ABSTRACT
The α-arrestin domain containing protein 3 (ARRDC3) is a tumor suppressor in triple-negative breast carcinoma (TNBC), a highly metastatic subtype of breast cancer that lacks targeted therapies. Thus, understanding the mechanisms and targets of ARRDC3 in TNBC is important. ARRDC3 regulates trafficking of protease-activated receptor 1 (PAR1, also known as F2R), a G-protein-coupled receptor (GPCR) implicated in breast cancer metastasis. Loss of ARRDC3 causes overexpression of PAR1 and aberrant signaling. Moreover, dysregulation of GPCR-induced Hippo signaling is associated with breast cancer progression. However, the mechanisms responsible for Hippo dysregulation remain unknown. Here, we report that the Hippo pathway transcriptional co-activator TAZ (also known as WWTR1) is the major effector of GPCR signaling and is required for TNBC migration and invasion. Additionally, ARRDC3 suppresses PAR1-induced Hippo signaling via sequestration of TAZ, which occurs independently of ARRDC3-regulated PAR1 trafficking. The ARRDC3 C-terminal PPXY motifs and TAZ WW domain are crucial for this interaction and are required for suppression of TNBC migration and lung metastasis in vivo. These studies are the first to demonstrate a role for ARRDC3 in regulating GPCR-induced TAZ activity in TNBC and reveal multi-faceted tumor suppressor functions of ARRDC3.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Hippo pathway, PAR1, PAR2, Thrombin, S1P, LPA
Summary: The α-arrestin ARRDC3 is a multi-functional tumor suppressor that distinctly regulates GPCR trafficking and TAZ-dependent Hippo signaling in metastatic triple negative breast cancer.
INTRODUCTION
Basal-like triple-negative breast cancer (TNBC) is highly aggressive, lacks targeted treatment options and exhibits therapeutic resistance (Foulkes et al., 2010; Gong et al., 2017). Arrestin-domain containing protein 3 (ARRDC3) is an emerging tumor suppressor for highly metastatic breast cancer, with diminished expression in basal-like TNBC due to gene deletion or epigenetic silencing (Adelaide et al., 2007; Lin et al., 2020; Soung et al., 2014). The loss of ARRDC3 expression correlates with increased breast cancer metastasis, tumor recurrence and poor prognosis (Adelaide et al., 2007). Conversely, ARRDC3 overexpression in TNBC reverses epithelial-to-mesenchymal transition and reduces chemo-resistance (Soung et al., 2019). However, the mechanism by which ARRDC3 exerts its tumor suppressor function in basal-like metastatic breast cancer is poorly understood.
The α-arrestin ARRDC3 is structurally similar to the multi-functional β-arrestin scaffold protein and possesses arrestin-like N and C domains but differs by the presence of C-terminal PPXY motifs. The ARRDC3 PPXY motifs are known to mediate interaction with WW domains (Rauch and Martin-Serrano, 2011). In basal-like breast carcinoma, ARRDC3 was previously reported to modulate trafficking of integrin β4 (Draheim et al., 2010), a protein marker for poor prognosis enriched in TNBC (Bierie et al., 2017). Our group showed that ARRDC3 regulates trafficking of protease-activated receptor 1 (PAR1, also known as F2R; Arakaki et al., 2018a; Dores et al., 2015), a G-protein-coupled receptor (GPCR) implicated in breast cancer progression. PAR1 expression is markedly increased in breast cancer biopsies and correlates with metastasis and poor prognosis (Arakaki et al., 2018b; McAuley et al., 2019). Overexpression of PAR1 also occurs in TNBC, owing, in part, to defective lysosomal trafficking, resulting in persistent signaling, cellular invasion and tumor growth (Arakaki et al., 2018a; Arora et al., 2008; Booden et al., 2004). Intriguingly, PAR1 expression is high and ARRDC3 expression is low or absent in TNBC (Arakaki et al., 2018a,b; Draheim et al., 2010). Moreover, re-expression of ARRDC3 is sufficient to rescue PAR1-defective lysosomal trafficking in TNBC and thereby attenuates PAR1-mediated persistent signaling and invasion (Arakaki et al., 2018a). These studies indicate that ARRDC3 tumor suppressor function is linked to regulation of receptor trafficking, but whether ARRDC3 displays other tumor suppressor functions to control GPCR signaling in TNBC is not known.
The Hippo pathway is dysregulated in many cancers and triggers tumorigenesis, metastasis and drug resistance (Ma et al., 2019; Piccolo et al., 2014). Core components of the Hippo pathway include the MST1 and MST2 kinases (also known as STK4 and STK3, respectively), which phosphorylate LATS1 and LATS2 kinases that directly phosphorylate the transcriptional co-activators yes-associated protein (YAP, also known as YAP1) and WW domain-containing transcription regulator protein 1 (TAZ), key effectors of the Hippo pathway. Phosphorylation of YAP and TAZ promotes cytoplasmic retention, whereas dephosphorylation triggers nuclear translocation and gene expression (Ma et al., 2019). Hippo signaling is dynamically regulated by soluble factors that act mainly through GPCRs, including PAR1, lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) receptors (Mo et al., 2012; Nag et al., 2018; Yu et al., 2012). Activation of PAR1 with the peptide agonist TRAP6 causes YAP and TAZ dephosphorylation and nuclear translocation, both of which are required for induction of gene expression (Mo et al., 2012). While YAP and TAZ have previously been shown to function largely redundantly (Plouffe et al., 2018), recent studies suggest an emerging role for TAZ in invasive breast cancer. Increased TAZ expression and activity is associated with high-grade human breast cancers and metastasis, and correlates with poor prognosis (Bartucci et al., 2015; Chan et al., 2008; Cordenonsi et al., 2011). Although GPCRs activate both YAP and TAZ, it is not known whether YAP and TAZ have distinct or overlapping functions in Hippo pathway signaling induced by GPCRs in TNBC.
Here, we have examined whether YAP and TAZ activation exert redundant or distinct functions in GPCR-stimulated Hippo signaling, and whether ARRDC3 regulates GPCR-induced Hippo signaling in TNBC. Our studies demonstrate that TAZ, and not YAP, is the major effector of GPCR-stimulated Hippo signaling in TNBC. We further show that ARRDC3 binds to and suppresses TAZ and not YAP activation, resulting in reduction of gene expression, breast carcinoma migration and metastasis in vivo. Thus, ARRDC3 inhibits GPCR-induced TAZ activity independently of receptor trafficking, indicating that ARRDC3 is a multifunctional tumor suppressor protein in TNBC.
RESULTS
Hippo pathway activation induced by thrombin requires PAR1
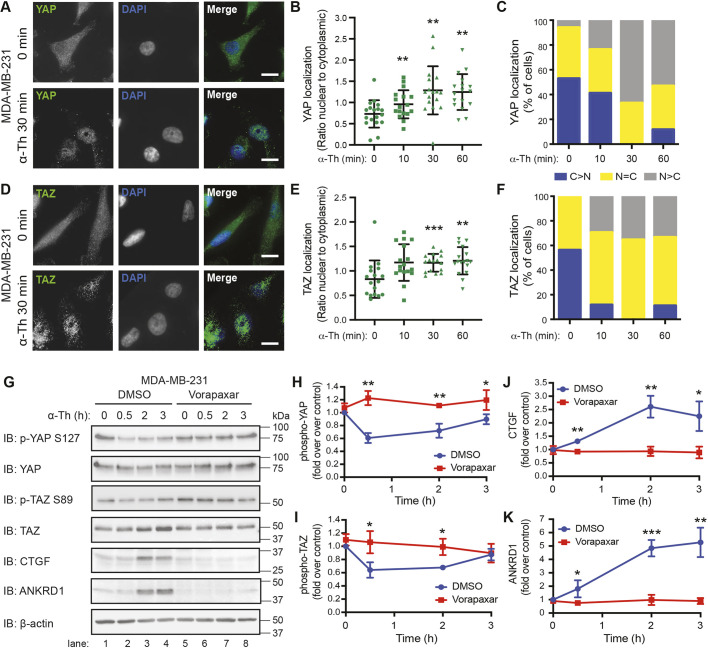
To define the role of PAR1 in Hippo signaling induced by its natural ligand thrombin, nuclear translocation and dephosphorylation of YAP and TAZ transcriptional co-activators were examined in TNBC MDA-MB-231 cells. Serum-starved MDA-MB-231 cells displayed Hippo signaling activity based on the detection of YAP and TAZ phosphorylation in unstimulated cells (Fig. 1G, lanes 1 and 5). However, immunofluorescence confocal microscopy detected a larger fraction of YAP and TAZ localized in cytoplasm compared with the nucleus in serum-starved cells not treated with agonist (Fig. 1A–F), with representative images shown in Fig. 1A,D. Incubation with thrombin for 30 min caused significant translocation and nuclear accumulation of YAP (Fig. 1A–C) and TAZ (Fig. 1D–F), as assessed by immunofluorescence confocal microscopy. The major initiator of YAP and TAZ nuclear translocation is dephosphorylation, and this was assessed next. Thrombin promoted a rapid and significant decrease in phosphorylation of YAP S127 and TAZ S89 at 30 min that was sustained for 2 h in serum-starved cells (Fig. 1G, lanes 1–5; Fig. 1H,I). However, in cells preincubated with vorapaxar, a PAR1 selective antagonist, thrombin-induced YAP S127 and TAZ S89 dephosphorylation was significantly inhibited (Fig. 1G–I). Thrombin also promoted a significant increase in connective tissue growth factor (CTGF, also known as CCN2) and ankyrin repeat domain 1 (ANKRD1) expression, major effectors of Hippo signaling in MDA-MB-231 cells (Fig. 1G,J,K), which was significantly reduced by vorapaxar (Fig. 1G,J,K). These results indicate that PAR1 is required for thrombin-induced Hippo signaling in TNBC.
Fig. 1.
Thrombin activates the Hippo pathway in metastatic breast cancer cells through PAR1. (A–F) Subcellular localization of YAP (A–C) and TAZ (D–F) in MDA-MB-231 cells. Representative images of YAP (A) and TAZ (D) (green), and DAPI (blue) basally and after incubation with 10 nM α-thrombin (α-Th). Scale bars: 10 μm. (B,E) Ratio of nuclear-to-cytoplasmic localization of YAP (B) or TAZ (E) at the indicated times. Data are mean±s.d. Statistical significance was determined by one-way ANOVA (with Tukey′s post hoc test) of each time point compared with 0 min (n=18, six fields of view from three biological replicates). (C,F) Percentage of cells displaying greater cytoplasmic (blue; C>N), nuclear (gray; N>C) or equal (yellow; N=C) staining of YAP (C) or TAZ (F) at the indicated time points. (G–K) MDA-MB-231 cells were pretreated with DMSO or Vorapaxar (10 µM) then treated with 10 nM α-thrombin for the indicated times. Representative immunoblotting (IB) using the indicated antibodies is shown (G). β-actin expression was used as a control. Results are represented as the fold-change in YAP phosphorylation (H), TAZ phosphorylation (I), CTGF expression (J) and ANKRD1 expression (K) relative to 0 min DMSO control. Data are mean±s.d., n=3. Statistical significance was determined using an unpaired t-test, comparing DMSO to Vorapaxar treatment at each time point. *P<0.05; **P<0.01; ***P<0.001.
TAZ, but not YAP, is the major effector of GPCR-stimulated Hippo signaling
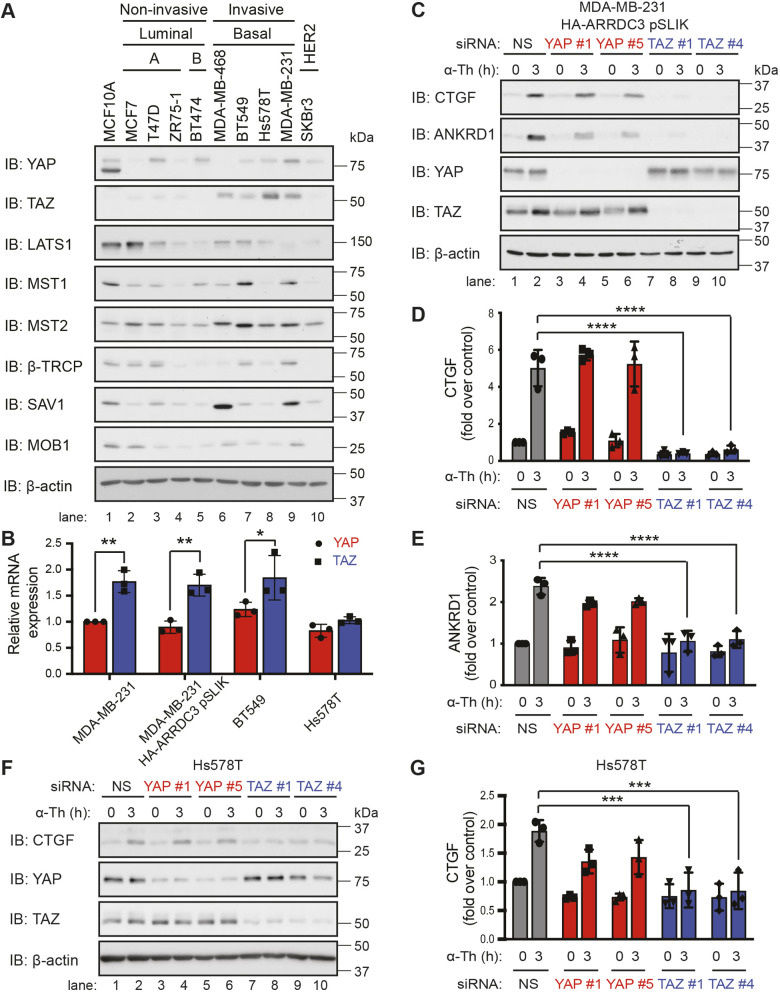
High TAZ expression and activity has been implicated in breast cancer progression (Chan et al., 2008; Cordenonsi et al., 2011; Diaz-Martin et al., 2015). To determine whether expression of TAZ alone, or along with other Hippo pathway components, correlates with invasiveness, we profiled a panel of basal invasive TNBCs that exhibit high PAR1 expression and luminal non-invasive breast carcinoma that display low PAR1 expression (Arakaki et al., 2018a; Booden et al., 2004). In contrast to variable expression of Hippo pathway components, including YAP, observed in both invasive and non-invasive breast carcinoma, high expression of TAZ was detected primarily in TNBC (Fig. 2A, lanes 6–9) and not in luminal non-invasive or HER2-positive breast carcinoma (Fig. 2A, lanes 1–5 and 10). The relative expression of YAP and TAZ mRNA transcript abundance was also determined by qPCR. There was no significant difference in TAZ and YAP mRNA transcript abundance in Hs578T TNBC cells (Fig. 2B). However, a significant but modest increase in TAZ mRNA transcripts compared with YAP was detected in parental MDA-MB-231 and BT549 cells (Fig. 2B). A similar difference in TAZ versus YAP mRNA abundance was observed in MDA-MB-231 cells stably expressing doxycycline-regulated HA–ARRDC3 single lentivector for inducible knockdown (pSLIK) (Fig. 2B), a cell system optimized to interrogate ARRDC3 function (Arakaki et al., 2018a). Thus, transcriptional and post-transcriptional processes likely regulate YAP and TAZ protein expression, resulting in higher TAZ expression in TNBC compared with luminal breast carcinoma. PAR1 expression is similarly high in TNBC, whereas ARRDC3 expression is low or absent in this subtype (Arakaki et al., 2018a; Booden et al., 2004).
Fig. 2.
TAZ, but not YAP, is required for thrombin-mediated activation of the Hippo pathway in invasive breast cancer cell lines. (A) Equivalent amounts (20 µg) of cell lysates from various breast cancer cell lines were immunoblotted (IB) for Hippo pathway components: YAP, TAZ, LATS1, MST1, MST2, β-TRCP (also known as BTRC), SAV1 and MOB1 expression. β-actin expression was used as a control. (B) YAP (red) and TAZ (blue) mRNA expression in invasive breast cancer cell lines was quantified by qPCR. Results are normalized to S18 mRNA expression and are represented as the fold-change relative to YAP expression in parental MDA-MB-231. Data are mean±s.d., n=3. Statistical significance was determined by two-way ANOVA with Tukey′s post hoc test. MDA-MB-231 HA–ARRDC3 pSLIK (C–E) and Hs578T (F,G) cells were transfected with the indicated siRNAs and stimulated with 10 nM α-thrombin (α-Th). Results are represented as the fold increase in CTGF expression (D,G) and ANKRD1 expression (E) relative to 0 min non-specific siRNA (NS)-transfected control. Data are mean±s.d., n=3. Statistical significance was determined by two-way ANOVA with Tukey′s post hoc test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
YAP and TAZ are thought to function redundantly in most cell types and are both expressed in TNBC (Fig. 2A) (Plouffe et al., 2018). To determine whether YAP and TAZ function redundantly in GPCR-stimulated Hippo signaling, induction of CTGF and ANKRD1 expression by thrombin-activated PAR1 was assessed in YAP and TAZ siRNA depleted MDA-MB-231 HA–ARRDC3 pSLIK cells. Knockdown of YAP expression with two different siRNAs failed to significantly affect thrombin-induced CTGF and ANKRD1 expression compared with non-specific siRNA control cells (Fig. 2C–E). Conversely, depletion of TAZ by siRNA caused significant inhibition of thrombin-stimulated expression of CTGF and ANKRD1 compared with non-specific siRNA control cells (Fig. 2C–E), suggesting that YAP and TAZ have distinct functions. TAZ, but not YAP, also emerged as the major effector of thrombin-induced Hippo pathway activation in parental MDA-MB-231 cells (Fig. S1A,B). Next, we used Hs578T cells to determine whether YAP and TAZ differentially regulate thrombin-promoted Hippo pathway activation in other TNBC. Similar to MDA-MB-231 cells, thrombin-stimulated CTGF expression was blocked by depletion of TAZ but not YAP in Hs578T cells (Fig. 2F,G). Thrombin failed to induce ANKRD1 expression in Hs578T cells and was not examined (Fig. S1C). These data indicate that TAZ, but not YAP, is the major effector of thrombin-induced Hippo signaling in TNBC.
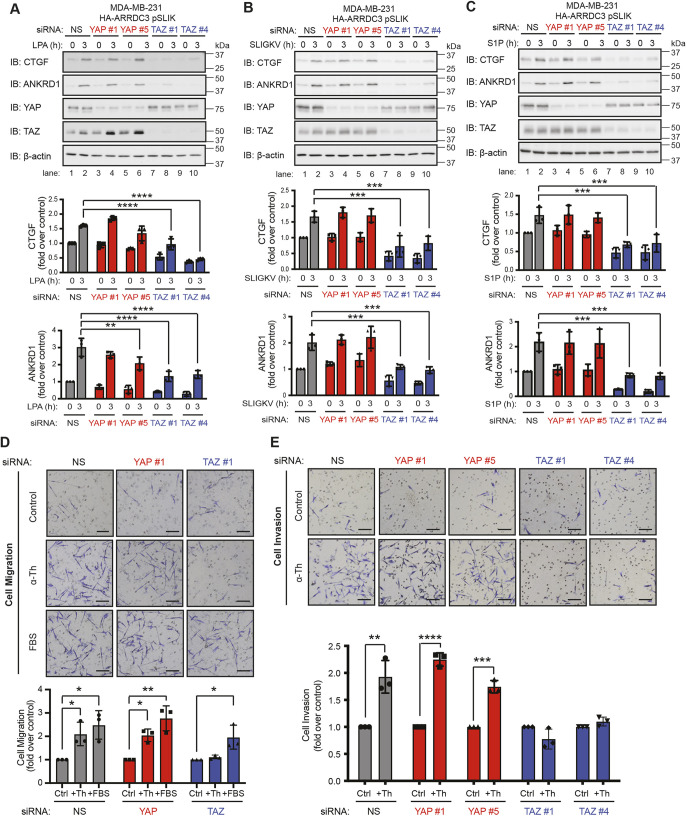
To determine whether YAP and TAZ differentially function in Hippo signaling stimulated by other GPCRs in TNBC, we examined signaling by the LPA receptors (LPARs), by PAR2 (also known as F2RL1) and by S1P receptors (S1PRs) in MDA-MB-231 HA–ARRDC3 pSLIK cells. Similar to thrombin, activation of LPA receptors with their cognate ligand induced a significant increase in both CTGF and ANKRD1 expression in non-specific siRNA cells that was significantly inhibited in TAZ- but not YAP-knockdown cells (Fig. 3A). Similarly, cells treated with either SLIGKV, a PAR2 selective peptide agonist, or S1P also showed a marked increase in CTGF and ANKRD1 expression in the non-specific siRNA-transfected cells, and this increase in expression was significantly inhibited in TAZ- but not YAP-deficient cells (Fig. 3B,C). These findings indicate that TAZ, but not YAP, functions as the main effector of GPCR-stimulated Hippo signaling in TNBC.
Fig. 3.
TAZ, but not YAP, is required for GPCR-mediated activation of the Hippo pathway and for thrombin-mediated migration and invasion. (A–C) MDA-MB-231 HA–ARRDC3 pSLIK cells were transfected with the indicated siRNAs and stimulated with 100 nM lysophosphatidic acid (LPA, A), 1 µM SLIGKV peptide agonist (B) or 100 nM sphingosine-1-phosphate (S1P, C). Samples were immunoblotted (IB) with antibodies against the indicated proteins. Results are represented as the fold-increase in CTGF and ANKRD1 expression relative to 0 h non-specific siRNA (NS)-transfected control. Data are mean±s.d., n=3. Statistical significance was determined by two-way ANOVA with Tukey′s post hoc test. (D,E) MDA-MB-231 HA–ARRDC3 pSLIK cells were transfected with the indicated siRNAs and incubated in transwells, with or without 100 pM α-thrombin (α-Th) or 0.5% FBS as control for migration assay (D), or with or without 1 pM α-thrombin for the invasion assay (E). Images are representative of three independent experiments. Scale bars: 20 μm. Results were quantified and are represented as the fold change over untreated control cells. Data are mean±s.d., n=3. Statistical significance was determined by one-way ANOVA (with Tukey′s post hoc test) for migration assay and unpaired t-test for invasion assay. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
The specific roles of YAP versus TAZ in thrombin-stimulated cell migration and invasion are not known and were examined in TNBC. Thrombin stimulated a significant increase in migration of control non-specific siRNA-transfected cells, comparable with fetal bovine serum (FBS)-induced migration observed in MDA-MB-231 HA–ARRDC3 pSLIK cells (Fig. 3D). In contrast, thrombin-induced cell migration was significantly inhibited in TAZ-depleted cells (Fig. 3D), whereas FBS-induced cell migration remained intact (Fig. 3D). Cell migration induced by thrombin or FBS was not impaired in YAP-deficient cells (Fig. 3D), consistent with a role for TAZ and not YAP. Moreover, thrombin caused a significant increase in invasion of control non-specific siRNA cells (Fig. 3E) that was similar in YAP-depleted cells (Fig. 3E). However, loss of TAZ expression significantly inhibited thrombin-stimulated breast carcinoma cell invasion (Fig. 3E). Thus, YAP and TAZ differ in their capacity to regulate thrombin-induced breast carcinoma cell migration and invasion, with TAZ emerging as the key effector of GPCR-stimulated Hippo signaling in TNBC.
ARRDC3 suppresses GPCR-stimulated Hippo signaling independently of receptor trafficking
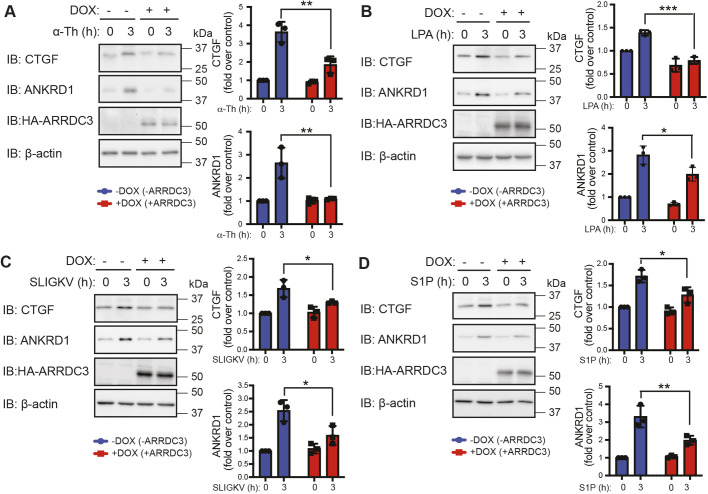
The loss of ARRDC3 tumor suppressor function is essential for PAR1-promoted breast cancer progression (Arakaki et al., 2018a), raising the possibility that ARRDC3 may regulate Hippo pathway signaling induced by PAR1. As ARRDC3 expression is lost or suppressed in TNBC (Adelaide et al., 2007; Arakaki et al., 2018a; Draheim et al., 2010), MDA-MB-231 cells stably expressing HA–ARRDC3 pSLIK were used to allow doxycycline-inducible expression of HA–ARRDC3. In control MDA-MB-231 HA–ARRDC3 pSLIK cells not treated with doxycycline and deficient in ARRDC3, incubation with thrombin caused a marked increase in CTGF and ANKRD1 expression (Fig. 4A). However, thrombin-stimulated CTGF and ANKRD1 expression was significantly inhibited in doxycycline-induced ARRDC3-expressing cells (Fig. 4A), suggesting that ARRDC3 suppresses Hippo signaling. Similar to thrombin-activated PAR1, ARRDC3 inhibited induction of CTGF and ANKRD1 expression by ligands acting at the LPARs, PAR2 and S1PRs (Fig. 4B–D). Thus, ARRDC3 regulates Hippo signaling induced by multiple GPCR ligands in TNBC.
Fig. 4.
ARRDC3 re-expression suppresses GPCR activation of the Hippo pathway. (A–D) ARRDC3 expression was induced by doxycycline (DOX) treatment in MDA-MB-231 HA–ARRDC3 pSLIK cells, then stimulated with 10 nM α-thrombin (α-Th; A), 100 nM LPA (B), 1 µM SLIGKV (C) or 100 nM S1P (D). Samples were immunoblotted (IB) with antibodies against the indicated proteins. Results are the fold increase in CTGF and ANKRD1 expression relative to 0 min −DOX control. Data are mean±s.d., n=3. Statistical significance was determined by two-way ANOVA with Tukey′s post hoc test. *P<0.05; **P<0.01; ***P<0.001.
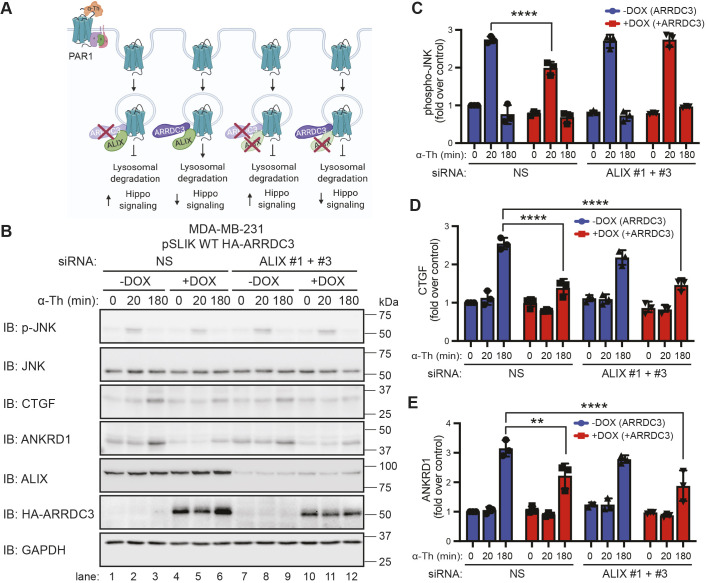
ARRDC3 functions as a tumor suppressor by facilitating PAR1 lysosomal trafficking through an ALIX-dependent pathway in invasive breast carcinoma (Fig. 5A). ARRDC3 and ALIX (also known as PDCD6IP) are both required for activated PAR1 lysosomal trafficking (Fig. 5A) (Arakaki et al., 2018a; Dores et al., 2015), and loss of either ARRDC3 or ALIX function blocks lysosomal degradation. We also recently showed that both ARRDC3 and ALIX are required to prevent activated PAR1 recycling to the plasma membrane and to attenuate Gα12/13 -mediated JNK signaling (Arakaki et al., 2018a). To determine whether ARRDC3 suppresses activated PAR1-stimulated Hippo signaling independent of receptor trafficking, we examined whether blockade of activated PAR1 lysosomal trafficking by siRNA knockdown of ALIX results in decreased Hippo signaling in the presence of ARRDC3 (Fig. 5A). MDA-MB-231 HA–ARRDC3 pSLIK cells transfected with non-specific siRNA or ALIX-specific siRNAs were treated with or without doxycycline to induce ARRDC3 expression followed by thrombin stimulation. MDA-MB-231 cells transfected with non-specific siRNA expressing ALIX but not ARRDC3 were stimulated with thrombin and showed a significant increase in JNK1 phosphorylation, as well as induction of CTGF and ANKRD1 expression (Fig. 5B, lanes 1–3; Fig. 5C–E). As expected, re-expression of ARRDC3 in cells expressing ALIX significantly reduced thrombin-stimulated JNK1 phosphorylation (Fig. 5B, lanes 4–6; Fig. 5C–E), which has been attributed to restoration of PAR1 lysosomal trafficking (Arakaki et al., 2018a), and further attenuated induction of CTGF and ANKRD1 expression (Fig. 5B, lanes 4–6; Fig. 5C–E). In cells lacking both ALIX and ARRDC3 expression, thrombin caused a significant increase in JNK1 phosphorylation and induction of CTGF and ANKRD1 expression (Fig. 5B, lanes 7–9; Fig. 5C–E). However, in cells lacking ALIX, re-expression of ARRDC3 retained the capacity to suppress thrombin-induced CTGF and ANKRD1 expression, despite blockade of PAR1 lysosomal trafficking that resulted in enhanced JNK1 phosphorylation (Fig. 5B, lanes 10–12; Fig. 5C–E). These findings indicate that ARRDC3 regulates PAR1-stimulated Hippo signaling via a mechanism that is independent of ALIX and receptor trafficking in TNBC.
Fig. 5.
ARRDC3 regulates Hippo signaling independently of its function on PAR1 trafficking and degradation. (A) Illustration of ARRDC3 and ALIX requirement for facilitating lysosomal degradation and hypothesis of the effect of ARRDC3 on Hippo signaling. Illustration created with BioRender.com. (B–E) MDA-MB-231 HA–ARRDC3 pSLIK cells were transfected with the indicated siRNAs (NS, non-specific control siRNA) and treated with doxycycline (DOX) for ARRDC3 overexpression. Cells were then stimulated with 10 nM α-thrombin (α-Th) for the indicated times, and samples were immunoblotted (IB) using antibodies against the indicated proteins. Results are the fold-increase in JNK phosphorylation (C), CTGF expression (D) and ANKRD1 expression (E) relative to 0 min −DOX, NS-transfected control. Data are mean±s.d., n=3. Statistical significance was determined by two-way ANOVA with Tukey′s post hoc test. **P<0.01; ****P<0.0001.
ARRDC3 suppresses thrombin-induced dephosphorylation and nuclear translocation of TAZ but not YAP
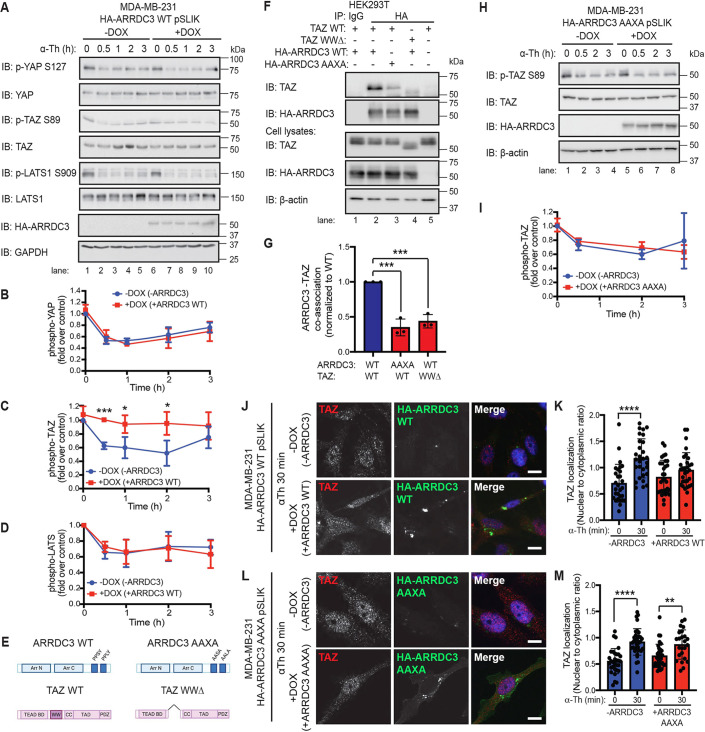
Hippo pathway activation occurs via a core kinase cascade that phosphorylates and activates LATS1 and LATS2 kinases, which phosphorylate and inactivate YAP and TAZ; but how ARRDC3 integrates into the Hippo pathway is not known and was examined in MDA-MB-231 HA–ARRDC3 pSLIK cells. In cells with or without ARRDC3 expression, basal phosphorylation of LATS1, YAP and TAZ was observed in unstimulated cells (Fig. 6A, lanes 1 and 5), indicating that Hippo signaling activity is turned on. After thrombin incubation, a significant decrease in phosphorylation of LATS1 as well as YAP and TAZ was detected in cells lacking ARRDC3 expression (Fig. 6A, lanes 1–5; Fig. 6B–D). However, in ARRDC3-expressing cells, thrombin-stimulated TAZ dephosphorylation was significantly blocked, with no difference in LATS and YAP dephosphorylation kinetics compared with control cells (Fig. 6D, lanes 6–10; Fig. 6B–D), suggesting that ARRDC3 regulates thrombin-induced Hippo signaling by controlling TAZ activity in invasive breast carcinoma. Notably, ARRDC3 expression had no significant effect on the basal expression of YAP or TAZ determined at 0 min (Fig. S2A). However, ARRDC3 suppressed thrombin-induced TAZ expression (Fig. S2A), whereas there was no significant effect of ARRDC3 on YAP expression.
Fig. 6.
ARRDC3 re-expression blocks thrombin-mediated TAZ dephosphorylation and nuclear localization through co-association between ARRDC3 and TAZ. (A–D,H,I) MDA-MB-231 wild-type (WT: A–D) and AAXA mutant (H,I) HA–ARRDC3 pSLIK cells were treated with doxycycline (DOX) to induce ARRDC3 expression and stimulated with 10 nM α-thrombin (α-Th) for various times. Samples were immunoblotted (IB) using antibodies against the indicated proteins. Results are represented as the fold-change in YAP phosphorylation (B), TAZ phosphorylation (C,I), and LATS phosphorylation (D) relative to 0 min −DOX control. Data are mean±s.d., n=3. Statistical significance was determined by unpaired t-test at each time point. (E) Illustration of constructs of ARRDC3 and TAZ, with domains indicated (BD, binding domain; CC, coiled-coils; TAD, topologically associating domain; PDZ, PDZ domain; WW domain deletion, WWΔ). Illustration created with BioRender.com. (F,G) HEK293T cells transiently expressing wild-type and mutant constructs were immunoprecipitated (IP) with HA antibody to pulldown HA–ARRDC3. Non-specific IgG IP was used as a control. Immunoprecipitate and cell lysate input were analyzed by immunoblotting, as indicated. Results are quantified by densitometry, and co-association of ARRDC3–TAZ (G) is represented as fold over wild-type control. Statistical significance determined using one-way ANOVA with Tukey′s post hoc test. Data are mean±s.d., n=3. (J–M) TAZ subcellular localization following thrombin treatment was determined by immunofluorescence staining of endogenous TAZ (red) in MDA-MB-231 wild-type (J,K) and AAXA mutant (L,M) HA–ARRDC3 pSLIK cells; HA (green) was stained to detect ARRDC3, and DAPI (blue) for cell nuclei. Scale bars: 10 μm. (K,M) Quantification of the ratio nuclear to cytoplasmic TAZ localization. Data are mean±s.d. Statistical significance was determined by one-way ANOVA (with Tukey′s post hoc test) of each time point compared with 0 min (n=27, nine fields of view from three biological replicates). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
TAZ contains a single WW domain that mediates protein–protein interaction with PPXY motifs, whereas ARRDC3 contains two C-terminal PPXY motifs that bind to WW domains (Fig. 6E). However, it is not known whether ARRDC3 and TAZ co-associate, and this was examined in HEK293T cells. Wild-type TAZ and HA–ARRDC3 showed robust co-association in anti-HA co-immunoprecipitates (co-IPs) but not in IgG control (Fig. 6F, lanes 1,2). Neither ARRDC3 nor TAZ was detected in co-IPs in cells expressing only TAZ (Fig. 6F, lane 5), indicating a specific interaction. In contrast, wild-type TAZ showed a significant decrease in co-association with the HA–ARRDC3 AAXA double mutant, where the crucial prolines (P) and tyrosine (Y) of the C-terminal PPXY motifs were converted to alanine (A), compared with wild-type ARRDC3 (Fig. 6E; Fig. 6F, lanes 2 and 3; Fig. 6G), suggesting that the PPXY motifs are crucial for ARRDC3–TAZ interaction. Moreover, deletion of the single WW domain of TAZ also resulted in a significant loss of interaction with wild-type ARRDC3 (Fig. 6E; Fig. 6F, lanes 2 and 4; Fig. 6G). Thus, ARRDC3 and TAZ interaction occurs via a WW domain interaction with PPXY motifs.
To determine whether ARRDC3 regulates TAZ function via the PPXY motifs, thrombin-induced TAZ dephosphorylation and nuclear translocation were examined in MDA-MB-231 cells expressing wild-type HA–ARRDC3 or the AAXA mutant. In contrast to wild-type ARRDC3, which suppresses TAZ dephosphorylation (Fig. 6A,C), thrombin-induced TAZ dephosphorylation was not affected in ARRDC3 AAXA mutant-expressing cells (Fig. 6H, lanes 5–8; Fig. 6I). As expected, ARRDC3 AAXA expression failed to affect thrombin-stimulated YAP dephosphorylation (Fig. S2B). These results suggest that the interaction of ARRDC3 with TAZ suppresses dephosphorylation induced by thrombin. Nuclear localization of TAZ stimulated by thrombin was also significantly inhibited by wild-type ARRDC3 (Fig. 6J,K; Fig. S2C,D). In contrast to wild-type ARRDC3, the ARRDC3 AAXA mutant failed to block thrombin-stimulated TAZ nuclear translocation (Fig. 6L,M). Neither wild-type ARRDC3 nor AAXA mutant expression had any effect on thrombin-stimulated nuclear translocation of YAP (Fig. S2E–H).
ARRDC3–TAZ interaction inhibits TAZ–TEAD binding and is required for suppression of thrombin-induced CTGF and ANKRD1 expression and cell migration
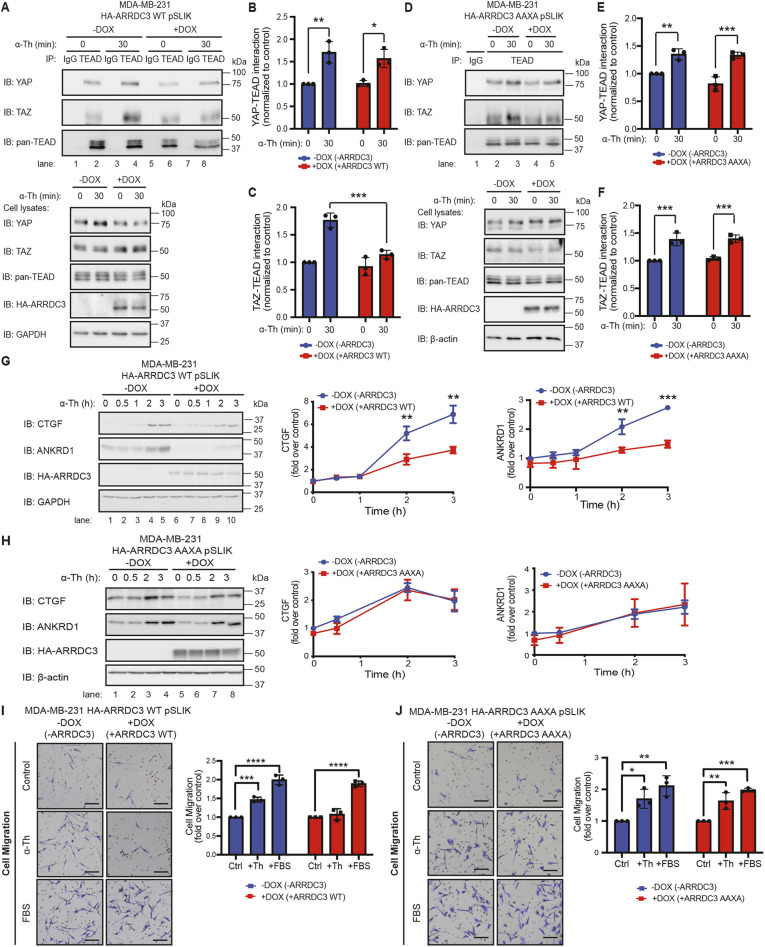
After dephosphorylation and translocation to the nucleus, YAP and TAZ, which lack DNA-binding motifs, interact with the TEA domain (TEAD) family members 1–4 DNA-binding transcription factors to regulate gene transcription (Lamar et al., 2012; Zhao et al., 2007). To determine whether ARRDC3 regulates YAP or TAZ activity, TEAD binding to YAP and TAZ was examined in MDA-MB-231 HA–ARRDC3 pSLIK cells by co-IP. In cells lacking ARRDC3 expression, thrombin-induced a significant increase in YAP–TEAD and TAZ–TEAD interaction (Fig. 7A, lanes 1–4; Fig. 7B,C). Although the thrombin-induced YAP–TEAD interaction was retained in wild-type ARRDC3-expressing cells (Fig. 7A, lanes 5–8; Fig. 7B), the TAZ–TEAD interaction was significantly inhibited by wild-type ARRDC3 expression (Fig. 7A, lanes 5–8; Fig. 7C). Contrary to wild-type ARRDC3, however, expression of AAXA mutant ARRDC3 failed to alter thrombin-induced TAZ–TEAD interaction and, as expected, had no effect on the YAP–TEAD interaction (Fig. 7D–F). These findings suggest that the ARRDC3–TAZ interaction mediated by PPXY motifs prevents the thrombin-stimulated TAZ–TEAD interaction.
Fig. 7.
ARRDC3 re-expression inhibits TAZ–TEAD binding and attenuates downstream Hippo signaling and thrombin-induced migration, dependent on the PPXY motifs of ARRDC3. (A–H) MDA-MB-231 wild-type (WT; A–C,G) and AAXA mutant (D–F,H) HA–ARRDC3 pSLIK cells were treated with doxycycline (DOX) to induce ARRDC3 expression and stimulated with 10 nM α-thrombin (α-Th) for indicated times. (A–F) Cells were lysed and immunoprecipitated (IP) with anti-TEAD antibody or anti-IgG control. IP samples and cell lysates were immunoblotted (IB) with antibodies against the indicated proteins. Results are quantified, and co-association of YAP–TEAD (B,E) and TAZ–TEAD (C,F) is represented as fold over −DOX 0 min control. Data are mean±s.d., n=3. Statistical significance determined using two-way ANOVA with Tukey′s post hoc test. (G,H) Results are the fold-change in CTGF and ANKRD1 expression relative to 0 min −DOX control. Data are mean±s.d., n=3. Statistical significance was determined by unpaired t-test at each time point. (I,J) Migration assay with MDA-MB-231 wild-type (I) and AAXA mutant (J) HA–ARRDC3 pSLIK cells treated with doxycycline to induce ARRDC3 expression and incubated with or without 100 pM α-thrombin or 0.5% FBS. Images shown are representative of three independent experiments. Scale bars: 20 μm. Results are the fold change over untreated control cells. Data are mean±s.d., n=3. Statistical significance was determined by one-way ANOVA with Tukey′s post hoc test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Next, the functional consequences of ARRDC3 regulation of TAZ were examined in MDA-MB-231 HA–ARRDC3 pSLIK cells. As shown previously, thrombin induced significant expression of the YAP/TAZ-targeted genes CTGF and ANKRD1 in cells lacking ARRDC3 expression (Fig. 7G, lanes 1–5), whereas expression of ARRDC3 resulted in significant inhibition of thrombin-induced CTGF and ANKRD1 expression (Fig. 7G, lanes 6–10). In contrast, expression of the ARRDC3 AAXA mutant failed to block thrombin-induced CTGF and ANKRD1 expression (Fig. 7H, lanes 5–8), compared with the response observed in cells lacking ARRDC3 (Fig. 7H, lanes 1–4). Thus, the suppression of TAZ activation by ARRDC3 is sufficient to block CTGF and ANKRD1 gene expression, indicating that YAP function is not necessary for thrombin-induced Hippo signaling in invasive breast carcinoma.
The effect of ARRDC3–TAZ interaction on breast carcinoma cell migration was also examined using MDA-MB-231 HA–ARRDC3 AAXA mutant pSLIK cells. In cells lacking ARRDC3 induction, thrombin stimulated a significant increase in cellular migration (Fig. 7I), which was similarly observed in cells incubated with FBS (Fig. 7I). In wild-type ARRDC3-expressing cells, thrombin-induced cell migration was markedly reduced compared with FBS-promoted cellular migration, which remained intact in cells expressing ARRDC3 (Fig. 7I). In contrast, expression of the ARRDC3 AAXA mutant defective in TAZ binding failed to block thrombin-stimulated cell migration (Fig. 7J), whereas FBS-induced cell migration remained intact and was comparable with that observed in ARRDC3-deficient cells (Fig. 7J). These results indicate that ARRDC3 suppresses thrombin-induced TAZ-dependent breast carcinoma cell migration.
The ARRDC3–TAZ interaction is required for suppression of breast carcinoma metastasis
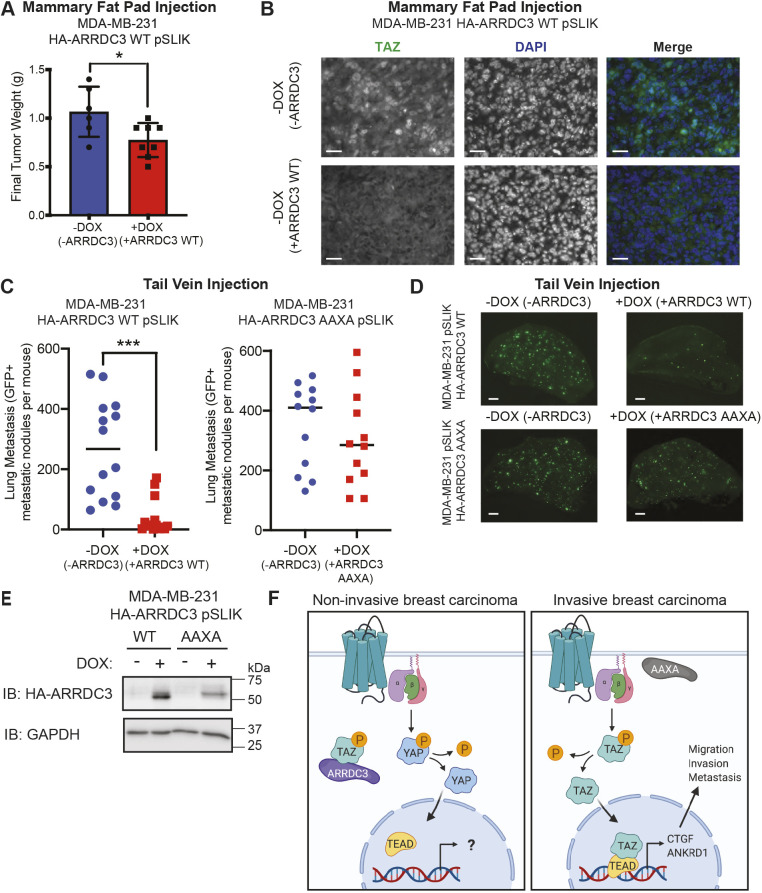
To investigate the effect of ARRDC3 on TAZ activity in vivo, we orthotopically injected MDA-MB-231 wild-type HA–ARRDC3 pSLIK cells into the mammary fat pad of immunocompromised NOD scid gamma (NSG) mice. After mammary fat pad injection, mice were randomly assigned and fed normal chow or doxycycline chow for ARRDC3 expression. As previously reported (Draheim et al., 2010), ARRDC3 re-expression reduced tumor growth compared with tumors lacking ARRDC3, determined 6 weeks post-implantation (Fig. 8A). The effect of ARRDC3 re-expression on TAZ activity was then examined by immunohistochemistry. In tumors of mice fed normal chow lacking ARRDC3 expression, nuclear TAZ localization was detected, suggesting high TAZ activity (Fig. 8B). However, tumors from mice treated with doxycycline to induce ARRDC3 expression in MDA-MB-231 cells showed reduced tumor volume and diminished TAZ localization in the nucleus (Fig. 8B). Together with our in vitro findings, these results suggest that ARRDC3 functions by inhibiting TAZ activity in vivo.
Fig. 8.
ARRDC3 re-expression blocks in vivo breast cancer growth and metastasis, dependent on the PPXY motifs of ARRDC3. (A,B) MDA-MB-231 wild-type (WT) HA–ARRDC3 pSLIK cells were injected in the mammary fat pad of NSG mice fed with or without doxycycline (DOX). (A) Final tumor weight 6 weeks post-implantation. Statistical significance determined by unpaired t-test (mean±s.d.; n=6 mice in −DOX group, n=8 mice in +DOX group). (B) Representative images of immunohistochemistry of mammary fat pad tumors stained for TAZ (green) and DAPI for nuclei (blue). Scale bars: 25 µm. (C,D) GFP-labeled MDA-MB-231 wild-type or AAXA mutant HA–ARRDC3 pSLIK cells were injected into the tail vein of NSG mice. (C) Quantification of GFP-positive metastatic nodules in the lungs of the mice collected 2 weeks after injection. Line indicates the median. Statistical significance determined by unpaired t-test with Welch's correction (wild type, n=14 mice per group; AAXA, n=12 mice per group). (D) Representative fluorescence images of GFP-positive metastatic lesions in the lungs of mice. GFP signal indicates tumor cell extravasation, seeding, growth and colonization in the lung. Scale bars: 1 mm. (E) Verification of HA–ARRDC3 wild-type or HA–ARRDC3 AAXA re-expression in MDA-MB-231 pSLIK cells collected prior to tail-vein injection. Lysates immunoblotted (IB) for HA–ARRDC3 and GAPDH expression. (F) ARRDC3 is highly expressed in normal mammary epithelial cells or luminal non-invasive breast carcinoma cells, and co-associates with TAZ, leading to its cytoplasmic retention and attenuated GPCR-mediated Hippo pathway signaling. However, when ARRDC3 is absent, e.g. in invasive basal-like breast carcinoma, or is lacking PPXY motifs and thus functionally inactive, GPCR signaling activates TAZ through TAZ dephosphorylation, promoting its nuclear localization, binding to the TEAD family of transcriptional co-activators, and inducing CTGF and ANKRD1 expression, leading to cell migration, invasion and metastasis. P, phosphorylation. Model created with BioRender.com. *P<0.01; ***P<0.001.
The role of ARRDC3 in breast tumor metastasis is not known and was examined using a tail-vein injection model. MDA-MB-231 HA–ARRDC3 wild-type and AAXA mutant pSLIK cells transduced with GFP and pre-treated with or without doxycycline for 48 h in vitro were injected into the tail vein of immunocompromised NSG mice. Mice injected with pre-treated doxycycline cells were fed doxycycline chow to induce ARRDC3 expression, while mice injected with non-treated cells were fed normal chow; lung metastasis was quantified by immunofluorescence microscopy. After 2 weeks, a high metastatic tumor burden was observed with a large number of GFP-positive nodules detected in the lung tissue of mice injected with control HA–ARRDC3 wild-type pSLIK cells and not treated with doxycycline (Fig. 8C,D). Conversely, the number of detected metastatic nodules was significantly reduced in doxycycline-treated mice injected with wild-type ARRDC3 pSLIK cells (Fig. 8C,D), suggesting that ARRDC3 suppresses metastasis. Control mice injected with HA–ARRDC3 AAXA mutant pSLIK cells and not treated with doxycycline also exhibited high tumor burden (Fig. 8C,D). However, unlike mice injected with wild-type HA–ARRDC3 pSLIK cells, mice injected into the tail-vein with ARRDC3 AAXA mutant pSLIK cells and treated with doxycycline formed abundant metastatic foci (Fig. 8C,D), suggesting that the ARRDC3 AAXA mutant fails to suppress breast carcinoma metastasis. Induction of wild-type HA–ARRDC3 and AAXA mutant expression in MDA-MB-231 pSLIK cells was confirmed by immunoblotting for HA–ARRDC3 in the pool of cells collected before injection that were treated with doxycycline (Fig. 8E). Collectively, these in vivo metastasis results combined with the cellular and biochemical data above indicate that ARRDC3 functions specifically to inhibit TAZ and not YAP activity induced by GPCRs, resulting in suppression of Hippo-mediated induction of CTGF and ANKRD1 expression, cell migration and breast carcinoma metastasis in vivo (Fig. 8F).
DISCUSSION
Basal-like TNBC remains a crucial subtype contributing to breast cancer mortality due to its high metastatic potential and lack of molecular targets (Foulkes et al., 2010; Gong et al., 2017). GPCRs, including PAR1, play significant roles in breast cancer progression yet are currently underused as therapeutic targets (Hamilton and Trejo, 2017; Insel et al., 2018; Nag et al., 2018). Hippo signaling, which normally prevents YAP and TAZ activation, is turned off predominantly by GPCRs, including PAR1, LPARs, PAR2 and S1PRs, to promote proliferation and invasion (Mo et al., 2012; Yu et al., 2012). However, the mechanisms responsible for dysregulation of the Hippo signaling induced by GPCRs in TNBC is not known. Here, we show that the transcriptional co-activator TAZ, but not YAP, is the major effector of GPCR-induced Hippo signaling in TNBC, promoting cell migration and invasion. We further demonstrate that ARRDC3 suppresses GPCR-induced Hippo signaling through TAZ, which occurs independently of ARRDC3 regulation on receptor trafficking. The ARRDC3 C-terminal PPXY motifs mediate interaction with the WW domain of TAZ, resulting in TAZ cytoplasmic retention and inhibition of Hippo signaling. Our study also indicates that the capacity of ARRDC3 to suppress breast carcinoma migration and metastasis in vivo is dependent on ARRDC3 engagement with TAZ. Thus, ARRDC3 exhibits multiple tumor suppressor functions, including regulation of receptor trafficking and control of GPCR-induced activity of TAZ in TNBC.
GPCRs preferentially signal via TAZ, but not YAP, in TNBC
Although YAP and TAZ are largely functionally redundant, TAZ has emerged as an important driver of breast cancer progression. Both overexpression of TAZ and nuclear localization, which are indicative of high TAZ activity, are correlated with high-grade metastatic breast cancer and poor prognosis (Chan et al., 2008; Cordenonsi et al., 2011). In addition, TAZ overexpression promotes breast carcinoma proliferation, migration, invasion and epithelial-to-mesenchymal transition (Chan et al., 2008), whereas loss of TAZ expression impairs migration and invasion, metastatic colonization and chemoresistance (Bartucci et al., 2015; Chan et al., 2008). A previous study showed that combined depletion of both YAP and TAZ inhibited breast carcinoma invasion induced by TRAP6 – a synthetic peptide agonist that activates both PAR1 and PAR2 (Lerner et al., 1996; Mo et al., 2012). Here, we show that activation of PAR1 with thrombin, its natural agonist, stimulates both YAP and TAZ dephosphorylation, and is blocked by the PAR1-specific antagonist vorapaxar, indicating that PAR1 triggers Hippo signaling in TNBC. Moreover, loss of TAZ but not YAP expression in TNBC is sufficient to block PAR1-stimulated CTGF and ANKRD1 gene expression, migration and invasion. TAZ, but not YAP, was also shown to be the major effector for gene induction stimulated by other GPCR agonists, including LPA, SLIGKV and S1PR1 in TNBC (Fig. 3). A dominant role for TAZ has been similarly demonstrated for the G-protein-coupled estrogen receptor (GPER) in invasive ductal carcinoma (Zhou et al., 2015). In that study, activation of GPER was shown to induce gene expression, migration, invasion and tumor growth through a TAZ-dependent pathway in estrogen receptor (ER)+ breast carcinoma (Zhou et al., 2015). These findings suggest that TAZ plays a pivotal role in Hippo signaling induced by GPCRs in both TNBC and ER+ breast carcinoma.
ARRDC3 is a multi-functional tumor suppressor in invasive breast carcinoma
ARRDC3 expression is low or absent in basal-like breast carcinoma, including TNBC, resulting from gene deletion or epigenetic silencing (Adelaide et al., 2007; Soung et al., 2014). However, the mechanisms by which ARRDC3 exerts its tumor suppressor functions are poorly understood. Given that the α-arrestin ARRDC3 shares structural homology with the multi-faceted β-arrestin scaffolds (Qi et al., 2014), ARRDC3 likely also exhibits multiple functions. ARRDC3 was shown to regulate trafficking of integrin β4, and suppressed migration, invasion and tumor growth of TNBC (Draheim et al., 2010). We previously demonstrated that ARRDC3 is both necessary and sufficient for regulating lysosomal trafficking of PAR1, and suppresses persistent signaling and invasion of TNBC (Arakaki et al., 2018a). Here, we report that ARRDC3 displays an additional tumor suppressor function through direct regulation of GPCR-stimulated Hippo signaling in TNBC, resulting in inhibition of metastasis in vivo. We found that ARRDC3 suppressed Hippo-mediated CTGF and ANKRD1 expression induced by several GPCR agonists acting through PAR1, LPARs, PAR2 and S1PRs (Fig. 4), indicating that ARRDC3 functions broadly to control Hippo signaling. Moreover, ARRDC3-mediated suppression of Hippo signaling occurs independently of receptor trafficking, as blockade of PAR1 trafficking by depletion of ALIX was not sufficient to enhance Hippo signaling in the presence of ARRDC3 (Fig. 5). PAR1 and a subset of other GPCRs use a non-canonical ALIX and ARRDC3 pathway for lysosomal sorting (Dores et al., 2012, 2016, 2015), which is distinct from the canonical ubiquitin-dependent lysosomal sorting pathway used by most classical GPCRs. In addition, ARRDC3 regulates Hippo signaling induced by activated PAR2 (Fig. 4), while PAR2 traffics to lysosomes via the canonical ubiquitin-mediated pathway independently of ALIX and ARRDC3 (Dores et al., 2012; Hasdemir et al., 2007). The LPARs and SPR1 also use the canonical ubiquitin-driven lysosomal pathway for degradation (Rosen et al., 2008; Zhao et al., 2016). Yet LPA- and S1P-induced Hippo signaling is suppressed by ARRDC3. Together, these studies support our findings that ARRDC3 functions on the Hippo pathway independently of its role on GPCR trafficking.
ARRDC3 suppresses GPCR-stimulated Hippo signaling by sequestering TAZ in the cytoplasm, thereby preventing gene induction, migration and metastasis of TNBC
A previous study in Drosophila reported that Leash, a homolog of ARRDC3, interacts with Yorkie, the homolog of YAP and TAZ, and inhibits Hippo signaling by facilitating Yorkie degradation (Kwon et al., 2013; Verghese and Moberg, 2020). This discovery gave credence to the idea that mammalian ARRDC3 might regulate Hippo signaling through interactions with YAP and TAZ. In addition, ARRDC3 contains C-terminal PPXY motifs that directly interact with WW domains, conserved regions that fold into a triple-stranded β-sheet present in both YAP and TAZ. In colorectal cancer cells, ARRDC3 co-associates with YAP and enhances YAP degradation, thereby suppressing tumorigenesis and chemotherapy sensitization (Shen et al., 2018). Similarly, in renal cell carcinoma, ARRDC3 and YAP were shown to interact via the ARRDC3 PPXY motifs and WW domains of YAP, resulting in YAP degradation and suppression of tumor progression (Xiao et al., 2018). Here, we report that ARRDC3 interacts with TAZ via the PPXY motifs of ARRDC3 and the single WW domain of TAZ. Unlike studies of YAP in colorectal and renal carcinoma, ARRDC3 does not regulate the stability of TAZ in TNBC. Instead, ARRDC3 suppresses PAR1-induced TAZ dephosphorylation and nuclear translocation in TNBC (Fig. 6), but fails to modulate YAP activity. Moreover, ARRDC3 interaction with TAZ is required for suppression of PAR1-stimulated CTGF and ANKRD1 gene expression and cell migration, as well as in vivo metastasis of TNBC (Figs 6–8). The precise mechanism by which ARRDC3 prevents PAR1-induced dephosphorylation of TAZ is not known, but may involve the specificity and stability of the ARRDC3–TAZ versus ARRDC3–YAP interaction, as YAP contains several structural features not present in TAZ, including an additional WW domain, a SH3-binding motif and an N-terminal proline-rich region (Plouffe et al., 2018). In addition, post-translational modifications, such as phosphorylation and ubiquitylation, may influence ARRDC3–TAZ interaction and function (Batista et al., 2020; Lei et al., 2008; Meng et al., 2016; Wagner et al., 2012), and are an important future area of investigation.
GPCRs are highly targeted therapeutically, currently representing 34% of all FDA-approved treatments (i.e. 475 drugs); however, only eight drugs targeting GPCRs are in use for oncology (Wu et al., 2019). Despite the fact that GPCRs are widely dysregulated in cancer and contribute to tumorigenesis by promoting proliferation, invasion and evasion of the immune system, this receptor class remains under-used as drug targets in oncology (Wu et al., 2019). Thus, ongoing investigations aimed at unraveling GPCR function at the molecular and cellular level in invasive breast cancer may reveal new targets or combination of targets for the development of new therapeutic strategies for the treatment of TNBC. In summary, our study reveals an unanticipated role for TAZ in GPCR-mediated gene induction, migration and invasion of TNBC, and a multi-functional role for ARRDC3 as a tumor suppressor in regulation of GPCR-stimulated TAZ activity. These studies further indicate that TAZ could be used as a drug target due to its crucial role in TNBC migration, invasion and metastasis driven by dysregulated GPCRs.
MATERIALS AND METHODS
Reagents and antibodies
Human α-thrombin was from Enzyme Research Technologies. SLIGKV peptide agonist was synthesized at Tufts Core Facility. Lysophosphatidic acid (#3854) and shingosine-1-phosphate (#1370) were purchased from Tocris Bioscience. Vorapaxar (SCH530348, #1755) was purchased from Axon Medchem and tetracycline-free FBS (#631101) was from Takara Bio. Dabigatran (an α-thrombin inhibitor) was from Boehringer Ingelheim Pharma. Rabbit anti-phospho-YAP S127 (#4911, 1:1000), rabbit anti-YAP (#14074, 1:1000), rabbit anti-phospho-TAZ S89 (#59971, 1:1000), rabbit anti-TAZ (#4883, 1:1000), rabbit anti-MST1 (#3682, 1:1000), rabbit anti-MST2 (#3952, 1:1000), rabbit anti-SAV1 (#13301, 1:1000), rabbit anti-MOB1 (#13730, 1:1000), rabbit anti-β-TRCP (#4394, 1:1000), rabbit anti-phospho-JNK1/2 (#9251, 1:1000), mouse anti-JNK1 (#3708, 1:1000), rabbit anti-phospho-LATS1 S909 (#9157, 1:1000), rabbit anti-LATS1 (#3477, 1:1000), mouse HA-tag Alexa Fluor 488 Conjugate (#2350, 1:500), rabbit anti-pan-TEAD (#13295, 1:1000), rabbit anti-HA (#3724, 1:1000) and normal rabbit IgG (#2729, 1:1000) antibodies were purchased from Cell Signaling Technology. Goat anti-rabbit secondary antibody conjugated to Alexa Fluor 594 (#A-21244, 1:1000), goat anti-rabbit secondary antibody conjugated to Alexa Fluor 488 (#A-11008, 1:1000), DAPI (#D-106), ProLong Gold Antifade Mountant (#P36930) and doxycycline chow (200 mg/kg doxycycline pellets, #14727450) were purchased from ThermoFisher Scientific. Mouse monoclonal anti-CTGF E-5 (#sc-365970, 1:1000) and monoclonal anti-ALIX antibody (#sc-53538, 1:1000) were from Santa Cruz Biotechnology, and rabbit polyclonal anti-ANKRD1 antibody (#11427-1-AP, 1:1000) was purchased from Proteintech. Monoclonal anti-β-actin antibody (#A5316, 1:30,000), rat tail collagen (#C3897) and anti HA-peroxidase (3F10, 1:1000) were purchased from Sigma-Aldrich. Goat anti-mouse (#170-6516) and goat anti-rabbit (#170-6515) secondary antibodies conjugated to horseradish peroxidase (HRP) were purchased from Bio-Rad. Doxycycline hydrochloride (#J67043) was purchased from Alfa Aesar. Mouse monoclonal anti-HA antibody (HA.11) (#MMS-101R, 1:1000) was purchased from Covance. Anti-GAPDH antibody (#GTX627408, 1:30,000) was purchased from GeneTex. Protein A-Sepharose CL-4B beads were from GE Healthcare. Mouse IgG (#010-0102, 1:1000) was purchased from Rockland Immunochemicals. Mouse anti-rabbit IgG, light chain specific (#211-032-171, 1:1000) and goat anti-mouse IgG, light chain specific (#115-035-174, 1:1000) antibodies were purchased from Jackson Immuno Research Laboratories.
Cell culture and ARRDC3 induction
MDA-MB-231 cells were maintained in Leibowitz-15 medium (#11415064, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (v/v). MDA-MB-231 HA–ARRDC3 wild-type and AAXA pSLIK cells were generated as previously described (Arakaki et al., 2018a) and cultured in Leibowitz-15 medium supplemented with 10% tetracycline-free FBS (#631101 from Takara Bio USA). All cell lines were purchased from ATCC and grown according to ATCC instructions. Cells were routinely tested for mycoplasma contamination. To induce ARRDC3 expression, MDA-MB-231 cells were treated with doxycycline for 48 h prior to the experiments.
Transfection with siRNA
siRNA transfections were performed using Oligofectamine (Life Technologies) according to the manufacturer's instructions. Signaling, migration and invasion assays described were performed 48 h after transfection. All single siRNAs were purchased from Qiagen: siRNA sequences and concentrations were as follows: non-specific (ns), 5′-CUACGUCCAGGAGCGCACC-3′ (25 nM); YAP #1 target sequence, 5′-AAGACATCTTCTGGTCAGAGA-3′ (25 nM); YAP #5 target sequence, 5′-CAGGTGATACTATCAACCAAA-3′ (25 nM); TAZ #1 target sequence, 5′-CTGCGTTCTTGTGACAGATTA-3′ (25 nM); TAZ #4 target sequence, 5′-ACAGTAGTACCAAATGCTTTA-3′ (25 nM); ALIX #1 target sequence, 5′-AAGTACCTCAGTCTATATTGA-3′ (12.5 nM); and ALIX #3 target sequence, 5′-AATCGAGACGCTCCTGAGATA-3′ (12.5 nM). For all conditions, cells were transfected with one siRNA, with the exception of ALIX, which was treated with ALIX #1 and ALIX #3 siRNAs.
Immunofluorescence confocal microscopy
Cells were plated at a density of 3×105 cells per well on fibronectin-coated glass coverslips placed in a 12-well dish, grown overnight and serum starved overnight to yield ∼80% confluence. Cells were treated with 2 mM leupeptin for 1 h at 37°C, stimulated with α-thrombin, and processed and imaged as previously described (Arakaki et al., 2018a). Slides were immunostained with anti-HA Alexa Fluor 488 conjugate antibody, with DAPI to stain nuclei and with either anti-YAP or anti-TAZ antibody. Confocal images were collected using an Olympus IX81 DSU spinning confocal microscope fitted with a Plan Apo 60× oil objective and a Hamamatsu ORCAER digital camera using Metamorph 7.7.4.0 software (Molecular Devices). The ratio of nuclear to cytoplasmic YAP and TAZ localization was quantified using the ImageJ intensity ratio nuclei cytoplasm tool on six to ten fields of view for each condition, from three biological independent replicates. When quantifying the percentage of cells displaying equal nuclear and cytoplasmic staining (N=C), cells showing 45–55% ratio nuclear:cytoplasmic were considered equal.
Signaling assays and immunoblotting
Signaling assays were performed essentially as described previously (Arakaki et al., 2018a). Briefly, 1.2×105 cells were seeded in a 24-well plate and grown overnight. Cells were then serum-starved overnight to yield ∼80% confluence then treated with agonist for the indicated times (Arakaki et al., 2018a). Whole-cell lysates were collected directly in 2× Laemmli sample containing 200 mM DTT and immunoblotted as previously described (Arakaki et al., 2018a). Samples were immunoblotted on separate gels to detect phospho-proteins and total proteins. Membranes were reprobed to assess protein expression of loading controls for normalization during quantification. Immunoblots were quantified by densitometry using ImageJ software.
Quantitative RT-PCR
Cells were plated in 6 cm dishes and grown overnight to yield ∼80% confluence. RNA was extracted using Direct-zol RNA Miniprep Plus kit (#R2072, Zymo Research) and the SuperScript IV VILO Master Mix with ezDNase enzyme kit (#11766050, ThermoFisher Scientific) was used for reverse transcription PCR, according to the manufacturer's instructions. Total RNA (750 ng) was used for RT-PCR. Quantitative PCR was performed using the TaqMan Fast Advanced Master Mix (#4444964, ThermoFisher Scientific) in technical triplicates. Three biological independent replicates were performed, and YAP and TAZ expression were normalized to S18 expression and compared with YAP expression in MDA-MB-231 cells. TaqMan Gene Expression Assay probes (Thermo Fisher Scientific) were as follows: YAP1, Hs00902712_g1; WWTR1 (TAZ), Hs00210007_m1; and 18S, Hs03003631_g1.
Immunoprecipitation
To assess the interaction of TEAD with YAP and TAZ, MDA-MB-231 pSLIK cells were plated in 10 cm dishes at 1.7×106 cells per dish. Cells were serum-starved overnight and treated with 10 nM α-thrombin for 30 min at ∼80% confluence. To assess the interaction of ARRDC3 with TAZ, HEK293T cells were plated in 6 cm dishes at 6.4×105 cells per dish and transiently transfected. After thrombin stimulation for MDA-MB-231 pSLIK or 48 h after transfection for HEK293T cells, cells were lysed with NP-40 lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, 10 μg/ml leupeptin, aprotinin, trypsin protease inhibitor, pepstatin, 100 μg/ml benzamide, 1 mM PMSF, 50 mM β-glycerophosphate, 20 mM NaF, 1 mM Na3VO4 and 20 mM N-ethylmaleimide]. Cell lysates were homogenized using a needle and syringe, and cleared by centrifugation. Supernatants were pre-cleared with Protein A-Sepharose beads and protein concentrations determined using a BCA assay (Thermo Fisher Scientific). Equal amounts of normalized lysates were immunoprecipitated with appropriate antibodies overnight at 4°C followed by incubation of BSA-blocked Protein-A beads (GE Healthcare) for 2 h. Immunoprecipitates were washed and eluted in 2× Laemmli sample buffer containing 200 mM DTT. Cell lysates and immunoprecipitates were analyzed by immunoblotting.
Migration and invasion assays
For ARRDC3 re-expression experiments, cells were seeded at 2×105 cells in 12-well dishes, grown overnight and serum-starved overnight to yield ∼80% confluence. Migration and invasion assays were performed essentially as previously described (Arakaki et al., 2018a). Cells were dissociated using Cellstripper solution (#25-056-Cl, Corning) and seeded onto rat tail collagen-coated migration chambers (#353097 from Corning) or BioCoat Matrigel invasion chambers (Corning) with or without α-thrombin or 0.5% FBS added. Cells were allowed to migrate or invade for 5 h at 37°C, fixed and stained with 0.5% Crystal Violet in ethanol. Membranes were dried overnight and cells that had migrated or invaded through the membrane were imaged using a Leica DMi1 inverted microscope. Cell migration and invasion were quantified by cell count in nine fields of view at 10× magnification for each condition, from three biological independent replicates.
Orthotopic mammary fat pad xenograft and a tail vein injection mouse model of metastasis
NSG mice were obtained from the University of California (San Diego, USA) in-house breeding colony and maintained in a pathogenic-free environment. For orthotopic mammary fat pad xenografts, 6- to 8-week-old female NSG mice received an orthotopic injection consisting of 1×106 MDA-MB-231 HA–ARRDC3 wild-type pSLIK cells resuspended in 50 µl PBS mixed with an equal volume of Matrigel into the inguinal mammary fat pad. Mice were randomized into two groups: one group was fed normal chow (control) and the other doxycycline chow (200 mg/kg doxycycline pellets, Fisher Scientific, #14727450) to induce ARRDC3 expression. The growth of mammary fat pad tumors was measured weekly with callipers. Tumors were excised from euthanized mice 6 weeks post-implantation, formalin-fixed and paraffin wax-embedded for sectioning. The UCSD Moores Cancer Center Tissue Technology Shared Resource performed the embedding, sectioning and Hematoxylin and Eosin (H&E) staining. For the tail-vein injection model, GFP-transduced MDA-MB-231 HA–ARRDC3 wild-type (4×105 cells in 50 µl PBS) or HA–ARRDC3 AAXA (3×105 cells in 50 µl PBS) pSLIK cells were pre-treated with or without doxycycline in vitro for 48 h then injected into the lateral tail vein of 5- to 8-week-old female NSG mice. Mice injected with cells that were pretreated with doxycycline were given doxycycline chow; mice not treated were given normal chow. Mice were euthanized 2 weeks post-injection; lungs were imaged and metastatic nodules counted with a Zeiss Axio Zoom.V16 microscope. All procedures were performed according to an animal protocol approved by the UC San Diego Institutional Animal Care and Use Committee.
Immunohistochemistry
Tumors formed from orthotopic mammary fat pad xenograft were formalin-fixed, paraffin-embedded and sectioned by the UCSD Moores Cancer Center Tissue Technology Shared Resource. Slides were baked at 50°C overnight then deparaffinated and rehydrated. Antigen retrieval was performed with Tris-EDTA (pH 9) containing 0.05% Tween-20 and permeabilization with 0.2% Triton X-100 in PBS. Slides were blocked with 2.5% normal horse serum and incubated with TAZ antibody (#72804 Cell Signaling Technology, 1:200 dilution) overnight at 4°C. Bound antibody was detected with Alexa Fluor 488-conjugated secondary antibody (1:500) and counterstained with 1 µM DAPI in PBS. Slides were mounted with Vectashield antifade mounting medium (H-1000, Vector Laboratories) and imaged on Nikon E800 microscope with Zeiss AxioCam HRm. Representative images of two tumors (four separate mice in total) are shown for each experimental condition.
Data analysis
Statistical significance was determined using an unpaired t-test, one-way ANOVA or two-way ANOVA with Tukey′s post hoc tests using Prism 8.0 software (GraphPad). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Supplementary Material
Acknowledgements
We thank all members of the Trejo laboratory for comments and advice. We are especially thankful to Dr Jing Yang (UC San Diego) for her advice in the experimental design for mouse xenograft experiments. We also thank Dr Richard Daneman for use of his lab's dissecting fluorescent microscope.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.K.S.A., W.-A.P., J.T.; Methodology: A.K.S.A., W.-A.P.; Validation: A.K.S.A., W.-A.P., H.W.; Formal analysis: A.K.S.A., W.-A.P., H.W., I.R.-M., L.C.; Investigation: A.K.S.A., W.-A.P., H.W., I.R.-M., L.C.; Resources: A.K.S.A., T.S.G., J.T.; Data curation: A.K.S.A.; Writing - original draft: A.K.S.A.; Writing - review & editing: A.K.S.A., W.-A.P., H.W., J.T.; Visualization: A.K.S.A., W.-A.P.; Supervision: A.K.S.A., T.S.G., J.T.; Project administration: A.K.S.A., J.T.; Funding acquisition: A.K.S.A., T.S.G., J.T.
Funding
This work was supported by National Institutes of Health (R35 GM127121 to J.T.) and The Ben and Catherine Ivy Foundation (to T.S.G.). A.K.S.A. was supported by a Howard Hughes Medical Institute Gilliam Pre-doctoral Fellowship and by the National Cancer Institute of the National Institutes of Health under Award Number T32CA080416. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/DOI/10.1242/jcs.254888/
References
- Adelaide, J., Finetti, P., Bekhouche, I., Repellini, L., Geneix, J., Sircoulomb, F., Charafe-Jauffret, E., Cervera, N., Desplans, J., Parzy, D.et al. (2007). Integrated profiling of basal and luminal breast cancers. Cancer Res. 67, 11565-11575. 10.1158/0008-5472.CAN-07-2536 [DOI] [PubMed] [Google Scholar]
- Arakaki, A. K. S., Pan, W.-A., Lin, H. and Trejo, J. (2018a). The α-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein–coupled receptor lysosomal sorting and signaling. J. Biol. Chem. 293, 3350-3362. 10.1074/jbc.RA117.001516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki, A. K. S., Pan, W.-A. and Trejo, J. (2018b). GPCRs in cancer: protease-activated receptors, endocytic adaptors and signaling. Int. J. Mol. Sci. 19, 1886. 10.3390/ijms19071886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, P., Cuevas, B. D., Russo, A., Johnson, G. L. and Trejo, J. (2008). Persistent transactivation of EGFR and ErbB2/HER2 by protease-activated receptor-1 promotes breast carcinoma cell invasion. Oncogene 27, 4434-4445. 10.1038/onc.2008.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartucci, M., Dattilo, R., Moriconi, C., Pagliuca, A., Mottolese, M., Federici, G., Benedetto, A. D., Todaro, M., Stassi, G., Sperati, F.et al. (2015). TAZ is required for metastatic activity and chemoresistance of breast cancer stem cells. Oncogene 34, 681-690. 10.1038/onc.2014.5 [DOI] [PubMed] [Google Scholar]
- Batista, T. M., Dagdeviren, S., Carroll, S. H., Cai, W., Melnik, V. Y., Noh, H. L., Saengnipanthkul, S., Kim, J. K., Kahn, C. R. and Lee, R. T. (2020). Arrestin domain-containing 3 (Arrdc3) modulates insulin action and glucose metabolism in liver. Proc. Natl. Acad. Sci. USA 117, 6733-6740. 10.1073/pnas.1922370117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierie, B., Pierce, S. E., Kroeger, C., Stover, D. G., Pattabiraman, D. R., Thiru, P., Liu Donaher, J., Reinhardt, F., Chaffer, C. L., Keckesova, Z.et al. (2017). Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. USA 114, E2337-E2346. 10.1073/pnas.1618298114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booden, M. A., Eckert, L. B., Der, C. J. and Trejo, J. (2004). Persistent signaling by dysregulated thrombin receptor trafficking promotes breast carcinoma cell invasion. Mol. Cell. Biol. 24, 1990-1999. 10.1128/MCB.24.5.1990-1999.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. W., Lim, C. J., Guo, K., Ng, C. P., Lee, I., Hunziker, W., Zeng, Q. and Hong, W. (2008). A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 68, 2592-2598. 10.1158/0008-5472.CAN-07-2696 [DOI] [PubMed] [Google Scholar]
- Cordenonsi, M., Zanconato, F., Azzolin, L., Forcato, M., Rosato, A., Frasson, C., Inui, M., Montagner, M., Parenti, A. R., Poletti, A.et al. (2011). The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759-772. 10.1016/j.cell.2011.09.048 [DOI] [PubMed] [Google Scholar]
- Díaz-Martín, J., López-García, M. A., Romero-Pérez, L., Atienza-Amores, M. R., Pecero, M. L., Castilla, M. A., Biscuola, M., Santón, A. and Palacios, J. (2015). Nuclear TAZ expression associates with the triple-negative phenotype in breast cancer. Endocr. Relat. Cancer 22, 443-454. 10.1530/ERC-14-0456 [DOI] [PubMed] [Google Scholar]
- Dores, M. R., Chen, B., Lin, H., Soh, U. J. K., Paing, M. M., Montagne, W. A., Meerloo, T. and Trejo, J. (2012). ALIX binds a YPX3L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J. Cell Biol. 197, 407-419. 10.1083/jcb.201110031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores, M. R., Lin, H., Grimsey, N. J., Mendez, F. and Trejo, J. (2015). The α-arrestin ARRDC3 mediates ALIX ubiquitination and G protein-coupled receptor lysosomal sorting. Mol. Biol. Cell 26, 4660-4673. 10.1091/mbc.E15-05-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores, M. R., Grimsey, N. J., Mendez, F. and Trejo, J. (2016). ALIX regulates the ubiquitin-independent lysosomal sorting of the P2Y1 purinergic receptor via a YPX3L motif. PLoS ONE 11, e0157587. 10.1371/journal.pone.0157587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draheim, K. M., Chen, H.-B., Tao, Q., Moore, N., Roche, M. and Lyle, S. (2010). ARRDC3 suppresses breast cancer progression by negatively regulating integrin β4. Oncogene 29, 5032-5047. 10.1038/onc.2010.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes, W. D., Smith, I. E. and Reis-Filho, J. S. (2010). Triple-negative breast cancer. N. Engl. J. Med. 363, 1938-1948. 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- Gong, G., Kwon, M. J., Han, J., Lee, H. J., Lee, S. K., Lee, J. E., Lee, S.-H., Park, S., Choi, J.-S., Cho, S. Y.et al. (2017). A new molecular prognostic score for predicting the risk of distant metastasis in patients with HR+/HER2- early breast cancer. Sci. Rep. 7, 45554. 10.1038/srep45554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, J. R. and Trejo, J. (2017). Challenges and opportunities in protease-activated receptor drug development. Annu. Rev. Pharmacol. Toxicol. 57, 349-373. 10.1146/annurev-pharmtox-011613-140016 [DOI] [PubMed] [Google Scholar]
- Hasdemir, B., Bunnett, N. W. and Cottrell, G. S. (2007). Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) mediates post-endocytic trafficking of protease-activated receptor 2 and calcitonin receptor-like receptor. J. Biol. Chem. 282, 29646-29657. 10.1074/jbc.M702974200 [DOI] [PubMed] [Google Scholar]
- Insel, P. A., Sriram, K., Wiley, S. Z., Wilderman, A., Katakia, T., McCann, T., Yokouchi, H., Zhang, L., Corriden, R., Liu, D.et al. (2018). GPCRomics: GPCR expression in cancer cells and tumors identifies new, potential biomarkers and therapeutic targets. Front. Pharmacol. 9, 431. 10.3389/fphar.2018.00431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, Y., Vinayagam, A., Sun, X., Dephoure, N., Gygi, S. P., Hong, P. and Perrimon, N. (2013). The Hippo signaling pathway interactome. Science 342, 737-740. 10.1126/science.1243971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar, J. M., Stern, P., Liu, H., Schindler, J. W., Jiang, Z.-G. and Hynes, R. O. (2012). The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. USA 109, E2441-E2450. 10.1073/pnas.1212021109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Q.-Y., Zhang, H., Zhao, B., Zha, Z.-Y., Bai, F., Pei, X.-H., Zhao, S., Xiong, Y. and Guan, K.-L. (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28, 2426-2436. 10.1128/MCB.01874-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, D. J., Chen, M., Tram, T. and Coughlin, S. R. (1996). Agonist recognition by proteinase-activated receptor 2 and thrombin receptor. Importance of extracellular loop interactions for receptor function. J. Biol. Chem. 271, 13943-13947. 10.1074/jbc.271.24.13943 [DOI] [PubMed] [Google Scholar]
- Lin, S., Zhang, G., Zhao, Y., Shi, D., Ye, Q., Li, Y. and Wang, S. (2020). Methylation and serum response factor mediated in the regulation of gene ARRDC3 in breast cancer. Am. J. Transl. Res. 12, 1913-1927. [PMC free article] [PubMed] [Google Scholar]
- Ma, S., Meng, Z., Chen, R. and Guan, K.-L. (2019). The Hippo pathway: biology and pathophysiology. Annu. Rev. Biochem. 88, 577-604. 10.1146/annurev-biochem-013118-111829 [DOI] [PubMed] [Google Scholar]
- McAuley, J. R., Bailey, K. M., Ekambaram, P., Klei, L. R., Kang, H., Hu, D., Freeman, T. J., Concel, V. J., Hubel, N. E., Lee, J.-Y. L.et al. (2019). MALT1 is a critical mediator of PAR1-driven NF-κB activation and metastasis in multiple tumor types. Oncogene 38, 7384-7398. 10.1038/s41388-019-0958-4 [DOI] [PubMed] [Google Scholar]
- Meng, Z., Moroishi, T. and Guan, K.-L. (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1-17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, J.-S., Yu, F.-X., Gong, R., Brown, J. H. and Guan, K.-L. (2012). Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs). Genes Dev. 26, 2138-2143. 10.1101/gad.197582.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, J. K., Rudina, T., Maoz, M., Grisaru-Granovsky, S., Uziely, B. and Bar-Shavit, R. (2018). Cancer driver G-protein coupled receptor (GPCR) induced β-catenin nuclear localization: the transcriptional junction. Cancer Metastasis Rev. 37, 147-157. 10.1007/s10555-017-9711-z [DOI] [PubMed] [Google Scholar]
- Piccolo, S., Dupont, S. and Cordenonsi, M. (2014). The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94, 1287-1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Plouffe, S. W., Lin, K. C., Moore, J. L., III, Tan, F. E., Ma, S., Ye, Z., Qiu, Y., Ren, B. and Guan, K.-L. (2018). The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J. Biol. Chem. 293, 11230-11240. 10.1074/jbc.RA118.002715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, S., O'Hayre, M., Gutkind, J. S. and Hurley, J. H. (2014). Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3). J. Biol. Chem. 289, 4743-4752. 10.1074/jbc.M113.527473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch, S. and Martin-Serrano, J. (2011). Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J. Virol. 85, 3546-3556. 10.1128/JVI.02045-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, H., Gonzalez-Cabrera, P., Marsolais, D., Cahalan, S., Don, A. S. and Sanna, M. G. (2008). Modulating tone: the overture of S1P receptor immunotherapeutics. Immunol. Rev. 223, 221-235. 10.1111/j.1600-065X.2008.00645.x [DOI] [PubMed] [Google Scholar]
- Shen, X., Sun, X., Sun, B., Li, T., Wu, G., Li, Y., Chen, L., Liu, Q., Cui, M. and Zhou, Z. (2018). ARRDC3 suppresses colorectal cancer progression through destabilizing the oncoprotein YAP. FEBS Lett. 592, 599-609. 10.1002/1873-3468.12986 [DOI] [PubMed] [Google Scholar]
- Soung, Y. H., Pruitt, K. and Chung, J. (2014). Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Sci. Rep. 4, 3846. 10.1038/srep03846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung, Y. H., Chung, H., Yan, C., Ju, J. and Chung, J. (2019). Arrestin domain containing 3 reverses epithelial to mesenchymal transition and chemo-resistance of TNBC cells by up-regulating expression of miR-200b. Cells 8, 692. 10.3390/cells8070692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese, S. and Moberg, K. (2020). Roles of membrane and vesicular traffic in regulation of the Hippo pathway. Front. Cell Dev. Biol. 7, 384. 10.3389/fcell.2019.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, S. A., Beli, P., Weinert, B. T., Schölz, C., Kelstrup, C. D., Young, C., Nielsen, M. L., Olsen, J. V., Brakebusch, C. and Choudhary, C. (2012). Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics 11, 1578-1585. 10.1074/mcp.M112.017905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, V., Yeerna, H., Nohata, N., Chiou, J., Harismendy, O., Raimondi, F., Inoue, A., Russell, R. B., Tamayo, P. and Gutkind, J. S. (2019). Illuminating the Onco-GPCRome: Novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 294, 11062-11086. 10.1074/jbc.REV119.005601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J., Shi, Q., Li, W., Mu, X., Peng, J., Li, M., Chen, M., Huang, H., Wang, C., Gao, K.et al. (2018). ARRDC1 and ARRDC3 act as tumor suppressors in renal cell carcinoma by facilitating YAP1 degradation. Am. J. Cancer Res. 8, 132-143. [PMC free article] [PubMed] [Google Scholar]
- Yu, F.-X., Zhao, B., Panupinthu, N., Jewell, J. L., Lian, I., Wang, L. H., Zhao, J., Yuan, H., Tumaneng, K., Li, H.et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780-791. 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Wei, X., Li, W., Udan, R. S., Yang, Q., Kim, J., Xie, J., Ikenoue, T., Yu, J., Li, L.et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747-2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Wang, S., Wang, Z., Feng, X., Liu, P., Lv, X.-B., Li, F., Yu, F.-X., Sun, Y., Yuan, H.et al. (2015). Estrogen regulates Hippo signaling via GPER in breast cancer. J. Clin. Investig. 125, 2123-2135. 10.1172/JCI79573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Wei, J., Dong, S., Bowser, R. K., Zhang, L., Jacko, A. M. and Zhao, Y. (2016). Destabilization of Lysophosphatidic acid receptor 1 reduces cytokine release and protects against lung injury. EBioMedicine 10, 195-203. 10.1016/j.ebiom.2016.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.