Abstract
Antimicrobial peptides (AMPs) are ubiquitously present small peptides, which play a critical function in the innate immune system. The defensin class of AMPs represented an evolutionarily ancient family containing cationic cysteine residue and frequently expressed in epithelial or neutrophils cells. It plays myriad functions in host innate immune responses against various infection. Defensin has a broad spectrum of antimicrobial activities, including anti-bacteria, anti-viruses (AVPs), anti-fungi, anti-cancers, and also overcoming bacterial drug resistance. In this review, we compiled the progress on defensin, particularly incorporating the mechanism of action, their application as an antiviral agent, prospects in different areas, and limitations to be solved as an antiviral peptide. Defensins were explored, in particular, their capacity to stimulate innate and adaptive immunity by trigging as anti-coronavirus (COVID-19) peptides. The present review summarised its immunomodulatory and immunoenhancing properties and predominantly focused on its promising therapeutic adjuvant choices for combat against viral infection.
Keywords: Human defensins, Adjuvants vaccine, Immunomodulation, antiviral agent, antimicrobial peptide, Innate immunity
1. Introduction
Antimicrobial peptides (AMPs) are observed in almost all organisms. These are oligopeptides with short positively charged, which manifest a heterogeneity of structures and functions. AMPs are the integral element of the innate immune system and are produced against several infections in a wide range of organs. Most of them are generated as abundant polyprotein precursors, and post-processing delivers active peptides. These peptides were classified into several classes. Almost all the AMPs bestowed to hold antimicrobial activity, but only a few of them demonstrated antiviral features, and defensins are one of the significant class. Here we report on the application of human defensins in treating viral infections, with particular emphasis on SARS. In 2002, Severe Acute Respiratory Syndrome (SARS) virus originated in Guangdong, China, that infected 8422, leading to 916 deaths worldwide [1,2]. After a decade, in 2012, the MERS-CoV virus emerged as a zoonotic disease in the Middle East from bats to humans. MERS-CoV infects the non-ciliated respiratory tract epithelial cells via dipeptidyl peptidase 4 and CD26 receptors cause 806 deaths [3].
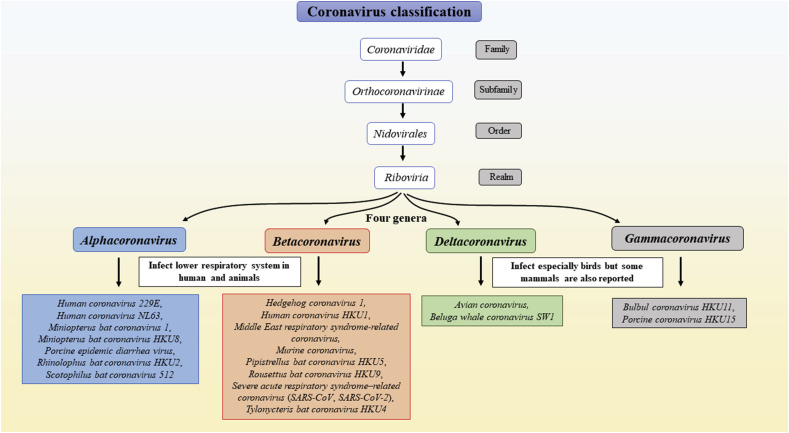
While scientists were looking for the mechanism of pathogenicity of SARS-CoV, and MERS-CoV, another highly contagious Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus pandemic outbreak, occurred in late December 2019 [4]. Bioinformatics analysis revealed that the SARS-CoV-2 virus belongs to the Coronaviridae family, and it shows 50% and 79% sequence similarity to its close relative MERS virus and SARS virus, respectively (Fig. 1 ).
Fig. 1.
The detailed classification of the coronavirus family for humans and birds.
The virus attacks the respiratory tract via ciliated epithelial cells using angiotensin-converting enzyme 2 (ACE2) & TMPRSS2 receptors [4]. The SARS-CoV-2 expanding pandemic has become globally challenging because of the high rate of mutations in its positive-stranded RNA genome and evolved spike proteins [5]. However, no specific drugs or treatment availability aggravated the situation [[6], [7], [8], [9]]. The pandemic severity could be estimated because, within few months, the novel SARS-CoV-2 virus surpassed the SARS and MERS-related severity. World researchers are searching for specific therapeutic strategies to prevent the COVID19-pandemic outbreak. [1,8]; https://www.worldometers.info/coronavirus/).
1.1. Virus structure
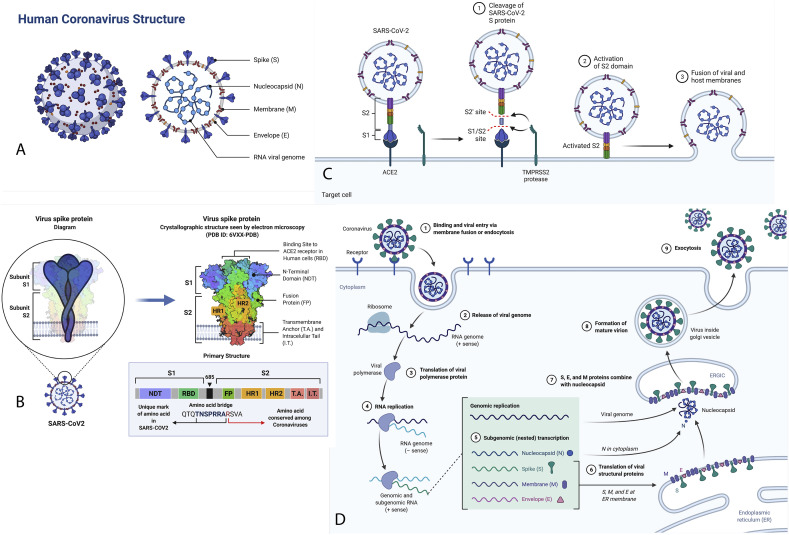
Homology study found that the SARS-CoV-2 and SARS-CoV virus has typical receptors and share 96% identity for primary protease enzyme (Fig. 2 ). SARS-CoV-2 virus (125 nm-sized) shares a 96.3% identity with bat-coronavirus RaTG13. The SARS-CoV-2 spike gene shows a ~75% sequence identity with human SARS-CoV and bat SL-CoVZXC21 [8]. The enveloped virus has a non-segmented phosphorylated nucleocapsid covered with 26–32 kb long positive-sense single-stranded RNA genomes (GeneBank ID MN908947). The genome encodes 9860 amino acids which include S-proteins, nucleocapsid proteins, membrane proteins, proteases (Mpro) and chymotrypsin-like protease (3CLpro), replication (pp1a and pp1ab), helicase (Non-structural protein-13, Nsp13), RNA-dependent RNA polymerase (Nsp12), and translational enzymes [6,8]. Nsp12 (3.1 Å) polymerase enzyme binds to Nsp7 and Nsp8 proteins to form a ~160 kDa functional RNA-synthesis machinery platform [4]. The virus majorly affects the respiratory and gastrointestinal tract. It causes several symptoms, including dry cough, common cold, pneumonia, headache, fever, sore throat, dyspnea, lungs oedema, renal dysfunctions, fatigue, hyperplasia, diarrhoea, haemorrhage, multiple patchy shadows, interstitial infiltrations, multiple patchy shadows in chest CT scan, and altered humoral & adaptive immunity. The numerous complications result in organ failure, leading to the patient's death [2,6,10] (Fig. 3 ).
Fig. 2.
A. The structure composition of COVID19. B. The S glycoprotein of the newly discovered SARS-CoV2 is composed of two subunits, S1 and S2, and is commonly represented as a sword-like spike. The Protein Data Bank (PDB) model of this glycoprotein reveals how the subunits comprise different regions that are fundamental to the infection process. S1 and S2 are linked together by a polybasic amino acid bridge, which may be necessary for studying viral targeting. C. Mechanism of SARS-CoV-2 Viral Entry. D. This pathway overview the coronavirus replication cycle. The figure depicts viral development from initial binding and release of the viral genome to eventual exocytosis of the mature virion.
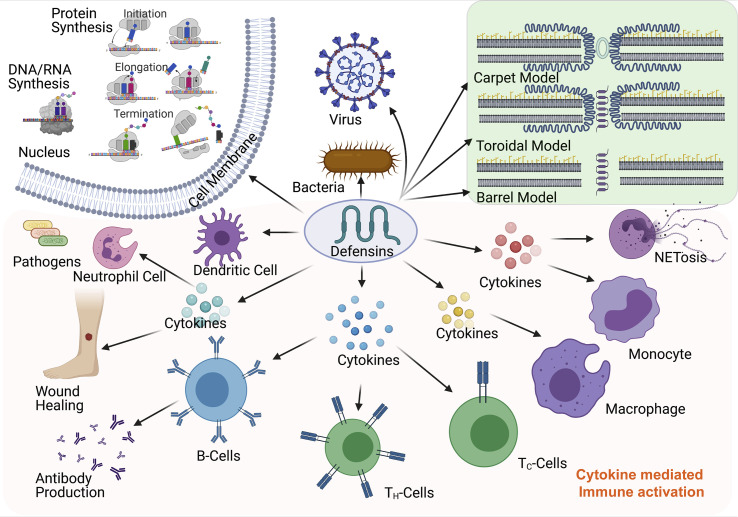
Fig. 3.
The illustration for the mechanism of actions of defensin peptides against pathogens and related activation of cytokines and cell types.
SARS-CoV-2 has an incubation period between 2 and 11 days (on average, within five days, the symptom could develop), and infected people have a recovery rate of 12–32 days. Diabetic and overage, people have more risk of getting SARS-CoV-2 infection. The non-symptomatic viral RNA could for remain in the blood up to 29 days. The drugs like Hydroxychloroquine (HCQ) and Azithromycin antibiotic treatment showed lowering the viral load through cell alkalinity action. The best strategy to prevent infection is a complete quarantine that benefits with no asymptomatic transfer [2,6]. COVID19 infected patients reported altered humoral immunity with the cytotoxic pattern, leukocytopenia, and lymphopenia than the non-infected person. The reports suggest that host antimicrobial peptides and immune cells play an essential role in lowering the virus load.
1.2. Virus host interaction
Coronavirus S-glycoprotein (S1 & S2) binds to cellular receptors (ACE2), leads to penetration of cells, and resulting in the initiation of disassembly of viral nucleocapsid to release viral RNA genome [6,11]. Other viral proteins like membrane, envelope, replicates, and protease help packs new viral particles. Trimeric S-protein consists of 1160–1400 amino acids, converted into S1 & S2 type-1 transmembrane glycoproteins by viral protease enzyme. The S-protein sequence 318–510 was reported as highly antigenic and modulating immune systems [3,6,12]. The S1-protein with 685 amino-acids sequence act as a receptor-binding domain (RBD) (Fig. 2).
It assists virus interactions with host cell receptors such as ACE-2, carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1), dipeptidyl peptidase-4 (DPP4), aminopeptidase-N (APN), and carbohydrates. Another S2-protein contains 588 amino acids with two fusion core subunits, heptad repeats-1 (HR1) and HR2, which form a hydrophobic six-helix bundle core result union the host cell membrane [3].
Predominantly, the ACE2 receptor present in the respiratory and intestinal wall has emerged as the most potential receptors for SARD-CoV entry. The protease activity on ACE2 from respiratory epithelial cell make ADAM17 a moving target for COVID19 [13]. The molecular weight ranges from 105 kDa to 120 kDa by proteolytic activity of ADAM17. International collaboration will open new avenues for the understanding of its proteomics structures [14]. Transcriptomics and proteomics studies from different tissues within the same individuals have a different cellular functional environment. The viral genome CpG Islands are targeted by the nucleotide-binding domain of the zinc finger antiviral protein (ZAP). The tissues specific communication between the CpG nucleotides and ZAPs recruit different nucleases to degrade viral genomes. Therefore evolved viruses majorly modified their CpG content in a manner to tissues particular infections. It has been reported that SARS-CoV-2 genomes have a significantly less amount of CpG content, suggesting their evolved mechanism to skip ZAP's actions [15].
1.3. Defensins
The mammals possess an immune surveillance system that protects them from pathogens. The defence system includes monocytes, macrophages, B-cells, T-cells, antigen-presenting cells such as neutrophils, dendritic cells, and immunostimulators cytokines & chemokines. These immune cells recognise pathogens by PPRs (pattern recognition receptors such as TLRs, RIG-I, lectin, and NOD) and PAMPs (pathogen-associated molecular patterns) [[16], [17], [18]]. The first line of the defence system protects the mammalian body from exposure to several airway microorganisms by producing nonspecific host defence antimicrobial peptides (HDPs) or antimicrobial peptides (AMPs) [19]. HDPs are produced by the invertebrate's tracheal mucosa lining, lungs, intestine, skins, mammary gland, and reproductive organs.
Mammalian AMPs are broadly classified into two principal categories, Cathelicidins (LL-37) and Defensins. The cathelicidin family embraces small cationic peptides, synthesised and stored in neutrophils and macrophages. They are part of the innate immune system and are generally proteolytically active proteins. Cathelicidin LL-37, an α-helical AMP, binds to viral glycoproteins and prevents virus cell entry [20]. However, defensin is a family of AMPs consisting of 6 cysteine residues that form 3 disulfide bonds and contain three subfamilies: α-defensins, β-defensins, and θ-defensins. All of these three differ from each other in the arrangement of disulfide bonds. These are found in tissues involved in the host immune response against microbial infections and are abundant in leukocyte granules. The α-defensins are present in several mammalian groups. Defensins are small cysteine-rich cationic peptides with a broad range of antimicrobial activity against bacteria, viruses, mycoplasma, tumour, and fungi. Defensin acts on the membrane or envelopes wall by its amphipathic nature [[16], [17], [18]].
1.4. General mechanism of actions
Defensins are antimicrobial peptide that takes part within the innate immunity of hosts. Humans have constitutively and/or inducible expression of α- and β-defensins known for their antiviral and antibacterial activities. The innate immune reaction incorporates the generation of complements proteins and interferons (IFNs). AMPs are vital for controlling contagious diseases and actuating adaptive immunity. It also acts as multifunctional peptides that participate in the disposal of pathogenic microorganisms, including bacteria, viruses, and fungi. In specific, defensins, which are significant AMP families in warm-blooded animals, contribute to the innate antimicrobial response by disrupting the cell wall of pathogens. The classical mechanistic step of cationic AMPs, such as defensins, is the disturbance of the anionic bacterial membrane [21]. That way, bacterial destruction occurs by the interaction between the electrostatic forces of positively charged amino acids within defensin(s) and the negatively charged on the cell surface [22]. Since the external surface of all microbes contains a negative charge (due to the presence of teichoic acids and/or lipopolysaccharides), positively charged and hydrophobic AMPs (in specific, defensins) nonspecifically “accumulate” on the surface of both gram-negative and gram-positive microorganisms. The antibacterial action of defensins is believed to be related to the membrane permeabilisation of microorganisms [23]. Nevertheless, a few AMPs have been found to use alternative components of antimicrobial activity [[24], [25], [26], [27]]. It has appeared that HNP-1 can hinder the synthesis of the bacterial cell wall by interacting with the forerunner of lipid II [28].
In a more elaborated way, bacterial layers are wealthy in negatively charged phospholipids, such as phosphatidylserine (PS), phosphatidyl-glycerol (PG), and cardiolipin (CL), which are further stabilised by divalent cations such as Ca2+ and Mg2+ ions. However, Gram-negative bacteria have an external membrane rich in an extra lipopolysaccharide, which stands as a barrier to the cytoplasmic layer. On the other hand, human cells are wealthy in neutrally charged phospholipids, such as phosphatidylcholine (PC), phosphatidylethanolamine (PE), and sphingomyelin (SM). This difference in membrane composition between humans and microbes makes AMPs exceedingly particular against microbes [29].
Three electrostatic models explain the action of AMPs on bacterial membranes: (1) the carpet model, which proposes that peptides are retained parallelly in the bilayers and, after accomplishing an adequate coverage, generate a cleanser effect and devastate the membrane; (2) the barrel stave model, in which peptides would be introduced perpendicularly within the bilayers, coalescing together to create a pore; and (3) the toroidal pore model, which suggests that peptides are presented perpendicularly in the lipid bilayer and produce a territorial membrane curvature, where a pore is shaped by both peptides and phospholipid head bunches [30,31]. For instance, the HNP-1 mechanism of action against E. coli contrasts against S. aureus (destruction of cell layer vs confinement of bacterial wall forerunner lipid II). HD-5 is successful in killing both gram-positive and -negative bacteria by increasing bacterial membrane penetrability. It has moreover been reported that HD-5 can bind to DNA and restrain cell replication. One component of the activity of HD-6 is the formation of nanonets and the capture of bacteria before they make physiological contact with epithelial cells, which anticipates bacterial attack [29]. Simultaneously, two mechanisms have been reported for hBD-3, the devastation of bacterial membrane and interaction with lipid II forerunner. A decrease of hBD-1 disulfide bridges increases its activity against pathogens. The recently detailed mechanism of action proposes that it forms extracellular traps (web-like structures containing AMPs) with neutrophils that capture and annihilate bacteria. hBD-2 acts by binding to negatively charged film phospholipids, inducing efflux of intracellular components and leading to cell death [32].
Although the expression of defensin genes is tightly controlled by cytokines as a candidate of the host defence and is suppressed by different virulent components of pathogens [33], it is critical to note that recently defensins were reported to regulate adaptive immunity by activating the recruitment and enactment of immune cells through various pathways connected with innate immunity [34]. For example, HBDs are chemotactic for immature dendritic cells (DCs) and memory T cells to the location of pathogen invasion by association with CCR6 and advance the adaptive resistant reaction by enlisting resistant cells [35,36].
1.5. Defensins: The antiviral specific function and mechanism of action
Defensins mainly act by removing the pathogens through membrane depolarisation, opsonisation of immune cells, cytokines stimulations, and inhibiting replication by inhibiting nuclear enzymes [[16], [17], [18]]. Six α-defensins (HNP1, HNP2, HNP3, HNP4, HD5, and HD6) and ~37 β-defensins are reported in humans. In that HNP1, HNP2, HNP3, HNP4 are produced by granulocytes, and HD5 and HD6 are reported in intestinal tissues. α-defensin consists of three exons encoding for UTR regions, signal peptide, and mature functional peptide. α-defensins attract the antigen-presenting cells at the site of infection by enhancing the expression of IL-10, ICAM-1, CD11b, CD11c, cytokines, and chemokines. These physiological altered cytokines and chemokines activate the immature or non-functional T-cells, natural killer cells, neutrophils, and dendritic cells, eliminating the viral pathogens by inducing humoral and adaptive immunity [37,38]. β-defensin consists of two exons encoding for the signal peptide and mature functional peptide. Both α and β defensins are primarily produced as pre-propeptide, later enzymatically cleaved to generate mature active peptides [37,39]. High concentrations of beta-defensins are associated with psoriasis, autoimmunity, and cytotoxicity [40]. It was found that using the bio-computational analysis of antimicrobial peptides databases has seven AMPs AP00180, AP00223, AP00225, AP00549, AP00729, AP00744, and AP00764, which bind most potentially to the MERS-CoV spike proteins and could appear biomarkers for the early diagnosis of its infections [3].
However, defensin's action on the virus is not directly related to membrane depolarisation as in bacteria. It acts as a linkage between immune surveillance and viral infections. Human beta-defensin 2 (HBD2) and HBD3 peptides are induced by inflammatory cytokines upon the pathogens invading (Fig. 4 ). HDP initially is reported as an antiviral peptide against the Herpes simplex virus (HSV). The constitutive expression of broad-spectrum of nonspecific HDPs in the invertebrates is a natural barrier for preventing bacteria, viruses, and other pathogens.
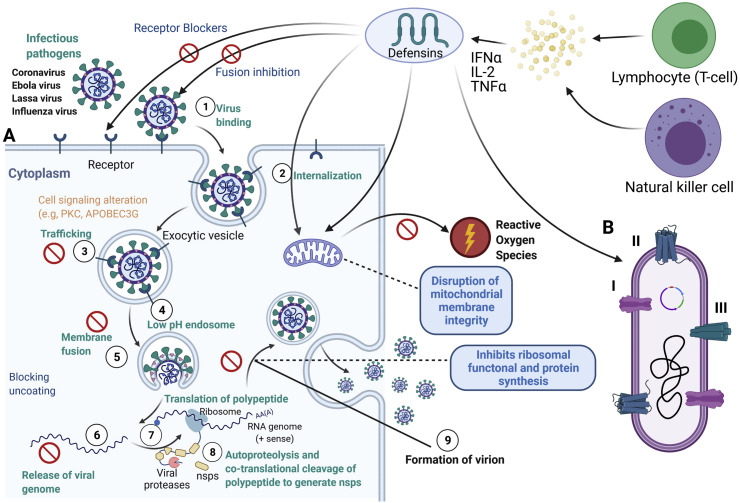
Fig. 4.
A. Antiviral action of defensins and how it involves in the inhibition of the enveloped and non enveloped viral based infections. B. Different modes of action as an antimicrobial peptide in the bacterial cell.
Nonetheless, even though the defensins are present, we still get infections; this raises the question of whether the body needs to produce more defensins or needs supportive adjuvants/vaccines to eliminate pathogens? [40]. In this regards, the recent studies on subunit vaccines report that defensin or peptides act as an excellent adjuvant to boost humoral and cellular immunity. It assists in eliminating the invading pathogens when supplied along with the vaccine. HBD 2-conjugated S-protein receptor-binding domain (S-RBD) elevated the expression of antiviral cytokines such as IFN-β, IFN-γ, PKR, RNase L, TNF-α, IL-1β IL-6, and NOD2. It also induces the activation of immune cells such as natural killer cells, T-cells, dendritic cells, macrophages, granulocytes, and higher S-protein neutralising antibodies than alone S-RBD.
HNP1-3 defensins inactivate the different categories of viruses like herpes simplex virus-1 (HSV-1), HSV-2, IAV, and vesicular stomatitis virus in physiological conditions based on in-vitro assays [41]. The results showed that HNP1 and HD5 lead to aggregation of BK polyomavirus and restrain its early replication [42]. hBD-1 and hBD-3 show intense antimicrobial activity against respiratory infections [43,44]. TNF-α- and IFN-γ induced HBD2, and HBD3 antimicrobial peptides bind to the CCR6 receptor of APCs cells and directly link innate and adaptive immune systems. In atopic dermatitis (AD) and cystic fibrosis (CF) diseases, HBD2 and HBD3 expressions are inhibited by IL-4 and IL-13 induced STAT-6 mediated suppressors of cytokine signalling-1 (SOCS-1) and SOCS-3 pathways [45].
Other organisms like equine beta-defensins 1 (eBD1), eBD2, and eBD3 are induced in the respiratory tract in response to equine herpes virus infections [46]. Porcine β-defensin 2 affects bacteria and pseudorabies virus [47]. P9 peptide derived from mouse β-defensin-4 makes aggregation with viral glycoproteins, which prevent virus RNA release, and P9 has reported a broad range of antiviral properties against various respiratory viruses, including MERS-CoV, SARS-CoV, H7N9, IAV, H1N1, H5N1, and H3N2 [48]. The rat beta-defensin-2 and IL-22 emerged biomarkers for the multidrug-resistant Klebsiella pneumonia bacterial infection [49]. The primates (non-human) θ-defensins mimic synthetic retrocyclins such as hapivirins, and diprovirins peptides neutralise the viruses, including H3N2, HIV-1, IAV, H1N1, HSV-1, and SARS-CoV [50] (Fig. 4A).
There are different modes and determinants of defensin binding to viruses. First, defensins interact with lipid bilayers, which is encouraged by the presence of adversely charged phospholipids. Four of the α-defensins (HNP1-3 and HD5) and HBD3 are lectins capable of binding to glycoproteins and glycolipids. Moreover, defensins can interact with protein–DNA or protein-protein interactions. Since they are cationic and amphipathic, defensins can bind with ligands through both charge–charge and hydrophobic interactions. Defensin oligomerisation, particularly for α-defensins, and conformational stability conferred by disulfide bonds further impact binding. Each of these interactions contributes to the antiviral action of defensins, and their relative significance depends on the specific virus/defensin combine beneath examination (Fig. 4B).
Moreover, direct interactions between defensins and structural components of the viruses, especially the lipid bilayer of encompassed viruses, may devastate or destabilise the virus and render it non-infectious. In contrast, numerous defensin generates multimeric structures, which have been illustrated in both crystal structures and arrangement. Interactions between defensin peptides to neighbouring viruses will cause virions to be aggregate. For instance, it has been demonstrated for IAV by HNP1 and HNP2 and for HAdV-5 and BKV by HD5 [51]. Likewise, defensin binding to viral proteins could disrupt receptor interactions critical for viral entry into the cell. HNP1-3, HD5, and HBD3, for example, bind with recombinant viral glycoprotein (gB) of both HSV-1 and HSV-2 to prevent adhesion. Also, HNP4 and HD6 inhibit HSV-1 and HSV-2 infection but could not interact with viral glycoproteins [52]. HNP4 and HD6 bind to heparan sulfate, the receptor for attachment, as well as other glycosaminoglycans. HBD3 is the only defensin capable of interacting with both host cell receptors and viral glycoproteins. Additionally, HNP1-4 interacts with HIV-1's gp41 and gp120 and the cell surface receptor CD4 [53,54].
Nevertheless, defensins have the opportunity to neutralisation the virus even after entry or post-entry. Infection is not completed by just penetrating the host cell membrane. Viral transcription, protein production, assembly, and departure must all happen to achieve a replicative cycle. These steps allow defensins to block viral infection by focusing on the virus, mainly targeting the cell. For instance, HNP2 and HD5 can interact with HSV-2 DNA and conjecture that this seems to contribute to inhibition by blocking gene expression, even though a post-transcription block by an unknown mechanism [55]. Moreover, numerous types of papillomaviruses are sensitive to HNP1-4 and HD5. The only known step that is currently hampered is the nuclear localisation of the HPV-16 genome, the final phase in the virus entry pathway [56]. Unlike the case for HAdV-C neutralisation, the HPV genome is exposed beneath HNP1 and HD5 neutralisation conditions, implying that virus uncoating occurs in the presence of the defensins [56,57]. Defensin's antiviral mechanisms are targeting the host cell by blocking fusion by cross-linking host proteins. Instead of focusing on viral fusion proteins to block, the enveloped virus fused with the host cell. Whereas the HBD3 and an engineered human θ-defensin called retrocyclin 2 appeared to repress IAV fusion cross-linking host glycoproteins [58]. Retrocyclin 2 and HBD3 binding restrain the mobility of host surface proteins within the early viral fusion pore region, limiting its maturation to complete fusion.
Numerous viruses regulate the protein kinase C (PKC) signalling amid entry and infection: HIV-1 requires phosphorylated PKC for viral fusion, transcription, integration, and gathering [59,60]. IAV requires PKC for endosomal elude and nuclear entry [61,62]. HSV requires PKC for cell entry and nuclear departure of the viral capsid [63,64]. As HNP1-3 is known to hinder the action of PKC in vitro [65], modifying or interfering with this cellular signalling pathway may be another defending mediated antiviral mechanism, which clarifies the post-entry block to infection watched for some viruses. HNP1 treatment of cells earlier to or during infection with either IAV or HIV-1 diminishes the levels of phosphorylated PKC.
Cell signalling pathways interceded by the chemokine receptor CCR6 moreover play a part in defensin-mediated HIV inhibition. HBD2 is known to bind CCR6 and has been appeared to initiate expression of host restriction factor APOBEC3G (apolipoprotein B mRNA-editing chemical, catalytic polypeptide-like 3G) that has antiviral action against HIV [66,67]. Thus, defensins can both repress cellular pathways required for viral infection and enact intracellular antiviral mechanisms. The importance of defensins in viral pathogenesis in vivo has been demonstrated earlier. For instance, administration of rhesus θ-defensin protected mice from lethal SARS coronavirus challenge without affecting lung viral titers, likely due to a decrease in immunopathology within the treated animals [68]. There is no illustration of a direct role for endogenous defensins in blocking virus infection in vivo; in huge part, the lack of a complete defensin knockout animal demonstrates model generation [69].
1.6. Defensins against SARS-CoV
The lectin-like human α defensin 5 (HD5) and HD6 peptides protect intestinal infections. Lectins are proteins or glycoproteins that specifically bind to carbohydrates present on the glycoproteins surface. 2019-nCoV S-proteins and its intestinal receptor ACE2 both are glycosylated proteins. HD5 binds to the ligand-binding domain (LBD) of ACE2 glycoproteins and masks its availability to enter the 2019-nCoV virus. This interruption between viral glycoproteins and cellular receptors activates the adaptive immune antigen-presenting phagocytes cells and interferes with nuclear enzymes that prevent viral cellular replication (Park et al., 2018). Human neutrophil peptide-4, bovine beta-defensin 1, HD5, HD6, rat neutrophil peptide 1 (RNP-1), RNP-2, chicken avian beta-defensin 5 (AvBD5), AvBD6, and channel catfish beta-defensin peptide have the antiviral activity against the Middle East Respiratory Syndrome Coronavirus [3]. HNP1-6, HD5-6, and HBD2-3 defensins remove the viruses by getting in the way of viral entry, membrane destabilisation, immune-modulating, downregulating receptors and ligands such as ACE2, CD4, CCR6 CXCR4, CXCL & CXCL5, lysosome fusion, inhibiting essential proteases, central dogma enzymes, and post assembly of virion particles [37,70].
Bio-computational designed Brilacidin is a synthetic non-peptide defensin mimic drug that destabilises the viral membrane by its amphipathic & hydrophobic nature, and its immunomodulatory property influence the expression of IL-1β, IL-6, TNF-α cytokines, and cAMP& PDE4/PDE3 pathways which are associated with bronchodilator and anti-inflammatory effects of COVID-19 disease. The smaller size, more effective antimicrobial activity, bioavailability, low enzymatic degradation and toxicity, natural and low-cost production make the Brilacidin a most potent drug against SARS-CoV-2 than the known antiviral drugs. Dalbavancin, Telavancin, Oritavanci, and Roflumilast are other Brilacidin like drugs used for MERS, SARS, and SARS-CoV2 viral inhibitions [71]. Rhesus macaque's leukocytes express θ-defensin1 (RTD-1) and RTD-2 antimicrobial peptides in response to viral infections. HD5-6, HNP1-6, HBD2-6, and retrocyclin-2 inactivate the various enveloped and non-enveloped virion particles by interrupting virus pre and post-entry steps, chemokines, and cytokines immunomodulation, chemotaxis of immune cells [72].
1.7. Secondary infection or co-infections
The secondary infection or co-infections of other harmful pathogens have reported respiratory system infecting viruses such as SARS, MERS, and Spanish influenza. Coronaviruses majorly affect the respiratory and intestinal systems and thus cause the suppression of immunity. The chance of secondary bacterial or fungal infections increases when the patient suffering from low immunity, especially individuals with hepatitis or diabetes, could face dangerous high complications like ketoacidosis and inflammations [[73], [74], [75]].
The Spanish influenza pandemic was reported for co-infections of severe bacterial species. The significant co-infections of negative bacilli bacteria and Candida fungus in the lower respiratory tract were reported in the SARS virus infection [75]. As the SARS-CoV-2 pandemic spread across the world and caused hundreds of thousands of infections and death cases, it has been predicted that more substantial secondary infection threats are lurking behind SARS-CoV-2 infections [76]. The respiratory COVID-19 lung infection predisposes to bacterial and viral infections such as Streptococcus pneumoniae, S. aureus, Escherichia coli Klebsiella pneumonia, Enterococcus, and Influenza [77]. Another risk of hidden threat is increasing the multidrug resistance in the secondary infections causing bacterial species.
In this regard, defensins are known for their broad range of antimicrobial activities, immune stimulations, and non-resistance to drugs. Porcine IL-22 (pIL-22) induces the expression of the beta-defensin 2, IL-18, and IFN-l against invading porcine epidemic diarrhoea virus (PEDV), porcine rotavirus (PoRV), and enteric coronaviruses infections [78]. RBD conjugated HBD2 influences innate immunity and enhanced adaptive immunity by increasing the expression of proinflammatory cytokines such as IFNs and nod2 in the case of MERS-Cov respiratory viral infections [79]. RBD specific REGN3048 and REGN3051 antibodies inhibit the MERS-CoV infection [80]. Temporins AMPs have MERS-CoV binding properties [81]. Defensin 4, bactericidal-permeability increasing protein (Bpifa1), and cathelicidins antimicrobial peptide activate the phagocytic macrophages and dendritic immune cells in the IAV infections [82].
The recognition of PAMPs by TLRs guides the synthesis of cytokines, chemokines, and defensin AMPs by activation of MAP kinase and NFk cascades. In this way, the increased expression of defensin recruits the adaptive immune cells at the site of infections [83]. The direct association of HNP1 and HNP5 alfa-defensins with pVI capsid protein inhibits the disassembly of adenovirus. HNPs affect the influenza viral replication and immune system by interfering with the host protein kinase C pathway and the hemagglutination-inhibiting activity of surfactant protein D. HNPs has also been reported for their involvement in the aggregation of IAVs and bacteria, which increase their uptake by immune cells [[84], [85], [86], [87]]. Nine human defensins have been reported to interfere with herpes simplex viral cellular entry [88]. HD5 defensin directly binds to the BK virus (BKV) and involve in the virion aggregation uptake by immune cells the immune cells' virion aggregation uptake [42]. Theta-defensin decreases the ICP4 expression and VP6 protein transport of HSV [89]. hBD2 and hBD3 affect the early transcription of X4 and R5 HIV viruses [90]. The lectin binding domains of HNP1 & HNP4 binds to the O- and N-glycosylated HIV-gp120 protein receptors and interferes with the nuclear protein kinase C signalling for the synthesis of cytokines and chemokines [72]. Defensin adjuvant with HIV peptide interacts with CCR6 receptors induces the recruitment and proliferation of T-cells, dendritic cells, and macrophages [91]. The combination of inactivated fowl virus, Poly di-sodium carboxylatoethylphenoxyphosphazene (PCEP), and avian beta-defensin have shown active and more extended antibody response in chicken [92,93]. Similarly, murine beta-defensin 2 with a combination of hemagglutinin and nucleoprotein induce active immunisation against highly pathogenic H5N1 influenza lung infection [94]. Bacterial titer and infection increase in the trachea region of the mBD1- deficient mice [95].
1.8. Defensins based treatment options
There is an urgent requirement to find out well-choreographed drugs or vaccines by bioinformatically and wet-lab experimentally [8]. Computational homology and molecular docking analysis found that orf1ab, ORF3a, ORF8, ORF10, and surface proteins bind to the heme-porphyrin ring and beta chain-1 haemoglobin. It could affect the less haemoglobin availability for oxygen transportation which results in inflammation in the respiratory tract. Few viral drugs such as Chloroquine and Favipiramir were found to prevent orf1ab, ORF3a, ORF7, ORF10, surface proteins, and envelope proteins binding to the heme group relieving from respiratory distress [96]. Most of the viruses have similar kinds of functional protein machinery for viral replication and transcription. Several reported drugs inhibit viral entry, replication, transcription, translation, metabolism, virion assembly, and new viron releasing from infected cells. The reported viral inhibitors are Remdesivir [97,98] lopinavir [97,[99], [100], [101]], ritonavir [100,101], nelfinavir [102,103], Chloroquine, arbidol [104,105]. Baricitinib [106], Amantadine [107], Hexachlorophene, Hydroxychloroquine, Ribavirin [108], Zanamivir, Efavirenz [109], Oseltamivir [100], 2′-Deoxy-2′-fluorocytidine [110], Peramivir [111], Darunavir [101], Umifenovir [101], Favipiravir [101], Dolutegravir, Acyclovir [112], Ganciclovir [112], Atazanavir [113], Piscidin, caerin, maculatin [114], and Rifampicin [115] could be effective against SARS-CoV-2 virus. British pharmaceutical company GlaxoSmithKline (GSK), a British pharmaceutical company, and Coalition for Epidemic Preparedness Innovations (CEPI) collaboratively are working on adjuvants vaccine for SARS-CoV2 [49]. The available SARS-CoV-2 genomes in public databases (MN975262.1, MN985325.1, MN938384.1, MN997409.1, MN994467.1, MN994468.1, MN985325.1, MN988668.1, MN988669.1, MN988713.1, MN908947.3, and NC_045512.2) could be used for immunoinformatic designing of B-cell and T-cell multi-epitopes with adjuvants for the development of the viral vaccine, drugs [[116], [117], [118]].
Serine endoprotease furin disrupts S-protein and RNAi targeting structural genes. Notably, Remdesivir & Chloroquine inhibit the viral replication-transcriptional events by interfering with nucleic acid metabolisms. The DRACO (dsRNA-activated caspaseoligomerize) induce virus-infected cell apoptosis, naphthalene inhibitors targeting Coronavirus proteases, a monoclonal antibody against S1-RBD motif, and inhibitors against S2 C-terminal membrane fusion could be important targets for the anti-SARS-CoV viral drugs [119,120]. Human defensin HD5, hBD1-3, and P9-peptide (mouse β-defensin 4) have immunomodulatory and immunoenhancing properties make them attractive broad range antiviral therapeutic adjuvant choice for the development of a viral vaccine against enveloped and non-enveloped viruses such as human immunodeficiency virus (HIV), dengue virus (DENV), influenza A virus (IAV), respiratory syncytial virus (RSV), MERS-CoV, SARS-CoV, and 2019-CoV-2 [3,37].
1.9. Advantages of defensin-mediated viral defence over other antiviral drugs and vaccines
Over the past years, only limited new antibacterial and antiviral drugs have arrived in the pharmaceutical industry [121]. Despite worldwide inventiveness for developing a superfluity of synthetic and semi-synthetic drugs, drug resistance is still endured as one of the premier health problems and constitutes challenges for prosperous combat against most of the pathogenic infections. Consequently, there is a growing requirement for identifying and characterising new potential drugs and therapeutic targets from natural mechanisms. Contrarily gradual increment in antimicrobial resistance leads to diminished treatment effectiveness and increment within the duration of treatment, an increment in mortality, and budgetary costs on treatment [122,123]. For illustration, 19,000 individuals pass on every year from diseases caused by methicillin-resistant strains of Staphylococcus aureus (MRSA) within the United States [124], whereas the yearly monetary costs on the treatment of this infection contain $3 billion. Agreeing to the most recent report from the Centers for Disease Control and Avoidance (USA), the money-related burden related to increasing microbial resistance comprises almost $55 Billion and 8 Million extra bed days [125]. It is evaluated that by 2050, more than 10 million individuals will pass on annually from diseases caused by safe strains, and by that time, the worldwide economy will lose about US $100 trillion due to this issue [126].
The development of resistance occurs due to different causes and molecular processes. The widespread antimicrobial resistance is two factors—mutations and horizontal gene exchange [127]. This is also a characteristic of the frequent natural evolutionary process of adaptation of microorganism contact with substances with antimicrobial properties [128]. Since the human body is in continuous contact with a vast number of pathogenic and non-pathogenic microorganisms, natural systems have carried on with evolution over thousands of years to thwart numerous pathogenic microbes. Within the preparation of advancement, defence mechanisms have formed that permit, to begin with, to identify the pathogen and, if required, work out adequately to control microorganism advance entrance and spread. These errands are fulfilled through innate immunity, which can immediately recognise and destroy infectious operators of different nature [129]. However, the main imperative component of innate immunity is antimicrobial peptides (AMPs). For instance, Defensins are innate defence molecules of the ancient root that can be followed back to living beings from approximately 500 million a long time ago [[130], [131], [132]].
AMPs have a broad range of antimicrobial action against different infectious operators: bacteria, viruses, parasites, and protozoa. Among the six kingdoms (archaea, microbes, protists, fungi, animals, and plants), more than 3000 AMPs have been recognised until now [133]. Among AMPs of incredible intrigued are human defensins: human beta-defensin-1 (hBD-1), human neutrophil peptide-1 (HNP-1), and human beta-defensin-3 (hBD-3), since they have a wide range of antimicrobial action [134,135].
1.10. Limitation
Many specified mechanisms make defensins a great candidate for future medications, but certain drawbacks delay their development as genuine pharmaceuticals. These problems include the rapid turnover within the human body, high cost of generation, osmotic sensitivity, and nonspecific haemolytic activity on human cells; the last affects directly clinical trials, which need vast amounts of material. These physicochemical properties have delayed the introduction of defensins and other AMPs as medicine. Before utilised in clinical applications, transportation, steadiness within the human body, targeted delivery, controlled release, and immunogenicity of these peptides must be improved. To overcome these challenges, much effort has been put into progressing the deep plasma proteomics, peptides’ half-life using different approaches, such as by design of peptidomimetics, counting cyclisation, lipidation, an amalgamation of hybrid peptides, or utilise of nanocarriers [29,[136], [137], [138]].
Although, insertion of native AMPs into clinical use as a monotherapy for bacterial diseases features several constraints: for instance, synthesis expense, cytotoxicity for macroorganism, immunogenicity, hemolytic activity, and pharmacokinetic specifics [139,140]. Other methods should be used to understand these issues: such as modifying provincial AMPs or designing primitive antimicrobial peptides [139] and utilising local AMPs at minimum dosages in combination with conventional antimicrobial synthetic drugs [135,141].
2. Conclusion
Defensins act as short cationic broad range immunostimulators to activate innate and adaptive immunity in response to infectious diseases. In respiratory viral infections, defensin abolishes viruses by inhibiting their entry, interfering with replications, and producing interleukins, cytokines, and chemokines. Also, stimulate to down-regulate the receptors essential for viral survival leading to the death of virus particles. Respiratory viral infection invites secondary bacterial infections, and the involvement of both viral and bacterial attacks makes the body's immune system weak. In that context, defensins might act as a helpful adjuvant when conjugated with viral proteins such as HBD2-S-RBD immunogens raised the expression of antiviral cytokines and immune cells than alone S-RBD. HD5-LBD of ACE2 glycoproteins could be adjuvant to raise an antibody against the 2019-nCoV virus. Several human defensins were reported for their antiviral activity against enveloped and non-enveloped viruses. These characteristics make them a promising adjuvant for the development of viral vaccines against COVID-19.
Funding
Any organisation does not explicitly fund the work. The author declares that there is no conflict of interest in this work.
Author contributions
SSS, SAA, and PS have drafted the manuscript, SAA has contributed to scientific idea/editing and supervised the whole project, PK and MSS have contributed to the manuscript arrangement. All authors contributed to the article and approved the submitted version.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Subhash Singh Solanki, Parul Singh, Poonam Kashyap, Manish Singh Sansi, Syed Azmal Ali.
Acknowledgment
We would also like to thank the Indian Council of Agriculture Research-National Dairy Research Institute (NDRI) Director, India, for providing the necessary facilities to carry out research work. SSS is thankful to the Department of Biotechnology (DBT) for the SRF fellowship. SAA is grateful to the Indian Council of Medical Research (ICMR) for the SRF fellowship.
References
- 1.Yap C.K. Antiviral compounds from marine bivalves for evaluation against SARS-CoV-2. J. PeerSci. 2020;2(2) [Google Scholar]
- 2.Hui K.P., Cheung M.C., Perera R.A., Ng K.C., Bui C.H., Ho J.C., Poon L. L. s. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8(7):687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mustafa S., Balkhy H., Gabere M. Peptide-protein interaction studies of antimicrobial peptides targeting middle east respiratory syndrome coronavirus spike protein: an in silico approach. Adv. Bioinfo. 2019 doi: 10.1155/2019/6815105. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12(2):244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virk J.S., Ali S.A., Kaur G. Recent update on COVID-19 in India: is locking down the country enough? MedRxiv. 2020 Jan 1 doi: 10.1101/2020.04.06.20053124. [DOI] [Google Scholar]
- 6.Jain N., Shankar U., Majee P., Kumar A. BioRxiv; 2020. Scrutinising the SARS-CoV-2 Protein Information for the Designing an Effective Vaccine Encompassing Both the T-Cell and B-Cell Epitopes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karmakar S., Kumar S., Katiyar V. 2020. Comparative Domain-fold Analysis of the SARS-CoV-2 ORF1ab Polyprotein: Insight into Co-evolution, Conservation of Folding Patterns, Potential Therapeutic Strategies, and the Possibility of Reemergence. [Google Scholar]
- 8.Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., Ciccozzi M. The 2019‐new coronavirus epidemic: evidence for virus evolution. J. Med. Virol. 2020;92(4):455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehman H.M., Mirza M.U., Saleem M., Froeyen M., Ahmad S., Gul R., Bhinder M.A. 2020. A Putative Prophylactic Solution for COVID-19: Development of Novel Multiepitope Vaccine Candidate against SARS‐COV‐2 by Comprehensive Immunoinformatic and Molecular Modelling Approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T., Xie J., He Y., Fan H., Baril L., Qiu Z., Zuo Y. Long-term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PloS One. 2006;1(1):e24. doi: 10.1371/journal.pone.0000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkar B., Ullah M.A., Johora F.T., Taniya M.A., Araf Y. BioRxiv; 2020. The Essential Facts of Wuhan Novel Coronavirus Outbreak in China and Epitope-Based Vaccine Designing against 2019-nCoV. [Google Scholar]
- 12.Zakhartchouk A.N., Sharon C., Satkunarajah M., Auperin T., Viswanathan S., Mutwiri G., Cameron C. Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: implications for a subunit vaccine. Vaccine. 2007;25(1):136–143. doi: 10.1016/j.vaccine.2006.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palau V., Riera M., Soler M.J. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol. Dial. Transplant. 2020;35(6):1071–1072. doi: 10.1093/ndt/gfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida A.M., Ali S.A., Ceciliani F., Eckersall P.D., Hernandez-Castellano L.E., Han R., Hodnik J.J., Jaswal S., Lippolis J.D., McLaughlin M., Miller I. Domestic animal proteomics in the 21st century: a global retrospective and viewpoint analysis. J. Proteom. 2021 Apr 7:104220. doi: 10.1016/j.jprot.2021.104220. [DOI] [PubMed] [Google Scholar]
- 15.Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol. Biol. Evol. 2020;37(9):2699–2705. doi: 10.1093/molbev/msaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh P.K., Jia H.P., Wiles K., Hesselberth J., Liu L., Conway B.A.D., Tack B.F. Production of β-defensins by human airway epithelia. Proc. Natl. Acad. Sci. Unit. States Am. 1998;95(25):14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semple F., Dorin J.R. β-Defensins: multifunctional modulators of infection, inflammation and more? J. Innate Immun. 2012;4(4):337–348. doi: 10.1159/000336619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasad S.V., Fiedoruk K., Daniluk T., Piktel E., Bucki R. Expression and function of host defense peptides at inflammation sites. Int. J. Mol. Sci. 2020;21(1):104. doi: 10.3390/ijms21010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar R., Ali S.A., Singh S.K., Bhushan V., Mathur M., Jamwal S., Mohanty A.K., Kaushik J.K., Kumar S. Antimicrobial peptides in farm animals: an updated review on its diversity, function, modes of action and therapeutic prospects. Vet. Sci. 2020 Dec;7(4):206. doi: 10.3390/vetsci7040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y., Cooper C.L., Wang G., Morwitzer M.J., Kota K., Tran J.P., Luo L.G. Iscience; 2020. Engineered Human Cathelicidin Antimicrobial Peptides Inhibit Ebola Virus Infection; p. 100999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. 2014;7:545–594. doi: 10.3390/ph7050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilhelmelli F., Vilela N., Albuquerque P., et al. Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front. Microbiol. 2013;4:353. doi: 10.3389/fmicb.2013.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wimley W.C., Hristova K. The mechanism of membrane permeabilisation by peptides: still an enigma. Aust. J. Chem. 2020;73(3):96. doi: 10.1071/CH19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oren Z., Shai Y. Mode of action of linear amphipathic a-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 25.Mor A., Nicolas P. The NH2-terminal alpha-helical domain 1-18 of dermaseptin is responsible for antimicrobial activity. J. Biol. Chem. 1994;269:1934–1939. [PubMed] [Google Scholar]
- 26.Chan D.I., Prenner E.J., Vogel H.J. Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim. Biophys. Acta. 2006;1758(9):1184–1202. doi: 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki K., Shioyama T., Okamura E., Umemura J., Takenaka T., Takaishi Y., Fujita T., Miyajima K. A comparative study on interactions of a-aminoisobutyric acid containing antibiotic peptides, trichopolyn I and hypelcin A with phosphatidylcholine bilayers. Biochim. Biophys. Acta Biomembr. 1991;1070(2):419–428. doi: 10.1016/0005-2736(91)90082-J. [DOI] [PubMed] [Google Scholar]
- 28.De Leeuw E., Li C.C., Zeng P., Li C.C., De Buin M.D., Lu W.Y.W., Breukink E., Lu W.Y.W. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2010;584(8):1543–1548. doi: 10.1016/j.febslet.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amerikova Meri, Ivanka Pencheva El-Tibi, Maslarska Vania, Bozhanov Stanislav, Tachkov Konstantin. Antimicrobial activity, mechanism of action, and methods for stabilisation of defensins as new therapeutic agents. Biotechnol. Biotechnol. Equip. 2019;33(1):671–682. doi: 10.1080/13102818.2019.1611385. [DOI] [Google Scholar]
- 30.Jin G., Weinberg A. Human antimicrobial peptides and cancer. Semin. Cell Dev. Biol. 2019;88:156–162. doi: 10.1016/j.semcdb.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Midura-Nowaczek K., Markowska A. Antimicrobial peptides and their analogs: searching for new potential therapeutics. Perspect. Med. Chem. 2014;6:73–80. doi: 10.4137/PMC.S13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pachon-Ibanez M.E., Smani Y., Pach_on J., et al. Perspectives for clinical use of engineered human host defense antimicrobial peptides. FEMS Microbiol. Rev. 2017;41:323–342. doi: 10.1093/femsre/fux012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diamond G., Beckloff N., Weinberg A., Kisich K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharmaceut. Des. 2009;15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oppenheim J.J., Yang D. Alarmins: chemotactic activators of immune responses. Curr. Opin. Immunol. 2005;17:359–365. doi: 10.1016/j.coi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Yang D., Chertov O., Bykovskaia S.N., Chen Q., Buffo M.J., Shogan J., Anderson M., Schröder J.M., Wang J.M., Howard O.M., Oppenheim J.J. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 36.Chertov O., Michiel D.F., Xu L., Wang J.M., Tani K., Murphy W.J., Longo D.L., Taub D.D., Oppenheim J.J. Identification of defensin-1, defensin-2, and CAP37/azurocidin as T-cell chemoattractant proteins released from interleukin-8-stimulated neutrophils. J. Biol. Chem. 1996;271:2935–2940. doi: 10.1074/jbc.271.6.2935. [DOI] [PubMed] [Google Scholar]
- 37.Biragyn A. Defensins-non-antibiotic use for vaccine development. Curr. Protein Pept. Sci. 2005;6(1):53–60. doi: 10.2174/1389203053027601. [DOI] [PubMed] [Google Scholar]
- 38.Giacalone V.D., Margaroli C., Mall M.A., Tirouvanziam R. Neutrophil adaptations upon recruitment to the lung: new concepts and implications for homeostasis and disease. Int. J. Mol. Sci. 2020;21(3):851. doi: 10.3390/ijms21030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holly M.K., Diaz K., Smith J.G. Defensins in viral infection and pathogenesis. Ann. Rev. Virol. 2017;4:369–391. doi: 10.1146/annurev-virology-101416-041734. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C., Yang M. The role and potential application of antimicrobial peptides in autoimmune diseases. Front. Immunol. 2020;11:859. doi: 10.3389/fimmu.2020.00859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daher K.A., Selsted M.E., Lehrer R.I. Direct inactivation of viruses by human granulocyte defensins. J. Virol. 1986;60(3):1068–1074. doi: 10.1128/jvi.60.3.1068-1074.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dugan A.S., Maginnis M.S., Jordan J.A., Gasparovic M.L., Manley K., Page R., Atwood W.J. Human α-defensins inhibit BK virus infection by aggregating virions and blocking binding to host cells. J. Biol. Chem. 2008;283(45):31125–31132. doi: 10.1074/jbc.M805902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishimoto H., Mukae H., Date Y., Shimbara T., Mondal M.S., Ashitani J., Nakazato M. Identification of hBD-3 in respiratory tract and serum: the increase in pneumonia. Eur. Respir. J. 2006;27(2):253–260. doi: 10.1183/09031936.06.00105904. [DOI] [PubMed] [Google Scholar]
- 44.Cruz Díaz L.A., Gutiérrez Ortega A., Chávez Álvarez R.D.C., Velarde Félix J.S., Prado Montes de Oca E. Regulatory SNP rs5743417 impairs constitutive expression of human β‐defensin 1 and has high frequency in Africans and Afro‐Americans. Int. J. Immunogenet. 2020;47(4):332–341. doi: 10.1111/iji.12475. [DOI] [PubMed] [Google Scholar]
- 45.Albanesi C., Fairchild H.R., Madonna S., Scarponi C., De Pità O., Leung D.Y., Howell M.D. IL-4 and IL-13 negatively regulate TNF-α-and IFN-γ-induced β-defensin expression through STAT-6, suppressor of cytokine signaling (SOCS)-1, and SOCS-3. J. Immunol. 2007;179(2):984–992. doi: 10.4049/jimmunol.179.2.984. [DOI] [PubMed] [Google Scholar]
- 46.Cleemput J.V., Poelaert K.C., Laval K., Vanderheijden N., Dhaenens M., Daled S., Boyen F., Pasmans F., Nauwynck H.J. Alphaherpesvirus exploits antimicrobial β-1 defensins to initiate respiratory tract infection. J. Virol. 2020;94(8) doi: 10.1128/JVI.01676-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J., Qi Y., Wang A., Huang C., Liu X., Yang X., Zhou R. Porcine β-defensin 2 inhibits proliferation of pseudorabies virus in vitro and in transgenic mice. Virol. J. 2020;17(1):1–7. doi: 10.1186/s12985-020-1288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K.M., Zhang A.J.X. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan J., Luo Y., Qin Y., Wu C., Han X., Ouyang H., Li N. The expression of β-Defensin-2, IL-22, IL-22R1 and IL-10R2 in rat model of Klebsiella pneumonia and their correlation with histological grades. Exp. Lung Res. 2020;46(5):109–116. doi: 10.1080/01902148.2020.1725690. [DOI] [PubMed] [Google Scholar]
- 50.Doss M., Ruchala P., Tecle T., Gantz D., Verma A., Hartshorn A., Hartshorn K.L. Hapivirins and diprovirins: novel θ-defensin analogs with potent activity against influenza A virus. J. Immunol. 2012;188(6):2759–2768. doi: 10.4049/jimmunol.1101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gounder A.P., Wiens M.E., Wilson S.S., Lu W., Smith J.G. Critical determinants of human alpha-defensin 5 activity against non-enveloped viruses. J. Biol. Chem. 2012;287:24554–24562. doi: 10.1074/jbc.M112.354068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hazrati E., Galen B., Lu W., Wang W., Ouyang Y., Keller M.J., Herold B.C. Human α-and β-defensins block multiple steps in herpes simplex virus infection. J. Immunol. 2006;177(12):8658–8666. doi: 10.4049/jimmunol.177.12.8658. [DOI] [PubMed] [Google Scholar]
- 53.Wang W., Owen S.M., Rudolph D.L., Cole A.M., Hong T., Waring A.J., et al. Activity of alpha- and theta-defensins against primary isolates of HIV-1. J. Immunol. 2004;173:515–520. doi: 10.4049/jimmunol.173.1.515. [DOI] [PubMed] [Google Scholar]
- 54.Demirkhanyan L.H., Marin M., Padilla-Parra S., Zhan C., Miyauchi K., Jean-Baptiste M., Melikyan G.B. Multifaceted mechanisms of HIV-1 entry inhibition by human α-defensin. J. Biol. Chem. 2012;287(34):28821–28838. doi: 10.1074/jbc.M112.375949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hazrati E., Galen B., Lu W., Wang W., Ouyang Y., Keller M.J., et al. Human alpha- and beta-defensins block multiple steps in herpes simplex virus infection. J. Immunol. 2006;177:8658–8666. doi: 10.4049/jimmunol.177.12.8658. [DOI] [PubMed] [Google Scholar]
- 56.Buck C.B., Day P.M., Thompson C.D., Lubkowski J., Lu W., Lowy D.R., et al. Human alpha-defensins block papillomavirus infection. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith J.G., Nemerow G.R. Mechanism of adenovirus neutralisation by human alpha-defensins. Cell Host Microbe. 2008;3:11–19. doi: 10.1016/j.chom.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Leikina E., Delanoe-Ayari H., Melikov K., Cho M.S., Chen A., Waring A.J., et al. Carbohydrate-binding molecules inhibit viral fusion and entry by cross-linking membrane glycoproteins. Nat. Immunol. 2005;6:995–1001. doi: 10.1038/ni1248. [DOI] [PubMed] [Google Scholar]
- 59.Yu G., Shen F.S., Sturch S., Aquino A., Glazer R.I., Felsted R.L. Regulation of HIV-1 gag protein subcellular targeting by protein kinase C. J. Biol. Chem. 1995;270:4792–4796. doi: 10.1074/jbc.270.9.4792. [DOI] [PubMed] [Google Scholar]
- 60.Contreras X., Mzoughi O., Gaston F., Peterlin M.B., Bahraoui E. Protein kinase C-delta regulates HIV-1 replication at an early post-entry step in macrophages. Retrovirology. 2012;9:37. doi: 10.1186/1742-4690-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sieczkarski S.B., Brown H.A., Whittaker G.R. Role of protein kinase C betaII in influenza virus entry via late endosomes. J. Virol. 2003;77:460–469. doi: 10.1128/JVI.77.1.460-469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Root C.N., Wills E.G., McNair L.L., Whittaker G.R. Entry of influenza viruses into cells is inhibited by a highly specific protein kinase C inhibitor. J. Gen. Virol. 2000;81:2697–2705. doi: 10.1099/0022-1317-81-11-2697. [DOI] [PubMed] [Google Scholar]
- 63.Constantinescu S.N., Cernescu C.D., Popescu L.M. Effects of protein kinase C inhibitors on viral entry and infectivity. FEBS Lett. 1991;292:31–33. doi: 10.1016/0014-5793(91)80826-o. [DOI] [PubMed] [Google Scholar]
- 64.Leach N.R., Roller R.J. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology. 2010;406:127–137. doi: 10.1016/j.virol.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charp P.A., Rice W.G., Raynor R.L., Reimund E., Kinkade J.M., Ganz T., et al. Inhibition of protein kinase C by defensins, antibiotic peptides from human neutrophils. Biochem. Pharmacol. 1988;37:951–956. doi: 10.1016/0006-2952(88)90187-6. [DOI] [PubMed] [Google Scholar]
- 66.Lafferty M.K., Sun L., DeMasi L., Lu W., Garzino-Demo A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood. 2010;115:1564–1571. doi: 10.1182/blood-2009-06-226423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu G., Shen F.S., Sturch S., Aquino A., Glazer R.I., Felsted R.L. Regulation of HIV-1 gag protein subcellular targeting by protein kinase C. J. Biol. Chem. 1995;270:4792–4796. doi: 10.1074/jbc.270.9.4792. [DOI] [PubMed] [Google Scholar]
- 68.Wohlford-Lenane C.L., Meyerholz D.K., Perlman S., Zhou H., Tran D., Selsted M.E., et al. Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J. Virol. 2009;83:11385–11390. doi: 10.1128/JVI.01363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson S.S., Wiens M.E., Smith J.G. Antiviral mechanisms of human defensins. J. Mol. Biol. 2013;425(24):4965–4980. doi: 10.1016/j.jmb.2013.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Demirkhanyan L.H., Marin M., Padilla-Parra S., Zhan C., Miyauchi K., Jean-Baptiste M., et al. Multifaceted mechanisms of HIV-1 entry inhibition by human alpha-defensin. J. Biol. Chem. 2012;287:28821–28838. doi: 10.1074/jbc.M112.375949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Innovation Pharmaceuticals . 2020. Mechanism of Action, Pre/Clinical Data and Academic Literature Supporting the Development of Brilacidin as a Potential Novel Coronavirus (COVID-19) Treatment. [Google Scholar]
- 72.Ding J., Chou Y.Y., Chang T.L. Defensins in viral infections. J. Innate Immun. 2009;1(5):413–420. doi: 10.1159/000226256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu L., She Z.G., Cheng X., Qin J.J., Zhang X.J., Cai J., Li H. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metabol. 2020;31(6):1068–1077.e3. doi: 10.1016/j.cmet.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chee Y.J., Ng S.J.H., Yeoh E. Diabetic ketoacidosis precipitated by Covid-19 in a patient with newly diagnosed diabetes mellitus. Diabetes Res. Clin. Pract. 2020;164 doi: 10.1016/j.diabres.2020.108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou P., Liu Z., Chen Y., Xiao Y., Huang X., Fan X.G. Bacterial and fungal infections in COVID-19 patients: a matter of concern. Infect. Contr. Hosp. Epidemiol. 2020:1–2. doi: 10.1017/ice.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwon W.J., Li G., Zheng M., Kaur H., Magbual N., Dalai S., Kwon W.J., Li G., Zheng M., Kaur H., Magbual N., Dalai S. 2020. (2020). Superinfections and Co-infections in COVID-19. Medpage Today. [Google Scholar]
- 77.MacIntyre C.R., Chughtai A.A., Barnes M., Ridda I., Seale H., Toms R., Heywood A. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a (H1N1) pdm09. BMC Infect. Dis. 2018;18(1):637. doi: 10.1186/s12879-018-3548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xue M., Zhao J., Ying L., Fu F., Li L., Ma Y., Liu P. IL-22 suppresses the infection of porcine enteric coronaviruses and rotavirus by activating STAT3 signal pathway. Antivir. Res. 2017;142:68–75. doi: 10.1016/j.antiviral.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J., Yang Y.L., Jang Y.S. Human β-defensin 2 is involved in CCR2-mediated Nod2 signal transduction, leading to activation of the innate immune response in macrophages. Immunobiology. 2019;224(4):502–510. doi: 10.1016/j.imbio.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mustafa S., Balkhy H., Gabere M.N. Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J. Infect. Publ. Health. 2018;11(1):9–17. doi: 10.1016/j.jiph.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marimuthu S.K., Nagarajan K., Perumal S.K., Palanisamy S., Subbiah L. Insilico alpha-helical structural recognition of temporin antimicrobial peptides and its interactions with Middle East respiratory syndrome-coronavirus. Int. J. Pept. Res. Therapeut. 2020;26(3):1473–1483. doi: 10.1007/s10989-019-09951-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.LeMessurier K.S., Lin Y., McCullers J.A., Samarasinghe A.E. Antimicrobial peptides alter early immune response to influenza A virus infection in C57BL/6 mice. Antivir. Res. 2016;133:208–217. doi: 10.1016/j.antiviral.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Froy O. Microreview: regulation of mammalian defensin expression by Toll‐like receptor‐dependent and independent signalling pathways. Cell Microbiol. 2005;7(10):1387–1397. doi: 10.1111/j.1462-5822.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- 84.Salvatore M., García-Sastre A., Ruchala P., Lehrer R.I., Chang T., Klotman M.E. β-defensin inhibits influenza virus replication by cell-mediated mechanism (s) JID (J. Infect. Dis.) 2007;196(6):835–843. doi: 10.1086/521027. [DOI] [PubMed] [Google Scholar]
- 85.Hartshorn K.L., White M.R., Tecle T., Holmskov U., Crouch E.C. Innate defense against influenza A virus: activity of human neutrophil defensins and interactions of defensins with surfactant protein D. J. Immunol. 2006;176(11):6962–6972. doi: 10.4049/jimmunol.176.11.6962. [DOI] [PubMed] [Google Scholar]
- 86.Doss M., White M.R., Tecle T., Gantz D., Crouch E.C., Jung G., Hartshorn K.L. Interactions of α-, β-, and θ-defensins with influenza A virus and surfactant protein D. J. Immunol. 2009;182(12):7878–7887. doi: 10.4049/jimmunol.0804049. [DOI] [PubMed] [Google Scholar]
- 87.Tecle T., White M.R., Gantz D., Crouch E.C., Hartshorn K.L. Human neutrophil defensins increase neutrophil uptake of influenza A virus and bacteria and modify virus-induced respiratory burst responses. J. Immunol. 2007;178(12):8046–8052. doi: 10.4049/jimmunol.178.12.8046. [DOI] [PubMed] [Google Scholar]
- 88.Hazrati E., Galen B., Lu W., Wang W., Ouyang Y., Keller M.J., et al. Human alpha- and beta-defensins block multiple steps in herpes simplex virus infection. J. Immunol. 2006;177:8658–8666. doi: 10.4049/jimmunol.177.12.8658. [DOI] [PubMed] [Google Scholar]
- 89.Yasin B., Wang W., Pang M., Cheshenko N., Hong T., Waring A.J., Lehrer R.I. θ defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J. Virol. 2004;78(10):5147–5156. doi: 10.1128/JVI.78.10.5147-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun L., Finnegan C.M., Kish-Catalone T., Blumenthal R., Garzino-Demo P., Maggiore G.M.L.T., Lu W. Human β-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J. Virol. 2005;79(22):14318–14329. doi: 10.1128/JVI.79.22.14318-14329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mohan T., Sharma C., Bhat A.A., Rao D.N. Modulation of HIV peptide antigen specific cellular immune response by synthetic α-and β-defensin peptides. Vaccine. 2013;31(13):1707–1716. doi: 10.1016/j.vaccine.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 92.Sarfraz M., Suleman M., Tikoo S.K., Wheler C., Potter A.A., Gerdts V., Dar A. Immune responses to in ovo vaccine formulations containing inactivated fowl adenovirus 8b with poly [di (sodium carboxylatoethylphenoxy)] phosphazene (PCEP) and avian beta defensin as adjuvants in chickens. Vaccine. 2017;35(6):981–986. doi: 10.1016/j.vaccine.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 93.Zhang H.H., Yang X.M., Xie Q.M., Ma J.Y., Luo Y.N., Cao Y.C., Bi Y.Z. The potent adjuvant effects of chicken β-defensin-1 when genetically fused with infectious bursal disease virus VP2 gene. Vet. Immunol. Immunopathol. 2010;136(1–2):92–97. doi: 10.1016/j.vetimm.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 94.Vemula S.V., Pandey A., Singh N., Katz J.M., Donis R., Sambhara S., Mittal S.K. Adenoviral vector expressing murine β-defensin 2 enhances immunogenicity of an adenoviral vector based H5N1 influenza vaccine in aged mice. Virus Res. 2013;177(1):55–61. doi: 10.1016/j.virusres.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ryan L.K., Wu J., Schwartz K., Yim S., Diamond G. β-Defensins coordinate in vivo to inhibit bacterial infections of the trachea. Vaccines. 2018;6(3):57. doi: 10.3390/vaccines6030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu W., Li H. COVID-19: attacks the 1-beta chain of hemoglobin and captures the porphyrin to inhibit human heme metabolism. Preprint. 2020;10 04. [Google Scholar]
- 97.Choy K.T., Wong A.Y.L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Peiris M. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang C., Wang S., Li D., Zhao X., Han S., Wang T., Wang L. bioRxiv; 2020. Lectin-like Intestinal Defensin Inhibits 2019-nCoV Spike Binding to ACE2. [Google Scholar]
- 99.Yao T.T., Qian J.D., Zhu W.Y., Wang Y., Wang G.Q. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus—a possible reference for coronavirus disease‐19 treatment option. J. Med. Virol. 2020;92(6):556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. J. Biomol. Struct. Dyn. 2020:1–6. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 101.Costanzo M., De Giglio M.A.R., Roviello G.N. SARS-CoV-2: recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Curr. Med. Chem. 2020;27(27):4536–4541. doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- 102.Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J., Tamamura H. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004;318(3):719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Musarrat F., Chouljenko V., Dahal A., Nabi R., Chouljenko T., Jois S.D., Kousoulas K.G. The anti‐HIV drug nelfinavir mesylate (viracept) is a potent inhibitor of cell fusion caused by the SARS‐CoV‐2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID‐19 infections. J. Med. Virol. 2020;92(10):2087–2095. doi: 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deng L., Li C., Zeng Q., Liu X., Li X., Zhang H., Xia J. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: a retrospective cohort study. J. Infect. 2020;81(1):e1–e5. doi: 10.1016/j.jinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking the trimerisation of viral spike glycoprotein? Int. J. Antimicrob. Agents. 2020:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Favalli E.G., Biggioggero M., Maioli G., Caporali R. Baricitinib for COVID-19: a suitable treatment? Lancet Infect. Dis. 2020;20(9):1012–1013. doi: 10.1016/S1473-3099(20)30262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Griffin S.D., Beales L.P., Clarke D.S., Worsfold O., Evans S.D., Jaeger J.…Rowlands D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535(1–3):34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 108.Morgenstern B., Michaelis M., Baer P.C., Doerr H.W., Cinatl J., Jr. Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Resear. Commun. 2005;326(4):905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rismanbaf A. Potential treatments for COVID-19; a narrative literature review. Arch. Acad. Emerg. Med. 2020;8(1) [PMC free article] [PubMed] [Google Scholar]
- 110.Kumaki Y., Day C.W., Smee D.F., Morrey J.D., Barnard D.L. In vitro and in vivo efficacy of fluorodeoxycytidine analogs against highly pathogenic avian influenza H5N1, seasonal, and pandemic H1N1 virus infections. Antivir. Res. 2011;92(2):329–340. doi: 10.1016/j.antiviral.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anderson P.O. Breastfeeding and respiratory antivirals: coronavirus and influenza. Breastfeed. Med. 2020;15(3) doi: 10.1089/bfm.2020.29149.poa. 128-128. [DOI] [PubMed] [Google Scholar]
- 112.Dunn D.L., Gillingham K.J., Kramer M.A., Schmidt W.J., Erice A.L.E.J.O., Balfour H.H., Jr., Payne W.D. A prospective randomised study of acyclovir versus ganciclovir plus human immune globulin prophylaxis of cytomegalovirus infection after solid organ transplantation. Transplantation. 1994;57(6):876–884. doi: 10.1097/00007890-199403270-00019. [DOI] [PubMed] [Google Scholar]
- 113.Fintelman-Rodrigues N., Sacramento C.Q., Lima C.R., da Silva F.S., Ferreira A., Mattos M., Miranda M. bioRxiv; 2020. Atazanavir Inhibits SARS-CoV-2 Replication and Proinflammatory Cytokine Production. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hu H., Guo N., Chen S., Guo X., Liu X., Ye S., He Q. Antiviral activity of Piscidin 1 against pseudorabies virus both in vitro and in vivo. Virol. J. 2019;16(1):95. doi: 10.1186/s12985-019-1199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Petric Domina . 2020. What is the connection between rifampicin and coronaviruses? [Google Scholar]
- 116.Lau S.Y., Wang P., Mok B.W.Y., Zhang A.J., Chu H., Lee A.C.Y., Chen Z. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microb. Infect. 2020;9(1):837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vac. 2020;5:18. doi: 10.1038/s41541-020-0170-0. 2020, Published, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mirza M.U., Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J. Pharmaceut. Anal. 2020;10(4):320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nyanguile O.F. Peptide antiviral strategies as an alternative to treat lower respiratory viral infections. Front. Immunol. 2019;10:1366. doi: 10.3389/fimmu.2019.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Hilgenfeld R. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Andrei S., Droc G., Stefan G. FDA approved antibacterial drugs: 2018–2019. Discoveries. 2019;7(4):e102. doi: 10.15190/d.2019.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rolain J.-M., Abat C., Jimeno M.-T., Fournier P.-E., Raoult D. Do we need new antibiotics? Clin. Microbiol. Infect. Dis. 2016;22(5):408–415. doi: 10.1016/j.cmi.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 123.Fair R.J., Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fischbach M.A., Walsh C.T. Antibiotics for emerging pathogens. Science. 2009;325(5944):1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Us C.D.C. Centers for Disease Control and Prevention; Atlanta: 2019. Antibiotic Resistance Threats in the United States. [Google Scholar]
- 126.O'Neill J. WHO; Geneva: 2016. Review on Antimicrobial Resistance. Tackling Drug-Resistant Infections Globally. [Google Scholar]
- 127.Martinez J.L., Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 2000;44(7):1771–1777. doi: 10.1128/AAC.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Martinez J.L., Fajardo A., Garmendia L., Hernandez A., Linares J.F., Martínez-Solano L., Sánchez M.B. A global view of antibiotic resistance. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2009;33(1):44–65. doi: 10.1111/j.1574-6976.2008.00142.x. [DOI] [PubMed] [Google Scholar]
- 129.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16(4):343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Phoenix D.A., Dennison S.R., Harris F. Antimicrobial Peptides. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2013. Antimicrobial peptides: their history, evolution, and functional promiscuity; pp. 1–38. [Google Scholar]
- 131.Erwin D.H., Davidson E.H. The last common bilaterian ancestor. Development. 2002;129:3021–3032. doi: 10.1242/dev.129.13.3021. [DOI] [PubMed] [Google Scholar]
- 132.Zhu S., Gao B. Evolutionary origin of β-defensins. Dev. Comp. Immunol. 2013;39:79–84. doi: 10.1016/j.dci.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 133.Wang G., Li X., Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pachón-Ibáñez M.E., Smani Y., Pachón J., Sánchez-Céspedes J. Perspectives for clinical use of engineered human host defense antimicrobial peptides. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2017;41(3):323–342. doi: 10.1093/femsre/fux012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bolatchiev A. Antibacterial activity of human defensins against Staphylococcus aureus and Escherichia coli. PeerJ. 2020;8 doi: 10.7717/peerj.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kaur G., Poljak A., Ali S.A., Zhong L., Raftery M.J., Sachdev P. Extending the depth of human plasma proteome coverage using simple fractionation techniques. J. Proteome Res. 2021 Jan 20;20(2):1261–1279. doi: 10.1021/acs.jproteome.0c00670. [DOI] [PubMed] [Google Scholar]
- 137.Mittal D., Kaur G., Singh P., Yadav K., Ali S.A. Nanoparticle-based sustainable agriculture and food science: recent advances and future outlook. Front. Nanotechnol. 2020 Dec 4;2(10) [Google Scholar]
- 138.Ali S.A., Mittal D., Kaur G. In-situ monitoring of xenobiotics using genetically engineered whole-cell-based microbial biosensors: recent advances and outlook. World J. Microbiol. Biotechnol. 2021 May;37(5):1–24. doi: 10.1007/s11274-021-03024-3. [DOI] [PubMed] [Google Scholar]
- 139.Lei J., Sun L.C., Huang S., Zhu C., Li P., He J., Mackey V., Coy D.H., He Q.Y. The antimicrobial peptides and their potential clinical applications. Am. J. Tourism Res. 2019;11(7):3919–3931. [PMC free article] [PubMed] [Google Scholar]
- 140.Moravej H., Moravej Z., Yazdanparast M., Heiat M., Mirhosseini A., Moosazadeh Moghaddam M., Mirnejad R. Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 2018;24(6):747–767. doi: 10.1089/mdr.2017.0392. [DOI] [PubMed] [Google Scholar]
- 141.Zharkova M.S., Orlov D.S., Golubeva O.Y., Chakchir O.B., Eliseev I.E., Grinchuk T.M., Shamova O.V. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics: a novel way to combat antibiotic resistance? Front. Cell. Infect. Microbiol. 2019;9:8027. doi: 10.3389/fcimb.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]