver cancer prevention.
Of particular interest is that the down-regulation of RACGAP1 and RAC1 oncogenes by butyrate-containing STLs is mediated by cytosine DNA and histone H3K27 promoter methylation. This finding corresponded to a recent report showing that demethylation of histone H3K4me2, the functionally opposite to H3K27me3 histone modification, at the RACGAP1 promoter decreased expression of RACGAP1 and was accompanied by reduced HCC cell migration and inhibition of tumor growth in vivo [52]. This indicates that targeting the RACGAP1 and RAC1 oncogenes, using dietary factors with the unique epigenome modulating abilities, may be a promising strategy to prevent HCC development. Consistent with this suggestion, it has been shown that S-adenosylmethionine, a universal methyl donor for all cellular methylation reactions, inhibits proliferation, adhesion, migration, and invasion of activated hepatic stellate cells by RAC1 promoter cytosine DNA methylation [53, 54].
In conclusion, the results of this study demonstrate that the inhibitory effect of butyrate-containing STLs on rat liver carcinogenesis is associated with the epigenetically-mediated inhibition of the RACGAP1/RAC1 oncogenic signaling pathway and EMT markers, which is one of the key events in the development of HCC.
Supplementary Material
Funding:
This work did not receive any specific grants from the funding agencies in the public, commercial or non-profit sectors.
Footnotes
The views expressed in this manuscript do not necessarily represent those of the U.S. Food and Drug Administration.
5. References
- [1].Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvessi M, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol 2017;67: 302–9. [DOI] [PubMed] [Google Scholar]
- [2].Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15: 599–616. [DOI] [PubMed] [Google Scholar]
- [3].Stewart BW, Wild CP, editors. World Cancer Report 2014, International Agency for Research on Cancer, Lyon, 2014. [Google Scholar]
- [4].Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett 2014;342: 223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zucman-Rossi J, Villanueva A, Nault J-C, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149: 1226–39. [DOI] [PubMed] [Google Scholar]
- [6].The Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169: 1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bupathi M, Kaseb A, Meric-Bernstam F, Naing A. Hepatocellular carcinoma: where there is unmet need. Mol Oncol 2015; 9: 1501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Singh S, Singh PP, Roberts LR, Sanchez W. Chemopreventive strategies in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2014;11: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol 2018;68: 526–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Semple-Roberts E, Hayes MA, Armstrong D, Becker RA, Racz WJ, Farber E. Alternative methods of selecting rat hepatocellular nodules resistant to 2-acetylaminofluorene. Int J Cancer 1987;40: 643–5. [DOI] [PubMed] [Google Scholar]
- [11].Petrelli A, Perra A, Cora D, Sulas P, Menegon S, Manca C, et al. MicroRNA/gene profiling unveils early molecular changes and nuclear factor erythroid related factor 2 (NRF2) activation in a rat model recapitulating human hepatocellular carcinoma (HCC). Hepatology 2014;59: 228–41. [DOI] [PubMed] [Google Scholar]
- [12].Heidor R, de Conti A, Ortega JF, Furtado KS, Silva RC, Tavares PELM, et al. The chemopreventive activity of butyrate-containing structured lipids in experimental rat hepatocarcinogenesis. Mol Nutr Food Res 2016;60: 420–9. [DOI] [PubMed] [Google Scholar]
- [13].Uehara T, Pogribny IP, Rusyn I. The DEN and CCl4-induced mouse model of fibrosis and inflammation-associated hepatocellular carcinoma. Curr Protoc Pharmacol 2014;66: 14.30.1–14.30.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].de Conti A, Dreval K, Tryndyak V, Orisakwe OE, Ross SA, Beland FA, et al. Inhibition of the cell death pathway in nonalcoholic steatohepatitis (NASH)-related hepatocarcinogenesis is associated with histone H4 lysine 16 deacetylation. Mol Cancer Res 2017;15: 1163–72. [DOI] [PubMed] [Google Scholar]
- [15].Fang H, Harris SC, Su Z, Chen M, Qian F, Shi L, et al. ArrayTrack: an FDA and public genomic tool. Methods Mol Biol 2009;563: 379–98. [DOI] [PubMed] [Google Scholar]
- [16].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. Stat Methodol 1995;57: 289–300. [Google Scholar]
- [17].Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, et al. g:Profiler˗a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 2016;44: W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13: 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Merico D, Isserlin R, Bader GD. Visualizing gene-set enrichment results using the Cytoscape plug-in enrichment map. Methods Mol Biol 2011;781: 257–77. [DOI] [PubMed] [Google Scholar]
- [20].Kucera M, Isserlin R, Arkhangorodsky A, Bader GD. AutoAnnotate: a Cytoscape app for summarizing networks with semantic annotations. F1000Research 2016;5: 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hall A Rho family GTPases. Biochem Soc Trans 2012;40: 1378–82. [DOI] [PubMed] [Google Scholar]
- [22].Kazaniets MG, Caloca MJ. The Rac GTPase in cancer: from old concepts to new paradigms. Cancer Res 2017;77: 5445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lawson CD, Der CJ. Filling GAPs in our knowledge: ARHGAP11A and RACGAP1 act as oncogenes in basal-like breast cancers. Small GTPases 2018;9: 290–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sabra H, Brunner M, Mandati V, Wehrle-Haller B, Lallemand D, Ribba A-S, et al. β1 integrin-dependent Rac/group I PAK signaling mediates YAP activation of Yes-associated protein 1 (YAP1) via NF2/merlin. J Biol Chem 2017;292: 19179–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 2005;122: 421–34. [DOI] [PubMed] [Google Scholar]
- [26].Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell 2016;29: 783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of Hippo/YAP pathway in liver physiology and cancer. J Hepatol 2015;63: 1491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Park JH, Shin JE, Park HW. The role of Hippo pathway in cancer stem cell biology. Mol Cells 2018;41: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jolly MK, Somarelli JA, Sheth M, Biddle A, Tripathi SC, Armstrong AJ, et al. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol Ther 2019; 194: 161–84. [DOI] [PubMed] [Google Scholar]
- [30].Espinoza I, Miele L. Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett 2013;341: 41–5. [DOI] [PubMed] [Google Scholar]
- [31].Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T, et al. SOX2 promotes tumor metastasis by stimulating epithelial-to-mesenchymal transition via regulation of WNT/β-catenin signal network. Cancer Lett 2013;336: 379–89. [DOI] [PubMed] [Google Scholar]
- [32].Basu S, Cheriyamundath S, Ben-Ze’ev A. Cell-cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Research 2018;7: 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bachmann VA, Bister K, Stefan E. Interplay of PKA and Rac. Fine-tuning of Rac localization and signaling. Small GTPases 2013;4: 247–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang C, Wang W, Liu Y, Yong M, Yang Y, Zhou H. Rac GTPase activating protein 1 promotes oncogenic progression of epithelial ovarian cancer. Cancer Sci 2018;109: 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang X-M, Cao X-Y, He P, Li J, Feng M-X, Zhang Y-L, et al. Overexpression of Rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing Hippo signaling to promote cytokinesis. Gastroenterology 2018;155: 1233–49. [DOI] [PubMed] [Google Scholar]
- [36].de Conti A, Tryndyak V, Doerge DR, Beland FA, Pogribny IP. Irreversible down-regulation of miR-375 in the livers of Fischer 344 rats after chronic furan exposure. Food Chem Toxicol 2016;98: 2–10. [DOI] [PubMed] [Google Scholar]
- [37].World Cancer Research Fund/American Institute for Cancer Research. Diet, Nutrition, Physical Activity and Cancer: a Global Perspective. A summary of the Third Expert Report. 2018. [Google Scholar]
- [38].Meerzaman D, Dunn BK, Lee M, Chen Q, Yan C, Ross S. The promise of omics-based approaches to cancer prevention. Semin Oncol 2016;43: 36–48. [DOI] [PubMed] [Google Scholar]
- [39].Zhou Z, Li Y, Hao H, Wang Y, Zhou Z, Wang Z, et al. Screening Hub genes as prognostic biomarkers of hepatocellular carcinoma by bioinformatics analysis. Cell Transplant 2019; 28: 76S–86S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Qi J, Zhou J, Tang X-Q, Wang Y. Gene biomarkers derived from clinical data of hepatocellular carcinoma. Interdiscip Sci 2020; (in press). [DOI] [PubMed] [Google Scholar]
- [41].He Q-L, Qin S-Y, Tao L, Ning H-J, Jiang H-X. Prognostic value and prospective molecular mechanism of miR-100-5p in hepatocellular carcinoma: a comprehensive study based on 1,258 samples. Oncol Lett 2019; 18: 6126–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li J, Zhang S, Hu Q, Zhang K, Jin J, Zheng X, et al. The NKD1/Rac1 feedback loop regulates the invasion and migration ability of hepatocarcinoma cells. Sci Rep 2016;6, 26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang SM, Ooi LLPJ, Hui KM. Upregulation of Rac GTPase-activating protein 1 is significantly associated with early recurrence of human hepatocellular carcinoma. Clin. Cancer Res 2011;17: 6040–51. [DOI] [PubMed] [Google Scholar]
- [44].Lin C, Hu Z, Lei B, Tang B, Yu H, Qiu X, et al. Overexpression of Yes-associated protein and its association with clinicopathological features of hepatocellular carcinoma: a meta-analysis. Liver Int 2017;37: 1675–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, et al. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol 2014;61: 1088–96. [DOI] [PubMed] [Google Scholar]
- [46].Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, et al. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 2014;147: 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nishio M, Sugimachi K, Goto H, Wang J, Morikawa T, Miyachi Y, et al. Dysregulated YAP1/TAZ and TGF-β signaling mediate hepatocarcinogenesis in Mob1a/1b-deficient mice. Proc Natl Acad Sci USA 2016;113: E71–E80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Patel SH, Camargo FD, Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology 2017;152: 533–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Guignandon A, Faure C, Neutelings T, Rattner A, Mineur P, Linossier M-T, et al. Rac1 GTPase silencing counteracts microgravity-induced effects on osteoblastic cells. FASEB J 2014;28: 4077–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jiang Z-B, Ma B-Q, Liu S-G, Li J, Yang G-M, Hou Y-B, et al. miR-365 regulates liver cancer stem cells via RAC1 pathway. Mol Carcinog 2018;58: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Humphries-Bickley T, Castillo-Pichardo L, Hernandez-O’Farrill E, Borrero-Garcia LD, Forestier-Roman I, Gerena Y, et al. Charcterization of a dual Rac/Cdc42 inhibitor MBQ-167 in metastatic cancer. Mol Cancer Ther 2017;16: 805–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pu J, Wang J, Wei H, Lu T, Wu X, Wu Y, et al. lncRNA MAGI2-AS3 prevents the development of HCC via recruiting KDM1A and promoting H3K4me2 demethylation of the RACGAP1 promoter. Mol Ther Nucleic Acids 2019; 18: 351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhang F, Zhuge Y-Z, Li Y-J, Gu J-X. S-adenosylmethionine inhibits the activated phenotype of human hepatic stellate cells via Rac1 and matrix metalloproteinases. Int Immunopharmacol 2014;19: 193–200. [DOI] [PubMed] [Google Scholar]
- [54].Bian K, Zhang F, Wang T, Zou X, Duan X, Chen G, et al. S-Adenosylmethionine suppresses the expression of Smad3/4 in activated human hepatic stellate cells via Rac1 promoter methylation. Mol Med Rep 2016;13: 3867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


