Abstract
The development of recombinant COVID-19 vaccines has resulted from scientific progress made at an unprecedented speed during 2020. The recombinant spike glycoprotein monomer, its trimer, and its recombinant receptor-binding domain (RBD) induce a potent anti-RBD neutralizing antibody response in animals. In COVID-19 convalescent sera, there is a good correlation between the antibody response and potent neutralization. In this review, we summarize with a critical view the molecular aspects associated with the interaction of SARS-CoV-2 RBD with its receptor in human cells, the angiotensin-converting enzyme 2 (ACE2), the epitopes involved in the neutralizing activity, and the impact of virus mutations thereof. Recent trends in RBD-based vaccines are analyzed, providing detailed insights into the role of antigen display and multivalence in the immune response of vaccines under development.
Short abstract
SARS-CoV-2 receptor-binding domain (RBD)-specific antibodies are crucial for neutralizing efficacy. Emerging mutations, the quality of the IgG response, and the benefit of multivalent display are analyzed here for RBD subunit vaccines.
1. Introduction
Viral infections are initiated with the binding of viral particles to the host’s surface cellular receptors, a process that defines the virus’s cellular and tissue tropism. In SARS-CoV-2, this process is mediated by the viral spike (S) glycoprotein trimer on the virion surface through its receptor-binding domain (RBD).
The S-glycoprotein is a 1273-amino acid polypeptide with 22 N-glycans, a class I fusion protein,1 which forms trimers on the virus surface. Each trimer has three main topological domains: head, stalk, and cytoplasmic tail. The head contains the S1 subunit with two domains: the N-terminal domain (NTD) and the receptor-binding domain (RBD), where the receptor-binding motif (RBM) is responsible for direct interaction with its receptor in human cells, the angiotensin-converting enzyme 2 (ACE2).2
SARS-CoV-2 uses the same host-cell entry receptor as SARS-CoV, ACE2,3 which is expressed, among others, in specific subsets of human respiratory epithelial cells in nasal passages, airways, and alveoli.4 S-Glycoprotein trimers have most of the time all RBDs in a hidden conformation, the “down” conformation, which seeks to evade immune recognition but, at the same time, cannot interact with ACE2. A structural transition occurs from the “down” to the “up” RBD conformation, achieving high-affinity binding to ACE2. Following RBD–ACE2 binding, the S-glycoprotein is cleaved by host proteases, allowing membrane fusion and entry of the virus.5
Knowledge on the SARS-CoV-2–ACE2 interaction and its neutralization by antibodies has been achieved at an unprecedented speed. Recombinant protein subunit vaccines in clinical development include recombinant SARS-CoV-2 proteins as active components: (i) S-glycoprotein monomer,6 (ii) S-glycoprotein trimer,7,8 and (iii) RBD-based immunogens with several forms of antigen display (monomeric, dimeric, or multivalent, vide infra). All induce anti-RBD neutralizing antibodies in laboratory animals, and some are in evaluation in humans. In this paper, we show key molecular aspects regarding virus–host cell interaction, mechanisms of virus neutralization by antibodies blocking such interaction, and the impact of virus mutation on transmissibility and escape from neutralizing antibodies. Attention is paid to how these aspects relate to vaccine development, as well as to the different types of RBD displays in vaccine candidates.
2. Structure of SARS-CoV-2 RBD and Its Interaction with the hACE2 Receptor
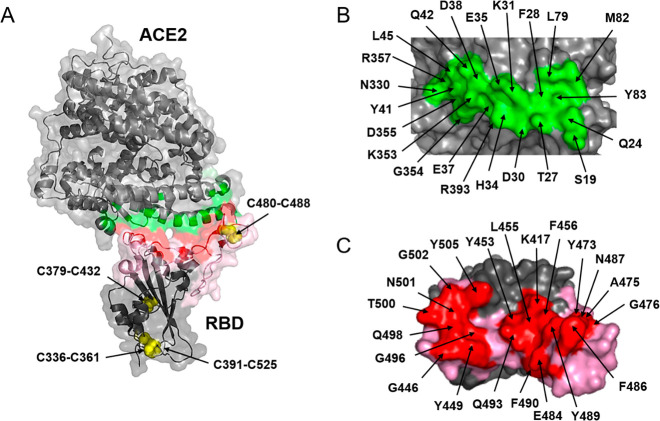
SARS-CoV-2 RBD comprises 193 amino acid residues (from Thr333 to Pro527), including the core and the RBM covering residues 438–506 (Figure 1, in pink). The core is structured around a twisted, five-strand antiparallel β-sheet with short connecting helices and loops and is stabilized by three disulfide bridges: Cys336–Cys361, Cys379–Cys432, and Cys391–Cys525. A fourth disulfide bridge, Cys480–Cys488, connects the loops at the distal ends of the RBM.9
Figure 1.
RBD–ACE2 interaction. (A) Transparent surface and ribbon representation of the SARS-CoV-2 RBD (residues Arg319–Phe541) complexed to the ACE2 N-terminal peptidase domain (residues Ser19–Asp615). The interface ACE2–RBD is colored in green and red, respectively, and the RBM in pink. The arrows indicate four different disulfide bridges able to stabilize the RBD. (B) Amino acids of ACE2 directly interacting with RBD (green). (C) Amino acids of RBD directly interacting with ACE2 (red).
In terms of glycosylation, the two domains (NTD and RBD) in the S-glycoprotein head are very different. NTD is highly glycosylated (7 N-linked carbohydrates on Asn 61, 74, 122, 149, 165, 234, 282), while RBD has one N-linked carbohydrate chain on Asn343 located in the core with no glycosylation at RBM. The N-linked carbohydrate chain on Asn331 is located between the two domains.10 The glycosylation pattern shows a higher prevalence of complex N-glycans compared with the most common oligomannoside-type glycans11 exposed on viruses infecting humans12 (e.g., HIV-1 and Ebola). In the spike trimer, the highest carbohydrate density in the NTD surrounds the head’s surface, helping camouflage the RBD, especially in the “down” conformation (vide infra).
ACE2 is the cellular receptor for three coronaviruses: NL-63, SARS-CoV, and SARS-CoV-2.13 This receptor is a zinc-dependent carboxypeptidase that cleaves one residue from the C-terminus of angiotensin peptides. It participates in blood pressure regulation14 and is also associated with protection from severe acute respiratory failure. During SARS-CoV and SARS-CoV-2 infection, lung injury is associated with down regulation of ACE2.15 Binding to the hACE2 receptor is a critical initial step in SARS-CoV16 and SARS-CoV-2 infection.17−20 SARS-CoV-2 RBD binds in vitro to hACE2 with an affinity in the low nanomolar range.21 This high ACE2-binding affinity is due to the large interacting surfaces: 864 Å on RBD,2 involving 21 amino acids (Figure 1C, in red) and 823 Å on the hACE2 receptor,2 involving 22 amino acids (Figure 1B, in green). A network of hydrophilic and hydrophobic interactions is established at the RBD–ACE2 interface with 13 hydrogen bonds and two salt bridges (K417-D30 and E484-K31).17−20 For SARS-CoV-2, the crystal structures of RBD complexed with hACE2 have been determined (recombinant in Hi5 insect cells, PDB codes 6M0J,96VW1,16 and 6LZG(2)).
Epidemiologic and biochemical studies have shown that the infectivity of different SARS-CoV-2 strains is proportional to the binding free energy (BFE) between the strain-specific RBD and ACE2 in the host cell.22 RBM mutations resulting in an affinity increase also lead to enhanced virus transmissibility,23 favoring the spread of the mutated virus. The probability that novel variants emerge during human to human transmission is favored by the increasing number of infected individuals, attaining >100 million confirmed cases as of 28 January 2021. The low number of novel mutations reaching high frequency in sequenced SARS-CoV-2 genomes24 is compensated by the number of individuals infected. In the COVID-19 prevaccine era, the increased affinity for the receptor was probably the main driving force for mutation, which is associated with a faster spread and in some cases leads to fast and complete replacement of the original strain.
The following single mutations of SARS-CoV-2 illustrate their impact on affinity and viral spreading: N501Y, S477N, E484K, L452R, and K417N/T (Figure 2A). Mutation N501Y at the RBM is probably the best known and the leading mutation in the new lineage B.1.1.7, isolated in the U.K. in September 202025 and rapidly spreading to more than 75 countries by mid-February 2021. New intermolecular contacts (between Tyr501 of RBD and ACE2,26Figure 2B) through hydrophobic (ACE2, Tyr41 aromatic ring, and Lys353 aliphatic chain) and polar (ACE2, ε-amino group of Lys353) interactions contribute to increased affinity compared to the wild type virus.27,28 The higher affinity increases the rate of transmission in about 60%.29 The fast spreading and, consequently, the high level of circulation may promote epistatic mutation such as Q498R, which would additionally increase the affinity through a more favorable interaction with L45 and Q42 at hACE2 (Figure 2C). If this event takes places, we should expect an increment in the infectivity.24 The binding affinity to hACE2 of RBD mutations selected by nature, S477N, E484K, and N501Y, strongly correlate to their frequency: in a decreasing order: E484K/N501Y double mutant (KDapp 126 pM), N501Y (KDapp 455 pM), and S477N (KDapp 710 pM) (value for WT: 1600 pM).24
Figure 2.
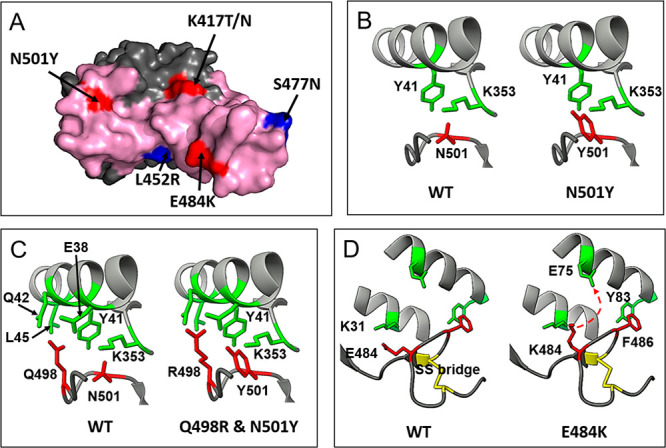
Selected mutations in RBD. (A) Location of each mutated amino acid with respect to the RBM surface (pink). Residues belonging to the ACE2 epitope are highlighted in red, and those without contact with ACE2 are colored in blue. (B) and (D) New interactions associated with N501Y and E484K mutations, respectively. (C) Potential epistatic mutation Q498R associated with N501Y.
Mutation E484K usually appears together with N501Y, slightly increasing the affinity for ACE2.30 Two lineages have this double mutation in common: lineage B.1.351, that emerged after the first epidemic wave in the most affected metropolitan area within the Eastern Cape Province31 in South Africa (and spread to more than 30 countries), and lineage B.1.1.28, initially described in Rio de Janeiro, and currently widely spread in Brazil32 and other countries. Recently, mutations N501Y and E484K appeared together in the U.K. following the spread of lineage B.1.1.7. E484K suppresses the salt bridge RBD E484–hACE2 K31, abolishing this important RBD–hACE2 charge attraction, and predicting a reduction in the affinity, especially at the new K484–K31 interaction.33 Molecular dynamics simulation identified the potential formation of a new salt bridge RBD K484–hACE2 E75 as a consequence of the flexibility in the RBD 475–487 loop region.34 This allows a transient ion pairing to be established between RBD K484–hACE2 E75 and a new salt bridge (Figure 2D).
3. SARS-CoV-2 S-Glycoprotein Trimers and Conformational Changes on the Virus Surface
SARS-CoV-2 S-glycoprotein trimers are exposed on the virus surface. Each of the three RBDs may adopt two conformations, called “down” and “up”. A fully exposed, accessible RBD is possible only in the “up” conformation, where its RBM can interact with cellular hACE2 receptors (Figure 1).35 The cryoelectron microscopy (CEM) of SARS-CoV-2 S-glycoprotein reveals a predominance of the “down” conformation,36 which is ineffective for binding the receptor.
The S1 glycoprotein domain, containing the RBD and NTD, is the protein’s most flexible part. Interactions between neighboring monomers show significant energetic differences for “up–down” and “down–down” conformations.37 Molecular dynamics simulations38 have shown that the RBD switches from the “down” to the “up” position through a semiopen intermediate that reduces the free energy barrier between the closed (“down”) and open (“up”) states. This intermediate may already bind to hACE2. In addition, interchain interactions are stronger and more numerous in “up–down” than in “down–down” conformations.
Studies using CEM confirmed that S-glycoprotein trimers protrude from the virion surface. An individual virion contains 25 ± 9 trimers in a highly dynamic structure where the opening of the RBD is stochastic. On average, approximately 41% of trimers have no RBD “up” (i.e., all their RBDs are “down”), while one RBD is “up” in 45% of trimers and two RBDs are “up” in only 13% of trimers.39 Structural analysis revealed that the D614G mutation that emerged in Europe in February 2020 increased the RBD “up” proportion from 41% to 82%.40 This new mutation seriously impacted transmissibility, and, as a consequence of higher viral load and shedding, it became dominant in the pandemic by April 2020.
When a single RBD adopts the “up” conformation, its RBM becomes accessible for binding to hACE2 but also becomes more accessible to neutralizing antibodies. On the contrary, in the “down” conformation, the RBM is camouflaged due to the shielding effect of glycans covering ∼35% of its surface,41 thus preventing binding to hACE2 but also reducing effective recognition by the immune system. The mutation D614G that increases transmissibility also exposes better the RBD and makes the virus more susceptible to neutralization by anti-RBD monoclonal antibodies.42 The RBD core is camouflaged in both the “up” and “down” conformations42 by the shielding effect of N-glycans (at Asn343, Asn331, Asn234, and Asn165).
4. SARS-CoV-2 RBD as a Target for Neutralizing Antibodies
Many SARS-CoV-2-binding antibodies have been identified in convalescent COVID-19 patients’ sera, but only a few led to virus neutralization and protection from the disease.43 Recent studies have shown that RBD-binding antibodies are responsible for 90% of the neutralizing activity in convalescent human sera.44 To date, almost 500 (476 and increasing) neutralizing antibodies (NAbs) targeting the SARS-CoV-2 RBD have been screened from B-cells of COVID-19 patients. They are regularly updated in the coronavirus antibody database (Cov-AbDab).45
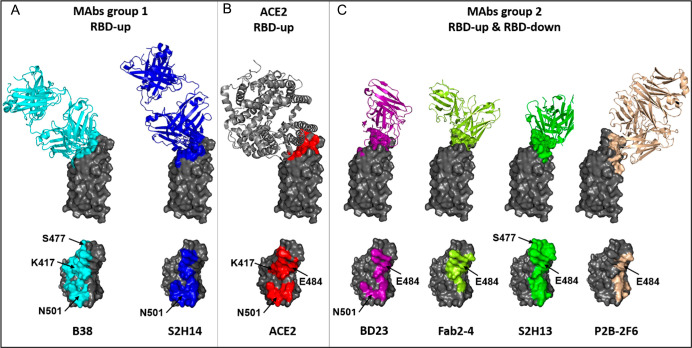
Human monoclonal NAbs have led to a better understanding of RBD epitopes and the angle of approach necessary for binding and neutralization. NAbs can be clustered in several groups based on their interaction with distinct RBD-binding sites. Here we consider only the most potent neutralizing antibodies, which are associated with RBM binding and clustered in two main groups. The target for type 1 antibodies (different classifications can be found elsewhere) is the fully exposed surface in the RBD “up” conformation of the trimer (open state, Figure 3C).44,46 They block directly the RBM interaction with hACE2 (Figure 3B). An important number of the most efficient NAbs have their heavy chains encoded by IGVH3-53 or IGVH3-66 with a short CDRH3 length (9–12 amino acids) and light chains often encoded by IGLV1-9/3-20. These antibodies interact with an epitope overlapping the hACE2 interacting surface (Figure 3A,B). The paired heavy-light chain signatures contribute to potent SARS-CoV-2 neutralization in convalescent antibody responses.47
Figure 3.
RBD epitopes of NAbs and location of the important mutations. (A) Example of antibodies of type 1. NAbs B38 and S2H14 interact with their respective epitopes overlapping the ACE2-binding region. (B) RBD surface (red) delimited by residues interacting with ACE2. (C) Example of antibodies of type 2. NAbs BD23, Fab2-4, S2H13, and P2B-2F6 interact with epitopes exposed in the RBD-down and RBD-up conformations.
Type 2 NAbs interact with the RBD-binding surface accessible in both “up” and “down” conformations (open/closed S states), thus blocking ACE2–RBD binding without functionally mimicking ACE2.48 Their various pairs IGHV–IGLV are responsible for the diversity of recognized RBD epitopes and angles of approach to RBD (Figure 3C).
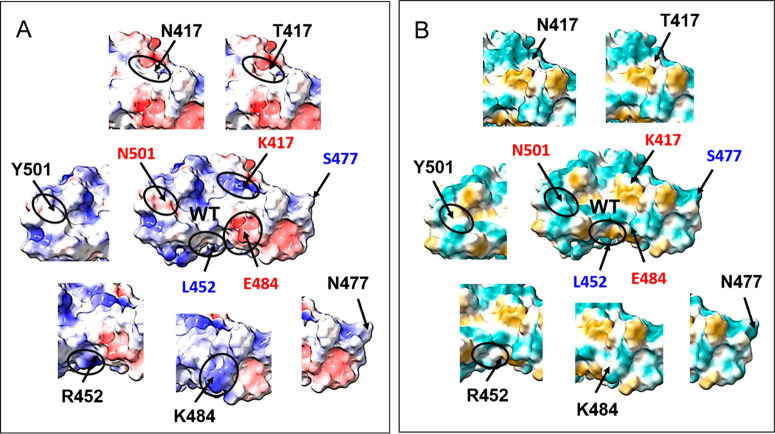
The human neutralizing polyclonal response is a combination of type 1 and type 2 antibodies, with different angles of approach to RBD epitopes, mostly involving residues of the RBM. This diversity is important for avoiding mutants’ escape from neutralization. In the naïve population, SARS-CoV-2 is evolving for increasing its infectivity and affinity for ACE2, as previously described. However, in the immune population, this evolution is driven by the adaptation to the host immune system through a selection of escaping mutants. The best example of such mutation is E484K, found in vitro as a response to virus incubation with convalescent plasma.49 The E484 position bear a negative charge (glutamate) in the WT RBD (Figure 4), but after the E484K mutation, the position changes to a positive charge at Lys. This perturbation has an important impact on the epitopes, mainly for type 2 antibodies (Figure 3C). This mutation reduces significantly both the naturally acquired and vaccine-induced immunity in lineages B.1.351, B.1.1.28, and in a double mutant derived from B.1.1.7. Lineage B.1.351 raised concern by its potential to escape from therapeutically relevant monoclonal antibodies50−52 and by reducing 6–10 times the neutralization titer in convalescent and vaccinated sera.51 However, this reduction is vaccine-dependent; for example, in the serum of a subject vaccinated with a dimeric RBD, the average neutralization titer is reduced only from 106 to 67.53
Figure 4.
(A) Electrostatic potential (red −10, +10 blue) and (B) hydrophobicity (green −20, +20 gold) maps represented on surfaces of the WT RBD and selected mutated RBD. Small circles show the biggest changes induced by mutation.
Another mutation impacting the electrostatic potential is K417N/T, which suppresses the positive charge at Lys. (Figure 4). Its impact is not as marked as E484K but is relevant enough to abolish neutralization by some antibodies binding to this region.54 Other individual mutations are less effective. For example, N501Y only induces a small increase in hydrophobicity (yellow) and a small decrease in the potential (red surface). As a result, the reduction of neutralization observed for the N501Y strain in convalescent sera is rather small.55 In the WT strain, position L452 neither binds to hACE2 nor interacts with antibodies. Yet, the recently reported mutation L452R, appearing in California,56 modifies the surface hydrophobicity (Figure 4). However, so far it is not clear whether this mutation can affect the affinity for hACE2 or the antibodies’ neutralization capacity.
5. Antigen Orientation and Multivalent Display in RBD-Based Vaccines
Once the immunodominance of SARS-CoV-2 RBD was proven,44 with this domain being the target of most NAbs,47,57,58 the question was whether this individual protein per se would become an effective vaccine immunogen.59,60 The high quality and functionality of anti-RBD antibodies, along with the good correlation between their presence and the neutralization capacity of COVID-19 convalescent sera,61,62 support the RBD potential as vaccine antigen. Nevertheless, a great effort has been devoted to determining whether this protein alone or a multivalent RBD construct is a better class of immunogen. Here we analyze some evidence based on reports from vaccine developers.
5.1. Monomer
The use of monomeric RBD as antigen is indeed the most straightforward approach, and some groups have developed immunogens based on RBD expressed in insect cells,63 yeast (Pichia pastoris),64 and mammalian cells (CHO and HEK293).65,66 In general, such versions of the antigen have induced a good response of NAbs in various animal models, including mice,63,65 rabbits,63 rodents,66 and nonhuman primates (NHPs), these latter showing a notable protection against an in vivo challenge with SARS-CoV-2.63 One of these reports revealed that the antibody response was dose-dependent and that the immune sera—rather than splenic T cells—was responsible for the protective immunity.63 To our knowledge, two of these monomeric RBD vaccine candidates have advanced to phase 1 and 2 clinical evaluation.67 In addition, one mRNA vaccine, namely, ARCoV,68 expressing monomeric RBD in the host cells has advanced to phase 1 after proving high titers of NAbs and a Th1-biased cellular response in mice and NHPs. Nonetheless, recent studies have shown that multivalent RBD versions have a greater and higher quality antibody response than the monomer versions in laboratory animals.61,69−71
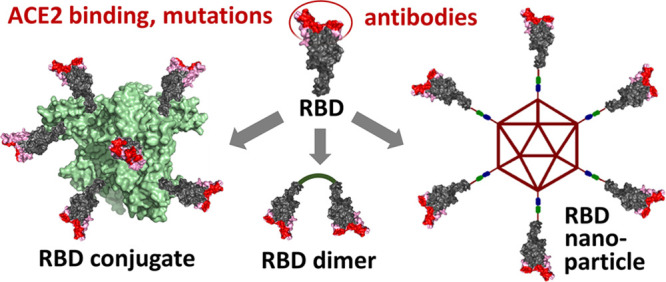
As shown in Figure 5A, a rationale for the improved immune response of multimeric RBD is its capacity to enhance B-cell activation by cross-linking their receptors,70,71 in contrast to the typically poorer B-cell response achieved by the monomeric antigen. To achieve this, new trends in vaccine development are focusing on oligomeric, conjugate, and nanoparticle RBD immunogens featuring multivalent display of this antigen. The key to succeed with such constructions is not antigen multimerization per se but its combination of a suitable RBD orientation/presentation seeking to enhance the neutralizing response. This needs to be done in such a way that the RBM is exposed better than other epitopes of the RBD core, thus directing most of the IgG response toward the motif that actually interacts with the hACE2 receptor and initiates virus internalization. This design should lead to a neutralizing/binding antibody ratio similar or—ideally—higher than that of convalescent sera, resulting in a protective vaccine.
Figure 5.
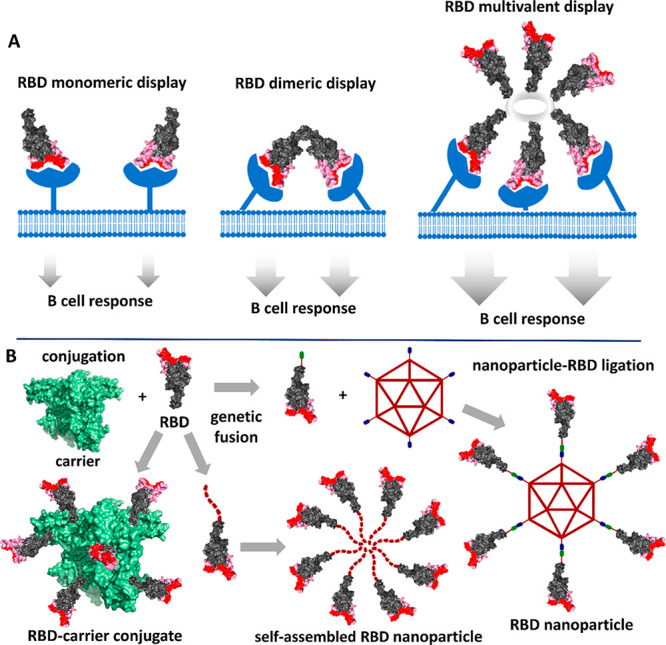
(A) Comparison of the B-cell response with different types of RBD immunogens. The multivalent RBD display permits cross-linking B-cell receptors leading to a more intense signaling. (B) Representation of the constructions of multivalent-displayed RBD immunogens by chemical conjugation, self-assembling, and ligation processes.
5.2. Dimer
The design of dimeric RBD is a clear bet for antigen multimerization. Deep analysis of the RBD structure reveals that both the N- and C-terminal regions are far away from the RBM and are therefore suitable sites for multimerization or ligation. This logic has been followed for the design of dimeric RBD vaccines70,72,73 in which two RBDs are connected by their C-terminal tails. This design may allow not only cross-linking of B-cell receptors (Figure 5A), but at the same time it can provide the correct exposure and orientation of the RBM, thus guiding the antibody response toward this motif. Consequently, it has been proven that RBD dimers elicited NAb titers 10–100-fold higher than those produced by the monomeric antigen.70 This suggests that the immunodominant RBM epitopes are more accessible than the RBD core, as compared with the monomer where the entire RBD surface is fully exposed to immune recognition. For example, the vaccine candidate named ZF200170,73 is a SARS-CoV-2 RBD dimer designed with a tandem repeat single chain (sc-dimer) that connects the two protein domains.70 ZF2001 completed phase 1 and 2 clinical trials proving the elicitation of very high titers of NAbs. In parallel, our group developed a dimeric SARS-CoV-2 RBD vaccine named ‘Soberana01’, also under clinical evaluation,72 in which a disulfide bond connects the two RBDs at Cys538.
Other vaccine platforms have also focused on RBD oligomers, as seen in DNA74 and mRNA75 vaccines expressing RBD trimers to improve immunogenicity. Such nucleic acid vaccines express RBD sequences incorporating a foldon trimerization tag at the C-terminus, which triggers protein trimerization upon expression in the host cells.
5.3. Multivalent Conjugate
An effective multivalent display of the RBD antigen can be achieved using the conjugate vaccine technology. In this platform relying on conjugation chemistry, multiple copies of the RBD antigen are chemically attached to an immunogenic carrier protein seeking both to activate multiple B-cell receptors (Figure 5B) and to take advantage of the carrier-specific pre-existing T helper cells to improve antibody production. In the context of COVID-19 vaccines, the challenge is to carry out a precise RBD conjugation to the usually large carrier without affecting the RBD neutralizing epitopes in the chemical reaction. Our group has developed a vaccine candidate, namely, Soberana02, in which multiple RBDs are site-selectively conjugated to the tetanus toxoid using a Cys residue placed at the C-terminus, which is far away from the relevant RBM region.76 In this design, the exposure of the neutralizing epitopes is maximized, while the immunogenic carrier favors the production of CD4+ T helper cells required for a potent antibody response. In animal models, this vaccine candidate elicited a robust neutralizing response.76 Phase 1 and 2 clinical trials77,78 showed good neutralizing seroconversion in humans, and the candidate advanced to phase 3 clinical evaluation as of March 2021.
5.4. Protein Nanoparticle
This is a technology69,79 that combines the advantages of antigen presentation in a virus-like particle of nanometric size with those of multivalent display to enhance B-cell activation. Indeed, the antigen display achieved with this type of self-assembling nanoparticle (NP) can mimic that in SARS-CoV-2. Nevertheless, a favorable RBD display needs to be attained during NP assembly to favor the spatial presentation of RBM. As depicted in Figure 5B, two approaches can be followed to successfully display multiple RBDs on the surface of immunogenic particles resembling the natural virus. The first approach consists of the expression of RBD genetically fused to another protein fragment or tail, enabling its self-assembling either alone or with another protein. An example of this construction is a hybrid nanoparticle71 with an RBD fused at its N-terminus (also distant from the RBM, not blocking the neutralizing epitopes) to protein I53-50A. Such a fusion protein self-assembles with protein I53-50B into two-component nanoparticles displaying about 60 RBD copies on the surface. These multivalent nanoparticles elicited NAbs titers markedly higher than those elicited by the RBD monomer and up to 10-fold higher than the prefusion-stabilized S ectodomain trimer. They also exhibited a neutralizing/binding ratio higher than that in convalescent human sera.71 Fusion of the RBD to Helicobacter pylori ferritin has also led to immunogenic, self-assembling nanoparticles in which the RBDs form radial multivalent projections that correctly expose their neutralizing epitopes.80
A second approach is the use of a protein ligation technology81 to conjugate several RBDs to the surface of preassembled nanoparticles. Examples are the construction of bacteriophage AP205 capsid-like particles decorated with multiple RBD copies82 and the assembly of multicomponent ferritin-based nanoparticles83 by ligating both the SARS-CoV-2 RBD and heptad repeat (HR) antigens to the ferritin nanocore. In these cases, the RBD was genetically fused to a ligation tag either at its N- or C-terminus, thus eluding any steric hindrance at the crucial RBM epitopes that need to be exposed at the surface. RBD-decorated mosaic nanoparticles displaying not only multiple SARS-CoV-2 RBDs but also RBDs of zoonotic coronaviruses have also been obtained84 by a “plug-and-display” strategy.81 The RBD-displaying nanoparticles herein discussed provide typically monodisperse and stable formulations, which elicited robust neutralizing immune responses in mice, ferrets, and even NHP. It remains to be proven whether such protective efficacy can be replicated in humans, as done for other antiviral nanoparticle vaccines.69,79
6. Conclusions
The RBD–ACE2 interaction is essential for SARS-CoV-2 infectivity. For virus neutralization, antibodies blocking this interaction are fundamental both in naturally acquired and vaccine-induced immunity. Understanding this process is essential for vaccine improvement and for fighting emerging SARS-CoV-2 variants. Virus mutation driven either by ACE2 affinity enhancement—increasing infectivity—or by escaping from acquired or vaccine-induced immunity poses a new challenge. Vaccine candidates based on the RBD monomer have proven success in preclinical evaluation, and they can most likely afford sufficient protection in humans to become approved. However, compared with monomeric RBD immunogens, those featuring multivalent RBD display (i.e., RBD dimers, RBD-carrier conjugates and protein nanoparticles) show greater promise due to the enhanced B-cell response and likely longer-lasting immunity. Evidence suggests that vaccine candidates based on multimeric RBD constructs benefit from both antigen multivalency and a maximized exposure of RBM epitopes to improve not only the IgG response but also neutralizing efficacy in comparison with monovalent antigens. The induction of an improved immunity and neutralizing response, along with the generalization of platforms that incorporate the emerging RBD mutants, should place RBD vaccines at a competitive position to provide a fast response against the COVID-19 pandemic.
7. Methods
Structural analyses were performed from PDB: 6M0J for the complex ACE2–RBD,9 PDB: 7BZ5 for B38,85 PDB: 7BYR for BD23,86 PDB: 6XEY for Fab2-4,87 PDB: 7BWJ for P2B-2F6,88 PDB: 7JV2 for S2H13 and PDB: 7JX3 for S2H1444 antibodies complexed with RBD or the full trimeric spike structure using the PyMOL Molecular Graphics System.89 Virtual mutations and molecular surface by electrostatic and molecular hydrophobicity potential were realized with UCSF ChimeraX 1.1.90 PyMOL and ChimeraX were also used for structural renderings for figures.
Acknowledgments
We are grateful to Fondo de Ciencia e Innovación (FONCI, Cuba) for financial support (Project-2020-20). We thank Gail Reed for editing English grammar.
Author Contributions
∥ Y.V.-B. and D.S.-M. contributed equally to this paper.
The authors declare the following competing financial interest(s): Y.V.-B., D.S.-M, S.F., L.R., B.S.-R., K.L., D.G.-R., D.G.R., and V.V.B. are co-inventors on provisional SARS-CoV-2 vaccine patents including results covered here.
References
- Zhang Y.; Zhao W.; Mao Y.; Chen Y.; Wang S.; Zhong Y.; Su T.; Gong M.; Du D.; Lu X.; Cheng J.; Yang H. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol. Cell Proteomics 2021, 20, 100058. 10.1074/mcp.RA120.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Zhang Y.; Wu L.; Niu S.; Song C.; Zhang Z.; Lu G.; Qiao C.; Hu Y.; Yuen K. Y.; Wang Q.; Zhou H.; Yan J.; Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181 (4), 894–904. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.; Yang X. L.; Wang X. G.; Hu B.; Zhang L.; Zhang W.; Si H. R.; Zhu Y.; Li B.; Huang C. L.; Chen H. D.; Chen J.; Luo Y.; Guo H.; Jiang R. D.; Liu M. Q.; Chen Y.; Shen X. R.; Wang X.; Zheng X. S.; Zhao K.; Chen Q. J.; Deng F.; Liu L. L.; Yan B.; Zhan F. X.; Wang Y. Y.; Xiao G. F.; Shi Z. L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579 (7798), 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muus C.; Luecken M. D.; Eraslan G.; Waghray A.; Heimberg G.; Sikkema L.; Kobayashi Y.; Vaishnav E. D.; Subramanian A.; Smilie C.; Jagadeesh K.; Duong E. T.; Fiskin E.; Triglia E. T.; Ansari M.; Cai P.; Lin B.; Buchanan J.; Chen S.; Shu J.; Haber A. L.; Chung H.; Montoro D. T.; Adams T.; Aliee H.; Samuel J.; Andrusivova A. Z.; Angelidis I.; Ashenberg O.; Bassler K.; Bécavin C.; Benhar I.; Bergenstråhle J.; Bergenstråhle L.; Bolt L.; Braun E.; Bui L. T.; Chaffin M.; Chichelnitskiy E.; Chiou J.; Conlon T. M.; Cuoco M. S.; Deprez M.; Fischer D. S.; Gillich A.; Gould J.; Guo M.; Gutierrez A. J.; Habermann A. C.; Harvey T.; He P.; Hou X.; Hu L.; Jaiswal A.; Jiang P.; Kapellos T.; Kuo C. S.; Larsson L.; Leney-Greene M. A.; Lim K.; Litviňuková M.; Lu J.; Ludwig L. S.; Luo W.; Maatz H.; Madissoon E.; Mamanova L.; Manakongtreecheep K.; Marquette C.-H.; Mbano I.; McAdams A. M.; Metzger R. J.; Nabhan A. N.; Nyquist S. K.; Penland L.; Poirion O. B.; Poli S.; Qi C.; Queen R.; Reichart D.; Rosas I.; Schupp J.; Sinha R.; Sit R. V.; Slowikowski K.; Slyper M.; Smith N.; Sountoulidis A.; Strunz M.; Sun D.; Talavera-López C.; Tan P.; Tantivit J.; Travaglini K. J.; Tucker N. R.; Vernon K.; Wadsworth M. H.; Waldman J.; Wang X.; Yan W.; Zhao W.; Ziegler C. G. K. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv 2020, 10.1101/2020.04.19.049254. [DOI] [Google Scholar]
- Coutard B.; Valle C.; de Lamballerie X.; Canard B.; Seidah N. G.; Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020, 176, 104742. 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Zheng Q.; Yu H.; Wu D.; Xue W.; Xiong H.; Huang X.; Nie M.; Yue M.; Rong R.; Zhang S.; Zhang Y.; Wu Y.; Wang S.; Zha Z.; Chen T.; Deng T.; Wang Y.; Zhang T.; Chen Y.; Yuan Q.; Zhao Q.; Zhang J.; Gu Y.; Li S.; Xia N. SARS-CoV-2 spike produced in insect cells elicits high neutralization titres in non-human primates. Emerging Microbes Infect. 2020, 9 (1), 2076–2090. 10.1080/22221751.2020.1821583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C.; Albert G.; Reed P.; Neal S.; Plested J. S.; Zhu M.; Cloney-Clark S.; Zhou H.; Patel N.; Frieman M. B.; Haupt R. E.; Logue J.; McGrath M.; Weston S.; Piedra P. A.; Cho I.; Robertson A.; Desai C.; Callahan K.; Lewis M.; Price-Abbott P.; Formica N.; Shinde V.; Fries L.; Linkliter J. D.; Griffin P.; Wilkinson B.; Smith G.; Glenn G. M. First-in-human trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. medRxiv 2020, 10.1101/2020.08.05.20168435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S.; Ozorowski G.; Turner H. L.; Antanasijevic A.; Huang D.; Wang X.; Torres J. L.; Diedrich J. K.; Tian J. H.; Portnoff A. D.; Patel N.; Massare M. J.; Yates J. R. 3rd; Nemazee D.; Paulson J. C.; Glenn G.; Smith G.; Ward A. B. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 2020, 370 (6520), 1089–1094. 10.1126/science.abe1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the hACE2 receptor. Nature 2020, 581 (7807), 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Casalino L.; Gaieb Z.; Goldsmith J. A.; Hjorth C. K.; Dommer A. C.; Harbison A. M.; Fogarty C. A.; Barros E. P.; Taylor B. C.; McLellan J. S.; Fadda E.; Amaro R. E. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020, 6 (10), 1722–1734. 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369 (6501), 330–333. 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Berndsen Z. T.; Raghwani J.; Seabright G. E.; Allen J. D.; Pybus O. G.; McLellan J. S.; Wilson I. A.; Bowden T. A.; Ward A. B.; Crispin M. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11 (1), 2688. 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015, 89 (4), 1954–64. 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y.; Kuba K.; Rao S.; Huan Y.; Guo F.; Guan B.; Yang P.; Sarao R.; Wada T.; Leong-Poi H.; Crackower M. A.; Fukamizu A.; Hui C. C.; Hein L.; Uhlig S.; Slutsky A. S.; Jiang C.; Penninger J. M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436 (7047), 112–6. 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhol F.; Sarlon G.; Deharo J. C.; Vaisse B. Downregulation of hACE2 induces overstimulation of the renin-angiotensin system in COVID-19: should we block the renin-angiotensin system?. Hypertens. Res. 2020, 43 (8), 854–856. 10.1038/s41440-020-0476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H.; Geng Q.; Auerbach A.; Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581 (7807), 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11 (1), 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Park Y. J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181 (2), 281–292. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Kruger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; Muller M. A.; Drosten C.; Pohlmann S. SARS-CoV-2 cell entry depends on hACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181 (2), 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Shang J.; Graham R.; Baric R. S.; Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94 (7), e00127–20 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Guo Y.; Pan Y.; Zhao Z. J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020, 525 (1), 135–40. 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Wang R.; Wang M.; Wei G. W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020, 432 (19), 5212–5226. 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradník J.; Marciano S.; Shemesh M.; Zoler E.; Chiaravalli J.; Meyer B.; Rudich Y.; Dym O.; Elad N.; Schreiber G. SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. bioRxiv 2021, 10.1101/2021.01.06.425392. [DOI] [Google Scholar]
- Li X.; Wang W.; Zhao X.; Zai J.; Zhao Q.; Li Y.; Chaillon A. Transmission dynamics and evolutionary history of 2019-nCoV. J. Med. Virol. 2020, 92 (5), 501–511. 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Investigation of novel SARS-CoV-2 variant of concern 202012/01. England Public Health 2021, technical briefing 3. [Google Scholar]
- Ahmed W.; Philip A. M.; Biswas K. H. Stable interaction of the UK B.1.1.7 lineage SARS-CoV-2 S1 spike N501Y mutant with hACE2 revealed by molecular dynamics simulation. bioRxiv 2021, 10.1101/2021.01.07.425307. [DOI] [Google Scholar]
- Starr T. N.; Greaney A. J.; Hilton S. K.; Ellis D.; Crawford K. H. D.; Dingens A. S.; Navarro M. J.; Bowen J. E.; Tortorici M. A.; Walls A. C.; King N. P.; Veesler D.; Bloom J. D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and hACE2 binding. Cell 2020, 182 (5), 1295–1310. 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan B.; Wang H.; Huynh T. Molecular mechanism of the N501Y mutation for enhanced binding between SARS-CoV-2’s spike protein and human hACE2 receptor. bioRxiv 2021, 10.1101/2021.01.04.425316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E.; Mishra S.; Mishra M.; Barrett J. C.; Johnson R.; Geidelberg L.; Hinsley W. R.; Laydon D. J.; Dabrera G.; O’Toole A.; Amato R.; Ragonnet-Cronin M.; Harrison I.; Jackson B.; Ariani C. V.; Boyd O.; Loman N. J.; McCrone J. T.; Gonçalves S.; Jorgensen D.; Myers R.; Hill V.; Jackson D. K.; Gaythorpe K.; Groves N.; Sillitoe J.; Kwiatkowski P.. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. Imperial College London 2020, Report 42. [Google Scholar]
- Liu Z.; VanBlargan L. A.; Bloyet L. M.; Rothlauf P. W.; Chen R. E.; Stumpf S.; Zhao H.; Errico J. M.; Theel E. S.; Liebeskind M. J.; Alford B.; Buchser W. J.; Ellebedy A. H.; Fremont D. H.; Diamond M. S.; Whelan S. P. J. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv 2020, 10.1101/2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H.; Wilkinson E.; Giovanetti M.; Iranzadeh A.; Fonseca V.; Giandhari J.; Doolabh D.; Pillay S.; San E. J.; Msomi N.; Mlisana K.; von Gottberg A.; Walaza S.; Allam M.; Ismail A.; Mohale T.; Glass A. J.; Engelbrecht S.; Van Zyl G.; Preiser W.; Petruccione F.; Sigal A.; Hardie D.; Marais G.; Hsiao M.; Korsman S.; Davies M.-A.; Tyers L.; Mudau I.; York D.; Maslo C.; Goedhals D.; Abrahams S.; Laguda-Akingba O.; Alisoltani-Dehkordi A.; Godzik A.; Wibmer C. K.; Sewell B. T.; Lourenço J.; Alcantara L. C. J.; Pond S. L. K.; Weaver S.; Martin D.; Lessells R. J.; Bhiman J. N.; Williamson C.; de Oliveira T. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020, 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Faria N. R.; Morales Claro I.; Candido D.; Moyses Franco L. A.; Andrade P. S.; Coletti T. M.; Silva C. A. M.; Sales F. C.; Manuli E. R.; Aguiar R. S.; Gaburo N.; da C. Camilo C.; Fraiji N. A.; Esashika Crispim M. A.; Carvalho M.; Rambaut A.; Loman N.; Pybus O. G.; Sabino E. C.. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. 2020, https://virological.org/t/586.
- Cheng M. H.; Krieger J. M.; Kaynak B.; Arditi M.; Bahar I. Impact of South African 501.V2 variant on SARS-CoV-2 spike infectivity and neutralization: a structure-based computational assessment. bioRxiv 2021, 10.1101/2021.01.10.426143. [DOI] [Google Scholar]
- Nelson G.; Buzko O.; Spilman P.; Niazi K.; Rabizadeh S.; Soon-Shiong P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv 2021, 10.1101/2021.01.13.426558. [DOI] [Google Scholar]
- Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367 (6485), 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C. L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367 (6483), 1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M. H.; Bastidas O. Static all-atom energetic mappings of the SARS-Cov-2 spike protein with potential latch identification of the down state protomer. bioRxiv 2020, 10.1101/2020.05.12.091090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur M.; Taka E.; Yilmaz S. Z.; Kilinc C.; Aktas U.; Golcuk M. Conformational transition of SARS-CoV-2 spike glycoprotein between its closed and open states. J. Chem. Phys. 2020, 153 (7), 075101 10.1063/5.0011141. [DOI] [PubMed] [Google Scholar]
- Ke Z.; Oton J.; Qu K.; Cortese M.; Zila V.; McKeane L.; Nakane T.; Zivanov J.; Neufeldt C. J.; Cerikan B.; Lu J. M.; Peukes J.; Xiong X.; Kräusslich H.-G.; Scheres S. H. W.; Bartenschlager R.; Briggs J. A. G. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498. 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D.; Alameh M. G.; de Silva T.; Collini P.; Hornsby H.; Brown R.; LaBranche C. C.; Edwards R. J.; Sutherland L.; Santra S.; Mansouri K.; Gobeil S.; McDanal C.; Pardi N.; Hengartner N.; Lin P. J. C.; Tam Y.; Shaw P. A.; Lewis M. G.; Boesler C.; Sahin U.; Acharya P.; Haynes B. F.; Korber B.; Montefiori D. C. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe 2021, 29 (1), 23–31. 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.; He Y.; Zhou Y.; Liu S.; Zheng B.-J.; Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7 (3), 226–236. 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Wan Y.; Luo C.; Ye G.; Geng Q.; Auerbach A.; Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (21), 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L.; Park Y. J.; Tortorici M. A.; Czudnochowski N.; Walls A. C.; Beltramello M.; Silacci-Fregni C.; Pinto D.; Rosen L. E.; Bowen J. E.; Acton O. J.; Jaconi S.; Guarino B.; Minola A.; Zatta F.; Sprugasci N.; Bassi J.; Peter A.; De Marco A.; Nix J. C.; Mele F.; Jovic S.; Rodriguez B. F.; Gupta S. V.; Jin F.; Piumatti G.; Lo Presti G.; Pellanda A. F.; Biggiogero M.; Tarkowski M.; Pizzuto M. S.; Cameroni E.; Havenar-Daughton C.; Smithey M.; Hong D.; Lepori V.; Albanese E.; Ceschi A.; Bernasconi E.; Elzi L.; Ferrari P.; Garzoni C.; Riva A.; Snell G.; Sallusto F.; Fink K.; Virgin H. W.; Lanzavecchia A.; Corti D.; Veesler D. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 2020, 183 (4), 1024–1042. 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C. O.; West A. P. Jr.; Huey-Tubman K. E.; Hoffmann M. A. G.; Sharaf N. G.; Hoffman P. R.; Koranda N.; Gristick H. B.; Gaebler C.; Muecksch F.; Lorenzi J. C. C.; Finkin S.; Hagglof T.; Hurley A.; Millard K. G.; Weisblum Y.; Schmidt F.; Hatziioannou T.; Bieniasz P. D.; Caskey M.; Robbiani D. F.; Nussenzweig M. C.; Bjorkman P. J. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 2020, 182 (4), 828–842. 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould M. I. J.; Kovaltsuk A.; Marks C.; Deane C. M. CoV-AbDab: the coronavirus antibody database. Bioinformatics 2020, btaa739. 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M.; Liu H.; Wu N. C.; Wilson I. A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021, 538, 192. 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banach B. B.; Cerutti G.; Fahad A. S.; Shen C.-H.; de Souza M. O.; Katsamba P. S.; Tsybovsky Y.; Wang P.; Nair M. S.; Huang Y.; Urdániz I. M. F.; Steiner P. J.; Gutiérrez-González M.; Liu L.; López Acevedo S. N.; Nazzari A.; Wolfe J. R.; Luo Y.; Olia A. S.; Teng I.-T.; Yu J.; Zhou T.; Reddem E. R.; Bimela J.; Pan X.; Madan B.; Laflin A. D.; Nimrania R.; Yuen K.-T.; Whitehead T. A.; Ho D. D.; Kwong P. D.; Shapiro L.; DeKosky B. J. Paired heavy and light chain signatures contribute to potent SARS-CoV-2 neutralization in public antibody responses. bioRxiv 2021, 10.1101/2020.12.31.424987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.; Xiang R.; Deng X.; Wang L.; Yu Z.; Tian S.; Liang R.; Li Y.; Ying T.; Jiang S. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5 (1), 212. 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E.; Piccini G.; Licastro D.; Casalino L.; Johnson N. V.; Paciello I.; Monego S. D.; Pantano E.; Manganaro N.; Manenti A.; Manna R.; Casa E.; Hyseni I.; Benincasa L.; Montomoli E.; Amaro R. E.; McLellan J. S.; Rappuoli R. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020, 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C. K.; Ayres F.; Hermanus T.; Madzivhandila M.; Kgagudi P.; Lambson B. E.; Vermeulen M.; van den Berg K.; Rossouw T.; Boswell M.; Ueckermann V.; Meiring S.; von Gottberg A.; Cohen C.; Morris L.; Bhiman J. N.; Moore P. L. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv 2021, 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- Tada T.; Dcosta B. M.; Samanovic-Golden M.; Herati R. S.; Cornelius A.; Mulligan M. J.; Landau N. R. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv 2021, 10.1101/2021.02.05.430003. [DOI] [Google Scholar]
- Wang P.; Liu L.; Iketani S.; Luo Y.; Guo Y.; Wang M.; Yu J.; Zhang B.; Kwong P. D.; Graham B. S.; Mascola J. R.; Chang J. Y.; Yin M. T.; Sobieszczyk M.; Kyratsous C. A.; Shapiro L.; Sheng Z.; Nair M. S.; Huang Y.; Ho D. D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. bioRxiv 2021, 10.1101/2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- Huang B.; Dai L.; Wang H.; Hu Z.; Yang X.; Tan W.; Gao G. F. Neutralization of SARS-CoV-2 VOC 501Y.V2 by human antisera elicited by both inactivated BBIBP-CorV and recombinant dimeric RBD ZF2001 vaccines. bioRxiv 2021, 10.1101/2021.02.01.429069. [DOI] [Google Scholar]
- Luan B.; Huynh T. Insights on SARS-CoV-2’s mutations for evading human antibodies: sacrifice and survival. bioRxiv 2021, 10.1101/2021.02.06.430088. [DOI] [PubMed] [Google Scholar]
- Collier D.; Meng B.; Ferreira I.; Datir R.; Temperton N.; Elmer A.; Kingston N.; Graves B.; McCoy L.; Smith K.; Bradley; Thaventhiram J.; Ceron-Gutierrez L.; Barcenas-Morales G.; Wills M.; Doffinger R.; Gupta R. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Davis B. D.; Chen S. S.; Martinez J. M. S.; Plummer J. T.; Vail E. Emergence of a novel SARS-CoV-2 strain in Southern California, USA. medRxiv 2021, 10.1101/2021.01.18.21249786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P. J. M.; Caniels T. G.; van der Straten K.; Snitselaar J. L.; Aldon Y.; Bangaru S.; Torres J. L.; Okba N. M. A.; Claireaux M.; Kerster G.; Bentlage A. E. H.; van Haaren M. M.; Guerra D.; Burger J. A.; Schermer E. E.; Verheul K. D.; van der Velde N.; van der Kooi A.; van Schooten J.; van Breemen M. J.; Bijl T. P. L.; Sliepen K.; Aartse A.; Derking R.; Bontjer I.; Kootstra N. A.; Wiersinga W. J.; Vidarsson G.; Haagmans B. L.; Ward A. B.; de Bree G. J.; Sanders R. W.; van Gils M. J. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369 (6504), 643–650. 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M. A.; Beltramello M.; Lempp F. A.; Pinto D.; Dang H. V.; Rosen L. E.; McCallum M.; Bowen J.; Minola A.; Jaconi S.; Zatta F.; De Marco A.; Guarino B.; Bianchi S.; Lauron E. J.; Tucker H.; Zhou J.; Peter A.; Havenar-Daughton C.; Wojcechowskyj J. A.; Case J. B.; Chen R. E.; Kaiser H.; Montiel-Ruiz M.; Meury M.; Czudnochowski N.; Spreafico R.; Dillen J.; Ng C.; Sprugasci N.; Culap K.; Benigni F.; Abdelnabi R.; Foo S. C.; Schmid M. A.; Cameroni E.; Riva A.; Gabrieli A.; Galli M.; Pizzuto M. S.; Neyts J.; Diamond M. S.; Virgin H. W.; Snell G.; Corti D.; Fink K.; Veesler D. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 2020, 370 (6519), 950–957. 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W.; He L.; Zhang X.; Pu J.; Voronin D.; Jiang S.; Zhou Y.; Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17 (6), 613–620. 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.; Du L.; Jiang S. Learning from the past: development of safe and effective COVID-19 vaccines. Nat. Rev. Microbiol. 2021, 19, 211. 10.1038/s41579-020-00462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.; Shang J.; Jiang S.; Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020, 11, 298. 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D. F.; Gaebler C.; Muecksch F.; Lorenzi J. C. C.; Wang Z.; Cho A.; Agudelo M.; Barnes C. O.; Gazumyan A.; Finkin S.; Hägglöf T.; Oliveira T. Y.; Viant C.; Hurley A.; Hoffmann H.-H.; Millard K. G.; Kost R. G.; Cipolla M.; Gordon K.; Bianchini F.; Chen S. T.; Ramos V.; Patel R.; Dizon J.; Shimeliovich I.; Mendoza P.; Hartweger H.; Nogueira L.; Pack M.; Horowitz J.; Schmidt F.; Weisblum Y.; Michailidis E.; Ashbrook A. W.; Waltari E.; Pak J. E.; Huey-Tubman K. E.; Koranda N.; Hoffman P. R.; West A. P.; Rice C. M.; Hatziioannou T.; Bjorkman P. J.; Bieniasz P. D.; Caskey M.; Nussenzweig M. C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584 (7821), 437–442. 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Wang W.; Chen Z.; Lu S.; Yang F.; Bi Z.; Bao L.; Mo F.; Li X.; Huang Y.; Hong W.; Yang Y.; Zhao Y.; Ye F.; Lin S.; Deng W.; Chen H.; Lei H.; Zhang Z.; Luo M.; Gao H.; Zheng Y.; Gong Y.; Jiang X.; Xu Y.; Lv Q.; Li D.; Wang M.; Li F.; Wang S.; Wang G.; Yu P.; Qu Y.; Yang L.; Deng H.; Tong A.; Li J.; Wang Z.; Yang J.; Shen G.; Zhao Z.; Li Y.; Luo J.; Liu H.; Yu W.; Yang M.; Xu J.; Wang J.; Li H.; Wang H.; Kuang D.; Lin P.; Hu Z.; Guo W.; Cheng W.; He Y.; Song X.; Chen C.; Xue Z.; Yao S.; Chen L.; Ma X.; Chen S.; Gou M.; Huang W.; Wang Y.; Fan C.; Tian Z.; Shi M.; Wang F.-S.; Dai L.; Wu M.; Li G.; Wang G.; Peng Y.; Qian Z.; Huang C.; Lau J. Y.-N.; Yang Z.; Wei Y.; Cen X.; Peng X.; Qin C.; Zhang K.; Lu G.; Wei X. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020, 586 (7830), 572–577. 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Pollet J.; Chen W. H.; Versteeg L.; Keegan B.; Zhan B.; Wei J.; Liu Z.; Lee J.; Kundu R.; Adhikari R.; Poveda C.; Mondragon M. V.; de Araujo Leao A. C.; Rivera J. A.; Gillespie P. M.; Strych U.; Hotez P. J.; Bottazzi M. E. SARS-CoV-2 RBD219-N1C1: a yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and t-cell immunity in mice. bioRxiv 2020, 10.1101/2020.11.04.367359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang J.; Gu C.; Zhou B.; Zhang C.; Yang Y.; Xu S.; Bai L.; Zhang R.; Deng Q.; Yuan Z.; Tang H.; Qu D.; Lavillette D.; Xie Y.; Huang Z. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discovery 2020, 6, 61. 10.1038/s41421-020-00199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan B. D.; Mou H.; Zhang L.; Guo Y.; He W.; Ojha A.; Parcells M. S.; Luo G.; Li W.; Zhong G.; Choe H.; Farzan M. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv 2020, 10.1101/2020.04.10.036418. [DOI] [Google Scholar]
- Clinical trial phase II NCT04718467, Clinical trial phase II RPCEC00000306, Clinical trial phase I NCT04522089.
- Zhang N. N.; Li X. F.; Deng Y. Q.; Zhao H.; Huang Y. J.; Yang G.; Huang W. J.; Gao P.; Zhou C.; Zhang R. R.; Guo Y.; Sun S. H.; Fan H.; Zu S. L.; Chen Q.; He Q.; Cao T. S.; Huang X. Y.; Qiu H. Y.; Nie J. H.; Jiang Y.; Yan H. Y.; Ye Q.; Zhong X.; Xue X. L.; Zha Z. Y.; Zhou D.; Yang X.; Wang Y. C.; Ying B.; Qin C. F. A thermostable mRNA vaccine against COVID-19. Cell 2020, 182 (5), 1271–1283. 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M. D.; Shukla S.; Chung Y. H.; Beiss V.; Chan S. K.; Ortega-Rivera O. A.; Wirth D. M.; Chen A.; Sack M.; Pokorski J. K.; Steinmetz N. F. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020, 15 (8), 646–655. 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- Dai L.; Zheng T.; Xu K.; Han Y.; Xu L.; Huang E.; An Y.; Cheng Y.; Li S.; Liu M.; Yang M.; Li Y.; Cheng H.; Yuan Y.; Zhang W.; Ke C.; Wong G.; Qi J.; Qin C.; Yan J.; Gao G. F. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell 2020, 182 (3), 722–733. 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Fiala B.; Schafer A.; Wrenn S.; Pham M. N.; Murphy M.; Tse L. V.; Shehata L.; O’Connor M. A.; Chen C.; Navarro M. J.; Miranda M. C.; Pettie D.; Ravichandran R.; Kraft J. C.; Ogohara C.; Palser A.; Chalk S.; Lee E. C.; Guerriero K.; Kepl E.; Chow C. M.; Sydeman C.; Hodge E. A.; Brown B.; Fuller J. T.; Dinnon K. H. 3rd; Gralinski L. E.; Leist S. R.; Gully K. L.; Lewis T. B.; Guttman M.; Chu H. Y.; Lee K. K.; Fuller D. H.; Baric R. S.; Kellam P.; Carter L.; Pepper M.; Sheahan T. P.; Veesler D.; King N. P. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 2020, 183 (5), 1367–1382. 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RPCEC00000332; RPCEC00000338; RPCEC00000349.
- Yang S.; Li Y.; Dai L.; Wang J.; He P.; Li C.; Fang X.; Wang C.; Zhao X.; Huang E.; Wu C.; Zhong Z.; Wang F.; Duan X.; Tian S.; Wu L.; Liu Y.; Luo Y.; Chen Z.; Li F.; Li J.; Yu X.; Ren H.; Liu L.; Meng S.; Yan J.; Hu Z.; Gao L.; Gao G. F. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD protein vaccine against COVID-19 in adults: pooled analysis of two randomized, double-blind, placebo-controlled, phase 1 and 2 trials. medRxiv 2020, 10.1101/2020.12.20.20248602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.; Tostanoski L. H.; Peter L.; Mercado N. B.; McMahan K.; Mahrokhian S. H.; Nkolola J. P.; Liu J.; Li Z.; Chandrashekar A.; Martinez D. R.; Loos C.; Atyeo C.; Fischinger S.; Burke J. S.; Slein M. D.; Chen Y.; Zuiani A.; Lelis F. J. N.; Travers M.; Habibi S.; Pessaint L.; Van Ry A.; Blade K.; Brown R.; Cook A.; Finneyfrock B.; Dodson A.; Teow E.; Velasco J.; Zahn R.; Wegmann F.; Bondzie E. A.; Dagotto G.; Gebre M. S.; He X.; Jacob-Dolan C.; Kirilova M.; Kordana N.; Lin Z.; Maxfield L. F.; Nampanya F.; Nityanandam R.; Ventura J. D.; Wan H.; Cai Y.; Chen B.; Schmidt A. G.; Wesemann D. R.; Baric R. S.; Alter G.; Andersen H.; Lewis M. G.; Barouch D. H. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369 (6505), 806–811. 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. J.; Lyke K. E.; Kitchin N.; Absalon J.; Gurtman A.; Lockhart S.; Neuzil K.; Raabe V.; Bailey R.; Swanson K. A.; Li P.; Koury K.; Kalina W.; Cooper D.; Fontes-Garfias C.; Shi P. Y.; Tureci O.; Tompkins K. R.; Walsh E. E.; Frenck R.; Falsey A. R.; Dormitzer P. R.; Gruber W. C.; Sahin U.; Jansen K. U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586 (7830), 589–593. 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Valdes-Balbin Y.; Santana-Mederos D.; Quintero L.; Fernandez S.; Rodriguez L.; Ramirez B. S.; Perez R.; Acosta C.; Méndez Y.; Ricardo M. G.; Hernandez T.; Bergado G.; Pi F.; Valdes A.; Ramirez U.; Oliva R.; Soubal J.-P.; Garrido R.; Cardoso F.; Landys M.; Gonzalez H.; Farinas M.; Enriquez J.; Noa E.; Suarez A.; Fang C.; Espinosa L. A.; Ramos Y.; González L. J.; Climent Y.; Rojas G.; Relova E.; Cabrera Y.; Losada S. L.; Boggiano T.; Ojito E.; Monzon K. L.; Chiodo F.; Paquet F.; Chen G.-W.; Rivera D. G.; Garcia-Rivera D.; Verez-Bencomo V. SARS-CoV-2 RBD-Tetanus toxoid conjugate vaccine induces a strong neutralizing immunity in preclinical studies. bioRxiv 2021, 10.1101/2021.02.08.430146. [DOI] [PubMed] [Google Scholar]
- RPCEC00000340.
- RPCEC00000347.
- Lopez-Sagaseta J.; Malito E.; Rappuoli R.; Bottomley M. J. Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 2016, 14, 58–68. 10.1016/j.csbj.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I.; Kim D.; Yu K.-M.; Seo H. D.; Lee S.-A.; Casel M. A. B.; Jang S.-G.; Kim S.; Jung W.; Lai C.-J.; Choi Y. K.; Jung J. U. Development of spike receptor-binding domain nanoparticle as a vaccine candidate against SARS-CoV-2 infection in ferrets. bioRxiv 2021, 10.1101/2021.01.28.428743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune K. D.; Leneghan D. B.; Brian I. J.; Ishizuka A. S.; Bachmann M. F.; Draper S. J.; Biswas S.; Howarth M. Plug-and-display: decoration of virus-like particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougeroux C.; Goksøyr L.; Idorn M.; Soroka V.; Myeni S. K.; Dagil R.; Janitzek C. M.; Søgaard M.; Aves K.-L.; Horsted E. W.; Erdoǧan S. M.; Gustavsson T.; Dorosz J.; Clemmensen S.; Fredsgaard L.; Thrane S.; Vidal-Calvo E. E.; Khalifé P.; Hulen T. M.; Choudhary S.; Theisen M.; Singh S. K.; Garcia-Senosiain A.; Van Oosten L.; Pijlman G.; Hierzberger B.; Domeyer T.; Nalewajek B. W.; Strøbæk A.; Skrzypczak M.; Andersson L. F.; Buus S.; Buus A. S.; Christensen J. P.; Dalebout T. J.; Iversen K.; Harritshøj L. H.; Mordmüller B.; Ullum H.; Reinert L. S.; de Jongh W. A.; Kikkert M.; Paludan S. R.; Theander T. G.; Nielsen M. A.; Salanti A.; Sander A. F. Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 2021, 12 (1), 324. 10.1038/s41467-020-20251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.; Zou F.; Yu F.; Li R.; Yuan Y.; Zhang Y.; Zhang X.; Deng J.; Chen T.; Song Z.; Qiao Y.; Zhan Y.; Liu J.; Zhang J.; Zhang X.; Peng Z.; Li Y.; Lin Y.; Liang L.; Wang G.; Chen Y.; Chen Q.; Pan T.; He X.; Zhang H. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity 2020, 53 (6), 1315–1330. 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. A.; Gnanapragasam P. N. P.; Lee Y. E.; Hoffman P. R.; Ou S.; Kakutani L. M.; Keeffe J. R.; Wu H. J.; Howarth M.; West A. P.; Barnes C. O.; Nussenzweig M. C.; Bjorkman P. J. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 2021, 371 (6530), 735–741. 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Wang F.; Shen C.; Peng W.; Li D.; Zhao C.; Li Z.; Li S.; Bi Y.; Yang Y.; Gong Y.; Xiao H.; Fan Z.; Tan S.; Wu G.; Tan W.; Lu X.; Fan C.; Wang Q.; Liu Y.; Zhang C.; Qi J.; Gao G. F.; Gao F.; Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368 (6496), 1274–1278. 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Su B.; Guo X.; Sun W.; Deng Y.; Bao L.; Zhu Q.; Zhang X.; Zheng Y.; Geng C.; Chai X.; He R.; Li X.; Lv Q.; Zhu H.; Deng W.; Xu Y.; Wang Y.; Qiao L.; Tan Y.; Song L.; Wang G.; Du X.; Gao N.; Liu J.; Xiao J.; Su X. D.; Du Z.; Feng Y.; Qin C.; Qin C.; Jin R.; Xie X. S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 2020, 182 (1), 73–84. 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Wang P.; Nair M. S.; Yu J.; Rapp M.; Wang Q.; Luo Y.; Chan J. F.; Sahi V.; Figueroa A.; Guo X. V.; Cerutti G.; Bimela J.; Gorman J.; Zhou T.; Chen Z.; Yuen K. Y.; Kwong P. D.; Sodroski J. G.; Yin M. T.; Sheng Z.; Huang Y.; Shapiro L.; Ho D. D. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584 (7821), 450–456. 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Ju B.; Zhang Q.; Ge J.; Wang R.; Sun J.; Ge X.; Yu J.; Shan S.; Zhou B.; Song S.; Tang X.; Yu J.; Lan J.; Yuan J.; Wang H.; Zhao J.; Zhang S.; Wang Y.; Shi X.; Liu L.; Zhao J.; Wang X.; Zhang Z.; Zhang L. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584 (7819), 115–119. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- De Lano W. L.Pymol; De Lano Scientific: South San Francisco, CA. 2002.
- Goddard T. D.; Huang C. C.; Meng E. C.; Pettersen E. F.; Couch G. S.; Morris J. H.; Ferrin T. E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27 (1), 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]