Abstract
In medical image segmentation tasks, deep learning-based models usually require densely and precisely annotated datasets to train, which are time-consuming and expensive to prepare. One possible solution is to train with the mixed-supervised dataset, where only a part of data is densely annotated with segmentation map and the rest is annotated with some weak form, such as bounding box. In this paper, we propose a novel network architecture called Mixed-Supervised Dual-Network (MSDN), which consists of two separate networks for the segmentation and detection tasks respectively, and a series of connection modules between the layers of the two networks. These connection modules are used to extract and transfer useful information from the detection task to help the segmentation task. We exploit a variant of a recently designed technique called ‘Squeeze and Excitation’ in the connection module to boost the information transfer between the two tasks. Compared with existing model with shared backbone and multiple branches, our model has flexible and trainable feature sharing fashion and thus is more effective and stable. We conduct experiments on 4 medical image segmentation datasets, and experiment results show that the proposed MSDN model outperforms multiple baselines.
Keywords: Mixed-supervised, Dual-network, Squeeze-and-excitation, Medical image segmentation
1. Introduction
Deep learning-based methods have achieved remarkable success in several important computer vision tasks, such as image classification (Krizhevsky et al., 2012; Simonyan and Zisserman, 2014; He et al., 2016; Huang et al., 2017), object detection (Ren et al., 2015; Redmon et al., 2016; He et al., 2017; Lin et al., 2017), and image segmentation (Long et al., 2015; Chen et al., 2017; Pohlen et al., 2017), especially for segmentation of anatomical structures from medical images (Ronneberger et al., 2015; Milletari et al., 2016; Litjens et al., 2017). Recently, such methods have been extensively applied to many medical image tasks, including segmentation of brain tumors(Shen et al., 2017), lung nodules (Jiang et al., 2018), and other structures (Chen et al., 2016; Fan et al., 2019). With the power of automatic representation learning and end-to-end training, deep learning methods greatly reduce the difficulty of feature extraction and outperforms the methods of hand-crafted feature engineering. However, all these methods require a large amount of training data with dense and accurate annotations for training, which is very expensive, time-consuming and laborious to prepare. In the area of medical image analysis, preparing annotations of training data usually requires not only strong medical background knowledge but also rich experience of diagnosis, thus the cost of preparing sufficient training data becomes even higher.
Therefore, weakly-supervised segmentation training with insufficient labels, e.g. image tags (Wang et al., 2018), points (Bearman et al., 2016) or bounding boxes (Rajchl et al., 2016) has attracted a lot of attention recently. Such techniques have also been applied to medical imaging. For example, Cai et al. (Cai et al., 2018) propose to train a 3D lesion volume segmentation model using annotated 2D slices in an iteratively slice-wise propagated fashion. Kervadec et al. (Kervadec et al., 2019) design a differentiable term to enforce inequality constraints directly in the loss function so that the weakly-labeled data can be leveraged. Although these works have made some progress, there still exists some gap in performance compared to the fully-supervised trained models. This makes it difficult to apply to medical scenario, where accurate segmentation is usually required for surgical planning, pathological analysis or disease diagnosis. Further, the training of weakly-supervised models becomes more complex as these models are usually trained in multi-step iteration mode between model learning and full label generation (Wang et al., 2018; Rajchl et al., 2016; Cai et al., 2018) or with additional constraint term in the loss function (Kervadec et al., 2019).
Another promising solution is training with mixed-supervised datasets, where only a part of data is annotated with dense segmentation map and the rest is labeled with some weak mode (for example, the bounding box which is considered in this paper). Typical methods handle such kind of datasets with multi-branch networks in a multi-task learning setting (Shah et al., 2018; Mlynarski et al., 2018; Bhalgat et al., 2018), where basic feature extractor (backbone) is shared and different branches are used for data with different kinds of annotation. Bhalgat et al. (Bhalgat et al., 2018) focus on the optimal balance between the number of annotations needed for different supervision types and presents a budget-based cost-minimization framework in a mixed-supervision setting. The insight of these models is that low level features are shared by different tasks while high level features differ. However, low-level features of different tasks may not be exactly identical, so simply sharing low-level features with joint multi-task training may suffer from limited or even inverse training effects when exploiting the weakly-labeled data.
In this paper, we propose a novel network architecture with dual-network for mixed-supervised medical image segmentation. We consider the bounding boxes as weak annotation, and take the segmentation task as the target task, which is augmented with object detection task (the auxiliary task). Different from the multi-branch network with shared backbone (Shah et al., 2018; Mlynarski et al., 2018), our new architecture contains two separate networks for each task so the features of the two tasks are decoupled and relatively independent. The two networks are linked by a series of connection modules that exist between the corresponding layers, which take the convolution features of detection network as input, squeeze them to extract useful information and transfer it to the segmentation network to help the training of the segmentation task. We exploit a variant of a recently designed feature attention technique called ‘Squeeze and Excitation’ (Hu et al., 2018; Roy et al., 2018, 2019) in the connection modules to boost the information transfer. Compared with existing model with shared backbone and multiple branches, our model has flexible and trainable feature sharing fashion and thus is more effective and stable. The proposed model is used for mixed-supervised segmentation, and contains two networks for each task, so we name it as Mixed-Supervised Dual-Network (MSDN). We perform evaluation on 4 medical image segmentation datasets (3 CT datasets and 1 RGB dataset): lung nodule, cochlea of inner ear, kidney and medical instrument, all of which are of great clinical significance for image-guided therapy and surgery. Experimental results show that our model outperforms multiple baselines in all the datasets.
This work has already been presented at the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2019) (Wang et al., 2019). We extend the conference paper as follows: First, we conduct experiments on two more public datasets and compare with more baseline models to further validate the effectiveness of the proposed method. Second, we include greater discussion about the settings of our method, such as the number of layers that contain SE block and the type of SE block. Further, we provide more related works and details to help the readers better understand our method.
The rest of the paper is organized as follows. In Section 2 we provide some background knowledge related to this paper, including the segmentation and detection model exploited in this paper and principle of SE module. In Section 3 we introduce in detail the architecture and training procedure of the proposed MSDN model. In Section 4 we detail the experimental settings, results and discussions about the model settings. Section 5 contains the conclusion and future works from this study.
2. Background
2.1. Deep learning-based segmentation
Recent deep learning-based segmentation methods exploit some variant of fully convolutional network (FCN) (Long et al., 2015) to predict the class labels of all the pixels in an image in parallel. Due to the pooling layers and upsampling operation, spatial information may be lost during prediction thus there exists some inaccuracy in the predicted segmentation map, especially in the sharp region such as boundaries. So the skip architecture is proposed in FCN (Long et al., 2015) to tackle this problem. Following this thought, Ronneberger et al. propose a very standard and popular framework for medical image segmentation called U-Net (Ronneberger et al., 2015), which contains symmetric Encoder and Decoder. The features of each Encoder layer are skip-connected to the corresponding Decoder layer to recover spatial information lost. Chen et al. (Chen et al., 2016) propose DCAN, which trains a FCN model jointly with additional contour supervision to further recover the inaccuracy in the contour region. Pohlen et al. (Pohlen et al., 2017) design a more elaborate segmentation model called FRRN based on ResNet to improve the effectiveness of skip connection. Since in this paper, we mainly focus on the model training with mixed-supervised dataset and compare it with fully-supervised manner and other multi-task learning method, we choose the standard U-Net as our basic segmentation model because it is very neat and easy to implement, but our method can be applied to any hierarchical network structure.
2.2. Deep learning-based detection
Current deep learning-based object detection models belong to either two-stage or one-stage approach. Typical two-stage object detectors include Faster RCNN (Ren et al., 2015) and Mask RCNN (He et al., 2017). They both include region proposal network (RPN) to automatically generate a group of candidate region proposals that may contain some objects as the first stage, followed by a classification head to recognize the object type for each proposed region as the second stage. Mask RCNN (He et al., 2017) exploits an additional object segmntation branch in parallel with the recognition branch. For one-stage approach, YOLO (Redmon et al., 2016) and RetinaNet (Lin et al., 2017) are two representative works. They propose to predict the bounding box and object type simultaneously in a single stage, so the forward procedure is much faster. However, due to the great imbalance between the positive (containing object) and negative (background) regions, direct training may force the model to focus too much on the background, so the detection accuracy may be lower than that of two-stage method. Lin et al. (Lin et al., 2017) propose the focal loss to tackle the imbalance problem to acquire both fast and accurate detection model. In this paper, we consider detection as the auxiliary task to assist the learning of segmentation task, so high accuracy in detection is not imperative. We follow the idea of one-stage RetinaNet and build a custom detection network in our own way, which borrows the idea of Anchor (Ren et al., 2015) and uses U-Net as backbone. Focal loss is also used to fix the imbalance between the positive and negative regions.
2.3. Squeeze and excitation (SE)
‘Squeeze-and-Excitation’ (SE) (Hu et al., 2018) is a recently-proposed mechanism to enhance the representational ability of networks, which exists between two CNN layers. The SE module first squeezes the output feature of previous layer by global pooling to capture the channel aggregation information. Then this vector is passed to the gating path to get the representation of channel-wise dependencies, which is used to rescale the feature map to indicate the importance of different channels, see Fig. 1(a). Roy et al. (Roy et al., 2018a) term the SE module in (Hu et al., 2018) as Spatial Squeeze and Channel Excitation (cSE) and design a different version called Channel Squeeze and Spatial Excitation (sSE). The sSE module squeezes the feature map in channel dimension by 1 × 1 convolution to preserve more spatial information, thus is more effective for image segmentation task, see Fig. 1(b). In this paper, we name these two SE modules as unary SE, because the squeeze and excitation are conducted on the same feature. Roy et al. (Roy et al., 2019) further propose a binary SE block where one feature map is squeezed and used to recalibrate another, and apply it to the few-shot segmentation problem. Since our work is more related to the binary SE module, we will introduce it in a more detailed way as follows.
Fig. 1.
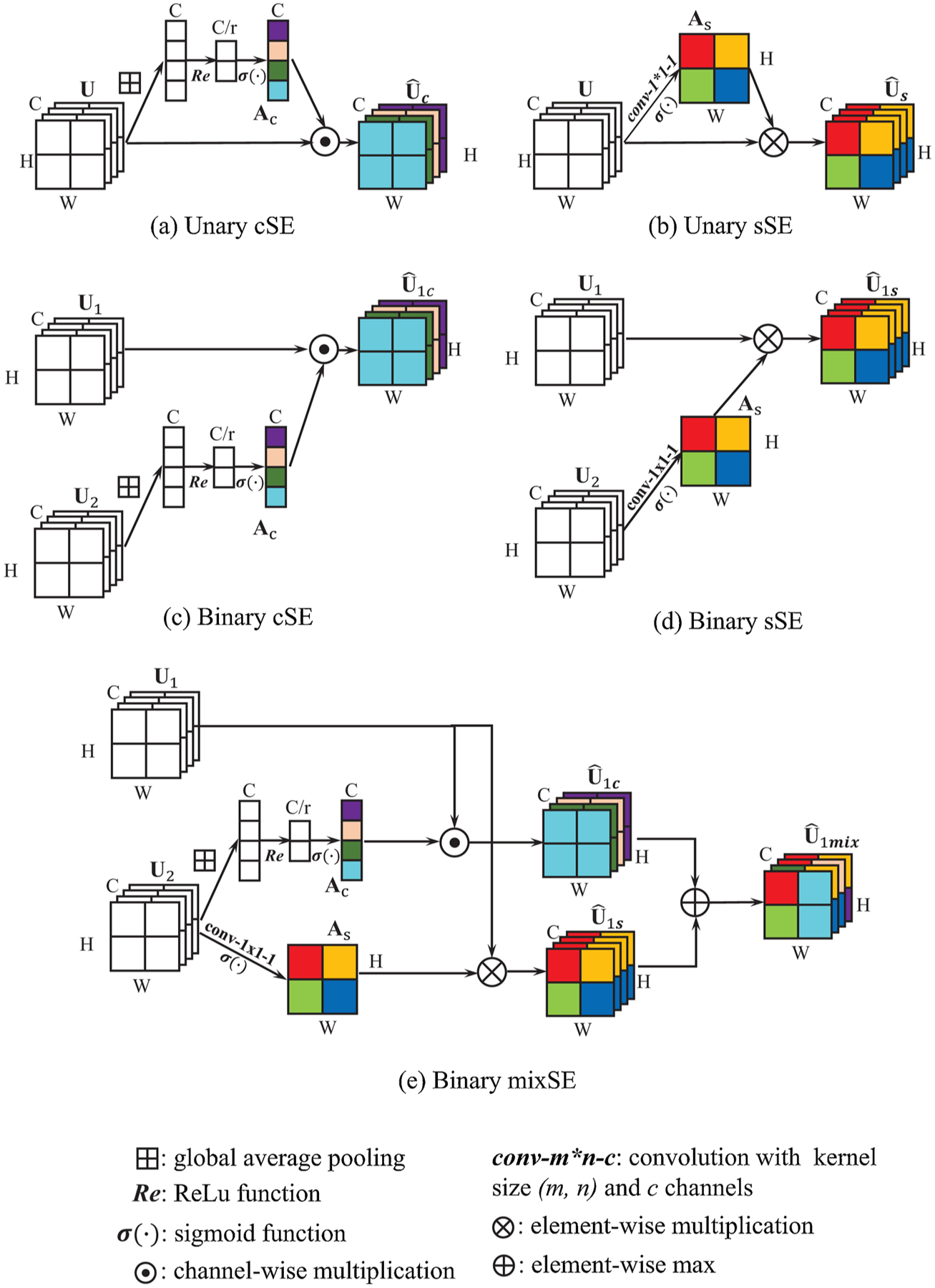
Illustration of different forms of Squeeze and Excitation (SE) module. The main difference between the Unary and Binary form is recalibration weight comes from the input feature itself or another feature.
We consider the convolution feature maps and from two convolution layers as the input of the binary SE module and denotes its ith channel of feature n. For the Binary cSE, feature U2 is first squeezed to by global average pooling. Then the recalibration vector Ac is calculated by
| (1) |
where and are the weights of the two fully-connected (fc) layers. r is the reduction ratio. σ and Re denote the sigmoid and ReLU function. The recalibrated feature is calculated by
| (2) |
For binary sSE, feature U2 is squeezed by 1 × 1 convolution with kernel weight . The squeezed feature is then passed through sigmoid function to derive the recalibration weight ,
| (3) |
Then each feature channel of U1 is multiplied element-wise by As to get the spatially recalibrated feature as output
| (4) |
Here ∘ denotes the Hadamard product, * denotes the convolution operation and σ denotes the sigmoid function. For binary mixSE, both SE modules mentioned above are performed and the two recalibrated features are fused. Here we choose the max fusion as shown in (Roy et al., 2018) that results in the best performance. Thus, the mixed recalibrated feature is
| (5) |
In this paper, we propose to use the binary SE module as the connection between our dual-network architecture for information extraction and transfer. All the three forms of Binary SE module are evaluated and compared. Detailed experiment results can be found in Section 4.
3. Mixed-supervised dual-network
3.1. Backbone
The MSDN is made up of two separate subnetworks for the segmentation and detection tasks respectively (as shown in Fig. 2), both of which are based on the U-Net and contain 9 convolution stages, with 4 stages in the Encoder, 4 in the Decoder and 1 in the Bottleneck. Each convolution stage contains 2 dilated-convolution layers with kernel size 3 × 3, followed by batch normalization (BN) (Ioffe and Szegedy, 2015) and rectified linear unit (ReLU). The output feature of each stage in the Encoder is skip-connected to the corresponding Decoder stage to recover spatial information caused by maxpooling. Dilation factors in the 9 feature stages are set as [1,2,2,2,4,2,2,2,1] respectively to enlarge the receptive field. The stride and padding are chosen accordingly to make the size of the output feature identical to that of the input.
Fig. 2.
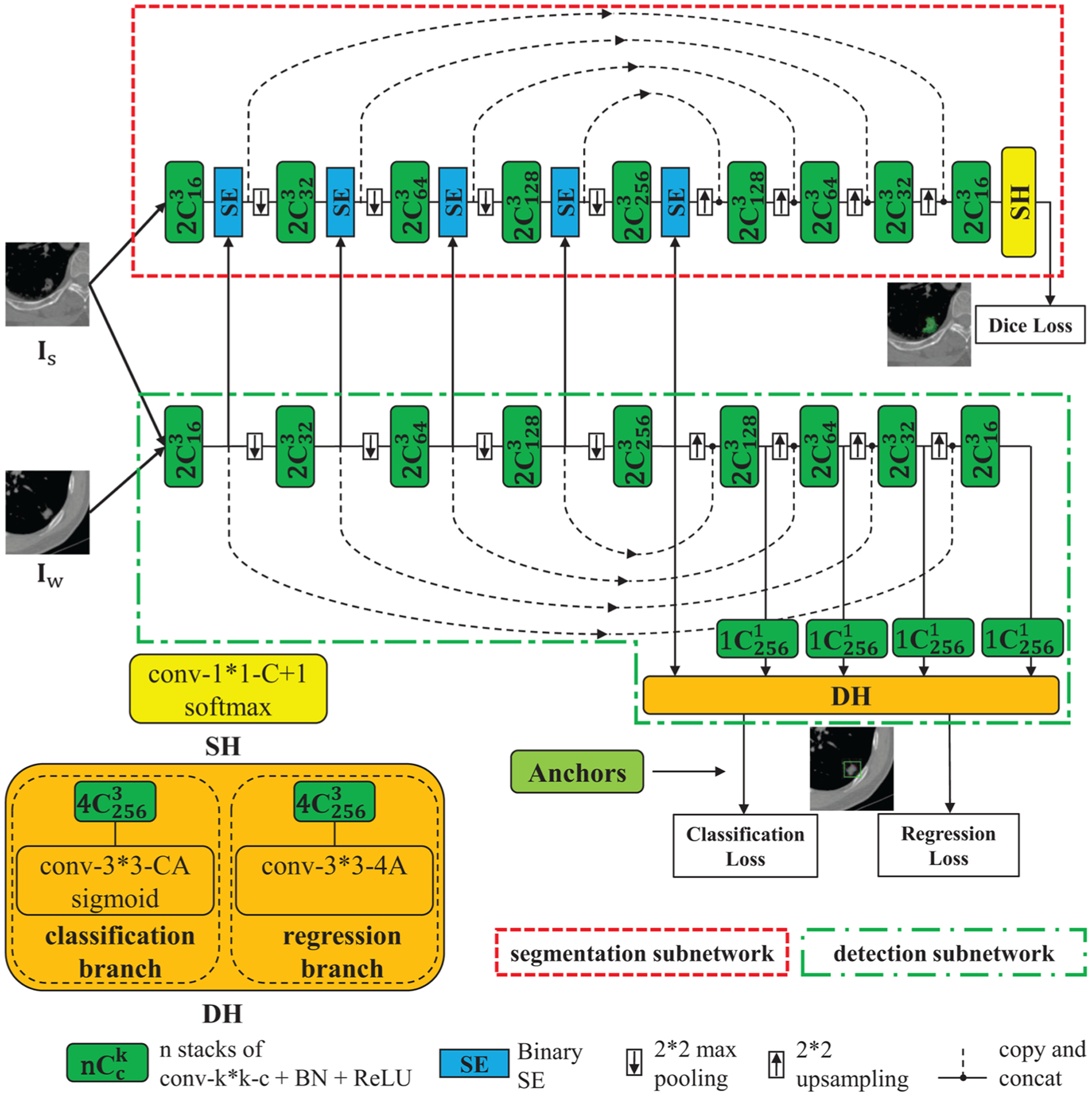
The structure of Mixed-Supervised Dual-Network (MSDN).
SE modules are added after each stage in the Encoder and Bottleneck of the segmentation subnetwork, which squeeze the detection feature and recalibrate the segmentation feature from the same stage. In this way, our model can extract useful information from the auxiliary detection subnetwork to facilitate the training of the segmentation subnetwork. We also try to add SE modules to different positions and this way gives the best performance. Detailed results can be found in Section 4.
3.2. Segmentation head
The Segmentation Head (SH) takes the feature map from the last convolution stage as input and pass it to a 1 × 1 convolution layer followed by a channel-wise softmax to output a C+1-channel dense segmentation map, where C is the number of segmentation classes and we treat the background as an additional class. Dice loss (Milletari et al., 2016) is minimized to train the segmentation subnetwork, that is
| (6) |
where and are the ith element of ground-truth and model predicted nodule segmentation map, respectively, a is a small value used for numerical stability and N is the number of training samples.
3.3. Detection head
We follow the idea of 1-stage object-detection model, similar to (Shah et al., 2018; Lin et al., 2017), to build the Detection Head (DH), which consists of a classification branch and a bounding box regression branch. The main difference is that here we use the U-Net model as backbone to extract features. The DH takes the features from the Bottleneck stage and all the Decoder stages (i.e. in total 5 feature levels) of the detection subnetwork as input to produce class predictions for C target classes and object locations via bounding boxes. We build anchors on the 5 feature levels with areas of 642, 322, 162, 82 and 42, respectively. At each position of each feature level, we build anchors at three aspect ratios 1:2, 1:1, 2:1 and three scales 20, 21/3, 22/3 of area, totally A = 9 anchors of different shapes and sizes as reference bounding boxes. We adopt a similar method to (Lin et al., 2017) to assign anchors to object boxes if the intersection-over-union (IoU) is above 0.5 (positive anchors) and to background if IoU is between [0,0.4) (negative anchors). The rest of anchors are ignored when computing training loss. Each positive anchor is assigned a C-length one-hot vector indicating the object type in it and a 4-length vector for box regression. The DH predicts the class label (C-length vector) of the object and the relative position (4-length vector) to the corresponding ground-truth bounding boxes for each of the A anchors at each spatial position. Thus, the classification branch takes as input the features from the detection subnetwork through 4 3 × 3 convolution layers with 256 filters and each followed by ReLU activation and one 3 × 3 convolution layer with C × A filters. A sigmoid function is used to scale the output of classification branch to [0, 1]. The structure of the regression branch is the same as the classification branch except that the last convolution layer is with 4 × A filters and no activation. Note that parameters of DH are shared across all feature levels and the number of input channels is set to that of the Bottleneck feature, so we add 1 × 1 convolution layers behind the features from other layers to make number the input channels identical.
Focal loss (Lin et al., 2017) is used in bounding box classification. We consider as the output of the classification branch, where element pn,c at (n, c) denotes the possibility that the nth box candidate containing object of c type. Define as
| (7) |
where yn,c = {0, 1} is the ground-truth label. The classification focal loss can by written as
| (8) |
Here γ is focusing parameter and is set to 2 for all the experiments.
For box regression, we use the standard parameterization of the 4 box coordinates and smooth L1 function following (Ren et al., 2015; Lin et al., 2017) to construct the loss function, denoted by Lreg.
3.4. Model training
We use the mixed-annotated dataset to train the model. The strongly- and weakly-annotated data are mixed and randomly shuffled. For each training data batch, the strongly-annotated data Is goes through the Encoder of the detection and segmentation subnetworks and the segmentation features are recalibrated by the detection features for the Decoder to claculate the segmentation loss. The weakly-annotated data Iw only goes through the detection subnetwork to get the detection loss. The weighted sum of the segmentation loss from the strongly-labeled data and the detection loss from weakly-labeled data is minimized to train the model:
| (9) |
where λ is the weight factor to balance the loss of the two tasks.
Remark Our model has a similar structure to (Roy et al., 2019), as both works design a dual architecture with two subnetworks and exploit SE modules as connection. However, Ref. (Roy et al., 2019) focuses on the few-shot segmentation problem. The two subnetworks are used for the same segmentation task and trained jointly in the meta-learning mode, i.e. training images go to the subnetwork in the below and their features are used to recalibrate those of testing images in the meta-dataset. SE modules exist in every feature stage of the base network. While, our model is designed for mixed-supervised segmentation problem, the two networks are used for different tasks and trained iteratively in different learning modes. The images with different annotation forms go through different paths. Because of that, the features of the two subnetworks in shallow layers may be relative to each other and those in deep layers may be task-specific. In the experiments we find that using SE in the shallow layers, specifically, in the Encoder and Bottleneck results in the best performance. Detailed results can be found in the following section.
4. Experiments
4.1. Datasets
We conduct experiments on 4 medical image segmentation datasets: lung nodule, cochlea of inner ear, kidney segmentation on CT images and medical instrument segmentation on an RGB dataset, all of which are of great clinical significance. The detail of each dataset is as follows:
4.1.1. Lung nodule dataset
The lung nodule dataset contains 320 non-contrast CT volumes that was acquired on a 64-detector CT system (GE Light Speed VCT or GE Discovery CT750 HD, GE Healthcare, Milwaukee, WI, USA) using the scan parameters: reconstruction interval, 1.25 mm; section width, 1.25 mm; display field of view (DFOV) ranged from 28 cm to 36 cm; pitch, 0.984; 120 kV; and 35 mA; matrix size, 512 × 512, pixel size ranged from 0.55mm to 0.7mm. We randomly select 160, 80 and 80 samples for training, validation and testing.
4.1.2. Cochlea of inner ear dataset
The inner ear dataset contains 146 non-contrast temporal bone CT volumes that was acquired on a Siemens Somatom scanner using the scan parameters: 120 kV; 167 mA, slice thickness, 1mm; matrix size, 512 × 512, and pixel size, 0.40625. 66, 40 and 40 samples are randomly chosen as training, validation and testing dataset.
4.1.3. Kidney dataset
Our kidney data is from the 2019 Kidney Tumor Segmentation (KiTS19) Challenge (Heller et al., 2019). The released dataset contains data of multi-phase CT imaging from 300 patients who underwent nephrectomy for kidney tumors, and 210 of them are randomly selected as training dataset and both the kidney and tumor are labeled. We split the 210 training CT volumes with labels into 150, 30 and 30 as training, validation and testing dataset.
4.1.4. Medical instrument dataset
The medical instrument dataset is obtained from the 2019 Robust Endoscopic Instrument Segmentation (ROBUST-MIS) Challenge. The training dataset is collected from the surgeries of 16 patients at the Heidelberg University Hospital, Department of Surgery and contains more than 4000 annotated images of instruments used during minimally invasive surgery, which include the grasper, scalpel, trocar, clip applicator, hooks, stapling device, suction and other instruments. Training with the whole dataset is very time-consuming, so in our experiments we pick images that only contain grasper, resulting in a sub-dataset with 1241 images. Among the 16 surgeries, we randomly select 14, 1 and 1 for training, validation and testing, with 697, 281 and 263 images respectively.
4.2. Experiment settings
Adam optimizer (Kingma and Ba, 2014) is exploited to train all the models. The initial learning rate is set to 0.0001 and is reduced by a factor of 0.8 if the mean validation Dice score doesn’t increase in 5 epochs. When the score does not increase by 20 epochs, we stop the training and perform testing. Dropout (Srivastava et al., 2014) with 0.1 is used to the output of each convolution stage to avoid overfitting. Training batch size is set to 4.
We extract a 2D slice from each 3D volume where the target structure takes up the largest area. The kidney dataset contains both the kidney and tumor. We only select the slices that contain kidney. For the lung and cochlea dataset, we first crop image patch with size 140 × 140 centered around the target structure and randomly crop to 128 × 128 during training. For kidney dataset, we first extract the central 468 × 468 patch and randomly crop to 448 × 448, then we downsample to 224 × 224 for training. For medical instrument dataset, the original images are of size 540 × 960. We first downsample the image to 135 × 240, then randomly crop to 128 × 224 for training. Both the annotations of segmentation and detection of all the 4 datasets are manually made by different experts.
For all the 4 datasets, we perform data augmentation through random horizontal and vertical flipping, adding Gaussian noise and random crop. For the medical instrument dataset, we also use color jitter. All images are normalized by subtracting the mean and dividing by the standard deviation of the training data. We test 5 different proportions of strongly-annotated data for the 3 CT datasets and 2 different proportions for medical instrument dataset. Note that we set the weight factor λ to 1.0. We apply the SE module to the Encoder and Bottleneck, and the type is chosen as Binary sSE in all the experiments. The impact of their different choices will be evaluated in Sections 4.4–4.6.
4.3. Results and analysis
To better evaluate our proposed method, we compare our MSDN with 7 baseline methods for all the 4 datasets, which are detailed as follows:
U-Net
U-Net + Unary sSE:
MS-Net
MS-Net + Unary sSE
MS-Net2
MS-Net2 + Unary sSE
MSDN−
For the first baseline model U-Net, we build it with the same number of convolution layers as the segmentation subnetwork of our model. For the U-Net+Unary sSE, Unary sSE module is added after every convolution stage of the U-Net model. For the MS-Net, we follow the idea of MS-Net (Shah et al., 2018) and implement it in our own way by building a multi-stream network based on U-Net, where all features from the Decoder are input to the detection head(DH) and the model is trained jointly in multi-task learning mode. For the MS-Net + Unary sSE, we add Unary sSE modules after every convolution stage of the MS-Net, as this model may benifit from both the sSE and mixed-supervision. The MS-Net2 and MS-Net2 + Unary sSE are another two variants of MS-Net (Shah et al., 2018), where only encoder path is shared and two decoder paths are built for different tasks. We also compare to a reduced version of our model called MSDN−, where we remove the detection part and only preserve the U-Net and the Binary sSE modules. Here we give the results of our MSDN with SE type of sSE, and sSE modules are added in the Encoder and Bottleneck part (see Fig. 2). The weight factor λ is set to 1.0. Detailed discussion about these hyper-parameters can be found in Sections 4.4 to 4.6. We run each experiment repeatly for 5 times and the mean dice score with 95% confidence interval is recorded.
One technical detail in our experiments is that we do not use Dropout to the models related to Unary sSE modules, as we surprisingly find that Unary sSE modules are not compatible with Dropout. In Table 1, we present the results of U-Net and U-Net+Unary sSE with/without Dropout on the Lung Nodule and Cochlea datasets. We can see that if we remove Dropout, the Dice Score of U-Net will decrease. However, the performance of U-Net+Unary sSE will be consistently improved. In this situation, U-Net+Unary sSE outperforms U-Net, demonstrating the effectiveness of Unary sSE modules. Based on these results, we can conclude that the Unary sSE module does not work well with Dropout. In all the experiments, we remove Dropout from the Unary sSE-related models for a fair comparison. Besides this, all the models are trained with the same settings described in Section 4.2.
Table 1.
Impact of dropout. Two strong-weak data splits of the lung nodule and cochlea datasets are tested.
| Lung nodule | Cochlea | |||
|---|---|---|---|---|
| 160-0 | 100-60 | 33-33 | 22-44 | |
| U-Net, w D | 84.04 ± 0.40 | 81.85 ± 0.31 | 86.55 ± 0.81 | 85.01 ± 0.39 |
| U-Net+Unary sSE, w D | 84.01 ± 0.11 | 81.35 ± 1.39 | 85.30 ± 0.28 | 84.38 ± 0.03 |
| U-Net, w/o D | 83.91 ± 0.65 | 81.47 ± 0.78 | 86.41 ± 0.38 | 84.84 ± 1.02 |
| U-Net+Unary sSE, w/o D | 84.85 ± 0.31 | 82.21 ± 0.59 | 86.71 ± 0.38 | 85.17 ± 0.31 |
The results are listed in Table 2–5. We can see that our model performs better than all the baselines in all the strong-weak data split. Compared with models trained in a fully-supervised manner, the performance is still comparable. When there are few strongly-annotated data for training, the performances of baselines may decrease dramatically (see the last column). However, the performance of our model still remains satisfying. The MS-Net may improve the results in some degree, but this is not always the case, and sometimes the improvement is marginal. In contrast, our model has better robustness. The MS-Net2, where a smaller part of the model is shared, performs better and more stable than MS-Net, and our MSDN that keeps the entire models of different tasks decoupled performs even better. This indicates that the fully sharing of features from different tasks may not always be the best way to exploit the weakly-labeled data and the flexible and trainable feature sharing fashion in our model is more effective and stable. The Unary sSE modules can improve the segmentation performance in most cases, but not as much as our model. Our MSDN outperforms MSDN−, which proves that our model benefits from exploiting the Binary sSE modules in the mixed-supervised training mode rather than the usage of Binary sSE modules only. Some qualitative results are shown in Fig. 3.
Table 2.
Test Dice Score (%) of lung nodule segmentation.
| Methods | Strong-weak data split (160 in total) | ||||
|---|---|---|---|---|---|
| 160-0 | 120-40 | 100-60 | 80-80 | 60-100 | |
| U-Net | 84.04 ± 0.40 | 82.25 ± 0.39 | 81.85 ± 0.31 | 80.51 ± 0.59 | 80.18 ± 0.97 |
| U-Net+Unary sSE | 84.85 ± 0.31 | 82.33 ± 0.81 | 82.21 ± 0.59 | 82.12 ± 0.84 | 80.77 ± 0.37 |
| MS-Net | – | 82.75 ± 1.04 | 82.38 ± 0.53 | 81.72 ± 1.19 | 79.80 ± 1.26 |
| MS-Net+Unary sSE | – | 83.01 ± 0.43 | 82.77 ± 0.55 | 82.61 ± 0.91 | 81.03 ± 0.61 |
| MS-Net2 | – | 83.07 ± 0.85 | 82.45 ± 0.63 | 82.24 ± 1.05 | 81.01 ± 0.79 |
| MS-Net2+Unary sSE | – | 83.34 ± 0.77 | 83.05 ± 0.53 | 82.95 ± 0.85 | 81.91 ± 0.79 |
| MSDN− | 84.90 ± 0.60 | 82.31 ± 1.14 | 82.17 ± 0.51 | 81.02 ± 1.10 | 80.50 ± 0.37 |
| MSDN | – | 83.58 ± 1.20 | 83.56 ± 0.52 | 83.01 ± 0.69 | 82.37±0.98 |
Table 5.
Test Dice Score (%) of medical instrument segmentation.
| Methods | Strong-weak data split(697 in total) | ||
|---|---|---|---|
| 697-0 | 348-349 (50%) | 174-523 (25%) | |
| U-Net | 70.60 ± 0.87 | 65.73 ±0.39 | 62.55 ±0.11 |
| U-Net+Unary sSE | 71.25 ± 0.76 | 67.31 ± 0.88 | 65.93 ± 1.08 |
| MS-Net | – | 67.61 ± 0.56 | 65.98 ±0.75 |
| MS-Net+Unary sSE | – | 68.59 ± 0.35 | 66.15 ± 0.75 |
| MS-Net2 | – | 68.51 ± 0.54 | 66.38 ±0.85 |
| MS-Net2+Unary sSE | – | 68.92 ± 0.47 | 66.55 ± 0.82 |
| MSDN− | 70.86 ±0.72 | 68.71 ± 1.33 | 62.22 ± 0.56 |
| MSDN | – | 69.05 ±0.65 | 66.89 ±0.89 |
Fig. 3.
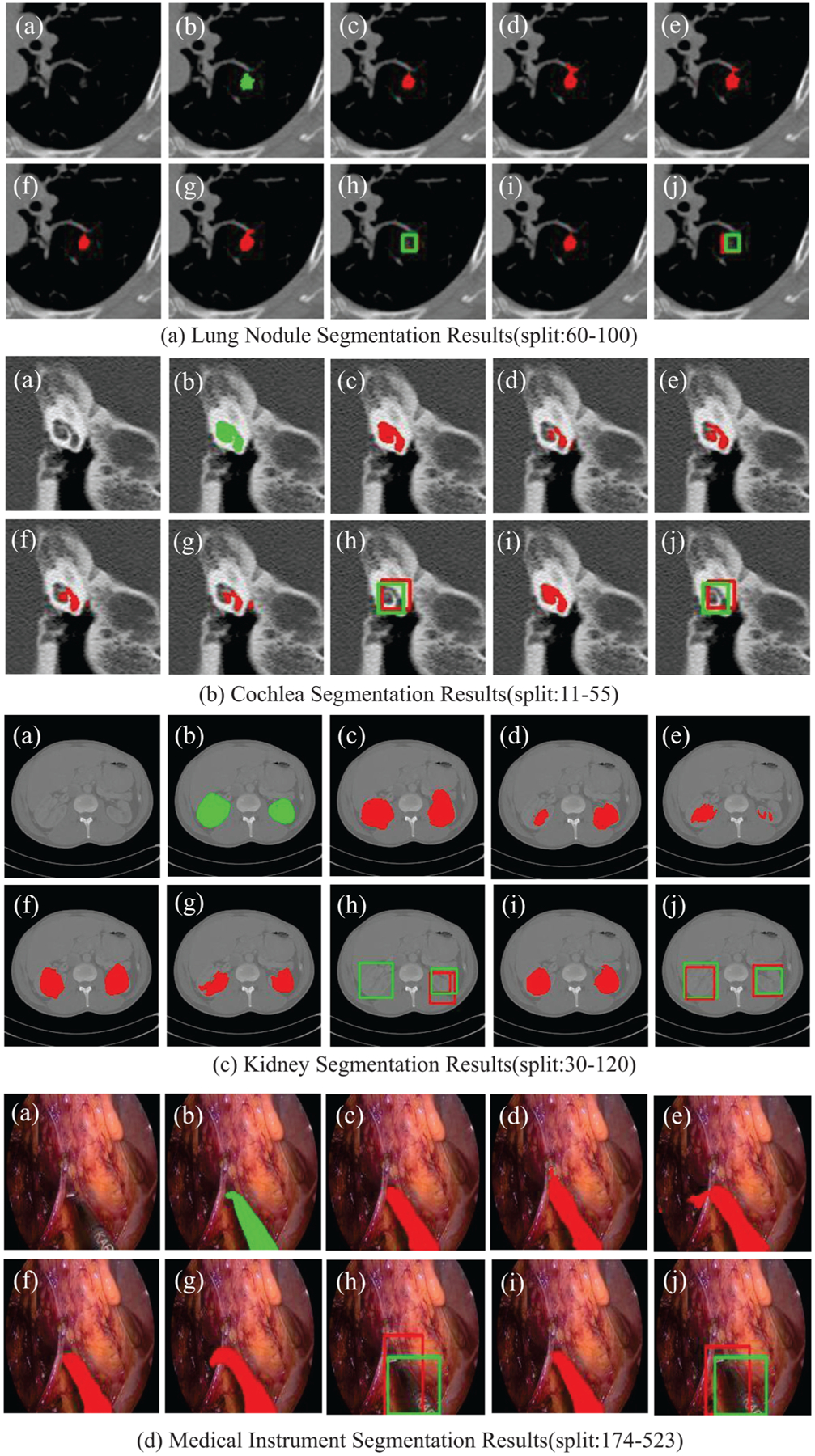
(a) Original image. (b) Ground truth. (c) U-Net trained in full-supervised manner. (d) U-Net trained with only strongly-annotated data. (e) U-Net+Unary sSE. (f) MSDN. (g) and (h) Segmentation and detection results of Variant MS-Net. (i) and (j) Segmentation and detection results of MSDN.
4.4. Impact of the weight λ
To evaluate the impact of the weight factor λ, we set λ to 0.5, 0.75, 1.0, 1.25, and 1.5, respectively, and conduct experiments with two proportions of strongly-annotated data for each of the four datasets. The results are listed in Table 6. We can draw the following conclusions. First, the performance of the proposed model does not vary too much with the weight factor from 0.5 to 1.25, which means that the model is not very sensitive to the weight factor within a certain range. Second, setting the weight factor to 0.75 or 1.0 yields the best results. Third, a large weight factor (i.e., 1.5) will remarkably damage the performance, as it will make the model focus too much on the auxiliary detection task and ignore the main segmentation task.
Table 6.
Test Dice Score (%) with different values of λ.
| Lung nodule | Cochlea | |||
|---|---|---|---|---|
| 80-80 | 60-100 | 22-44 | 11-55 | |
| 0.5 | 82.79 ± 0.68 | 82.12 ± 1.01 | 86.83 ± 0.33 | 85.32 ± 1.11 |
| 0.75 | 82.93 ± 0.61 | 82.44 ± 0.81 | 87.08 ± 0.33 | 85.47 ± 0.96 |
| 1.0 | 83.01 ± 0.69 | 82.37 ± 0.98 | 87.11 ± 0.28 | 85.60 ± 1.76 |
| 1.25 | 82.85 ± 0.57 | 82.17 ± 0.95 | 86.89 ± 0.44 | 85.38 ± 1.21 |
| 1.5 | 81.87 ± 0.57 | 81.55 ± 0.95 | 85.84 ± 0.37 | 84.93 ± 0.91 |
| Kidney | Medical instrument | |||
| 30-120 | 15-135 | 50% | 25% | |
| 0.5 | 87.25 ± 0.91 | 83.36 ± 0.95 | 68.92 ± 0.55 | 66.59 ± 1.07 |
| 0.75 | 87.44 ± 0.77 | 83.78 ± 1.21 | 69.01 ± 0.71 | 66.87 ± 0.91 |
| 1.0 | 87.34 ± 0.87 | 83.72 ± 1.58 | 69.05 ± 0.65 | 66.89 ± 0.89 |
| 1.25 | 87.17 ± 0.95 | 83.18 ± 1.19 | 68.95 ± 0.65 | 66.62 ± 0.88 |
| 1.5 | 86.40 ± 1.01 | 82.33 ± 1.03 | 67.71 ± 0.68 | 65.59 ± 0.87 |
4.5. Impact of SE module amount in MSDN
In this part, we explore the relationship between the amount of SE modules and the model performance. Different amounts of SE modules mean different degrees that the features of segmentation are recalibrated by object detection features. As is shown in Fig. 4, we test 4 SE amounts, denoted by N1 to N4. Ni means that SE modules are added to the position and all its previous layers so that these features of segmentation are recalibrated. In this section, we choose sSE modules for all the experiments. The results are listed in Table 7.
Fig. 4.
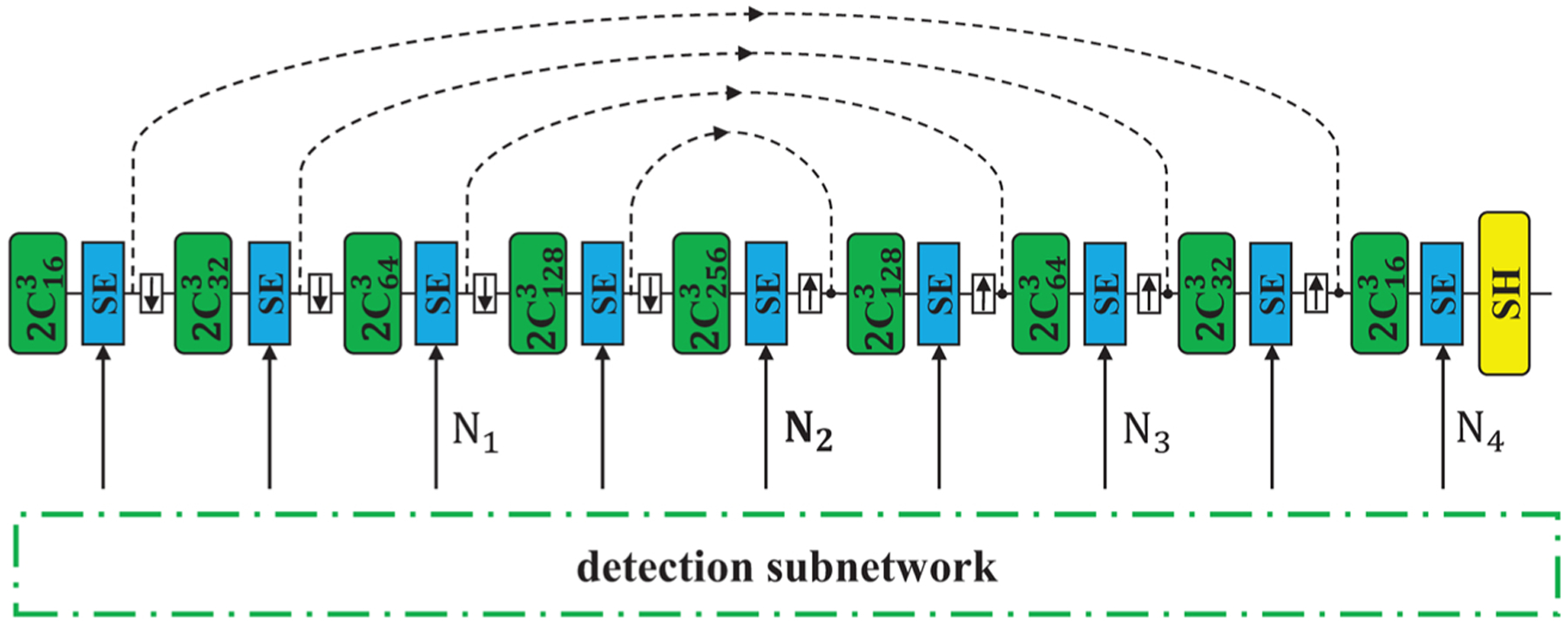
Illustration of different amount of SE modules used in MSDN. Ni means that SE modules are added to the position and all its previous layers so that these features of segmentation are recalibrated.
Table 7.
Test Dice Score (%) with different SE amounts.
| Lung nodule | Cochlea | |||
|---|---|---|---|---|
| 80-80 | 60-100 | 22-44 | 11-55 | |
| N1 | 81.59 ± 1.18 | 80.12 ± 1.31 | 86.63 ± 1.01 | 85.32 ± 1.11 |
| N2 | 83.01 ± 0.69 | 82.37 ± 0.98 | 87.11 ± 0.28 | 85.60 ± 1.76 |
| N3 | 81.56 ± 1.41 | 81.29 ± 1.61 | 86.74 ± 0.93 | 85.82 ± 1.62 |
| N4 | 81.89 ± 0.27 | 80.82 ± 0.95 | 85.74 ± 0.77 | 85.44 ± 0.91 |
| Kidney | Medical instrument | |||
| 30-120 | 15-135 | 50% | 25% | |
| N1 | 84.57 ± 0.98 | 80.36 ± 0.65 | 65.52 ± 0.52 | 64.59 ± 0.77 |
| N2 | 87.34 ± 0.87 | 83.72 ± 1.58 | 69.05 ± 0.65 | 66.89 ± 0.89 |
| N3 | 83.72 ± 2.28 | 81.68 ± 0.47 | 68.76 ± 1.18 | 66.04 ± 0.56 |
| N4 | 86.70 ± 1.14 | 80.68 ± 1.19 | 68.36 ± 0.45 | 65.61 ± 0.87 |
We can see from the results that in most cases using SE in the shallow layers, specifically, in the Encoder and Bottleneck results in the best performance. One exception is that for the Cochlea segmentation with split 11–55, adding sSE modules to two more layers performs the best. In short, adding SE modules to a part of former layers yields better results than adding SE modules to all the layers. This is because the two subnetworks are used for different tasks, so the features of the two subnetworks in shallow layers may be more relative to each other and those in deep layers may be more task-specific. Using task-specific features for recalibration may introduce feature bias to the segmentation task, thus may result in worse generalization. So we only use SE modules in the Encoder and Bottleneck layers, which is one of the differences from (Roy et al., 2019).
4.6. Impact of SE module type in MSDN
We compare 3 types of binary SE in our MSDN model, cSE (Fig. 1 (c)), sSE (Fig. 1 (d)) and mixSE (Fig. 1 (e)). One hyper-parameter of cSE and mixSE is the reduction ratio r. Based on the results from (Roy et al., 2018b), although r = 2 yields the best results, the difference of other choices of r is not very obvious. However, smaller r will result in more parameters in the model. Therefore, we set r to 8 in our experiments. The aggregation strategy is chosen as Max for mixSE in all the experiments because it works the best for the segmentation task. The results are shown in Table 8. We can see that mixSE performs the best in most cases. However, we find that training MSDN with cSE or mixSE modules cost much more time and GPU memory than that with sSE modules. This is because of the fully-connected layers in the feature squeeze path. The performance of sSE module isn’t too much worse than that of mixSE, and sometimes even better. Based on these consideration, we apply sSE module to our MSDN model in the experiments.
Table 8.
Test Dice Score (%) with different SE types.
| Lung nodule | Cochlea | |||
|---|---|---|---|---|
| 80-80 | 60-100 | 22-44 | 11-55 | |
| cSE | 82.09 ± 0.59 | 80.57 ± 0.77 | 86.61 ± 1.59 | 85.58 ± 1.97 |
| sSE | 83.01 ± 0.69 | 82.37 ± 0.98 | 87.11 ± 0.28 | 85.60 ± 1.76 |
| mixSE | 81.29 ± 1.62 | 79.75 ± 1.14 | 86.91 ± 1.49 | 86.28 ± 1.04 |
| Kidney | Medical instrument | |||
| 30-120 | 15-135 | 50% | 25% | |
| cSE | 84.55 ± 2.56 | 83.06 ± 1.64 | 66.57 ± 0.52 | 65.07 ± 0.72 |
| sSE | 87.34 ± 0.87 | 83.72 ± 1.58 | 69.05 ± 0.65 | 66.89 ± 0.89 |
| mixSE | 88.08 ± 0.82 | 85.37 ± 2.38 | 69.56 ± 0.57 | 68.96 ± 1.11 |
5. Conclusion and future work
In this paper, we propose a novel architecture called Mixed-Supervised Dual-Network (MSDN) for mixed-supervised medical image segmentation task. It consists of two separate networks for the detection and segmentation tasks respectively, and a series of sSE modules as connection between the two networks so that the useful information of the detection task can be extracted and transferred to facilitate the training of segmentation task. Compared with existing multi-task learning model with shared backbone and multiple branches, our model has flexible and trainable feature sharing fashion and thus is more effective and stable. We conduct experiments on four medical image datasets and the results show that our model outperforms multiple baselines. The limitation of our method is that it cannot be directly applied to the situation where there are more than two forms of annotations. So future works will follow the idea of some related works, such as (Lu et al., 2017; Wang et al., 2016), to extend our method to multi-task scenario.
Table 3.
Test Dice Score (%) of cochlea segmentation.
| Methods | Strong-weak data split (66 in total) | ||||
|---|---|---|---|---|---|
| 66-0 | 44-22 | 33-33 | 22-44 | 11-55 | |
| U-Net | 88.62 ± 0.08 | 87.41 ± 0.16 | 86.55 ± 0.81 | 85.01 ± 0.39 | 80.85 ± 0.42 |
| U-Net+Unary sSE | 89.05 ± 0.11 | 87.73 ± 0.45 | 86.71 ± 0.38 | 85.17 ± 0.31 | 83.17 ± 0.37 |
| MS-Net | – | 87.54 ± 0.36 | 86.03 ± 0.25 | 84.71 ± 0.53 | 82.60 ± 0.52 |
| MS-Net+Unary sSE | – | 87.73 ± 0.41 | 87.11 ± 0.43 | 85.05 ± 0.57 | 83.99 ± 0.45 |
| MS-Net2 | – | 87.66 ± 0.41 | 86.73 ± 0.73 | 86.41 ± 0.53 | 83.91 ± 0.92 |
| MS-Net2+Unary sSE | – | 87.78 ± 0.54 | 87.15 ± 0.93 | 86.73 ± 0.54 | 84.71 ± 0.83 |
| MSDN− | 88.73 ± 0.33 | 86.73 ± 1.02 | 85.68 ± 0.35 | 85.10 ± 0.15 | 80.81 ± 0.47 |
| MSDN | – | 87.91 ± 0.28 | 87.27 ± 1.08 | 87.11 ± 0.28 | 85.60 ± 1.76 |
Table 4.
Test Dice Score (%) of kidney segmentation.
| Methods | Strong-weak data split (150 in total) | ||||
|---|---|---|---|---|---|
| 150-0 | 90-60 | 60-90 | 30-120 | 15-135 | |
| U-Net | 89.09 ± 0.95 | 88.17 ± 0.68 | 88.01 ± 0.81 | 84.71 ± 0.37 | 77.79 ± 0.52 |
| U-Net+Unary sSE | 91.05 ± 1.25 | 88.31 ± 0.37 | 88.08 ± 0.93 | 86.02 ± 1.13 | 78.95 ± 0.91 |
| MS-Net | – | 88.75 ± 1.04 | 88.38 ± 1.53 | 83.72 ± 1.19 | 81.08 ± 1.26 |
| MS-Net+Unary sSE | – | 89.05 ± 0.91 | 88.61 ± 0.99 | 85.15 ± 0.79 | 82.61 ± 0.88 |
| MS-Net2 | – | 89.21 ± 0.87 | 88.57 ± 1.32 | 86.53 ± 1.35 | 82.98 ± 1.01 |
| MS-Net2+Unary sSE | – | 89.47 ± 0.94 | 88.72 ± 0.55 | 87.13 ± 0.74 | 83.40 ± 0.81 |
| MSDN− | 90.69 ± 1.04 | 89.47 ± 1.51 | 88.41 ± 1.02 | 83.02 ± 1.32 | 78.09 ± 1.75 |
| MSDN | – | 89.61 ± 0.79 | 89.06 ± 0.97 | 87.34 ± 0.87 | 83.72 ± 1.58 |
Acknowledgments
This project was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health through Grant Numbers P41EB015898, R01EB025964 and R01DK119269, and China Scholarship Council (CSC). Unrelated to this publication, Jayender Jagadeesan owns equity in Navigation Sciences,Inc. He is a co-inventor of a navigation device to assist surgeons in tumor excision that is licensed to Navigation Sciences. Dr. Jagadeesan’s interests were reviewed and are managed by BWH and Partners HealthCare in accordance with their conflict of interest policies.
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
References
- Bearman A, Russakovsky O, Ferrari V, Fei-Fei L, 2016. What’s the point: semantic segmentation with point supervision. European Conference on Computer Vision 549–565. [Google Scholar]
- Bhalgat Y, Shah M, Awate S, 2018. Annotation-Cost Minimization for Medical Image Segmentation Using Suggestive Mixed Supervision Fully Convolutional Networks arXiv preprint arXiv:1812.11302.
- Cai J, Tang Y, Lu L, Harrison AP, Yan K, Xiao J, Yang L, Summers RM, 2018. Accurate weakly-supervised deep lesion segmentation using large-scale clinical annotations: slice-propagated 3d mask generation from 2d recist. International Conference on Medical Image Computing and Computer-Assisted Intervention 396–404. [Google Scholar]
- Chen H, Qi X, Yu L, Heng P-A, 2016. Dcan: deep contour-aware networks for accurate gland segmentation. Proceedings of the IEEE conference on Computer Vision and Pattern Recognition 2487–2496. [Google Scholar]
- Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL, 2017. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell 40 (4), 834–848. [DOI] [PubMed] [Google Scholar]
- Fan G, Liu H, Wu Z, Li Y, Feng C, Wang D, Luo J, Wells W, He S, 2019. Deep learning-based automatic segmentation of lumbosacral nerves on CT for spinal intervention: a translational study. Am. J. Neuroradiol 40 (6), 1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 770–778. [Google Scholar]
- He K, Gkioxari G, Dollár P, Girshick R, 2017. Mask R-CNN. Proceedings of the IEEE International Conference on Computer Vision 2961–2969. [Google Scholar]
- Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, Rosenberg J, Blake P, Rengel Z, Oestreich M, et al. , 2019. The KiTS19 Challenge Data: 300 Kidney Tumor Cases With Clinical Context, CT Semantic Segmentations, and Surgical Outcomes arXiv preprint arXiv:1904.00445.
- Hu J, Shen L, Sun G, 2018. Squeeze-and-excitation networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 7132–7141. [Google Scholar]
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ, 2017. Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 4700–4708. [Google Scholar]
- Ioffe S, Szegedy C, 2015. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift arXiv preprint arXiv:1502.03167.
- Jiang J, Hu Y-C, Liu C-J, Halpenny D, Hellmann MD, Deasy JO, Mageras G, Veeraraghavan H, 2018. Multiple resolution residually connected feature streams for automatic lung tumor segmentation from CT images. IEEE Trans. Med. Imaging 38 (1), 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervadec H, Dolz J, Tang M, Granger E, Boykov Y, Ayed IB, 2019. Constrained-cnn losses for weakly supervised segmentation. Med. Image Anal 54, 88–99. [DOI] [PubMed] [Google Scholar]
- Kingma DP, Ba J, 2014. Adam: A Method for Stochastic Optimization arXiv preprint arXiv:1412.6980.
- Krizhevsky A, Sutskever I, Hinton GE, 2012. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst 1097–1105. [Google Scholar]
- Lin T-Y, Goyal P, Girshick R, He K, Dollár P, 2017. Focal loss for dense object detection. Proceedings of the IEEE International Conference on Computer Vision 2980–2988. [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. , 2017. A survey on deep learning in medical image analysis. Med. Image Anal 42, 60–88. [DOI] [PubMed] [Google Scholar]
- Long J, Shelhamer E, Darrell T, 2015. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 3431–3440. [DOI] [PubMed] [Google Scholar]
- Lu Y, Kumar A, Zhai S, Cheng Y, Javidi T, Feris R, 2017. Fully-adaptive feature sharing in multi-task networks with applications in person attribute classification. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 5334–5343. [Google Scholar]
- Milletari F, Navab N, Ahmadi S-A, 2016. V-net: fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV) 565–571. [Google Scholar]
- Mlynarski P, Delingette H, Criminisi A, Ayache N, 2018. Deep Learning with Mixed Supervision for Brain Tumor Segmentation arXiv preprint arXiv:1812.04571. [DOI] [PMC free article] [PubMed]
- Pohlen T, Hermans A, Mathias M, Leibe B, 2017. Full-resolution residual networks for semantic segmentation in street scenes. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 4151–4160. [Google Scholar]
- Rajchl M, Lee MC, Oktay O, Kamnitsas K, Passerat-Palmbach J, Bai W, Damodaram M, Rutherford MA, Hajnal JV, Kainz B, et al. , 2016. Deepcut: object segmentation from bounding box annotations using convolutional neural networks. IEEE Trans. Med. Imaging 36 (2), 674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmon J, Divvala S, Girshick R, Farhadi A, 2016. You only look once: unified, real-time object detection. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 779–788. [Google Scholar]
- Ren S, He K, Girshick R, Sun J, 2015. Faster R-CNN: towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst 91–99. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U -net: convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention 234–241. [Google Scholar]
- Roy AG, Navab N, Wachinger C, 2018a. Concurrent spatial and channel ‘squeeze & excitation’ in fully convolutional networks. International Conference on Medical Image Computing and Computer-Assisted Intervention 421–429. [Google Scholar]
- Roy AG, Navab N, Wachinger C, 2018b. Recalibrating fully convolutional networks with spatial and channel ”squeeze and excitation” blocks. IEEE Trans. Med. Imaging 38 (2), 540–549. [DOI] [PubMed] [Google Scholar]
- Roy AG, Siddiqui S, Pölsterl S, Navab N, Wachinger C, 2019. ‘Squeeze & Excite’ Guided Few-Shot Segmentation of Volumetric Images arXiv preprint arXiv: 1902.01314. [DOI] [PubMed]
- Shah MP, Merchant S, Awate SP, 2018. Ms-net: mixed-supervision fully-convolutional networks for full-resolution segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention 379–387. [Google Scholar]
- Shen H, Wang R, Zhang J, McKenna SJ, 2017. Boundary-aware fully convolutional network for brain tumor segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention 433–441. [Google Scholar]
- Simonyan K, Zisserman A, 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition arXiv preprint arXiv:1409.1556.
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R, 2014. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res 15 (1), 1929–1958. [Google Scholar]
- Wang J, Cheng Y, Schmidt Feris R, 2016. Walk and learn: facial attribute representation learning from egocentric video and contextual data. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2295–2304. [Google Scholar]
- Wang X, You S, Li X, Ma H, 2018. Weakly-supervised semantic segmentation by iteratively mining common object features. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 1354–1362. [Google Scholar]
- Wang D, Li M, Ben-Shlomo N, Corrales CE, Cheng Y, Zhang T, Jayender J, 2019. Mixed-Supervised Dual-Network for Medical Image Segmentation arXiv preprint arXiv:1907.10209. [DOI] [PMC free article] [PubMed]


