Abstract
Background
To study the protein profile of the serum exosomes of patients with coronary artery aneurysms (CAA) caused by Kawasaki disease (KD).
Methods
Two-dimensional electrophoresis (2-DE) was used to identify proteins from the exosomes of serum obtained from children with CAA caused by KD, as well as healthy controls. Differentially expressed proteins were identified using matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF MS) analysis.
Results
Thirty two differentially expressed proteins were identified (18 up-regulated and 14 downregulated) from serum exosomes of children with CAA and were compared to healthy controls. The expression levels of 4 proteins (TN, RBP4, LRG1, and APOA4) were validated using Western blotting. Classification analysis and protein–protein network analysis showed that they are associated with multiple functional groups, including host immune response, inflammation, apoptotic process, developmental process, and biological adhesion process.
Conclusions
These findings establish a comprehensive proteomic profile of serum exosomes from children with CAA caused by KD, and provide additional insights into the mechanisms of CAA caused by KD.
Keywords: Kawasaki disease, coronary aneurysm, exosome, proteomics
Introduction
Kawasaki disease (KD) is the most prevalent inflammatory coronary artery disease with an unknown etiology [1]. Eighty percent of patients are younger than 5 years old [2]. It is characterized by the development of coronary artery aneurysms (CAAs) [3]. CAAs may cause ruptures, thrombosis, stenosis, or myocardial ischemia if there is a failure or delay in treatment. It is therefore important to make an accurate diagnosis and choose the appropriate therapy during early stages of the disease.
Several scoring systems have been established to identify the risk factors for CAA [4, 5]. Duration of fever has been reported as a potent risk factor. Male sex, younger patient age, and delayed diagnosis and treatment have also been found to be correlated with the development of CAA. Laboratory-detected indexes such as thrombocytopenia, leukocytosis, reduced hemoglobin, and reduced serum albumin are also prominent risk factors [6]. However, no specific laboratory test exists for CAA, and it is hard to establish a diagnosis, especially in early stages. The methods for diagnosing CAA are primarily imaging systems such as echocardiography [7], computed tomography (CT), and magnetic resonance imaging (MRI) [3]. However, these methods have a relatively low sensitivity. The incidence of CAA has decreased as a result of treatments with intravenous immunoglobulin (IVIG); however, up to 5% of treated patients still develop CAA compared to up to 25% of untreated patients [2]. Therefore, it is crucial to understand the molecular pathogenesis of CAA to improve diagnosis and therapy of CAA in patients with KD.
Exosomes, 30–100 nm membrane vesicles of endocytic origin that are secreted by most cell types, are important intercellular communicators [8, 9]. As exosomes can transmit many molecules intercellularly, they are considered a potential source of new biomarkers. Exosomes may act as mediators for the activation of signaling mechanisms in the target cell, intercellular communication, transfer of proteins and miRNAs, or the disposal of cellular components [10]. Thus, exosomes carry physiological and pathological biomarker information, and changes in serum exosome protein composition may reflect the pathological processes of systemic diseases. Proteomics analysis is currently considered to be a powerful tool for the global evaluation of protein expression and has been widely applied in studying biomarkers and the molecular pathogenesis of diseases [11].
In the current study, we employed two-dimensional electrophoresis (2-DE) and matrix-assisted laser desorption ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF MS) to compare the proteomes of exosomes from serum samples collected from healthy children and from children with CAA caused by KD. Western blotting was used to confirm the results of the 2-DE analysis. According to available research, this is the first study to perform a proteomic analysis in the context of pathogenesis of CAA caused by KD, providing a comprehensive atlas of the serum exosome proteome, and providing clues for further diagnostic and therapeutic strategies for CAA caused by KD.
Methods
Preparation of serum samples
This study was approved by the Ethical Committee of Guangzhou Women and Children’s Medical Center (permit [2013]077); the legal guardians of all children enrolled in the study provided written informed consent. Blood samples from 5 patients with CAA caused by KD were randomly recruited according to the criteria of the American Heart Association and the Japanese Ministry of Health and Welfare [12]. The diagnoses of all patients were validated by more than 2 pediatric cardiologists, and all other possible diseases were ruled out. Blood samples from 5 healthy children were used as a control group. Blood samples were separated by centrifugation at 1,000 × g for 10 min. Serum aliquots were collected and stored at −80°C. The serum obtained was further processed for exosome isolation.
Precipitation of serum exosomes
Exosomes were separated from the sera of all participants using ExoQuick precipitation (System Biosciences Inc., Palo Alto, CA, USA) according to the manufacturer instructions [13, 14].
Exosome characterization
Serum exosomes were precipitated using the exosome extraction reagent. The extracts were centrifuged at 1,500 × g for 10 min at 4°C. The exosome pellet was suspended in phosphate-buffered saline (PBS) in 4 × the volume of serum. A copper mesh was placed on a wax plate, then 100 μL exosome suspension was added for 4 min. The copper mesh was removed and placed in 2% phosphotungstic acid for 5 min. The mesh was then laid on filter paper for drying. The morphological features of the exosomes were observed using transmission electron microscopy (TEM).
Exosome characterization using Western blot analysis
The exosome pellet was dissolved in protein lysis buffer, and a Bradford protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA) was used to determine protein concentration. Samples were separated by one-dimensional (1-D) SDS-PAGE, then transferred to a PVDF membrane. The membrane was incubated in anti-CD63 (1:1000), -HSP90α (1:1000), and -Flotillin (1:1000) antibodies at 4°C overnight, followed by incubation with secondary antibodies at room temperature for 1 h. Specific protein bands were visualized using the SuperSignal enhanced chemiluminescence (ECL) system (Thermo Fisher Scientific, Waltham, MA, USA) and imaged on x-ray film.
Protein extraction of serum exosome
The exosome samples were grouped into two pooled samples consisting of equal amounts of the 5 experimental samples from each group (CAA and control groups) before proteomic analysis. The Bradford protein assay kit was subsequently used to determine the final protein concentration of the exosomes according to the manufacturer instructions.
2-DE
According to a protocol published previously [15], 2-DE analysis was performed with slight modifications. In brief, the prepared pooled protein samples (120 mg protein on analytical gels, or 600 mg protein on preparative gels) were mixed with rehydration buffer to a volume of 450 μL. IPG strips (pH 4–7, 24 cm, GE Healthcare, Little Chalfont, UK) were used for the first dimension to isolate the altered proteins, and the following parameters were used: 20°C, 300 V for 30 min, 700 V for 30 min, 1,500 V for 1.5 h, 9,900 V for 3 h, 9,900 V for 6.5 h, 600 V for 20 h, 8,000 V constant for a total of 56 000 Vh. After completion of the isoelectric focusing (IEF) program, the strips were equilibrated in two steps: 15 min in an IPG equilibration buffer (6 M urea, 2% SDS, 30% glycerol, 0.375M Tris, 20 mg/mL DTT, and a trace of bromophenol blue), and then alkylated for 15 min. Subsequently, a 12.5% 2-D SDS-PAGE was performed. Electrophoresis was carried out at 20 mA per gel for 40 min and then at 30 mA per gel until the dye front reached the bottom. The protein spots were visualized by either Coomassie Brilliant Blue G-250 staining or silver staining.
Image analysis
The gels were analyzed using the Image-Master 2D Platinum software (GE Healthcare). The normalized protein amount for each spot was calculated as the ratio of the volume on each spot to the total spot volume on the gel. Protein spots with significant differences in abundance (increase or decrease of > 1.5-fold) were selected for further analysis.
In-gel digestion
Differentially expressed protein spots were manually excised from the 2-DE gels. Each gel piece was washed twice in deionized water, destained with 50% methanol for 30 min at 37°C, and dehydrated in 100 μL acetonitrile (ACN) for 20 min at room temperature. Next, the samples were swollen in 50 μL 100 mM NH4HCO3, dehydrated a second time, and incubated in 1 μg/50 μL trypsin (Promega, Madison, WI, USA) for 30 min at 4°C. Then, coverage solution (50 mM NH4HCO3, 10% ACN, deionized water) was added to the samples and incubated for 16 h at 37°C. After removing the coverage solution by aspiration, the peptide mixtures were extracted with 2.5% trifluoroacetic acid (TFA)/90% ACN for 30 min at room temperature and vacuum dried.
Protein identification
The material was dissolved in 1.5 μL solution containing deionized water, 0.1% TFA, and 50% ACN after vacuum drying. Then, 0.8 μL of the mixture was loaded onto a MALDI sample target plate using 0.5 μL HCCA (5 mg/mL a-Cyano-4-hydro-xycinnamic acid) matrix, dried at room temperature and analyzed using the ABI MALDI-TOF/TOF Proteomics Analyzer mass spectrometer (Applied Biosystems, USA). The UV laser was operated at a 200 Hz repetition rate at a wavelength of 355 nm and an accelerated voltage of 20 kV.
Database searching
The experimental MS data were matched to a corresponding virtual peptide mass database derived from GPS Explorer v 3.6, Mascot, and the UniProt database under the taxonomy of Homo sapiens. Protein identification was carried out by peptide mass fingerprint (PMF) with Mascot software (http://www.matrixscience.com). The search parameters used in PMF were as follows: species: Homo sapiens; enzyme: trypsin; fixed modifications: carbamidomethylation; variable modifications: oxidation (M). The gene name, function, and Gene Ontology (GO) category of each protein was determined with the Mascot v 2.1 protein database search engine and the UniProt Homo sapiens protein database.
Western blot analysis
Protein extracts from the serum exosomes of 5 KD patients with CAA and 5 healthy children were separated by SDS-PAGE (11–15% acrylamide). After being transferred to PVDF membranes and blocked overnight, primary antibody was added for 1 h, followed by washing in PBS, application of a secondary HRP-conjugated antibody, and development using an ECL system (Thermo Fisher Scientific). Anti-tetranectin (TN) (1:1000), anti-Leucine-rich alpha-2-glycoprotein (LRG1) (1:1000), anti-Retinol-binding protein 4 (RBP4) (1:1000), and anti-Apolipoprotein A-IV (APOA4) (1:1000) were used as primary antibodies.
Protein categorization
Differentially expressed proteins were classified using Protein Analysis Through Evolutionary Relationships (PANTHER; http://www.pantherdb.org). PANTHER ontology is a highly controlled vocabulary categorized according to molecular function, biological process, and protein class.
Protein network construction
An interaction map of the differentially expressed proteins was established using Pathway Studio 5.0 (Ariadne Genomics, Rockville, MD, USA), a text-mining tool that can construct protein interaction networks and pathways. It includes pathway components, protein–protein interactions, functional classes, proteins, and their cellular processes. In this study, the shortest path analysis was selected.
Results
Exosome isolation and validation
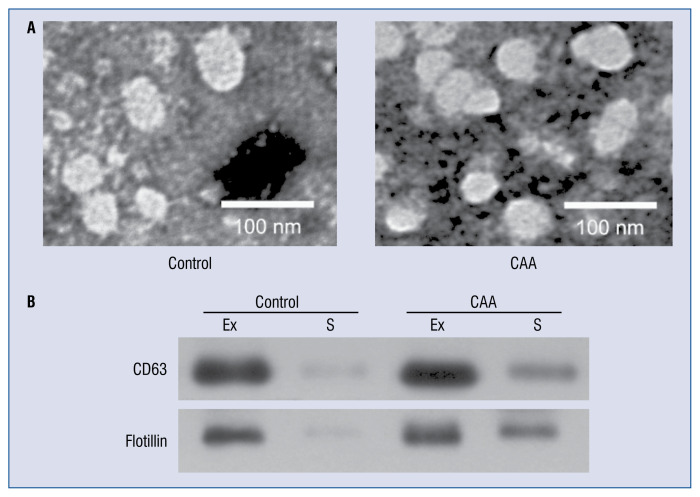
Microvesicles isolated from the sera of children with CAA caused by KD and healthy controls were examined by TEM and Western blotting. Spherical structures 30–100 nm in diameter were observed by TEM (Fig. 1A). It was validated that the microvesicles were exosomes through Western blot analysis using antibodies against 3 exosomal markers: HSP90α, CD63, and Flotillin. The expression levels of these 3 markers were markedly higher in the microvesicle fraction than in the serum fraction (Fig. 1B). These results validated that the isolated microvesicles were exosomes.
Figure 1.
Characterization of exosomes in serum samples from patients with coronary artery aneurysm (CAA) caused by Kawasaki disease and healthy children. A. Morphological characterization of exosomes by transmission electron microscopy; B. Molecular characterization of exosomes by Western blotting. Protein extracts prepared from sera (S) or exosomes (Ex) were examined using antibodies against exosomal protein markers (CD63, HSP90α, and flotillin).
Identification of differential proteins between CAA patients and healthy controls by proteomic analysis
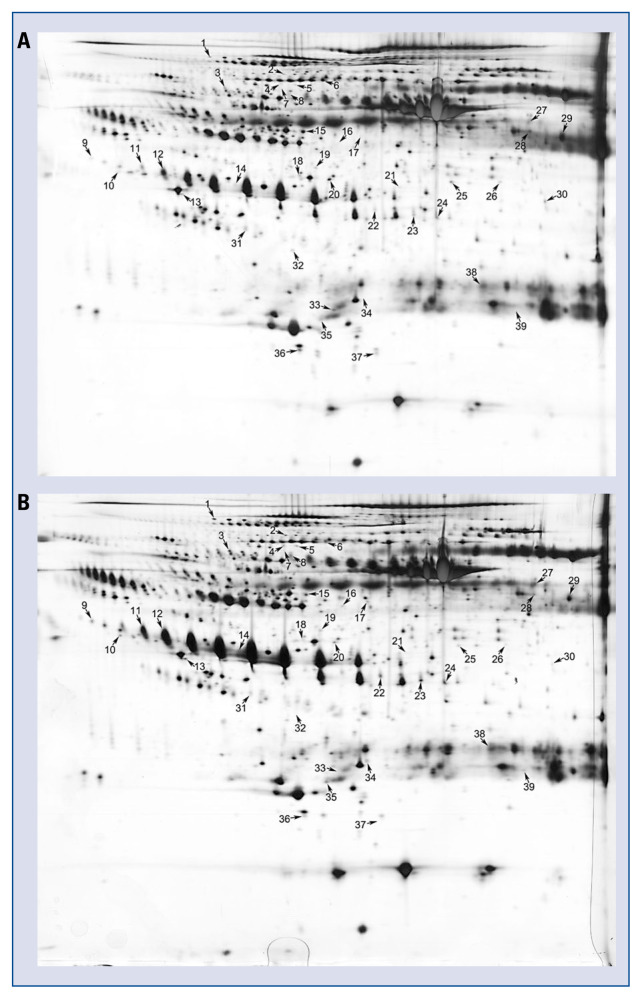
Pooled serum exosome proteins from 5 KD patients with CAA and 5 healthy controls were separated by 2-DE (Fig. 2A–B). After visual review, 39 differential protein spots with a minimum of 1.5-fold difference in expression between the two groups were selected for MALDI-TOF/TOF MS analysis. Differentially expressed proteins were picked from gels and identified using MALDI-TOF/TOF MS. In total, 32 differentially expressed proteins (18 up-regulated and 14 down-regulated) were successfully identified (Table 1). Some proteins appeared as multiple spots in 2-DE, likely due to their isoforms and/or post-translational modifications.
Figure 2.
Two-dimensional electrophoresis analysis of the proteome of serum exosomes. Gels were visualized by silver staining. Arrows indicate differentially expressed proteins in Kawasaki disease (KD) patients with coronary artery aneurysm (CAA) and healthy controls. Spot numbers correspond to the proteins in Table 1. A. Healthy children; B. KD patients with CAA.
Table 1.
Differentially expressed proteins between Kawasaki disease patients with coronary artery aneurysm and healthy children.
| Spot No.* | Protein | Accession no. | Protein MW (Da) | Protein pI | CAA/C | Sequence coverage | Protein score |
|---|---|---|---|---|---|---|---|
| 1 | Alpha-1-antitrypsin | P01009 | 44280 | 5.37 | 1.95588 | 32% | 319 |
| 2 | Complement C4-B | P0C0L5 | 194216 | 6.89 | 1.67713 | 13% | 255 |
| 3 | Inter-alpha-trypsin inhibitor heavy chain H4 | Q14624 | 103536 | 6.64 | 2.05879 | 18% | 251 |
| 4 | Afamin | P43652 | 70963 | 5.64 | 1.90293 | 23% | 287 |
| 5 | Inter-alpha-trypsin inhibitor heavy chain H4 | Q59FS1 | 76971 | 5.72 | 2.36148 | 23% | 183 |
| 6 | Complement C4-A | P0C0L4 | 84758 | 5.33 | −1.96522 | 26% | 229 |
| 7 | Insulin-like growth factor-binding protein complex acid labile subunit | Q8TAY0 | 66735 | 6.33 | 1000000 | 26% | 325 |
| 8 | Alpha-1B-glycoprotein | P04217 | 52479 | 5.65 | 3.7226 | 26% | 185 |
| 9 | Leucine-rich alpha-2-glycoprotein | P02750 | 38382 | 6.45 | 2.34081 | 31% | 464 |
| 10 | Complement C4-B | P0C0L5 | 40795 | 5.19 | 1.50231 | 26% | 137 |
| 11 | Haptoglobin | P00738 | 38868 | 6.26 | 8.13111 | 29% | 269 |
| 12 | Haptoglobin | P00738 | 38868 | 6.26 | 3.62451 | 20% | 141 |
| 13 | Complement C3 | P01024 | 40204 | 4.79 | 1.71764 | 48% | 278 |
| 14 | Apolipoprotein A-IV | P06727 | 45371 | 5.28 | 2.06449 | 27% | 158 |
| 15 | Antithrombin-III | P01008 | 49350 | 5.95 | −1.65022 | 40% | 476 |
| 16 | CFI protein | Q8WW88 | 44285 | 8.49 | 2.18167 | 7% | 74 |
| 17 | CFI protein | Q8WW88 | 44285 | 8.49 | 2.9125 | 9% | 128 |
| 18 | CD5 antigen-like | O43866 | 39603 | 5.28 | −1.86353 | 43% | 246 |
| 19 | Adipocyte plasma membrane-associated protein | Q9HDC9 | 46622 | 5.82 | 1.63727 | 26% | 230 |
| 20 | Sex hormone-binding globulin | P04278 | 28876 | 5.32 | −1.75668 | 46% | 392 |
| 21 | Sex hormone-binding globulin | P04278 | 28876 | 5.32 | −1.57588 | 35% | 88 |
| 22 | Haptoglobin-related protein | P00739 | 39300 | 6.67 | 2.302 | 20% | 156 |
| 23 | Haptoglobin | P00738 | 39496 | 6.42 | 2.93725 | 28% | 76 |
| 24 | Complement factor H | P08603 | 36506 | 5.97 | 1.72789 | 39% | 152 |
| 25 | Complement factor H-related protein 1 | Q03591 | 38766 | 7.38 | −2.13194 | 29% | 130 |
| 26 | Ig mu chain C region | P01871 | 50117 | 6.4 | −1.7426 | 21% | 136 |
| 27 | Alpha-2-macroglobulin | P01023 | 71321 | 5.47 | 1.87957 | 7% | 115 |
| 28 | Serum albumin | P02768 | 58513 | 5.96 | −1.80408 | 40% | 244 |
| 29 | Beta-2-glycoprotein 1 | D9IWP9 | 37485 | 8.37 | −2.1629 | 28% | 221 |
| 30 | Serotransferrin | P02787 | 37241 | 6.49 | −1.72825 | 39% | 291 |
| 31 | Clusterin | P10909 | 36997 | 5.74 | −1.7058 | 14% | 162 |
| 32 | Transthyretin | P02766 | 12789 | 5.26 | −1000000 | 68% | 155 |
| 33 | Ig kappa chain V-I region Rei | P01607 | 23779 | 8.75 | −1000000 | 44% | 120 |
| 34 | Ig lambda chain V-IV region Hil | P01717 | 23020 | 6.69 | 1000000 | 32% | 103 |
| 35 | Ig kappa chain C region | P01834 | 23523 | 5.72 | 1.59839 | 42% | 159 |
| 36 | Retinol-binding protein 4 | Q5VY30 | 21287 | 5.27 | −2.64472 | 33% | 130 |
| 37 | Tetranectin | P05452 | 22951 5 | .52 | −1000000 | 42% | 295 |
| 38 | Immunoglobulin lambda-like polypeptide 5 | B9A064 | 22819 | 5.79 | 1.71135 | 42% | 85 |
| 39 | Ig kappa chain C region | P01834 | 24352 | 6.99 | 1000000 | 39% | 118 |
Spot numbers correspond to those in Figure 2.
MW — molecular weight; pI — isoelectric point; CAA — coronary artery aneurysm; C — control
Validation of differential proteins by Western blotting
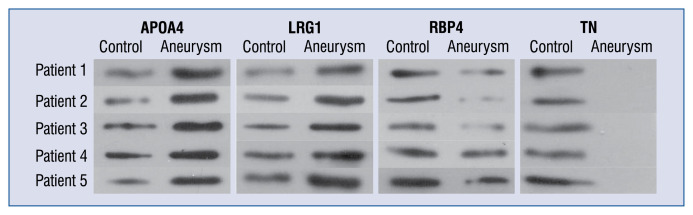
To further confirm the proteomic analysis results, Western blotting was performed to examine the expression levels of APOA4, LRG1, RBP4, and TN in protein extracts from the serum exosomes of 5 patients with CAA caused by KD and 5 healthy controls. The expression levels of APOA4 and LRG1 were significantly higher in the CAA group than in the control group (Fig. 3). The expression levels of RBP4 and TN were significantly lower in the CAA group than in the control group (Fig. 3). This result was consistent with the results of proteomic analysis.
Figure 3.
Western blot analysis showing the expression of TN, APOA4, RBP4, and LRG1 in 5 Kawasaki disease patients with coronary artery aneurysm and 5 healthy children.
Classification analysis of identified proteins
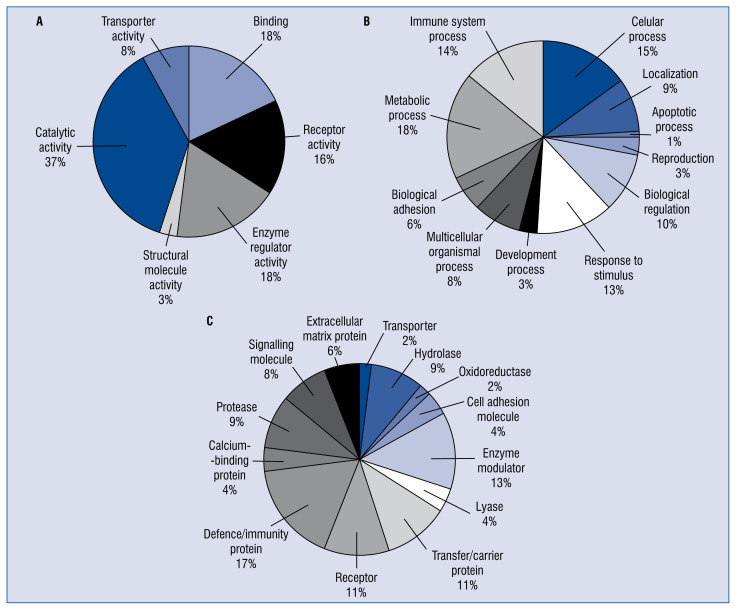
According to GO cellular functions and processes, the proteins were distributed into categories based on molecular function, biological processes, and protein classes using the PANTHER classification system (Fig. 4). Most of the identified proteins played roles in the metabolic process (18.0%) and immune system process (14.0%). Other biological processes that the proteins were involved in included response to stimulus (13.0%), biology regulation (10.0%), and localization (9.0%). In terms of protein class, most of the proteins were classified as defense/immunity protein (17.0%), enzyme modulator (13.0%), or transfer/carrier protein (11.0%); others belonged to receptor (11%), hydrolase (9.0), signaling molecule (8.0%), and others. As for molecular function, the most dominant function that the identified proteins were involved in was catalytic activity (37.0%), followed by enzyme regulator activity (18.0%), binding (18.0%), receptor activity (16.0%), transporter activity (8.0%), and structural molecule activity (3.0%).
Figure 4.
Classification analysis of differentially expressed proteins identified between Kawasaki disease patients with coronary artery aneurysm (CAA) and healthy controls. Categorization was based on information that was obtained from the online Protein Analysis Through Evolutionary Relationships (PANTHER) classification system in terms of (A) molecular function, (B) biological process, and (C) protein class.
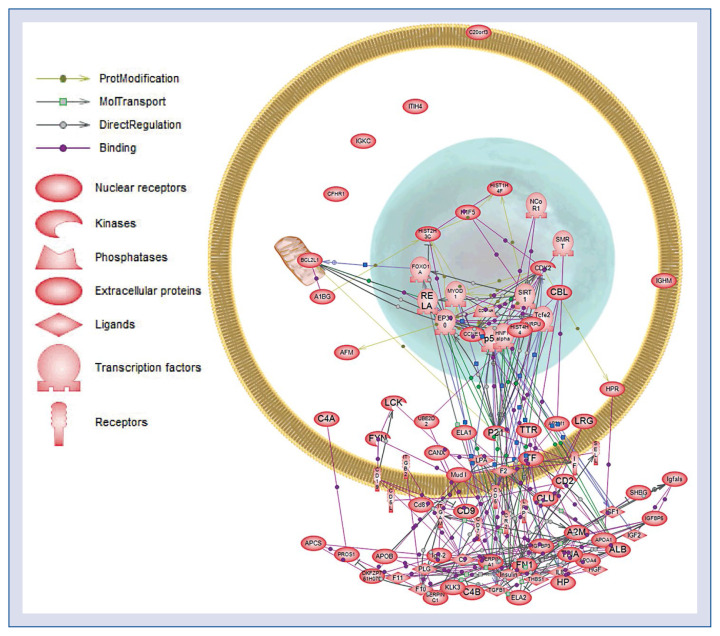
Network modeling of differentially expressed proteins
To explore the associations between differentially expressed proteins, a protein network was constructed with Pathway Studio 5.0. Complex interactions between mitochondrial, nuclear, cytoplasmic, and extracellular proteins were present in the prediction network (Fig. 5). Further research is needed to explore the role of these proteins and their interaction pathways in the pathogenesis of CAA due to KD.
Figure 5.
Protein network of the identified proteins constructed using Pathway Studio 5.0. The differentially expressed proteins in Table 1 were imported into Pathway Assist, and an interaction model was generated using the shortest path algorithm. The legend of the interaction network is summarized on the right of the figure. Each node represents either a protein entity or a control mechanism of the interaction.
Discussion
Cardiovascular complications are a major cause of morbidity in KD; the primary concern is CAA, which develops in 15–25% of untreated children [16]. Importantly, there is no specific modality to establish a diagnosis, especially in early stages. Treatment with a high dose of IVIG is proven to be effective in reducing the prevalence of CAA to less than 5%. However, about 10% of patients do not respond to this treatment, which represents an important clinical dilemma owing to the increased risk of developing CAA [17]. Therefore, it is critical to understand the molecular pathogenesis of CAA in order to improve the effectiveness of diagnosis and therapy for CAA caused by KD. Protein profiles from patients at specific stages can be more accurate in reflecting the status of disease progression.
In this study, 2-DE and MALDI-TOF/TOF MS were used to analyze proteomic changes in the serum exosomes of patients with CAA caused by KD. 32 differentially expressed proteins were identified in serum exosome samples, offering a spectrum of the proteomic changes caused by CAA. The differential expression of 4 proteins (TN, LRG1, RBP4, and APOA4) was then validated by Western blotting. Finally, these differentially expressed proteins were analyzed using a bioinformatics approach. The majority of differentially expressed proteins were involved in the host immune response, inflammation, apoptotic process, developmental process, and biological adhesion process. This largely favors molecular diagnosis and therapy for patients with CAA caused by KD.
Tetranectin, a plasminogen kringle 4 binding C-type lectin [18], is a plasma protein secreted by monocytes, neutrophils, and macrophages. The biological function of TN has not been fully elucidated, though it is thought to play an important role in the regulation of proteolytic and fibrinolysis processes by binding to plasminogen. It is also believed to play a critical role in mineralization during osteogenesis and in myogenesis during embryonic development [19]. Furthermore, TN has been implicated as a potential biomarker in conditions ranging from cancer to Parkinson’s disease [20]. The present results, detected by 2-DE and confirmed by Western blotting, showed that the expression of TN in the CAA samples was significantly lower compared to healthy controls. Although validation with a larger sample size is necessary, these preliminary results have provided the first indication, according to available research, that TN potentially participates in the pathogenesis of CAA caused by KD, and that it is a potential CAA biomarker. The function and role of TN in CAA caused by KD still remains elusive and requires further investigation.
LRG1 is a protein from the leucine-rich repeat (LRR) family. LRG1 is expressed during granulocyte differentiation and might play an important role in the immune response when the host clears infections. Previous research has shown that LRG1 is involved in important pathological and biological processes, such as cell adhesion, signal transduction, and protein–protein interactions [21]. Recently, LRG1 was found to be elevated in the serum of patients with lung cancer, liver cancer, and pancreatic adenocarcinoma [22, 23]. In the present study, it was found that LRG1 was up-regulated in CAA serum exosomes, suggesting that it may be involved in the development of CAA, and that the immune response to infection might contribute to the formation of CAA in KD.
RBP4 functions to deliver retinol to tissues [24]. RBP4 has been correlated with cardiometabolic risk in humans and is related to the degree of carotid intima–media thickness. Recent studies on aortic rings have shown the vasodilatory effect of RBP4 results from increased nitric oxide production in vascular endothelial cells. In addition, decreased levels of RBP4 have been found in men during the acute phase of myocardial infarction and in patients with familial hyper-cholesterolemia at high risk of an ischemic event; this suggests that decreased RBP4 levels lead to an increased susceptibility to ischemic events, and that RBP4 is a potential protective factor against cardiovascular events [25, 26]. In the present study, for the first time, it was shown that RBP4 was down-regulated in CAA serum exosomes compared to healthy controls. It seems that the down-regulation of RBP4 plays a role in the development of CAA due to KD.
APOA4, a 46-kDa glycoprotein, is produced primarily in the small intestine and is discharged into the mesenteric lymph [27]. It enters the plasma compartment as a structural protein of very low-density lipoproteins (VLDL), high-density lipoproteins (HDL), chylomicrons, or in the lipoprotein-free fraction [28]. Currently, the physiological function of APOA4 is not fully understood. Several studies have shown that it plays a role in reverse cholesterol transport. Some studies have reported that APOA4 has antioxidant and anti-atherogenic properties. A meaningful change in the serum APOA4 concentration has been found in patients with several disorders including renal disease, malabsorption syndrome, and chronic pancreatitis. In the present study, it was found that APOA4 was up-regulated in the serum exosome samples of patients with CAA. It seems that APOA4 may participate in the development of CAA due to KD.
Conclusions
In conclusion, a set of proteins were identified that were differentially expressed in CAA serum exosomes compared to healthy control samples. Of them, TN, RBP4, LRG1, and APOA4 were validated by Western blotting. Although none of the above-mentioned differentially expressed proteins is specifically correlated with CAA caused by KD, this preliminary study clearly demonstrated that the levels of these proteins were significantly changed in patients with CAA compared to healthy controls. Preliminary results suggest that these proteins may participate in the pathogenesis of CAA caused by KD. Future studies are necessary to elucidate the molecular role and mechanisms of these proteins in CAA caused by KD.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Project No. 81500275 to H.-L.J.), and the Natural Science Foundation of Guangdong Province (Project No. 2016A030313080 to H.-L.J.).
Footnotes
Conflict of interest: None declared
References
- 1.Takahashi K, Oharaseki T, Yokouchi Y. Pathogenesis of Kawasaki disease. Clin Exp Immunol. 2011;164(Suppl 1):20–22. doi: 10.1111/j.1365-2249.2011.04361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uehara R, Belay ED. Epidemiology of Kawasaki disease in Asia, Europe, and the United States. J Epidemiol. 2012;22(2):79–85. doi: 10.2188/jea.JE20110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duarte R, Cisneros S, Fernandez G, et al. Kawasaki disease: a review with emphasis on cardiovascular complications. Insights Imaging. 2010;1(4):223–231. doi: 10.1007/s13244-010-0035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabharwal T, Manlhiot C, Benseler SM, et al. Comparison of factors associated with coronary artery dilation only versus coronary artery aneurysms in patients with Kawasaki disease. Am J Cardiol. 2009;104(12):1743–1747. doi: 10.1016/j.amjcard.2009.07.062. [DOI] [PubMed] [Google Scholar]
- 5.McCrindle BW, Li JS, Minich LL, et al. Coronary artery involvement in children with Kawasaki disease: risk factors from analysis of serial normalized measurements. Circulation. 2007;116(2):174–179. doi: 10.1161/CIRCULATIONAHA.107.690875. [DOI] [PubMed] [Google Scholar]
- 6.Leung DYM, Meissner HC, Shulman ST, et al. Prevalence of superantigen-secreting bacteria in patients with Kawasaki disease. J Pediatr. 2002;140(6):742–746. doi: 10.1067/mpd.2002.123664. [DOI] [PubMed] [Google Scholar]
- 7.Baddour LM, Wilson WR, Bayer AS, et al. Infective endocarditis: diagnosis, antimicrobial therapy, and management of complications: a statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils on Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association endorsed by the Infectious Diseases Society of America. Circulation. 2005;111(23):e394–e434. doi: 10.1161/CIRCULATIONAHA.105.165564. [DOI] [PubMed] [Google Scholar]
- 8.Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18(5):199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson RJ, Jensen SS, Lim JWE. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8(19):4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 10.Vlassov AV, Magdaleno S, Setterquist R, et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Jia HL, Huang JM, et al. Identification of biomarkers for lymph node metastasis in early-stage cervical cancer by tissue-based proteomics. Br J Cancer. 2014;110(7):1748–1758. doi: 10.1038/bjc.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayusawa M, Sonobe T, Uemura S, et al. Revision of diagnostic guidelines for Kawasaki disease (the 5th revised edition) Pediatr Int. 2005;47(2):232–234. doi: 10.1111/j.1442-200x.2005.02033.x. [DOI] [PubMed] [Google Scholar]
- 13.Rekker K, Saare M, Roost AM, et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47(1–2):135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Wu SC, Yang JCS, Rau CS, et al. Profiling circulating micro-RNA expression in experimental sepsis using cecal ligation and puncture. PLoS One. 2013;8(10):e77936. doi: 10.1371/journal.pone.0077936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Li, Jia HL, Huang WM, et al. Monitoring of the serum proteome in Kawasaki disease patients before and after immunoglobulin therapy. Biochem Biophys Res Commun. 2014;447(1):19–25. doi: 10.1016/j.bbrc.2014.03.108. [DOI] [PubMed] [Google Scholar]
- 16.Cohen P, O’Gara P. Coronary Artery Aneurysms. Cardiology in Review. 2008;16(6):301–304. doi: 10.1097/crd.0b013e3181852659.. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Oharaseki T, Yokouchi Y, et al. Kawasaki disease as a systemic vasculitis in childhood. Ann Vasc Dis. 2010;3(3):173–181. doi: 10.3400/avd.sasvp01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemmensen I, Petersen LC, Kluft C. Purification and characterization of a novel, oligomeric, plasminogen kringle 4 binding protein from human plasma: tetranectin. Eur J Biochem. 1986;156(2):327–333. doi: 10.1111/j.1432-1033.1986.tb09586.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang ES, Sun Y, Guo JG, et al. Tetranectin and apolipoprotein A-I in cerebrospinal fluid as potential biomarkers for Parkinson’s disease. Acta Neurol Scand. 2010;122(5):350–359. doi: 10.1111/j.1600-0404.2009.01318.x. [DOI] [PubMed] [Google Scholar]
- 20.Obrist P, Spizzo G, Ensinger C, et al. Aberrant tetranectin expression in human breast carcinomas as a predictor of survival. J Clin Pathol. 2004;57(4):417–421. doi: 10.1136/jcp.2003.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Donnell LC, Druhan LJ, Avalos BR. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol. 2002;72(3):478–485. [PubMed] [Google Scholar]
- 22.Kakisaka T, Kondo T, Okano T, et al. Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2DDIGE): up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;852(1–2):257–267. doi: 10.1016/j.jchromb.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okano T, Kondo T, Kakisaka T, et al. Plasma proteomics of lung cancer by a linkage of multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis. Proteomics. 2006;6(13):3938–3948. doi: 10.1002/pmic.200500883. [DOI] [PubMed] [Google Scholar]
- 24.Quadro L, Blaner WS, Salchow DJ, et al. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999;18(17):4633–4644. doi: 10.1093/emboj/18.17.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calò LA, Maiolino G, Pagnin E, et al. Increased RBP4 in a human model of activated anti-atherosclerotic and antiremodelling defences. Eur J Clin Invest. 2014;44(6):567–572. doi: 10.1111/eci.12270. [DOI] [PubMed] [Google Scholar]
- 26.Bobbert T, Raila J, Schwarz F, et al. Relation between retinol, retinol-binding protein 4, transthyretin and carotid intima media thickness. Atherosclerosis. 2010;213(2):549–551. doi: 10.1016/j.atherosclerosis.2010.07.063. [DOI] [PubMed] [Google Scholar]
- 27.Steinmetz A, Barbaras R, Ghalim N, et al. Human apolipoprotein A-IV binds to apolipoprotein A-I/A-II receptor sites and promotes cholesterol efflux from adipose cells. J Biol Chem. 1990;265(14):7859–7863. [PubMed] [Google Scholar]
- 28.Omori M, Watanabe M, Matsumoto K, et al. Impact of serum apolipoprotein A-IV as a marker of cardiovascular disease in maintenance hemodialysis patients. Ther Apher Dial. 2010;14(3):341–348. doi: 10.1111/j.1744-9987.2010.00809.x. [DOI] [PubMed] [Google Scholar]