Abstract
Backgroud:
To analyze the correlation between gene polymorphisms of 5,10- methylenetetrahydrofolate reductase (MTHFR) and risk of unexplained recurrent pregnancy loss (URPL) in Chinese women.
Methods:
Eligible studies were searched in Pubmed, Embase, Web of Science, Wanfang, and China National Knowledge Infrastructure (CNKI) databases. Established inclusion criteria were used to screening articles, subsequently evaluate the quality of the included studies, Stata 16.0 PM and RevMan 5.3 software were conducted for meta-analysis. The pooled odds ratio (OR) with 95% confidence interval (CI) was determined to assess the relationship between MTHFR and risk of URPL in Chinese women.
Results:
For MTHFR C677T, fifty studies were included, involving 6677 URPL cases and 8111 controls. The overall results showed that MTHFR C677T was significantly correlated with URPL risk, especially in the homozygous model (TT vs CC; OR 3.06; 95% CI 2.56–3.66). For MTHFR A1298C, twenty-first studies were included, involving 3439 URPL cases and 3155 controls. The results showed that MTHFR A1298C was also significantly correlated with URPL risk in recessive (CC vs AC + AA; OR 1.55; 95% CI 1.25–1.93) and homozygous (CC vs AA; OR 1.53; 95% CI 1.22–1.91) models. In addition, sub-group results showed that no significant difference between north and south China populations in the MTHFR gene polymorphisms and URPL risk. Of note, the patients carrying MTHFR C677T and MTHFR A1298C joint mutants had no synergistic effect (OR 2.71; 95% CI 0.84–8.70) on the occurrence of URPL compared with the wild-type homozygous genotype (MTHFR 677CC/ MTHFR 1298AA).
Conclusion:
Studies included in this meta-analysis suggested that MTHFR 677T allele and 677TT genotype and MTHFR 1298CC genotype were both associated with URPL; testing MTHFR C677T gene polymorphism was a more appropriate target compared with other mutations in the prediction of URPL.
Keywords: 5, 10 – methylenetetrahydrofolate reductase, unexplained recurrent pregnancy loss, meta-analysis
1. Introduction
Recurrent pregnancy loss (RPL) was defined as two or more failed clinical pregnancies by American Society for Reproductive Medicine.[1] A total of 1-–5% of women in reproductive age will experience RPL.[2,3] Although abnormalities of the genital tract and the uterine structure, autoimmune diseases, genetic disorders, and inherited thrombophilia had been identified as risk factors for RPL, nearly 50% of RPL cases were still classified as URPL.[4] MTHFR reduced 5,10-methylenetetrahydrofolate (5,10-MTHF) to 5-methylenetetrahydrofolate (5-MTHF), and 5-MTHF participated in the conversion of homocysteine to methionine. The reduction of MTHFR resulted in the increased level of homocysteine in the blood.[5] High level blood homocysteine was a significant risk factor for URPL and detected in about 30% URPL cases.[6,7] MTHFR gene polymorphisms were widely believed to play a key role in the risk of URPL, MTHFR C677T and MTHFR A1298C of which were the two most-investigated single nucleotide polymorphisms.[8–11]
Numerous case-control studies and meta-analyses reported that MTHFR C677T was significantly associated with URPL.[10–12] The frequencies of the MTHFR 677T allele and 677TT genotype increased along the south-north direction, and showed significantly geographical variations among Chinese population.[13–15] However, to our best knowledge, no meta-analysis compared the relationship between MTHFR C677T geographical distribution and the risk of URPL by meta-analysis.
The correlations between MTHFR A1298C and the risk of URPL were also reported by lots of meta-analyses. However, these results were controversial.[8–11] Three meta-analyses indicated MTHFR A1298C polymorphisms were significantly associated with URPL,[9,11,16] whereas others showed no significant correlation between them.[8,10] Considered that a series of novel case-control studies about Chinese women's URPL had been published, an updated meta-analysis needed to further validate the association between MTHFR A1298C and the risk of URPL. The frequencies of MTHFR 1298C allele and 1298CC genotype decreased along the southern-northern direction, and also showed significantly geographical variations.[13,14] Hence, it is also important to clarify the correlation between MTHFR A1298C geographical distributions and the risk of URPL by meta-analysis.
The MTHFR 677T allele and the 677TT genotype reduced the activity of MTHFR, and the MTHFR 1298C allele and the 1298CC genotype increased folate level in the blood.[9] To our best knowledge, no meta-analysis investigates the association between MTHFR C677T and MTHFR A1298C joint mutation and the risk of URPL.
Therefore, we performed a comprehensive and updated meta-analysis to drive a precise estimation of association between MTHFR C677T or MTHFR A1298C gene polymorphisms and the risk of URPL in Chinese women, to unravel the effects of geographical variations of MTHFR C677T or MTHFR A1298C gene polymorphisms on the risk of URPL, and to analyze the association between MTHFR C677T and MTHFR A1298C joint mutation and the risk of URPL. The findings of this meta-analysis may help predict the risk of URPL in Chinese women from the angle of MTHFR C677T or / and MTHFR A1298C gene polymorphisms.
2. Materials and methods
The meta-analysis was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)[17] This review has been registered on the PROSPERO website as No. CRD42020173815 (To enable PROSPERO to focus on COVID-19 registrations during the 2020 pandemic, this registration record was automatically published exactly as submitted. The PROSPERO team has not checked eligibility).
2.1. Search strategy
A systematic literature search for studies published up to March 25, 2020, was performed. Relevant studies were identified by searching Pubmed, Embase, Web of Science, Wanfang, and CNKI databases with the following key words: ‘Chinese’ or ‘China’, ‘methylenetetrahydrofolate reductase’ or ‘MTHFR’, ‘recurrent miscarriage’ or ‘recurrent abortion’ or ‘spontaneous abortion’ or ‘recurrent pregnancy loss’ or ‘recurrent fetal loss’. Only studies that had been published in English or Chinese were included.
2.2. Inclusion and exclusion criteria
Studies were selected according to the following criteria:
-
1.
case-control studies;
-
2.
all the participants were Chinese women. URPL was defined as at least two miscarriages, and the controls were women with at least one live birth;
-
3.
MTHFR C677T and/or MTHFR A1298C gene polymorphisms were detected and sufficient data regarding genotype distributions were provided; and
-
4.
articles were published in English or Chinese.
For duplicates, only the studies with more complete data were included. Case reports, comments, review papers were excluded. The full-text screening according to the inclusion and exclusion procedures was summarized in Figure 1.
Figure 1.
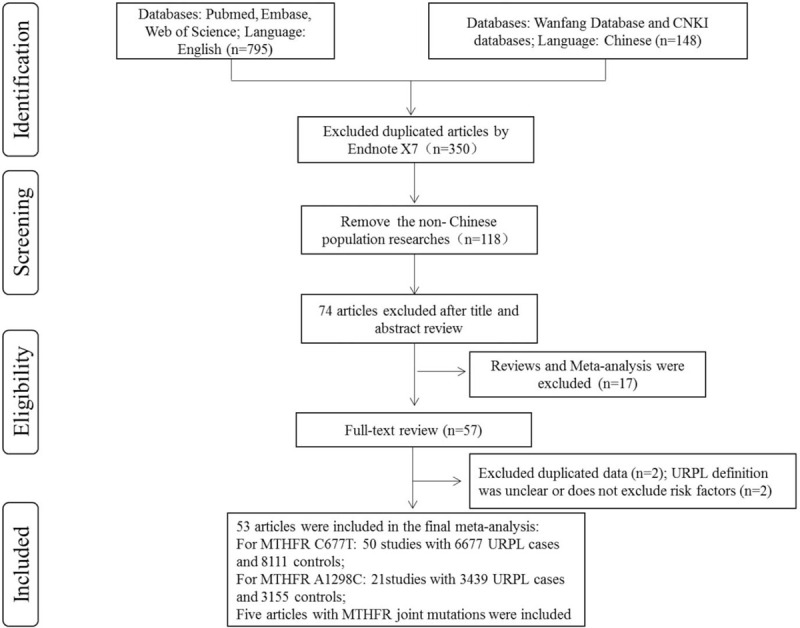
Flowchart of the literature search.
2.3. Data extraction
The following information from each eligible publication was recorded: first name of the first author, year of publication, province, sample size, number of cases and controls with genotypes, the genotype frequencies of control group were evaluated for Hardy–Weinberg equilibrium (HWE), and the quality of included studies was estimated by the 9-star Newcastle-Ottawa Scale (NOS).[18] The stars of 0–3, 4–6, and 7–9 were considered of low, moderate, and high quality, respectively.
2.4. Data analysis
All statistical analyses were performed using the STATA 16.0PM and RevMan 5.3 software. The two-tailed Student t test was employed to analyze the geographical distribution data, P < .05 was considered statistically significant. The strength of the association between the MTHFR gene polymorphisms (C677T and A1298C) and URPL risk in the Chinese women was assessed by the odds ratios (ORs) with 95% confidence intervals (CIs). For MTHFR C677T, the pooled ORs were calculated for the dominant model (TT + CT vs CC), recessive model (TT vs CT + CC), heterozygous model (CT vs CC), homozygous model (TT vs CC), and an allele model (T vs C); for MTHFR A1298C, the pooled ORs were also calculated for the dominant model (CC + AC vs AA), recessive model (CC vs AC + AA), heterozygous model (AC vs AA), homozygous model (CC vs AA), and an allele model (C vs A). χ2 and Higgins I2 statistics were performed to assess heterogeneity between studies. If I2 > 50% and P < .01, the pooled ORs were analyzed using the random effects model (REM), or else, the fixed effects model (FEM) was used. Publication bias was estimated by Egger's linear regression test. If P < .05, a significant publication bias was considered.
3. Results
3.1. Main characteristics and quality assessment of the included studies
Overall, fifty-three articles were included in the final meta-analysis. For MTHFR C677T, fifty studies with 6677 URPL cases and 8111 controls were included.[19–68] The stars of NOS ranged from 5-9, and only two studies were classed as moderate quality.[24,51] The main characteristics of all included studies were shown in Table 1. Twenty-one articles demonstrated the association of MTHFR A1298C polymorphism with risk of URPL involving 3439 URPL cases and 3155 controls.[23,24,33–35,37,39,42,43,46,47,56,59,63,64,66–71] All studies were classed as high quality according to the NOS score. The main characteristics were shown in Table 2. Five studies investigated the relationship between the MTHFR joint mutations and the risk of URPL,[23,56,59,66,68] all studies were classed as high quality, and the main characteristics were listed in Table 3.
Table 1.
Characteristics of all selected studies included on MTHFR C677T.
| Cases No. | Control No. | |||||||||||||
| Author | Provence ∗ | Publication year | Pregnancy loss(times) | Total | CC | CT | TT | Total | CC | CT | TT | HWE † | P values ‡ | NOS score § |
| Wang et al., [19] | Shanxi | 2002 | ≥2 | 62 | 13 | 33 | 16 | 119 | 43 | 53 | 23 | Y | .36 | 7 |
| Song et al., [20] | Guangdong | 2003 | ≥2 | 50 | 36 | 2 | 12 | 56 | 40 | 12 | 4 | N | .04 | 8 |
| Li et al, [21] | Shandong | 2004 | ≥2 | 57 | 16 | 32 | 9 | 50 | 25 | 20 | 5 | Y | .74 | 7 |
| Guan et al, [22] | Shandong | 2005 | ≥3 | 127 | 13 | 59 | 55 | 117 | 19 | 73 | 25 | N | .01 | 8 |
| Wang et al, [23] | Shanghai | 2006 | ≥2 | 147 | 49 | 78 | 20 | 82 | 43 | 34 | 5 | Y | .61 | 8 |
| Ren et al, [24] | Shanxi | 2007 | ≥2 | 71 | 9 | 40 | 22 | 93 | 29 | 38 | 26 | Y | .08 | 5 |
| Wan et al, [25] | Shandong | 2007 | ≥2 | 80 | 6 | 46 | 28 | 60 | 19 | 33 | 8 | Y | .28 | 9 |
| Xu et al, [26] | Shandong | 2007 | ≥2 | 112 | 21 | 48 | 43 | 100 | 32 | 50 | 18 | Y | .84 | 8 |
| Ma et al, [27] | Shanxi | 2008 | ≥2 | 60 | 12 | 32 | 16 | 60 | 19 | 34 | 7 | Y | .16 | 8 |
| Zhang et al, [29] | Jilin | 2009 | ≥2 | 56 | 12 | 25 | 19 | 50 | 20 | 22 | 8 | Y | .64 | 7 |
| Wang et al, [28] | Jiangsu | 2009 | ≥2 | 50 | 36 | 2 | 12 | 125 | 89 | 27 | 9 | N | .00 | 7 |
| Zhong et al, [30] | Ningxia | 2010 | ≥3 | 141 | 72 | 53 | 16 | 160 | 114 | 43 | 3 | Y | .65 | 9 |
| Wang et al, [31] | Shandong | 2011 | ≥2 | 159 | 18 | 82 | 59 | 127 | 28 | 78 | 21 | N | .01 | 8 |
| Han et al, [32] | Beijing | 2012 | ≥2 | 71 | 10 | 35 | 26 | 58 | 25 | 15 | 18 | N | .00 | 8 |
| Hu et al, [34] | Guangdong | 2014 | ≥3 | 52 | 29 | 14 | 9 | 16 | 11 | 4 | 1 | Y | .47 | 8 |
| Cao et al, [33] | Shanghai | 2014 | ≥2 | 166 | 53 | 83 | 30 | 82 | 29 | 43 | 10 | Y | .33 | 9 |
| Luo et al, [37] | Zhejing | 2015 | ≥2 | 125 | 40 | 70 | 15 | 135 | 60 | 65 | 10 | Y | .18 | 9 |
| Zhu et al, [39] | Henan | 2015 | ≥2 | 118 | 60 | 40 | 18 | 174 | 100 | 72 | 2 | N | .01 | 9 |
| Wang et al, [38] | Zhejiang | 2015 | ≥2 | 125 | 40 | 70 | 15 | 905 | 374 | 471 | 60 | N | .00 | 9 |
| Guo et al, [36] | Guangdong | 2015 | ≥2 | 62 | 15 | 29 | 18 | 59 | 31 | 16 | 11 | N | .00 | 9 |
| Gao et al, [35] | Henan | 2015 | ≥2 | 378 | 130 | 185 | 63 | 423 | 224 | 160 | 39 | Y | .18 | 7 |
| Tang et al, [41] | Guizhou | 2016 | ≥2 | 100 | 38 | 37 | 25 | 50 | 25 | 16 | 9 | N | .04 | 8 |
| Wang et al, [42] | Jiangsu | 2016 | ≥2 | 190 | 97 | 64 | 29 | 180 | 103 | 75 | 2 | N | .00 | 9 |
| Yue et al, [44] | Shanxi | 2016 | ≥2 | 130 | 30 | 68 | 32 | 130 | 32 | 70 | 28 | Y | .37 | 8 |
| Shang et al, [40] | Henan | 2016 | ≥2 | 349 | 79 | 150 | 120 | 421 | 220 | 175 | 26 | Y | .25 | 9 |
| Xie et al, [43] | Tianjin | 2016 | ≥2 | 244 | 31 | 94 | 119 | 116 | 23 | 62 | 31 | Y | .42 | 8 |
| Huang et al, [47] | Guangdong | 2017 | ≥2 | 83 | 19 | 39 | 25 | 90 | 30 | 48 | 12 | Y | .29 | 9 |
| Shen et al, [50] | Henan | 2017 | ≥2 | 100 | 10 | 40 | 50 | 100 | 14 | 54 | 32 | Y | .25 | 8 |
| Hua et al, [46] | Shanghai | 2017 | ≥2 | 140 | 32 | 72 | 36 | 143 | 51 | 71 | 21 | Y | .64 | 8 |
| Wang et al, [54] | Zhejiang | 2017 | ≥3 | 79 | 20 | 43 | 16 | 280 | 116 | 122 | 42 | Y | .29 | 8 |
| Zhan et al, [55] | Anhui | 2017 | ≥2 | 120 | 31 | 48 | 41 | 98 | 55 | 32 | 11 | Y | .07 | 9 |
| Wang et al, [52] | Zhejiang | 2017 | ≥2 | 100 | 15 | 45 | 40 | 50 | 19 | 21 | 10 | Y | .35 | 7 |
| Wang et al, [53] | Zhejiang | 2017 | ≥2 | 50 | 11 | 21 | 18 | 50 | 20 | 24 | 8 | Y | .86 | 7 |
| Jiang et al, [48] | Guangxi | 2017 | ≥2 | 152 | 76 | 60 | 16 | 313 | 197 | 96 | 20 | Y | .08 | 9 |
| Ding et al, [45] | Hubei | 2017 | ≥2 | 100 | 20 | 48 | 32 | 100 | 34 | 48 | 18 | Y | .88 | 7 |
| Li et al, [49] | Shanxi | 2017 | ≥2 | 50 | 7 | 25 | 18 | 50 | 22 | 13 | 15 | N | .00 | 7 |
| Shi et al, [51] | Shandong | 2017 | ≥2 | 69 | 8 | 35 | 26 | 169 | 46 | 92 | 31 | Y | .21 | 6 |
| Zhang et al, [56] | Zhejiang | 2017 | ≥2 | 50 | 14 | 24 | 12 | 10 | 4 | 6 | 0 | Y | .18 | 7 |
| Zhu et al, [59] | Beijing | 2018 | ≥2 | 370 | 166 | 157 | 47 | 144 | 66 | 59 | 19 | Y | .32 | 9 |
| Sun et al, [58] | Anhui | 2018 | ≥2 | 108 | 26 | 60 | 22 | 181 | 53 | 91 | 37 | Y | .86 | 9 |
| Li et al, [57] | Beijing | 2018 | ≥2 | 100 | 18 | 44 | 38 | 100 | 35 | 41 | 24 | Y | .09 | 9 |
| Lin et al, [63] | Guangdong | 2019 | ≥2 | 403 | 213 | 153 | 37 | 342 | 253 | 78 | 11 | Y | .11 | 9 |
| Xu et al, [67] | Henan | 2019 | ≥2 | 218 | 26 | 87 | 105 | 264 | 40 | 122 | 102 | Y | 0.72 | 9 |
| Li et al, [62] | Gansu | 2019 | ≥2 | 264 | 64 | 124 | 76 | 381 | 118 | 209 | 54 | N | 0.01 | 8 |
| Xu et al, [66] | Zhejiang | 2019 | ≥2 | 108 | 38 | 41 | 29 | 140 | 69 | 53 | 18 | Y | 0.13 | 8 |
| Bai et al, [60] | Zhejiang | 2019 | ≥2 | 72 | 27 | 28 | 17 | 116 | 48 | 54 | 14 | Y | 0.84 | 9 |
| Wu et al, [65] | Henan | 2019 | ≥2 | 109 | 14 | 49 | 46 | 487 | 91 | 242 | 154 | Y | 0.81 | 8 |
| Cai et al, [61] | Fujian | 2019 | ≥2 | 150 | 32 | 69 | 49 | 120 | 65 | 47 | 8 | Y | 0.90 | 8 |
| Liu et al, [64] | Jiangsu | 2019 | ≥2 | 170 | 38 | 84 | 48 | 170 | 55 | 86 | 29 | Y | 0.64 | 8 |
| Xu et al, [68] | Henan | 2020 | ≥2 | 230 | 29 | 90 | 111 | 264 | 40 | 122 | 102 | Y | 0.72 | 9 |
Table 2.
Characteristics of all selected studies included on MTHFR A1298C.
| Cases No. | Control No. | |||||||||||||
| Author | Province ∗ | Publication year | Pregnancy loss(times) | Total | AA | AC | CC | Total | AA | AC | CC | HWE † | P values ‡ | NOS score § |
| Li et al, [69] | Shandong | 2003 | ≥2 | 57 | 33 | 21 | 3 | 50 | 29 | 18 | 3 | Y | .93 | 8 |
| Wang et al, [23] | Shanghai | 2006 | ≥2 | 148 | 103 | 35 | 10 | 82 | 60 | 20 | 2 | Y | .83 | 7 |
| Ren et al, [24] | Shanxi | 2007 | ≥2 | 71 | 49 | 20 | 2 | 93 | 69 | 23 | 1 | Y | .54 | 7 |
| Chen et al, [70] | Hainan | 2013 | ≥2 | 59 | 24 | 29 | 6 | 87 | 38 | 44 | 5 | Y | .09 | 8 |
| Hu et al, [34] | Guangdong | 2014 | ≥3 | 52 | 33 | 12 | 7 | 16 | 12 | 3 | 1 | Y | .25 | 8 |
| Cao et al, [33] | Shanghai | 2014 | ≥2 | 166 | 132 | 31 | 3 | 82 | 49 | 31 | 2 | Y | .25 | 8 |
| Luo et al, [37] | Zhejiang | 2015 | ≥2 | 125 | 82 | 40 | 3 | 135 | 78 | 54 | 3 | Y | .07 | 9 |
| Zhu et al, [39] | Henan | 2015 | ≥2 | 118 | 48 | 58 | 12 | 174 | 76 | 88 | 10 | N | .02 | 8 |
| Gao et al, [35] | Henan | 2015 | ≥2 | 378 | 180 | 118 | 80 | 423 | 210 | 165 | 48 | Y | .08 | 7 |
| Li et al, [71] | Jiangsu | 2015 | ≥2 | 60 | 31 | 21 | 8 | 150 | 84 | 61 | 5 | Y | .12 | 7 |
| Wang et al, [42] | Jiangsu | 2016 | ≥2 | 190 | 77 | 93 | 19 | 180 | 79 | 91 | 10 | N | .01 | 9 |
| Xie et al, [43] | Tianjin | 2016 | ≥2 | 244 | 165 | 74 | 5 | 116 | 82 | 29 | 5 | Y | .25 | 8 |
| Huang et al, [47] | Guangdong | 2017 | ≥2 | 83 | 52 | 28 | 3 | 90 | 64 | 23 | 3 | Y | .60 | 9 |
| Hua et al, [46] | Shanghai | 2017 | ≥2 | 140 | 100 | 36 | 4 | 143 | 86 | 53 | 4 | Y | .21 | 9 |
| Zhang et al, [56] | Zhejiang | 2017 | ≥2 | 50 | 30 | 19 | 1 | 10 | 5 | 4 | 1 | Y | .88 | 7 |
| Zhu et al, [59] | Guangdong | 2018 | ≥2 | 370 | 243 | 114 | 13 | 144 | 83 | 56 | 5 | Y | .23 | 8 |
| Lin et al, [63] | Guangxi | 2019 | ≥2 | 403 | 231 | 144 | 28 | 342 | 221 | 102 | 19 | Y | .12 | 9 |
| Xu et al, [67] | Henan | 2019 | ≥2 | 218 | 155 | 58 | 5 | 264 | 214 | 44 | 6 | N | .05 | 9 |
| Xu et al, [66] | Zhejiang | 2019 | ≥2 | 108 | 61 | 37 | 10 | 140 | 83 | 46 | 11 | Y | .21 | 8 |
| Liu et al, [64] | Jiangsu | 2019 | ≥2 | 170 | 119 | 49 | 2 | 170 | 114 | 53 | 3 | Y | .26 | 8 |
| Xu et al, [68] | Henan | 2020 | ≥2 | 230 | 156 | 67 | 7 | 264 | 214 | 44 | 6 | N | .05 | 9 |
Table 3.
Characteristics of all selected studies included on MTHFR joint mutations.
| Case No. | Control No. | |||||||||
| Author | Provence | Publication year | Pregnancy loss(times) | Joint mutation | No mutation | Joint mutation | No mutation | HWE ∗ | P value † | NOS score ‡ |
| Wang et al, [23] | Shanghai | 2006 | ≥2 | 23 | 27 | 7 | 28 | Y | .61 | 8 |
| Zhang et al, [56] | Zhejiang | 2017 | ≥2 | 12 | 6 | 2 | 1 | Y | .18 | 7 |
| Zhu et al, [59] | Beijing | 2018 | ≥2 | 48 | 88 | 23 | 28 | Y | .32 | 9 |
| Xu et al, [66] | Zhejiang | 2019 | ≥2 | 41 | 4 | 28 | 18 | Y | .72 | 9 |
| Xu et al, [68] | Henan | 2020 | ≥2 | 49 | 4 | 28 | 18 | Y | .72 | 9 |
3.2. Geographical distributions of MTHFR gene polymorphisms
The prevalence of the two gene polymorphisms varied significantly among different populations and showed apparent geographical gradients. For MTHFR C677T, the MTHFR 677TT genotype frequency was significantly higher in URPL group compared with control group (P < .001), and the frequency increased along the south-north direction. For MTHFR A1298C, the MTHFR 1298CC genotype frequency was slightly higher in URPL group compared with control (not statistically significant), and the frequency decreased along the south-north direction (Table 4).
Table 4.
Distribution frequencies under different MTHFR gene polymorphisms.
| Groups | No. | Genotype frequencies [n (%)] | ||||
| Wild type | Heterozygous type | Homozygous type | ||||
| C677T | Total | Cases | 6677 | 1919 (28.84) | 2957 (44.48) | 1801 (26.69) ∗ |
| Control | 8111 | 3318 (41.03) | 3592 (44.26) | 1201 (14.72) | ||
| North | Cases | 3725 | 874 (23.46) | 1673 (44.91) | 1178 (31.26) | |
| Control | 4217 | 1444 (34.24) | 1952 (46.29) | 821 (19.47) | ||
| South | Cases | 2952 | 1045 (35.40) | 1284 (43.50) | 623 (21.10) | |
| Control | 3894 | 1874 (48.13) | 1640 (42.12) | 380 (9.76) | ||
| A1298C | Total | Cases | 3439 | 2206 (61.18) | 1139 (32.10) | 241 (6.72) |
| Control | 3155 | 2010 (61.81) | 1072 (33.34) | 155 (4.85) | ||
| North | Cases | 1686 | 1029 (61.03) | 530 (31.44) | 127 (7.53) | |
| Control | 1528 | 977 (63.94) | 467 (30.56) | 84 (5.50) | ||
| South | Cases | 1753 | 1075 (61.32) | 574 (32.74) | 104 (5.93) | |
| Control | 1627 | 973 (59.80) | 585 (35.96) | 69 (4.24) | ||
3.3. MTHFR C677T and/or MTHFR A1298C gene polymorphisms and URPL risk
The ORs and 95% CIs of the association between MTHFR gene polymorphisms and URPL risk were considered under different models (Figure S1 and S2 ). Main meta-analyzed results of the association of MTHFR C677T and URPL risk were summarized in Table 5. The overall pooled results indicated significant associations between all the MTHFR C677T gene polymorphisms and the risk of URPL, especially homozygous model (TT vs CC; OR 3.06; 95% CI 2.56–3.66). When excluding studies that deviated from HWE, significant associations were also found in all these models (Table 5). The overall results of MTHFR A1298C polymorphism also showed significant association in the recessive (CC vs AC + AA; OR 1.55; 95% CI 1.25–1.93) and homozygous (CC vs AA; OR 1.53; 95% CI 1.22–1.91) models. When excluding studies that deviated from HWE, significant associations were still found in these two models (Table 6). Through linkage analysis of the MTHFR C677T and A1298C loci, our results found that the patients carrying these two MTHFR mutants had no synergistic effect (OR 2.71; 95% CI 0.84–8.70) on the occurrence of URPL compared with the individuals carrying the wild-type homozygous genotype (677CC/1298AA) (Figure 2).
Table 5.
ORs and 95% Cis for the MTHFR C677T and URPL under different models.
| Contrast | Group | No. | I2(%) | Model ∗ | Pooled OR(95% CI) | Egger (P value) | P value † |
| TT + CT vs CC | Overall | 50 | 51.9 | REM | 1.91 (1.70–2.15) | .61 | |
| HWE: yes | 38 | 49.7 | FEM | 2.01 (1.84–2.19) | .75 | ||
| Distribution: North | 25 | 59.6 | REM | 1.99 (1.65–2.39) | .57 | ||
| Distribution: South | 25 | 43.0 | FEM | 1.86 (1.67–2.07) | |||
| TT vs CT + CC | Overall | 50 | 57.0 | REM | 2.24 (1.93–2.60) | .03 | |
| HWE: yes | 38 | 58.5 | REM | 2.16 (1.83–2.56) | .11 | ||
| Distribution: North | 25 | 69.1 | REM | 2.16 (1.74–2.68) | .47 | ||
| Distribution: South | 25 | 31.2 | FEM | 2.24 (1.92–2.61) | |||
| CT vs CC | Overall | 50 | 50.7 | REM | 1.59 (1.40–1.80) | .84 | |
| HWE: yes | 38 | 25.5 | FEM | 1.70 (1.55–1.87) | .44 | ||
| Distribution: North | 25 | 53.9 | REM | 1.63 (1.36–1.96) | .98 | ||
| Distribution: South | 25 | 49.4 | FEM | 1.60 (1.42–1.80) | |||
| TT vs CC | Overall | 50 | 57.8 | REM | 3.06 (2.56–3.66) | .06 | |
| HWE: yes | 38 | 63.9 | REM | 2.95 (2.38–3.67) | .28 | ||
| Distribution: North | 25 | 70.3 | REM | 3.10 (2.33–4.11) | .91 | ||
| Distribution: South | 25 | 31.9 | FEM | 2.95 (2.48–3.49) | |||
| T vs C | Overall | 50 | 61.3 | REM | 1.74 (1.60–1.90) | .70 | |
| HWE: yes | 38 | 69.2 | REM | 1.75 (1.57–1.95) | .99 | ||
| Distribution: North | 25 | 69.8 | REM | 1.74 (1.53–1.97) | .92 | ||
| Distribution: South | 25 | 49.3 | FEM | 1.73 (1.60–1.87) |
Table 6.
ORs and 95% CIs for the MTHFR A1298C and URPL under different models.
| Contrast | Group | No. | I2(%) | Model ∗ | Pooled OR(95% CI) | Egger (P value) | P value † |
| CC + AC vs AA | Overall | 21 | 54.5 | REM | 1.07 (0.90–1.26) | .06 | |
| HWE: yes | 17 | 43.1 | FEM | 0.99 (0.88–1.12) | .64 | ||
| Distribution: North | 8 | 58.2 | REM | 1.24 (0.95–1.56) | .10 | ||
| Distribution: South | 13 | 50.9 | REM | 0.97 (0.78–1.24) | |||
| CC vs AA + AC | Overall | 21 | 0.0 | FEM | 1.55 (1.25–1.93) | .04 | |
| HWE: yes | 17 | 1.6 | FEM | 1.54 (1.21–1.97) | .08 | ||
| Distribution: North | 8 | 9.6 | FEM | 1.63 (1.21–2.19) | .64 | ||
| Distribution: South | 13 | 0.0 | FEM | 1.47 (1.06–1.93) | |||
| AC vs AA | Overall | 21 | 56.5 | REM | 1.02 (0.85–1.21) | .75 | |
| HWE: yes | 17 | 39.3 | FEM | 0.92 (0.81–1.05) | .97 | ||
| Distribution: North | 8 | 67.9 | REM | 1.17 (0.87–1.58) | .15 | ||
| Distribution: South | 13 | 45.6 | FEM | 0.95 (0.82–1.11) | |||
| CC vs AA | Overall | 21 | 0.0 | FEM | 1.53 (1.22–1.91) | .06 | |
| HWE: yes | 17 | 0.0 | FEM | 1.49 (1.16–1.91) | .10 | ||
| Distribution: North | 8 | 0.0 | FEM | 1.57 (1.15–2.13) | .82 | ||
| Distribution: South | 13 | 0.0 | FEM | 1.49 (1.07–2.07) | |||
| C vs A | Overall | 21 | 52.7 | REM | 1.10 (0.97–1.26) | .29 | |
| HWE: yes | 17 | 49.9 | FEM | 1.07 (0.97–1.18) | .29 | ||
| Distribution: North | 8 | 51.1 | REM | 1.22 (1.01–1.47) | .08 | ||
| Distribution: South | 13 | 51.9 | REM | 1.02 (0.85–1.23) |
Figure 2.
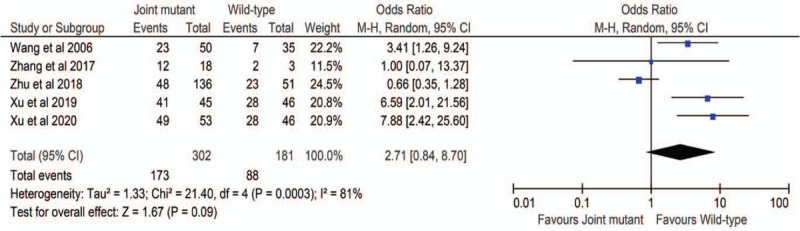
Random effect forest plot of MTHFR joint mutant vs the wild-type homozygous genotype.
3.4. Relationship between MTHFR C677T or MTHFR A1298C geographical distribution and the risk of URPL
We also performed a sub-group analysis stratified by north and south China (Table 5 and 6). For MTHFR C677T, twenty-five studies with 3725 URPL cases and 4217 controls were included in the north China and twenty-five studies with 2952 URPL cases and 3894 controls were included in the south China. Significant associations between MTHFR C677T and URPL risk were found in both north and south China in all the models. However, no significant difference was observed in all the genotype groups between north and south China in the risk of UPRL (Table 5). For MTHFR A1298C, eight studies with 1686 URPL cases and 1528 controls were included in the north China and thirteen studies with 1753 URPL cases and 1627 controls were included in the south China. Sub-group analyzed results still showed significant association in the recessive and homozygous models of MTHFR A1298C polymorphism in north and south China. In addition, we also found a significant association in the allele model (C vs A; OR 1.22; 95% CI 1.01–1.47) in north China. There was also no significant difference in all the genotype groups between north and south of China in the risk of UPRL (Table 6).
3.5. Sensitivity analysis
We conducted sensitivity analysis to ascertain the primary origin of the heterogeneity. Through sensitivity analysis, the present study showed that no individual study had affected the pooled ORs (see Figure S1 and S2 , Supplemental Content, which illustrates the sensitivity analysis results).
3.6. Publication bias
Egger's test was performed to evaluate funnel plot symmetry statistically (see Figure S3 and S4 , Supplemental Content, which illustrates the publication bias results). For MTHFR C677T, the egger's regression asymmetry test showed significant publication bias (P = .03) in the recessive model (Table 5). For MTHFR A1298C, significant publication bias (P = .04) was also found in the recessive model (Table 6). However, after excluding non-HWE studies, no publication bias was found on the recessive model of both MTHFR C677T and MTHFR A1298C mutations (Table 5 and 6).
4. Discussion
In our meta-analysis, we collected 50 studies with 6677 URPL cases and 8111 controls to investigate the association between MTHFR C677T gene polymorphism and the risk of URPL. The results revealed that all the C677T mutations of MTHFR were significantly associated with the risk of URPL in the Chinese population. The MTHFR 677 T allele and TT genotype may increase the risk of URPL. These results were expected because all the individual studies included in our meta-analysis presented these trends in their populations. The results were also consistent with previous meta-analysis results among Chinese population reported by Chen et al and Ren et alet alet al,[8,72] and among Asian population reported by Parveen et al, Cao et al, and Wu et al,[73–75]. For MTHFR A1298C, a total of 21 studies involving 3439 URPL cases and 3155 controls were included. A significant association between MTHFR A1298C mutations (recessive [CC vs AC + AA; OR 1.55; 95% CI 1.25–1.93] and homozygous [CC vs AA; OR 1.53; 95% CI 1.22–1.91] models) and URPL was observed. Among the related 21 studies, two studies showed that the MTHFR 1298CC genotype occurred more significantly frequent in the URPL population than in the control group,[35,71] and eleven studies described the same trend,[23,24,34,39,42,63,66–68,70] while eight studies showed the opposite trend.[33,37,43,46,56,59,64,69] Our present meta-analysis showed significantly positive results, which were similar with the meta-analyzed ones reported by Yang et al and Zhang et al,[9,11], but were inconsistent with previous meta-analyzed results reported by Chen et al, Rai et al, and Cao et al, [8,74,76]. We hypothesize that part of the reasons for these controversial results might be related to negative results occupied high weights and different ethnic background in their meta-analyses.[9] The relationship between MTHFR C677T and MTHFR A1298C joint mutation and the risk of URPL were investigated by five studies and our result showed no significant association among Chinese women (OR 2.71; 95% CI 0.84–8.70). Further analysis revealed that among the five studies, one study presented MTHFR C677T and MTHFR A1298C joint mutation more frequent in the control than in the URPL group and has the highest weight (24.52%).[59] And this might be the reason why we did not get statistically significant result.
The association of geographical distributions of the MTHFR C677T and MTHFR A1298C gene polymorphisms with URPL was not clear among Chinese population. Our results showed that geographical distributions of the MTHFR C677T or MTHFR A1298C gene polymorphisms were not associated with URPL.
Some potential limitations of this meta-analysis should be addressed. One of the limitations was high heterogeneity. It influenced the interpretation of the meta-analysis result. Sample size, racial difference, and deviations of allele distributions from the HWE law played important roles.[77] After excluded non HWE studies, the results still played high heterogeneity in the recessive, homozygous, and additive comparisons in MTHFR C677T group. Our further analysis showed the high heterogeneity was caused by Shang et al, whose pooled OR value and sample size were high.[40] After we removed this study, all the I2 values decreased to less than 50%. While for MTHFR A1298C, no significant heterogeneity was found in all the comparisons after remove non HWE articles. The other limitation was publication bias. Positive results interested only by researchers and journals might be the possible reason for publication bias.[78,79] However, in our present study, deviation from HWE law was an indication of potential publication bias. After excluding the non HWE studies, all the P values increased to over.05 in both MTHFR C677T and MTHFR A1298C groups. In addition, for the association between MTHFR C677T and MTHFRA1298C joint mutation and the risk of URPL, only five studies were involved, so heterogeneity and publication bias were not investigated. Therefore, additional studies were needed to reevaluate the risk of MTHFR C677T and MTHFR A1298C joint mutation in URPL. In addition, although we performed a broad search in five different databases to find studies for inclusion criteria, it is impossible to confirm that all available studies were included, which may exhibit another limitation of our meta-analysis.
In conclusion, this meta-analysis suggested that MTHFR 677T allele and 677TT genotype and MTHFR 1298CC genotype may be risk factors for the development of URPL. Moreover, geographical distribution of the MTHFR C677T or MTHFR A1298C gene polymorphisms was not associated with the risk of URPL for Chinese women. We suggested that MTHFR C677T and MTHFR A1298C mutants should be tested in Chinese pregnant women.
Author contributions
Data curation: Xiaoying Wang, Qiang Sun, Baiqian Xing.
Investigation: Zhongdong Li.
Methodology: Genzhu Wang, Xiaoying Wang, Qiang Sun.
Resources: Zhaohui Lin, Zhikun Xun.
Software: Genzhu Wang, Qiang Sun, Baiqian Xing.
Supervision: Zhongdong Li.
Validation: Zhaohui Lin, Xiaoying Wang, Zhikun Xun, Baiqian Xing.
Writing – original draft: Genzhu Wang.
Writing – review & editing: Zhaohui Lin, Zhongdong Li.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: CI = confidence interval, CNKI = China National Knowledge Infrastructure, FEM = fixed effects model, HWE = Hardy–Weinberg equilibrium, MTHFR = 5,10-methylenetetrahydrofolate reductase, 5,10-MTHF = 5,10-methylenetetrahydrofolate, 5-MTHF = 5-methylenetetrahydrofolate, OR = odds ratio, PRISMA = the Preferred Reporting Items for Systematic Reviews and Meta-Analyses, RPL = recurrent pregnancy loss, NOS = Newcastle-Ottawa Scale, REM = random effects model, URPL = unexplained recurrent pregnancy loss.
How to cite this article: Wang G, Lin Z, Wang X, Sun Q, Xun Z, Xing B, Li Z. The association between 5, 10 – methylenetetrahydrofolate reductase and the risk of unexplained recurrent pregnancy loss in China: A Meta-analysis. Medicine. 2021;100:17(e25487).
Compliance with ethical standards
The authors have no funding to disclose.
The authors have no conflicts of interest to disclose.
Ethical approval: All data utilized in our meta-analysis are extracted from publicly available material; therefore, ethical approval is waived.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental digital content is available for this article.
MTHFR = 5,10-methylenetetrahydrofolate reductase, HWE = Hardy–Weinberg equilibrium.
Bold for north China, regular for south China.
Y: genotype frequencies of control group were meet HWE criteria; N: genotype frequencies of control group were not meet HWE criteria.
P value for HWE in control group.
Scores estimated by the 9-star Newcastle–Ottawa Scale.
MTHFR = 5,10-methylenetetrahydrofolate reductase, HWE = Hardy–Weinberg equilibrium.
Bold for north China, regular for south China.
Y: genotype frequencies of control group were meet HWE criteria; N: genotype frequencies of control group were not meet HWE criteria.
P value for HWE in control group.
Scores estimated by the 9-star Newcastle–Ottawa Scale.
MTHFR = 5,10-methylenetetrahydrofolate reductase, HWE = Hardy–Weinberg equilibrium, No. = number.
Y: genotype frequencies of control group were meet HWE criteria; N: genotype frequencies of control group were not meet HWE criteria.
P value for HWE in control group.
Scores estimated by the 9-star Newcastle–Ottawa Scale.
∗P < .001 vs control, MTHFR = 5,10-methylenetetrahydrofolate reductase, No. = number.
OR = odds ratio, CI = confidence interval, MTHFR = 5,10-methylenetetrahydrofolate reductase, URPL = unexplained recurrent pregnancy loss, No. = number, HWE = Hardy–Weinberg equilibrium, NOS = Newcastle–Ottawa Scale.
REM: random effects model; FEM: fixed effects model.
P value for north vs south.
OR = odds ratio, CI = confidence interval, MTHFR = 5,10-methylenetetrahydrofolate reductase URPL = unexplained recurrent pregnancy loss, No. = number, HWE = Hardy–Weinberg equilibrium.
REM: random effects model; FEM: fixed effects model.
P value for north vs south.
References
- [1].PCotASfR Medicine. Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril 2012;98:1103–11. [DOI] [PubMed] [Google Scholar]
- [2].von Eye Corleta H. It is time to respect the American Society for Reproductive Medicine definition of recurrent pregnancy loss. Fertil Steril 2010;94:e61. [DOI] [PubMed] [Google Scholar]
- [3].Hong Li Y, Marren A. Recurrent pregnancy loss: a summary of international evidence-based guidelines and practice. Aust J Gen Prac 2018;47:432–6. [DOI] [PubMed] [Google Scholar]
- [4].Ticconi C, Pietropolli A, Di Simone N, et al. Endometrial immune dysfunction in recurrent pregnancy loss. Int J Mol Sci 2019;20: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Goyette P, Sumner JS, Milos R, et al. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet 1994;7:195–200. [DOI] [PubMed] [Google Scholar]
- [6].Gueant JL, Candito M, Andres E, et al. Familial pernicious anaemia with hyperhomocysteinaemia in recurrent early pregnancy loss. Throm Haemost 2004;92:1147–9. [PubMed] [Google Scholar]
- [7].Steegers-Theunissen RP, Boers GH, Blom HJ, et al. Hyperhomocysteinaemia and recurrent spontaneous abortion or abruptio placentae. Lancet (London, England) 1992;339:1122–3. [DOI] [PubMed] [Google Scholar]
- [8].Chen H, Yang X, Lu M. Methylenetetrahydrofolate reductase gene polymorphisms and recurrent pregnancy loss in China: a systematic review and meta-analysis. Arch Obstet Gynaecol 2016;293:283–90. [DOI] [PubMed] [Google Scholar]
- [9].Yang Y, Luo Y, Yuan J, et al. Association between maternal, fetal and paternal MTHFR gene C677T and A1298C polymorphisms and risk of recurrent pregnancy loss: a comprehensive evaluation. Arch Obstet Gynaecol 2016;293:1197–211. [DOI] [PubMed] [Google Scholar]
- [10].Du B, Shi X, Yin C, et al. Polymorphisms of methalenetetrahydrofolate reductase in recurrent pregnancy loss: an overview of systematic reviews and meta-analyses. J Assist Reprod Genet 2019;36:1315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang Y, He X, Xiong X, et al. The association between maternal methylenetetrahydrofolate reductase C677T and A1298C polymorphism and birth defects and adverse pregnancy outcomes. Prenat Diagn 2019;39:03–9. [DOI] [PubMed] [Google Scholar]
- [12].Pi T, Liang YQ, Xia HY, et al. Prevalence of the methylenetetrahydrofolate reductase 677C > T polymorphism in the pregnant women of Yunnan Province, China. Medicine 2020;99:e22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang X, Fu J, Li Q, et al. Geographical and ethnic distributions of the MTHFR C677T, A1298C and MTRR A66G gene polymorphisms in Chinese populations: a meta-analysis. PLoS one 2016;11:e0152414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang B, Liu Y, Li Y, et al. Geographical distribution of MTHFR C677T, A1298C and MTRR A66G gene polymorphisms in China: findings from 15357 adults of Han nationality. PLoS one 2013;8:e57917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guo X, Xing H, Li Y, et al. The relation between the MTHFR gene and recurrent spontaneous abortion and to analyze the regional distribution. Chin J Birth Health Hered 2016;24:29–31. [Google Scholar]
- [16].Zhang Y, Zhan W, Du Q, et al. Variants c.677 C > T, c.1298 A > C in MTHFR, and c.66 A > G in MTRR affect the occurrence of recurrent pregnancy loss in Chinese women. Genet Test Mol Biomarkers 2020;24:717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shamseer L, Moher D, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ (Clin Res ed) 2015;350:g7647. [DOI] [PubMed] [Google Scholar]
- [18].Wells G, Shea B, O’Connell D, et al. The Newcastle–Ottawa Scale (NOS) For Assessing The Quality of Nonrandomised Studies In Meta-Analyses. 2011;http://www.ohri.ca/programs/clinical_epidemiology/oxford.asphttp://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 2020.04.23. [Google Scholar]
- [19].Wang Y, Li F, Li Y, et al. The study on the relationship between the methylenetetrahydofolate reductase 677 C→T mutation and unexplained recurrent pregnancy loss. Chin J Prac Gynecol Obstet 2002;18:291–3. [Google Scholar]
- [20].Song L, Qi Q, Tian G, et al. The relationship between methylenetetrahydrofolate reductase and risk of recurrent pregnancy loss. JMBE 2003;9:246–7. [Google Scholar]
- [21].Li X, Zhang Y, Xu Y, et al. Study on the relationship of MTHFR polymorphisms with unexplained recurrent spontaneous abortion. Chin J Med Gen 2004;21:39–42. [PubMed] [Google Scholar]
- [22].Guan L, Du X, Wang J, et al. Association of genetic polymorphisms in plasminogen activator inhibitor-1 gene and 5,10-methylenetetrahydrofolate reductase gene with recurrent early spontaneous abortion. Chin J Med Gen 2005;22:330–3. [PubMed] [Google Scholar]
- [23].Wang X, Ma Z, Lin Q. Inherited thrombophilia in recurrent spontaneous abortion among Chinese women. Int J Gynaecol Obstet 2006;92:264–5. [DOI] [PubMed] [Google Scholar]
- [24].Ren J, Han X, Liu Xe, et al. Methylenetetrahydrofolate reductase genepolymorphism in women with recurrent pregnancy loss. Chin J Perinat Med 2007;10:80–4. [Google Scholar]
- [25].Wan J, Liu X, Zhang H, et al. Analyzes of susceptible gene in women with recurrent spontaneous abortions. Chin J Birth Health Hered 2007;15:09–10. [Google Scholar]
- [26].Xu L, Liu X, Zhang H, et al. Relationship between three thrombophilic gene mutations and unexplained recurrent early spontaneous abortion. Chin JObstet Gynecol 2007;42:180–3. [PubMed] [Google Scholar]
- [27].Ma S, Zheng M. The Relation of MTHFRC677T Gene Polymorphism to Recurrent Abortion. Health Med Res Prac High Inst 2008;5:04–6. [Google Scholar]
- [28].Wang S, Shi X, Shen R, et al. The role of folate metabolism related enzymes gene polymorphisms on recurrent early spontaneous abortion. C J Birth Health Hered 2009;17:12–3. [Google Scholar]
- [29].Zhang J, Zuo W, Wu G, et al. Association between MTHFR gene C677T polymorphism and habitual abortion. J Jilin Univ (Medicine edition) 2009;35:698–701. [Google Scholar]
- [30].Zhong H, Liu J, Zhu Y, et al. Study on correlation between MTHFR polymorphisms and repeated spontaneous abortion. Ningxia Med J 2010;32:675–6. [Google Scholar]
- [31].Wang S, Jia Y, Yang D, et al. Correlation between MTHFR gene C677T polymorphism and unexplained recurrent spontaneous abortion. Matern Child Health Care China 2011;29:1385–7. [Google Scholar]
- [32].Han H, Shen H, Wang Y, et al. Association of C677T Methylenetetrahy Hydrofolate Reductase (MTHFR) polymorphism and homocysteine with recurrent pregnancy loss. Reprod Contracep 2012;32:486–9. [Google Scholar]
- [33].Cao Y, Zhang Z, Zheng Y, et al. The association of idiopathic recurrent early pregnancy loss with polymorphisms in folic acid metabolism-related genes. Genes Nutr 2014;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hu X, Liang P, Xi L, et al. The association of methylenetetrahydrofolate reductase gene mutation with unexplained recurrent miscarriage. Chin J Birth Health Hered 2014;22:87–9. [Google Scholar]
- [35].Gao J, Wang T, Xiao H, et al. MTHFR and MTRR gene polymorphisms involved in folate metabolism and their relations to unexplained recurrent spontaneous abortion in Henan province. Clin Med 2015;35:01–4. [Google Scholar]
- [36].Guo B. The association between gene polymorphisms of 5, 10-methylenetetrahydrofolate reductase and the risk of unexplained recurrent pregnancy loss. Clin Res 2015;23:153–4. [Google Scholar]
- [37].Luo L, Chen Y, Wang L, et al. Polymorphisms of genes involved in the folate metabolic pathway impact the occurrence of unexplained recurrent pregnancy loss. Reprod Sci (Thousand Oaks, Calif) 2015;22:845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang X, Luo L, Wang L, et al. MTHFR C677T polymorphism contributes to unexplained recurrent spontaneous abortion. Chin J Lab Med 2015;38:243–6. [Google Scholar]
- [39].Zhu L. Polymorphisms in the methylene tetrahydrofolate reductase and methionine synthase reductase genes and their correlation with unexplained recurrent spontaneous abortion susceptibility. Genet Mol Res 2015;14:8500–8. [DOI] [PubMed] [Google Scholar]
- [40].Shang P. Analysis of the relationship between MTHFR C677T polymorphism and unknown reason Embryo arrest by gene chip. J Chin Phys 2016;18:702–5. [Google Scholar]
- [41].Tang D, Wu Z, Jin Y, et al. The relationship between gene polymorphisms of MTHFR C677T, homocysteine and recurrent pregnancy loss. Chin J Birth Health Hered 2016;24:15–7. [Google Scholar]
- [42].Wang K. Correlation analysis of SNP polymorphisms of MTHFR and MTRR with recurrent miscarriage of Han Chinese pregnant women. Chin J Wom Child Health 2016;27:1195–7. [Google Scholar]
- [43].Xie X, Zhang Y, Xin L, et al. The relationship of the folate metabolism related gene polymorphisms of MTHFR and MTRR with unexplained recurrent spontaneous abortion. Tianjin Med J 2016;44:1243–6. [Google Scholar]
- [44].Yue H, Huang J, Wang X, et al. Effect of PA1-1, FV, F II and MTHFR polymorphisms on women with recurrent miscarriage. Chin J Birth Health Hered 2016;24:14–6. [Google Scholar]
- [45].Ding X. Relationship between MTHFRC677T gene polymorphism and RSA and URSA. Med J Nat Defen Forces Southwest China 2017;27:37–9. [Google Scholar]
- [46].Hua Z, Huang C, Lu Y, et al. Study on the correlation between MTHFR gene polymorphism and unexplained recurrent spontaneous abortion. Int J Lab Med 2017;38:16–7. [Google Scholar]
- [47].Huang S, Tang G, Liu Q, et al. The relationship between MTHFR gene polymorphism and unexplained recurrent spontaneous abortion. J Int Rep Health/Fam Plan 2017;36:382–4. [Google Scholar]
- [48].Jiang W, Lu Y, Li Y, et al. Association between MTHFR C677T polymorphism and unexplained recurrent spontaneous abortion in Nanning Han patients. J Guangxi Med Univ 2017;34:1728–30. [Google Scholar]
- [49].Li H, Huang J. Association of methylenetetrahydrofolate reductase gene 677 polymorphism with recurrent spontaneous abortion. Shaanxi Med J 2017;46:855–7. [Google Scholar]
- [50].Shen H, Feng X, Luo S, et al. The relationship between MTHFR C677T gene polymorphisms, homocysteine level and recurrent pregnancy loss. Chin J Birth Health Hered 2017;25:07–8. [Google Scholar]
- [51].Shi C, Yan Q, Jiang L. Study on the relationship between the polymorphism of MTHFR gene C677T locus and unexplained recurrent spontaneous abortion in Yantai. China Contin Med Edu 2017;9:75–6. [Google Scholar]
- [52].Wang C, Ding C. Clinical study on the correlation between MTHFR gene polymorphism and traditional Chinese medical syndromes in recurrent spontaneous abortion. Zhejiang J Trad Chin Med 2017;52:625–6. [Google Scholar]
- [53].Wang C, Ding C, Yang X, et al. Relationship between HCV levels and the polymorphism of MTHFR gene with traditional Chinese medical type of kidney deficiency and blood stasis in recurrent spontaneous abortion patients. Zhejiang J Integ Trad Chin West Med 2017;27:470–3. [Google Scholar]
- [54].Wang M, Li X, Tang S. Relationship between MTHFR gene polymorphism and spontaneous abortion. Mat Child Health Care China 2017;32:92–4. [Google Scholar]
- [55].Zhan Q, He L. Correlation analysis of MTHFR and PAI-1 polymorphisms with recurrent spontaneous abortion. Acta Acad Med Wannan 2017;36:38–40. [Google Scholar]
- [56].Zhang J, Zeng Y. Analysis of mutations in methylenetetrahydrofolate reductase gene in patients with unexplained recurrent miscarriage. J Prev Med 2017;29:202–5. [Google Scholar]
- [57].Li Q, Chang L, Liu P, et al. Association between maternal and fetal MTHFR polymorphism (C677T) and risk of unexplained recurrent spontaneous abortion. J Rep Med 2018;27:981–4. [Google Scholar]
- [58].Sun Y, Zhu S, Wang C, et al. The relationship between MTHFR C677 polymorphism and recurrent spontaneous abortion. Chin J Birth Health Hered 2018;26:28–30. [Google Scholar]
- [59].Zhu Y, Wu T, Ye L, et al. Prevalent genotypes of methylenetetrahydrofolate reductase (MTHFR) in recurrent miscarriage and recurrent implantation failure. J Assis Rep Gen 2018;35:1437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bai W, Tang S. Relationship between MTHFR C677T gene polymorphism and recurrent spontaneous abortion detected by PCR-fluorescence probe method. J Math Med 2019;32:868–9. [Google Scholar]
- [61].Cai X, Fan Z, Yang A. Polymorphism of methylenetetrahydrofolate reductase gene C677T and plasminogen activator inhibitor gene 4G/5G and unexplained recurrent miscarriage. Chin J Clin Ration Drug Use 2019;12:151–2. [Google Scholar]
- [62].Li A, Niu L, Zhang Y. Association between MTHFR gene polymorphism and recurrent spontaneous abortion in Qingyang area. For Med Sci (Section of Medgeography) 2019;40:30–2. [Google Scholar]
- [63].Lin Z, Li Q, Sun Y, et al. Interactions between genetic variants involved in the folate metabolic pathway and serum lipid, homocysteine levels on the risk of recurrent spontaneous abortion. Lipids Health Dis 2019;18:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu Y, Liu M, Xiang J, et al. Correlation between MTHFR gene polymorphism and unexplained recurrent spontaneous abortion. Chin J Birth Health Hered 2019;27:06–8. [Google Scholar]
- [65].Wu J. Polymorphism analysis of MTHFR gene C677T locus in women with unexplained recurrent miscarriage in Luoyang. Chin J Birth Health Hered 2019;27:956–7. [Google Scholar]
- [66].Xu J, Liang L, Chen X, et al. Relationships between MTHFR gene polymorphism and threatened abortion, recurrent spontaneous abortion. Matern Child Heal Care China 2019;34:1326–8. [Google Scholar]
- [67].Xu Y, Ban Y, Ran L, et al. Relationship between unexplained recurrent pregnancy loss and 5,10-methylenetetrahydrofolate reductase) polymorphisms. Fertil Steril 2019;111:597–603. [DOI] [PubMed] [Google Scholar]
- [68].Xu Y, Ban Y, Ran L, et al. Relationship between unexplained recurrent spontaneous abortion and MTHFR gene polymorphism. J Zhengzhou Univ (Medical Sciences) 2020;55:112–5. [Google Scholar]
- [69].Li X, Xu Y, Jiang S. Study on correlation between MTHFR polymorphisms and unexplained repeated spontaneous abortion. J Prac Obst Gynecol 2003;19:179–80. [Google Scholar]
- [70].Chen H, Tang L, Chen C, et al. Unexplained recurrent spontaneous abortion and methylenetetrahydrofolate reductase and methionine synthase reductase gene polymorphism. Chin J Bir Health Her 2013;21:29–31. [Google Scholar]
- [71].Li X, Chen L, Guo H, et al. Study on polymorphism of folate metabolism enzyme gene and susceptibility to repeated spontaneous abortion. Contemp Med 2015;21:01–2. [Google Scholar]
- [72].Ren A, Wang J. Methylenetetrahydrofolate reductase C677T polymorphism and the risk of unexplained recurrent pregnancy loss: a meta-analysis. Fertil Steril 2006;86:1716–22. [DOI] [PubMed] [Google Scholar]
- [73].Parveen F, Tuteja M, Agrawal S. Polymorphisms in MTHFR, MTHFD, and PAI-1 and recurrent miscarriage among North Indian women. Arch Gynecol Obstet 2013;288:1171–7. [DOI] [PubMed] [Google Scholar]
- [74].Cao Y, Xu J, Zhang Z, et al. Association study between methylenetetrahydrofolate reductase polymorphisms and unexplained recurrent pregnancy loss: a meta-analysis. Gene 2013;514:105–11. [DOI] [PubMed] [Google Scholar]
- [75].Wu X, Zhao L, Zhu H, et al. Association between the MTHFR C677T polymorphism and recurrent pregnancy loss: a meta-analysis. Genet Test Mol Biomarkers 2012;16:806–11. [DOI] [PubMed] [Google Scholar]
- [76].Rai V. Methylenetetrahydrofolate reductase gene A1298C polymorphism and susceptibility to recurrent pregnancy loss: a meta-analysis. Cell Mol Biol (Noisy-le-grand) 2014;60:27–34. [PubMed] [Google Scholar]
- [77].Thakkinstian A, McElduff P, D’Este C, et al. A method for meta-analysis of molecular association studies. Stat Med 2005;24:1291–306. [DOI] [PubMed] [Google Scholar]
- [78].Vickers A, Goyal N, Harland R, et al. Do certain countries produce only positive results?. a systematic review of controlled trials. Control Clin Trials 1998;19:159–66. [DOI] [PubMed] [Google Scholar]
- [79].Matsuura Y, Takazawa Wlch N, Sakai T, et al. Clinical trial registration, and publication in acupuncture studies: a systematic review. Integ Med Res 2020;9:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


