Abstract
Regulation of the anti-apoptotic BCL2 protein determines cell survival and is frequently abnormal in B cell lymphomas. An evolutionarily conserved post-translational mechanism for over-expression of BCL2 in human B cell lymphomas and the BCL2 paralogue CED-9 in Caenorhabditis elegans results from loss-of-function mutations in human FBXO10 and its C.elegans paralogue DRE-1, a BCL2/CED-9-binding subunit of the SKP-CULLIN-FBOX (SCF) ubiquitin ligase. Here, we tested the role of FBXO10 in BCL2 regulation by producing mice with two different CRISPR/Cas9-engineered Fbxo10 mutations: an Asp54Lys (E54K) missense mutation in the FBOX domain and a Cys55SerfsTer55 frameshift (fs) truncating mutation. Mice homozygous for either mutant allele were born at the expected Mendelian frequency and appeared normal in body weight and appearance as adults. Spleen B cells from homozygous mutant mice did not have increased BCL2 protein, nor were the numbers of mature B cells or germinal centre B cells increased as would be expected if BCL2 was increased. Other lymphocyte subsets that are also regulated by BCL2 levels also displayed no difference in frequency in homozygous Fbxo10 mutant mice. These results support one of two conclusions: either FBXO10 does not regulate BCL2 in mice, or it does so redundantly with other ubiquitin ligase complexes. Possible candidates for the latter include FBXO11 or ARTS-XIAP. The difference between the role of FBXO10 in regulating BCL2 protein levels in C. elegans and in human DLBCL, relative to single-gene deficient mouse leukocytes, should be further investigated.
Introduction
Survival of many cells, notably mature B-lymphocytes, is promoted by and depends upon the Bcl2 gene encoding an essential inhibitor of apoptosis [1–5]. The B-cell leukemia-lymphoma-2 (BCL2) gene was discovered because hybrid BCL2-Immunoglobulin Heavy chain (IGH) fusion transcripts [6–8] resulting in aberrantly high BCL2 protein expression [9] are often created by a t(14; 18) chromosomal translocation that occurs in 85% of human follicular B cell lymphomas [10,11] and 34% of germinal centre (GC)-type diffuse large B-cell lymphomas (DLBCL) [12]. While expressed in other mature B cell subsets, BCL2 is absent in normal GC B cells due to BCL6-mediated transcriptional suppression [13,14], but this regulation is disrupted by t(14;18) that brings BCL2 under control of the constitutively active IGH promoter [12,15]. BCL2 over-expression due to 18q21 amplification or activated NF-κB signalling often occurs in activated B cell (ABC)-type DLBCL [16]. Missense BCL2 point mutations are also frequently observed, associated with activation-induced cytidine deaminase (AID)-mediated somatic hypermutation (SHM) and exhibiting significant negative selection against BCL2 loss-of-function mutations [17]. Together, translocations, amplifications and missense mutations make BCL2 the second most highly mutated gene in DLBCL [18].
BCL2 is a moderately long-lived protein with a 10-hour half-life in mature B cells [19,20]. Stability of BCL2 and its anti-apoptotic paralogues, relative to the even longer-lived pro-apoptotic BAX and BAK proteins, is a key determinant of anti-apoptotic potency [21,22]. Despite the importance of BCL2 regulation for normal and neoplastic lymphocytes, remarkably little is known about mechanisms controlling BCL2 protein accumulation and turnover [23]. Protein ubiquitination resulting in proteasomal degradation is an important mechanism determining protein stability. An important family of protein ubiquitin ligases comprise the S phase kinase-associated protein 1 (SKP1)–cullin 1 (CUL1)–F-box protein (SCF) complexes [24,25]. Specific protein substrates for ubiquitination by a given SCF complex are recognised by diverse domains in the 69 different FBOX proteins. The FBOX domain itself mediates interaction with SKP1, which in turn binds CUL1 and RBX1 to activate the E2 ubiquitin ligase.
In a genetic screen in Caenorhabditis elegans, Chiorazzi et al. [26] identified a strain with a recessive S275L missense mutation in the F-box domain of the product of gene dre-1 that prevented apoptosis of the tail spike cell. Two additional dre-1 alleles had a similar effect and complementation confirmed the variant as causal, whilst transgenic over-expression of dre-1 resulted in an opposing effect of increased apoptosis. The DRE-1 protein bound weakly to the C. elegans BCL2 homologue, CED-9, and strong epistasis occurred between a weak loss-of-function dre-1 mutation and a weak loss-of-function Ced-9 mutation. The phenotypic effects of the dre-1 mutation were recapitulated by RNA interference (RNAi) against C. elegans SKP-Cullin complex proteins skr-1 and cul-1, and expression followed by co-immunoprecipitation showed the dre-1 S275L FBOX domain mutation diminished DRE-1 binding to the C. elegans SKP1 paralogue.
The C. elegans DRE-1 protein most closely resembles two human proteins, FBXO11 and FBXO10, with FBXO11 being the closest homologue [27]. Only FBXO10 and FBXO11 have the same combination of F-box and a Carbohydrate-binding proteins And Sugar Hydrolases (CASH) domain as DRE-1, but FBOX11 is confined to the nucleus where it controls BCL6 protein levels [28] whereas BCL2 is cytoplasmic [26]. Using over-expression and RNAi experiments, Chiorazzi et al. demonstrated that FBXO10 is the BCL2-binding subunit of an SCF cytoplasmic ubiquitin ligase complex that ubiquitinates BCL2 to trigger proteasomal degradation in DLBCL. The relevance of this process was supported by infrequent FBXO10 partial loss-of-function somatic mutations and frequently reduced mRNA expression in DLBCL samples from their cohort [26]. Low FBXO10 mRNA resulting in high BCL2 also appears to drive accumulation of mantle cell lymphomas (MCL) [29] derived from marginal zone or memory B cells [30].
Mice expressing BCL2 under the control of the IGH enhancer (Eμ) have increased accumulation of BCL2 protein in B cells, dramatically increased numbers of mature B cells and GC B cells, and develop low-incidence pre-B lymphomas, immunoblastic lymphomas and plasmacytomas [31–34]. Constitutive over-expression of BCL2 in all hematopoietic lineages, in transgenic mice where the human BCL2 gene is fused to the Vav gene promoter, has a potent effect on the survival, development and maturation of many blood cell types [35] and results in increased incidence of follicular lymphoma [36]. We therefore hypothesized that mice with germline Fbxo10 loss-of-function mutations would have increased BCL2 protein in B cells and correspondingly increased B cell and GC B cell accumulation, and increased BCL2 and dysregulated survival in other blood cell types. Here, we tested this hypothesis by analysing mice with either a CRISPR/Cas9-engineered germline deletion in Fbxo10 or partial loss-of-function E54K missense mutation in the F-box domain.
Results
Germline Fbxo10E54K and Fbxo10frameshift mutant mice appear at Mendelian frequencies, present with no visible clinical phenotype and age normally
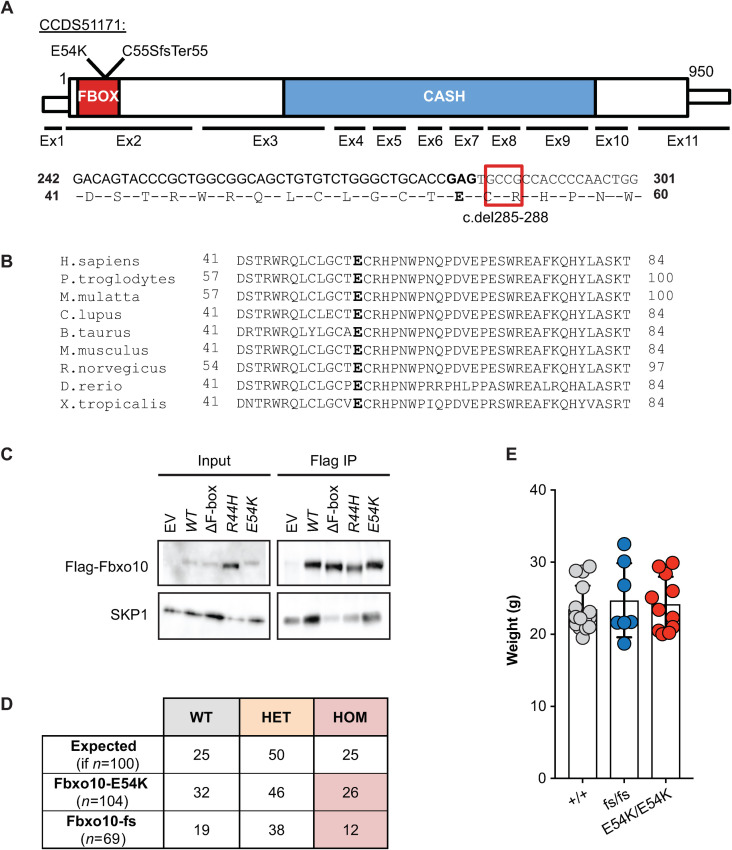
Our interest in FBXO10 was stimulated by the identification of a very rare, predicted damaging, missense variant E54K inherited in homozygous state from healthy heterozygous parents in a child with multiple autoimmune diseases and possible learning difficulties (unpublished data). We have since discovered compound heterozygous TNFAIP3 mutations explaining the child’s autoimmunity, but it was notable that E54K had been independently found as a heterozygous de novo mutation in a child with autism spectrum disorder [37]. The E54 residue lies within the F-box superfamily domain (SSF81383, FBXO10 residues 6–80) required for SCF complex assembly, is strictly conserved from fish to humans, and the substitution from glutamic acid to lysine represents a non-conservative charge reversal (Fig 1A and 1B). When FLAG-tagged FBXO10 was expressed in HEK293T cells, the E54K substitution decreased immunoprecipitation of endogenous SKP1 to a similar extent as the partial loss-of-function R44H FBOX mutation (Fig 1C) previously characterised in a human lymphoma [26].
Fig 1. Viable mice with homozygous germline Fbxo10E54K or Fbxo10fs mutations.
(A) Schematic of mouse Fbxo10 mRNA CCDS51171, showing position of exons, location of mutations, FBOX (SSF81383) and three tandem CASH (SM00722) domains, and the four cDNA nucleotides deleted in the Fbxo10C55SfsTer55 (fs) allele. (B) Alignment of the FBOX10 amino acid sequence from the indicated species: E54 in bold. (C) Expression vectors, either empty (EV) or encoding FLAG-tagged human FBXO10 wildtype (WT) or with the indicated missense mutations or deleting the F-box domain (ΔF-box) to completely abolish interaction with endogenous SKP1 [26], were transfected into HEK293T cells and lysates or anti-FLAG immunoprecipitates western blotted with antibodies to FLAG or SKP1. (D) Expected and observed numbers of offspring of the indicated genotypes from intercrossed heterozygous parents. Statistical analysis by Chi-Square test with n = 2 degrees of freedom, testing for differences relative to a 1WT:2HET:1HOM expected Mendelian ratio (p = 0.61 and p = 0.55 for Fbxo10E54K or Fbxo10fs respectively). (E) Body weight of Fbxo10+/+, Fbxo10fs/fs and Fbxo10E54K/E54K mice 9–20 weeks old (p = 0.51 and p = 0.62 for Fbxo10E54K or Fbxo10fs, respectively). Each dot represents an individual mouse of the indicated genotype. Statistical comparison between each mutant and wild-type group was performed by t-test corrected for multiple comparisons using the Holm-Sidak method.
To explore E54K as a candidate mutation, Fbxo10E54K mice were produced by CRISPR/Cas9 gene editing in mouse embryos following established molecular and animal husbandry techniques [38]. Two independent alleles were engineered and propagated in C57BL/6J mice (Fig 1A): a point mutation in exon 2 changing the Glutamate 54 codon to Lysine (E54K), or a 4 nucleotide deletion in codons 55 and 56 within exon 2 changing the Cysteine 55 codon to Serine and creating a reading frame shift and premature stop codon after 55 codons (c.del285_288 or p.Cys55SerfsTer55; abbreviated as fs). Both alleles were confirmed by Sanger sequencing and genotyping that readily identified heterozygotes and homozygotes for each allele (S1 Fig). The fs deletion does not create a new splice donor site and there is no evidence of alternate splice forms of Fbxo10 that skip exon 2 in mouse or human. It is therefore likely to create a null allele, although we lack suitable antibodies to test for a protein remnant in primary mouse cells. When heterozygous animals were intercrossed, neither E54K nor the frameshift mutation resulted in altered frequencies of heterozygous or homozygous mutant mice relative to expected Mendelian ratios (Fig 1D). Adult homozygous mutant animals up to 50 weeks old appeared normal and healthy, and had no significant difference in body weight from wild-type littermates (Fig 1E).
FBXO10 deletion or missense mutation had no detectable effect on B cells in bone marrow or spleen
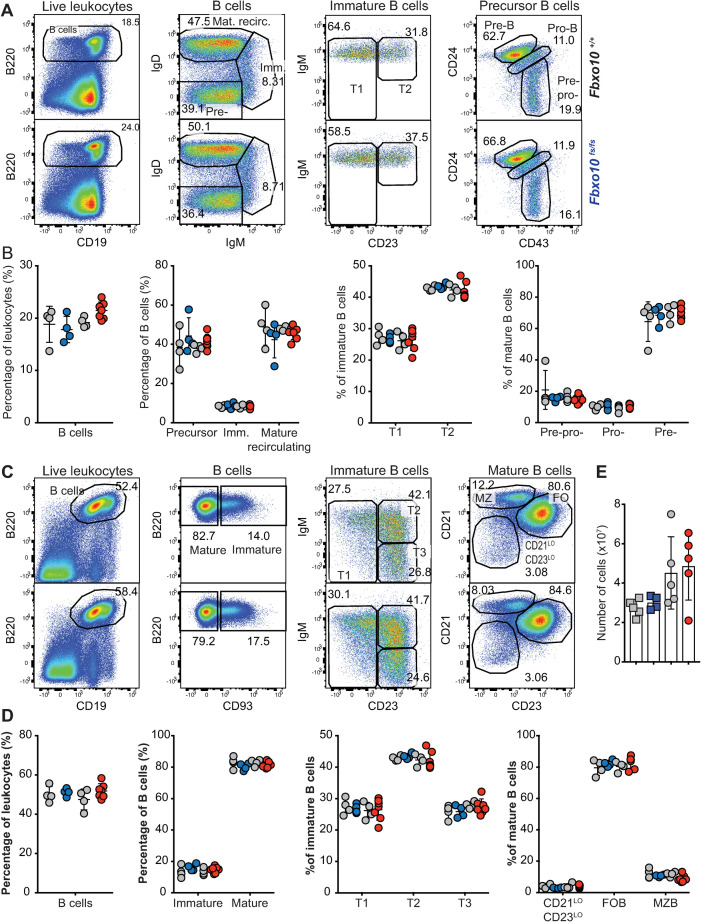
Flow cytometric analysis of early B cell development in the bone marrow revealed no discernable difference between Fbxo10 wild-type and mutant mice in the frequencies of bone marrow B cells, nor in the subsets of mature recirculating B cells, immature B cells, or the different stages of precursor B cell differentiation (Fig 2A and 2B). Congruent with this, there were no detectable differences in expression of B220, CD19, CD93, IgM, IgD, CD21, CD23 or CD86 in these various subsets (S2A Fig).
Fig 2. Fbxo10 frameshift or missense mutation do not discernably affect B cell subsets in the bone marrow or spleen.
(A) Representative flow cytometric gating strategy to delineate B cell developmental subsets in the bone marrow. Numbers denote cells in the gate as percentage of parent population. (B) Frequency of indicated B cell subsets in the bone marrow in Fbxo10fs/fs and Fbxo10+/+ littermate control mice (blue and left set of grey circles, respectively) and in Fbxo10E54K/$54K and Fbxo10+/+ littermate control mice (red and right-hand grey circles, respectively). (C) Representative flow cytometric gating strategy to delineate splenic B cell subsets. (D) B cell subsets in the spleen of Fbxo10+/+, Fbxo10fs/fs and Fbxo10E54K/E54K mice. (E) Spleen cellularity in Fbxo10fs/fs and littermate control mice (blue and grey circles, respectively) and in Fbxo10E54K/$54K and Fbxo10+/+ littermate control mice (red and grey circles, respectively). (B, D, E) Each dot represents data from an individual animal. Data are representative of n = 2 experiments on mice 40–50 weeks old, and similar results observed in n = 2 experiments on mice 10–20 weeks old. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wild-type controls with p < 0.05.
Neither mutation resulted in any changes in distribution of B cell maturation subsets in the spleen (Fig 2C and 2D), nor in any detectable changes in surface expression by splenic B cell subsets of the markers listed above (S2B Fig). Inguinal lymph node and spleen cellularity were also unaffected (Fig 2E). Once more, the lack of a visible effect of Fbxo10 mutation or deletion, even in elderly mice, indicates that FBXO10 plays no role or a functionally redundant role in B cell early development and splenic B cell maturation.
FBXO10 deletion or missense mutation had no visible effect on the magnitude or quality of a polyclonal GC B cell response to SRBC immunisation
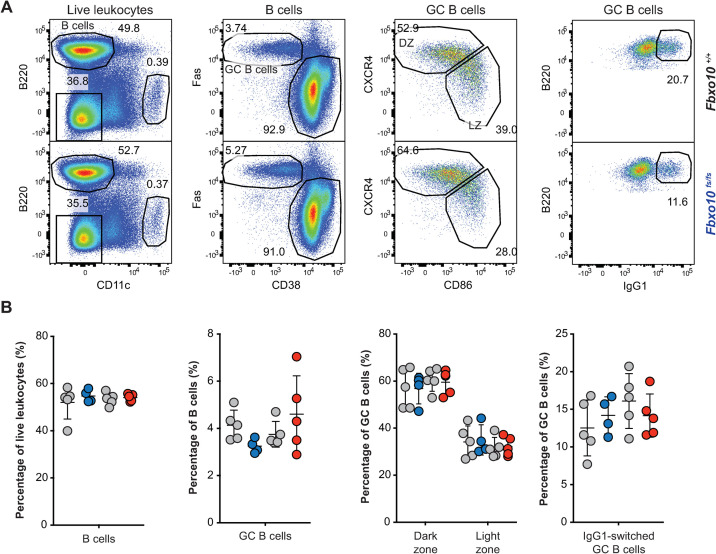
FBXO10 is particularly highly expressed by GC B cells and appears to be most important for regulating BCL2 protein levels in GCB-type DLBCL, based on the reduced FBXO10 mRNA expression and low frequency heterozygous FBXO10 hypomorphic missense alleles in DLBCL and the high FBXO10 mRNA expression in GC B cells [26]. We therefore tested for increased accumulation of GC B cells in Fbxo10E54K and Fbxo10fs mice in a T cell-dependent response following sheep red blood cell (SRBC) immunisation. Sacrifice of Fbxo10fs/fs, Fbxo10E54K/E54K and wild-type mice 7 days post SRBC-immunisation demonstrated no significant difference in the magnitude of the GC response, nor the dark zone/light zone distribution or the fraction of IgG1 class-switched GC B cells (Fig 3A and 3B).
Fig 3. Fbxo10 frameshift or missense mutation do not visibly expand or alter the GC response to SRBC immunisation.
(A) Representative flow cytometric gating strategy to delineate GC B cells responding to SRBC immunisation. Numbers denote cells in the gate as a percentage of parent population. (B) Frequency of B cells, GC B cells, light zone/dark zone distribution and IgG1-class-switched GC B cells in the spleen of Fbxo10+/+ (grey circles matched with mutant siblings) Fbxo10fs/fs (blue circles), and Fbxo10E54K/E54K (red circles) mice day 7 post-immunisation. B: each dot represents an individual biological replicate. Data are representative of n = 2 experiments on mice 10–20 weeks old. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wildtype controls with p < 0.05.
FBXO10 deletion or missense mutation had no visible effect on B or T cell expression of putative FBXO10 targets
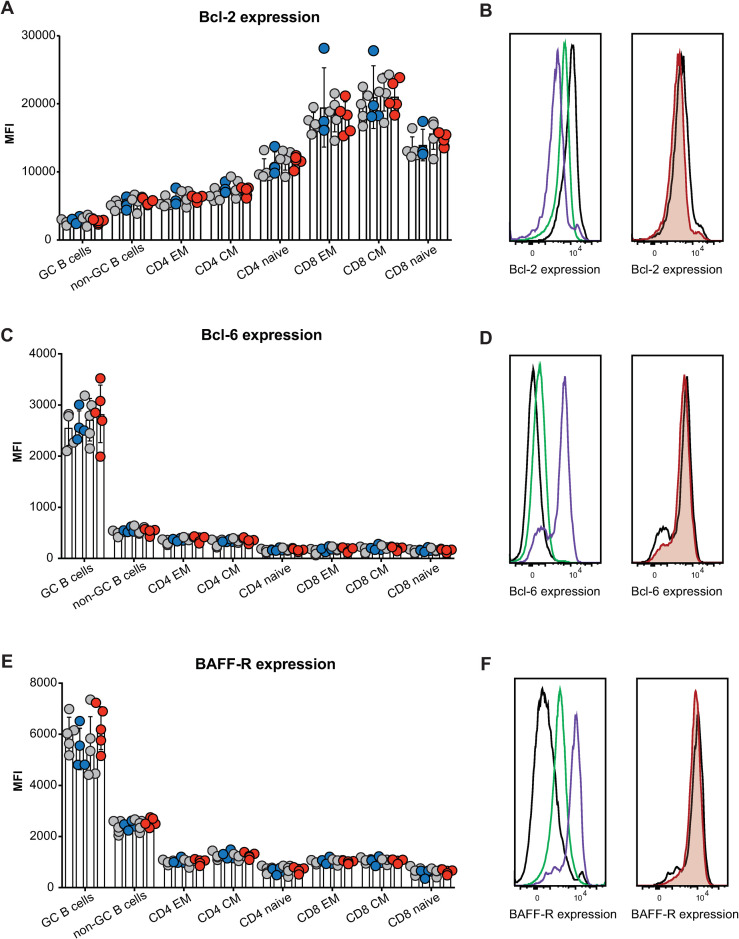
Given the lack of any increase in mature B cells or GC B cells, as would be expected if FBXO10 deficiency resulted in increased BCL2 protein accumulation, we measured BCL2 protein levels in single B cells by intracellular antibody staining followed by flow cytometric analysis. We expected to observe an increase in BCL2 protein consistent with the results of shRNA knockdown of FBXO10 in human B lymphoma cells [26]. By contrast, we observed no differences in expression of BCL2 between wild-type and mutant GC B cells (Fig 4A and 4B) The same was true for BCL6 (Fig 4C and 4D) and BAFF-R that is an FBXO11 target in lymphoma (Fig 4E and 4F). The same was true for non-GC B cells as well as for effector memory, central memory and naïve CD4 and CD8 T cells (Fig 4A–4F). Importantly, the well-validated changes in expression of BAFF-R, BCL-6, and BCL-2 between lymphoid subsets, such as increased BCL-6 expression in GC B cells relative to non-GC B cells or increased BCL-2 expression in effector memory T cells relative to naïve T cells, provided a useful internal control to validate successful staining in terms both of specificity and sensitivity (Fig 4B, 4D and 4F).
Fig 4. Fbxo10 frameshift or missense mutation do not discernably alter the protein expression levels of putative Fbxo10 targets in lymphoma.
(A,C,E) Mean fluorescence intensity (MFI) for BCL2, BCL6 or BAFF-R expression, respectively, in splenic B and T cell subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 10–20 weeks old, day 7 post-immunisation with SRBCs. (B,D,F) Left panel: representative histogram overlay of fluorescent antibody staining for intracellular BCL-2 or BCL-6 or cell surface BAFF-R in T cells (black), non-GC B cells (green) or GC B cells (purple). Right panel: representative histogram overlay for BCL2, BCL6 or BAFF-R expression in Fbxo10+/+ (black) versus Fbxo10E54K/E54K (red) GC B cells. Each dot represents an individual animal, with genotypes as in Fig 2. Data are representative of n = 2 experiments on mice 10–20 weeks old, 7 days post-immunisation with SRBC, and similar results obtained for n = 2 experiments on un-immunised mice 40–50 weeks old. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wildtype controls with p < 0.05.
FBXO10 deletion or missense mutation had no detectable effect on T cell thymic development or T cell splenic maturation
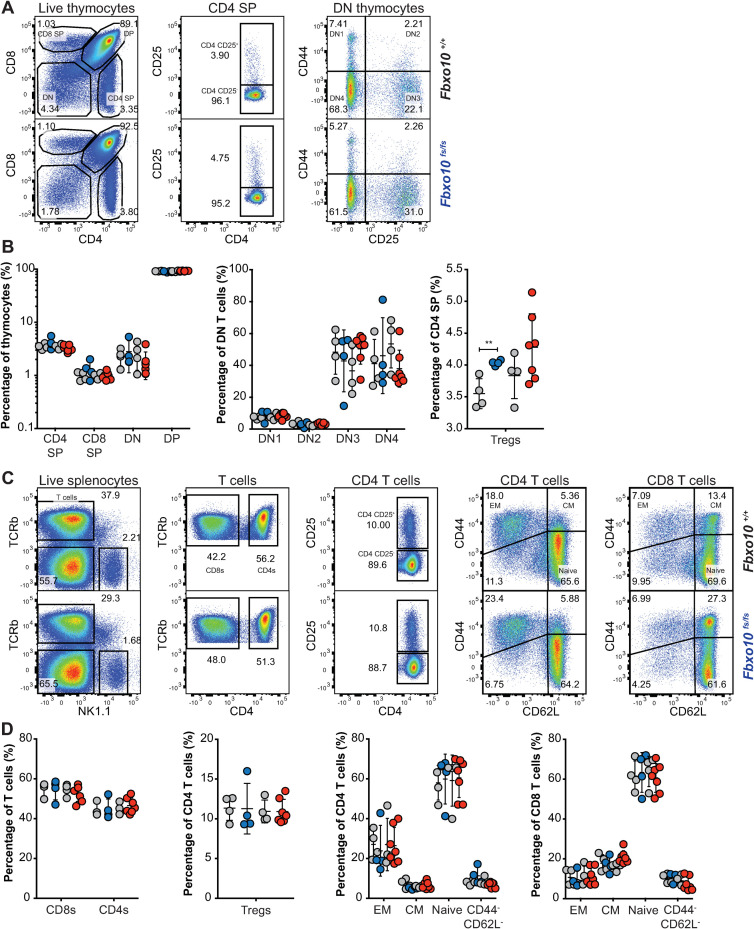
Because expression of the Vav-BCL2 transgene in mice also causes a marked elevation of T lymphocytes and altered relative abundances of developing CD4- CD8- double negative (DN), CD4+ CD8+ double positive (DP) and single positive (SP) thymocytes [35], we investigated T cell development and maturation in Fbxo10E54K or Fbxo10fs mutant mice. Our analysis revealed no significant difference in fractions of thymic DN, DP, CD4 and CD8 SP T cells in elderly (40–50 week old) Fbxo10fs and Fbxo10E54K mutant mice, relative to their wild-type littermate controls, nor in the fractions of early developing DN1-DN4 thymocytes (Fig 5A and 5B). There were also no detectable changes in expression of CD25, CD44, CD69, PD1 and CD62L by these thymic subsets (S3A and S3B Fig). In our hands, the only significant effect of Fbxo10 deletion or missense mutation was a very slight increase in frequency of Tregs in the thymus (Fig 5A and 5B) that was a consistent trend in different cohorts. Splenic T cell subsets were also not significantly affected by Fbxo10 mutations, as the percentage of T cells, CD4:CD8 ratio, fraction of Tregs and of CD4 and CD8 effector memory, central memory and naïve subsets were comparable in mutant relative to wild-type mice (Fig 5C and 5D). Similarly, no changes in expression of maturation/activation markers were detected in these various subsets between wild-type and mutant mice (S3C Fig).
Fig 5. Fbxo10 frameshift or missense mutation do not discernably alter thymic or spleen T cell subsets.
(A) Representative flow cytometric gating strategy to delineate T cell developmental populations in the thymus. Numbers denote cells in gate as percentage of parent population. (B) T cell developmental subsets in the thymus of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice. (C) Representative flow cytometric gating strategy to delineate splenic T cell subsets. (D) T cell subsets in the spleen of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice. B, D: each dot represents data from an individual mouse: Fbxo10+/+ in grey, Fbxo10fs/fs in blue, Fbxo10E54K/E54K in red as in Fig 2. Results representative of n = 2 experiments on mice 40–50 weeks old, and similar results observed in n = 2 experiments on mice 10–20 weeks old. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method, ** p < 0.01, all other differences were not significant.
The lack of any observable effects in young or elderly mice, despite the common exacerbation of underlying immune defects with age in mice and humans, indicates that FBXO10 plays no or a redundant role in murine T cell development and maturation, at least in un-immunised mice. Further analysis using antigen-specific challenge models may reveal a context-specific role for FBXO10 in T cells. Fbxo10 expression has for example been shown to increase in Jurkat cells upon cellular stress, downstream of LEDGF signalling [39]. Nevertheless, we can conclude that FBXO10 alone is not required for the development, differentiation or survival of T cells in mice.
FBXO10 deletion or missense mutation had no detectable effect on the distribution of murine lymphoid and myeloid leukocyte subsets in the bone marrow or spleen
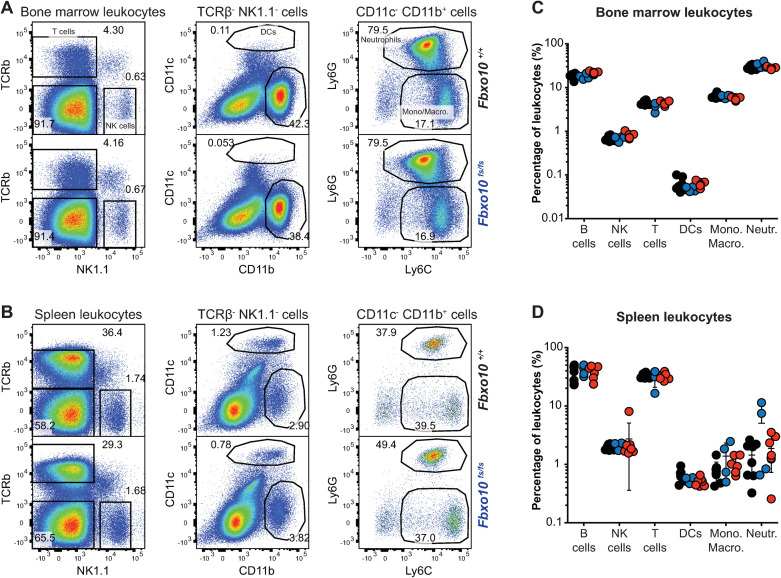
Similarly, despite the changes in lymphoid and myeloid subsets in Vav-BCL2 transgenic mice [35], the distribution of NK cells, dendritic cells, monocytes/macrophages and dendritic cells in the bone marrow (Fig 6A and 6C) and spleen (Fig 6B and 6D) was not significantly different between Fbxo10 mutant or wild-type mice (Fig 6A–6D). No consistent changes in size, granularity or expression of CD11b, CD11c, Ly6G, Ly6C, CD44, CD62L proteins were detected in any leukocyte subset in the bone marrow (S4A Fig) or spleen (S4B Fig) of Fbxo10-mutant mice. As many of these proteins are well-validated markers of activation and differentiation of myeloid cells, we may infer that development and maturation of myeloid cells are largely unaffected by Fbxo10E54K and Fbxo10fs.
Fig 6. Fbxo10 frameshift or missense mutation do not cause discernable differences in myeloid and lymphoid leukocytes within the spleen or bone marrow.
(A) Representative flow cytometry gating strategy to delineate leukocyte populations in the bone marrow. Numbers denote cells in gate as percentage of parent population. (B) Representative flow cytometric gating strategy to delineate leukocyte populations in the spleen. (C) Leukocyte subsets in the bone marrow of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice. (D) Leukocyte subsets in the spleen of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice. C, D: each dot represents an individual biological replicate. Results representative of n = 2 experiments on mice 40–50 weeks old, and similar results observed in n = 2 experiments on mice 10–20 weeks old. Fbxo10+/+ in black, Fbxo10fs/fs in blue, Fbxo10E54K/E54K in red. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wildtype controls with p < 0.05.
Discussion
Based on somatic mutations in DLBCL, germline mutations in C elegans, and experimental overexpression and RNAi knockdown experiments in human DLBCL cells, we hypothesised that germline loss of function Fbxo10 mutations in mice would cause increased BCL2 protein accumulation in mature B cells and GC B cells and corresponding increased B cell accumulation. This hypothesis was not supported here by characterisation of C57BL/6J mice with a germline missense mutation or frameshift mutation in Fbxo10. No visible morphological or immune cellular phenotype resulted from FBXO10 loss-of-function in mice. Mutant mice presented with normal breeding frequencies (Fig 1), spleen cellularity (Fig 2), unchanged B and T cell early development and splenic maturation (Figs 2 and 5), normal frequencies of leukocyte subsets in the bone marrow or spleen (Fig 6), and identical expression of protein markers associated with development, differentiation, activation and migration in all of these various leukocyte subsets (S2–S4 Figs).
Mutation or loss of Fbxo10 did not affect the magnitude of a polyclonal GC B cell response to SRBC immunisation, nor the GC dark zone/light zone distribution or frequency of IgG1 class-switched cells (Fig 3). Finally, wild-type and mutant lymphoid subsets presented with identical expression of BCL2 (Fig 4). These observations were made not only in 10–12 week old adult mice, but also in elderly 40–50 week old mice where one can often observe exacerbation of underlying immune defects over time. Of 59 mice aged to 30–50 weeks old, no Fbxo10-mutant (or wild-type) mouse developed a solid organ or lymphoid malignancy. Thus, Fbxo10 hypomorphic mutation or deletion results in no visible changes in expression of FBXO10 target BCL2 in mice, even in the GC (nor of FBXO11 target BCL6). The importance of FBXO10 function to BCL2 expression and survival of DLBCL [26,28] may be associated with a concomitant loss of redundant or compensatory mechanisms in these cells.
Another role identified for FBXO10 in human lymphoma cell lines is in the negative regulation of BCR signalling via BCR signalling-induced membrane re-localisation followed by degradation of human germinal-centre associated lymphoma (HGAL, also called GCET2) protein levels [40]. HGAL is GC B cell-specific, enhances BCR signalling by increasing activation of Syk downstream effectors and human HGAL-transgenic mice develop lymphoid hyperplasia in elderly mice [41]. Notably however, deletion of HGAL (also called M17) had no effect on the GC response in mice [42]. The normal GC responses observed in mice with frameshift or missense FBXO10 do not support a critical role for FBXO10 in degrading HGAL in mice, although we have not measured HGAL levels in mutant GC B cells.
The related protein, FBXO11, may theoretically compensate for FBXO10 loss of function mutations. In the gnoMAD database analysing 124,000 adult human exomes or genomes, FBXO11 has a pLI = 1.0, due to much lower than expected occurrence of heterozygous stop gain or frameshift mutations. This is consistent with evidence for human FBXO11 haploinsufficiency, with heterozygous germline de novo loss-of-function alleles found recurrently in children with neurodevelopmental disorders [37,43,44], and with high frequency heterozygous loss-of-function somatic mutations in human B cell lymphoma [28]. In mice, an Fbxo11 missense mutation in the CASH domain causes a heterozygous developmental disorder of the ear and homozygous lethal dysmorphism [45], while homozygous conditional Fbxo11 deletion in GC B cells increases their number and BCL6 protein levels [46]. By contrast, FBXO10 has a pLI = 0 in gnoMAD indicating that heterozygous null mutations occur at the expected frequency in the adult human population. Evidence against FBXO11 as a redundant paralogue for BCL2 regulation comes from FBXO11 siRNA knockdown in human B cell lymphoma cells, which dramatically enhanced BCL6 protein stability after protein translation was pharmacologically blocked, but did not enhance BCL2 protein stability analysed in the same cell lysates [28].
Another candidate compensatory BCL2-regulator is ARTS (gene name SEPT4), which serves as an adapter to promote BCL2 ubiquitination by the XIAP ubiquitin ligase in apoptotic cells [47]. ARTS-deficient B cells in mice nevertheless develop and accumulate in normal numbers, suggesting that ARTS is also unnecessary or redundant for regulating BCL2-dependent B cell survival [48]. However, ARTS deficiency does promote exaggerated mature B cell accumulation in Emu-MYC transgenic B cells where MYC is dysregulated and promotes apoptosis, and this effect is abolished in ARTS-XIAP double-deficient B cells [48]. Given the importance of balanced BCL2 and BIM protein levels for controlling normal B cell survival and suppressing B cell lymphoma [1], it would not be surprising that BCL2 protein turnover be governed by multiple, redundant ubiquitin ligases.
To our knowledge, our results constitute the first characterisation of mice with homozygous loss-of-function mutations in FBXO10. They highlight the importance of investigating the functional redundancy/synergy of FBXO10 loss-of-function with mutations in other pathways and with loss-of-function of SCF complex members such as FBXO11. The incongruity between the role of FBXO10 in inducing cell death of the C. elegans tail spike cell and of human DLBCL cells relative to leukocyte subsets in the mouse should be further investigated.
Materials and methods
Mice
Mice were bred, using heterozygous by heterozygous pairs, at Australian BioResources (MossVale, NSW, Australia) and kept in specific pathogen-free conditions at the Garvan Institute (Sydney, Australia). All animal studies were approved and conducted in compliance with the guidelines set by the Garvan/St.Vincent’s Animal Ethics Committee. All the mice in this study were monitored weekly for weight changes, signs of discomfort or poor health–no signs of deteriorating health were detected in any of these mice. 4–10 mice were analysed per genotype per independent experiment, to ensure statistical robustness of resulting data.
Fbxo10E54K and Fbxo10KO mice were produced by the Mouse Engineering Garvan/ABR (MEGA) Facility using CRISPR/Cas9 gene targeting in mouse embryos following established molecular and animal husbandry techniques [49]. The single guide RNA (sgRNA) was based on a target site exon 2 of Fbxo10 (CCAGTTGGGGTGGCGGCACTCGG) (protospacer-associated motif = PAM italicised and underlined) and was microinjected into the nucleus and cytoplasm of C57BL/6J zygotes together with polyadenylated S.pyogenes Cas9 mRNA and a 150 base single-stranded, anti-sense, deoxy-oligonucleotide homologous recombination substrate carrying the E54K (GAG>AAG) mutation and a PAM-inactivating silent mutation in the T53 codon (ACC>ACA). A founder mouse heterozygous for both substitutions was obtained and used to establish the Fbxo10E54K line. An additional founder carrying a 4bp frame shift mutation after the first base of the C55 codon was bred to establish the Fbxo10Δ line. Both lines were maintained on an inbred C57BL/6J background. All experiments were approved by the Garvan/St Vincent’s Animal Ethics Committee. Mice were bred and housed in specific pathogen-free conditions at Australian BioResources (Moss Vale) and the Garvan Institute Biological Testing Facility.
Flow cytometric analysis
Mouse organs were harvested into FACS buffer (PBS/1% BSA/0.02% sodium azide) and single cell suspensions passed through a 70 μm cell strainer (Falcon, Corning, NY, USA). In analysis of spleen or blood immune subsets, red blood cell (RBC) lysis was performed using lysis buffer solution (0.8% ammonium chloride, 0.08% sodium bicarbonate, 0.04% EDTA disodium salt, pH 7.3).
Single cell suspensions were stained with antibodies targeting cell-surface (B220, BAFF-R, IgM, IgD, IgG1, Ly-6C, Ly-6G, PD-1, TCRβ, CD3, CD11b, CD11c, CD19, CD21/35, CD23, CD24, CD25, CD28, CD38, CD43, CD44, CD62L, CD69, CD86, CD93, CD95, CD278) or intracellular proteins (BCL-2, BCL-6, CD152) and cells were acquired on an LSR II analyser (BD Pharmingen), followed by flow cytometric analysis using the FlowJo Software (FlowJo LLC, Ashland, OR, USA).
Immunoprecipitation
Expression vectors, either empty or encoding FLAG-tagged human FBXO10 wild-type or with the indicated mutations, were transfected into HEK293T cells and lysates or anti-FLAG immunoprecipitates western blotted with antibodies to FLAG or SKP1. Immunoprecipitations were performed as previously described [26], using FLAG (Sigma F3165) and SKP1 (Santa Cruz sc-5281) antibodies. Construction of cDNA of FLAG-tagged FBXO10, ΔFBXO10 and FBXO10 R44H in a retroviral vector and transfection into HEK293T cells were also performed as previously described [26]. FBXO10 E54K was generated by site-directed mutagenesis (Stratagene 200521–5) using the following primers:
Fbxo10_E54K_F: GTCTGGGCTGCACCGAGTGCCGCCACCCCAACTGG
Fbxo10_E54K_R: CCAGTTGGGGTGGCGGCACTCGGTGCAGCCCAGAC
Statistical analysis
GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA) was used for analysis of flow cytometry or ELISA data. For comparisons between genotypes, the variance was approximately equal between samples and comparisons were made using a Student’s t-test, and corrected for multiple comparisons using the Holm-Sidak method. For these tests, p < 0.05 was considered statistically significant. In all flow cytometry summary figures, each data point represents an individual mouse. Error bars indicate the mean and standard distribution. *p<0.05; **p<0.01; ***p<0.001.
Supporting information
CRISPR/Cas9-engineered mice were produced as described in the materials and methods. (A-C) Sanger sequencing traces of a portion of the Fbxo10 gene encoding amino acids 49 to 64 of FBXO10: (A) wild type sequence showing the target site of the sgRNA used and its associated PAM and highlighting the E54 codon (B) sequence from a mouse heterozygous for the E54K mutation (together with the PAM inactivating mutation in the T53 codon) (C) sequence from a mouse heterozygous for a four base pair deletion (GCCG, highlighted) resulting in a reading frame shift after the E54 codon. Note that the Sanger sequencing read is from right to left, and the two frameshifted sequences that are superimposed downstream of the deletion are shown above. (D) High resolution melt curve analysis plots of DNA from wild-type and mutant mice, showing the alternative fluorescence profiles resulting from heterozygous or homozygous E54K (left) or C55SfsTer55 (right) mutations.
(TIF)
(A) Mean fluorescence intensity (MFI) for IgM, IgD, CD93, CD86 expression in splenic B cell subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. (B) MFI for IgM, IgD, CD24, CD43 expression in bone marrow B cell subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. Each dot represents an individual biological replicate in Fbxo10+/+ (black), Fbxo10E54K/E54K (red) or Fbxo10fs/fs (blue) mice. Similar results were obtained for multiple protein markers (CD19, CD21/35, CD23, CD24, CD43, CD86, etc.). Results are representative of n = 2 experiments on un-immunised mice 40–50 weeks old and similar results were obtained for n = 2 experiments on mice 10–20 weeks old, 7 days post-immunisation with SRBC. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wildtype controls with p < 0.05.
(TIF)
(A,B) Representative histogram overlays of Fbxo10+/+ (grey fill) or Fbxo10fs/fs (blue fill) thymocyte subsets relative to Fbxo10+/+ control thymocytes (black line) for CD44 (A) or for CD69 (B). Results are representative of results obtained for other markers: CD25, CD69, PD1, CD3, etc. (C) Representative histogram overlays of Fbxo10+/+ (grey fill) or Fbxo10fs/fs (blue fill) splenic T cells relative to Fbxo10+/+ control cells (black line) showing CD62L (left 3 panels) or CD44 (right 3 panels). Results are representative of results obtained for other markers: CD25, CD62L, PD1, CD3, etc.
(TIF)
(A) Mean fluorescence intensity (MFI) for CD11b, Ly6G, CD62L, CD44 expression in spleen leukocyte subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. (B) Mean fluorescence intensity (MFI) for CD11b, Ly6G, CD62L, CD44 expression in bone marrow leukocyte subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. Each dot represents an individual biological replicate in Fbxo10+/+ (black), Fbxo10E54K/E54K (red) or Fbxo10fs/fs (blue) mice. Similar results were obtained for multiple protein markers (NK1.1, Ly6G, FSC, SSC-A, MHC II, etc.). Results are representative of n = 2 experiments on un-immunised mice 40–50 weeks old and similar results were obtained for n = 2 experiments on mice 10–20 weeks old, 7 days post-immunisation with SRBC. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wildtype controls with p < 0.05.
(TIF)
(TIF)
Acknowledgments
We thank the Garvan Institute ABR, GMG and Flow Cytometry facilities for expert animal husbandry, genotyping and cell sorting.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by NHMRC Grants APP1113904, APP1081858 and APP1108800. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bouillet P, Cory S, Zhang LC, Strasser A, Adams JM. Degenerative disorders caused by Bcl-2 deficiency prevented by loss of its BH3-only antagonist Bim. Dev Cell. 2001;1(5):645–53. 10.1016/s1534-5807(01)00083-1 . [DOI] [PubMed] [Google Scholar]
- 2.Nakayama K, Nakayama K, Negishi I, Kuida K, Sawa H, Loh DY. Targeted disruption of Bcl-2 alpha beta in mice: occurrence of gray hair, polycystic kidney disease, and lymphocytopenia. Proc Natl Acad Sci U S A. 1994;91(9):3700–4. 10.1073/pnas.91.9.3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S, et al. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci U S A. 1991;88(19):8661–5. 10.1073/pnas.88.19.8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335(6189):440–2. 10.1038/335440a0 . [DOI] [PubMed] [Google Scholar]
- 5.Veis DJ, Sorenson CM, Shutter JR, Korsmeyer SJ. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 1993;75(2):229–40. 10.1016/0092-8674(93)80065-m . [DOI] [PubMed] [Google Scholar]
- 6.Bakhshi A, Jensen JP, Goldman P, Wright JJ, McBride OW, Epstein AL, et al. Cloning the chromosomal breakpoint of t(14;18) human lymphomas: clustering around JH on chromosome 14 and near a transcriptional unit on 18. Cell. 1985;41(3):899–906. 10.1016/s0092-8674(85)80070-2 . [DOI] [PubMed] [Google Scholar]
- 7.Cleary ML, Smith SD, Sklar J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell. 1986;47(1):19–28. 10.1016/0092-8674(86)90362-4 . [DOI] [PubMed] [Google Scholar]
- 8.Tsujimoto Y, Croce CM. Analysis of the structure, transcripts, and protein products of bcl-2, the gene involved in human follicular lymphoma. Proc Natl Acad Sci U S A. 1986;83(14):5214–8. 10.1073/pnas.83.14.5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngan BY, Chen-Levy Z, Weiss LM, Warnke RA, Cleary ML. Expression in non-Hodgkin’s lymphoma of the bcl-2 protein associated with the t(14;18) chromosomal translocation. N Engl J Med. 1988;318(25):1638–44. 10.1056/NEJM198806233182502 . [DOI] [PubMed] [Google Scholar]
- 10.Weiss LM, Warnke RA, Sklar J, Cleary ML. Molecular analysis of the t(14;18) chromosomal translocation in malignant lymphomas. N Engl J Med. 1987;317(19):1185–9. 10.1056/NEJM198711053171904 . [DOI] [PubMed] [Google Scholar]
- 11.Yunis JJ, Oken MM, Kaplan ME, Ensrud KM, Howe RR, Theologides A. Distinctive chromosomal abnormalities in histologic subtypes of non-Hodgkin’s lymphoma. N Engl J Med. 1982;307(20):1231–6. 10.1056/NEJM198211113072002 . [DOI] [PubMed] [Google Scholar]
- 12.Iqbal J, Sanger WG, Horsman DE, Rosenwald A, Pickering DL, Dave B, et al. BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma. Am J Pathol. 2004;165(1):159–66. 10.1016/s0002-9440(10)63284-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ci W, Polo JM, Cerchietti L, Shaknovich R, Wang L, Yang SN, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113(22):5536–48. 10.1182/blood-2008-12-193037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito M, Novak U, Piovan E, Basso K, Sumazin P, Schneider C, et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2009;106(27):11294–9. 10.1073/pnas.0903854106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–47. 10.1056/NEJMoa012914 . [DOI] [PubMed] [Google Scholar]
- 16.Iqbal J, Neppalli VT, Wright G, Dave BJ, Horsman DE, Rosenwald A, et al. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. J Clin Oncol. 2006;24(6):961–8. 10.1200/JCO.2005.03.4264 . [DOI] [PubMed] [Google Scholar]
- 17.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109(10):3879–84. 10.1073/pnas.1121343109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017;171(2):481–94 e15. 10.1016/j.cell.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merino R, Ding L, Veis DJ, Korsmeyer SJ, Nunez G. Developmental regulation of the Bcl-2 protein and susceptibility to cell death in B lymphocytes. EMBO J. 1994;13(3):683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed JC. A day in the life of the Bcl-2 protein: does the turnover rate of Bcl-2 serve as a biological clock for cellular lifespan regulation? Leuk Res. 1996;20(2):109–11. 10.1016/0145-2126(95)00135-2 . [DOI] [PubMed] [Google Scholar]
- 21.Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, Hilton AA, Ellis S, et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128(6):1173–86. 10.1016/j.cell.2007.01.037 . [DOI] [PubMed] [Google Scholar]
- 22.Rooswinkel RW, van de Kooij B, de Vries E, Paauwe M, Braster R, Verheij M, et al. Antiapoptotic potency of Bcl-2 proteins primarily relies on their stability, not binding selectivity. Blood. 2014;123(18):2806–15. 10.1182/blood-2013-08-519470 . [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen TN, McKee A, Wang M, Kushnir E, White J, Refaeli Y, et al. Bim and Bcl-2 mutually affect the expression of the other in T cells. J Immunol. 2007;179(6):3417–24. 10.4049/jimmunol.179.6.3417 . [DOI] [PubMed] [Google Scholar]
- 24.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–67. 10.1146/annurev.cellbio.15.1.435 . [DOI] [PubMed] [Google Scholar]
- 25.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14(6):369–81. 10.1038/nrm3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiorazzi M, Rui L, Yang Y, Ceribelli M, Tishbi N, Maurer CW, et al. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proc Natl Acad Sci U S A. 2013;110(10):3943–8. 10.1073/pnas.1217271110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fielenbach N, Guardavaccaro D, Neubert K, Chan T, Li D, Feng Q, et al. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12(3):443–55. 10.1016/j.devcel.2007.01.018 . [DOI] [PubMed] [Google Scholar]
- 28.Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481(7379):90–3. 10.1038/nature10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Bouchlaka MN, Wolff J, Grindle KM, Lu L, Qian S, et al. FBXO10 deficiency and BTK activation upregulate BCL2 expression in mantle cell lymphoma. Oncogene. 2016;35(48):6223–34. 10.1038/onc.2016.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh SH, Rosenquist R. Immunoglobulin gene analysis of mature B-cell malignancies: reconsideration of cellular origin and potential antigen involvement in pathogenesis. Med Oncol. 2005;22(4):327–41. 10.1385/MO:22:4:327 . [DOI] [PubMed] [Google Scholar]
- 31.McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57(1):79–88. 10.1016/0092-8674(89)90174-8 . [DOI] [PubMed] [Google Scholar]
- 32.Strasser A, Harris AW, Vaux DL, Webb E, Bath ML, Adams JM, et al. Abnormalities of the immune system induced by dysregulated bcl-2 expression in transgenic mice. Curr Top Microbiol Immunol. 1990;166:175–81. 10.1007/978-3-642-75889-8_22 . [DOI] [PubMed] [Google Scholar]
- 33.McDonnell TJ, Korsmeyer SJ. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14; 18). Nature. 1991;349(6306):254–6. 10.1038/349254a0 . [DOI] [PubMed] [Google Scholar]
- 34.Strasser A, Harris AW, Cory S. E mu-bcl-2 transgene facilitates spontaneous transformation of early pre-B and immunoglobulin-secreting cells but not T cells. Oncogene. 1993;8(1):1–9. . [PubMed] [Google Scholar]
- 35.Ogilvy S, Metcalf D, Print CG, Bath ML, Harris AW, Adams JM. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci U S A. 1999;96(26):14943–8. 10.1073/pnas.96.26.14943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egle A, Harris AW, Bath ML, O’Reilly L, Cory S. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 2004;103(6):2276–83. 10.1182/blood-2003-07-2469 . [DOI] [PubMed] [Google Scholar]
- 37.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–50. 10.1038/nature10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc. 2014;9(8):1956–68. 10.1038/nprot.2014.134 . [DOI] [PubMed] [Google Scholar]
- 39.Xu X, Powell DW, Lambring CJ, Puckett AH, Deschenes L, Prough RA, et al. Human MCS5A1 candidate breast cancer susceptibility gene FBXO10 is induced by cellular stress and correlated with lens epithelium-derived growth factor (LEDGF). Mol Carcinog. 2014;53(4):300–13. 10.1002/mc.21977 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo F, Luo Y, Jiang X, Lu X, Roberti D, Lossos C, et al. Recent BCR stimulation induces a negative autoregulatory loop via FBXO10 mediated degradation of HGAL. Leukemia. 2020;34(2):553–66. 10.1038/s41375-019-0579-5 . [DOI] [PubMed] [Google Scholar]
- 41.Romero-Camarero I, Jiang X, Natkunam Y, Lu X, Vicente-Duenas C, Gonzalez-Herrero I, et al. Germinal centre protein HGAL promotes lymphoid hyperplasia and amyloidosis via BCR-mediated Syk activation. Nat Commun. 2013;4:1338. 10.1038/ncomms2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schenten D, Egert A, Pasparakis M, Rajewsky K. M17, a gene specific for germinal center (GC) B cells and a prognostic marker for GC B-cell lymphomas, is dispensable for the GC reaction in mice. Blood. 2006;107(12):4849–56. 10.1182/blood-2005-10-4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritzen D, Kuechler A, Grimmel M, Becker J, Peters S, Sturm M, et al. De novo FBXO11 mutations are associated with intellectual disability and behavioural anomalies. Hum Genet. 2018;137(5):401–11. 10.1007/s00439-018-1892-1 . [DOI] [PubMed] [Google Scholar]
- 44.Gregor A, Sadleir LG, Asadollahi R, Azzarello-Burri S, Battaglia A, Ousager LB, et al. De Novo Variants in the F-Box Protein FBXO11 in 20 Individuals with a Variable Neurodevelopmental Disorder. Am J Hum Genet. 2018;103(2):305–16. 10.1016/j.ajhg.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardisty-Hughes RE, Tateossian H, Morse SA, Romero MR, Middleton A, Tymowska-Lalanne Z, et al. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum Mol Genet. 2006;15(22):3273–9. 10.1093/hmg/ddl403 . [DOI] [PubMed] [Google Scholar]
- 46.Schneider C, Kon N, Amadori L, Shen Q, Schwartz FH, Tischler B, et al. FBXO11 inactivation leads to abnormal germinal-center formation and lymphoproliferative disease. Blood. 2016;128(5):660–6. 10.1182/blood-2015-11-684357 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edison N, Curtz Y, Paland N, Mamriev D, Chorubczyk N, Haviv-Reingewertz T, et al. Degradation of Bcl-2 by XIAP and ARTS Promotes Apoptosis. Cell Rep. 2017;21(2):442–54. 10.1016/j.celrep.2017.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Fernandez M, Kissel H, Brown S, Gorenc T, Schile AJ, Rafii S, et al. Sept4/ARTS is required for stem cell apoptosis and tumor suppression. Genes Dev. 2010;24(20):2282–93. 10.1101/gad.1970110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Staudt LM. Protein ubiquitination in lymphoid malignancies. Immunol Rev. 2015;263(1):240–56. 10.1111/imr.12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CRISPR/Cas9-engineered mice were produced as described in the materials and methods. (A-C) Sanger sequencing traces of a portion of the Fbxo10 gene encoding amino acids 49 to 64 of FBXO10: (A) wild type sequence showing the target site of the sgRNA used and its associated PAM and highlighting the E54 codon (B) sequence from a mouse heterozygous for the E54K mutation (together with the PAM inactivating mutation in the T53 codon) (C) sequence from a mouse heterozygous for a four base pair deletion (GCCG, highlighted) resulting in a reading frame shift after the E54 codon. Note that the Sanger sequencing read is from right to left, and the two frameshifted sequences that are superimposed downstream of the deletion are shown above. (D) High resolution melt curve analysis plots of DNA from wild-type and mutant mice, showing the alternative fluorescence profiles resulting from heterozygous or homozygous E54K (left) or C55SfsTer55 (right) mutations.
(TIF)
(A) Mean fluorescence intensity (MFI) for IgM, IgD, CD93, CD86 expression in splenic B cell subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. (B) MFI for IgM, IgD, CD24, CD43 expression in bone marrow B cell subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. Each dot represents an individual biological replicate in Fbxo10+/+ (black), Fbxo10E54K/E54K (red) or Fbxo10fs/fs (blue) mice. Similar results were obtained for multiple protein markers (CD19, CD21/35, CD23, CD24, CD43, CD86, etc.). Results are representative of n = 2 experiments on un-immunised mice 40–50 weeks old and similar results were obtained for n = 2 experiments on mice 10–20 weeks old, 7 days post-immunisation with SRBC. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wildtype controls with p < 0.05.
(TIF)
(A,B) Representative histogram overlays of Fbxo10+/+ (grey fill) or Fbxo10fs/fs (blue fill) thymocyte subsets relative to Fbxo10+/+ control thymocytes (black line) for CD44 (A) or for CD69 (B). Results are representative of results obtained for other markers: CD25, CD69, PD1, CD3, etc. (C) Representative histogram overlays of Fbxo10+/+ (grey fill) or Fbxo10fs/fs (blue fill) splenic T cells relative to Fbxo10+/+ control cells (black line) showing CD62L (left 3 panels) or CD44 (right 3 panels). Results are representative of results obtained for other markers: CD25, CD62L, PD1, CD3, etc.
(TIF)
(A) Mean fluorescence intensity (MFI) for CD11b, Ly6G, CD62L, CD44 expression in spleen leukocyte subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. (B) Mean fluorescence intensity (MFI) for CD11b, Ly6G, CD62L, CD44 expression in bone marrow leukocyte subsets of Fbxo10+/+, Fbxo10fs/fs, Fbxo10E54K/E54K mice 40–50 weeks old. Each dot represents an individual biological replicate in Fbxo10+/+ (black), Fbxo10E54K/E54K (red) or Fbxo10fs/fs (blue) mice. Similar results were obtained for multiple protein markers (NK1.1, Ly6G, FSC, SSC-A, MHC II, etc.). Results are representative of n = 2 experiments on un-immunised mice 40–50 weeks old and similar results were obtained for n = 2 experiments on mice 10–20 weeks old, 7 days post-immunisation with SRBC. Statistical analysis: t-test corrected for multiple comparisons using the Holm-Sidak method yielded no evidence for significant differences between mutants and wildtype controls with p < 0.05.
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.