Abstract
The release of small extracellular vesicles (sEVs) has recently been reported, but knowledge of their function in neuron development remains limited. Using LC-MS/MS, we found that sEVs released from developing cortical neurons in vitro obtained from mice of both sexes were enriched in cytoplasm, exosome, and protein-binding and DNA/RNA-binding pathways. The latter included HDAC2, which was of particular interest, because HDAC2 regulates spine development, and populations of neurons expressing different levels of HDAC2 co-exist in vivo during the period of spine growth. Here, we found that HDAC2 levels decrease in neurons as they acquire synapses and that sEVs from HDAC2-rich neurons regulate HDAC2 signaling in HDAC2-low neurons possibly through HDAC2 transfer. This regulation led to a transcriptional decrease in HDAC2 synaptic targets and the density of excitatory synapses. These data suggest that sEVs provide inductive cell-cell signaling that coordinates the development of dendritic spines via the activation of HDAC2-dependent transcriptional programs.
SIGNIFICANCE STATEMENT A role of small extracellular vesicles (sEVs; also called exosomes) in neuronal development is of particular interest, because sEVs could provide a major signaling modality between developing neurons when synapses are not fully functional or immature. However, knowledge of sEVs on neuron, and more precisely spine development, is limited. We provide several lines of evidence that sEVs released from developing cortical neurons regulate the development of dendritic spines via the regulation of HDAC2 signaling. This paracrine communication is temporally restricted during development because of the age-dependent decrease in sEV release as neurons mature and acquire spines.
Keywords: cortex, development, exosome, HDAC, spine, synapse
Introduction
Every cell in the body, including neurons, releases nanosized material that has the potential to impact surrounding cells. The released nanomaterial that has drawn the most attention is small extracellular vesicles (sEVs), especially exosomes, that form when intraluminal vesicles in multivesicular bodies (MVBs) are released into the extracellular milieu on MVB fusion with the plasma membrane (Valadi et al., 2007; Lee et al., 2012; Lawson et al., 2017). sEVs carry a cargo rich in proteins and RNAs and have emerged as a powerful mechanism of intercellular communication in all cell types (Valadi et al., 2007; Lee et al., 2012; Zappulli et al., 2016; Jeppesen et al., 2019). With respect to neuronal cells, sEVs have been shown to be released and transferred between cortical neurons (Fauré et al., 2006; Lachenal et al., 2011; Laulagnier et al., 2017). There is also accumulating evidence that sEVs have several roles during brain development (for review, see Budnik et al., 2016; Zappulli et al., 2016), including contributing to retrograde signaling at the neuromuscular junction (Korkut et al., 2009, 2013) and in cultured mouse and human neurons (Lee et al., 2018; Sharma et al., 2019). A role of sEVs in neuronal development, including the growth of dendritic spines, is of particular interest because sEVs could provide a major signaling modality between developing neurons when synapses are not fully functional or immature. However, knowledge of sEVs on neuron, and more precisely spine development, is limited.
To identify a potential role of sEVs in neuron development, we performed a protein screen using sEVs isolated from the medium of cultured developing cortical neurons. We identified the presence of the epigenetic regulator HDAC2, which was of particular interest because of its negative regulation of spine maturation (Akhtar et al., 2009; Guan et al., 2009). In addition, although HDAC2 is ubiquitously expressed in neurons, its levels are development dependent (Jones et al., 2009). In particular, in the neocortex, HDAC2 levels decrease as neurons mature, leading to the coexistence of neuronal populations with different HDAC2 levels (Jones et al., 2009; Gräff et al., 2012). This led us to examine whether sEVs could affect the development of dendritic spines via regulation of HDAC2 signaling. We performed a series of experiments in vitro that confirms the decrease in HDAC2 expression as neurons mature, the presence of HDAC2 in sEVs, and a role for sEVs in regulating the levels of HDAC2-dependent synaptic players and dendritic spine development.
Materials and Methods
Animals
Research protocols were approved by the Yale University Institutional Animal Care and Use Committee. All experiments were performed on CD-1 (Charles River) mice of either sex.
Neuronal culture
Cerebral cortices from embryonic day (E)15 mice were isolated after removing meninges and placed in ice-cold Hibernate E (Invitrogen, #A12476-01) supplemented with B27 (Invitrogen, #17504-044). The cortices were then incubated in prewarmed and activated papain digestion solution (Worthington, #LK003150). After a 15-min digestion, neurons were dissociated and plated in MEM medium (Invitrogen, #11095-080) supplemented with 5% fetal bovine serum (FBS; Invitrogen, #16000) and 0.6% glucose (Sigma, # G8769). Plating medium was replaced with neurobasal medium (Invitrogen, #21103-049) supplemented with B27 and GlutaMax-1 (Invitrogen, #35050-061) after neurons have attached. The medium was changed every 3 d.
Western blotting
sEVs were lysed in reducing sample buffer [0.25 m Tris–HCl (pH 6.8), 40% glycerol, 8% SDS, 5% 2-mercaptoethanol, and 0.04% bromophenol blue] or non-reducing sample buffer (without 2-mercaptoethanol, for CD9 and CD63) and then boiled for 5 min. Primary neurons were homogenized in radioimmunoprecipitation assay buffer, 1× Halt Protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific), 5 mm EDTA, and 20 units/ml DNase I (Roche). All lysates were run on Tris-glycine gradient gels (Bio-Rad; #456-1084). Proteins were transferred to polyvinylidene difluoride and blocked in 5% milk and incubated with primary antibodies in 0.25% bovine serum albumin (BSA; concentrations of primary antibody are listed in Table 1). Horseradish peroxidase-conjugated anti-rabbit or anti-mouse were used as secondary antibodies. All density measurements were performed by using NIH ImageJ software.
Table 1.
List of antibodies
| Antibodies | Company | Catalog number | Dilution |
|---|---|---|---|
| Primary antibodies | |||
| Acetyl-histone H2B | Cell Signaling Technology | 12799 | 1:5000 |
| Acetyl-histone H3 | Cell Signaling Technology | 8173 | 1:5000 |
| Alix | Cell Signaling Technology | 2171 | 1:3000 |
| Calnexin | Cell Signaling Technology | 2679 | 1:3000 |
| Santa Cruz Biotechnology | sc-11397 | 1:1000 | |
| Calreticulin | Cell Signaling Technology | 12238 | 1:3000 |
| CD9 | Santa Cruz Biotechnology | sc-9148 | 1:1000 |
| CD63 | Novus | NBP2-36567 | 1:1000 |
| Santa Cruz Biotechnology | sc-15363 | 1:1000 | |
| FLT1 | Cell Signaling Technology | 3253 | 1:3000 |
| GAPDH | Santa Cruz Biotechnology | sc-25778 | 1:5000 |
| GFP | Cell Signaling Technology | 2956 | 1:5000 |
| GM130 | Cell Signaling Technology | 12480 | 1:3000 |
| Histone H2B | Cell Signaling Technology | 12364 | 1:5000 |
| Histone H3 | Cell Signaling Technology | 4499 | 1:5000 |
| HA-Tag | Cell Signaling Technology | 3724 and 2367 | 1:5000 (both) |
| HDAC2 | Cell Signaling Technology | 5113 | 1:5000 |
| Homer1 | Cell Signaling Technology | 8231 | 1:5000 |
| PSD-95 | Cell Signaling Technology | 3450 | 1:5000 |
| SHANK | Cell Signaling Technology | 12218 | 1:5000 |
| Santa Cruz Biotechnology | sc-23543 | 1:1000 | |
| TuJ1 | Cell Signaling Technology | 5568 | 1:1000 (IF) |
| Secondary antibody | |||
| Anti-rabbit | Cell Signaling Technology | 7074 | 1:5000 |
| Anti-mouse | Cell Signaling Technology | 7076 | 1:5000 |
| 488 anti-chicken | ThermoFisher | A-11039 | 1:1000 (IF) |
| 633 anti-rabbit | ThermoFisher | A-21070 | 1:1000 (IF) |
Isolation of sEVs and quantification
We used sequential centrifugation procedures and filtration. Conditioned medium was first centrifuged at 300 × g for 10 min at 4°C to remove detached cells, followed by a 0.22-μm filtration, an ultracentrifugation at 120,000 × g for 70 min at 4°C using a fixed-angle bucket rotor (Beckman), and a washing step in PBS to eliminate contaminating proteins. Pellets were re-suspended in 50-µl PBS by syringing through a sterile 27-gauge needle to prevent clumping together. The isolated sEV fraction was immediately characterized or stored at −80°C for later use (no more than one week). The quantification of sEVs was performed using EXOCET Exosome Quantitation kit by following the product manual (System Biosciences).
Fixation, immunofluorescence
Cultured neurons were fixed in prewarmed 4% paraformaldehyde (37°C) for 10 min, followed by treatment in PBS containing 0.1% Triton X-100 for 10 min. Culture neurons were then incubated in blocking solution (5% BSA in PBS) for 1 h at room temperature. Following the blocking step, primary antibodies were added to PBS supplemented with 1% BSA for overnight treatment at 4°C. Neurons were then washed using PBS and incubated in secondary antibodies at room temperature for 1 h, which was followed by PBS washing. Coverslips were mounted onto standard microscope slides using ProLong Gold antifade reagent (Invitrogen). Analysis of staining for cell count and cross-sectional area was performed using NIH ImageJ software.
Spine analyses
Spine analysis was performed blindly using the automated spine identifier in NeuronStudio (Rodriguez et al., 2008) on confocal Z-stack images (0.2-µm steps) acquired with a 60× oil Olympus Uplan SAPO (N.A. 1.35) objective. Spine density and shape, including thin, stubby, and mushroom, were analyzed on 100-μm-long dendritic segments on a secondary dendrite per neuron. Thin spines have a thin, long neck and a small bulbous head, whereas mushroom spines have a larger head. Stubby spines are devoid of a neck and are prominent during postnatal development (Rochefort and Konnerth, 2012). A mushroom type is defined as a spine with a head to neck ratio >1.1 and a head diameter >0.35 µm.
Nucleofection of primary neurons and plasmids
Mouse Neuron Nucleofector kit (Lonza, #VPG-1001) was used for nucleofection as previously described (Zhang et al., 2014). Briefly, following microdissection of the cortices and neuron dissociation, 4–6 × 106 cells were spun down and re-suspended in nucleofection solution with 3- to 5-µg plasmids. DNA was transfected by nucleofector program 0–005. Neurons were then plated in MEM medium supplemented with 5% FBS and 0.6% glucose and maintained in neurobasal medium supplemented with B27 and GlutaMax-1.
Plasmids included: pSico-Hdac2 with the sequence GACCGTCTCATTCCATAAA and pSico from Addgene #11578, pCMV-HDAC2-N-3xHA from GeneCopoeia EX-Mm02993-M06, CMV, N-terminal 3XHA, pCAG-GFP from Addgene #11150, and pCAG-tdTomato (Pathania et al., 2012).
sEV and drug treatment in vitro
We used a 6:1 ratio (i.e., cell culture density being sixfold higher for extracellular nanovesicle collection than for the density of recipient neurons) or a 3:1 ratio. Both treatment conditions gave similar results. The following drugs were purchased from Cayman Chemical, Trichostatin A (TSA; #89730) and SAHA (#10009929).
Electron microscopy
Electron microscopy (without additional staining) was performed by the imaging core facility in the Department of Physiology at Yale. Purified vesicles were resuspended in 4% wt/vol paraformaldehyde in phosphate buffered solution (pH 7.4) and embedded for 20 min at room temperature in a formvar-carbon-coated grid. The embedded vesicles were washed in PBS, fixed in 1% gluteraldehyde for 5 min, and stained with saturated aqueous uranyl oxalate. Samples were subsequently embedded in 0.4% wt/vol uranyl acetate and 1.8% wt/vol methylcellulose on ice for 10 min. Samples were dried at room temperature before visualization with a Carl Zeiss 910 electron microscope (Carl Zeiss Microscopy).
Sucrose gradient
Isolated extracellular nanovesicles were analyzed by sucrose gradient centrifugation as extracellular nanovesicles float at densities ranging from 1.15 to 1.19 g/ml on sucrose gradients. Vesicles from the endoplasmic reticulum float at 1.18–1.25 g/ml and those from the Golgi float at 1.05–1.12 g/ml. Most protein aggregates float at 1.22 g/ml (Quillin and Matthews, 2000; Théry et al., 2006). The sucrose gradient centrifugation was performed according to a published protocol (Crewe et al., 2018). Following nanovesicle isolation as detailed above, isolated extracellular nanovesicles were resuspended in sucrose (1 ml at 90% sucrose) at a final concentration of 82% and transferred to 13.2-ml tubes. The sucrose gradient was overlaid on top of the extracellular nanovesicle preparation by successively adding a specific 10–82% sucrose solution starting with the highest and finishing with the lowest sucrose concentration (1.002–1.34 g/ml calculated density). Following 16 h of centrifugation at 100,000 × g, six fractions (F) were collected from top to bottom that were composed of two layers each: 10–16% (F1), 22–28% (F2), 34–40% (F3), 46–52% (F4), 58–64% (F5), and 70–82% (F6). Each fraction was resuspended in PBS and centrifuged at 120,000 × g for 70 min. Western blottings were performed for all the pellets from all six fractions.
Proteinase K protection assay
Proteinase K protection assay was done as previously described (Wang et al., 2017). The isolated extracellular nanovesicles were resuspended in PBS and finally incubated in 1% saponin (Sigma-Aldrich, #47036), 1.67 µg/ml proteinase K (Thermo Fisher Scientific, #AM2546), or 1.67 µg/ml proteinase K together with 1% saponin for 1 h at 37°C. The samples were followed by Western blot analysis.
Proteomic acquisition and analysis
Sample preparation
Proteins were extracted from the isolated fraction containing extracellular nanovesicles using a chloroform:MeOH:water procedure (Wessel and Flügge, 1984). Precipitated proteins were dissolved in 20-µl 8 m urea, 400 mm NH4HCO3 before reduction using 2 µl of dithiothreitol (DTT) and incubated at 37°C for 20 min. Cysteines were then alkylated by the addition of 2 µl of iodoacetamide and incubated in the dark at room temperature for 30 min. The urea concentration was decreased to a final concentration of 2 m urea, 100 mm NH4HCO3 with the addition of 36 µl of water; 20 µl of 0.1 mg/ml Lys-C (Promega, sequencing grade) was added and the sample was digested at 37°C for 6 h, then 4 µl of 0.1 mg/ml trypsin (Promega, sequencing grade) was added and the samples were digested at 37°C for 16 h. Samples were stored at −20°C until just before LC-MS/MS analysis. Into each sample to be injected, a 1× of Pierce's peptide retention time (RT) calibration mixture containing 15 known heavy stable isotopes labeled peptides was added, making a final concentration 1/400th of the 0.5 pmol/µl stock solution.
LC-MS/MS on the LTQ orbitrap
The LTQ Orbitrap Elite is equipped with a Waters nanoAcquity UPLC system and uses a Waters Symmetry C18 180 µm × 20 mm trap column and a 1.7 µm, 75 µm × 250 mm nanoAcquity UPLC column (35°C) for peptide separation. Peptide separation was performed with a linear gradient over 160 min. UPLC run time was at a flow rate of 300 nl/min. Trapping was conducted for 3 min at 5 µl/min in 99% buffer A (0.1% FA in water) and 1% buffer B [0.075% FA in acetonitrile (ACN)] before eluting with linear gradients that will reach 30% B at 140 min, 40% B at 155 min, and 85% B at 160 min. Two blanks (first 100% ACN, second buffer A) will follow each injection to ensure against sample carry over. MS was acquired in the Orbitrap using one microscan and a maximum inject time of 900 ms followed by 15 data-dependent MS/MS acquisitions in the ion trap (with precursor ions threshold of >1000); the total cycle time for both MS and MS/MS acquisition was 2.4 s. Peaks were targeted for MS/MS fragmentation by collision-induced dissociation (CID) were first isolated with a 2-Da window followed by normalized collision energy of 35%. Dynamic exclusion was activated where former target ions were excluded for 30 s.
Data analyses
Feature extraction, chromatographic/spectral alignment, data filtering, and statistical analysis were performed using Nonlinear Dynamics Progenesis QI software (v. 2.0; www.nonlinear.com). First, the raw data files were imported into the program. A sample run was chosen as a reference (usually at or near the middle of all runs in a set), and all other runs were automatically aligned to that run to minimize RT variability between runs. No adjustments are necessary in the m/z dimension because of the high mass accuracy of the spectrometer (typically <3ppm). All runs were selected for detection with an automatic detection limit. On the order of 11,000–28,000 features were detected for each sample set. Features within RT ranges of 0–16 and 102–120 min were filtered out, as were features with charge ≥+8. A normalization factor was then calculated for each run to account for differences in sample load between injections. The experimental design was setup to group multiple injections from each run. The algorithm then calculates and tabulates raw and normalized abundances, max fold change, and ANOVA p values for each feature in the data set. The features were tagged in sets based on characteristics such as the number of MS/MS > 1, and p < 0.05. The MS and MS/MS collected for the experiment were filtered to exclude spectra with rank >10 or isotope >3 to ensure that the highest quality MS/MS spectral data are used for peptide assignments and subsequent protein ID. The remaining MS/MS were exported to an .mgf (Mascot generic file) for database searching. An .xml file of the Mascot search result was created, then imported into the Progenesis QI software, where search hits (peptides ID) are assigned to corresponding features, and protein quantitation was then calculated by the Progenesis QI software.
Database searching
The .mgf files created by the Progenesis QI were searched in-house using the Mascot algorithm (Hirosawa et al., 1993; version 2.2.0) for un-interpreted MS/MS spectra. The data were searched against the SWISSPROT mouse protein database (2015). Search parameters are as follows: type of search: MS/MS Ion Search; Enzyme: Trypsin/Lys-C; Variable modifications: Carbamidomethyl (Cys), Oxidation (Met), Acetyl (Lys), Phospho (ST), and Phospho (Y); Mass values: Monoisotopic; Protein mass: Unrestricted; Peptide mass tolerance: ± 10 ppm; Fragment mass tolerance: ± 0.5 Da; Charge: +7; Max missed cleavages: 3; Decoy: Yes; Instrument type: ESI-TRAP. Using the Mascot database search algorithm, proteins were considered identified when Mascot lists it as significant and more than two unique peptides match the same protein. The Mascot significance score (similar to the “confident scores” column in the progenesis QI protein features spreadsheet) match is based on a MOWSE score and relies on multiple matches to more than one peptide from the same protein. The Mascot search results were exported to an .xml file using a false discovery rate of 1% or less for the protein ID. The emPAI (exponentially modified protein abundance index; Ishihama et al., 2005) was used to estimate the protein level within the LC-MS/MS data and are provided in (Extended Data Fig. 2-1).
To perform gene enrichment analysis, the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/) was used (Huang da et al., 2009a,b). Three types of Gene Ontology (GO) term (molecular function, biological process, and cellular component) pathways were used for annotations. After analysis, the overrepresentation analysis p values and counts for each GO term pathway were obtained. The top 15 GO term pathways with the most significant p values were selected for the subsequently analysis.
Statistical analyses (except for the proteomic analysis)
Data were plotted in Prism 7 (GraphPad Software). Statistical significance was determined using unpaired two-tailed Student's t test and one-way ANOVA with Tukey's post hoc test with p < 0.05 for significance for all experiments. Data are presented as mean ± SEM including the data points in some of the plots. Each experiment was reproduced at least three times, e.g., three mice or three sets of culture. The Student's t tests were all two-sided and unpaired. The sample size calculation was performed using power analysis with G*Power 3 (Faul et al., 2007). For each set of experiments, the sample size was estimated for an effect size of 30–70% using SD calculated from the control population and a power at 0.9 (β = 0.1) and an α of 0.05. Normality tests were performed using the D'Agostino–Pearson omnibus test.
Results
Developing cortical neurons release sEVs enriched in protein and DNA/RNA binding molecules
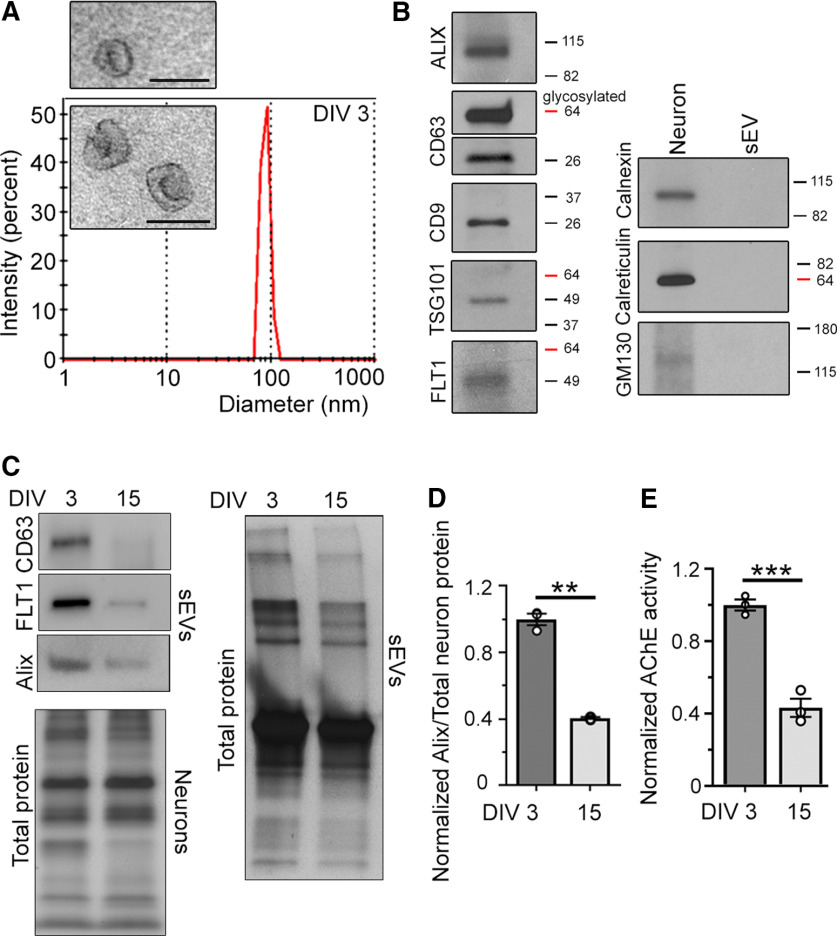
To identify molecules in sEVs that are known to regulate neuron development, we performed a protein screen using sEVs isolated from the culture medium of developing cortical neurons. sEVs were first isolated from the medium of neurons cultured for 3 d in vitro (DIV), which is before spine development. Spine formation starts around DIV9 and ends around DIV21 in our glia-free culture system. The culture medium was collected and subjected to filtration and sequential centrifugations to isolate sEVs (Zhang et al., 2018; Jeppesen et al., 2019). The isolated vesicles contained classical sEVs (or exosomes) based on their mean diameter (91 nm, range: 60–105 nm; Fig. 1A) that is smaller than the diameter of shedding microvesicles and apoptotic bodies (Couzin, 2005; Valadi et al., 2007; Théry et al., 2009; Lee et al., 2012; Dujardin et al., 2014; Janas et al., 2016; Jeppesen et al., 2019), their cup-like morphology assessed by electron microscopy (Fig. 1A, inset), the presence of markers of MVB-generated sEVs, including flotillin 1 (FLT1), CD63, and Alix, and the absence of endoplasmic reticulum proteins (Fig. 1B; El-Andaloussi et al., 2012; Jeppesen et al., 2019). Using a similar procedure, we isolated sEVs from more mature neurons at DIV15 during spine development. Remarkably, there was a decrease in the quantity of sEVs released by DIV15 compared with DIV3 neurons based on an observable decrease in Alix, CD63, and FLT1 levels and total proteins in sEVs (Fig. 1C). Quantification of Alix levels suggested a 56% decrease in sEVs released by DIV15 compared with DIV3 cortical neurons assuming that there were no changes in the level of Alix per sEV (Fig. 1D). We also assessed the quantity of sEVs released by DIV15 and DIV3 neurons (at similar density) using a commercially-available colorimetric assay that measures the enzymatic activity of acetylcholinesterase (AChE) known to be present in sEVs. We found a 57% decrease in AChE activity in DIV15 versus DIV3 sEVs (Fig. 1E). These data suggest a decrease in the quantity of sEV released from cultured cortical neurons as they mature.
Figure 1.
Developing cortical neurons release sEVs. A, Histogram of size distribution of nanovesicles determined by nanoparticle tracking analysis (NanoSight). Inset, Electron micrographs of purified nanovesicles. Scale bars: 125 nm. B, Immunoblots for sEV markers (left blots) and cytoplasmic organelles and apoptotic body markers (right). C, Immunoblots of Alix, FLT1, and CD63 in sEVs from the medium of DIV3 and DIV15 neurons and total protein from neurons at each time point. D, Quantification of Alix in DIV3 and DIV15 sEV divided by the total protein in neurons at each respective time point and normalized to DIV3. E, AChE enzymatic activity in DIV3 and DIV15 sEV normalized to DIV3. sEVs were collected from the medium of cultured neurons of similar density. Unpaired two-tail t test; ***p < 0.001, **p < 0.01.
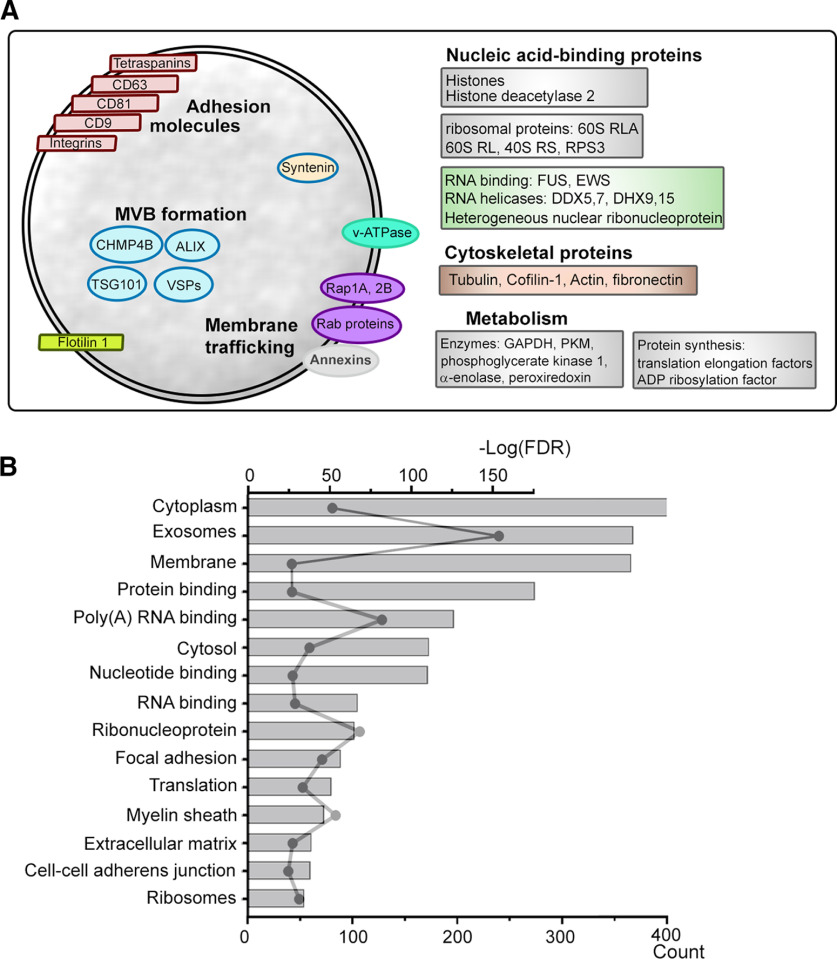
We then examined the content of sEVs from DIV3–DIV6 cortical neurons using LC-MS/MS analyses. We identified 660 proteins by at least one significant peptide (Extended Data Fig. 2-1). We found a combination of molecules reported to be markers of sEV while the marker of classical microvesicles, Annexin A1, was absent (Jeppesen et al., 2019). The markers of sEVs included: membrane proteins such as CD63, CD81, CD9, FLT1, EGFR, Integrin β1, and proteins associated with the biogenesis of sEVs such as Alix, synthenin-1, TSG101, VPS28, VPS37B (Fig. 2A). Isolated sEVs also contained proteins involved in the trafficking of synaptic vesicles (Takamori et al., 2006; Burré and Volknandt, 2007). Pathway analysis revealed that out of the top 15 most significant pathways, protein and DNA/RNA binding molecules were the most represented in addition to cytoplasm and exosome pathways (Fig. 2B; Extended Data Fig. 2-2).
Figure 2.
sEVs released by cortical neurons are enriched in protein and DNA/RNA binding molecules. A, Diagram illustrating some of the molecules known to be enriched in sEVs (e.g., CD63, CD9, FLT1). B, Pathway analysis. FDR, false discovery rate, from DIV3–DIV6 neurons. Scale bars: 100 nm (inset).
Excel spreadsheet listing the results of the proteomics data analysis. Download Figure 2-1, XLSX file (65.1KB, xlsx) .
Excel spreadsheet listing the results of the pathway analysis. Download Figure 2-2, XLSX file (120.3KB, xlsx) .
HDAC2 is present in sEVs released from cortical neurons and its levels decrease as neurons mature
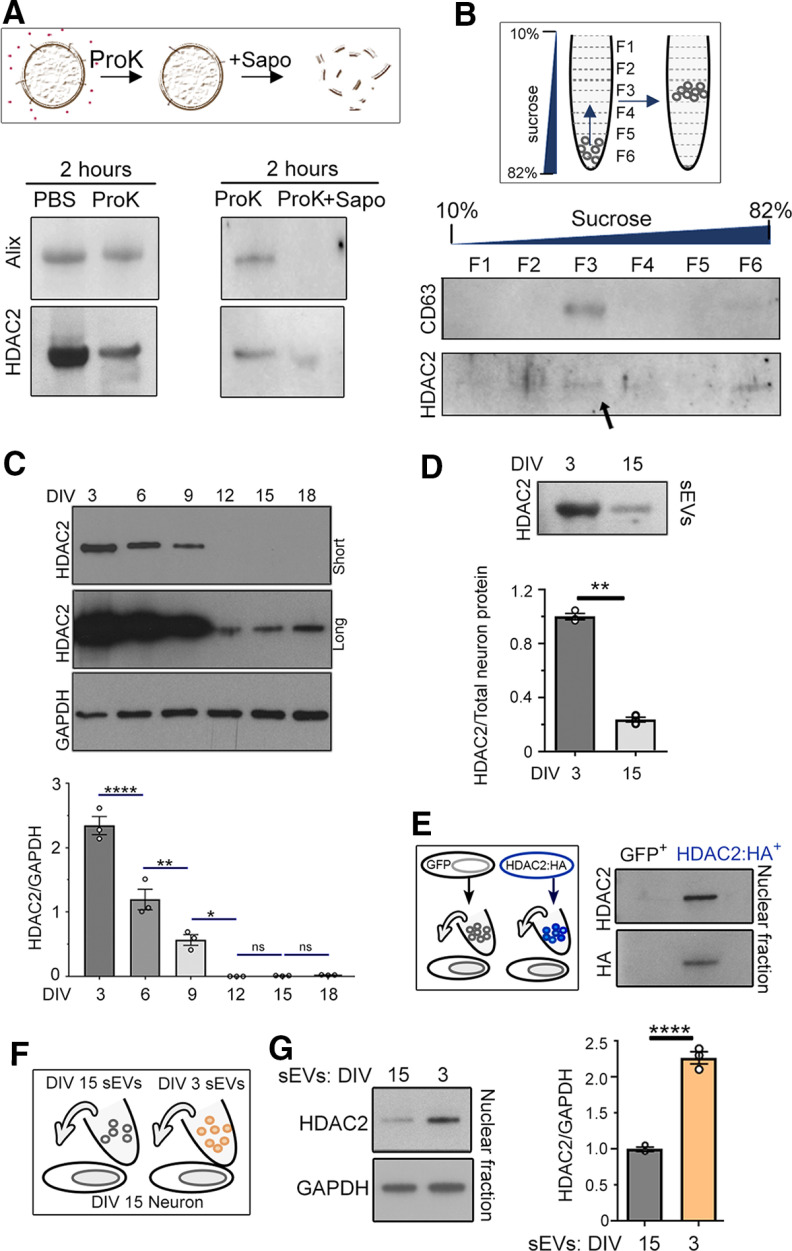
Among the DNA/RNA binding molecules, we identified the epigenetic regulator HDAC2, which is known to suppress dendritic spine development through transcriptional regulation of synaptic molecules (Akhtar et al., 2009; Guan et al., 2009). We thus performed a series of experiments to validate that HDAC2 was indeed released in sEVs collected from the medium of DIV3 cortical neurons. We first isolated sEVs and treated them for 2 h with proteinase K alone to degrade any extracellular proteins or proteinase K with saponin to degrade vesicles (Fig. 3A). Proteinase K treatment of isolated sEVs decreased, but did not abolish, the level of both Alix and HDAC2 detected by immunoblotting. The decrease in Alix and HDAC2 following a proteinase K treatment suggests the possibility of sEV degradation or the presence of free Alix or HDAC2 in the medium that would be digested by proteinase K. Nevertheless, both signals were eliminated with the addition of saponin (Fig. 3A). To validate that proteinase K was efficient at digesting proteins and that saponin itself did not degrade sEVs and their content, we examined whether HDAC2 persisted when sEV were treated with saponin only (for 30 min). HDAC2 was indeed detected following sEV treatment with saponin only and was almost eliminated following treatment with saponin plus proteinase K (30 min; data not shown). Together, these data suggest the presence of HDAC2 in sEVs. We next performed a sucrose gradient centrifugation that further separates sEVs (low density) from non-vesicular particles and debris (high density). The isolated fraction was resuspended into a 12 layer-sucrose gradient (10–82%). After centrifugation, two layers were collected at a time leading to six fractions used for HDAC2 and CD63 immunoblotting (Fig. 3B; Kowal et al., 2016). HDAC2 signal was found in the sEV fraction (F3, also expressing CD63).
Figure 3.
HDAC2 is present in sEV released from cortical neurons and its levels decrease as neurons mature. A, top, Diagram illustrating the effect of Proteinase K (ProK) and saponin (Sapo) on sEVs. Bottom, Western blots of Alix and HDAC2 in the sEV fraction following treatment with PBS or ProK, or 2 hours treatment with ProK or ProK + Sapo. B, Diagram of the sucrose gradient and western blots of CD63 and HDAC2 recovered following a sucrose gradient of sEVs isolated from DIV 3 neurons. C, Western blots and quantification for HDAC2 in cortical neurons at different DIV. D, Western blots for HDAC2 in sEV released from DIV 3 and DIV 15 from neurons and quantification of the data. The lysate was the same as the one used in Figure 1 and thus data normalization was done using total neuron protein as in Figure 1C. E, Diagram of the experimental paradigm (left) and western blots for HDAC2 and HA-tag in neurons treated with sEVs from neurons nucleofected with either GFP or HA-tagged HDAC2 (right). F, Diagram of the experimental paradigm. G, Immunoblots for HDAC2 and GAPDH in the nuclear fraction of DIV 15 neurons treated with sEVs from DIV 3 or 15 neurons and quantification of the blots. N = 3 sets of cultures for all experiments. Plots are mean ± SEM. Unpaired 2-tail t-test (D and G) and one-way ANOVA followed by Tukey post-hoc (C). ****p < 0.0001, **p < 0.01, *p < 0.05; ns, not significant.
An intriguing characteristic of HDAC2 is its developmentally regulated expression during cortical development, in particular, a decrease during spine development (Jones et al., 2009; Gräff et al., 2012). Consistent with this, we found that as neurons mature, HDAC2 levels decreased (DIV3 compared with DIV15; Fig. 3C). We also found that the level of HDAC2 in the sEV fraction significantly decreased (by 75%) as neurons mature (Fig. 3D). This decrease in HDAC2 in the sEV fraction could result from a decrease in sEV cargo packaging or a decrease in the quantity of released sEVs, or a combination of both. To then determine whether HDAC2 could be transferred between neurons or whether sEV treatment alters HDAC2 expression in recipient cells, we expressed an HA-tagged HDAC2 in cultured neurons (Fig. 3E). sEVs were isolated from the medium of transfected cultured neurons and applied to non-transfected neurons. Cortical neurons treated with sEVs from HA-tagged HDAC2-expressing neurons were positive for HA and displayed increased nuclear HDAC2 relative to controls (Fig. 3E). These data suggest that HA-tagged HDAC2 was transferred. To assess endogenous HDAC2 expression in recipient cells, we replaced the medium of DIV15 neurons with medium containing sEVs isolated from DIV3 or DIV15 neurons (used as control) considering the low level of HDAC2 in DIV15 compared with DIV3 neurons (Fig. 3F). The level of HDAC2 was assessed 1 d postmedium replacement. We found a significant increase in nuclear HDAC2 following treatment with sEVs from DIV3 versus DIV15 neurons (Fig. 3G). These data suggest that sEVs released from immature neurons increase HDAC2 expression in more mature neurons. This effect could result from either HDAC2 transfer or the transfer of molecules in sEV that alter HDAC2 expression in recipient neurons.
sEV treatment alters the levels of HDAC2 synaptic targets and spine development
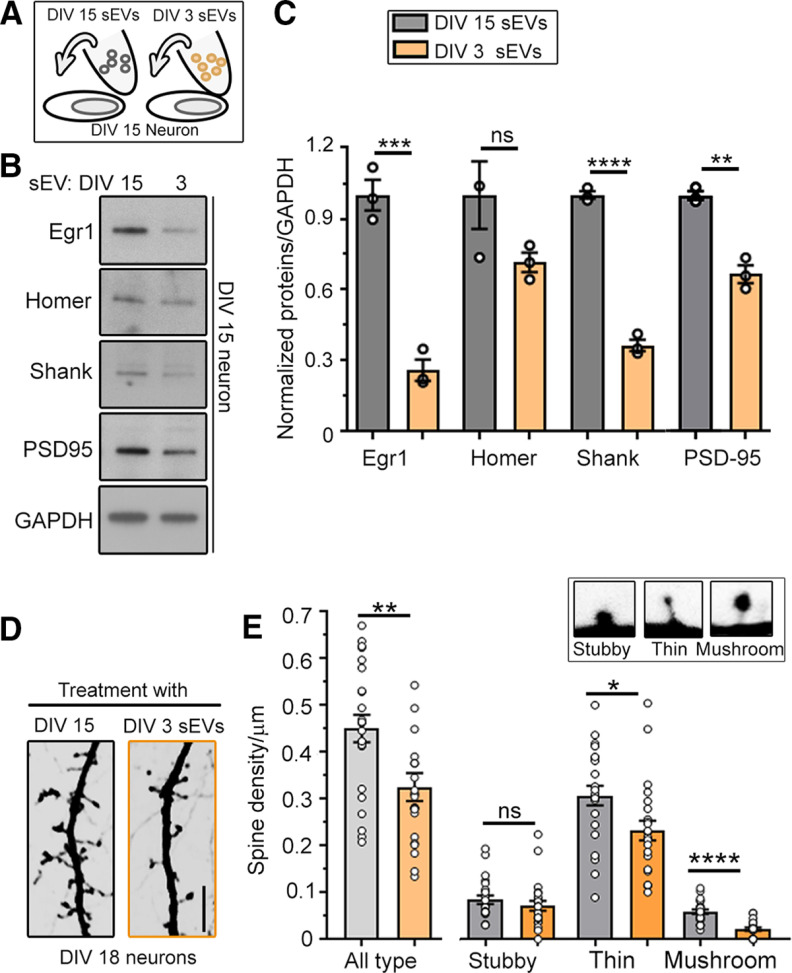
Considering the increase in HDAC2 expression in the recipient neurons following sEV treatment, we examined whether sEVs from the medium of DIV3 neurons would alter the expression of HDAC2-synaptic targets and spine development in DIV15 neurons. As described above, the medium of DIV15 neurons was replaced with medium containing sEVs isolated from DIV3 neurons (Fig. 4A). As control, the medium of DIV15 neurons was replaced with medium containing sEVs isolated from DIV15 neurons. At DIV18, Western blottings and spine properties were analyzed. Treatment of DIV15 neurons with medium containing sEVs from DIV3 neurons decreased the expression of the HDAC2-targets, Egr1, Shank, and PSD-95 (Fig. 4B,C; Gräff et al., 2012). This treatment also decreased the density of spines, including mushroom spines, in DIV15–DIV18 neurons compared with the control condition (Fig. 4D,E). These data indicate that sEVs released from developing cortical neurons transfer molecules that regulate HDAC2 signaling and inhibit spine development.
Figure 4.
sEV treatment alters the levels of HDAC2 synaptic targets and spine development. A, Diagram of the experimental paradigm. B, Western blottings for HDAC2-targeted synaptic proteins in the cytoplasmic fraction of neurons receiving medium replacement with sEVs from DIV3 or DIV15 neurons. C, Quantification of the blots shown in B (N = 3 sets of cultures). D, Confocal images of spines of DIV18 neurons that received medium replacement at DIV15 with medium containing sEVs from either DIV15 or DIV3 neurons. E, Bar graphs of spine density following treatment with sEVs from DIV3 (orange) or DIV15 (gray) neurons (N = 22–24 neurons per condition, three sets of DIV15 culture per condition). Inset, Images of the three different types of spine analyzed: stubby, thin, and mushroom. Unpaired two-tail t test for all plots (mean ± SEM); *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant.
sEVs control spine development via regulation of HDAC2 signaling
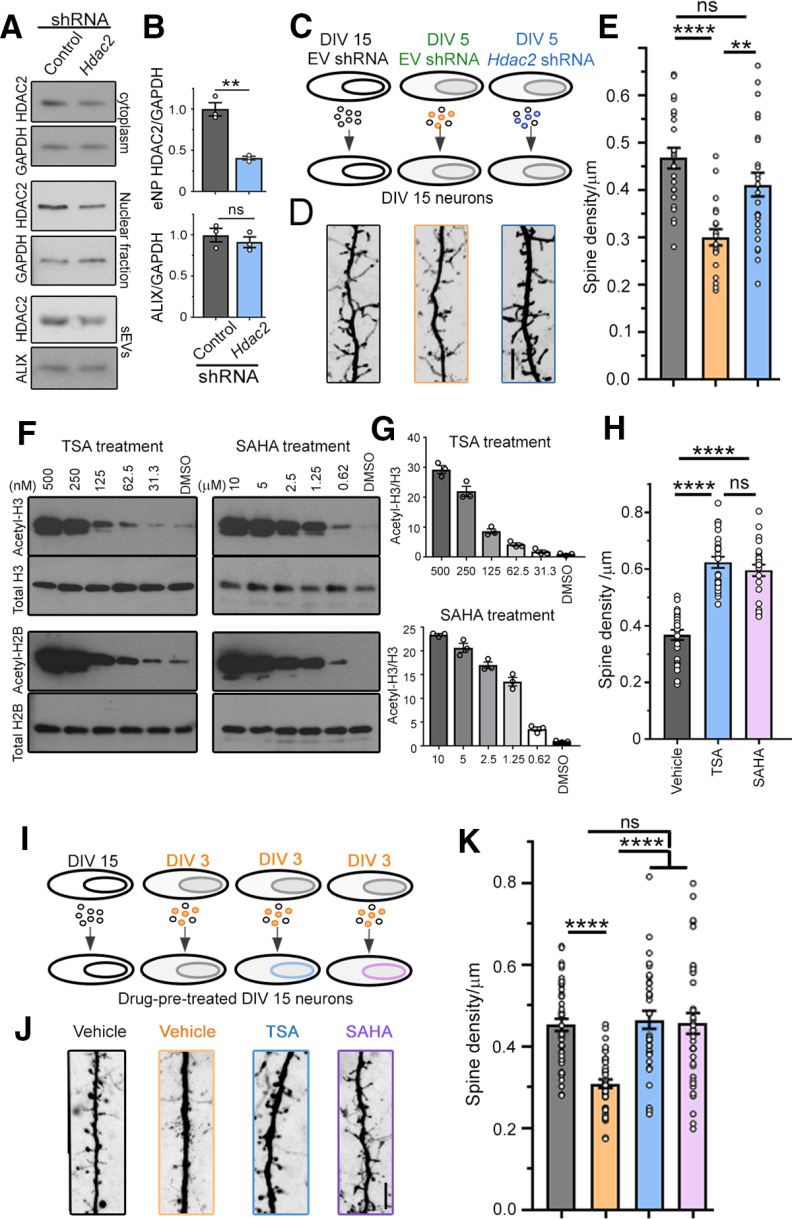
To assess whether increased HDAC2 expression in recipient neurons was involved in spine regulation following sEV treatment, we asked whether the effect of sEVs on spine development could be prevented by decreasing HDAC2 levels in the donor neurons and their released sEVs. Hdac2 and control shRNA were expressed using nucleofection at the time of plating. Expressing an shRNA against Hdac2 in cortical neurons led to a significant decrease in HDAC2 levels in the transfected neurons and in their released sEVs without affecting the quantity of released sEVs as assessed by Alix immunoblotting (Fig. 5A,B). Decreasing HDAC2 levels in DIV5 neurons significantly alleviated the decrease in spine density in DIV15–DIV18 neurons caused by sEV treatment from DIV5 neurons (DIV5 instead of DIV3 was used to allow efficient HDAC2 decrease in sEVs; Fig. 5C–E). Decreasing HDAC2 levels in neurons could potentially alter the content of sEVs and contribute to a rescue of spine properties independent of HDAC2 signaling. To rule out this possibility, we pretreated receiving neurons with HDAC blockers, TSA (250 nm) or SAHA (5 μm), which were confirmed to decrease acetyl-H3 and acetyl-H2B levels (quantification not shown for acetyl-H2B; Fig. 5F,G). Neurons were treated for 3 d before harvest. These blockers also significantly increased spine density in DIV18 neurons (Fig. 5H) as expected since HDAC2 is known to suppress dendritic spine development (Akhtar et al., 2009; Guan et al., 2009). Pretreatment of receiving neurons with TSA or SAHA prevented the decrease in spine density induced by treatment with sEVs from DIV3 neurons (Fig. 5I–K). These data suggest that sEVs contribute to the control of spine development through regulation of HDAC2 signaling in recipient neurons. This may result from direct transfer of HDAC2, but alternatives possibilities are discussed below.
Figure 5.
sEVs affect spine development via the regulation of HDAC2 signaling in recipient cells. A, HDAC2 and GAPDH Western blottings in the cytoplasm and nuclear fraction of DIV5 neurons, and HDAC2 and ALIX Western blottings in sEVs released from neurons containing control or Hdac2 shRNA. B, Quantification of the blots in A. C, Diagram of the experimental paradigm. D, E, Images of spines (D) and spine density (E) in DIV18 neurons posttreatment with sEVs from DIV15 neurons, or DIV5 neurons with or without Hdac2 shRNA. F, Western blottings for acetyl-H3 and acetyl-H2B, targets of HDAC2, examined in cortical neurons treated with different concentrations of TSA and SAHA. G, Quantification of the acetyl-H3/total H3 Western blotting signals shown in F. H, Spine density per micrometer in DIV18 neurons treated with TSA and SAHA for 3 d. I, Diagram of the experimental paradigm. J, K, Images of spines (J) and spine density (K) in DIV18 neurons pretreated with vehicle or HDAC2 blockers before sEV treatment. N = 20–24 neurons/condition, N = 3 culture sets for all experiments. Unpaired two-tail t test (B) and one-way ANOVA followed by Tukey's post hoc test (E, J). Plots are mean ± SEM; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05; ns, not significant. Scale bar: 5 µm.
Discussion
Here, we identified sEVs as signaling elements controlling dendritic spine development by regulating HDAC2 signaling in recipient neurons. To gain insight into a potential function of sEVs on developing cortical neurons, we performed a proteomic analysis that identified >600 proteins in sEVs. Many of these proteins are markers or have been previously reported in sEVs. We also identified proteins involved in synaptic vesicle release and recycling (e.g., SNARE complexes) that may participate in MVB signaling. We found that sEVs were enriched in DNA and RNA binding proteins, including HDAC2. HDAC2 is a well-known epigenetic regulator of the transcription of many genes, including genes encoding synaptic proteins, and it also controls dendritic spine development and the establishment of synapses (Akhtar et al., 2009; Guan et al., 2009). We thus focused on HDAC2 and validated its presence in sEVs as shown with the proteinase K treatment and the sucrose gradient. Recently, the presence of non-vesicular nanoparticles co-isolated with sEVs has been identified (Jeppesen et al., 2019). It has been proposed that a vesicular compartment, called amphisome, is generated by the fusion of MBV and autophagosomes, and co-releases sEVs and non-vesicular nanoparticles into the extracellular space on fusion with the plasma membrane (Papandreou and Tavernarakis, 2017; Palmulli and van Niel, 2018; Jeppesen et al., 2019). Considering the presence of HDAC2 in the non-vesicular fraction (F6) and the identification of non-vesicular nanoparticle markers using LC-MS/MS (e.g., histones, ribosomal protein S3, and fibronectin), it is possible that HDAC2 is in both sEVs and non-vesicular nanoparticles. However, this would need to be further investigated. We next examined whether endogenous HDAC2 could be transferred from newly plated cortical neurons, which are rich in HDAC2, to cortical neurons that are developing spines and express low levels of HDAC2. The levels of HDAC2 indeed decreased as cortical neurons mature in vitro, which recapitulates in vivo data (Jones et al., 2009; Gräff et al., 2012). Using immunoblotting, we detected increased HDAC2 expression in more mature neurons treated with sEVs isolated from newly plated neurons. This increase could result from either HDAC2 transfer via sEVs, but sEV cargo could also induce a change in the recipient neurons leading to HDAC2 increase without HDAC2 transfer. We did not further investigate this issue but rather asked whether the increase in HDAC2 expression was accompanied with a change in the expression of HDAC2 synaptic targets. We found that sEV treatment led to a decrease in the expression of synaptic proteins known to be regulated by HDAC2. Consistent with these data, treatment with sEVs from developing neurons decreased the density of spines in more mature cortical neurons. This effect resulted from an increase in HDAC2 signaling in the recipient neurons since it was prevented by expressing an shRNA against Hdac2 in donor neurons and by pretreating recipient neurons with a blocker of HDAC2. These data clearly show that sEVs provide a signal that increases HDAC2 signaling in recipient neurons that regulate spine development. Additional data showing a decrease in Alix and HDAC2 expression, AChE activity, and total protein in the DIV15 versus the DIV3 sEV fraction suggest that the quantity of released sEV decreases as neurons mature. It is also possible that the content of Alix, HDAC2, and AChE in sEV decreases over time. Either way, these data suggest that sEVs provide a strong signal during development that may decline as neurons mature. Collectively, these data show that sEVs provide a signal that regulates synaptogenesis between cortical neurons at different stages of development. Considering that HDAC2 is ubiquitously expressed in neurons, albeit with different levels during development, we expect that this sEV-based communication modality exerts significant influence on other neuronal networks.
Footnotes
This work was supported by the National Institutes of Health (NIH) Grant R01 NS086329, CURE Innovator Award, the National Alliance for Research on Schizophrenia and Depression Distinguished Investigator Award funded by the Hofmann Trust (A.B.), and the NIH Grant SIG RR031795 (to T.T.L.). We thank Jean Kanyo for her assistance with the proteomics data collection.
The authors declare no competing financial interests.
References
- Akhtar MW, Raingo J, Nelson ED, Montgomery RL, Olson EN, Kavalali ET, Monteggia LM (2009) Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci 29:8288–8297. 10.1523/JNEUROSCI.0097-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V, Ruiz-Cañada C, Wendler F (2016) Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci 17:160–172. 10.1038/nrn.2015.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burré J, Volknandt W (2007) The synaptic vesicle proteome. J Neurochem 101:1448–1462. 10.1111/j.1471-4159.2007.04453.x [DOI] [PubMed] [Google Scholar]
- Couzin J (2005) Cell biology: the ins and outs of exosomes. Science 308:1862–1863. 10.1126/science.308.5730.1862 [DOI] [PubMed] [Google Scholar]
- Crewe C, Joffin N, Rutkowski JM, Kim M, Zhang F, Towler DA, Gordillo R, Scherer PE (2018) An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell 175:695–708.e13. 10.1016/j.cell.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin S, Begard S, Caillierez R, Lachaud C, Delattre L, Carrier S, Loyens A, Galas MC, Bousset L, Melki R, Auregan G, Hantraye P, Brouillet E, Buee L, Colin M (2014) Ectosomes: a new mechanism for non-exosomal secretion of tau protein. PLoS One 9:e100760. 10.1371/journal.pone.0100760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Andaloussi S, Lee Y, Lakhal-Littleton S, Li J, Seow Y, Gardiner C, Alvarez-Erviti L, Sargent IL, Wood MJ (2012) Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc 7:2112–2126. 10.1038/nprot.2012.131 [DOI] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A (2007) G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 39:175–191. 10.3758/bf03193146 [DOI] [PubMed] [Google Scholar]
- Fauré J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B, Grange J, Schoehn G, Goldberg Y, Boyer V, Kirchhoff F, Raposo G, Garin J, Sadoul R (2006) Exosomes are released by cultured cortical neurones. Mol Cell Neurosci 31:642–648. 10.1016/j.mcn.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Gräff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, Su SC, Samiei A, Joseph N, Haggarty SJ, Delalle I, Tsai LH (2012) An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 483:222–226. 10.1038/nature10849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459:55–60. 10.1038/nature07925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa M, Hoshida M, Ishikawa M, Toya T (1993) MASCOT: multiple alignment system for protein sequences based on three-way dynamic programming. Comput Appl Biosci 9:161–167. 10.1093/bioinformatics/9.2.161 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009a) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009b) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272. 10.1074/mcp.M500061-MCP200 [DOI] [PubMed] [Google Scholar]
- Janas AM, Sapoń K, Janas T, Stowell MHB, Janas T (2016) Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. Biochim Biophys Acta 1858:1139–1151. 10.1016/j.bbamem.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ (2019) Reassessment of exosome composition. Cell 177:428–445.e18. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Overly CC, Sunkin SM (2009) The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci 10:821–828. 10.1038/nrn2722 [DOI] [PubMed] [Google Scholar]
- Korkut C, Ataman B, Ramachandran P, Ashley J, Barria R, Gherbesi N, Budnik V (2009) Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 139:393–404. 10.1016/j.cell.2009.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut C, Li Y, Koles K, Brewer C, Ashley J, Yoshihara M, Budnik V (2013) Regulation of postsynaptic retrograde signaling by presynaptic exosome release. Neuron 77:1039–1046. 10.1016/j.neuron.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C (2016) Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA 113:E968–E977. 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, Blot B, Haase G, Goldberg Y, Sadoul R (2011) Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci 46:409–418. 10.1016/j.mcn.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Laulagnier K, Javalet C, Hemming FJ, Sadoul R (2017) Purification and analysis of exosomes released by mature cortical neurons following synaptic activation. Methods Mol Biol 1545:129–138. 10.1007/978-1-4939-6728-5_9 [DOI] [PubMed] [Google Scholar]
- Lawson C, Kovacs D, Finding E, Ulfelder E, Luis-Fuentes V (2017) Extracellular vesicles: evolutionarily conserved mediators of intercellular communication. Yale J Biol Med 90:481–491. [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Shin SM, Zhong P, Kim HT, Kim DI, Kim JM, Do Heo W, Kim DW, Yeo CY, Kim CH, Liu QS (2018) Reciprocal control of excitatory synapse numbers by Wnt and Wnt inhibitor PRR7 secreted on exosomes. Nat Commun 9:3434. 10.1038/s41467-018-05858-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, El AS, Wood MJ (2012) Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet 21:R125–R134. 10.1093/hmg/dds317 [DOI] [PubMed] [Google Scholar]
- Palmulli R, van Niel G (2018) To be or not to be… secreted as exosomes, a balance finely tuned by the mechanisms of biogenesis. Essays Biochem 62:177–191. 10.1042/EBC20170076 [DOI] [PubMed] [Google Scholar]
- Papandreou ME, Tavernarakis N (2017) Autophagy and the endo/exosomal pathways in health and disease. Biotechnol J 12. [DOI] [PubMed] [Google Scholar]
- Pathania M, Torres-Reveron J, Yan L, Kimura T, Lin TV, Gordon V, Teng ZQ, Zhao X, Fulga TA, Van VD, Bordey A (2012) miR-132 enhances dendritic morphogenesis, spine density, synaptic integration, and survival of newborn olfactory bulb neurons. PLoS One 7:e38174. 10.1371/journal.pone.0038174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin ML, Matthews BW (2000) Accurate calculation of the density of proteins. Acta Crystallogr D Biol Crystallogr 56:791–794. 10.1107/s090744490000679x [DOI] [PubMed] [Google Scholar]
- Rochefort NL, Konnerth A (2012) Dendritic spines: from structure to in vivo function. EMBO Rep 13:699–708. 10.1038/embor.2012.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL (2008) Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One 3:e1997. 10.1371/journal.pone.0001997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Mesci P, Carromeu C, McClatchy DR, Schiapparelli L, Yates JR 3rd, Muotri AR, Cline HT (2019) Exosomes regulate neurogenesis and circuit assembly. Proc Natl Acad Sci USA 116:16086–16094. 10.1073/pnas.1902513116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, Müller SA, Rammner B, Gräter F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmüller H, Heuser J, et al. (2006) Molecular anatomy of a trafficking organelle. Cell 127:831–846. 10.1016/j.cell.2006.10.030 [DOI] [PubMed] [Google Scholar]
- Théry C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Théry C, Ostrowski M, Segura E (2009) Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9:581–593. 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Wang Y, Balaji V, Kaniyappan S, Krüger L, Irsen S, Tepper K, Chandupatla R, Maetzler W, Schneider A, Mandelkow E, Mandelkow EM (2017) The release and trans-synaptic transmission of tau via exosomes. Mol Neurodegener 12:5. 10.1186/s13024-016-0143-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143. 10.1016/0003-2697(84)90782-6 [DOI] [PubMed] [Google Scholar]
- Zappulli V, Friis KP, Fitzpatrick Z, Maguire CA, Breakefield XO (2016) Extracellular vesicles and intercellular communication within the nervous system. J Clin Invest 126:1198–1207. 10.1172/JCI81134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Bartley CM, Gong X, Hsieh LS, Lin TV, Feliciano DM, Bordey A (2014) MEK-ERK1/2-dependent FLNA overexpression promotes abnormal dendritic patterning in tuberous sclerosis independent of mTOR. Neuron 84:78–91. 10.1016/j.neuron.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, et al. (2018) Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 20:332–343. 10.1038/s41556-018-0040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel spreadsheet listing the results of the proteomics data analysis. Download Figure 2-1, XLSX file (65.1KB, xlsx) .
Excel spreadsheet listing the results of the pathway analysis. Download Figure 2-2, XLSX file (120.3KB, xlsx) .