ABSTRACT
Nuclear Ca2+ has emerged as one of the most potent mediators of the dialogue between neuronal synapses and the nucleus that regulates heterochromatin states, transcription factor activity, nuclear morphology and neuronal gene expression induced by synaptic activity. Recent studies underline the importance of nuclear Ca2+ signaling in long-lasting, activity-induced adaptation and maintenance of proper brain function. Diverse forms of neuroadaptation require transient nuclear Ca2+ signaling and cyclic AMP-responsive element-binding protein (CREB1, referred to here as CREB) as its prime target, which works as a tunable switch to drive and modulate specific gene expression profiles associated with memory, pain, addiction and neuroprotection. Furthermore, a reduction of nuclear Ca2+ levels has been shown to be neurotoxic and a causal factor driving the progression of neurodegenerative disorders, as well as affecting neuronal autophagy. Because of its central role in the brain, deficits in nuclear Ca2+ signaling may underlie a continuous loss of neuroprotection in the aging brain, contributing to the pathophysiology of Alzheimer's disease. In this Review, we discuss the principles of the ‘nuclear calcium hypothesis’ in the context of human brain function and its role in controlling diverse forms of neuroadaptation and neuroprotection. Furthermore, we present the most relevant and promising perspectives for future studies.
KEY WORDS: Alzheimer's disease, Autophagy, CREB signaling, Epigenetics, Lysosome, Nuclear calcium
Summary: This Review discusses the ‘nuclear calcium hypothesis’ in the context of human brain function, the role of nuclear Ca2+ in controlling diverse forms of neuroadaptation and neuroprotection, and its dysregulation in disease.
Introduction
Ca2+ ions influence numerous signaling pathways within the vast majority of cells and thus impact nearly every aspect of life (Berridge et al., 2000). In order to adapt to changing intra- and extra-cellular conditions, cells must induce specific signaling cascades to regulate and integrate crucial cellular processes, including proliferation, differentiation, migration and cell death (Carrión et al., 1999; Ivanova et al., 2017; Sammels et al., 2010). The spatiotemporal distribution of Ca2+ ions is tightly controlled and contributes to the versatility of this second messenger, allowing fast and efficient genomic responses that are required for the adaptation to continuously changing conditions (Berridge et al., 2000; Carrión et al., 1999; Ivanova et al., 2017; Newton et al., 2016; Sammels et al., 2010). Due to the underlying chemistry, the importance and versatility of Ca2+ ions influence local electrostatic fields, thus altering the structure of thousands of proteins upon forming stable complexes, as compared to monovalent ions such as Na+ or K+ (Clapham, 2007). Additionally, the large and complex electron shell of a Ca2+ ion allows the formation of tight and irregular coordination bonds that are characteristic of biomolecules, unlike smaller chemical relatives such as Mg2+ (Carafoli et al., 2001; Clapham, 2007). These Ca2+-driven conformational modifications of proteins enable its function as a second messenger and, depending on the source of Ca2+ release and its destination, can drive local or long distance changes in cellular processes (Hardingham et al., 1997). The role of nuclear Ca2+ is well documented in the nervous system, where its role in the regulation of processes responsible for neuroadaptations and neuronal homeostasis is crucial (Hardingham et al., 1997, 2001). In recent years, the focus has turned to the cell nucleus as a unique location of action for Ca2+ ions, and robust, genetically encoded tools for the measurement (using GCaMP–NLS, a Ca2+ sensor with a nuclear localization signal) and depletion (using the Ca2+/calmodulin-binding polypeptide CaMBP4 or parvalbumin targeted to the cell nucleus) of nuclear Ca2+ have been developed (Bading, 2013). Activity-driven changes in nuclear Ca2+ levels alter the neuronal gene expression program that is responsible for adaptive responses, including neuroprotection, chronic pain, memory formation and memory extinction, and allow cells to rapidly adjust to a constantly changing environment (Hemstedt et al., 2017; Limbäck-Stokin et al., 2004; Papadia et al., 2005; Simonetti et al., 2013; Weislogel et al., 2013; Zhang et al., 2009). The ‘nuclear calcium hypothesis’, first proposed 20 years ago, emphasizes the role of Ca2+ flux into the nucleus and stresses its importance as a modulator of evolutionarily conserved pathways that are often dysregulated in pathological states (Bading, 2000). In this Review, we aim to provide a summary of recent findings in the field of nuclear Ca2+ and strengthen the ‘nuclear calcium hypothesis’ in the context of a regulation of pivotal cellular processes in the nervous system.
Reservoirs and sources of nuclear Ca2+
Along with the large excess of Ca2+ in the extracellular space, subcellular compartments constitute a second main store of Ca2+ ions in the cellular environment (Berridge et al., 2000). Organelles, such as lysosomes, peroxisomes, mitochondria, endoplasmic reticulum (ER) and the Golgi, are well established as suppliers of Ca2+ in response to different stimuli (Joseph et al., 2019; Lloyd-Evans and Waller-Evans, 2020; Vandecaetsbeek et al., 2011; Wacquier et al., 2019). Transient sequestration inside these structures provides a dynamic spatiotemporal distribution of Ca2+ that facilitates efficient and specific signal transduction. In mammalian cells, Ca2+ is released from intracellular stores via specialized channels, including inositol (1,4,5)-trisphosphate receptors (IP3Rs), ryanodine receptors (RyRs), two-pore channels (TPCs), Na+/Ca2+ exchangers (NCXs) and sarcoendoplasmic reticulum Ca2+-ATPase pumps (SERCAs) (Blaustein and Lederer, 1999; Galione, 2019; Lanner et al., 2010; Prole and Taylor, 2019; Toyoshima, 2009; Vangeel and Voets, 2019). The shared lumen of the ER, the nucleoplasmic reticulum and the nuclear envelope (NE) constitutes the most abundant Ca2+ store and can release ions in response to different molecular triggers (Bootman and Bultynck, 2020). The NE does not represent a measurable physical barrier to shuttling of Ca2+ between the cytoplasm and nucleoplasm, thereby allowing efficient crosstalk to maintain synapse-to-nucleus communication (Bengtson et al., 2010; Eder and Bading, 2007). The IP3Rs and RyRs, located on the outer nuclear membrane (ONM), are involved in the regulation of nuclear Ca2+ levels (Echevarría et al., 2003; Humbert et al., 1996; Laflamme et al., 2002; Marius et al., 2006).
The NE exhibits invaginations that can reach deep into the nucleus and contribute to various cellular processes, such as genome organization, transcription and stress responses (Heessen and Fornerod, 2007; Ruault et al., 2008; Wittmann et al., 2009). These NE invaginations are a common feature of many cell types, including neurons (Jorgens et al., 2017; Malhas et al., 2011; Wittmann et al., 2009). Nuclear invaginations generally are divided into two types (I and II), which fulfill distinct functions in the crosstalk between the nucleus, the nucleoplasmic reticulum and the cytosol (Malhas et al., 2011). Type I nuclear invaginations are folds of the inner nuclear membrane (INM) that increase the surface area of the nucleoplasmic reticulum, whereas type II nuclear invaginations are lined by both membranes (INM and ONM); they increase the surface area of the cell nucleus, contain nuclear pore complexes (NPCs) and often connect to nucleoli to promote rRNA export (Bourgeois et al., 1979; Fricker et al., 1997; Schoen et al., 2017; Wittmann et al., 2009). In addition to rRNA and small molecules (such as ATP, nucleotides and ions), the prevailing functional model of passive transport (the ‘rigid barrier’ model) also defines macromolecules of size less than 40 kDa as able to enter or exit the nuclear space via passive diffusion (Knockenhauer and Schwartz, 2016; Timney et al., 2016). Larger macromolecules require specific transport sequences, namely a nuclear localization signal (NLS) or a nuclear export signal (NES), which are recognized by specific transport proteins responsible for shuttling the cargo macromolecules through the NPC (Cautain et al., 2015). Conversely, an interesting hypothesis has been posited based on observations of an unexpected mode of Ca2+ flux through NPCs that, rather than being dependent on the large central NPC lumen, is instead dependent on small peripheral pores that are sensitive to ATP and Ca2+ (Shahin et al., 2001). In the same study, blockage of passive transport through NPCs did not affect electrical conductance in isolated Xenopus laevis nuclei, and electron microscopy analyses revealed the formation of additional symmetrically arranged small pores around the main lumen of NPCs (Shahin et al., 2001). Although this mode of Ca2+ diffusion needs further validation, the presence of peripheral ATP-dependent pores in NPCs has been confirmed in human fibroblasts (Maimon et al., 2012), providing an additional level of complexity to our understanding of diffusion of Ca2+ into the nucleus (Gerasimenko and Gerasimenko, 2004; Rodrigues et al., 2009). Furthermore, synaptic activity leads to drastic nuclear morphology changes, creating a network of nuclear invaginations that penetrate deep into the nuclear interior, allowing for release of Ca2+ deep into the nucleus (Wittmann et al., 2009).
Additionally, other pumps and channels on the NE, such as big potassium channels (BKs) (Li et al., 2014) or the NCX that couples Ca2+ efflux and influx to the influx and efflux of Na+ (Blaustein and Lederer, 1999), have been implicated in nuclear Ca2+ signaling (Secondo et al., 2018, 2020). However, it is important to consider that NPCs span two lipid bilayers of the NE, allowing direct exchange between the cytosol and nucleoplasm, unlike transport through single-membrane gates in the NE formed by RyR or IP3R (Fig. 1B). Therefore, NPCs are considered to be the main site for Ca2+ entry into the nucleus (Bading, 2013).
Fig. 1.
Regulation of nuclear Ca2+-dependent gene expression in a neuronal cell. (A) Levels of cytosolic Ca2+ can be increased as a result of synaptic inputs such as glutamate. (I) Glutamate activates NMDARs, AMPA receptors and kainate-type glutamate receptors at the synapse, giving rise to an excitatory postsynaptic potential that may initiate an action potential (AP). (II) APs activate VGCCs in the axon, resulting in Ca2+ entry from the extracellular space into the cytosol. (III) Stimulation of GPCRs by glutamate triggers production of IP3, which in turn activates the release of Ca2+ from the ER through IP3Rs, resulting in a propagation of intercellular Ca2+ waves. (IV) Activation of RyRs additionally amplifies Ca2+ release from the ER or the Golgi through Ca2+-induced Ca2+ release. (B) Ca2+ enters the nucleus by two main routes, either directly from the ER or from the cytoplasm. Ca2+ ions from the ER may enter the nucleoplasm (for example via BK channels). The second route of Ca2+ ions depends on type II nuclear invaginations, which are composed of both INM and ONM. These invaginations are enriched in NPCs, allowing for effective entry of Ca2+ into the nucleus. While IP3R, BK and RyR channels allow efflux of Ca2+ from the ER into the cytosol, NPCs facilitate Ca2+ transport into the nucleus. Nuclear Ca2+ triggers activation of CaMKII and CaMKIV, which in turn modulate epigenetic and transcriptional programs, triggering defined gene expression crucial for proper neuronal function. Figure contains images adapted from Servier Medical Art (http://smart.servier.com) under the terms of a CC BY 3.0 license.
In parallel with the abovementioned mechanisms, proper Ca2+ flux is additionally controlled by the buffering action of the ER, lysosomes and mitochondria (Berridge et al., 2000; Raffaello et al., 2016). Importantly, mitochondrial Ca2+ overload is associated with neuronal death in a mouse model of Alzheimer's disease (AD) (Britti et al., 2020), and impaired mitochondrial Ca2+ efflux contributes to disease progression in multiple experimental models of neurodegenerative diseases as a result of the dysregulation of the mitochondrial Na+/Ca2+ exchanger NCLX (also known as SLC8B1; Jadiya et al., 2019; Kostic et al., 2015; Verma et al., 2017). Our understanding of the mechanisms underlying mitochondrial Ca2+ regulation in neurodegeneration is expanding quickly, and these mechanisms have been reviewed in detail elsewhere (Giorgi et al., 2018); however, a link to nuclear Ca2+ signaling has not yet been made, and this remains to be assessed in future studies.
The interplay of all these abovementioned channels maintains a high capacity for Ca2+ transport into the nucleus that can be seen as a buffering compartment providing rapid recovery from high Ca2+ concentrations in the ER, mitochondria or cytosol in order to protect cells from harmful Ca2+ overload (Annunziato et al., 2004; Carafoli, 1988). The term ‘calcium signaling toolkit’, which refers to all the Ca2+ channels, pumps, exchangers and signaling components, was coined two decades ago (Berridge et al., 2000). Based on more recent findings, in our opinion it seems reasonable to suggest that the ‘nuclear calcium signaling toolkit’ is an independent set of components dedicated exclusively to proteins that are involved in the regulation of nuclear Ca2+ (Bading, 2013). Nevertheless, this concept may appear somewhat simplified in the context of Ca2+ signaling, as it has been shown that the ‘calcium signaling toolkit’ differs between brain regions, especially in pathological conditions such as AD (Grolla et al., 2013).
Nuclear versus cytosolic Ca2+
An increase in cytosolic Ca2+ in response to synaptic activity can be achieved by several means and is known to be relayed to the nucleus (Fig. 1A; Box 1). Glutamate release from presynaptic terminals activates N-methyl-D-aspartate receptors (NMDARs) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors and kainate-type receptors on the postsynaptic site, which allow influx of Na+ and smaller amounts of Ca2+ (Fig. 1A, inset I). The influx of these ions evokes the so-called excitatory postsynaptic potential, a voltage that initiates action potentials if the depolarization at the axon hillock reaches a threshold (Bading, 2013; Bengtson et al., 2010). An action potential at the hillock propagates back to the axon and into the dendrites, causing opening of voltage-gated Ca2+ channels (VGCCs) and, consequently, Ca2+ influx into the cytoplasm (Fig. 1A, inset II; Bengtson et al., 2013).
Box 1. Nuclear Ca2+ in synaptic plasticity.
Proper neuronal signaling requires adequate Ca2+ flux, which is controlled by plasma membrane receptors and voltage-dependent ion channels (Bading, 2013; Brini et al., 2014; see Fig. 1). Whereas Na+ and K+ are used to generate short-lived action potentials (of ∼1 ms), Ca2+-based action potentials last far longer (beyond 100 ms) and are prone to Ca2+ buildup. Especially after prolonged and repeated stimulation, Ca2+ accumulates in the cytosol and the nucleus, driving gene transcription and contributing to the late phase of long-term potentiation or depression of synaptic efficacy, a process that may underlie learning and that is altered in neurodegenerative diseases (Bading, 2013). Epigenetic changes and activation of specific transcriptional programs caused by flux of Ca2+ ions into the nucleus (see Fig. 2) are two molecular processes thought to be involved in LTM, affecting long-term potentiation and long-term depression. Whereas long-term depression produces a long-lasting decrease in synaptic strength, long-term potentiation refers to a long-lasting (up to several days) increase in synaptic efficacy that involves relocalization of ion channels and transcriptional changes required for the consolidation of information storage and LTM formation (Cooke and Bliss, 2006). The development of molecular tools to specifically interfere with nuclear Ca2+ signaling, such as CaMBP4 (Wang et al., 1995) or a nuclear-targeted version of the Ca2+-binding protein parvalbumin, has led to key discoveries demonstrating that the late phase of long-term potentiation and formation of LTM, as well as memory extinction, require nuclear Ca2+ signaling in mice (Hemstedt et al., 2017; Limbäck-Stokin et al., 2004) and in Drosophila (Weislogel et al., 2013). These findings thus identify nuclear Ca2+ signaling as an evolutionarily conserved mechanism that controls memory consolidation and higher-order brain functions. In contrast, short-term memory (defined to last in the order of minutes) does not require gene transcription or nuclear Ca2+ signaling (Berridge, 2014; Gibbs et al., 1979; Hemstedt et al., 2017; Limbäck-Stokin et al., 2004).
Ca2+ release into the cytoplasm can also be achieved through the activation of G-protein-coupled receptors (GPCRs) that respond to a variety of agonists, described in detail elsewhere (Huang and Thathiah, 2015). GPCR stimulation results in the activation of phospholipase C (PLC), which hydrolyzes phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] located in the plasma membrane to generate two second messengers, inositol trisphosphate (IP3) and diacylglycerol (DAG). IP3 then activates IP3Rs to induce Ca2+ release into cytosol (Fig. 1A, inset III), which can be further amplified by Ca2+-induced Ca2+ release (CICR) through RyRs (Fig. 1A, inset IV). While it is widely accepted that nuclear Ca2+ transients elicit a biological response different to that elicited by cytosolic Ca2+ transients (Bading, 2013; Hardingham et al., 1997; Leite et al., 2003), it has been debated whether nuclear Ca2+ is regulated independently of cytosolic Ca2+ transients. For instance, it is known that the genomic events regulating the expression patterns of target genes in stimulated neurons are differentially controlled by nuclear versus cytosolic Ca2+ signals (Bading, 2013; Chawla et al., 1998; Mauceri et al., 2011). Differential gene expression in response to location of Ca2+ signal was first demonstrated by Hardingham et al. (1997), who showed that blocking nuclear Ca2+ with the non-diffusible Ca2+ chelator BAPTA and subsequent stimulation of L-type VGCCs results in expression of serum-response element (SRE) target genes but not those of cyclic AMP-responsive element-binding protein (CREB1, referred to here as CREB). These data suggest that SRE-mediated gene expression is controlled by an increase in cytosolic Ca2+, whereas CREB, which binds to the cAMP-response element (CRE), requires an increase in nuclear Ca2+ in order to stimulate its transcription-activating function (Hardingham et al., 1997). This study thus provided the first evidence for the role of nuclear Ca2+ as a regulator of gene expression through CREB, a major transcription factor in neurons that is implicated in neuronal plasticity and long-term memory (LTM) and whose loss has a role in the pathology of AD (Mayr and Montminy, 2001; Yin et al., 1995). CREB activity is regulated by multiple kinases, including protein kinase A (PKA) and Ca2+/calmodulin-dependent protein kinases (CaMKs), through phosphorylation on serine 133 and subsequent recruitment of the transcriptional co-activator CREB-binding protein (CBP) or its paralog p300 (also known as EP300) to CRE-containing promoters (Mayr and Montminy, 2001; Yin et al., 1994). Today, we know that nuclear Ca2+ can relay its signals through CaMKII and CaMKIV, which work together to mediate the effects of Ca2+ flux through L-type VGCCs and/or NMDARs in excitation–transcription coupling (Ma et al., 2014). CaMKIV is the principal regulator of gene transcription induced by synaptic activity (Bading, 2013). It is activated by a nuclear Ca2+–calmodulin complex leading to the phosphorylation of both CREB and CBP, which is required for activation of CREB and CBP target gene expression (Chawla et al., 1998; Hardingham et al., 1999; Hu et al., 1999; Impey et al., 2002). In addition to the classical view that nuclear Ca2+ influx occurs strictly by passive diffusion through NPCs (Fig. 1B), a role for Ca2+-mobilizing machinery in nucleoplasmic reticulum with a complex branched network of invaginations has been suggested for the generation of nuclear Ca2+ increases (Irvine, 2003; Malhas et al., 2011). Further findings indicating that the translocation of receptor tyrosine kinases (RTKs) following their activation can elicit nuclear Ca2+ transients suggest that nuclear Ca2+ signaling may be independent of cytosolic Ca2+ (De Miranda et al., 2019). In one study, the activation of c-Met (also known as MET) through its ligand hepatocyte growth factor (HGF) has been found to promote its nuclear translocation, which induces Ca2+ transients inside the nucleus (Gomes et al., 2008). Additionally, insulin receptor activation elicits a nuclear Ca2+ response through a similar mode of action in rat hepatocytes (Rodrigues et al., 2008). However, in both cases, it is possible that an initial Ca2+ increase is generated in the cytosol and subsequently transduced to the nucleus through NPCs. Generation of IP3 to induce nuclear Ca2+ release proceeds in the same manner as in the cytoplasm and is possibly due to availability of nuclear PLC isoforms, as well as PI(4,5)P2 and IP3R (Irvine, 2003). However, to our knowledge, compelling evidence for the localization of IP3R on the INM, for example based on immunoelectron microscopy studies, does not exist. Although a few studies suggest a self-sufficiency of nuclear Ca2+ regulation, there is thus far no convincing evidence for this claim. Conversely, a large body of work in healthy neurons has convincingly shown that influx of cytosolic Ca2+ transients into the nucleus directly regulates nuclear Ca2+ levels, which affect gene expression profiles through modifications of chromatin state, as discussed next.
Epigenetic changes in response to nuclear Ca2+ signaling
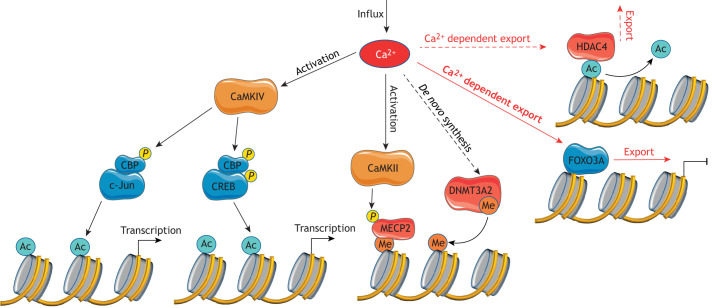
Over the past few decades, nuclear Ca2+ has been established as one of the main regulators of epigenetic changes in the cell. As a consequence of synaptic activity, elevated nuclear Ca2+ signaling induces chromatin remodeling, including DNA methylation and histone acetylation, which in turn regulate gene expression (Fig. 2), and has been linked to specific brain functions that are reviewed in detail elsewhere (Dulac, 2010; Fischle et al., 2003). One target of nuclear Ca2+ signaling is methyl CpG-binding protein 2 (MECP2, also known as MBD2), a protein that binds to methylated DNA and that is thought to act as a global chromatin modifier (Guy et al., 2011). Upon stimulation of synaptic activity, MECP2 is phosphorylated on a functionally relevant residue, serine 421. This phosphorylation event requires nuclear Ca2+ and is catalyzed by nuclear CaMKII (Buchthal et al., 2012). Although MECP2 binding to methylated DNA is traditionally associated with repression of transcription (Kinde et al., 2016; Nan et al., 1997), MECP2 has also been shown to act as a transcriptional activator, revealing a dual role as both a repressor and activator of gene expression (Chahrour et al., 2008; Horvath and Monteggia, 2018). Additionally, nuclear availability of the transcription factor forkhead box O3A (FOXO3A, also known as FOXO3), which plays a crucial role in cell death pathways, is also regulated through nuclear Ca2+-dependent export (Dick and Bading, 2010). As CBP interacts with many transcription factors (including CREB and c-Jun, also known as JUN) and regulatory proteins, nuclear Ca2+- and CaMKIV-driven modulation of CBP can influence a broad array of target genes in neurons, thereby regulating the proper functioning of the nervous system (Bading, 2000; Bedford et al., 2010; Cruzalegui et al., 1999). CBP contains a histone acetyltransferase (HAT) domain that is responsible for histone acetylation, which in turn initiates the induction of specific sets of target genes (Bading, 2000; Chawla et al., 1998; Vo and Goodman, 2001). Chromatin decondensation, which promotes gene expression, is additionally enhanced by the nuclear export of class IIa histone deacetylases (HDACs), which are directly regulated by a nuclear Ca2+-dependent mechanism in response to synaptic activity (Chawla et al., 2003; Schlumm et al., 2013). By removing acetyl residues from histones, which in most cases increases chromatin condensation leading to chromatin compaction, HDACs repress the expression of numerous genes and thus antagonize the HAT-dependent activity of CBP. Recently, two class IIa histone deacetylases, HDAC4 and HDAC5, have been shown to regulate expression of early response genes (ERGs) during associative learning, and this expression pattern is associated with proper synaptic architecture and regulation of learning and memory in mice (Zhu et al., 2019). HDAC4 is highly expressed in the brain and plays a pivotal role in the maintenance of cognitive functions (Bolger and Yao, 2005; Chawla et al., 2003; Darcy et al., 2010). There is growing interest in determining whether pharmacological inhibition of HDACs indeed correlates with an enhancement of LTM. There are reports that expression of a truncated, nuclear-restricted HDAC4 mutant results in deficits in LTM and hippocampal-dependent memory formation, indicating a possible non-nuclear role of HDAC4 in normal memory formation (Sando et al., 2012). This suggests that at least some genes required for LTM are subjected to HDAC4-mediated transcriptional repression in Drosophila (Fitzsimons et al., 2013) and in mouse models (Fitzsimons et al., 2013; Kim et al., 2012). In parallel to affecting histone acetylation and deacetylation, nuclear Ca2+ also contributes to epigenetic changes by inducing the expression of the de novo DNA methyltransferase, DNMT3A2, which triggers DNA methylation in response to neuronal activity (Oliveira et al., 2012). DNMT3A2 is linked to transcriptionally active euchromatin and has been shown to have a pivotal role in memory formation and consolidation by modulating numerous genes involved in synaptic plasticity and memory (Chen et al., 2002; Gulmez Karaca et al., 2020; Oliveira, 2016; Oliveira et al., 2016). A comprehensive transcriptome analysis has revealed that nuclear Ca2+ triggers the expression of a large pool of ∼185 genes (Zhang et al., 2009). This pool includes transcriptional regulators that control the following neuroadaptations: (1) memory, which is regulated by CREB, downstream regulatory element antagonist modulator (DREAM, also known as KCNIP3), CBP, MECP2 and class IIa HDACs; (2) acquired neuroprotection, which is regulated by CREB and class IIa HDACs; (3) addiction, which is regulated by CREB, CBP and MECP2; and (4) pain, which is regulated by CREB, CBP, MECP2 and DREAM. The target genes of these regulators are reviewed elsewhere (Bading, 2013; Zhang et al., 2009).
Fig. 2.
Epigenetic and transcriptional mechanisms triggered by nuclear Ca2+. Ca2+ influx into the nucleus modulates the activity and localization of several proteins that can influence chromatin organization, leading to transcriptional induction via CaMK activation (black arrows) or repression of specific target genes owing to Ca2+-dependent export of regulatory factors (red arrows). Transcriptional activity of chromatin is determined by the methylation (Me) and acetylation (Ac) of histones, which is accomplished by chromatin-modifying proteins, including CBP, DNMT3A2 and HDAC4. Ca2+-dependent activation of CaMKIV phosphorylates (P) CBP, which contains a histone acetyltransferase (HAT) domain, responsible for histone acetylation and thus chromatin decondensation. Histone acetylation can be enhanced by nuclear export of HDAC4, whose enzymatic activity antagonizes the action of CBP. Activated CBP can interact with transcription factors, such as c-Jun; this provides an additional mechanism to control transcription of target genes. Additionally, availability of the transcription factor FOXO3A, which is crucial for cell-death pathways, is also regulated through its nuclear Ca2+-dependent export. Nuclear Ca2+ also modulates DNA methylation that is linked to transcriptionally active euchromatin. CaMKII-dependent activity of DNMT3A2 or de novo synthesis of this protein, as well as phosphorylation of the transcriptional regulator MECP2, are strictly regulated by the level of nuclear Ca2+. Figure contains images adapted from Servier Medical Art (http://smart.servier.com) under the terms of a CC BY 3.0 license.
Furthermore, CREB has been found to regulate the induction of a variety of autophagic genes, including those encoding SESN2, unc-51-like autophagy activating kinase 1 (ULK1), autophagy related 7 (ATG7) and transcription factor EB (TFEB) (Lee et al., 2014; Seok et al., 2014). These findings suggest a functional link between nuclear Ca2+ and regulation of autophagy on a transcriptional level, as discussed below.
An emerging role for CREB in the regulation of cellular clearance
Cellular clearance is required for various physiological processes and its dysregulation is observed in many human diseases (Malik et al., 2019; Mizushima and Levine, 2020). Autophagy is a complex process that facilitates the constitutive turnover of intracellular components but also eliminates potentially damaging materials in response to stress conditions (Levine and Kroemer, 2019). Thus, the maintenance of cellular clearance driven by autophagy not only regulates crucial homeostatic and pro-survival mechanisms (He and Klionsky, 2009; Klionsky, 2007), but is also involved in dendritic retraction and synaptic pruning when hyperactive in pathological conditions (Levine and Kroemer, 2019). Among the numerous triggers of autophagy, damaged organelles, infection and periods of starvation are the most common (Kroemer et al., 2010; Lum et al., 2005). As autophagy is strictly dependent on lysosomal activity, which ultimately allows the degradation of cytosolic factors, the autophagosomal (auto)-lysosomal system is a major regulator of autophagic flux (Huber and Teis, 2016; Yim and Mizushima, 2020). As the pro-survival effect of autophagy can be dysregulated in pathological states, such as cancer and neurodegenerative diseases (Yang and Klionsky, 2010), pharmacological agents that can modulate autophagy in human pathophysiological conditions have been widely investigated (Rubinsztein et al., 2012). The postmitotic nature of mature neurons renders them particularly vulnerable to consequences of autophagy deregulation. As neurons and other terminally differentiated cells are unable to dilute toxic proteins and damaged organelles by cell division, they strictly depend on basal autophagy for the proper turnover of cytoplasmic contents (Hara et al., 2006). Autophagy is also crucial for both embryonic and adult neurogenesis, and is required for correct membrane turnover in the axons (Ha et al., 2017; Komatsu et al., 2007; Vázquez et al., 2012). While nuclear Ca2+ controls cellular clearance pathways largely at the transcriptional level, cytosolic Ca2+ regulates autophagy in a multitude of ways, and these have been recently reviewed elsewhere (Bootman et al., 2018). In general, high cytosolic Ca2+ stimulates autophagy, whereas Ca2+ chelators hamper autophagic flux (Brady et al., 2007; Gao et al., 2008; Høyer-Hansen et al., 2007; Sakaki et al., 2008).
Over the last few years, TFEB has emerged as a key regulator of autolysosomal clearance pathways. TFEB is regulated by the nutrient-sensing mechanistic target of rapamycin complex 1 (mTORC1) kinase complex that is located on lysosomal membranes (Inoki et al., 2003). When active, mTORC1 phosphorylates TFEB, allowing it to remain in the cytoplasm. During starvation, mTORC1 dissociates from lysosomal membranes and no longer phosphorylates TFEB, which now can translocate into the nucleus. Inside the nucleus, TFEB directly binds to promoter elements containing the so-called coordinated lysosomal expression and regulation (CLEAR) sequence, which regulates expression of lysosomal and autophagosomal genes (Sardiello et al., 2009; Settembre et al., 2011, 2012). Importantly, a more recently identified axis for the regulation of TFEB and autophagy activity in hepatic cells involving the farsenoid X receptor (FXR, also known as NR1H4) and CREB (FXR–CREB) has been shown to work independently of mTORC1 and to be slower and more persistent in the regulation of autophagy and activation of lysosomal gene expression than mTORC1 signaling (Lee et al., 2014; Seok et al., 2014). Notably, CREB upregulates crucial autophagy genes, such as ATG7, ULK1 and SESN2, whereas activated FXR opposes these effects by disrupting the functional complex between CREB and CREB-regulated transcription coactivator 2 (CRTC2) required for the CREB-dependent transcription (Seok et al., 2014). It is currently unknown whether a similar axis of autophagy regulation by CREB exists in cells of neuronal origin. Among the numerous studies highlighting the importance of CREB signaling and autophagy in neuroprotection, some reports suggest that induction of autophagy and CREB are strongly linked and are interdependent in a variety of murine models of neonatal hypoxia-ischemia brain injury and β-amyloid (Aβ)-induced neurodegeneration (Carloni et al., 2010; Singh et al., 2017). Although these studies attribute the observed effects of CREB to the expression of synaptic markers, neurotransmitter receptors and ion channels, rather than directly to autophagy, we expect that functional autophagy is induced under these conditions and drives neuroprotection through clearance of neurotoxic aggregates and defective organelles. Additional epistatic analyses are required to carefully assess the interplay between CREB and autophagy in these model systems. Overall, the above studies identify CREB as novel transcriptional activator of autophagy that may underlie its neuroprotective role and could constitute a promising pharmacological target for neurodegenerative diseases. However, it is important to consider that autophagy has a destructive potential and needs to be fine-tuned to avoid its hyperactivation leading to excessive synaptic pruning and worsening of the disease condition. To decipher the complex roles of nuclear Ca2+ and CREB in neuroprotection and conserved brain function, simpler model systems with a readily available genetic toolkit, such as Drosophila, offer clear advantages that are discussed below.
Nuclear Ca2+ signaling in long-term neuronal adaptations is evolutionarily conserved
Functional and morphological adaptations of synapses, dendrites and spines, as well as alterations in local and broader networks are essential features that underlie neuronal information processing and storage. In particular, adaptations underlying the formation of lasting memory are thought to involve functional and morphological changes within neuronal networks that result in a durable strengthening of certain communication pathways, while simultaneously depressing other pathways within the network (Bennett et al., 2018; Carasatorre and Ramirez-Amaya, 2012; Segal, 2017; Vogel-Ciernia and Wood, 2014). Neuronal adaptation is controlled by epigenetic changes and activation of specific transcriptional programs (Hsieh and Gage, 2005). The CREB family of transcription factors driven by nuclear Ca2+ is critical in controlling memory and neuronal adaptation in animals as divergent as mice, the sea slug Aplysia (Kandel, 2001) and Drosophila (Yin et al., 1994) (see Box 1). The suite of tools developed to control nuclear Ca2+ signaling in Drosophila in a temporally- and spatially-controlled manner has facilitated conceptual advances with regard to the function of nuclear Ca2+ transients (Guo et al., 2017; Jung et al., 2020; Pfeiffer et al., 2010). By restricting the expression of the nuclear Ca2+ blocker CaMBP4 to small subsets of distinct classes of neurons, termed Kenyon cells, within the mushroom body (MB) (Crittenden et al., 1998) – the region of the Drosophila brain that mediates learning and memory and contains ∼2000 Kenyon cells (Aso et al., 2014; de Belle and Heisenberg, 1994; Heisenberg et al., 1985; Modi et al., 2020) – the ∼600 γ-neurons (Aso et al., 2014) of the MB were found to be required for nuclear Ca2+ signaling (Weislogel et al., 2013). MB γ-neurons have recently been shown to play a major role in LTM consolidation by detecting spaced learning paradigms (as opposed to massed learning without spaces), as well as orchestrating activity and LTM-related gene expression in downstream neuronal networks (Awata et al., 2019). Furthermore, a pair of modulatory neurons, the anterior paired lateral neurons, which interact directly with MB γ-neurons to facilitate odor discrimination during olfactory learning (Lin et al., 2014), undergo synchronized synaptic pruning together with MB γ-neurons during brain development in Drosophila (Mayseless et al., 2018). This study showed that coordinated pruning of γ-neurons and anterior paired lateral neurons in the MB depends on the activity of MB γ-neurons and nuclear Ca2+ signaling, which controls expression of neuroprotective genes in the downstream anterior paired lateral neurons. Taken together, this work identifies nuclear Ca2+ as a key factor involved in coordinating changes to circuit architecture (Mayseless et al., 2018).
A recent study in Drosophila has reported that nuclear Ca2+ is depleted in neurons of the adult brain in the context of both aging and neurodegeneration induced by tau (also known as MAPT in mammals), and that nuclear Ca2+ depletion is a causal factor driving tau-induced neurotoxicity (for details see below; Mahoney et al., 2020). These findings add an important dimension to our fundamental understanding of brain aging and the ‘calcium hypothesis of AD’ (Khachaturian, 1984; Alzheimer's Association Calcium Hypothesis Workgroup, 2017). Future studies in Drosophila will no doubt contribute to a more complete understanding of the involvement of nuclear Ca2+ signaling in the neurophysiology and neuropathology.
Nuclear Ca2+-mediated neuroprotection and degeneration
The ‘calcium hypothesis of AD’, proposed in 1984 by Khachaturian, suggests a link between Ca2+ dyshomeostasis, neurodegeneration and AD pathophysiology (Khachaturian, 1984). Since then, numerous studies have investigated the role of Ca2+ in the pathogenesis of different neurological conditions, including AD, and these are reviewed elsewhere (Agostini and Fasolato, 2016; Galla et al., 2020; Kumar, 2020). The identification of nuclear Ca2+ signaling as a critical regulator of higher-order brain function points toward the potential involvement of dysregulated nuclear Ca2+ signaling in the etiology of diseases and mental disorders. While there is a vast literature focused on general Ca2+ dyshomeostasis in AD, the involvement of nuclear Ca2+ in the context of AD and related tauopathies has only recently been discovered (Mahoney et al., 2020; Reddy et al., 2016). These studies utilized genetically-encoded nuclear Ca2+ sensors (GCaMP3–NLS and GCaMP6–NLS) in induced pluripotent stem cell (iPSC)-derived neurons from patients with familial AD (FAD) (Reddy et al., 2016) and sporadic AD, as well as tau-transgenic Drosophila carrying human tau with an R406W mutation (Mahoney et al., 2020). iPSC-derived neurons with knockout of presenilin 1 (PS1) or with FAD-associated PS1 mutation show low levels of nuclear Ca2+, as well as reduced CaMKIV and CREB signaling (Reddy et al., 2016). Consequently, PS1-deficient cells have reduced expression of sestrin 2 (SESN2) (Reddy et al., 2016), a known target of nuclear Ca2+ signaling (Bading, 2013) and inhibitor of mTORC1 (Wolfson et al., 2016). These results causally link nuclear Ca2+ depletion, reduced CREB and SESN2 signaling to hyperactive mTORC1 activity in PS1-deficient cells, and also reveal that nuclear Ca2+ depletion aggravates autophagic and autolysosomal dysfunction as a consequence of mTORC1-driven inhibition of TFEB and its CLEAR gene network (Reddy et al., 2016). Furthermore, in human iPSC-derived neurons harboring an FAD PS1 mutation, SESN2 expression can be restored by pharmacologically elevating nuclear Ca2+, which in turn initiates TFEB-mediated clearance (Reddy et al., 2016). Other mechanisms through which PS1 modulates nuclear Ca2+ are possible and may involve its role as an ER-resident Ca2+ channel (Neely et al., 2011; Száraz et al., 2013; Tu et al., 2006). Importantly, TFEB activation effectively reduces aggregation of prion proteins, such as huntingtin, Aβ and phosphorylated tau (pTau), by enhancing lysosomal function, as shown in the HD N171-82Q mouse model of Huntington’s disease (Tsunemi et al., 2012), murine cortical primary astrocytes (Xiao et al., 2014) and in the rTg4510 mouse model of tauopathy (Polito et al., 2014). Additionally, in the mouse-model studies mentioned above, TFEB activity attenuates plaque pathogenesis and rescues behavioral deficits and neurodegeneration, making TFEB an attractive therapeutic target for treating AD and related tauopathies.
We have recently investigated the effects of pathogenic tau on nuclear Ca2+ signaling and CREB using a Drosophila model of tauopathy, as well as iPSC-derived neurons from patients with sporadic AD (Mahoney et al., 2020). RNA-sequencing analyses reveal a significant overrepresentation of CREB-regulated genes among differentially expressed transcripts (being either down- or up-regulated genes) in tau-transgenic Drosophila. Consistent with deregulation of CREB signaling in tau-expressing flies, we have observed that nuclear Ca2+ is depleted in neurons of tau-transgenic Drosophila and in iPSC-derived neurons from patients with sporadic AD, as determined by nuclear Ca2+ imaging. Tau-induced nuclear Ca2+ depletion appears to be a causal factor driving neurodegeneration, as further reduction of nuclear Ca2+ signaling through pan-neuronal expression of CaMBP4 significantly enhances tau-induced neurodegeneration in Drosophila (Mahoney et al., 2020). Interestingly, BK channels are thought to play a pivotal role in nuclear Ca2+ regulation, thus placing BK at the crossroads of nuclear Ca2+ and neuroprotection (Contet et al., 2016; Li and Gao, 2016). Indeed, we have found that pharmacological activation of BK channels elevates nuclear Ca2+ levels in brains of tau-transgenic Drosophila and significantly suppresses tau-induced neurotoxicity (Mahoney et al., 2020). These results suggest an involvement of BK channels in nuclear Ca2+ signaling (Li et al., 2014; Mahoney et al., 2020), timing and duration of K+ influx, and levels of local Ca2+ concentrations that impact neurotransmitter release (Contet et al., 2016; Wang et al., 2015a,b). Although the mechanism underlying tau-induced reduction of nuclear Ca2+ is unknown, we speculate that it could involve effects of pathogenic tau on nuclear architecture. Invaginations of the NE have been proposed to allow for targeted Ca2+ release to discrete regions of the nucleus (Marius et al., 2006), and neurons from postmortem human AD brains, brains of tau-transgenic Drosophila (Frost et al., 2016) and iPSC-derived neurons from patients with tauopathy (Paonessa et al., 2019) all feature such invaginations. In healthy neurons, synaptic activity induces dramatic changes in nuclear morphology that could be attributed to an increase in nuclear Ca2+ levels and associated signaling (Wittmann et al., 2009). In neurons from patients with AD, NE invaginations are lined with nuclear pores, suggesting that they could potentially deregulate nucleocytoplasmic trafficking of RNAs, proteins and other cellular factors, such as Ca2+ (Frost et al., 2016). Indeed, studies of tau-transgenic Drosophila reveal that tau-induced NE invaginations contain RNA, suggesting that they actively export and/or sequester RNA (Cornelison et al., 2019). Furthermore, altered nucleocytoplasmic transport in AD has been suggested to be a consequence of the direct binding of pathogenic forms of tau to the nucleoporin subunits of NPCs, thus disrupting their structural integrity (Eftekharzadeh et al., 2018). Based on these findings, we speculate that NE invaginations, which can be induced by synaptic activity in healthy neurons (Wittmann et al., 2009) or other means in disease conditions (Frost et al., 2016), lead to a redistribution of NPCs that could affect the nuclear Ca2+ dynamics. However, it is currently unknown whether the effects of tau on nuclear architecture are causally related to a disruption of nuclear Ca2+ signaling and to what extent low levels of nuclear Ca2+ drive AD pathology. To assess whether nuclear Ca2+ signaling plays a crucial role in the pathogenesis or progression of sporadic AD, CREB activity, target gene expression and nuclear morphology will need to be correlatively analyzed in human postmortem brain tissues from patients at different stages of the disease. In summary, we argue that a dysregulation of nuclear Ca2+ signaling is expected to trigger a cascade of events leading to the formation of known disease hallmarks and pathogenesis of neurological diseases (Fig. 3).
Fig. 3.
Health and disease conditions related to nuclear Ca2+ signaling. As a result of homeostatic synaptic and NMDAR activity, nuclear Ca2+ signaling promotes the expression of genes required for synapse maintenance, plasticity, molecular clearance and neuroprotection. In disease conditions, synaptic loss occurs as a result of dyshomeostatic activity, leading to decreased nuclear Ca2+ signaling. Reduced expression of nuclear Ca2+ target genes (including CREB, SESN2 and DREAM) leads to attenuated autophagic clearance and neuroprotection underlying memory impairment and neurodegeneration. Nuclear Ca2+ levels are likely to be reduced by other, synapse-independent mechanisms, such as a reduced permeability of nuclear pore complexes, that might be induced by accumulation of abnormal proteins or changes of nuclear morphology during cellular aging. Reduced nuclear Ca2+ signaling and subsequent expression of target genes further impairs autophagic lysosomal clearance and exacerbates the accumulation of abnormal proteins such as pTau, huntingtin or Aβ, and promotes further synaptic loss, leading to an aggravation of the pathological condition. ATG, autophagy-related protein; BECN1, Beclin-1; pCREB, phosphorylated CREB; ROS, reactive oxygen species; WIPI, human WD-repeat protein interacting with phosphoinositides.
Future perspectives
Recent studies exploring the roles of nuclear Ca2+ signaling in the brain bring us closer to deciphering its importance in proper brain function and its involvement in neurodegenerative diseases. In future studies, it will be important to work towards addressing a few key aspects of nuclear Ca2+ signaling, including the crosstalk between cytosolic and nuclear Ca2+ in health and disease. It is still not clear whether nuclear Ca2+ signaling can be fully autonomous and generated independently of activity-induced cytosolic Ca2+ transients. It is also currently unknown precisely how nuclear Ca2+ transients produce different epigenetic and transcriptional outcomes. While there are three obvious dimensions for such coding (i.e. space, time and strength), tools to monitor these parameters in vivo at high resolution are currently limited. Recent developments in Drosophila may provide more sensitive approaches for the imaging of nuclear events (Jung et al., 2020) that could potentially be combined with tools that allow quantification of Ca2+ transients in freely moving animals (Guo et al., 2017). The use of Drosophila and the many available disease-associated mutants to study nuclear Ca2+ at such high resolution will be helpful to assess the potential impact on disease onset and progression. However, as helpful as Drosophila and other models may be for the dissection of the mechanisms underlying (normal and altered) nuclear Ca2+ signaling and its downstream processes, these insights always need to be considered from an evolutionary perspective: apparent conservation of mechanisms between fly and man only suggests that a common ancestor of mammals and insects might have applied similar functional principles to solve a problem. How these ancient functional principles are implemented in modern animals after millions of years of independent evolution therefore requires careful analysis. For example, it will be important to investigate the etiology of the disrupted nuclear morphology present in AD-patient neurons and its relationship to dysregulated nuclear Ca2+ and CREB signaling; to uncover the mechanisms responsible for low levels of nuclear Ca2+, despite synaptic hyperactivity in early and mid-stages of AD; and to study the role of neurodegenerative disease-causing extrasynaptic NMDA receptors that antagonize the transcription-promoting activities of nuclear Ca2+ (Bading, 2017; Hardingham and Bading, 2010). Finally, the importance of nuclear Ca2+ signaling in many cellular processes may potentially be translated into therapies by targeting the key players responsible for its regulation, although suitable drugs that are able to increase nuclear Ca2+ levels are yet to be found.
Acknowledgements
Figures were created using Microsoft PowerPoint and images modified from Servier Medical Art (http://smart.servier.com). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License. We would like to thank Angelid Pabon, Anthony Williams, Viorica Raluca Contu and the other members of the laboratory for the discussion on figure design and current literature.
Footnotes
Competing interests
The authors declare no competing or financial interests.
References
- Agostini, M. and Fasolato, C. (2016). When, where and how? Focus on neuronal calcium dysfunctions in Alzheimer's Disease. Cell Calcium 60, 289-298. 10.1016/j.ceca.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Alzheimer's Association Calcium Hypothesis Workgroup (2017). Calcium Hypothesis of Alzheimer's disease and brain aging: a framework for integrating new evidence into a comprehensive theory of pathogenesis. Alzheimer's Demen. 13, 178-182.e17. 10.1016/j.jalz.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Annunziato, L., Pignataro, G. and Di Renzo, G. F. (2004). Pharmacology of brain Na+/Ca2+ exchanger: from molecular biology to therapeutic perspectives. Pharmacol. Rev. 56, 633-654. 10.1124/pr.56.4.5 [DOI] [PubMed] [Google Scholar]
- Aso, Y., Hattori, D., Yu, Y., Johnston, R. M., Iyer, N. A., Ngo, T.-T., Dionne, H., Abbott, L. F., Axel, R., Tanimoto, H.et al. (2014). The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577. 10.7554/eLife.04577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata, H., Takakura, M., Kimura, Y., Iwata, I., Masuda, T. and Hirano, Y. (2019). The neural circuit linking mushroom body parallel circuits induces memory consolidation in Drosophila. Proc. Natl. Acad. Sci. USA 116, 16080-16085. 10.1073/pnas.1901292116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading, H. (2000). Transcription-dependent neuronal plasticity: the nuclear calcium hypothesis. Eur. J. Biochem. 267, 5280-5283. 10.1046/j.1432-1327.2000.01565.x [DOI] [PubMed] [Google Scholar]
- Bading, H. (2013). Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 14, 593-608. 10.1038/nrn3531 [DOI] [PubMed] [Google Scholar]
- Bading, H. (2017). Therapeutic targeting of the pathological triad of extrasynaptic NMDA receptor signaling in neurodegenerations. J. Exp. Med. 214, 569-578. 10.1084/jem.20161673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford, D. C., Kasper, L. H., Fukuyama, T. and Brindle, P. K. (2010). Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics 5, 9-15. 10.4161/epi.5.1.10449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson, C. P., Freitag, H. E., Weislogel, J.-M. and Bading, H. (2010). Nuclear calcium sensors reveal that repetition of trains of synaptic stimuli boosts nuclear calcium signaling in CA1 pyramidal neurons. Biophys. J. 99, 4066-4077. 10.1016/j.bpj.2010.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson, C. P., Kaiser, M., Obermayer, J. and Bading, H. (2013). Calcium responses to synaptically activated bursts of action potentials and their synapse-independent replay in cultured networks of hippocampal neurons. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1833, 1672-1679. 10.1016/j.bbamcr.2013.01.022 [DOI] [PubMed] [Google Scholar]
- Bennett, S. H., Kirby, A. J. and Finnerty, G. T. (2018). Rewiring the connectome: evidence and effects. Neurosci. Biobehav. Rev. 88, 51-62. 10.1016/j.neubiorev.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge, M. J. (2014). Calcium regulation of neural rhythms, memory and alzheimer's disease. J. Physiol. 592, 281-293. 10.1113/jphysiol.2013.257527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge, M. J., Lipp, P. and Bootman, M. D. (2000). The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 1, 11-21. 10.1038/35036035 [DOI] [PubMed] [Google Scholar]
- Blaustein, M. P. and Lederer, W. J. (1999). Sodium/calcium exchange: its physiological implications. Physiol. Rev. 79, 763-854. 10.1152/physrev.1999.79.3.763 [DOI] [PubMed] [Google Scholar]
- Bolger, T. A. and Yao, T.-P. (2005). Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 25, 9544-9553. 10.1523/JNEUROSCI.1826-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman, M. D. and Bultynck, G. (2020). Fundamentals of cellular calcium signaling: a primer. Cold Spring Harb. Perspect. Biol. 12, a038802. 10.1101/cshperspect.a038802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman, M. D., Chehab, T., Bultynck, G., Parys, J. B. and Rietdorf, K. (2018). The regulation of autophagy by calcium signals: do we have a consensus? Cell Calcium 70, 32-46. 10.1016/j.ceca.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Bourgeois, C. A., Hemon, D. and Bouteille, M. (1979). Structural relationship between the nucleolus and the nuclear envelope. J. Ultrasructure Res. 68, 328-340. 10.1016/S0022-5320(79)90165-5 [DOI] [PubMed] [Google Scholar]
- Brady, N. R., Hamacher-Brady, A., Yuan, H. and Gottlieb, R. A. (2007). The autophagic response to nutrient deprivation in the hl-1 cardiac myocyte is modulated by Bcl-2 and sarco/endoplasmic reticulum calcium stores. FEBS J. 274, 3184-3197. 10.1111/j.1742-4658.2007.05849.x [DOI] [PubMed] [Google Scholar]
- Brini, M., Calì, T., Ottolini, D. and Carafoli, E. (2014). Neuronal calcium signaling: function and dysfunction. Cell. Mol. Life Sci. 71, 2787-2814. 10.1007/s00018-013-1550-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britti, E., Ros, J., Esteras, N. and Abramov, A. Y. (2020). Tau inhibits mitochondrial calcium efflux and makes neurons vulnerable to calcium-induced cell death. Cell Calcium 86, 102150. 10.1016/j.ceca.2019.102150 [DOI] [PubMed] [Google Scholar]
- Buchthal, B., Lau, D., Weiss, U., Weislogel, J.-M. and Bading, H. (2012). Nuclear calcium signaling controls methyl-CpG-binding protein 2 (MeCP2) phosphorylation on serine 421 following synaptic activity. J. Biol. Chem. 287, 30967-30974. 10.1074/jbc.M112.382507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli, E. (1988). Membrane transport of calcium: an overview. Methods Enzymol. 157, 3-11. 10.1016/0076-6879(88)57063-5 [DOI] [PubMed] [Google Scholar]
- Carafoli, E., Santella, L., Branca, D. and Brini, M. (2001). Generation, control, and processing of cellular calcium signals. Crit. Rev. Biochem. Mol. Biol. 36, 107-260. 10.1080/20014091074183 [DOI] [PubMed] [Google Scholar]
- Carasatorre, M. and Ramirez-Amaya, V. (2012). Network, cellular, and molecular mechanisms underlying long-term memory formation. In Neurogenesis and Neural Plasticity (ed. Belzung C. and Wigmore P.), pp. 73-115. Berlin Heidelberg: Springer. [DOI] [PubMed] [Google Scholar]
- Carloni, S., Girelli, S., Scopa, C., Buonocore, G., Longini, M. and Balduini, W. (2010). Activation of autophagy and Akt/CREB signaling play an equivalent role in the neuroprotective effect of rapamycin in neonatal hypoxia-ischemia. Autophagy 6, 366-377. 10.4161/auto.6.3.11261 [DOI] [PubMed] [Google Scholar]
- Carrión, A. M., Link, W. A., Ledo, F., Mellström, B. and Naranjo, J. R. (1999). DREAM is a Ca2+-regulated transcriptional repressor. Nature 398, 80-84. 10.1038/18044 [DOI] [PubMed] [Google Scholar]
- Cautain, B., Hill, R., De Pedro, N. and Link, W. (2015). Components and regulation of nuclear transport processes. FEBS J. 282, 445-462. 10.1111/febs.13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour, M., Jung, S. Y., Shaw, C., Zhou, X., Wong, S. T. C., Qin, J. and Zoghbi, H. Y. (2008). MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224-1229. 10.1126/science.1153252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla, S., Hardingham, G. E., Quinn, D. R. and Bading, H. (1998). CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science 281, 1505-1509. 10.1126/science.281.5382.1505 [DOI] [PubMed] [Google Scholar]
- Chawla, S., Vanhoutte, P., Arnold, F. J. L., Huang, C. L.-H. and Bading, H. (2003). Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85, 151-159. 10.1046/j.1471-4159.2003.01648.x [DOI] [PubMed] [Google Scholar]
- Chen, T., Ueda, Y., Xie, S. and Li, E. (2002). A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277, 38746-38754. 10.1074/jbc.M205312200 [DOI] [PubMed] [Google Scholar]
- Clapham, D. E. (2007). Calcium signaling. Cell 131, 1047-1058. 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- Contet, C., Goulding, S. P., Kuljis, D. A. and Barth, A. L. (2016). BK channels in the central nervous system. Int. Rev. Neurobiol . 128, 281-342. 10.1016/bs.irn.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, S. F. and Bliss, T. V. P. (2006). Plasticity in the human central nervous system. Brain 129, 1659-1673. 10.1093/brain/awl082 [DOI] [PubMed] [Google Scholar]
- Cornelison, G. L., Levy, S. A., Jenson, T. and Frost, B. (2019). Tau-induced nuclear envelope invagination causes a toxic accumulation of mRNA in Drosophila. Aging Cell 18, e12847. 10.1111/acel.12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden, J. R., Skoulakis, E. M. C., Han, K. A., Kalderon, D. and Davis, R. L. (1998). Tripartite mushroom body architecture revealed by antigenic markers. Learn. Mem. 5, 38-51. [PMC free article] [PubMed] [Google Scholar]
- Cruzalegui, F. H., Hardingham, G. E. and Bading, H. (1999). c-Jun functions as a calcium-regulated transcriptional activator in the absence of JNK/SAPK1 activation. EMBO J. 18, 1335-1344. 10.1093/emboj/18.5.1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcy, M. J., Calvin, K., Cavnar, K. and Ouimet, C. C. (2010). Regional and subcellular distribution of HDAC4 in mouse brain. J. Comp. Neurol. 518, 722-740. 10.1002/cne.22241 [DOI] [PubMed] [Google Scholar]
- de Belle, J. S. and Heisenberg, M. (1994). Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science 263, 692-695. 10.1126/science.8303280 [DOI] [PubMed] [Google Scholar]
- De Miranda, M. C., Rodrigues, M. A., De Angelis Campos, A. C., Faria, J. A. Q. A., Kunrath-Lima, M., Mignery, G. A., Schechtman, D., Goes, A. M., Nathanson, M. H. and Gomes, D. A. (2019). Epidermal growth factor (EGF) triggers nuclear calcium signaling through the intranuclear phospholipase Cδ-4 (PLCδ4). J. Biol. Chem. 294, 16650-16662. 10.1074/jbc.RA118.006961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, O. and Bading, H. (2010). Synaptic activity and nuclear calcium signaling protect hippocampal neurons from death signal-associated nuclear translocation of FoxO3a induced by extrasynaptic N-methyl-D-aspartate receptors. J. Biol. Chem. 285, 19354-19361. 10.1074/jbc.M110.127654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac, C. (2010). Brain function and chromatin plasticity. Nature 465, 728-735. 10.1038/nature09231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarría, W., Leite, M. F., Guerra, M. T., Zipfel, W. R. and Nathanson, M. H. (2003). Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat. Cell Biol. 5, 440-446. 10.1038/ncb980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder, A. and Bading, H. (2007). Calcium signals can freely cross the nuclear envelope in hippocampal neurons: somatic calcium increases generate nuclear calcium transients. BMC Neurosci. 8, 57. 10.1186/1471-2202-8-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekharzadeh, B., Daigle, J. G., Kapinos, L. E., Coyne, A., Schiantarelli, J., Carlomagno, Y., Cook, C., Miller, S. J., Dujardin, S., Amaral, A. S.et al. (2018). Tau protein disrupts nucleocytoplasmic transport in Alzheimer's Disease. Neuron 99, 925-940.e7. 10.1016/j.neuron.2018.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Wang, Y. and Allis, C. D. (2003). Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172-183. 10.1016/S0955-0674(03)00013-9 [DOI] [PubMed] [Google Scholar]
- Fitzsimons, H. L., Schwartz, S., Given, F. M. and Scott, M. J. (2013). The Histone Deacetylase HDAC4 regulates long-term memory in Drosophila. PLoS ONE 8, e83903. 10.1371/journal.pone.0083903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker, M., Hollinshead, M., White, N. and Vaux, D. (1997). Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J. Cell Biol. 136, 531-544. 10.1083/jcb.136.3.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, B., Bardai, F. H. and Feany, M. B. (2016). Lamin dysfunction mediates neurodegeneration in tauopathies. Curr. Biol. 26, 129-136. 10.1016/j.cub.2015.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galione, A. (2019). NAADP receptors. Cold Spring Harb. Perspect. Biol. 11, a035071. 10.1101/cshperspect.a035071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galla, L., Redolfi, N., Pozzan, T., Pizzo, P. and Greotti, E. (2020). Intracellular calcium dysregulation by the alzheimer's disease-linked protein presenilin 2. Int. J. Mol. Sci. 21, 770. 10.3390/ijms21030770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, W., Ding, W.-X., Stolz, D. B. and Yin, X.-M. (2008). Induction of macroautophagy by exogenously introduced calcium. Autophagy 4, 754-761. 10.4161/auto.6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimenko, O. and Gerasimenko, J. (2004). New aspects of nuclear calcium signalling. J. Cell Sci. 117, 3087-3094. 10.1242/jcs.01295 [DOI] [PubMed] [Google Scholar]
- Gibbs, M. E., Gibbs, C. L. and Ng, K. T. (1979). The influence of calcium on short-term memory. Neurosci. Lett. 14, 355-360. 10.1016/0304-3940(79)96174-3 [DOI] [PubMed] [Google Scholar]
- Giorgi, C., Marchi, S. and Pinton, P. (2018). The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 19, 713-730. 10.1038/s41580-018-0052-8 [DOI] [PubMed] [Google Scholar]
- Gomes, D. A., Rodrigues, M. A., Leite, M. F., Gomez, M. V., Varnai, P., Balla, T., Bennett, A. M. and Nathanson, M. H. (2008). c-Met must translocate to the nucleus to initiate calcium signals. J. Biol. Chem. 283, 4344-4351. 10.1074/jbc.M706550200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolla, A. A., Sim, J. A., Lim, D., Rodriguez, J. J., Genazzani, A. A. and Verkhratsky, A. (2013). Amyloid-β and Alzheimer's disease type pathology differentially affects the calcium signalling toolkit in astrocytes from different brain regions. Cell Death Dis. 4, e623-e627. 10.1038/cddis.2013.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulmez Karaca, K., Kupke, J., Brito, D. V. C., Zeuch, B., Thome, C., Weichenhan, D., Lutsik, P., Plass, C. and and Oliveira, A. M. M. (2020). Neuronal ensemble-specific DNA methylation strengthens engram stability. Nat. Commun. 11, 639. 10.1038/s41467-020-14498-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, F., Chen, X. and Rosbash, M. (2017). Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc. Natl. Acad. Sci. USA 114, E8780-E8787. 10.1073/pnas.1706608114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, J., Cheval, H., Selfridge, J. and Bird, A. (2011). The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 27, 631-652. 10.1146/annurev-cellbio-092910-154121 [DOI] [PubMed] [Google Scholar]
- Ha, S., Jeong, S.-H., Yi, K., Chung, K. M., Hong, C. J., Kim, S. W., Kim, E.-K. and Yu, S.-W. (2017). Phosphorylation of p62 by AMP-activated protein kinase mediates autophagic cell death in adult hippocampal neural stem cells. J. Biol. Chem. 292, 13795-13808. 10.1074/jbc.M117.780874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H.et al. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885-889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hardingham, G. E. and Bading, H. (2010). Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat. Rev. Neurosci. 11, 682-696. 10.1038/nrn2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham, G. E., Chawla, S., Johnson, C. M. and Bading, H. (1997). Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature 385, 260-265. 10.1038/385260a0 [DOI] [PubMed] [Google Scholar]
- Hardingham, G. E., Chawla, S., Cruzalegui, F. H. and Bading, H. (1999). Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron 22, 789-798. 10.1016/S0896-6273(00)80737-0 [DOI] [PubMed] [Google Scholar]
- Hardingham, G. E., Arnold, F. J. L. and Bading, H. (2001). Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat. Neurosci. 4, 261-267. 10.1038/85109 [DOI] [PubMed] [Google Scholar]
- He, C. and Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67-93. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heessen, S. and Fornerod, M. (2007). The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 8, 914-919. 10.1038/sj.embor.7401075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg, M., Borst, A., Wagner, S. and Byers, D. (1985). Drosophila mushroom body mutants are deficient in olfactory learning. J. Neurogenet. 2, 1-30. 10.3109/01677068509100140 [DOI] [PubMed] [Google Scholar]
- Hemstedt, T. J., Bengtson, C. P., Ramírez, O., Oliveira, A. M. M. and Bading, H. (2017). Reciprocal interaction of dendrite geometry and nuclear calcium–VEGFD signaling gates memory consolidation and extinction. J. Neurosci. 37, 6946-6955. 10.1523/JNEUROSCI.2345-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, P. M. and Monteggia, L. M. (2018). MeCP2 as an activator of gene expression. Trends Neurosci. 41, 72-74. 10.1016/j.tins.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyer-Hansen, M., Bastholm, L., Szyniarowski, P., Campanella, M., Szabadkai, G., Farkas, T., Bianchi, K., Fehrenbacher, N., Elling, F., Rizzuto, R.et al. (2007). Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Mol. Cell 25, 193-205. 10.1016/j.molcel.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Hsieh, J. and Gage, F. H. (2005). Chromatin remodeling in neural development and plasticity. Curr. Opin. Cell Biol. 17, 664-671. 10.1016/j.ceb.2005.09.002 [DOI] [PubMed] [Google Scholar]
- Hu, S.-C., Chrivia, J. and Ghosh, A. (1999). Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron 22, 799-808. 10.1016/S0896-6273(00)80738-2 [DOI] [PubMed] [Google Scholar]
- Huang, Y. and Thathiah, A. (2015). Regulation of neuronal communication by G protein-coupled receptors. FEBS Lett. 589, 1607-1619. 10.1016/j.febslet.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Huber, L. A. and Teis, D. (2016). Lysosomal signaling in control of degradation pathways. Curr. Opin. Cell Biol. 39, 8-14. 10.1016/j.ceb.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Humbert, J.-P., Matte, N., Artault, J.-C., Köppler, P. and Malviya, A. N. (1996). Inositol 1,4,5-trisphosphate receptor is located to the inner nuclear membrane vindicating regulation of nuclear calcium signaling by inositol 1,4,5-trisphosphate: Discrete distribution of inositol phosphate receptors to inner and outer nuclear membranes. J. Biol. Chem. 271, 478-485. 10.1074/jbc.271.1.478 [DOI] [PubMed] [Google Scholar]
- Impey, S., Fong, A. L., Wang, Y., Cardinaux, J.-R., Fass, D. M., Obrietan, K., Wayman, G. A., Storm, D. R., Soderling, T. R. and Goodman, R. H. (2002). Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron 34, 235-244. 10.1016/S0896-6273(02)00654-2 [DOI] [PubMed] [Google Scholar]
- Inoki, K., Li, Y., Xu, T. and Guan, K.-L. (2003). Rheb GTpase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829-1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine, R. F. (2003). Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 4, 349-361. 10.1038/nrm1100 [DOI] [PubMed] [Google Scholar]
- Ivanova, H., Kerkhofs, M., La Rovere, R. M. and Bultynck, G. (2017). Endoplasmic reticulum-mitochondrial Ca2+ fluxes underlying cancer cell survival. Front. Oncol. 7, 70. 10.3389/fonc.2017.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadiya, P., Kolmetzky, D. W., Tomar, D., Di Meco, A., Lombardi, A. A., Lambert, J. P., Luongo, T. S., Ludtmann, M. H., Praticò, D. and Elrod, J. W. (2019). Impaired mitochondrial calcium efflux contributes to disease progression in models of Alzheimer's disease. Nat. Commun. 10, 3885. 10.1038/s41467-019-11813-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgens, D. M., Inman, J. L., Wojcik, M., Robertson, C., Palsdottir, H., Tsai, W.-T., Huang, H., Bruni-Cardoso, A., López, C. S., Bissell, M. J.et al. (2017). Deep nuclear invaginations are linked to cytoskeletal filaments – integrated bioimaging of epithelial cells in 3D culture. J. Cell Sci. 130, 177-189. 10.1242/jcs.190967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, S. K., Booth, D. M., Young, M. P. and Hajnóczky, G. (2019). Redox regulation of ER and mitochondrial Ca2+ signaling in cell survival and death. Cell Calcium 79, 89-97. 10.1016/j.ceca.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, Y., Kennedy, A., Chiu, H., Mohammad, F., Claridge-Chang, A. and Anderson, D. J. (2020). Neurons that function within an integrator to promote a persistent behavioral state in Drosophila. Neuron 105, 322-333.e5. 10.1016/j.neuron.2019.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel, E. R. (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030-1038. 10.1126/science.1067020 [DOI] [PubMed] [Google Scholar]
- Khachaturian, Z. S. (1984). Towards theories of brain aging. In Handbook of Studies on Psychiatry and Old Age (ed. D. S. Kay and G. W. Burrows), pp. 7-30. Elsevier Science Publishers. [Google Scholar]
- Kim, M.-S., Akhtar, M. W., Adachi, M., Mahgoub, M., Bassel-Duby, R., Kavalali, E. T., Olson, E. N. and Monteggia, L. M. (2012). An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J. Neurosci. 32, 10879-10886. 10.1523/JNEUROSCI.2089-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde, B., Wu, D. Y., Greenberg, M. E. and Gabel, H. W. (2016). DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc. Natl. Acad. Sci. USA 113, 15114-15119. 10.1073/pnas.1618737114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J. (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931-937. 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- Knockenhauer, K. E. and Schwartz, T. U. (2016). The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162-1171. 10.1016/j.cell.2016.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, M., Wang, Q. J., Holstein, G. R., Friedrich, V. L., Iwata, J.-I., Kominami, E., Chait, B. T., Tanaka, K. and Yue, Z. (2007). Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA 104, 14489-14494. 10.1073/pnas.0701311104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic, M., Ludtmann, M. H. R., Bading, H., Hershfinkel, M., Steer, E., Chu, C. T., Abramov, A. Y. and Sekler, I. (2015). PKA phosphorylation of NCLX reverses mitochondrial calcium overload and depolarization, promoting survival of PINK1–deficient dopaminergic neurons. Cell Rep. 13, 376-386. 10.1016/j.celrep.2015.08.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer, G., Mariño, G. and Levine, B. (2010). Autophagy and the integrated stress response. Mol. Cell 40, 280-293. 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A. (2020). Calcium Signaling During Brain Aging and Its Influence on the Hippocampal Synaptic Plasticity. Adv. Exp. Med. Biol. 1131, 985-1012. 10.1007/978-3-030-12457-1_39 [DOI] [PubMed] [Google Scholar]
- Laflamme, K., Domingue, O., Guillemette, B. I. and Guillemette, G. (2002). Immunohistochemical localization of type 2 inositol 1,4,5-trisphosphate receptor to the nucleus of different mammalian cells. J. Cell. Biochem. 85, 219-228. 10.1002/jcb.10124 [DOI] [PubMed] [Google Scholar]
- Lanner, J. T., Georgiou, D. K., Joshi, A. D. and Hamilton, S. L. (2010). Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb. Perspect. Biol. 2, a003996. 10.1101/cshperspect.a003996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. M., Wagner, M., Xiao, R., Kim, K. H., Feng, D., Lazar, M. A. and Moore, D. D. (2014). Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516, 112-115. 10.1038/nature13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, M. F., Thrower, E. C., Echevarria, W., Koulen, P., Hirata, K., Bennett, A. M., Ehrlich, B. E. and Nathanson, M. H. (2003). Nuclear and cytosolic calcium are regulated independently. Proc. Natl. Acad. Sci. USA 100, 2975-2980. 10.1073/pnas.0536590100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, B. and Kroemer, G. (2019). Biological functions of autophagy genes: a disease perspective. Cell 176, 11-42. 10.1016/j.cell.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. and Gao, T. M. (2016). Functional Role of Mitochondrial and Nuclear BK Channels. Int. Rev. Neurobiol. 128, 163-191. 10.1016/bs.irn.2016.03.018 [DOI] [PubMed] [Google Scholar]
- Li, B., Jie, W., Huang, L., Wei, P., Li, S., Luo, Z., Friedman, A. K., Meredith, A. L., Han, M.-H., Zhu, X.-H.et al. (2014). Nuclear BK channels regulate gene expression via the control of nuclear calcium signaling. Nat. Neurosci. 17, 1055-1063. 10.1038/nn.3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbäck-Stokin, K., Korzus, E., Nagaoka-Yasuda, R. and Mayford, M. (2004). Nuclear Calcium/Calmodulin regulates memory consolidation. J. Neurosci. 24, 10858-10867. 10.1523/JNEUROSCI.1022-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, A. C., Bygrave, A. M., de Calignon, A., Lee, T. and Miesenböck, G. (2014). Sparse, decorrelated odor coding in the mushroom body enhances learned odor discrimination. Nat. Neurosci. 17, 559-568. 10.1038/nn.3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans, E. and Waller-Evans, H. (2020). Lysosomal Ca2+ homeostasis and signaling in health and disease. Cold Spring Harb. Perspect. Biol. 12, a035311. 10.1101/cshperspect.a035311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum, J. J., Bauer, D. E., Kong, M., Harris, M. H., Li, C., Lindsten, T. and Thompson, C. B. (2005). Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237-248. 10.1016/j.cell.2004.11.046 [DOI] [PubMed] [Google Scholar]
- Ma, H., Groth, R. D., Cohen, S. M., Emery, J. F., Li, B., Hoedt, E., Zhang, G., Neubert, T. A. and Tsien, R. W. (2014). γcaMKII shuttles Ca2+/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell 159, 281-294. 10.1016/j.cell.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, R., Ochoa Thomas, E., Ramirez, P., Miller, H. E., Beckmann, A., Zuniga, G., Dobrowolski, R. and Frost, B. (2020). Pathogenic Tau causes a toxic depletion of nuclear calcium. Cell Rep. 32, 107900. 10.1016/j.celrep.2020.107900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon, T., Elad, N., Dahan, I. and Medalia, O. (2012). The human nuclear pore complex as revealed by cryo-electron tomography. Structure 20, 998-1006. 10.1016/j.str.2012.03.025 [DOI] [PubMed] [Google Scholar]
- Malhas, A., Goulbourne, C. and Vaux, D. J. (2011). The nucleoplasmic reticulum: Form and function. Trends Cell Biol. 21, 362-373. 10.1016/j.tcb.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Malik, B. R., Maddison, D. C., Smith, G. A. and Peters, O. M. (2019). Autophagic and endo-lysosomal dysfunction in neurodegenerative disease. Mol. Brain 12, 100. 10.1186/s13041-019-0504-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marius, P., Guerra, M. T., Nathanson, M. H., Ehrlich, B. E. and Leite, M. F. (2006). Calcium release from ryanodine receptors in the nucleoplasmic reticulum. Cell Calcium 39, 65-73. 10.1016/j.ceca.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Mauceri, D., Freitag, H. E., Oliveira, A. M. M., Bengtson, C. P. and Bading, H. (2011). Nuclear Calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron 71, 117-130. 10.1016/j.neuron.2011.04.022 [DOI] [PubMed] [Google Scholar]
- Mayr, B. and Montminy, M. (2001). Transcriptional regulation by the phosphorylation-dependent factor creb. Nat. Rev. Mol. Cell Biol. 2, 599-609. 10.1038/35085068 [DOI] [PubMed] [Google Scholar]
- Mayseless, O., Berns, D. S., Yu, X. M., Riemensperger, T., Fiala, A. and Schuldiner, O. (2018). Developmental coordination during olfactory circuit remodeling in Drosophila. Neuron 99, 1204-1215.e5. 10.1016/j.neuron.2018.07.050 [DOI] [PubMed] [Google Scholar]
- Mizushima, N. and Levine, B. (2020). Autophagy in human diseases. N. Engl. J. Med. 383, 1564-1576. 10.1056/NEJMra2022774 [DOI] [PubMed] [Google Scholar]
- Modi, M. N., Shuai, Y. and Turner, G. C. (2020). The Drosophila mushroom body: from architecture to algorithm in a learning circuit. Annu. Rev. Neurosci. 43, 465-484. 10.1146/annurev-neuro-080317-0621333 [DOI] [PubMed] [Google Scholar]
- Nan, X., Campoy, F. J. and Bird, A. (1997). MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88, 471-481. 10.1016/S0092-8674(00)81887-5 [DOI] [PubMed] [Google Scholar]
- Neely, K. M., Green, K. N. and LaFerla, F. M. (2011). Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a γ-secretase-independent manner. J. Neurosci. 31, 2781-2791. 10.1523/JNEUROSCI.5156-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, A. C., Bootman, M. D. and Scott, J. D. (2016). Second messengers. Cold Spring Harb. Perspect. Biol. 8, a005926. 10.1101/cshperspect.a005926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, A. M. M. (2016). DNA methylation: A permissive mark in memory formation and maintenance. Learn. Mem. 23, 587-593. 10.1101/lm.042739.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, A. M. M., Hemstedt, T. J. and Bading, H. (2012). Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 15, 1111-1113. 10.1038/nn.3151 [DOI] [PubMed] [Google Scholar]
- Oliveira, A. M. M., Hemstedt, T. J., Freitag, H. E. and Bading, H. (2016). Dnmt3a2: a hub for enhancing cognitive functions. Mol. Psychiatry 21, 1130-1136. 10.1038/mp.2015.175 [DOI] [PubMed] [Google Scholar]
- Paonessa, F., Evans, L. D., Solanki, R., Larrieu, D., Wray, S., Hardy, J., Jackson, S. P. and Livesey, F. J. (2019). Microtubules deform the nuclear membrane and disrupt nucleocytoplasmic transport in Tau-mediated frontotemporal dementia. Cell Rep. 26, 582-593.e5. 10.1016/j.celrep.2018.12.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadia, S., Stevenson, P., Hardingham, N. R., Bading, H. and Hardingham, G. E. (2005). Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J. Neurosci. 25, 4279-4287. 10.1523/JNEUROSCI.5019-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, B. D., Ngo, T.-T. B., Hibbard, K. L., Murphy, C., Jenett, A., Truman, J. W. and Rubin, G. M. (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735-755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito, V. A., Li, H., Martini-Stoica, H., Wang, B., Yang, L., Xu, Y., Swartzlander, D. B., Palmieri, M., di Ronza, A., Lee, V. M.-Y.et al. (2014). Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 6, 1142-1160. 10.15252/emmm.201303671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prole, D. L. and Taylor, C. W. (2019). Structure and function of IP3 receptors. Cold Spring Harb. Perspect. Biol. 11, a035063. 10.1101/cshperspect.a035063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaello, A., Mammucari, C., Gherardi, G. and Rizzuto, R. (2016). Calcium at the center of cell signaling: interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem. Sci. 41, 1035-1049. 10.1016/j.tibs.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, K., Cusack, C. L., Nnah, I. C., Khayati, K., Saqcena, C., Huynh, T. B., Noggle, S. A., Ballabio, A. and Dobrowolski, R. (2016). Dysregulation of nutrient sensing and CLEARance in presenilin deficiency. Cell Rep. 14, 2166-2179. 10.1016/j.celrep.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, M. A., Gomes, D. A., Andrade, V. A., Leite, M. F. and Nathanson, M. H. (2008). Insulin induces calcium signals in the nucleus of rat hepatocytes. Hepatology 48, 1621-1631. 10.1002/hep.22424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, M. A., Gomes, D. A., Nathanson, M. H. and Leite, M. F. (2009). Nuclear calcium signaling: a cell within a cell. Braz. J. Med. Biol. Res. 42, 17-20. 10.1590/S0100-879X2008005000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruault, M., Dubarry, M. and Taddei, A. (2008). Re-positioning genes to the nuclear envelope in mammalian cells: impact on transcription. Trends Genet. 24, 574-581. 10.1016/j.tig.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Rubinsztein, D. C., Codogno, P. and Levine, B. (2012). Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709-730. 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki, K., Wu, J. and Kaufman, R. J. (2008). Protein kinase Cθ is required for autophagy in response to stress in the endoplasmic reticulum. J. Biol. Chem. 283, 15370-15380. 10.1074/jbc.M710209200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammels, E., Parys, J. B., Missiaen, L., De Smedt, H. and Bultynck, G. (2010). Intracellular Ca2+ storage in health and disease: A dynamic equilibrium. Cell Calcium 47, 297-314. 10.1016/j.ceca.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Sando, R., Gounko, N., Pieraut, S., Liao, L., Yates, J. and Maximov, A. (2012). HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151, 821-834. 10.1016/j.cell.2012.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello, M., Palmieri, M., di Ronza, A., Medina, D. L., Valenza, M., Gennarino, V. A., Di Malta, C., Donaudy, F., Embrione, V., Polishchuk, R. S.et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473-477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Schlumm, F., Mauceri, D., Freitag, H. E. and Bading, H. (2013). Nuclear calcium signaling regulates nuclear export of a subset of class iia histone deacetylases following synaptic activity. J. Biol. Chem. 288, 8074-8084. 10.1074/jbc.M112.432773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen, I., Aires, L., Ries, J. and Vogel, V. (2017). Nanoscale invaginations of the nuclear envelope: shedding new light on wormholes with elusive function. Nucleus 8, 506-514. 10.1080/19491034.2017.1337621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secondo, A., Esposito, A., Petrozziello, T., Boscia, F., Molinaro, P., Tedeschi, V., Pannaccione, A., Ciccone, R., Guida, N., Di Renzo, G.et al. (2018). Na+/Ca2+ exchanger 1 on nuclear envelope controls PTEN/Akt pathway via nucleoplasmic Ca2+ regulation during neuronal differentiation. Cell Death Discov. 4, 12. 10.1038/s41420-017-0018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secondo, A., Petrozziello, T., Tedeschi, V., Boscia, F., Pannaccione, A., Molinaro, P. and Annunziato, L. (2020). Nuclear localization of NCX: Role in Ca2+ handling and pathophysiological implications. Cell Calcium 86, 102143. 10.1016/j.ceca.2019.102143 [DOI] [PubMed] [Google Scholar]
- Segal, M. (2017). Dendritic spines: morphological building blocks of memory. Neurobiol. Learn. Mem. 138, 3-9. 10.1016/j.nlm.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Seok, S., Fu, T., Choi, S.-E., Li, Y., Zhu, R., Kumar, S., Sun, X., Yoon, G., Kang, Y., Zhong, W.et al. (2014). Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 516, 108-111. 10.1038/nature13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre, C., Di Malta, C., Polito, V. A., Arencibia, M. G., Vetrini, F., Erdin, S., Erdin, S. U., Huynh, T., Medina, D., Colella, P.et al. (2011). TFEB Links autophagy to lysosomal biogenesis. Science 332, 1429-1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre, C., Zoncu, R., Medina, D. L., Vetrini, F., Erdin, S., Erdin, S., Huynh, T., Ferron, M., Karsenty, G., Vellard, M. C.et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095-1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin, V., Danker, T., Enss, K., Ossig, R. and Oberleithner, H. (2001). Evidence for Ca2+ - and ATP-sensitive peripheral channels in nuclear pore complexes. FASEB J. 15, 1895-1901. 10.1096/fj.00-0838com [DOI] [PubMed] [Google Scholar]
- Simonetti, M., Hagenston, A. M., Vardeh, D., Freitag, H. E., Mauceri, D., Lu, J., Satagopam, V. P., Schneider, R., Costigan, M., Bading, H.et al. (2013). Nuclear calcium signaling in spinal neurons drives a genomic program required for persistent inflammatory pain. Neuron 77, 43-57. 10.1016/j.neuron.2012.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. K., Kashyap, M. P., Tripathi, V. K., Singh, S., Garg, G. and Rizvi, S. I. (2017). Neuroprotection through rapamycin-induced activation of autophagy and PI3K/Akt1/mTOR/CREB signaling against amyloid-β-induced oxidative stress, synaptic/neurotransmission dysfunction, and neurodegeneration in adult rats. Mol. Neurobiol. 54, 5815-5828. 10.1007/s12035-016-0129-3 [DOI] [PubMed] [Google Scholar]
- Száraz, P., Bánhegyi, G., Marcolongo, P. and Benedetti, A. (2013). Transient knockdown of presenilin-1 provokes endoplasmic reticulum stress related formation of autophagosomes in HepG2 cells. Arch. Biochem. Biophys. 538, 57-63. 10.1016/j.abb.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Timney, B. L., Raveh, B., Mironska, R., Trivedi, J. M., Kim, S. J., Russel, D., Wente, S. R., Sali, A. and Rout, M. P. (2016). Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215, 57-76. 10.1083/jcb.201601004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima, C. (2009). How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1793, 941-946. 10.1016/j.bbamcr.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Tsunemi, T., Ashe, T. D., Morrison, B. E., Soriano, K. R., Au, J., Roque, R. A. V., Lazarowski, E. R., Damian, V. A., Masliah, E. and La Spada, A. R. (2012). PGC-1α rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4, 142ra97. 10.1126/scitranslmed.3003799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, H., Nelson, O., Bezprozvanny, A., Wang, Z., Lee, S.-F., Hao, Y.-H., Serneels, L., De Strooper, B., Yu, G. and Bezprozvanny, I. (2006). Presenilins Form ER Ca2+ Leak Channels, a Function Disrupted by Familial Alzheimer's Disease-Linked Mutations. Cell 126, 981-993. 10.1016/j.cell.2006.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecaetsbeek, I., Vangheluwe, P., Raeymaekers, L., Wuytack, F. and Vanoevelen, J. (2011). The Ca2+ pumps of the endoplasmic reticulum and golgi apparatus. Cold Spring Harb. Perspect. Biol. 3, a004184. 10.1101/cshperspect.a004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangeel, L. and Voets, T. (2019). Transient receptor potential channels and calcium signaling. Cold Spring Harb. Perspect. Biol. 11, a035048. 10.1101/cshperspect.a035048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez, P., Arroba, A. I., Cecconi, F., de la Rosa, E. J., Boya, P. and De Pablo, F. (2012). Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy 8, 187-199. 10.4161/auto.8.2.18535 [DOI] [PubMed] [Google Scholar]
- Verma, M., Callio, J., Otero, P. A., Sekler, I., Wills, Z. P. and Chu, C. T. (2017). Mitochondrial calcium dysregulation contributes to dendrite degeneration mediated by PD/LBD-Associated LRRK2 mutants. J. Neurosci. 37, 11151-11165. 10.1523/JNEUROSCI.3791-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, N. and Goodman, R. H. (2001). CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276, 13505-13508. 10.1074/jbc.R000025200 [DOI] [PubMed] [Google Scholar]
- Vogel-Ciernia, A. and Wood, M. A. (2014). Neuron-specific chromatin remodeling: A missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology 80, 18-27. 10.1016/j.neuropharm.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacquier, B., Combettes, L. and Dupont, G. (2019). Cytoplasmic and mitochondrial calcium signaling: a two-way relationship. Cold Spring Harb. Perspect. Biol. 11, a035139. 10.1101/cshperspect.a035139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Campos, B., Kaetzel, M. A., Dedman, J. R. and Jamieson, G. A. (1995). Functional elimination of calmodulin within the nucleus by targeted expression of an inhibitor peptide. J. Biol. Chem. 270, 30245-30248. 10.1074/jbc.270.51.30245 [DOI] [PubMed] [Google Scholar]
- Wang, L., Kang, H., Li, Y., Shui, Y., Yamamoto, R., Sugai, T. and Kato, N. (2015a). Cognitive recovery by chronic activation of the large-conductance calcium-activated potassium channel in a mouse model of Alzheimer's disease. Neuropharmacology 92, 8-15. 10.1016/j.neuropharm.2014.12.033 [DOI] [PubMed] [Google Scholar]
- Wang, F., Zhang, Y., Wang, L., Sun, P., Luo, X., Ishigaki, Y., Sugai, T., Yamamoto, R. and Kato, N. (2015b). Improvement of spatial learning by facilitating large-conductance calcium-activated potassium channel with transcranial magnetic stimulation in Alzheimer's disease model mice. Neuropharmacology 97, 210-219. 10.1016/j.neuropharm.2015.05.027 [DOI] [PubMed] [Google Scholar]
- Weislogel, J.-M., Bengtson, C. P., Müller, M. K., Hörtzsch, J. N., Bujard, M., Schuster, C. M. and Bading, H. (2013). Requirement for nuclear calcium signaling in drosophila long-term memory. Sci. Signal. 6, ra33. 10.1126/scisignal.2003598 [DOI] [PubMed] [Google Scholar]
- Wittmann, M., Queisser, G., Eder, A., Wiegert, J. S., Bengtson, C. P., Hellwig, A., Wittum, G. and Bading, H. (2009). Synaptic activity induces dramatic changes in the geometry of the cell nucleus: interplay between nuclear structure, histone H3 phosphorylation, and nuclear calcium signaling. J. Neurosci. 29, 14687-14700. 10.1523/JNEUROSCI.1160-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson, R. L., Chantranupong, L., Saxton, R. A., Shen, K., Scaria, S. M., Cantor, J. R. and Sabatini, D. M. (2016). Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43-48. 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Q., Yan, P., Ma, X., Liu, H., Perez, R., Zhu, A., Gonzales, E., Burchett, J. M., Schuler, D. R., Cirrito, J. R.et al. (2014). Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 34, 9607-9620. 10.1523/JNEUROSCI.3788-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. and Klionsky, D. J. (2010). Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814-822. 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim, W. W.-Y. and Mizushima, N. (2020). Lysosome biology in autophagy. Cell Discov. 6, 6. 10.1038/s41421-020-0141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, J. C. P., Wallach, J. S., Del Vecchio, M., Wilder, E. L., Zhou, H., Quinn, W. G. and Tully, T. (1994). Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79, 49-58. 10.1016/0092-8674(94)90399-9 [DOI] [PubMed] [Google Scholar]
- Yin, J. C. P., Del Vecchio, M., Zhou, H. and Tully, T. (1995). CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in drosophila. Cell 81, 107-115. 10.1016/0092-8674(95)90375-5 [DOI] [PubMed] [Google Scholar]
- Zhang, S. J., Zou, M., Lu, L., Lau, D., Ditzel, D. A. W., Delucinge-Vivier, C., Aso, Y., Descombes, P. and Bading, H. (2009). Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 5, e1000604. 10.1371/journal.pgen.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Huang, M., Bushong, E., Phan, S., Uytiepo, M., Beutter, E., Boemer, D., Tsui, K., Ellisman, M. and Maximov, A. (2019). Class IIa HDACs regulate learning and memory through dynamic experience-dependent repression of transcription. Nat. Commun. 10, 3469. 10.1038/s41467-019-11409-0 [DOI] [PMC free article] [PubMed] [Google Scholar]