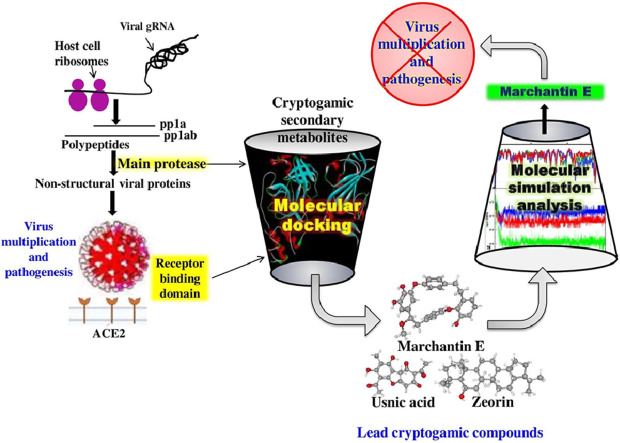
Abstract
The unprecedented quick spreading of newly emerged SARS-CoV-2, the virus responsible for causing COVID-19 has put the whole world in vast crisis. Several prophylactic interventions are being performed to discover the effective anti-COVID-19 agent. Thus, the present study aims to identify the cryptogamic secondary metabolites (CSMs) as potent inhibitors of two major targets of SARS-Cov2, namely 3-chymotrypsin-like protease (3CLpro) and receptor-binding domain (RBD) of spike glycoprotein (SGP), by implementing a computational approach. Molecular docking was carried out on Autodock 4.2 software with the 3CLpro (PDB ID:6LU7) and RBD of SGP (PDB ID:6W41) of the virus. Lopinavir and Arbidol were taken as positive controls to compare the efficacy of randomly selected 53 CSMs. The drug-likeness and pharmacokinetics properties of all metabolites were accessed to discern the anti-COVID 19 activity acting well at the physiological conditions. The docking results predicted that Marchantin E and Zeorin would potentially block the catalytic site of 3CLpro with the interaction energy values of -8.42 kcal/mol and -9.04 kcal/mol, respectively. In addition, Usnic acid revealed its ability to combat the interaction of RBD of SGP to angiotensin-converting enzyme-2 in docking analysis. To certify the potent metabolites for both targets of SARS-CoV-2, MD analysis was performed for 100 ns. The results confirmed that Marchantin E could inhibit SARS-CoV-2 3CLpro and RBD of SGP as well as reveals excellent pharmacokinetic properties. The present study suggests that the identified CSMs could be quickly positioned for further experimental validation to propose promising inhibitors of SARS-CoV-2.
Keywords: Molecular docking, Molecular dynamics, SARS-CoV-2, Main protease, Ribosomal binding domain, Cryptogamic secondary metabolites
Graphical abstract
1. Introduction
The virus, SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) discovered from the Wuhan city, Hebei province, China, in late 2019, causes a pneumonia-like syndrome known as COVID-19, currently bringing the global public health crisis and devastating the socioeconomic system of many countries [1], [2], [3]. Its transmission principle is human-to-human contact with infected body fluids such as cough droplets [3,4]. Therefore, the tally of COVID-19 diseased people is rising exponentially worldwide; more than 20 million infected people are recorded in more than 213 countries till 13th August 2020. SARS-CoV-2 is a positive-sense and single-stranded RNA virus that exhibits ~79% similarity to previously reported SARS-CoV, but the infectious ability of SARS-CoV-2 is much higher than SARS-CoV [5], [6], [7]. So, in March 2020, World Health Organization (WHO) affirmed that the COVID-19 is a pandemic.
As the scientific community has gotten acquainted with the pathogenesis of COVID-19, most of them have started racing to discover anti-COVID-19 agents and vaccines to curb the outbreak of SARS-CoV-2 [7]. Several drugs, such as ritonavir, lopinavir, hydroxychloroquine, chloroquine, azithromycin, and remdesivir are being repurposed to treat COVID-19 and their clinical trials in progress [8]. Nevertheless, we are still far away from a promising drug of COVID-19. Endless efforts, therefore, are needed to find effective and specific anti-COVID-19 drugs for fighting the deadly outbreak of SARS-CoV-2 across the globe.
Several macromolecules of SARS-CoV-2, including 3-chymotrypsin-like protease (3CLpro), endoribonuclease, RNA-dependent RNA polymerase, and 2-O methyltransferase, have been selected for designing and developing anti-COVID-19 drugs [7,9]. Among these, 3CLpro protein plays a key role in completing all stages of the virus life cycle, such as viral entry, viral protein maturation, and viral infection pathogenesis [10]. The 3CLpro inhibitors, therefore, can curb virus infection, and we speculated that 3CLpro would be an excellent therapeutic target protein to which novel anti-COVID-19 drugs would bind [11,12]. Only the homodimer form of 3CLpro has S1 pocket for substrate binding placed at a slit between domain I (residues 8–101) and II (residues 102–184) of its monomer. S1 pocket possesses the catalytic dyad (His41 and Cys145), essential residues for proteolysis activity [13]. Moreover, 3CLpro differs from human protease; its specific inhibitors will exhibit very low/negligible cytotoxicity. Another attractive target of SARS-CoV-2 is receptor-binding domain (RBD) of trimeric spike glycoprotein (SGP) of SARS-CoV-2 because this virus utilizes the SGPs to gain quick entry into bronchial epithelial cells by binding with a human receptor, Angiotensin-Converting Enzyme-II (ACE-II) on the host cells [9,14]. ACE-II receptor is located on many body organs, such as nasopharynx, lung, stomach, colon, bone marrow, liver, spleen, small intestine, lymph nodes, skin, thymus, kidney, brain, and oral and nasal mucosa [15,16]. After diagnosing the infected patient, it is essential to prevent the severity of virus infection on other organs. It is well reported that ACE-II receptor blockers exhibit adverse effect such as hyperkalaemia and angioedema [16]. Thus, blocking of the RBD of SGP, which interacts with amino acid residues of ACE-II could be an effective target domain for the treatment of COVID-19 infection and prevention of the risk of multi-organ failure.
Plant-derived bioactive secondary metabolites are of great interest in preventing all health-associated problems due to the absence of any adverse effects [17], [18], [19]. Cryptogams are groups of lower plants, including algae, mosses, lichens, and ferns that produce many bioactive secondary metabolites. Marchantins, a macrocyclic bisbibenzyls found in liverworts and usnic acid, a dibenzofuran derivative present in lichens have shown antiviral and other biological activities [20]. Thus, numerous phytochemicals belongs to different classes such as flavonoids, terpenoids, alkaloids and phenolics have been assessed for different targets of SARS-CoV2 using the computational tool because a large number of phytochemicals cannot be screened quickly using an in-vitro model [21,22]. Molecular docking and molecular simulations are not only cost effective but also time effective approach to identify a promising therapeutics agent of COVID-19.
We performed a high therapeutic screening of 53 cryptogamic secondary metabolites (CSMs) for two key targets of COVID-19; one is 3CLpro and the other is RBD of SGP. Docking studies revealed that marchantin E and zeorin would potentially block the catalytic site of 3CLpro, while usnic acid inhibited RBD of SGP. In addition, we calculated the pharmacokinetic properties of all phytochemicals to know their efficacy in vivo scale. MM-PBSA analysis was further carried out to refine and re-dock potential metabolites. The obtained results confirmed that marchantin E might inhibit SARS-CoV-2 3CLpro and RBD of SGP and reveal excellent pharmacokinetics. This effect of marchantin E needs to be validated through in vitro and in vivo experiments. To the best of our knowledge, this is the first report on the evaluation of CSMs for COVID-19.
2. Materials and methods
2.1. Ligand and protein structure preparation
The chemical structures of 53 CSMs were obtained from the chemical database of PubChem (https://pubchem.ncbi.nlm.nih.gov/) in the sdf format, which was transformed into pdb format for further ligand preparation using an online tool, SMILES converter (https://cactus.nci.nih.gov/translate/).
The three-dimensional (3D) structure of 3CLpro (PDB ID: 6LU7) and RBD of SGP (PDB ID: 6W41) were acquired from RCSB PDB (https://www.rcsb.org/). The retrieved 6LU7 was the homodimer of two chains (A and B), and both chains crystalized with a ligand, namely N3, and some water molecules, while 6W41 was bound with human antibody (chain H and L). The co-crystallized ligands and undesirable crystallographic water molecules from the 3D coordinate file of both receptors were excluded using UCSF Chimera ver 1.14, a freely available tool to visualize and analyze the molecular structure. Chain A of 6LU7 and chain C of 6W41 were selected for further analyses.
2.2. Molecular docking
Molecular docking analysis was accomplished on Autodock 4.2 tool to evaluate the target-ligand interaction energy and inhibition constant of CSMs. The structures of protein and ligand were optimized for docking analysis following Hassan et al. [23]. The autogrid was prepared on the entire macromolecule by screwing the grid size in x,y,z directions. The x, y, and z directions for 3CLpro were 12.723, 0.646 and 4.867, and RBD of SGP was −41.236, −45.024 and −17.888, respectively. Lamarckian genetic algorithm (GA 4.2) opted for the docking analysis with all default settings. The docked protein-ligand complex with the lowest energy cluster was selected from 10 hits for all. The binding free energy (ΔGbind) and inhibition constant (Ki) are expressed as kcal/mol and micromolar (µM), respectively. Discovery Studio Visualizer was used to assess the nature of interactions between ligand and protein.
2.3. Pharmacokinetics analysis
The absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis of all ligands was anticipated using the online tools; SwissADME (http://www.swissadme.ch/) and ProTox-II (http://tox.charite.de/protox_II/). The canonical SMILES of ligands were used as input for online servers.
2.4. Molecular dynamics (MD) simulation studies
The MD studies were performed using GROMACS version 5.1.1 (Groningen Machine for Chemical Simulations) by employing the GROMOS96 43a1 force field. A GROMOS96 compatible server, PRODRG, prepared ligand topology files. The protein-ligand complex files were prepared by combining the protein.gro and the ligand.gro files. The 3CLpro-Lopinavir, 3CLpro-Marchantin E, and 3CLpro-Zeorin complex consist of 3101, 3060, and 3043 atoms, respectively. While the RBD-Arbidol, RBD-Marchantin E, RBD-Zeorin and RBD-Usnic acid complex contains 2066, 2088, 2063, 2056 atoms, respectively. Periodic boundary conditions (PBC) were set at XYZ coordinates to ensure that the atoms stayed inside the simulation box, which was kept cubic in shape for both targets with at least 1 nm distance from the edge of the box. The X, Y, and Z coordinates for 3CLpro-Lopinavir/Marchantin E/Zeorin were 9.823 nm, 9.823 nm, and 9.823 nm, respectively, whereas these were 8.539 nm, 8.539 nm, and 8.539 nm for the RBD-Marchantin E/Zeorin/Usnic acid complex. The X, Y, and Z coordinates were 9.461, 9.461, and 9.461 nm for the RBD-Arbidol complex. Further, protein-ligand complexes' solvation was accomplished using simple point charge (SPC) waters in the simulation box. A total of 28,142, 20,665, and 20,682 water molecules were added to the simulation box of 3CLpro-Lopinavir, 3CLpro-Marchantin E, and 3CLpro-Zeorin, respectively. RBD-Arbidol, RBD-Marchantin E, RBD-Zeorin, and RBD-Usnic complex systems were solvated with 25,720, 19,669, 19,681, and 19,671 number of total water molecules. Four Na+in 3CLpro and two Cl−in RBD of SGP systems were added to neutralize the system. The energy of solvated system was minimized to remove stearic clashes between protein and water molecules using the steepest descent algorithm for 50,000 iteration steps and cut-off up to 1000 kJ/mol. Prior to MD simulations, two phases of equilibration were performed on an energy minimized solvated system by NVT (constant number of particles, volume, and temperature) and NPT (constant number of particles, pressure, and temperature) ensembles. Particle mesh Ewald (PME) was applied to treat the long-range coulombic interactions, with a Fourier grid spacing of 0.16 nm. All hydrogen bonds were constrained using the Linear Constraint Solver (LINCS) algorithm. MD simulation run was carried out for 100 ns with each step of 2 fs. Generated trajectories were further analyzed through a set of gromacs inbuilt tools such as g_rmsf, g_rmsf, g_hbond, g_covar and g_anaeig. The obtained results were analyzed using XMGRACE.
2.5. Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) calculation
MM-PBSA calculated the total interaction free energies of protein-ligand complexes prepared during simulations, a Gromacs supported approach. The program files were downloaded from the web address, http://rashmikumari.github.io/g_mmpbsa [24]. MM-PBSA methods were applied on snapshots of each simulated system which were extracted from the last 50 ns of the trajectory at the intervals of 10 ns to calculate various modules of interaction energy, including electrostatic interactions, Vander Waals interactions, non-polar solvation energy and polar solvation energy.
3. Results
3.1. Ligands and their druglikeness properties
Fifty-three CSMs belong to different structural skeletons, such as depsides, depsidones, benzofuran, xanthone, anthraquinone, and macrocyclic bisbibenzyl ether were deployed for this study (Table-S1). All metabolites were analyzed to assess if they followed the Lipinski rule of five and Veber's rule. These rules state that a ligand should not possess more than five hydrogen bond donors, logP (n-octanol and water partition coefficient), and not be more than ten hydrogen bond acceptors and rotatable bonds. The molecular weight of the ligand should not more than 500, and the total polar surface area must be less than or equal to 140 Ų [25]. These attributes of ligand structure govern their pharmacokinetics properties, such as ADMET. It is, therefore, necessary for an orally administrating drug to obey the Lipinski rule of five and Veber's rule. All selected CSMs followed both Lipinski rule and Veber's rule except fumaprotocetraric acid, galbinic acid, thamnolic acid, diploicin, and zeorin. These metabolites violated only one of the two rules. If a metabolite violates more than two drug-likeness rules, it is not considered a suitable candidate for oral administration. Lopinavir, selected as a positive control for anticatalytic activity of the 3CLpro violates two rules of drug-likeness: molecular weight and the other is t number of rotatable bonds. All other selected CSMs possessed good drug-likeness properties.
3.2. Molecular docking of CSMs to 3CLpro of SARS-CoV-2
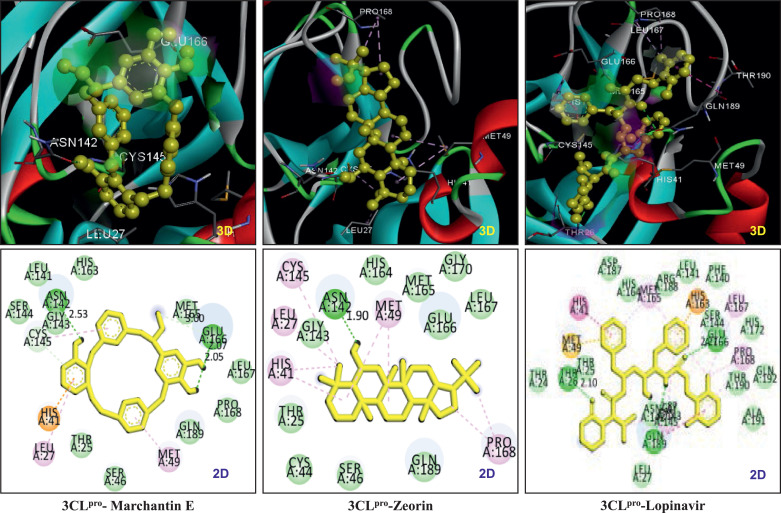
Molecular docking studies were performed with all selected CSMs and 3CLpro (6LU7) to find the potential inhibitory candidate of the catalytic site of the 3CLpro. Table-S2 represents the ΔGbind, Ki of all ligands, and the name of amino acid residues involved in stabilizing the protein-ligand complex. We found that out of a total, 34 compounds interacted with ΔGbind more than −7 kcal/mol, and 12 compounds showed ΔGbind between −7 to −8 kcal/mol. Only 4 compounds, zeorin, roccanin, marchantin E, and thiophannic acid, interacted with high affinity showing the ΔGbind below −8 kcal/mol. It was observed that roccanin and thiophannic acids were not found to be nestled to the S1 pocket of the 6LU7 during all ten hits of molecular docking. In contrast, zeorin and marchantin E firmly gripped the substrate-binding site of the 6LU7. The ΔGbind and Ki (IC50) of zeorin were −9.05 kcal/mol and 0.23 µM, respectively, which were almost similar to that of a known inhibitor of 3CLpro, lopinavir. The predicted ΔGbind and Ki (IC50) values of lopinavir were −9.16 kcal/mol and 0.697 µM, respectively. Zeorin interacted with Asn142 by hydrogen bonding and with Leu27, His41, Met49 Ser145, and Pro168 by pi-alkyl bonding. Ten other amino acid residues of the active site are involved in Van der Waal interaction to make a grip in the S1 pocket of the 6LU7 (Fig. 1 a). Marchantin E was second after zeorin in hitting the S1 pocket of 6LU7 with the implying least ΔGbind value of −8.4 Kcal/mol and IC50 value of 0.697 µM. The 6LU7-marchantin E complex was stabilized by three hydrogen bonds formed between hydroxyphenyl group of marchantin E and the amino acid residues, namely Asn142 and Glu166; other aromatic rings bound to Cys145 and Met49 by pi-alkyl interactions. The amino acid residues, Thr25, Leu27, His41, Ser46, Met49, Gly143, Ser144, His163, Met165, Leu167, Pro168, and Gln189 strengthened the 6LU7-marchantin E complex. Although, the 6LU7-lopinavir complex was observed with four hydrogen bonds, the length of hydrogen was higher than the 6LU7-Zeorine/marchantin E complex (Table 1 ). In conclusion, the binding mode of marchantin E and zeorine to 6LU7 indicates that both the metabolites can be effective against COVID-19 Fig. 2 .
Fig. 1.
3D and 2D images of interaction pattern of 3CLpro-Marchantin E, 3CLpro-Zeorin and 3CLpro-Lopinavir.
Table 1.
Binding interactions analysis of best docked CSMs with 3CLpro and RBD of SGP of SARS-CoV-2.
| Protein | Ligand | H-bond |
π-interactions |
||||
|---|---|---|---|---|---|---|---|
| Comp | Amino acid | Distance | Type | Amino acid | Distance (Å) | ||
| 3CLpro (PDBID:6LU7) | Marchantin E | C13-OH C1’-OH C6’-OH |
Asn142 Glu166 Glu166 |
2.53 2.05 2.07 |
π-Alkyl π-Cation π-Donor Hydrogen bond |
Leu27 Met49 Cys145 His41 Cys145 Glu166 |
6.92 4.23 7.68 5.31 6.28 5.66 |
| Zeorin | C6-OH | Asn142 | 1.90 | π-Alkyl | Leu27 Met49 His41 Cys145 Pro168 |
5.29 5.03, 6.44 4.55, 5.29 5.45 4.49, 3.59 |
|
| Lopinavir | NH5-CO10 CO29 NH27 CH17-OH18 |
Thr26 Glu166 Gln189 Gln189 |
4.04 3.39 4.39 5.49 |
π-Alkyl π-Sulfur π-Cation π-π T-shaped |
Cys145 Met165 Leu167 Pro168 Met49 His163 His41 Gln189 His163 |
6.26 4.72 6.44 5.17 4.26 6.44 4.26 4.49 6.44 |
|
| RBD of SGP (PDBID:6W41) |
Marchantin E | C1’-OH C6’-OH C1-O |
Leu492 Leu492 Phe490 |
5.69 5.84 4.45 |
π-Alkyl π-Anion π-π Stacked |
Tyr489 Leu455 Glu484 Phe486 |
4.45 5.72 5.00 4.97 |
| Zeorin | C6-OH C22-OH |
Gln493 Tyr449 |
4.26 5.03 |
π-Alkyl | Leu452 Leu455 Phe490 |
5.72 5.99 6.31 |
|
| Usnic acid | C11-O C7-OH C14-O |
Lys458 Gln474 Cys480 |
2.46 2.23 2.23 |
π-Anion π-Sigma |
Glu471 Tyr473 |
6.38 3.85 |
|
| Arbidol | C19-O-O C21 C28 C29 |
Phe490 Cys488 Glu484 Glu484 |
1.96 2.97 5.70 3.13 |
π-Alkyl π-Sulfur |
Leu455 Tyr489 Phe456 |
4.08 4.93 4.88 |
|
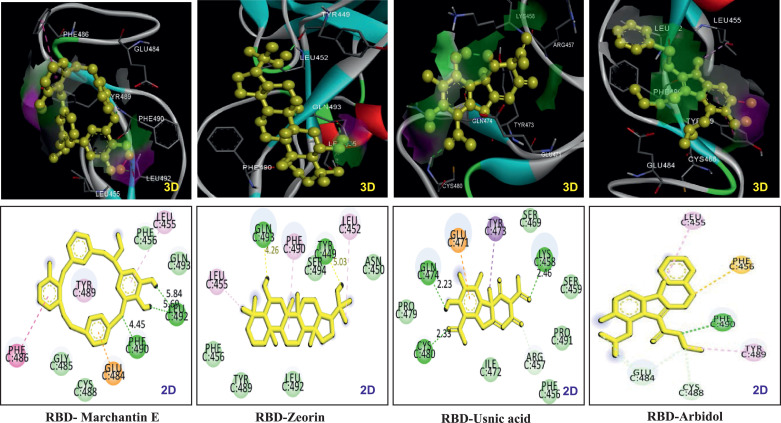
Fig. 2.
3D and 2D images of interaction pattern of RBD-Marchantin E, RBD-Zeorin, RBD-Usnic acid and RBD-Arbidol.
3.3. Molecular docking of CSMs to RBD of SGP of SARS-CoV2
The ΔGbind and Ki of all opted ligands for RBD of SGP of SARS-CoV2 revealed that eight compounds displayed the total ΔGbind values less than −7 Kcal/mol and Ki in between 1 and 7 µM (Table-S2). The binding affinity order of these eight compounds is granulatine = strepsilin (−7.04 Kcal/mol) < variolaric acid = scensidin (−7.17 Kcal/mol) < roccanin (−7.37 Kcal/mol) < lecanoric acid (−7.4 Kcal/mol) < usnic acid (−7.54 Kcal/mol) < marchantin E (−7.77 Kcal/mol) < zeorin (−7.8 Kcal/mol). An in-vitro study was concluded that arbidol, an anti-influenza agent, possesses efficiency to combat the SARS-CoV-2 virus entry. Therefore, arbidol was taken as comparable control for all selected ligands. Arbidol displayed ΔGbind value of −5.24 Kcal/mol and Ki value of 144.71 µM. We surmised that zeorin, marchantin E, and usnic acid might act as promising inhibitors to combat the COVID-19 infection based on binding affinity. Zeorin bound with an interaction energy value of −7.8 Kcal/mol and exhibited Ki value of 1.93 µM. Zeorin locked the ACE-II binding amino acid residues of RBD by forming two hydrogen bonds at amino acid residues, namely GLN493 and TYR449, and by interacting with other amino acids through Van der Waal bonding. The ΔGbind and Ki values for marchantin E were −7.77 Kcal/mol and 2.02 µM, respectively. These values were almost similar to zeorin and much higher than the positive control, arbidol. Marchantin E firmly interacted with SARS-CoV-2 RBD of SGP through three hydrogen bonds (two pi-pi stacking and one pi-anion) and four Van der Waal bonds (Table-1). Usnic acid was the third-best compound, encountered to RBD of SGP domain by forming three hydrogen bonds at the amino acid residues, Lys458, Gln474, and Cys480 (Table-1). These three complexes were further subjected to molecular dynamics studies to sight their stability.
3.4. Pharmacokinetics properties analysis
The determination of pharmacokinetic properties of drugs at the preliminary stages of drug discovery is essential to bring the drug up to clinical trial. The pharmacokinetic properties of any drug depend on its drug-likeness properties (Table-S1). Drugs showing the higher gastrointestinal absorption (GIA) property are considered the best candidate for oral administration. In our study, 38 compounds had high GIA efficiency. The topological polar surface area (TPSA) and molecular weight of a drug determine its permeability to the biological barrier. TPSA value of less than 140 Å2 is necessary for sufficient transmembrane penetration of a candidate, and a value less than 90 Å2 is required to cross the blood-brain barrier (BBB). Drugs that are developed for the central nervous system should come across the BBB [26]. If a syndrome is not associated with CNS, it is better to be a drug to exclude by BBB. Of fifty-three drug candidates, only five compounds, namely protolichesterinic acid, vulpinic acid, lichesterinic acid, olivetonide, and scensidin, were predicted to penetrate the BBB. The transportation of many drugs inside the cell is regulated by P-glycoprotein located in the plasma membrane, and therefore, the drugs possessing the ability to bind with P-glycoprotein cannot cross the plasma membrane [27]. Twelve compounds act as a substrate for p-glycoprotein (i.e. these compounds could not be easily reached up to the target). Except six compounds, such as fumaprotocetraric acid, thamnolic acid, gyrophoric acid, squamatic acid, alectorialic acid, and protocetraric acid showed good bioavailability score. In the metabolism study, granulatine, diffractaic acid, barbatic acid, usnic acid, galbinic acid, constictic acid, protocetraric acid, salazinic acid, marchantin E, zeorin, lecanoric acid, evernic acid, baeomycesic acid, ramalic acid, atranorin, gyrophoric acid, squamatic acid, ethyl orsellinate, alectorialic acid, and galapagin could be easily metabolized by cytochrome P450, a drug metabolizing enzyme. It could be due to the non-inhibitory effect of these compounds on the catalytic site of cytochrome P450 isoforms (i.e. CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4). The inhibition of these enzymes through therapeutic agents influences the drug clearance from the excretory system and resulted in the adverse effects of the drug. These 12 compounds, therefore, may not cause severe side effects. In the toxicity studies, scensidin, vulpinic acid, and thiophaninic acid were found to be hepatotoxic. It was also predicted that psoromic acid, galbinic acid, constictic acid, pannarin, diploicin, thiomelin, protocetraric acid, norstictic acid, menegazziaic acid, variolaric acid, physcion, salazinic acid, fumaprotocetraric acid, and stictic acid can be mutagenic. In addition, norstictic acid, stictic acid, menegazziaic acid, strepsilin, marchantin A, marchantin B, olivetonide, variolaric acid, and usnic acid could be carcinogenic in nature. The calculated lethal dose for the rat of all ligand except norlobaric acid and plagiochin A was in the range of 130 to 5000 mg/kg. The studied pharmacokinetic dimensions of all compounds are presented in Table-S4. After analyzing pharmacokinetic properties, we summarized that marchantin E possesses excellent pharmacokinetic properties compared to other compounds.
3.5. Molecular simulations study
Molecular simulations were employed to get insight into the rigidness of docked complexes for 10 ns using GROMACS 5.1 packages. The root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and number of hydrogen bond (H_bond) in docked complexes were analysed after simulations.
3.5.1. RMSD analysis
RMSD analysis assists in understanding the stability of docked protein-ligand complex during the simulation period. The calculated RMSD of the protein backbone and ligands during the 100 ns simulation is shown in Fig. 3 a and 3b. MPro-Zeorin complex showed deviations during the initial simulations. Still, over time, it was stable at around the RMSD value of 0.78 nm, whereas 3CLpro-Lopinavir complex was seemed to be steady at the RMSD value of 0.44 nm after the completion of the simulation period of 70 ns. 3CLpro-Marchantin E complex was found to deviate till 8 ns from 0.1 to 0.6 nm, and after that, it exhibited an almost constant RMSD value, which was 0.33 nm. The highest RMSD value was obtained for zeorin interactions with the 3CLpro in contrast to marchantin E and lopinavir. It indicates that zeorin showed less stability with 3CLpro, compared to marchantin E and lopinavir. When the backbone of the target was RBD of SGP, marchantin E exhibited a steady RMSD value of 0.79 nm after the 7 ns simulation periods. RBD-Zeorin complex showed variation till 4 ns; after that, it showed constant RMSD value at 1.1 nm. The sudden elevation in RMSD value was observed in the case of RBD-Usnic acid complex at 1 ns and was continue till 12 ns. After that, it was found in a range of 7.2 nm to 8.3 nm over the simulation periods. Similarly, RMSD value for RBD-arbidol complex was also deviated during the entire simulation period in the range of 1 to 1.8 nm and was too greater than marchantin E and zeorin. So, RBD-Marchantin E complex seems to be more stable as compared to RBD-Arabidol/Lopinavir.
Fig. 3.
RMSD graph of 3CLpro backbone (a) fixed with Marchantin E (blue line), Zeorin (green line) and Lopinavir (red line). RBD backbone of SGP (b) interacted with Marchantin E (blue line), Zeorin (green line), Usnic acid (purple line) and Arbidol (red line). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5.2. RMSF analysis
To evaluate the fluctuations of the targets in amino acid residues over the simulation period, the g_rmsf tool was applied (Fig. 4 a and 4b). The RMSF plots of all three 3CLpro-Zeorin/Marchantin E/Lopinavir complexes reflect the fluctuations at the amino acid residues of 220 to 287 because these residues were not involved in the interaction between target and ligands. All the above three complexes exhibit slight variation in the residues ranging from 1 to 180. Some deviations at the residues 97–102 and 182–197 were seen in the case of 3CLpro-Marchantin E and 3CLpro-Zeorin complex, respectively. In the 3CLpro-Lopinavir complex, the hiked fluctuation peak was observed at amino acids 47–49, suggesting that lopinavir interaction with Met49 is not durable. In the study of RBD-Marchantin E/Zeorin/Arbidol complexes, the RMSF plots displayed an almost similar trend of residues deviation profile indicating the equal binding affinity of marchantin E, zeorin, and arbidol with the 3CLpro. RMSF value for the residues of RBD-Usnic acid complex was found to be hiked in contrast to that of other complexes throughout the simulation periods. These results conclude that except for usnic acid, all studied ligands exhibited good interaction affinity with less flexibility to both targets.
Fig. 4.
RMSF graph of main protease (a) complexed with marchantin E (blue line), Zeorin (green line) and Lopinavir (red line). RBD of SGP (b) bound with Marchantin E (blue line), Zeorin (green line), Usnic acid (purple line) and Arbidol (red line). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5.3. Rg analysis
To evaluate the compactness of the molecular structure of the 3CLpro and RBD of SGP and ligands complex during 100 ns simulation, the “gyrate” tool was employed to generate the Rg plot. The compactness level of 3CLpro-MarchantinE/Zeorin/Lopinavir and RBD-MarchantinE/Zeorin/Usnic acid/Arbidol is displayed in Fig. 5 a and 5b, respectively. Lopinavir showed a constant Rg value at 2.14 nm after the 22 ns of simulation. In contrast, marchantin E exhibited strong fluctuation in Rg value till 40 ns, and after that, stable compactness with the Rg value of 2.12 nm was observed. Similarly, the 3CLpro-Zeorin complex was initially not found to be compact. The average Rg values for 3CLpro-Marchantin E and 3CLpro-Zeorin and 3CLpro-Lopinavir were 2.12, 2.11, and 2.14 nm, respectively. Similarly, the Rg values of RBD-Marchantin E, RBD-Zeorin, and RBD-Arbidol differed insignificantly, which were in the range of 1.80–1.81 nm, indicating that these all molecules fitted in the highly compact target domain. However, the RBD-Usnic acid complex attained slight loose compactness comparative to others; it exhibits the Rg value at 2.04 nm.
Fig. 5.
Rg graphs of 3CLpro (a) complexed with Marchantin E (blue line), Zeorin (green line) and Lopinavir (red line). RBD of SGP (b) bound with Marchantin E (blue line), Zeorin (green line), Usnic acid (purple line) and Arbidol (red line). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
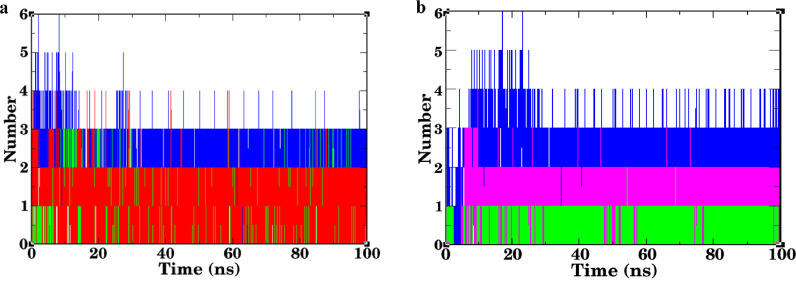
3.5.4. H_bond analysis
H_bond makes a significant contribution to stabilizing the protein-ligand complex. The g_hbond tool of GROMACS was used to determine the number of H_bond in the protein-ligand complex. The results revealed that the 3CLpro-Marchantin E complex interacted with three H_bonds in maximum hits during the MD simulation period, although the 5–6 H_bonds were observed at the initial stage of simulation (Fig. 6 a). Zeorin initially interacted with 3CLpro through a single H_bond, but over the 9 ns, the number of H_bond was increased, whereas strong H_bond interaction was seen in the case of the 3CLpro-Lopinavir complex. The average number of H_bond for the 3CLpro-Lopinavir complex was 2 (Fig. 6c). These data were consistent with the docking analysis of these three ligands. In the study of RBD-Marchantin E complex, it was observed that marchantin E was buried into RBD of SGP with 1–2 H_bonds till 5 ns over the simulation period. Thereafter, the number of H_bond was increased to an average of three (Fig. 6d). Similarly, in case of RBD-Usnic acid, no H_bond was detected till ~5 ns, after that, RBD-Usnic acid was found to be stabilized by 2 H_bonds in maximum hit (Fig. 6f). This suggests that the intensity of interaction was found to increase the time of the simulation. Poor H_bond interaction was seen in the case of the RBD-Zeorin complex over the simulation time (Fig. 6e). Likewise, the RBD-Arbidol complex formed 1–2 H_bonds during the complete simulation period (Fig. 6g). The obtained results concluded that the interaction of marchantin E and usnic acid with RBD was stronger than that of arbidol and zeorin. Likewise, the 3CLpro-Marchantin E complex seems to be more stable than 3CLpro-Lopinavir and 3CLpro-Zeorin complexes.
Fig. 6.
Plots of intramolecular H_bond between (a) 3CLpro-Marchantin E (blue)/3CLpro-Zeorin (green)/ 3CLpro-Lopinavir (red) and (b) RBD-Marchantin E (blue)/ RBD-Zeorin (green)/ RBD-Usnic acid (purple) and RBD-Arbidol (red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5.5. MM-PBSA analysis
The total ΔGbind was computed by the MM-PBSA approach after the MD simulation of the last 50 ns for all the complexes (Table 2 ). MM-PBSA approach aids in re-rank the docked complexes in terms of binding affinities. The computed total interaction energies were –46.11 ± 3.35 Kcal/mol, –17.98 ± 2.42 Kcal/mol and –62.69 ± 6.34 Kcal/mol, for the complex of 3CLpro-Marchantin E, 3CLpro-Zeorin, and 3CLpro-Lopinavir, respectively, suggesting that marchantin E interacted with SARS-CoV-2 3CLpro by utilizing less energy which was close to lopinavir. It is noticed that marchantin E exhibited a lower electrostatic energy value (−7.88 ± 2.39 Kcal/mol) compared to lopinavir (–3.22 ± 2.01 Kcal/mol). It is indicated that electrostatic interactions in the 3CLpro-Marchantin E complex were stronger than 3CLpro-Lopinavir. Zeorin was found to be superior compare to marchantin E and close to lopinavir in the molecular docking analysis. On the contrary, in the molecular simulation analysis, the obtained interaction energy was neither close to lopinavir nor marchantin E.
Table 2.
Contributions of various energy components to the ΔGbind (Kcal/mol), calculated by MM-PBSA method.
| Protein | Ligand | Electrostatic energy (kcal/mol) | Solvation energy (kcal /mol) | Van der Waal energy (kcal/mol) | SASA energy (kcal/mol) | Entropy (–TΔS) | Binding energy (kcal/mol) |
|---|---|---|---|---|---|---|---|
| 3CLpro (PDBID:6LU7) | Marchantin E | –7.88 ± 2.39 | 27.90 ± 3.16 | –61.68 ± 2.66 | –4.43 ± 0.22 | 35.76 | –46.11 ± 3.35 |
| Zeorin | –4.06 ± 2.86 | 12.4 ± 3.10 | –24.05 ± 2.98 | –2.33 ± 0.28 | 29.34 | –17.98 ± 2.42 | |
| Lopinavir | –3.22 ± 2.01 | 23.39 ± 3.87 | –76.80 ± 7.06 | –6.05 ± 0.58 | 59.6 | –62.69 ± 6.34 | |
| RBD of SGP (PDBID:6W41) | Marchantin E | –10.37 ± 3.16 | 20.03 ± 3.94 | –30.91 ± 3.18 | –2.83 ± 0.22 | 38.74 | –24.08 ± 3.05 |
| Zeorin | –0.17 ± 0.90 | 5.96 ± 1.79 | –30.11 ± 1.89 | –2.82 ± 0.21 | 38.74 | –27.24 ± 2.44 | |
| Usnic acid | –4.74 ± 2.24 | 12.84 ± 3.54 | –27.21 ± 4.92 | –2.34 ± 0.35 | 26.82 | –21.45 ± 3.89 | |
| Arbidol | –2.37 ± 1.21 | 5.70 ± 2.38 | –24.98 ± 2.83 | –2.37 ± 0.49 | 45.5 | –24.02 ± 2.69 |
SASA: solvent accessible surface area.
Each value represents the mean ± standard deviation calculated from 5 snapshots at 10 ns intervals of the last 50 ns MD run.
On the other hand, the RBD-Zeorin complex exhibited relatively lower ΔGbind (–24.08 ± 3.05 Kcal/mol) in contrast to RBD-Marchantin E (–27.24 ± 2.44 Kcal/mol), RBD-Usnic acid (–21.45 ± 3.89 Kcal/mol), RBD-Arbidol (–24.02 ± 2.69 Kcal/mol) (Table 2). It was observed that marchantin E and arbidol hit the RBD with the same interaction energy. Similar to the binding mode of marchantin E with the 3CLpro, the electrostatic interaction energy was found to be a major contributor to stabilizing the RBD-Marchantin E complex.
To get more insight the contribution of binding pocket residues to interaction of targets with ligands, the free energy decomposition per residue was employed (Fig. 7 a & 7b). The free energy decomposition analysis plot represents that the amino acid residues, Thr25, Leu27, His41, Met49, Leu141, Asn142, Ser144, Met165, Leu167, Pro168, Asp-187, and Gln189 energetically favor the binding stability of lopinavir to 3CLpro. In contrast, it was analysed that the amino acid residues, Thr25, Leu27, Met49, 50, Leu141, Asn142, Ser144, Cys145, Met165, Pro168, Asp187, and Gln189 were major residues interacting with the marchantinE and contributes to the total binding energy. However, Glu166 showed positive energy values suggesting that no favorable binding with marchantin E. Remarkably, it is noted that Met49 and Cys145 were found to be higher binding energy contributors with the value of −10.26 and −6.37 KJ/mol. In the binding with Zeorin with 3CLpro, no active site amino acid residues contribute to the stability of the 3CLpro Zeorin complex.
Fig. 7.
Per-residue energy decomposition analysis for 3CLpro (a) with Marchantin E (blue), Zeorin (green), and Lopinavir (red) and RBD (b) with Marchantin E (blue) Zeorin (green), RBD-Usnic acid, RBD-Arbidol. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In the analysis of RBD binding to arbidol, it was found that Glu484, Phe486, Gly485, Cys488, and Phe490 significantly contribute to the total energy. Similarly, the amino acids, Leu455, Tyr489, Phe490, Pro491, Leu492, Gln493, and Ser494 were considerably supported to marchantin E binding stability to RBD. However, the positive energy value of Glu484 indicates the unstable binding of marchantin E to Glu484. The binding of zeorin to RBD was mainly supported by the amino acids, Leu455, Phe456, Tyr473, Tyr489, Phe490, and Pro491, with the binding energy values of −5.33, −4.84, −8.26, −11.06, −1.95, and −6.33 KJ/mol, respectively. Per residue-free energy decomposition analysis of RBD-Usnic acid complex suggested Leu452, Leu455, Phe456, Glu484, Tyr489, Phe490, Pro491, Leu492, Gln493, and Ser494 energetically favor the binding of usnic acid.
4. Discussion
We performed in silico analysis of 53 CSMs to quickly determine the putative inhibitor of the prime targets, namely 3CLpro and RBD of SGP of SARS-CoV-2. This study presents the integration of two computational approaches, such as molecular docking and molecular dynamics, to strengthen the findings. The 3CLpro of coronaviruses facilitates the maturation of viral non-structural polyproteins (Nsp4-Nsp16), which regulate the viral replication, and viral invasion to host [28,29]. Therefore, we speculated that the 3CLpro would be a potential target for drug development. Earlier studies reported that the mutation at His41 and Cys145 amino acids completely diminished the catalytic activity of 3CLpro. In this study, the molecular docking analysis revealed that zeorin and marchantin E could efficiently block the catalytic dyad (His41 and Cys145) of 3CLpro with almost similar affinity to reference drug lopinavir. Lopinavir exhibits the binding energy value of −9.16 Kcal/mol, whereas zeorin and marchantin E show −9.04 Kcal/mol and −8.59 Kcal/mol, respectively. Our calculated binding energy value for lopinavir to 3CLpro, which is closed to the earlier reported value, −10.72 Kcal/mol. Marchantin E and lopinavir formed hydrogen bonds with Glu166 of 3CLpro, which aids in dimerizing two protomers and thereby maintains the correct orientation of S1 pocket of the substrate-binding site and functional enzyme [30]. i.e. Both compounds can alter the s1 pocket orientation, subsequently blocking the cleavage of the polypeptide necessary for the virus life cycle. Marchantin E, a macrocyclic bisbibenzyl ether, is commonly found in liverwort genera, Marchantia, and recently it was reported as a potential anti-influenza candidate [31,32]. Although we have also assessed other natural analogs of marchantin E, marchantin A and marchantin B. Marchantin E exhibited higher binding affinity with lower inhibition constant with 3CLpro than others. The structural-relationship analysis concluded that the hydroxyl group at C7′-position in marchantin E favors the binding to 3CLpro. Zeorin, a pentacyclic triterpenoid, is found in a few genera of lichen viz., Heterodermia, Rhinodina, and Diploschistes as well as of liverworts, such as Asterella, Reboulia, and Plagiochasma [33] . No earlier report on antiviral activity of zeorin is available. Likewise, Joshi et al., reported that two metabolites of lichens, namely, Calycin and Rhizocarpic acid display considerable affinity to 3CLpro. These metabolites also have not been earlier reported for antiviral activity [34].
SARS-CoV-2 takes first entry to host respiratory system by interacting with ACE-II of nasal epithelial through own RBD of SGP and then rapidly travels to lungs to cause the acute respiratory syndrome [16,35]. The crystal structure of the complex of human ACE-II and RBD of SGP revealed that the amino acid residues including Ser19, Gln24, Thr27, Phe28, Asp30, Lys31, His34, Glu35, Glu37, Asp38, Tyr41, Gln42, Leu45, Leu79, Met82, Tyr83, Asn330, Lys353, Gly354, Asp355, Arg357 and Arg393 of human ACE2 interacted with residues Gly446, Tyr449, Tyr453, Leu455, Phe456, Tyr473, Ala474, Ala475, Gly476, Asn487, Glu484 Phe486, Asn487, Tyr489, Phe490, Gln493, Gly496, Gln498, Asn501, Thr500, Gly502, Tyr505, respectively, of spike glycoprotein [2]. Strong viral binding with the human ACE-II is essential for virus entry.
To combat the severity of SARS-CoV2 viral infection, it is vital to obstruct the RBD interaction of SGP to ACE-II. The molecular docking analysis of 53 CSMs with RBD of SGP demonstrates that zeorin, marchantin E, and usnic acid have higher affinity to the amino acid residues of RBD of SGP, which involve in ACE-II binding as compared to reference anti-influenza drug, arbidol that combats the SARS-CoV-2 virus entry into host cells. Recently, arbidol has been approved for the clinical trial to be an anti-COVID-19 drug (https://clinicaltrials.gov/ct2/show/results/NCT01651663). We surmised that zeorin, marchantin E, and usnic acid may act as promising agents to combat the COVID-19 infection based on binding affinity. Usnic acid, a unique benzofuran, is major marker compound of genus Usnea and earlier reported for antiviral activity against influenza A virus subtype H1N1 [36].
As the molecular docking approach is applied to find the best dock pose of ligand based on binding energy, therefore, molecular simulation approach was performed to validate the docking and repositioning the best ligand by calculating the various measurements such H_bond, Rg, RMSD, and RMSF. RMSD analysis tells about the stability of docked protein-ligand complex during the simulation period. A higher RMSD value represents the complex's less stability, while a lower RMSD value represents a highly stable complex. Lower RMSD value of 3CLpro-Marchantin E and 3CLpro-Lopinavir complexes was observed compare to 3CLpro-zeorin, suggesting that 3CLpro-Marchantin E and 3CLpro-Lopinavir complexes are more stable compared to the 3CLpro-Zeorin complex. Less stability of the 3CLpro-Zeorin complex might be due to weak interaction of zeorin to 3CLpro. In docking analysis, zeorin stabilized by only one hydrogen bond, whereas Lopinavir and Marchantin E form more than two hydrogen bonds. The RMSD analysis of RBD-Usnic acid, RBD-Marchantin E, RBD-Zeorin, and RBD-Arabidol Complexes is studied was observed that RBD-Marchantin E complex deviates less (0.3 nm) as compared to others, suggesting that RBD-Marchantin E complex is highly stable. Like the main protease, the RBD-Marchantin E complex stabilizes by a higher H-bond and shows strong interaction throughout the simulation period. Although usnic acid also forms a similar number of hydrogen bonds to stabilize its complex, which exhibits a very high RMSD value after 1 ns, representing the ligand selected in docking analysis was not in good confirmation. Additionally, the characteristic of usnic acid is intramolecular H-bond formation, which might be responsible for unstable interaction during the entire simulation period [37].
Also, RMSF analysis gives an idea about the flexibility of amino acid residues in docked protein-ligand complex [38]. Lower RMSF value constrains residues' movement during the simulation period and specifies the stable interaction between protein and ligands [39]. The 3CLpro-Marchantin E complex displays the negligible loop change in RMSF values at the amino acid residues ranging from 1 to180 in contrast to 3CLpro-Lopinavir and 3CLpro-Zeorin, signifying that higher binding affinity of marchantin E to 3CLpro. This data exhibits consistency with RMSD analysis. Strikingly, a similar trend in residues deviation profile has been obtained for RBD-Marchantin E/Zeorin/Arbidol complexes except for RBD-Usnic acid complex, depicting that all complexes are having good interaction affinity with less flexibility to the virus RBD of SGP. When bound with usnic acid, higher fluctuation in RBD amino acids justified its unusual interaction and less stability of its complex as predicted by higher RMSD value.
Rg defines the compactness level of the docked protein-ligand complex in all dimensions [40]. Lower the value of Rg epitomizes the highly compact protein-ligand complex. The average Rg values for 3CLpro-Marchantin E, 3CLpro-Lopinavir, and 3CLpro-Zeorin complexes were almost similar for all. The 3CLpro-Lopinavir complex showed consistency with the earlier predicted study, in which the complex exhibited Rg value of ~2.1 nm [41]. Likewise, no significant change was observed in the Rg values of RBD-Marchantin E, RBD-Zeorin, and RBD-Arbidol complexes, indicating that these ligands fitted in the highly compact protein. However, the RBD-Usnic acid complex displays a higher Rg value, again indicating unfavorable interaction. Conclusively, targeted proteins exhibit the same behavior with different ligands except for usnic acid.
H_bond plays a crucial role in stabilizing the protein-ligand complex [28]. The average number of H_bond in 3CLpro-Marchantin E, 3CLpro-Zeorin, and 3CLpro-lopinavir complexes were 3, 1, and 2, respectively. These data were consistent with the docking analysis of these three ligands. Likewise, the 3CLpro-Marchantin E complex was more stable than 3CLpro-Lopinavir and 3CLpro-Zeorin complexes. It was also observed that RBD-Marchantin E complexes were strongly stabilized by hydrogen binding throughout the simulations as compared to arbidol, zeorin, and usnic acid.
When a ligand dock to a protein, various energy components, including electrostatic, Van der Waals, and Solvation energy, participate in their interactions and contribute to ligand's total binding affinity [23]. MM-PBSA analysis was executed to analyze the total binding energy of ligands. We have demonstrated that marchantin E interacts with the main protease by utilizing less energy close to lopinavir. Zeorin was more superior to marchantin E, and was close to lopinavir in the molecular docking analysis. On the contrary, in the molecular simulation analysis, its obtained interaction energy was neither close to lopinavir nor marchantin E. It might be due to zeorin's weak interaction to 3CLpro during the simulation period and the selection of less favorable confirmation of zeorin in molecular docking analysis. Interestingly, zeorin exhibits a higher binding affinity to RBD, whereas marchantin E and arbidol hit the RBD with the same interactions energy in molecular simulation analysis. Although Zeorin was stabilized by only one hydrogen bond during the simulation period, the higher contribution of hydrophobic interactions energy to total interaction energy brings it's in the first position. Taken together, marchantin E showed an excellent binding affinity with both targets of SARS-CoV-2 viz., 3CLpro, and RBD of SGP, while zeorin displayed only good affinity to the RBD of SGP. The study suggests that marchantin E of liverwort genera Marchantia may be a promising drug candidate to manage the COVID-19 pandemic and its consequences.
5. Conclusions
In the present study, 53 CSMs were screened for two principal targets of SARS-CoV-2, namely 3CLpro and RBD of SGP, using inexpensive and highly recommended computational algorithms during this crisis. Our molecular docking and molecular dynamics analysis conclude that Marchantin E might act as a good blocker of 3CLpro and RBD of SGP. At the same time, Zeorin would be an active inhibitor of RBD of SGP. Therefore, further in vitro and in vivo studies are required to propose these CSMs as potential drug candidates for controlling the COVID-19 pandemic.
Credit to authors
B.N.S. and S.K.B. conceived and designed the experiments. P. performed most of the experiments. P. and B.N.S. analysed the experimental data. P., B.N.S., S.K.B., A.K.A., and T.S.R. wrote the manuscript.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Dr. Brahma Nand Singh reports financial support was provided by DBT and CSIR and DBT, India. Dr. B.N. Singh reports a relationship with CSIR-NBRI, Lucknow that includes: employment.
Acknowledgement
The authors would like to acknowledge the Council of Science and Industrial Research (CSIR) and Department of Biotechnology (DBT; GAP-3439) India, to provide financial support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.130506.
Appendix. Supplementary materials
References
- 1.Lee P.I., Hsueh P.R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020;53:365–367. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/nejmoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Q. Phan, L.T., Nguyen, T.V., Luong, Q.C., Nguyen, T.V., Nguyen, H.T., Le, H.Q., Nguyen, T.T., Cao, T.M. and Pham, Importation and human-to-human transmission of a novel coronavirus in Vietnam, (2020) 2020–2022. [DOI] [PMC free article] [PubMed]
- 5.Lai C., Shih T., Ko W., Tang H., Hsueh P. International journal of antimicrobial agents severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui J. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 8.R. Wu, L. Wang, H.D. Kuo, A. Shannar, R. Peter, P.J. Chou, S. Li, R. Hudlikar, X. Liu, Z. Liu, G.J. Poiani, L. Amorosa, L. Brunetti, An update on current therapeutic drugs treating COVID-19, (2020) 56–70. [DOI] [PMC free article] [PubMed]
- 9.Pandey A., Nitin A., Basavaraj A., Mutalik S.P., Prassl R. Potential therapeutic targets for combating SARS-CoV-2 : drug repurposing, clinical trials and recent advancements. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan B., Huynh T., Cheng X., Lan G., Wang H.R. Targeting proteases for treating COVID-19. J. Proteome Res. 2020;19:4316–4326. doi: 10.1021/acs.jproteome.0c00430. [DOI] [PubMed] [Google Scholar]
- 11.P. Inhibitor, M. Hoffmann, H. Kleine-weber, S. Schroeder, M.A. Mu, C. Drosten, S. Po, M. Hoffmann, H. Kleine-weber, S. Schroeder, N. Kru, SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven article SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, (2020) 271–280. https://doi.org/10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed]
- 12.Huynh T., Wang H., Luan B. In Silico exploration of the molecular mechanism of clinically oriented drugs for possibly inhibiting SARS-CoV-2’s main protease. J. Phys. Chem. Lett. 2020;11:4413–4420. doi: 10.1021/acs.jpclett.0c00994. [DOI] [PubMed] [Google Scholar]
- 13.L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a -ketoamide inhibitors, 412 (2020) 409–412. [DOI] [PMC free article] [PubMed]
- 14.D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C. Hsieh, O. Abiona, B.S. Graham, J.S. Mclellan, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, 1263 (2020) 1260–1263. [DOI] [PMC free article] [PubMed]
- 15.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by. Nature. 2020;581 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh B.N., Shankar S., Srivastava R.K. Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms, perspectives and clinical applications Catechin backbone. Biochem. Pharmacol. 2011;82:1807–1821. doi: 10.1016/j.bcp.2011.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh B.N., Singh B.R., Singh R.L., Prakash D., Sarma B.K., Singh H.B. Antioxidant and anti-quorum sensing activities of green pod of Acacia nilotica L. Food Chem. Toxicol. 2009;47:778–786. doi: 10.1016/j.fct.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Singh B.N., Singh B.R., Singh R.L., Prakash D., Dhakarey R., Upadhyay G., Singh H.B. Oxidative DNA damage protective activity, antioxidant and anti-quorum sensing potentials of Moringa oleifera. Food Chem. Toxicol. 2009;47:1109–1116. doi: 10.1016/j.fct.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 20.D.H. Cornelissen JHC, Lang S.I., Soudzilovskaia N.A., Comparative cryptogam ecology : a review of bryophyte and lichen traits that drive biogeochemistry, (2007) 987–1001. https://doi.org/10.1093/aob/mcm030. [DOI] [PMC free article] [PubMed]
- 21.Yu R., Chen L., Lan R., Shen R., Li P. International Journal of Antimicrobial Agents Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6 doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan M., Shahzadi S., Seo S.Y., Alashwal H., Zaki N., Moustafa A.A. Molecular docking and dynamic simulation of AZD3293 and solanezumab effects against BACE1 to treat alzheimer's disease. Front. Comput. Neurosci. 2018;12:1–11. doi: 10.3389/fncom.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumari R., Kumar R., Lynn A. G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 25.Khanna V., Ranganathan S. Physiochemical property space distribution among human metabolites, drugs and toxins. BMC Bioinformatics. 2009;10:1–18. doi: 10.1186/1471-2105-10-S15-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shityakov S., Neuhaus W., Dandekar T., Förster C. Analysing molecular polar surface descriptors to predict blood-brain barrier permeation. Int. J. Comput. Biol. Drug Des. 2013;6:146–156. doi: 10.1504/IJCBDD.2013.052195. [DOI] [PubMed] [Google Scholar]
- 27.Amin M.L. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights. 2013;2013:27–34. doi: 10.4137/DTI.S12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. Dai, W., Zhang, B., Jiang, X.M., Su, H., Li, J., Zhao, Y., Xie, X., Jin, Z., Peng, J., Liu, F. Li, C. Dai, W., Zhang, B., Jiang, X.M., Su, H., Li, J., Zhao, Y., Xie, X., Jin, Z., Peng, J., Liu, F. Li, Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease, 1335 (2020) 1331–1335. [DOI] [PMC free article] [PubMed]
- 29.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Rolf Hilgenfeld M.D. Center for Structural and Cell Biology in Medicine, University of, BioRxiv; 2020. Substrate Specificity Profiling of SARS-CoV-2 M Pro Protease Provides Basis For anti-COVID-19 Drug Design Institute of Biochemistry. March 8. [Google Scholar]
- 30.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 31.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020 doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwai Y., Murakami K., Gomi Y., Hashimoto T., Asakawa Y., Okuno Y., Ishikawa T., Hatakeyama D., Echigo N., Kuzuhara T. Anti-influenza activity of marchantins, macrocyclic bisbibenzyls contained in liverworts. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0019825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarasova V.N., Obabko R.P., Himelbrant D.E., Boychuk M.A., Stepanchikova I.S., Borovichev E.A. Diversity and distribution of epiphytic lichens and bryophytes on aspen (Populus tremula) in the middle boreal forests of Republic of Karelia (Russia) Folia Cryptogam. Est. 2017;54:125–142. doi: 10.12697/fce.2017.54.16. [DOI] [Google Scholar]
- 34.T. Joshi, P. Sharma, T. Joshi, H. Pundir, S. Mathpal, S. Chandra, Structure-Based Screening of Novel Lichen Compounds for SARS Coronavirus Main protease (Mpro) and Angiotensin-Converting Enzyme 2 (ACE2) inhibitory potentials as multi-target inhibitors of COVID-19, 2 (2020). doi: 10.21203/rs.3.rs-26207/v1. [DOI]
- 35.I. Hamming, W. Timens, M.L.C. Bulthuis, A.T. Lely, G.J. Navis, H. Van Goor, Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus . a first step in understanding SARS pathogenesis, (2004) 631–637. https://doi.org/10.1002/path.1570. [DOI] [PMC free article] [PubMed]
- 36.Sokolov D.N., Zarubaev V.V., Shtro A.A., Polovinka M.P., Luzina O.A., Komarova N.I., Salakhutdinov N.F., Kiselev O.I. Anti-viral activity of (−)-and (+)-usnic acids and their derivatives against influenza virus A (H1N1) 2009. Bioorg. Med. Chem. Lett. 2012;22:7060–7064. doi: 10.1016/j.bmcl.2012.09.084. [DOI] [PubMed] [Google Scholar]
- 37.Green T.K., Lane C.A. Usnic acid and the intramolecular hydrogen bond. A computational experiment for the organic laboratory. J. Chem. Educ. 2006;83:1046–1048. doi: 10.1021/ed083p1046. [DOI] [Google Scholar]
- 38.Khan R.J., Jha R.K., Amera G.M., Jain M., Singh E., Pathak A., Singh R.P., Muthukumaran J., Singh A.K. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J. Biomol. Struct. Dyn. 2020;0:1–14. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan M.T., Ali A., Wang Q., Irfan M., Khan A., Zeb M.T., Zhang Y.J., Chinnasamy S., Wei D.Q. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2—A molecular dynamic study. J. Biomol. Struct. Dyn. 2020;0:1–11. doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peele K.A., Potla Durthi C., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., Indira M., Reddy A.R., Venkateswarulu T.C. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Informatics Med. Unlocked. 2020;19 doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2020;0:1–12. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.