Abstract
Glycolytic metabolism is closely involved in physiological homeostasis and pathophysiological states. Among glycolytic enzymes, phosphoglycerate mutase (PGAM) has been reported to exert certain physiological role in vitro, whereas its impact on glucose metabolism in vivo remains unclear. Here, we report the characterization of Pgam1 knockout mice. We observed that homozygous knockout mice of Pgam1 were embryonic lethal. Although we previously reported that both PGAM-1 and -2 affect global glycolytic profile of cancers in vitro, in vivo glucose parameters were less affected both in the heterozygous knockout of Pgam1 and in Pgam2 transgenic mice. Thus, the impact of PGAM on in vivo glucose metabolism is rather complex than expected before.
Introduction
Glucose is utilized as energy source, from bacterias to higher eukaryotes. Glycolysis provides energy supply in the form of ATP through oxidation of carbon atoms in glucose. As it also supports the synthesis of macromolecules [1], glycolysis is evolutionally conserved as one of vitally essential metabolisms.
Dysregulation in glycolysis, either the pathological enhancement or impairment, is closely related to human disease states. For example, most of cancer cells display enhanced glycolytic feature, known as the Warburg effect. On the other hand, impaired glycolysis is much involved in dysfunction in various tissues and degenerative disorders [2,3]. In humans and other vertebrates, glucose is delivered to various tissues through circulating blood. Even during prolonged fasting (no calorie feeding), blood sugar levels are minimally maintained, which enables various tissues and cells, e.g. neurons in brain, to utilize glucose from circulating blood. The patients with impaired uptake of glucose into cells display higher glucose levels in blood, known as diabetes or impaired glucose metabolism. Indeed, diabetes are observed both in human glycolytic enzyme deficiencies [4,5] and in knockout (KO) mice for glycolytic enzymes [6–9].
Glycolytic enzyme phosphoglycerate mutase (PGAM) converts 3-phosphoglycerate into 2-phosphoglycerate as an isomerase [10]. Two isoforms of PGAM (PGAM1 and PGAM2) distribute to overlapping but distinct tissues in vivo. However, both isoforms display the significant similarity in their sequences (79% identity) and enzymatic activities in vitro [11–13].
Besides its role in glycolysis, PGAM would play a key role connecting glycolysis to physiological homeostasis. PGAM is post-translationally regulated by several modification, including the phosphorylation, acetylation and ubiquitination by environmental stimuli or under stress conditions [13–15]. Moreover, PGAM activity modulates mitochondrial function [16] and the pentose phosphate pathway [17], implicating its involvement in the defense against oxidative stress [12]. Noteworthy, both PGAM-1 and -2 support cancerous cell growth through its nonenzymatic function. PGAM interacts with Chk1 kinase under oncogenic Ras expression, followed by boosts of glycolytic profiles [18].
However, the global impacts of PGAM in vivo is still unclear. Here we characterized the profiles of glucose metabolisms in mice with ablation of Pgam1 or global overexpression of Pgam2 in vivo. Homozygous knockout of Pgam1 is embryonic lethal, while its global heterozygous knockout mice displayed comparable glucose parameters in vivo, compared to those in wild type. Moreover, global overexpression of Pgam2 unlikely affects the in vivo profile for glucose metabolisms. Thus, the impact of PGAM on in vivo glucose metabolism is rather complex than expected before.
Materials and methods
Ethical statements
All procedures for animal experiments were approved and conducted in accordance with the principles and guidelines of the Animal Care and Use Committees of Kyoto University Graduate School of Medicine (Med Kyo 15556 and Med Kyo 15558).
Generation of mouse models
Pgam2-Tg mice were generated previously [13]. Pgam2-Tg is a strain of transgenic C57BL/6 mice that overexpresses the Pgam2 with a 3xFLAG tag under the cytomegalovirus immediate-to-early enhancer element and chicken β-actin promoter (CAG) [19].
Generation of the conditional knockout mouse of Pgam1 is previously reported [18]. For the study of whole body Pgam1 knockout mice, Pgam1+/- mice were generated by crossing between Pgam1flox/+ mice and CAG-Cre Tg mice, which ubiquitously express Cre recombinase under the CAG promoter. Subsequently, Pgam1+/-; CAG-Cre micewere crossed with wild-type mice and the genotypes of progenitor mice were analyzed by using PCR. Resulting Pgam1+/- mice without CAG-Cre transgene were applied for further experiment.
Mouse experiments
Intraperitoneal glucose tolerance test (IPGTT) was performed as previously described [20]. After 16 h of fasting, 1.5 g/kg body weight glucose was intraperitoneally injected into the mice. The blood glucose level of the mice was measured at 0, 15, 30, 60, and 120 min using a Glutest sensor (Sanwa Kagaku Kenkyusyo Co., Ltd. Aichi, Japan). For the high-fat diet study, 8-week-old male mice were fed diets with 60% of calories from fat (high-fat diet, Research Diets D12492) for 10 weeks. Body weight was measured every week. Finally, IPGTT was performed when the mice 18 weeks old. Blood tests for several biological markers were performed by Unitech Co. (Kashiwa, Japan). Muscle muss and body fat mass in mice were measured using magnetic resonance imaging (Latheta LCT-100, Hitachi).
Measurement of glycolytic enzyme activity
PGAM enzymatic activity was measured as previously described [18]. Briefly, mice tissues were homogenized by BioMasher II (Nippi, Tokyo, Japan) in lysis buffer (50 mM Tris–HCl [pH 8.0], 2 mM DTT, 2 mM EDTA, and 1% Triton X-100). Tissue lysates were incubated in reaction buffer (100 mM Tris–HCl [pH 8.0], 100 mM KCl, 0.5 mM EDTA, 2 mM MgCl2, 0.2 mM NADH, 3 mM ADP, and 10 μM 2,3-diphosphoglycerate) with enzyme mixture (0.6 U lactate dehydrogenase, 0.5 U pyruvate kinase, and 0.1 U enolase). The levels of NADH were monitored at 37°C, from adding to 1 mM 3-phosphoglyceric acid. Enzymatic activity was measured as NAD+ release.
RNA analysis
Total RNA was extracted with TRIzol (Invitrogen). cDNA pools were prepared by the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). Real-time quantitative PCR was carried out using Thunderbird SYBR qPCR mix (Toyobo). The level of expression of target gene was monitored by the Thermal Cycler Dice Real-Time system (Takara Bio., Kusatsu, Japan) and. Gene expression levels were normalized to Rpl13a mRNA and presented as values relative to wild-type mice.
The primer sequences are shown as below. Rpl13a:Fw 5’- TGC TGC TCT CAA GGT TGT TCG -3’, Re 5’-GCC TTT TCC TTC CGT TTC TCC-3’, Hk1:Fw 5’-AAG AAT GGC CTC TCC CGG-3’, Re 5’-CGC CGA GAT CCA GTG CAA TG-3’, Hk2:Fw 5’-ATA TGG TTG CCT CAT CTT GG-3’, Re 5’-CTC CCT CCC TCC CAA TG-3’, Hk3:Fw 5’-ATT CCT GGA TGC ATA CCC CGT -3’, Re 5’-GCC GCT GCA CCT AAA ACC TTT -3’, Gpi:Fw 5’-CCA ATG CAG AGA CAG CAA AGG-3’, Re 5’-CAC TTT GGC CGT GTT CGT AGA-3’, Pfkl:Fw 5’-GCT GCA ATG GAG TTG TG-3, Re 5’-GTA GCC AGG TAGC CAC AG-3’, Pfkm:Fw 5’-TGG AGC GAC TTG CTG AAT GAT -3’, Re 5’-TCA TTG TCG ATT GAG CCA ACC -3’, Aldo a:Fw 5’-CTG GCC ATC ATG GAA AAT GC-3’, Re 5’-TCA AGT CAT GGT CCC CAT CAG-3’, Aldo b:Fw 5’-ATC GGC GGA GTG ATC CTT TT-3’, Re 5’-TCC AAC TTG ATG CCC ACC A -3’, Tpi:Fw 5’-TGC CAA ACA ATG AGC ACT GC-3’, Re 5’-ATC AGA AGC ATG TGA CCG GTG-3’, Gapdh:Fw 5’-AGC CTC GTC CCG TAG ACA AAA-3’, Re 5’-TGG CAA CAA TCT CCA CTT TGC-3’, Pgk1:Fw 5’-TTT GGA CAA GCT GGA CGT GAA-3’, Re 5’-GCT TGG AAC AGC AGC CTT GAT-3’, Pgam1:Fw 5’-GTT GCG AGA TGC TGG CTA TGA-3’, Re 5’-CAC ATC TGG TCA ATG GCA TCC-3’, Pgam2:Fw 5’-TGG AAT GAG GAG ATC GCA CCT -3’, Re 5’-TCG GAC ATC CCT TCC AGA TGT -3’,Eno1:Fw 5’-TAT TGC GCC TGC TCT GGT TAG-3’, Re 5’-GGA TGG CAT TTG CAC CAA AT-3’, Eno3:Fw 5’-GGA GAA GAA GGC CTG CAA TTG -3’, Re 5’-CCC AGC CAT TAG ATT GTG CAA -3’,Pkm1:Fw 5’-CTG TTT GAA GAG CTT GTG GCG -3’, Re 5’-CTG CTA AAC ACT TAT AAG AGG CC -3’.
Statistical analysis
All data are expressed as the mean ± standard error of the mean (SEM) Comparisons between two independent groups were analyzed using an unpaired Student’s two-tailed t-test.
Results
Homozygous knockout mice of Pgam1 were embryonic lethal
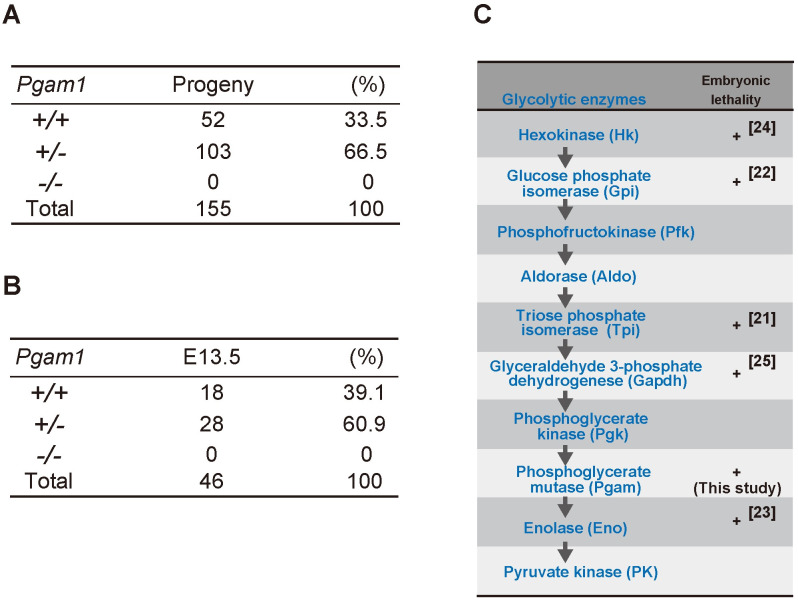
To analyze the physiological impact of PGAM inactivation in vivo, conditional KO mice for Pgam1 were established [18]. Global Pgam1 heterozygous KO (Pgam1+/−) mice were generated through crosses between CAG-Cre Tg mice and Pgam1flox/+ mice. Among 155 progenies obtained after mating between Pgam1+/− mice, the ratio of wild-type; heterozygous KO; and homozygous KO was 52:103:0 (Fig 1A). Thus, Pgam1 homozygous KO (Pgam1−/−) mice were embryonic lethal, consistent with the lack of observed Pgam1−/− embryos at embryonic day 13.5 (E13.5) (Fig 1B). The early embryonic lethality of Pgam1−/− mice validated the vital significance of PGAM in vivo, like the other glycolytic enzymes [21–25] (Fig 1C).
Fig 1. Embryonic lethality of homozygous knockout mice of Pgam1.
(A) Summary of Pgam1 genotypes for 155 progenies from crosses between Pgam1+/− mice. +/+; wild-type, +/−; heterozygous KO, −/−; homozygous KO. (B) Summary of Pgam1 genotypes for embryo from crossing between Pgam1+/− mice at embryonic day 13.5 (E13.5). (C) Schematic diagram of glycolytic pathway. (+) indicates embryonic lethality in homozygous knockout mice, as reported previously (ref. [21–25]).
Heterozygous Pgam1 knockout mice were viable with reduced PGAM acitvity
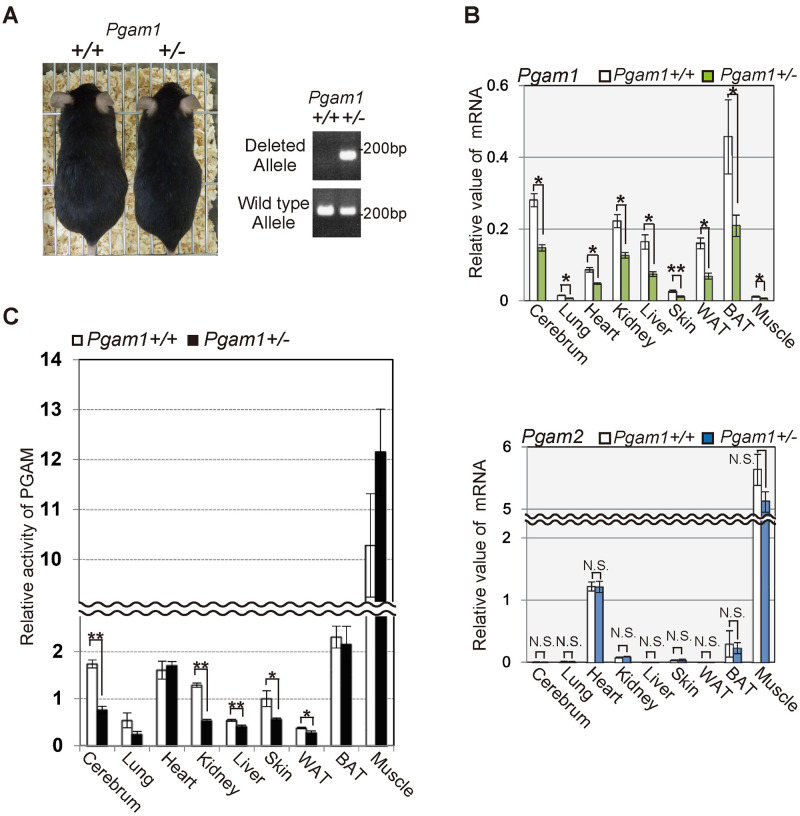
Although Pgam1 homozygous KO mice were embryonic lethal, Pgam1+/− mice are viable (Fig 2A). The mRNA levels of Pgam1 were decreased by approximately 50% in several tissues of Pgam1+/− mice (Fig 2B, upper panel), whereas the levels of Pgam2 mRNA were not affected in the examined tissues from these mice (Fig 2B, lower panel). PGAM1 and PGAM2 exhibit overlapping but distinct tissue distributions; PGAM1 is dominantly expressed in several tissues including the liver, white adipose tissue, aorta, and brain, whereas PGAM2 is found mainly in the muscle. The other tissues including the lung, heart, skin, and bone express both isoforms [11,13]. Consistently, the enzymatic activities of PGAM in Pgam1+/− mice were significantly decreased in several tissues including the cerebrum, kidney, and skin, or modestly decreased in the lung, liver, and white adipose tissue (WAT), but not in the heart, brown adipose tissue (BAT), and muscle, compared to those in control (Fig 2C). As several posttranslational modification for PGAM is heavily operating [26], unknown compensatory mechanisms by such modification may preserve its enzymatic activity in Pgam1 heterozygous knockout mice.
Fig 2. Heterozygous Pgam1 knockout mice were viable.
(A) Pgam1+/− mice are viable. Representative images of wild-type and Pgam1+/− mice at 35 weeks (left panel). Genomic PCR in indicated mice was performed to verify the wild-type or deleted alleles for Pgam1 (right panels). (B and C) Assessment of PGAM expression levels and enzymatic activity in various tissues from wild-type (n = 5) and Pgam1+/− mice (n = 5) at 35 weeks. (B) Pgam1+/− mice displayed approximately 50% reduction for Pgam1 mRNA levels, compared to those from wild-type mice (upper panel), while Pgam2 mRNA levels did not show significant difference (lower panel). (C) The PGAM activity were measured in indicated tissues. Each data is shown as relative values against those in control. +/+; wild-type, +/-; heterozygous knockout.
In vivo parameters for glucose metabolism were less affected by heterozygous Pgam1 knockoout
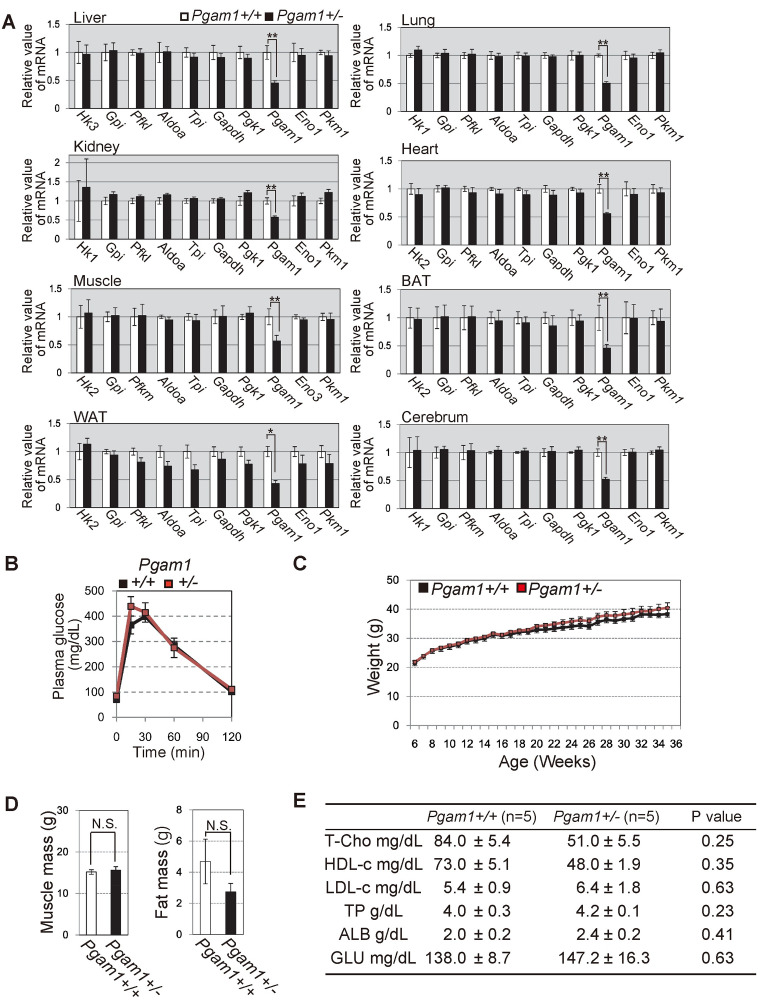
Recently we reported that PGAM status deeply affects glycolytic profiles in cancer cells with oncogenic Ras expression, but not in standard cells in vitro [18]. We examined the profiles of glycolytic mRNAs in the tissues of Pgam1+/− mice. We observed no significant difference in the profiles of glycolytic mRNAs between wild-type and heterozygous knockout of Pgam1 mice (Fig 3A).
Fig 3. Characterization of Pgam1 heterozygous KO mice.
(A) The comparison of glycolytic mRNAs profiles between wild-type and Pgam1+/− mice. The extracts from indicated tissues were tested. (B) Intraperitoneal glucose test (IPGTT) was performed in wild-type (n = 5) and Pgam1+/− mice (n = 4). (C-E) Several physiological parameters in wild-type (n = 5) and Pgam1+/− mice (n = 5). (C) Body weight were measured every week for 30 weeks. (D) Measurements of muscle mass and visceral-fat mass by computerized tomography at 35 weeks. (E) Total cholesterol (T-Cho), high-density lipoprotein cholesterol (HDL-c), low-density lipoprotein cholesterol (LDL-c), Total protein, (TP), albumin (ALB) and blood glucose (Glu) were determined in the blood plasma of 35 weeks mice. +/+; wild-type, +/-; heterozygous knockout. Data represent the mean ± SEM. Single-asterisks (*) and double-asterisk (**) indicate statistical significance of p<0.05 and p<0.005, respectively, Student’s t-test.
It was reported that brain-specific knockout of Glut4 in vivo, glucose transporter in glycolysis, displayed impaired glucose tolerance and insulin sensitivity [27]. As glycolysis is essential energy source in all tissues, it is possible that the impaired glycolysis in the limited organs could affect glucose metabolisms in vivo [28]. To examine this possibility, we next examined glucose metabolism of Pgam1+/− mice in vivo, as mice with one allele of glucokinase displayed diabetic phenotype [8]. In comparison to wild-type mice, Pgam1+/− mice displayed similar profiles for in vivo glucose metabolism (intraperitoneal glucose tolerance test (IPGTT) and serum glucose level), in addition to body weight, visceral-fat weight, muscle mass, total-cholesterol, and other biochemical blood markers (Fig 3B–3E). Collectively, in vivo parameters for glucose metabolism were less affected by heterozygous Pgam1 knockout.
Global overexpression of Pgam2 does not affect glucose tolerance in vivo
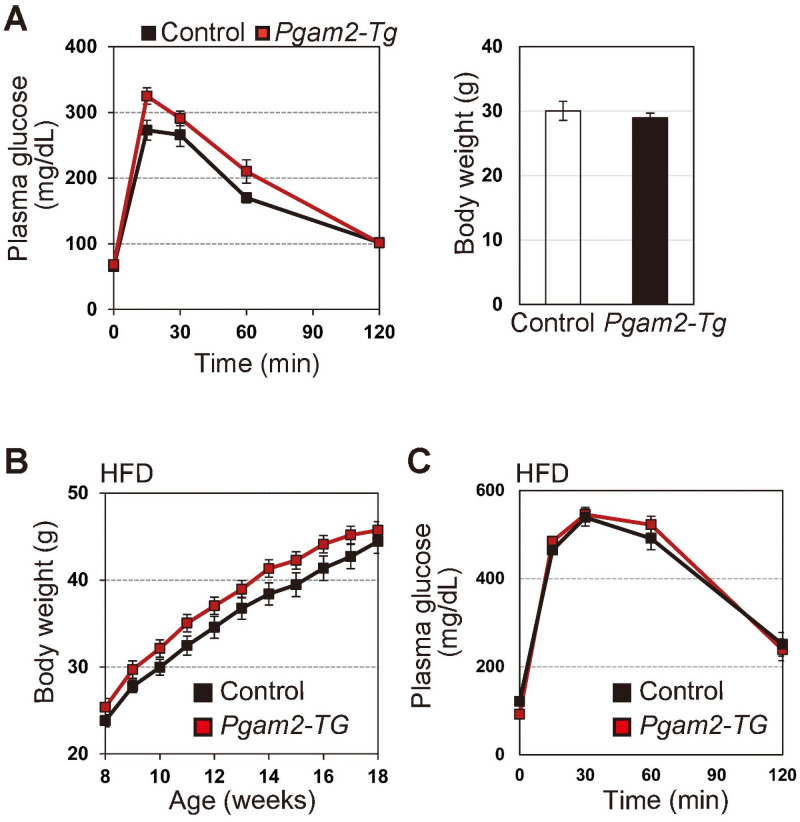
Next, we evaluated the impact of PGAM overexpression in vivo. We previously reported that heart-specific Pgam2-transgenic (Tg) mice displayed almost normal glycolytic features in the heart [16]. However, it remains unclear whether global PGAM overexpression affects glycolytic profiles in vivo. As it has been demonstrated that the overexpression of either PGAM isoform confers similar physiological impact [11–13], we evaluated the in vivo parameters of glucose metabolism in Pgam2-Tg mice, in which a Pgam2-FLAG transgene is driven by the CAG promoter. Pgam2-Tg mice exhibited a significant increase of PGAM protein in the whole body [18]. Pgam2-Tg mice also displayed their similar features with respect to body weight and IPGTT both under physiological (Fig 4A) and high fat-induced obesity conditions (Fig 4B and 4C), compared with those of wild-type mice. Thus, the parameters for glucose metabolism are less affected by PGAM overexpressing status in vivo under physiological conditions.
Fig 4. Glycolytic profiles in PGAM2-Tg mice.
(A) Intraperitoneal glucose test (IPGTT) was performed in control (wild-type) (n = 10) and Pgam2-Tg mice (n = 8) (Left panel). Body weight was measured at 18 weeks (Right panel). (B and C) High-fat diet (HFD) protocol was performed to induce obesity in control (n = 9) and Pgam2-Tg mice (n = 7). (B) Body weight was monitored during the protocol (left panel). (C) IPGTT was performed in HFD-fed obese mice at 18 weeks (right panel). Data represent the mean ± SEM.
Discussion
Here we reported on the profiles of in vivo glucose parameters in genetic models of glycolytic enzyme PGAM. Consistent to the findings in the ablation of other glycolytic enzymes, the embryonic lethality of homozygous knockout of Pgam1 supports the notion that glycolysis is vitally essential metabolism in vivo. Unexpectedly, however, Pgam1+/− and Pgam2 -Tg mice displayed comparable profiles of in vivo glucose parameters, as far as we tested.
Several genetic mutations of PGAM with its impaired enzymatic activity, have been reported in human PGAM deficient patients [29–31]. However, little is reported on the link between human PGAM deficiency and diabetes, except one case report on the adult patient of PGAM deficiency (51 years old) accompanied with type 2 diabetes [32]. Thus, it is still controversial whether human PGAM deficiency causes diabetes or not. Here, we observed that the in vivo parameters for glucose metabolism are less affected in Pgam1+/− mice. Moreover, Pgam2 -Tg in normal and high-fat diet conditions displayed the normal glucose tolerance. However, as Pgam1 +/- mice retained substantial PGAM enzymatic activity in several tissue including liver, muscle, WAT and BAT, it would be worthy to investigate tissue-specific Pgam1 homozygous knockout or Pgam2 knockout model in the future.
Different from our observations in genetic model mice for PGAM, several reports suggest that impaired glycolysis results in diabetes or impaired glucose metabolism both in human patients [4,5] and in knockout (KO) mice for the other glycolytic enzymes [8,9]. The discrepancy between our study and the inactivation of the other glycolytic enzymes could be possibly due to the different impact of individual glycolytic enzymes on glycolytic pathway. Interestingly, the catalytic reaction of PGAM is not rate-limiting step during glycolytic pathway under standard conditions, even when PGAM activity is reduced by approximately half of normal mean value both in human PGAM deficient patient [32] and in Pgam1 heterozygous knockout mice (this study) [18]. As PGAM enzyme activity is most prominent among glycolytic enzymes [30], the residual PGAM activity may be sufficient to maintain glycolysis in human or mice PGAM impairment. Interestingly, overall glycolysis is much affected if majority of PGAM proteins were degraded via ubiquitin-dependent proteolysis of PGAM under senescence-inducing stress [13], rather consistent to the embryonic lethality of Pgam1−/− mice (in this study). In this setting, PGAM would be rate-limiting in glycolysis. Moreover, we recently reported that the global enhancement of glycolytic profiles in cancerous cells, the Warburg effect, is provoked in vitro by the cooperation of PGAM with Chk1 kinase under oncogenic Ras expression [18]. Besides cancers, it is well known that the active proliferation of T cells after immunological stimuli or stress is accompanied by the enhanced glycolysis [33]. Pgam1 deficient T cells are impaired in maintenance of such glycolytic enhancement, followed by immunological dysfunction [34]. Thus, the impact of PGAM on glycolytic regulation would be cellular- or tissue-context dependent. Indeed, the boosts of glyclolytic mRNAs were observed in Pgam2-Tg mice under chemical carcinogenic protocol [18], while those in Pgam2-Tg mice under standard conditions were comparable to control [18]. It is possible that the modulation of PGAM proteins would be required in vivo for its physiological impact, as several post-translational regulations of PGAM were reported in vitro [26].
Further investigation might provide valuable clue to understand the impact of PGAM on glucose metabolisms in vivo.
Supporting information
(XLSX)
Acknowledgments
We thank all the staff of Geriatric unit and of the Department of Diabetes, Endocrinology and Nutrition at Kyoto University for their cooperation; Dr. Jun-ichi Miyazaki for the pCAGGS vector. The animal experiments were performed at the Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University.
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
This work was supported in part by grants from the Japan Society for the Promotion of Science (Grants No. 26310103 to HK and No. 15K19283 to HK), and by the Japan Agency for Medical Research and Development (AMED), Core Research for Evolutional Science and Technology (CREST JP17gm0610002h0306 to HK). HK; Hiroshi Kondoh.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor M, Wallhaus TR, Degrado TR, Russell DC, Stanko P, et al. An evaluation of myocardial fatty acid and glucose uptake using PET with [18F]fluoro-6-thia-heptadecanoic acid and [18F]FDG in Patients with Congestive Heart Failure. J Nucl Med. 2001;42(1):55–62. [PubMed] [Google Scholar]
- 3.Goyal MS, Vlassenko AG, Blazey TM, Su Y, Couture LE, et al. Loss of Brain Aerobic Glycolysis in Normal Human Aging. Cell Metab. 2017;26(2):353–360 e353. 10.1016/j.cmet.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ristow M, Vorgerd M, Mohlig M, Schatz H, Pfeiffer A. Deficiency of phosphofructo-1-kinase/muscle subtype in humans impairs insulin secretion and causes insulin resistance. J Clin Invest. 1997;100(11):2833–2841. 10.1172/JCI119831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Njolstad PR, Sovik O, Cuesta-Munoz A, Bjorkhaug L, Massa O, et al. Neonatal diabetes mellitus due to complete glucokinase deficiency. N Engl J Med. 2001;344(21):1588–1592. 10.1056/NEJM200105243442104 [DOI] [PubMed] [Google Scholar]
- 6.Guillam MT, Hummler E, Schaerer E, Yeh JI, Birnbaum MJ, et al. Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2. Nat Genet. 1997;17(3):327–330. 10.1038/ng1197-327 [DOI] [PubMed] [Google Scholar]
- 7.Richard AM, Webb DL, Goodman JM, Schultz V, Flanagan JN, et al. Tissue-dependent loss of phosphofructokinase-M in mice with interrupted activity of the distal promoter: impairment in insulin secretion. Am J Physiol Endocrinol Metab. 2007;293(3):E794–801. 10.1152/ajpendo.00168.2007 [DOI] [PubMed] [Google Scholar]
- 8.Grupe A, Hultgren B, Ryan A, Ma YH, Bauer M, et al. Transgenic knockouts reveal a critical requirement for pancreatic beta cell glucokinase in maintaining glucose homeostasis. Cell. 1995;83(1):69–78. 10.1016/0092-8674(95)90235-x [DOI] [PubMed] [Google Scholar]
- 9.Abel ED, Peroni O, Kim JK, Kim YB, Boss O, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409(6821):729–733. 10.1038/35055575 [DOI] [PubMed] [Google Scholar]
- 10.Rodwell VW, Towne JC, Grisolia S. The kinetic properties of yeast and muscle phosphoglyceric acid mutase. J Biol Chem. 1957;228(2):875–890. [PubMed] [Google Scholar]
- 11.Zhang J, Yu L, Fu Q, Gao J, Xie Y, et al. Mouse phosphoglycerate mutase M and B isozymes: cDNA cloning, enzyme activity assay and mapping. Gene. 2001;264(2):273–279. 10.1016/s0378-1119(00)00597-7 [DOI] [PubMed] [Google Scholar]
- 12.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65(1):177–185. [PubMed] [Google Scholar]
- 13.Mikawa T, Maruyama T, Okamoto K, Nakagama H, Lleonart ME, et al. Senescence-inducing stress promotes proteolysis of phosphoglycerate mutase via ubiquitin ligase Mdm2. J Cell Biol. 2014;204(5):729–745. 10.1083/jcb.201306149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalom-Barak T, Knaus UG. A p21-activated kinase-controlled metabolic switch up-regulates phagocyte NADPH oxidase. J Biol Chem. 2002;277(43):40659–40665. 10.1074/jbc.M206650200 [DOI] [PubMed] [Google Scholar]
- 15.Xu Y, Li F, Lv L, Li T, Zhou X, et al. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014;74(13):3630–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuda J, Niizuma S, Shioi T, Kato T, Inuzuka Y, et al. Persistent overexpression of phosphoglycerate mutase, a glycolytic enzyme, modifies energy metabolism and reduces stress resistance of heart in mice. PLoS One. 2013;8(8):e72173. 10.1371/journal.pone.0072173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22(5):585–600. 10.1016/j.ccr.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikawa T, Shibata E, Shimada M, Ito K, Ito T, et al. Phosphoglycerate Mutase Cooperates with Chk1 Kinase to Regulate Glycolysis. iScience. 2020;23(7):101306. 10.1016/j.isci.2020.101306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. 10.1016/0378-1119(91)90434-d [DOI] [PubMed] [Google Scholar]
- 20.Luo J, Sobkiw CL, Hirshman MF, Logsdon MN, Li TQ, et al. Loss of class IA PI3K signaling in muscle leads to impaired muscle growth, insulin response, and hyperlipidemia. Cell Metab. 2006;3(5):355–366. 10.1016/j.cmet.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 21.Merkle S, Pretsch W. Characterization of triosephosphate isomerase mutants with reduced enzyme activity in Mus musculus. Genetics. 1989;123(4):837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West JD. A genetically defined animal model of anembryonic pregnancy. Hum Reprod. 1993;8(8):1316–1323. 10.1093/oxfordjournals.humrep.a138249 [DOI] [PubMed] [Google Scholar]
- 23.Couldrey C, Carlton MB, Ferrier J, Colledge WH, Evans MJ. Disruption of murine alpha-enolase by a retroviral gene trap results in early embryonic lethality. Dev Dyn. 1998;212(2):284–292. [DOI] [PubMed] [Google Scholar]
- 24.Heikkinen S, Pietila M, Halmekyto M, Suppola S, Pirinen E, et al. Hexokinase II-deficient mice. Prenatal death of homozygotes without disturbances in glucose tolerance in heterozygotes. J Biol Chem. 1999;274(32):22517–22523. 10.1074/jbc.274.32.22517 [DOI] [PubMed] [Google Scholar]
- 25.Pretsch W, Favor J. Genetic, biochemical, and molecular characterization of nine glyceraldehyde-3-phosphate dehydrogenase mutants with reduced enzyme activity in Mus musculus. Mamm Genome. 2007;18(10):686–692. 10.1007/s00335-007-9055-z [DOI] [PubMed] [Google Scholar]
- 26.Mikawa T, LL ME, Takaori-Kondo A, Inagaki N, Yokode M, et al. Dysregulated glycolysis as an oncogenic event. Cell Mol Life Sci. 2015;72(10):1881–1892. 10.1007/s00018-015-1840-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reno CM, Puente EC, Sheng Z, Daphna-Iken D, Bree AJ, Routh VH, et al. Brain GLUT4 Knockout Mice Have Impaired Glucose Tolerance, Decreased Insulin Sensitivity, and Impaired Hypoglycemic Counterregulation. Diabetes. 2017;66(3):587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerich JE. Role of the kidney in normal glucose homeostasis and in the hyperglycaemia of diabetes mellitus: therapeutic implications. Diabet Med. 2010;27(2):136–42. 10.1111/j.1464-5491.2009.02894.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadjigeorgiou GM, Kawashima N, Bruno C, Andreu AL, Sue CM, et al. Manifesting heterozygotes in a Japanese family with a novel mutation in the muscle-specific phosphoglycerate mutase (PGAM-M) gene. Neuromuscul Disord. 1999;9(6–7):399–402. 10.1016/s0960-8966(99)00039-5 [DOI] [PubMed] [Google Scholar]
- 30.Naini A, Toscano A, Musumeci O, Vissing J, Akman HO, et al. Muscle phosphoglycerate mutase deficiency revisited. Arch Neurol. 2009;66(3):394–398. 10.1001/archneurol.2008.584 [DOI] [PubMed] [Google Scholar]
- 31.de Atauri P, Repiso A, Oliva B, Vives-Corrons JL, Climent F, et al. Characterization of the first described mutation of human red blood cell phosphoglycerate mutase. Biochim Biophys Acta. 2005;1740(3):403–410. 10.1016/j.bbadis.2004.11.023 [DOI] [PubMed] [Google Scholar]
- 32.Kawashima N, Mishima M, Shindo R, Hirano M, Kuwabara S, et al. Partial deficiency of phosphoglycerate mutase with diabetic polyneuropathy: the first Japanese patient. Intern Med. 1996;35(10):799–802. 10.2169/internalmedicine.35.799 [DOI] [PubMed] [Google Scholar]
- 33.Radu CG, Shu CJ, Shelly SM, Phelps ME, Witte ON. Positron emission tomography with computed tomography imaging of neuroinflammation in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2007;104(6):1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toriyama K, Kuwahara M, Kondoh H, Mikawa T, Takemori N, et al. T cell-specific deletion of Pgam1 reveals a critical role for glycolysis in T cell responses. Commun Biol. 2020;3(1):394. 10.1038/s42003-020-01122-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting information files.