Abstract
Schizophrenia is a serious neuropsychiatric disorder with abnormal age-related neurodevelopmental (or neurodegenerative) trajectories. Although an accelerated aging hypothesis of schizophrenia has been proposed, the quantitative study of the disruption of the physiological trajectory caused by schizophrenia is inconclusive. In this study, we employed 3 “epigenetic clock” methods to quantify the epigenetic age of a large sample size of whole blood (1069 samples from patients with schizophrenia vs 1264 samples from unaffected controls) and brain tissues (500 samples from patients with schizophrenia vs 711 samples from unaffected controls). We observed significant positive correlations between epigenetic age and chronological age in both blood and brain tissues from unaffected controls and patients with schizophrenia, as estimated by 3 methods. Furthermore, we observed that epigenetic age acceleration was significantly delayed in schizophrenia from the whole blood samples (aged 20–90 years) and brain frontal cortex tissues (aged 20–39 years). Intriguingly, the genes regulated by the epigenetic clock also contained schizophrenia-associated genes, displaying differential expression and methylation in patients with schizophrenia and involving in the regulation of cell activation and development. These findings were further supported by the dysregulated leukocyte composition in patients with schizophrenia. Our study presents quantitative evidence for a neurodevelopmental model of schizophrenia from the perspective of a skewed “epigenetic clock.” Moreover, landmark changes in an easily accessible biological sample, blood, reveal the value of these epigenetic clock genes as peripheral biomarkers for schizophrenia.
Keywords: schizophrenia, epigenetic clocks, epigenetic age acceleration, methylation, immune cell
Introduction
Schizophrenia, usually strikes in early adulthood, is a complex multifactorial chronic disorder with an unknown causative pathophysiology.1,2 Some studies have suggested that schizophrenia is a disorder of brain development, such as abnormalities in structural brain connectivity3; others have pointed out that it is connected with the phenotype of premature aging, such as cerebral cortex atrophy4 and telomere shortening.5 The neurodevelopmental model of schizophrenia, an emerging convergence cognition, posits that the genetic and epigenetic risk factors for schizophrenia converge on early brain development to perturb neurodevelopmental trajectories.2 However, most of the studies in this area are cross-sectional and qualitative and fail to quantitatively shed light on the progressive developmental changes of age-related development (aging) during the disease course. Therefore, it has been a challenge to verify the hypothesis that schizophrenia causes stunted growth of tissues and organs and decelerated or accelerated aging.
Recent studies based on DNA methylation data have revealed clock-like patterns of epigenetic changes in the human life cycle, which offers promise for addressing this challenge. These “epigenetic clocks” measure the cumulative effect of an epigenetic maintenance system, reflecting the dynamic landscape of interaction between genetic and environmental factors.6 They can accurately estimate the biological age of any tissue to measure physiological homeostasis and degradation throughout the entire life course, which enables the addressing of a host of questions in developmental biology and biological senescence research.7 Three extensively accepted “epigenetic clocks” are available.6,8,9 The “Hannum Clock” was developed using 71 cytosine-phosphate-guanine (CpG) sites from the whole blood samples, aged 19–101 years8; the “Multi-tissue Predictor” identified 353 CpG sites through training on different tissue types derived from thousands of individuals, including children and adults6; and the “DNAm PhenoAge” was recently developed based on 513 CpG sites from the blood samples of adults that incorporated 10 clinical indicators.9 The ability of these “epigenetic clocks” to predict health imbalances caused by psychiatric disorders has been verified in studies into autistic disorder,10 bipolar disorder,11 and depression symptomatology.12 Some scholars have pointed out that epigenetic age acceleration is related to the borderline clinically significant internalizing of problems by adolescent individuals, especially problems related to anxiety/depression and withdrawal, as well as affective and thought problems.13 Tracing the effects of schizophrenia on the epigenetic age of different organs and tissues across the life span can also be used to effectively measure the development process or the speed of aging. However, current studies have not found that schizophrenia can significantly accelerate or decelerate the epigenetic age, and there is a lack of analysis of different age groups.14–16 This may be due to insufficient sample sizes, and such studies mostly tend to use a single “epigenetic clock” without comparing the results of different predictors.
Here, we used a large collection of samples (2333 whole blood and 1211 brain tissues) from 13 independent DNA methylation datasets to analyze the epigenetic age of different tissues from individuals with schizophrenia or unaffected controls. It is worth mentioning that 3 different predictors6,8,9 were implemented to evaluate their estimation capabilities to obtain accurate results. This study provided some evidence that clarified the effects of schizophrenia on the developmental trajectories of different tissues in humans.
Methods
Three “epigenetic clock” described previously by Horvath,6 Hannum et al,8 and Levine et al9 were used to quantify the epigenetic age of a large sample size of whole blood (1069 samples from patients with schizophrenia vs 1264 samples from unaffected controls) and brain tissues (500 samples from patients with schizophrenia vs 711 samples from unaffected controls). Differential methylation analysis was examined using the limma17 package in R Studio. We obtained schizophrenia-associated differentially expressed genes from 2 published papers including B-cell transformed lymphoblastoid cell line (LCL)18 and postmortem brain tissues of PsychENCODE consortium.19 Six proportions of cell types (CD8+ T cells, CD4+ T cells, natural killer cells, B cells, monocytes, and granulocytes) for each of the blood samples were estimated as previously described.20 More detailed materials and methods are described in supplementary files.
Results
Epigenetic Age Acceleration Is Decreased in Patients With Schizophrenia
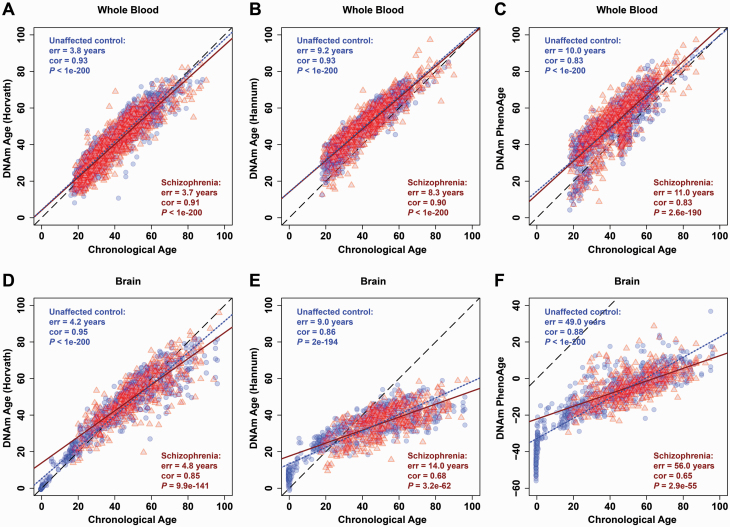
To calculate epigenetic age (also referred to as DNAm age), we employed 3 predictors to analyze DNA methylation data from whole blood or postmortem brain tissues of individuals with schizophrenia or unaffected controls (supplementary tables 1 and 2). We observed that the DNAm age of unaffected controls, as calculated by the 3 predictors, was significantly correlated with the corresponding chronological age based on whole blood DNA (blue dotted lines in figures 1A–C) and postmortem brain (blue dotted lines in figures 1D–F) methylation data. In particular, the “Multi-tissue Predictor” developed by Horvath6 showed a prominent advantage, with minimal errors (err) and higher correlation coefficients (cor) in both blood (err = 3.8 years, cor = 0.93, P < 1e-200; figure 1A) and postmortem brain datasets (err = 4.2 years, cor = 0.95, P < 1e-200; figure 1D) of unaffected controls. However, the regression lines calculated by Hannum et al8 and Levine et al9 showed a significant deviation from the diagonal in the postmortem brain (figures 1E and 1F). This may be because both predictors were trained on whole blood methylation data without a strong applicability in brain tissues. Similar results were observed in patients with schizophrenia (red solid lines in figure 1). Thus, “Multi-tissue Predictor” developed by Horvath6 was used in the subsequent analysis.
Fig. 1.
Correlation between epigenetic age and chronological age. The figure shows scatter plots of epigenetic age (y-axis) against chronological age (x-axis) in whole blood (A–C) and brain tissue (D–F) measured by the 3 “epigenetic clocks” described by Horvath6 (A, D), Hannum et al8 (B, E), and Levine et al9 (C, F). The black dashed line is the diagonal where epigenetic age equals chronological age. The regression of epigenetic age on chronological age was shown with blue dotted line for unaffected controls or with red solid line for schizophrenia patients. The prediction error (err), coefficient (cor), and P value of Pearson correlation analysis of epigenetic age with chronological age from unaffected controls (blue circles) or schizophrenia (red triangles) are shown within each panel.
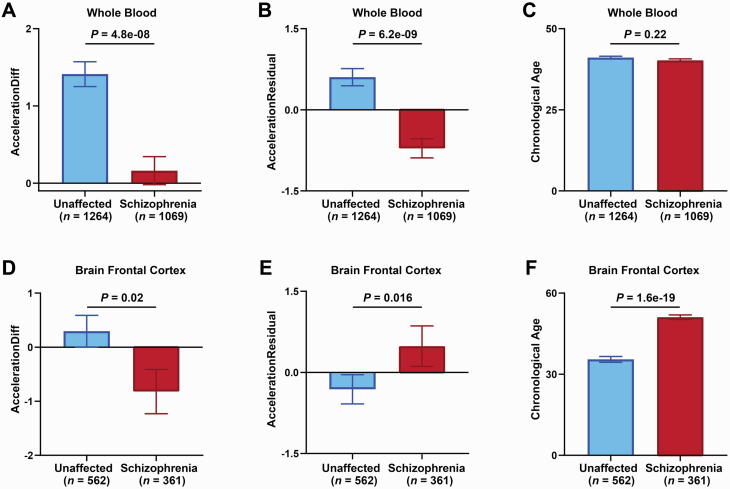
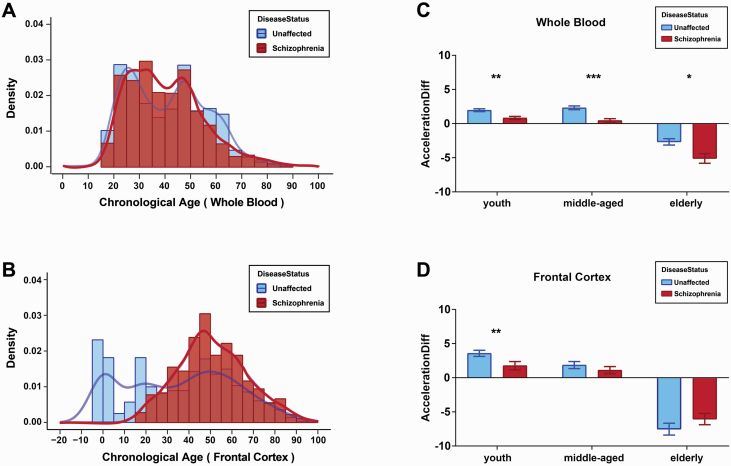
To examine the effect of schizophrenia on the development (aging) rate, we calculated the epigenetic age acceleration by calculating AccelerationDiff and AccelerationResidual6 and determined that epigenetic age is higher (AccelerationDiff or AccelerationResidual > 0) or lower (AccelerationDiff or AccelerationResidual < 0) than its chronological age in whole blood, as well as in 4 postmortem brain regions: frontal cortex, cerebellum, hippocampus, and striatum (supplementary tables 3 and 4). In whole blood, we observed that the AccelerationDiff value (decreased by 1.25 years; P = 4.8e-08, figure 2A) and AccelerationResidual (P = 6.2e-09, figure 2B) values displayed significant decreases in schizophrenia patients compared with unaffected controls; chronological age (P = .22, figure 2C) did not show any differences between schizophrenia patients and unaffected controls. In the brain frontal cortex, we observed that the AccelerationDiff value displayed a significant decrease (decreased by 1.12 years, P = .02; figure 2D), but AccelerationResidual values displayed a significant increase (P = .016, figure 2E) in schizophrenia patients compared with unaffected controls. This may be ascribed to highly significant differences in chronological age between the 2 groups (P = 1.6e-19, figure 2F). In the brain cerebellum, hippocampus, and striatum, we did not observe any significant differences in AccelerationDiff, AccelerationResidual, or chronological age between schizophrenia patients and unaffected controls (supplementary figure 1). Next, we investigated the variation pattern of epigenetic age acceleration in different age groups. We observed that the chronological age distribution of schizophrenia patients was similar to that of unaffected controls in the ages from 20 to 90 years of both whole blood and frontal cortex (figures 3A and 3B). We thus divided the subjects into 3 groups: youth group (20~39 years old), middle-aged group (40~59 years old), and elderly group (60~90 years old) and observed that the epigenetic age acceleration decreased significantly in schizophrenia patients compared with unaffected controls of all 3 age groups from the whole blood (figure 3C). In the brain frontal cortex, we only observed significantly decreased age acceleration in the youth group and a trend level in the middle-aged group of schizophrenia compared with unaffected controls (figure 3D). Altogether, the epigenetic age acceleration was delayed in schizophrenia, especially, in youth and middle-age stages.
Fig. 2.
Comparison of epigenetic age acceleration and chronological age between unaffected controls and schizophrenia patients. Epigenetic age accelerationDiff (A, D) and accelerationResidual (B, E) or chronological age (C, F) from unaffected control (blue) or schizophrenia (red) are shown with columns with standard error indicated by bars from whole blood (A–C) or brain frontal cortex (D–F). Significant differences in epigenetic age accelerationDiff and accelerationResidual or chronological age between unaffected control and schizophrenia by Kruskal-Wallis tests were shown with P < .05 above each panel. The epigenetic age acceleration was determined by calculating AccelerationDiff (ie, DNAm age minus chronological age) and AccelerationResidual (the residual resulting from linear regressing DNAm age on the chronological age), and a positive (negative) value of AccelerationDiff or AccelerationResidual indicates that the DNAm age of a given tissue is higher (or lower) than chronological age.
Fig. 3.
Change in epigenetic age acceleration with age by 3 stages. (A and B) The chronological age density distribution map of whole blood samples (A) and frontal cortex samples (B). (C and D) Comparison of epigenetic age acceleration in whole blood (C) and frontal cortex samples (D) between unaffected controls and schizophrenia patients in 3 physiological stages of youth (20~39 years old), middle-aged (40~59 years old), and elderly (60~90 years old). *P < .05; **P < .01; ***P < .001.
Schizophrenia-Associated Genes in the Epigenetic Clock Are Involved in the Regulation of Cell Activation and Growth
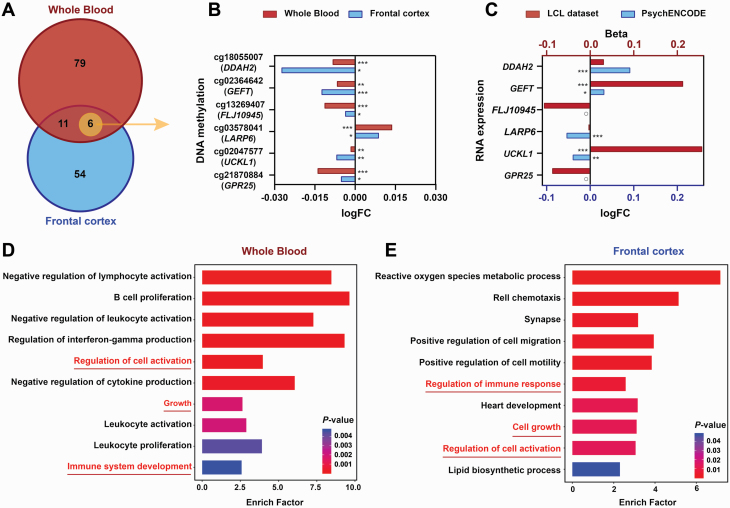
To explore the signs of decreased epigenetic age acceleration in schizophrenia, we performed differential methylation analysis and differentially expressed gene analysis of 353 CpG-annotated genes involved in the epigenetic clock between schizophrenia patients and unaffected controls. We first identified 96 schizophrenia-associated differential methylation probes (DMPs) (adjusted P-value < .05) from the whole blood dataset (supplementary table 5) and 71 DMPs (adjusted P-value < .05) from the brain frontal cortex dataset (supplementary table 6); further, we identified 17 DMPs coexisting in both blood and frontal cortex tissues (figure 4A), 6 of which were located in DDAH2, GEFT, FLJ10945, LARP6, UCKL1, and GPR25, showing consistent methylation alterations in blood with the frontal cortex (figure 4B). Intriguingly, 70%~80% of the DMPs were located within the vicinity of the promoter regions, suggesting that methylation was endowed with substantial capacity to regulate gene expression.
Fig. 4.
Differences in DNA methylation and mRNA expression between the 2 groups and enrichment analysis. (A) Venn diagram of differentially methylated sites (adjusted P-value < .05) selected from whole blood (red) and frontal cortex (blue). The 6 DMPs in the intersection showed a consistent change in methylation between the whole blood and brain frontal cortex. (B) Bar graph indicates the methylation alterations of 6 DMPs from whole blood (red) and frontal cortex (blue) between unaffected controls and schizophrenia patients. The x-axis represents methylation differences between 2 groups (log2-transformed fold change of case over control). *adjusted P-value < .05, **adjusted P-value < .01, and ***adjusted P-value < .001. (C) Bar graph indicates mRNA expression changes of the 6 DMP-annotated genes from the lymphoblastoid cell line (LCL) (red) and brain (blue) between unaffected controls and schizophrenia patients. The upper x-axis represents the mRNA expression differences between the 2 groups identified in the LCLs data (beta > 0 indicates upregulation in schizophrenia and beta < 0 indicates downregulation). The lower x-axis represents the mRNA expression differences between the 2 groups in the brain postmortem tissue of PsychENCODE dataset (fold change of case over control in log2 conversion). The circle represents that the gene was not detected in this dataset. (D, E) Gene ontology analysis of the 96 DMP-annotated genes in whole blood (D) and the 71 DMPs in the frontal cortex (E).
We then obtained schizophrenia-associated differentially expressed genes from 2 published papers, B-cell-transformed LCL18 and brain postmortem tissues of PsychENCODE consortium19 to identify whether genes containing disease-associated DMPs displayed differential gene expression in schizophrenia. Of the 96 blood DMPs, 39 genes in LCLs and 31 genes in brain tissues displayed significant expression alterations in schizophrenia, and 7 genes displayed consistent expression alterations in both LCLs and brain tissues from schizophrenia (supplementary table 5). Of the 71 frontal cortex DMPs, 32 genes in LCLs and 24 genes in brain tissues displayed significant expression alterations in schizophrenia, and 6 genes displayed consistent expression alterations in both LCLs and brain tissues from schizophrenia (supplementary table 6). Intriguingly, of 6 genes showing consistent schizophrenia-associated methylation alterations in both blood and brain tissues, DDAH2 messenger RNA (mRNA) levels were significantly increased in brain tissues from schizophrenia patients and displayed a similar trend in LCLs; GEFT (also named ARHGEF25) mRNA level was significantly increased in both LCLs and brain tissues of schizophrenia patients; LARP6 mRNA levels were significantly reduced in brain tissues from schizophrenia patients and displayed a similar trend in LCLs; UCKL1 mRNA level in LCLs was significantly increased but was reduced in brain tissues from schizophrenia patients; FLJ10945 (also named PRR34) and GPR25 mRNA levels were both reduced in LCLs from schizophrenia patients (figure 4C; supplementary tables 5 and 6). Similar or opposite alterations between DNA methylation and mRNA expression in schizophrenia further suggested the regulatory role of DMPs located in the enhancer or silencer regions. More importantly, DDAH2 is reported to map to a region (rs114115252 in chr 6: 30931418, P = 8e-20) that was previously found to be associated with schizophrenia based on a genome-wide association study (GWAS).21 In addition, DDAH2 (rs142536810, P = 5e-20),22GEFT (rs61937595, P = 6e-12),23 and FLJ10945/PRR34 (rs35245641, P = 2e-6)24 are also marked as “enhancers” by mapping GWAS SNPs to GeneHancer enhancers based on the GWAS catalog and GeneHancer database25 in the Genecards website (https://www.genecards.org). Altogether, these results further support that schizophrenia-associated genes are regulated by epigenetic clocks involved in the schizophrenia-associated decreased epigenetic age acceleration.
We further performed functional enrichment analysis on the annotated genes represented by differentially methylated CpG sites in the blood and frontal cortex. Gene ontology analysis revealed the significant enrichment of 96 DMP-annotated genes in whole blood in terms related to “regulation of cell activation,” “the proliferation and differentiation of immune cells,” “regulation of immune system process,” “growth and development,” “proliferation and differentiation,” and others (figure 4D, supplementary table 7). Intriguingly, we observed that “regulation of cell activation,” “cell migration and motility,” “tissue development,” and “biosynthetic and metabolic processes of macromolecules”-related terms were significantly enriched in Gene Ontology analysis of 71 DMPs identified in the frontal cortex (figure 4E; supplementary table 8).
Cell Composition Is Altered in Individuals With Schizophrenia
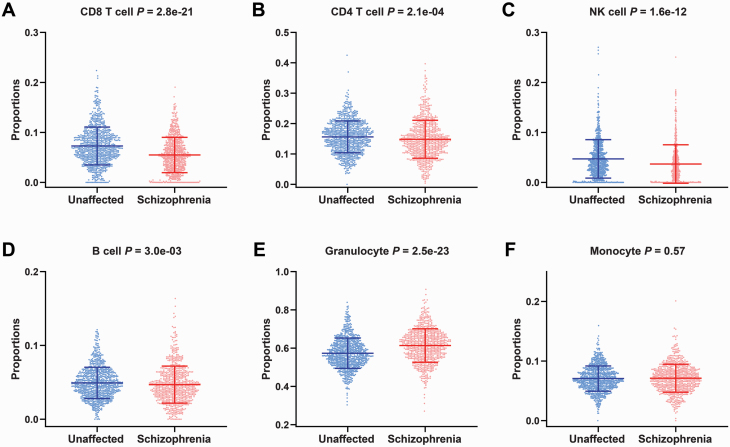
Functional annotation of schizophrenia-associated methylation alterations of epigenetic clock points to the regulation of cell activation and immune system process, and empirical studies show that epigenetic age acceleration has a relative correlation with blood cell composition26; therefore, we then examined whether blood cell composition was any different between patients who have schizophrenia and unaffected controls. We estimated the proportion of 6 immune cell types (supplementary table 3) using a previously described method.20 The results show that the patients who have schizophrenia had decreased proportions of CD8+ T cells (P = 2.8e-21, figure 5A), CD4+ T cells (P = 2.1e-04, figure 5B), natural killer cells (P = 1.6e-12, figure 5C), B cells (P = 3.0e-03, figure 5D), an increased proportion of granulocytes (P = 2.5e-23, figure 5E), and a similar trend line for the proportion of monocytes (P = .57, figure 5F) compared with unaffected controls.
Fig. 5.
Comparison of estimated cellular proportions between unaffected controls and schizophrenia patients. (A–-F) The y-axis is each of the estimated cellular proportions of CD8+ T cells (A), CD4+ T cells (B), natural killer cells (C), B cells (D), granulocytes (E), and monocytes (F). The P-value from Kruskal-Wallis tests is shown above each panel.
Discussion
To our knowledge, this study is the largest sample study to date of epigenetic age to quantify the effects of schizophrenia on the biological age of thousands of whole blood and brain tissues based on the “epigenetic clock” method developed by Horvath.6 Our findings first demonstrated that epigenetic age acceleration is delayed in patients who have schizophrenia. Although the accelerated aging hypothesis of schizophrenia has been proposed, the quantitative study of the displacement of the physiological aging trajectory caused by schizophrenia is inconclusive. Our observations were that DNAm age acceleration was reduced in patients who have schizophrenia, which was concluded from a large sample size of genome-wide methylation datasets from 2333 whole blood and 1211 brain tissues; this conclusion contradicts the accelerated aging hypothesis of schizophrenia. In fact, several previous studies also reported no accelerated brain or blood aging or moderate reduction of epigenetic age acceleration in relatively small sample sizes or uneven chronological age distribution between schizophrenia and control.14–16 This reduction in epigenetic age acceleration in patients who have schizophrenia that was observed in our study, together with observations that age-dependent DNA methylation alterations are disrupted in patients who have schizophrenia,27–29 supports the idea that schizophrenia is a neurodevelopmental disorder.
A skew in the epigenetic clock in schizophrenia was further supported by the evidence that the group of genes regulated by the epigenetic clock contains schizophrenia-associated genes. We demonstrated that some of the genes regulated by the epigenetic clock displayed schizophrenia-associated mRNA expression and DNA methylation alterations in blood and/or postmortem brain tissues or are reported to map to schizophrenia GWAS regions or enhancers. Intriguingly, 70%–80% of the schizophrenia-associated DMPs are annotated within the vicinity of the promoter regions. The biological functions of reduced epigenetic age acceleration underlying the development of schizophrenia might be mediated through the effects on gene expression. For example, the regions around the DDAH2 transcription start site and the first exon of GEFT displayed hypomethylation in both blood and brain tissues of schizophrenia patients, and their mRNA expression was upregulated in both LCLs and brain tissues of schizophrenia. Overexpression of DDAH2, which encodes dimethylarginine dimethylaminohydrolase 2 and functions in nitric oxide generation in islets, has been reported to activate secretagogin expression.30 Secretagogin was also found to be increased in serum from first-onset antipsychotic-naïve schizophrenia patients.31 Another schizophrenia-associated DMP, the TNFSF13B promoter, displayed hypomethylation in the whole blood dataset of schizophrenia, and its mRNA expression was upregulated in the LCLs and postmortem brain tissues of schizophrenia.32,33 TNFSF13B is reported to be involved in the stimulation of B- and T-cell function and the regulation of humoral immunity.32Moreover, cell activation, proliferation, migration, and motility as well as cytokine-, synapse-, and immune-related pathways were enriched for the schizophrenia-associated DMPs, further indicating that immune dysregulation and neurological dysplasia in schizophrenia may be the primary cause of the decreased epigenetic age acceleration.
Immune dysregulation in schizophrenia was further demonstrated by altered leukocyte composition. Leukocyte composition variations herald aberrant immune-inflammatory processes in most chronic medical conditions.34 The proportions of lymphocytes, including T cells, B cells, and NK cells, were reduced, while the proportions of lymphomyeloid cells, including granulocytes, were increased in the blood of patients who have schizophrenia relative to those of unaffected controls, which are consistent with previous studies.34,35 A reduced B cell proportion is reported to be associated with the progression of several neuroimmunologic disorders.33 This may be a new hint regarding the disruption of immune homeostasis in schizophrenia. Our recent study also demonstrated the presence of a mixed inflammatory response, including both pro- and anti-inflammatory responses, in schizophrenia depending on the duration or state of disease.36
There are some limitations in our research that need further exploration. First, although we had the largest sample size and covered the widest age range compared with relevant studies, it will still require larger samples to further verify these results, especially when making conclusions about different brain regions. Second, there is an insistent demand for detailed clinical characteristics of schizophrenia, such as medication and hospitalization. Indeed, antipsychotics may impact the DNA methylation status and levels of lymphocytes and various cytokines.37,38 Third, several potential confounders that may affect epigenetic age should be considered, including gender, diet, exercise, unhealthy habits (alcohol and tobacco abuse), education, and lifestyle factors.39,40
In summary, our study presents quantitative evidence for a neurodevelopmental model of schizophrenia from the perspective of a skewed “epigenetic clock,” resulting in a decreased epigenetic age acceleration and altered leukocyte composition, thus providing evidence for developmental delay and immune disorders in schizophrenia. Moreover, landmark changes in an easily accessible biological sample, blood, reveal the values of epigenetic age and may lead to translation of these epigenetic clock genes as peripheral biomarkers for schizophrenia.
Funding
We thank the National Natural Science Foundation of China [grant number 32000419 and 81671333] and the Guangdong Science and Technology Foundation [grant number 2019B030316032] for providing financial supports.
Supplementary Material
Acknowledgments
We gratefully acknowledge the many researchers who made their DNA methylation datasets publicly available and responded to my email requests. This study would not have been possible without the valuable data from the NCBI Gene Expression Omnibus database.
References
- 1. Millan MJ, Andrieux A, Bartzokis G, et al. Altering the course of schizophrenia: progress and perspectives. Nat Rev Drug Discov. 2016;15(7):485–515. [DOI] [PubMed] [Google Scholar]
- 2. Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017;18(12):727–740. [DOI] [PubMed] [Google Scholar]
- 3. Okazaki S, Otsuka I, Numata S, et al. Epigenetic clock analysis of blood samples from Japanese schizophrenia patients. npj Schizophr. 2019;5(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haren VMNE. Changes in cortical thickness during the course of illness in schizophrenia. Arch Gen Psychiatry. 2011;68(9):871–880. [DOI] [PubMed] [Google Scholar]
- 5. Wolkowitz OM, Jeste DV, Martin AS, et al. Leukocyte telomere length: effects of schizophrenia, age, and gender. J Psychiatr Res. 2017;85:42–48. [DOI] [PubMed] [Google Scholar]
- 6. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guevara EE, Lawler RR. Epigenetic clocks. Evol Anthropol. 2018;27(6):256–260. [DOI] [PubMed] [Google Scholar]
- 8. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu X, Chen W, Lin F, et al. DNA methylation profile is a quantitative measure of biological aging in children. Aging (Albany NY). 2019;11(22):10031–10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fries GR, Bauer IE, Scaini G, et al. Accelerated hippocampal biological aging in bipolar disorder. Bipolar Disord. 2020;22:498–507. [DOI] [PubMed] [Google Scholar]
- 12. Starnawska A, Tan Q, Soerensen M, et al. Epigenome-wide association study of depression symptomatology in elderly monozygotic twins. Transl Psychiatry. 2019;9(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suarez A, Lahti J, Czamara D, et al. The epigenetic clock and pubertal, neuroendocrine, psychiatric, and cognitive outcomes in adolescents. Clin Epigenetics. 2018;10(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voisey J, Lawford BR, Morris CP, et al. Epigenetic analysis confirms no accelerated brain aging in schizophrenia. npj Schizophr. 2017;3(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKinney BC, Lin H, Ding Y, Lewis DA, Sweet RA. DNA methylation evidence against the accelerated aging hypothesis of schizophrenia. npj Schizophr. 2017;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKinney BC, Lin H, Ding Y, Lewis DA, Sweet RA. DNA methylation age is not accelerated in brain or blood of subjects with schizophrenia. Schizophr Res. 2018;196:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health. New York, NY: Springer; 2005:397–420. [Google Scholar]
- 18. Duan J, Göring HHH, Sanders AR, et al. ; MGS . Transcriptomic signatures of schizophrenia revealed by dopamine perturbation in an ex vivo model. Transl Psychiatry. 2018;8(1):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gandal MJ, Zhang P, Hadjimichael E, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018;362(6420):eaat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Autism Spectrum Disorders Working Group of The Psychiatric Genomics C. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism 2017;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ikeda M, Takahashi A, Kamatani Y, et al. Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr Bull. 2019;45(4):824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pardiñas AF, Holmans P, Pocklington AJ, et al. ; GERAD1 Consortium; CRESTAR Consortium . Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goes FS, McGrath J, Avramopoulos D, et al. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet B Neuropsychiatr Genet. 2015;168(8):649–659. [DOI] [PubMed] [Google Scholar]
- 25. Fishilevich S, Nudel R, Rappaport N, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212(10):1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang T, Zong L, Zhou L, et al. Variation in global DNA hydroxymethylation with age associated with schizophrenia. Psychiatry Res. 2017;257:497–500. [DOI] [PubMed] [Google Scholar]
- 28. Li S, Yang Q, Hou Y, et al. Hypomethylation of LINE-1 elements in schizophrenia and bipolar disorder. J Psychiatr Res. 2018;107:68–72. [DOI] [PubMed] [Google Scholar]
- 29. Li S, Zong L, Hou Y, et al. Altered DNA methylation of the AluY subfamily in schizophrenia and bipolar disorder. Epigenomics 2019;11(6):581–586. [DOI] [PubMed] [Google Scholar]
- 30. Erzin G, Topçuoğlu C, Bayram Ş, et al. Secretagogin may not be a new neuroendocrine biomarker in schizophrenia while levels may reflect clinical severity. Psychiat Clin Psychoph. 2019;29(4):394–398. [Google Scholar]
- 31. Guest PC, Schwarz E, Krishnamurthy D, et al. Altered levels of circulating insulin and other neuroendocrine hormones associated with the onset of schizophrenia. Psychoneuroendocrinology 2011;36(7):1092–1096. [DOI] [PubMed] [Google Scholar]
- 32. Nocturne G, Mariette X. B cells in the pathogenesis of primary Sjögren syndrome. Nat Rev Rheumatol. 2018;14(3):133–145. [DOI] [PubMed] [Google Scholar]
- 33. Han J, Sun L, Fan X, et al. Role of regulatory b cells in neuroimmunologic disorders. J Neurosci Res. 2016;94(8):693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karpiński P, Samochowiec J, Frydecka D, Sąsiadek MM, Misiak B. Further evidence for depletion of peripheral blood natural killer cells in patients with schizophrenia: a computational deconvolution study. Schizophr Res. 2018;201:243–248. [DOI] [PubMed] [Google Scholar]
- 35. Kinoshita M, Numata S, Tajima A, et al. Aberrant DNA methylation of blood in schizophrenia by adjusting for estimated cellular proportions. Neuromolecular Med. 2014;16(4):697–703. [DOI] [PubMed] [Google Scholar]
- 36. Ni C, Jiang W, Wang Z, et al. LncRNA-AC006129.1 reactivates a SOCS3-mediated anti-inflammatory response through DNA methylation-mediated CIC downregulation in schizophrenia [published online ahead of print February 3, 2020]. Mol Psychiatry. doi: 10.1038/s41380-020-0662-3. [DOI] [PubMed] [Google Scholar]
- 37. de Witte L, Tomasik J, Schwarz E, et al. Cytokine alterations in first-episode schizophrenia patients before and after antipsychotic treatment. Schizophr Res. 2014;154(1-3):23–29. [DOI] [PubMed] [Google Scholar]
- 38. Melas PA, Rogdaki M, Ösby U, Schalling M, Lavebratt C, Ekström TJ. Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J. 2012;26(6):2712–2718. [DOI] [PubMed] [Google Scholar]
- 39. Quach A, Levine ME, Tanaka T, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY). 2017;9(2):419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu X, Huang Q, Javed R, Zhong J, Gao H, Liang H. Effect of tobacco smoking on the epigenetic age of human respiratory organs. Clin Epigenetics. 2019;11(1):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.