Abstract
Background
The main challenge in the study of schizophrenia is its high heterogeneity. While it is generally accepted that there exist several biological mechanisms that may define distinct schizophrenia subtypes, they have not been identified yet. We performed comprehensive gene expression analysis to search for molecular signals that differentiate schizophrenia patients from healthy controls and examined whether an identified signal was concentrated in a subgroup of the patients.
Methods
Transcriptome sequencing of 14 superior temporal gyrus (STG) samples of subjects with schizophrenia and 15 matched controls from the Stanley Medical Research Institute (SMRI) was performed. Differential expression and pathway enrichment analysis results were compared to an independent cohort. Replicability was tested on 6 additional independent datasets.
Results
The 2 STG cohorts showed high replicability. Pathway enrichment analysis of the down-regulated genes pointed to proteasome-related pathways. Meta-analysis of differential expression identified down-regulation of 12 of 39 proteasome subunit genes in schizophrenia. The signal of proteasome subunits down-regulation was replicated in 6 additional datasets (overall 8 cohorts with 267 schizophrenia and 266 control samples, from 5 brain regions). The signal was concentrated in a subgroup of patients with schizophrenia.
Conclusions
We detected global down-regulation of proteasome subunits in a subgroup of patients with schizophrenia. We hypothesize that the down-regulation of proteasome subunits leads to proteasome dysfunction that causes accumulation of ubiquitinated proteins, which has been recently detected in a subgroup of schizophrenia patients. Thus, down-regulation of proteasome subunits might define a biological subtype of schizophrenia.
Keywords: gene expression, postmortem brain samples, integrated analysis, ubiquitin proteasome system
Introduction
Schizophrenia affects 1% of the population and has a complex pathophysiology that is far from being fully understood. The main challenge is its high genetic and clinical heterogeneity.1 While for years several subtypes definitions were in scientific and clinical use, the DSM-5 has omitted them after concluding that they do not predict the course of illness.2 However, it is generally accepted that there exist several mechanisms that may define distinct schizophrenia subtypes, which have not been identified yet.
Recently, the ubiquitin proteasome system (UPS), governing protein degradation, has been associated with schizophrenia at both transcript3–6 and protein levels,7,8 with tendency for down-regulation in schizophrenia brain samples. On the genomic level, UPS pathways were enriched with schizophrenia-associated copy number variants,9 and the proteasome pathway was enriched in schizophrenia susceptibility genes.10
Recent findings suggest a more pronounced role of the UPS in schizophrenia. Accumulation of ubiquitinated proteins has been identified in brain samples of a subgroup of schizophrenia patients in the STG, frontal cortex, and prefrontal cortex samples.11 Another study detected elevated ubiquitinated protein levels in the orbitofrontal cortex of schizophrenia patients.12 While ubiquitin binds to proteins (which become “ubiquitinated”), targeting them for proteasome degradation, proteasome dysfunction can cause accumulation of ubiquitinated proteins,13 as has been detected in schizophrenia. Recent studies of proteasome activity in schizophrenia have, however, yielded inconsistent results.12,14 Thus, while the elevation of ubiquitinated protein levels seems to play a role in schizophrenia, it is not clear whether this is caused by dysfunction of the proteasome.
Two studies7,14 have examined protein levels of proteasome subunits in schizophrenia, with 3 regulatory subunits found to be decreased in both (Table 2). Several studies reported down-regulation of proteasome subunits genes,4,6,15,16 but only 2 subunits were found to be down-regulated in more than a single study (Table 2). Thus, while there is evidence for down-regulation of both transcript and protein levels of proteasome subunits in schizophrenia, the results are currently sporadic.
Table 2.
Proteasome Subunits Differential Gene and Protein Level Expression, in Previous Studies and in Our Meta-Analysis
| # | Proteasome Subunit Genes | Previous Gene Expression Studies | Previous Protein Level Studies7,14 | Our Meta-Analysis (SMRI+MSSM) | SMRI+ MSSM Meta-Analysis Summary Measure [Confidence Interval] |
|---|---|---|---|---|---|
| Structural subunits | |||||
| 20S core α subunits | |||||
| 1 | PSMA1 (also named 20S α1) | Down-regulated in 2 studies4,35 | Not measured | Unchanged | −0.37 [−0.97, 0.21] |
| 2 | PSMA2 (20S α2) | Not measured | Down-regulation | −1.13 [−1.68, −0.59] | |
| 3 | PSMA3 (20S α3) | Not measured | Unchanged | −0.63 [−1.67, 0.4] | |
| 4 | PSMA4 (20S α4) | Not measured | Unchanged | −0.43 [−0.9, 0.07] | |
| 5 | PSMA5 (20S α5) | Not measured | Down-regulation | −0.61 [−1.13, −0.09] | |
| 6 | PSMA6 (20S α6) | Unchanged in ref.14 | Down-regulation | −0.63 [−1.15, −0.12] | |
| 7 | PSMA7 (20S α7) | Not measured | Down-regulation | −0.79 [−1.32, −0.27] | |
| Catalytic subunits | |||||
| 20S core β subunits | |||||
| 8 | PSMB1 (20S β1) | Not measured | Unchanged | −0.17 [−0.73, 0.37] | |
| 9 | PSMB2 (20S β2) | Down-regulation trend7 (P = .08); unchanged in ref.14 | Down-regulation | −0.62 [−1.13, −0.11] | |
| 10 | PSMB3 (20S β3) | Not measured | Unchanged | −0.25 [−0.75, 0.25] | |
| 11 | PSMB4 (20S β4) | Not measured | Unchanged | −0.03 [−0.53, 0.46] | |
| 12 | PSMB5 (20S β5) | Unchanged7,14 | Down-regulation | −0.73 [−1.28, −0.18] | |
| 13 | PSMB6 (20S β6) | Not measured | Unchanged | −0.13 [−1.25, 0.97] | |
| 14 | PSMB7 (20S β7) | Not measured | Unchanged | −0.37 [−0.87, 0.13] | |
| Immunoproteasome β subunit genes | |||||
| PSMB8 (20S β5i) | Unchanged7,14 | unchanged in SMRI; absent in MSSM | |||
| 15 | PSMB9 (20S β1i) | Unchanged7 | Unchanged | −0.04 [−0.45, 0.54] | |
| 16 | PSMB10 (20S β2i) | Unchanged7,14 | Unchanged | 0.16 [−0.38, 0.71] | |
| Regulatory subunits | |||||
| 19S AAA-ATPase subunits (Rpt) | |||||
| 17 | PSMC1 (19S Rpt2) | Unchanged in ref.7; Down-regulated in ref.14 | Unchanged | 0.02 [−0.48, 0.52] | |
| 18 | PSMC2 (19S Rpt1) | Down-regulated in 2 studies7,14 | Down-regulation | −0.93 [−1.46, −0.4] | |
| 29 | PSMC3 (19S Rpt5) | Unchanged in ref.7; Down-regulated in ref.14 | Unchanged | −0.11 [−0.62, 0.38] | |
| 20 | PSMC4 (19S Rpt3) | Down-regulated in 2 studies7,14 | Down-regulation | −0.67 [−1.19, −0.15] | |
| 21 | PSMC5 (19S Rpt6) | Down-regulated in 2 studies7,14 | Unchanged | 0.02 [−0.48, 0.52] | |
| 22 | PSMC6 (19S Rpt4) | Down-regulated in 2 studies4,6 | Unchanged7; Down-regulated in ref.14 | Down-regulation | −0.83 [−1.36, −0.3] |
| 19S non-ATPase subunits (Rpn) | |||||
| 23 | PSMD1 (19S Rpn2) | Not measured | Unchanged | 0.07 [−0.42, 0.58] | |
| 24 | PSMD2 (19S Rpn1) | Not measured | Unchanged | −0.03 [−1.49, 1.41] | |
| 25 | PSMD3 (19S Rpn3) | Not measured | Unchanged | −0.34 [−0.87, 0.18] | |
| 26 | PSMD4 (19S Rpn10) | Unchanged7 | Unchanged | 0.09 [−0.41, 0.59] | |
| 27 | PSMD5 | Not measured | Unchanged | 0.17 [−0.69, 1.05] | |
| 28 | PSMD6 (19S Rpn7) | Not measured | Down-regulation | −0.62 [−1.13, −0.1] | |
| 29 | PSMD7 (19S Rpn8) | Not measured | Unchanged | −0.07 [−1.02, 0.86] | |
| 30 | PSMD8 (19S Rpn12) | Not measured | Unchanged | −0.39 [−0.9, 0.11] | |
| 31 | PSMD9 (19S Rpn4) | Not measured | Unchanged | −0.39 [−1.42, 0.63] | |
| 32 | PSMD10 | Not measured | Unchanged | 0.13 [−0.36, 0.63] | |
| 33 | PSMD11 (19S Rpn6) | Unchanged7 | Down-regulation | −0.73 [−1.25, −0.21] | |
| 34 | PSMD12 (19S Rpn5) | Not measured | Unchanged | −0.23 [−0.74, 0.26] | |
| 35 | PSMD13 (19S Rpn9) | Not measured | Unchanged | 0.05 [−0.58, 0.69] | |
| 36 | PSMD14 (19S Rpn11) | Unchanged7 | Down-regulation | −0.89 [−1.42, −0.37] | |
| 11S subunits | |||||
| 37 | PSME1 (11S α) | Down-regulated in ref.7; unchanged in ref.14 | Unchanged | −0.33 [−0.84, −0.16] | |
| 38 | PSME2 (11S β) | Unchanged7,14 | Unchanged | 0.29 [−0.65, 1.24] | |
| 39 | PSME3 (11S gamma) | Unchanged7 | Unchanged | −0.07 [−0.85, 0.7] |
Note: Previous gene expression studies’ results were listed only for genes which were detected as differentially expressed in more than one study. Down-regulation findings are boldfaced. In the meta-analysis, a gene is defined as down-regulated if its summary measure is lower than zero and the confidence interval does not cross zero.
A basic limitation of gene expression studies of schizophrenia is the fact that brain samples are usually composed of a mixture of cell types, which might dilute authentic changes. In addition, schizophrenia is highly heterogeneous1 and typical changes in gene expression are modest (fold change range 1.03–1.3317), which are thus difficult to detect. A relatively simple way to deal with these limitations is to perform a systematic comparison between independent datasets. Here we performed RNA-sequencing of STG samples from 14 schizophrenia and 15 control subjects from the Stanley Medical Research Institute (SMRI). We applied pathway enrichment analysis to the list of genes detected as differentially expressed. We then used an independent cohort from the Mount Sinai School of Medicine (MSSM) to test the replicability of our results. A systematic meta-analysis of the SMRI and MSSM was applied to a subgroup of 39 inter-connected genes, which showed a tendency for down-regulation in schizophrenia. Six additional cohorts of different brain regions were used to further examine the robustness of our results. One of the 6 datasets was from the same patients as the SMRI data described above, from a different brain region. Finally, we checked whether the signal characterizes a subgroup of the patients.
Methods
SMRI Subjects
STG postmortem tissues from 15 subjects with schizophrenia and 15 healthy controls were obtained from the SMRI using approved protocols for tissue collection and informed consent.18 Samples were examined by a neuropathologist to exclude cerebral pathologies.19 Diagnoses were performed independently by 2 psychiatrists according to DSM-IV, and matched by age, gender, postmortem interval (PMI) and pH (Table 1). RNA-sequencing was applied to 29 out of the 30 STG samples (one sample did not pass quality control – see below).
Table 1.
Subjects’ Characteristics
| Characteristics | Schizophrenia | Control | P-value |
|---|---|---|---|
| SMRI subjects | |||
| Number of subjects | 14 | 15 | |
| Gender (M/F) | 9/5 | 9/6 | 1 |
| Age (y) | 43.6 (13) | 48.1 (10.6) | .32 |
| Brain pH | 6.2 (0.3) | 6.3 (0.2) | .35 |
| RIN | 6.2 (0.5) | 6.4 (0.5) | .17 |
| PMI (min) | 2052 (900) | 1424 (596) | .03 |
| MSSM subjects | |||
| Number of subjects | 19 | 14 | |
| Gender (M/F) | 14/5 | 5/9 | .04 |
| Age (y) | 77.4 (10.9) | 82.4 (12.7) | .23 |
| Brain pH | 6.4 (0.2) | 6.6 (0.3) | .08 |
| PMI (min) | 814 (499) | 460 (429) | .04 |
Note: Average values (SD). To compare schizophrenia and controls 2-sided t-test P-values were calculated for the continuous variables and Fisher’s exact test P-value was calculated for M/F ratio.
MSSM Subjects
STG samples of 19 schizophrenia and 14 healthy controls were obtained from the Brain Bank of the Department of Psychiatry of the MSSM (Table 1). All cortical dissections and sample preparation were described previously20–22; see also the Supplementary Information. Gene expression was measured using Affymetrix HG-U133A microarrays.
RNA-Sequencing
Brain regions were dissected at SMRI and delivered to Israel, where total RNA was isolated using the Trizol method. The concentration of total RNA and RNA Integrity Number (RIN) were measured. Samples with concentration ≥ 10 ng/μl and RIN ≥5 were selected for sequencing (one schizophrenia sample was excluded). The mean RIN was 6.3 (± 0.5), and the mean ratio of 260/280 was 1.6 (±0.14). The mean total RNA yield was 15.4 μg (± 9.7). See Supplementary Methods for a description of the libraries preparation protocol. For raw RNA-sequencing data description see Supplementary Table 1S.
Mapping, Quantification of Gene Expression Levels and Pre-processing
We used standard software tools for mapping fragments to the genome and for quantification of gene expression levels. See Supplementary Methods for full description. Pre-processing: Lowess correction was calculated.23 Then expression threshold was set to 6 (log scale) to reduce noise. Filtering: Genes with expression values below 6 in at least 80% of the samples were excluded from the analysis, leaving 16 482 genes after filtering (out of 23 715). We compared this method to filtering by the coefficient of variation (CV). CV was calculated for each of the 23 715 genes. A cutoff of CV = 0.73 passed 16,482 genes. The 2 lists of 16,482 genes had 16 265 in common. Therefore the specific filtering method used did not affect our conclusions.
MSSM Microarray Pre-processing
MAS-5 algorithm was used for normalization. Lowess correction was then applied, expression levels below 20 were set to 20 and log2-transformation was applied. Probe-sets without assigned gene symbols were removed. 12 033 probe-sets were left for the rest of the analysis after filtering (out of 22 283), representing 8542 gene symbols. Probe sets of the same gene were combined. For full details, see Supplementary Methods.
Differential Gene Expression Analysis
A linear model was fitted to each gene by a stepwise procedure,24 using the MATLAB function stepwiselm with default parameters. As pH did not differ significantly between schizophrenia and controls (Table 1), at first age, gender and PMI were included as covariates. Later we added pH as well (see Discussion). The model was then refitted using only the selected variables, including diagnosis. Finally, for each gene, the diagnosis coefficient was statistically tested for being nonzero, implying an effect for schizophrenia, beyond any other effect of the covariates. This produced a t-statistic and a corresponding P-value, which were adjusted for multiple hypothesis testing using the false discovery rate (FDR) procedure.25 As the differentially expressed genes are subjected to further pathway enrichment analysis, a non-stringent FDR threshold of 15% was used. A standard 2-sample t-test was also performed; the results were very similar (Supplementary Figure 1S).
Pathway Enrichment Analysis Using GeneAnalytics
GeneAnalytics tool26 was used for pathway enrichment analysis. GeneAnalytics leverages PathCards (http://pathcards.genecards.org/), which clusters thousands of pathways from multiple sources into Superpathways, in order to improve inferences and reduce redundancy. Superpathways are scored by log2-transformation of the binomial P-value, which is equivalent to a corrected P-value with significance level <0.05.
Differential Expression STRING Database Network View
Network Creation.
Given a list of genes, a network is built. A network consists of genes (nodes) and genes’ co-expression relations (edges). The co-expression relations data was downloaded from the STRING database, version 10.5.27 Each such connection has a score between 0 and 1 that indicates the estimated likelihood that a given interaction is “biologically meaningful, specific and reproducible”.27 Only edges with STRING score greater than 0.1 are included in our network.
Differential Expression Network View.
Given a network and gene expression data, of both patients and controls, the following steps are taken, for each gene:
The mean expression and standard deviation values, Mc and Sc, are calculated using the control samples only.
The mean expression, Mp, is calculated using the patients’ samples.
Mp-Mc is calculated, the difference in the expression means between the 2 groups of samples.
The deviation from the control group is calculated by: (Mp-Mc)/Sc
Finally, the network is displayed as an undirected graph, with each node colored according to the deviation described above, (Mp-Mc)/Sc. The edges represent co-expression relations. Only genes that have co-expression relations with other genes in the network are displayed.
Results
UPS-Related Pathways are Enriched in the Group of Genes Which are Down-Regulated in SMRI STG Samples of Individuals With Schizophrenia
Differential expression analysis was performed, yielding 881 up-regulated and 986 down-regulated genes. In order to examine possible connection to antipsychotic medications, alcohol or substance use, we performed correlation analyses between the expression pattern of the differentially expressed genes and Fluphenazine equivalent dosage, substance use, and alcohol use measures. Correlation analyses for Fluphenazine equivalent dosage and alcohol use did not reveal any significant association with differential expression. Correlation analysis for substance use detected 2 down-regulated genes (out of 986) with statistically significant correlated expression (Supplementary Methods and Supplementary Figures 2S–4S).
Pathway enrichment analysis was applied separately to the up and down-regulated genes. Results are presented in Supplementary Tables 3S–4S for the up-regulated and down-regulated genes, respectively. Out of 49 pathways enriched in the down-regulated genes, 5 are directly UPS related (marked in Supplementary Table 4S). While several pathways have higher enrichment scores, we focus on the UPS and proteasome-related pathways, since 5 such pathways were enriched, and 11 closely related additional pathways were also enriched (Supplementary Table 4S). One of these pathways is Class I MHC Mediated Antigen Processing and Presentation, where proteins degraded by the proteasome are a major source of peptides presented by MHC class I molecules.28
The UPS Signal is Highly Replicated in the MSSM STG Samples
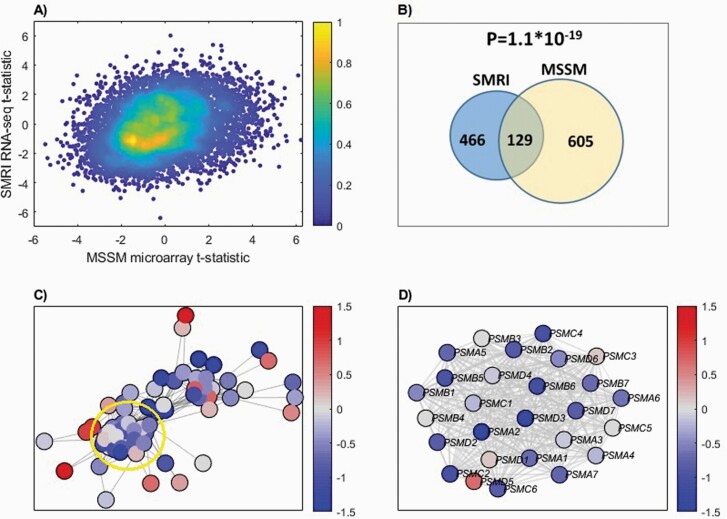
Our findings are replicated in the STG of the independent MSSM cohort of elderly subjects. We first examined whether the 2 datasets are comparable. Though microarrays differ from RNA-seq in their captured features, there was a significant positive correlation of the t-statistics (schizophrenia vs controls) between SMRI and MSSM across 7498 genes common to both platforms (Figure 1A).
Fig. 1.
(A) Binned density scatter plot comparing the t-statistics for case versus control differential expression between the independent MSSM replication cohort assayed on microarrays and the SMRI RNA-seq data; correlation between the statistics is .28 (P = 4.7 × 10−133). The colorbar represents the density in each cell, calculated by voronoi procedure48 and normalized to values between 0 (minimal density) and 1 (maximal density). (B) Hypergeometric P-value calculation for the intersection between SMRI and MSSM down-regulated genes. The 986 SMRI and 794 MSSM down-regulated genes were intersected with the 7498 genes that are present in both cohorts, yielding 595 SMRI and 734 MSSM down-regulated genes, with 129 shared genes. (C) SMRI Differential expression network view for Ubiquitin-Proteasome Dependent Proteolysis superPathway. A node’s color corresponds to the deviation of expression from the control samples group, in terms of standard deviation units. The edges represent STRING database co-expression relations. Only genes that have co-expression relations with other genes in the network are displayed. A subgroup of highly interconnected genes, coding for proteasome subunits, is circled (D) Zoom in on proteasome subunits. The same plot as in (C), for a subgroup of highly interconnected genes coding for proteasome subunits (circled in C).
We next repeated the differential expression and pathway enrichment analyses in the MSSM cohort. 919 genes and 794 genes were found to be up-regulated and down-regulated in schizophrenia, respectively. MSSM and SMRI differentially expressed genes significantly overlap (hypergeometric P-values: 9.8 × 10–7, 1.1 × 10–19 for the up-regulated and down-regulated genes, respectively; see Figure 1B).
Pathway enrichment analysis yielded 27 and 48 enriched pathways in up and down-regulated genes, respectively (results are listed in Supplementary Tables 5S and 6S). Intersecting SMRI 49 enriched pathways with 48 from MSSM in the down-regulated genes yields 30 shared pathways; see Supplementary Table 4S (hypergeometric P-value: 2.5 × 10–36). Four out of the 5 SMRI enriched UPS pathways were enriched also in the MSSM. A similar analysis of the up-regulated genes yields a hypergeometric P-value of 1.03 × 10–6. Interestingly, one of the pathways that were enriched in the MSSM up-regulated genes is Metabolism of Proteins, which contains UPS-related genes. Thus, while a subgroup of the pathway genes is up-regulated, another is down-regulated. For further details, see Supplementary Information.
A Network View of the UPS Identifies Down-Regulation of a Tightly Connected Cluster of Proteasome Subunits
To further explore the UPS differential expression, we used differential expression network view for SMRI (see Methods). It was applied to the Ubiquitin-Proteasome Dependent Proteolysis GeneAnalytics “superpathway”,26 which is representative of the UPS and was significantly enriched in both SMRI and MSSM (Supplementary Table 4S). The network view includes all 69 pathway genes for which network data was available from STRING,29 and not only those 27 genes that were found to be down-regulated. As can be seen in Figure 1C, there is a cluster of tightly inter-connected genes which are mostly down-regulated in schizophrenia (bluish colors of the nodes). Interestingly, this cluster is composed of proteasome subunits, as shown in Figure 1D. The same analysis of the MSSM yields a similar view (Supplementary Figure 5S).
Meta-Analysis of SMRI and MSSM Datasets Identifies Down-Regulation of Multiple Proteasome Subunits in STG Samples of Subjects With Schizophrenia
We performed a meta-analysis of the expression of each of the 39 proteasome subunit genes, whose expression has been measured by both SMRI and MSSM (see Supplementary Methods). The list of proteasome subunits genes, meta-analysis results and comparisons to previous gene expression and protein-level studies are summarized in Table 2. Overall 12 out of 39 subunit genes were found to be down-regulated.
Down-Regulation Signal of Proteasome Subunits in Schizophrenia is Replicated in 6 Independent Datasets of 5 Different Brain Regions
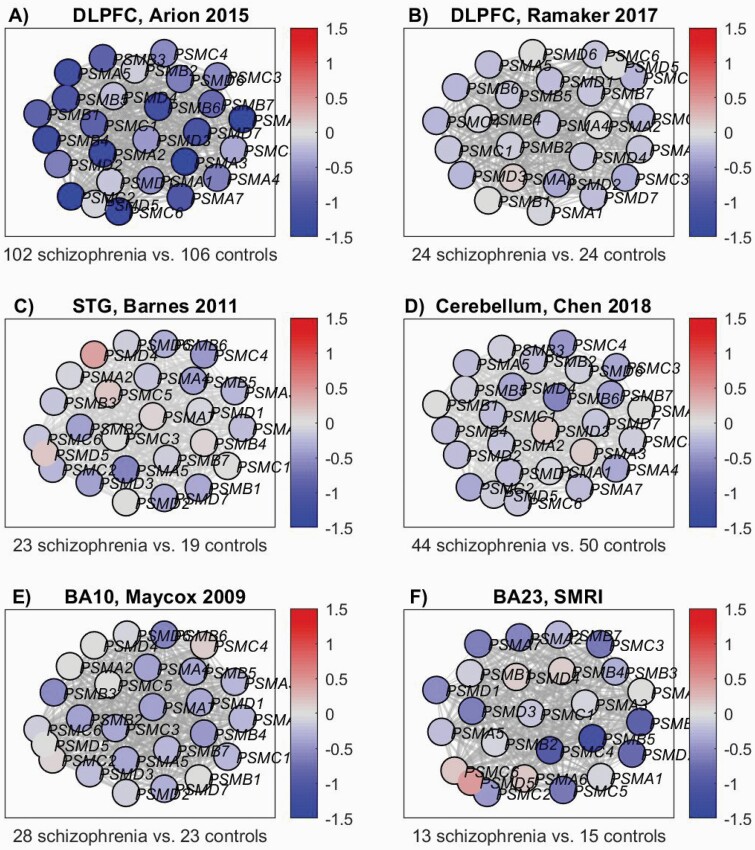
To examine whether down-regulation of proteasome subunits is specific to the STG we repeated the differential expression network analysis of the 39 proteasome subunit genes using 6 additional datasets (fully described in the Supplementary Information): dorso-lateral prefrontal cortex (DLFPC) samples from Arion 20156 and from Ramaker 2017,30 STG samples from Barnes 2011,31 Cerebellum samples from Chen 2018,32 Brodmann area 23 (BA23) samples from the SMRI cohort, and Brodmann area 10 (BA10) samples from Mycox 2009.33 The results are presented in Figure 2. The DLPFC samples of Arion 2015 (Figure 2A) exhibit pronounced down-regulation, while in the DLPFC samples of Ramaker 2017 (Figure 2B) the signal is weaker, though present in most of the genes; the binomial P-value for the number of genes with (even slightly) reduced expression versus the control group is P = 6.4 × 10−6). Interestingly, while Ramaker 201730 used brain samples composed of mixture of cells, Arion 20156 used laser microdissection to capture pyramidal neurons. Thus, the difference in down-regulation might be due to dilution of the signal, caused by the mixture of cell types used in Ramaker 2017. The Cerebellum samples from Chen 201832 (Figure 2D), BA10 samples from Mycox 2009 (Figure 2E) and BA23 SMRI (Figure 2F) show clear tendency for down-regulation (binomial P-values 6.9 × 10−7, 1.2 × 10−5, and .04, respectively), with modest magnitude (mostly less than 1 SD). STG samples of Barnes 201131 (Figure 2C) show a similar pattern. Down-regulation might be specific to neurons or subtypes of neurons; as the brain samples in these datasets are of mixture of cells, the signal might be diluted. Overall, this analysis replicates the signal of down-regulation of multiple proteasome subunits, in both the STG and additional 4 brain regions.
Fig. 2.
Proteasome subunits differential expression network view: The nodes’ colors correspond to the deviation from the group of the control samples, in terms of SD units. The edges represent STRING database co-expression relations. Only genes that have co-expression relations with other genes in the network are displayed. (A) DLPFC, Arion 2015 dataset.6 (B) DLPFC, Ramaker 2017 dataset.30 (C) STG, Barnes 2011 dataset.31 (D) Cerebellum, Chen 2018.32 (E) BA10, Mycox 2009 dataset.33 (F) BA23, SMRI dataset.
Down-Regulation of Proteasome Subunits in Schizophrenia is Concentrated in a Subgroup of the Patients
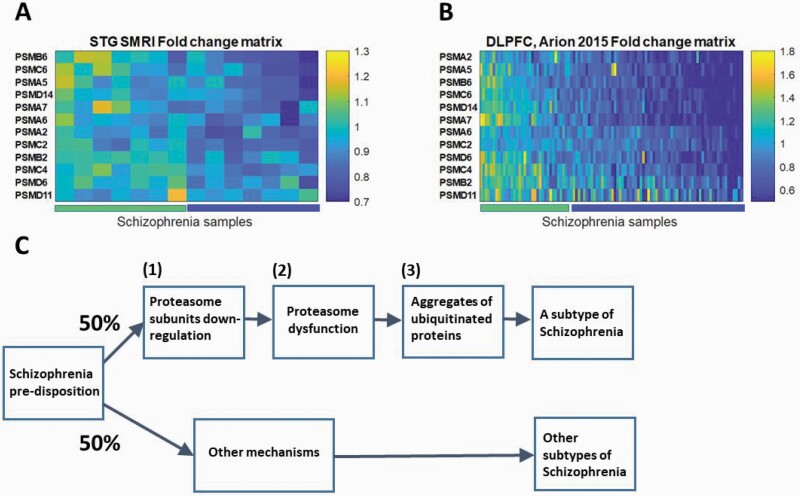
To explore whether the signal is concentrated in a subgroup of the patients, we applied fold change analysis of the 12 down-regulated proteasome subunits (listed in Table 2) to each of the SMRI schizophrenia samples. The results are plotted in Figure 3A. Half of the patients (7/14, “Group 2”; marked blue along the x-axis) show down-regulation tendency (bluish colors) of most of the 12 proteasome subunits genes, while the others (“Group 1”; marked green) show fold change values closer to 1, for most genes. The same analysis of the Arion 2015 dataset, of microdissected pyramidal neurons, yields even more pronounced distinction (Figure 3B). A similar picture emerges for the other 6 datasets (Supplementary Figure 7S). Support for this observation comes from a recent study,34 where transcriptomics analysis of 189 DLPFC samples of schizophrenia patients vs 206 healthy controls identified 2 molecular subtypes of schizophrenia. In “Type 1” (about half of the patients) 4 differentially expressed genes (schizophrenia vs controls) were detected, and in “Type 2” more than 3000. When examining the list of differentially expressed genes (Supplementary Table 3B), 28 proteasome subunits were differentially expressed, all down-regulated, in “Type 2,” while no proteasome subunit genes were differentially expressed in “Type 1.”
Fig. 3.
(A) SMRI STG schizophrenia samples fold change matrix of proteasome subunits genes. Each row represents one of the 12 proteasome subunits genes that were found to be down-regulated in schizophrenia in the meta-analysis of the SMRI and MSSM datasets. Each column represents one of the SMRI schizophrenia samples. The color code represents the fold change, ie, the expression value of the proteasome subunit gene in the specific sample, divided by its mean expression in the 15 control samples. Samples and genes locations were sorted by the SPIN tool.36 The left half of the samples, “Group 1,” are marked by the left bar along the x-axis and the right half, “Group 2,” by the right bar. (B) DLPFC Arion 2015 schizophrenia samples fold change matrix of proteasome subunits genes. The same plot as in (A) for the DLPFC Arion 2015 dataset. (C) A schematic preliminary model for a biological mechanism based division of schizophrenia patients into subtypes.
We then applied a similar analysis as in, ref.34 and compared each of “Group 1” and “Group 2” samples to the controls, separately. Differential expression analysis was applied to the 47 SMRI measured proteasome subunit genes, in each of the 2 groups. While in “Group 1” no differentially expressed genes were found, in “Group 2” 23 proteasome subunits were found to be differentially expressed (FDR < 15%; Supplementary Table 8S). We conclude that proteasome subunits down-regulation characterizes about half of the patients with schizophrenia.
Discussion
The main finding of our study is a global down-regulation of multiple proteasome subunits in post mortem brain samples of individuals with schizophrenia. Although several scenarios may be possible, a reasonable model (Figure 3C) is that given a predisposition to schizophrenia, certain (unknown) factors lead to (1) down-regulation of multiple proteasome subunits in about half the patients. This in turn leads to (2) proteasome dysfunction which causes (3) accumulation of ubiquitinated proteins. We discuss below the evidence relevant to each of the hypotheses (1)–(3) of our model.
Hypothesis (1) is supported by our main finding, which was replicated in 8 datasets of 5 different brain regions. We observed that the signal characterizes about half the patients. This observation is supported by,34 where 2 molecular subtypes of schizophrenia were detected, one (“Type 2”) with 28 down-regulated proteasome subunits genes, and another (“Type 1”), without dysregulation of these genes. In “Type 2” more than 3000 genes (about 25% of those measured) were dysregulated (up- and down-regulated ratio close to 1:1). Thus, the fact that 28 proteasome subunits genes are dysregulated is somewhat less surprising. However, as all the 28 were down-regulated, it makes the concordance with our results significant.
The fact that several studies identified decreased protein levels of proteasome subunit genes (Table 2) supports hypothesis (2), of proteasome dysfunction in schizophrenia. However, it was not established whether the lower protein levels are caused by lower expression of the coding genes. Moreover, previous studies of proteasome activity in schizophrenia yielded inconsistent results. While in ref.14 intra-cellular compartment-specific dysfunction in STG samples was found, no change has been detected in neither blood or brain in ref.12. A possible explanation of this inconsistency is that the signal is specific not only to a subgroup of the patients, but also to neurons or subtypes of neurons, and thus diluted. This is supported by our analysis of Arion 2015 dataset, of laser microdissected neurons,6 where higher magnitude of down-regulation was detected (Figure 2). This could also explain why the signal has not been detected by many previous relevant gene expression studies. Actually, if we look at some of the datasets (eg, Figure 2B–E), each proteasome subunit is not pronouncedly down-regulated. Only the analysis of the proteasome subunits as a group, measured in multiple datasets, enabled the detection of the global down-regulation signal.
Hypothesis (3), of accumulation of ubiquitinated proteins in schizophrenia, comes from 2 recent studies. In, ref.11 accumulation of ubiquitinated proteins has been identified for about half the patients in the STG, frontal cortex and prefrontal cortex samples. In, ref.12 ubiquitinated protein levels were found to be elevated in the orbitofrontal cortex of schizophrenia patients. While the fact that dysfunction of proteasome can cause accumulation of ubiquitinated proteins13 suggests a causative connection between hypotheses (2) and (3), this link was not examined in schizophrenia.
Interestingly, lower pH was detected in brains with accumulation of ubiquitinated proteins in ref.11 and was also associated with elevated ubiquitinated protein levels in ref.12. As pH did not differ significantly between schizophrenia and controls in neither SMRI nor MSSM (Table 1), it was not included as a covariate in the differential expression analysis. However, it may have more delicate associations with proteasome subunit genes’ expression, possibly in a subgroup of the patients. To examine this we performed a correlation analysis between pH levels and mean fold change of the 12 down-regulated proteasome subunits (listed in Table 2), in 7 datasets (for which pH information was available) and found a clear tendency for positive correlation (Supplementary Figure 8S). This is concordant with the association between lower pH and accumulation of ubiquitinated proteins shown in ref.11,12 and gives indirect support to our hypothesis that this accumulation is caused by down-regulation of the proteasome subunits.
In order to further explore the association between pH and proteasome subunits expression, we repeated the step-wise linear regression for both SMRI and MSSM for the 12 down-regulated proteasome subunits, including pH as a covariate (with age, PMI, and gender) for both cohorts and RIN was included for the SMRI (Supplementary Tables 9S–10S). This linear regression analysis gave moderate results in terms of the magnitude and statistical significance of the genes’ down-regulation, when compared to standard 2-sided t-test (Supplementary Figure 9S). However, the clear tendency for down-regulation remained, with statistical significance. We thus conclude that pH, or the other included potential conounding factors, cannot solely explain our observed decreased expression of the proteasome subunits.
In order to further explore the notion of subtypes of schizophrenia, we compared the mean fold change (FC) of the 12 down-regulated proteasome subunits to the polygenic risk score (PRS) of the STG SMRI samples (Supplementary Information and Supplementary Figure 10S). No statistically significant correlation was found (P-value .56); but when a single outlier sample (with largest PRS and smallest FC) was omitted, we did get a significant Pearson correlation of 0.67 (P-value .03), supporting our hypothesis of association between proteasome FC and clinical characteristics. However, since correlation was calculated from only 11 samples, this needs further investigation.
As described in, ref.11 the accumulated ubiquitinated proteins were enriched with nervous system development related pathways, suggesting its possible relation to disease pathogenesis through disruption of relevant pathways. In addition, clinical symptoms were correlated with 2 ubiquitin conjugation genes’ expression in patients’ peripheral blood.37 These findings might suggest that our hypothesized model defines a biological and clinical subtype of schizophrenia. In this context we note that Bortezomib, a proteasome inhibitor used in the treatment of cancer, is not known to cause psychosis when given to glioblastoma patients (where the brain-blood-barrier is disrupted).38–40 This seemingly suggests that proteasome dysfunction is not the cause of the symptoms seen in schizophrenia. In addition, pathways that apparently are not connected to the proteasome/UPS were found to be dysregulated, both by us and by,34 suggesting there are other mechanisms that underlie the pathogenesis of schizophrenia. However, interestingly, 4 of 6 pathways repeatedly found as dysregulated in schizophrenia (reviewed in ref.41), involve the UPS: presynaptic function,42 signaling,43 oxidative stress44 and cellular immune mechanisms.45 In addition, it was recently shown that antipsychotics modulate UPS-related protein levels in oligodendrocytes.46 However, it is still not clear whether the UPS has a causal role in schizophrenia and further study is needed to decipher this connection.
Our study is limited by several features. Every postmortem study represents only a snapshot at the end of life. This is especially relevant in schizophrenia, as its pathogenesis is probably rooted in early development.47 The fact that we compare independent cohorts of both relatively young and elderly subjects strengthens the validity of the results, but does not fully overcome this limitation. There is also the question of pharmacotherapy, as exposure to antipsychotics might affect gene expression. We found no significant correlation between Fluphenazine equivalent dose and expression levels. In addition, the fact that the subjects of the cohorts significantly differ in age suggests that duration of exposure to antipsychotics is unlikely to influence proteasome subunits expression substantively. The replication of the detected signal in 8 cohorts from 5 brain regions significantly increases the validity and generalizability of this signal. As gene expression does not always correlate with the levels of the coded proteins, the fact that we measure gene expression alone is a serious limitation, which causes difficulties in making definitive conclusions regarding the biological consequences of the results. While several studies detected decreased protein levels of proteasome subunits,7,14 the results were not fully consistent and the recent proteasome activity studies in schizophrenia were not consistent either, as described above. Thus, further study is needed in order to decipher the consequences of the global down-regulation of proteasome subunits we detect in schizophrenia, in terms of protein levels and proteasome activity.
Overall, we detect global down-regulation of proteasome subunits in schizophrenia, which characterizes about half of the patients. Based on ours and others’ recent findings we present a hypothesized model for a mechanism that defines a biological, and maybe also clinical, subtype of schizophrenia. This has the potential to lead to a better understanding of the biological and clinical subtypes of schizophrenia and to finding novel diagnostic and therapeutic tools.
Supplementary Material
Acknowledgments
We thank Professor Shai Izraeli for his advice and for providing the conditions for RNA isolation and quality control measurements. We thank Professor Harker Rhodes for his advice and insight regarding the interpretation of the results, and for sharing the results of the analysis he performed. We thank Professor Yifat Merbl for her advice and insight regarding the interpretation of the results. We also thank Maree Webster from the SMRI for her kind advice regarding the process of RNA isolation and Tian Ge for his helpful advice regarding the calculation of the PRS. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
Previous presentation: the paper has been uploaded to a preprint server, BioRxiv, doi: https://doi.org/10.1101/853226
Funding
This work was supported by a 2014 Brain and Behavior Research Foundation Young Investigator Grant, the Leir Charitable Foundation, the Veterans Administration Mental Illness Research, Education, and Clinical Center, and the National Institutes of Health Neurobiobank (HHSN271201300031).
References
- 1. Tsuang MT, Stone WS, Faraone SV. Genes, environment and schizophrenia. Br J Psychiatry Suppl. 2001;40:s18–s24. [DOI] [PubMed] [Google Scholar]
- 2. Mattila T, Koeter M, Wohlfarth T, et al. Impact of DSM-5 changes on the diagnosis and acute treatment of schizophrenia. Br J Psychiatry Suppl. 2015;41:637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22(7):2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altar CA, Jurata LW, Charles V, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58(2):85–96. [DOI] [PubMed] [Google Scholar]
- 5. Bousman CA, Chana G, Glatt SJ, et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arion D, Corradi JP, Tang S, et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry. 2015;20(11):1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scott MR, Rubio MD, Haroutunian V, Meador-Woodruff JH. protein expression of proteasome subunits in elderly patients with schizophrenia. Neuropsychopharmacology. 2016;41(3):896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rubio MD, Wood K, Haroutunian V, Meador-Woodruff JH. Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology. 2013;38(10):1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pescosolido MF, Gamsiz ED, Nagpal S, Morrow EM. Distribution of disease-associated copy number variants across distinct disorders of cognitive development. J Am Acad Child Adolesc Psychiatry. 2013;52(4):414–430.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang X, Lima L de A, Liu Y, et al. Common and rare genetic risk factors converge in protein interaction networks underlying schizophrenia. Front Genet. 2018;9:1–10. 10.3389/fgene.2018.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nucifora LG, MacDonald ML, Lee BJ, et al. Increased protein insolubility in brains from a subset of patients with schizophrenia. Am J Psychiatry. 2019;176(9):730–743. [DOI] [PubMed] [Google Scholar]
- 12. Bousman CA, Luza S, Mancuso SG, et al. Elevated ubiquitinated proteins in brain and blood of individuals with schizophrenia. Sci Rep. 2019;9(1):2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keller JN, Huang FF, Markesbery WR. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience. 2000;98(1):149–156. [DOI] [PubMed] [Google Scholar]
- 14. Scott MR, Meador-Woodruff JH. Intracellular compartment-specific proteasome dysfunction in postmortem cortex in schizophrenia subjects. Mol Psychiatry. 2020;25(4):776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vawter MP, Barrett T, Cheadle C, et al. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55(5):641–650. [DOI] [PubMed] [Google Scholar]
- 16. Chu TT, Liu Y, Kemether E. Thalamic transcriptome screening in three psychiatric states. J Hum Genet. 2009;54(11):665–675. [DOI] [PubMed] [Google Scholar]
- 17. Fromer M, Roussos P, Sieberts SK, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44(2):151–155. [DOI] [PubMed] [Google Scholar]
- 19. Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44(2):151–155. [DOI] [PubMed] [Google Scholar]
- 20. Hakak Y, Walker JR, Li C, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia, 2001/04/11. Proc Natl Acad Sci U S A 2001;98:4746–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katsel PL, Davis KL, Haroutunian V. Large-scale microarray studies of gene expression in multiple regions of the brain in schizophrenia and Alzheimer’s disease, 2005/03/31. Int Rev Neurobiol. 2005;63:41–82. [DOI] [PubMed] [Google Scholar]
- 22. Katsel P, Davis KL, Gorman JM, Haroutunian V. Variations in differential gene expression patterns across multiple brain regions in schizophrenia, 2005/06/01. Schizophr Res. 2005;77:241–252. [DOI] [PubMed] [Google Scholar]
- 23. Ballman KV, Grill DE, Oberg AL, Therneau TM. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics. 2004;20(16):2778–2786. [DOI] [PubMed] [Google Scholar]
- 24. Pope PT, Webster JT. The Use of an F -statistic in stepwise regression procedures. Technometrics. 1972;14:327–340. [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300. [Google Scholar]
- 26. Ben-Ari Fuchs S, Lieder I, Stelzer G, et al. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20(3):139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stoltze L, Nussbaum AK, Sijts A, Emmerich NP, Kloetzel PM, Schild H. The function of the proteasome system in MHC class I antigen processing. Immunol Today. 2000;21(7):317–319. [DOI] [PubMed] [Google Scholar]
- 29. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramaker RC, Bowling KM, Lasseigne BN, et al. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017;9(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barnes MR, Huxley-Jones J, Maycox PR, et al. Transcription and pathway analysis of the superior temporal cortex and anterior prefrontal cortex in schizophrenia. J Neurosci Res. 2011;89(8):1218–1227. [DOI] [PubMed] [Google Scholar]
- 32. Chen C, Meng Q, Xia Y, et al. The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci Transl Med. 2018;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maycox PR, Kelly F, Taylor A, et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry. 2009;14(12):1083–1094. [DOI] [PubMed] [Google Scholar]
- 34. Bowen EFW, Burgess JL, Granger R, Kleinman JE, Rhodes CH. DLPFC transcriptome defines two molecular subtypes of schizophrenia. Transl Psychiatry. 2019;9(1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vawter MP, Barrett T, Cheadle C, et al. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55(5):641–650. [DOI] [PubMed] [Google Scholar]
- 36. Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O, Notterman DA, Domany E. Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics. 2005;21(10):2301–2308. [DOI] [PubMed] [Google Scholar]
- 37. Bousman CA, Chana G, Glatt SJ, et al. Positive symptoms of psychosis correlate with expression of ubiquitin proteasome genes in peripheral blood. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(7):1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Odia Y, Kreisl TN, Aregawi D, Innis EK, Fine HA. A phase II trial of tamoxifen and bortezomib in patients with recurrent malignant gliomas. J Neurooncol. 2015;125(1):191–195. [DOI] [PubMed] [Google Scholar]
- 39. Raizer JJ, Chandler JP, Ferrarese R, et al. A phase II trial evaluating the effects and intra-tumoral penetration of bortezomib in patients with recurrent malignant gliomas. J Neurooncol. 2016;129(1):139–146. [DOI] [PubMed] [Google Scholar]
- 40. Bota DA, Eroglu Z, Reardon DA, et al. Phase II clinical trial of bortezomib and bevacizumab combination in recurrent glioblastoma. J Clin Oncol. 2011;29:2056–2056. [Google Scholar]
- 41. Kumarasinghe N, Tooney PA, Schall U. Finding the needle in the haystack: a review of microarray gene expression research into schizophrenia. Aust N Z J Psychiatry. 2012;46(7):598–610. [DOI] [PubMed] [Google Scholar]
- 42. Hegde AN. Proteolysis, synaptic plasticity and memory. Neurobiol Learn Mem. 2017;138:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6(3):231–242. [DOI] [PubMed] [Google Scholar]
- 44. Shang F, Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med. 2011;51(1):5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kammerl IE, Meiners S. Proteasome function shapes innate and adaptive immune responses. Am J Physiol Lung Cell Mol Physiol. 2016;311(2):L328–L336. [DOI] [PubMed] [Google Scholar]
- 46. Seabra G, de Almeida V, Reis-de-Oliveira G, Crunfli F, Antunes ASLM, Martins-de-Souza D. Ubiquitin–proteasome system, lipid metabolism and DNA damage repair are triggered by antipsychotic medication in human oligodendrocytes: implications in schizophrenia. Sci Rep. 2020;10:12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gogtay N, Vyas NS, Testa R, Wood SJ, Pantelis C. Age of onset of schizophrenia: perspectives from structural neuroimaging studies. Schizophr Bull. 2011;37(3):504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duyckaerts C, Godefroy G. Voronoi tessellation to study the numerical density and the spatial distribution of neurones. J Chem Neuroanat. 2000;20(1):83–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.