Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) mediated Coronavirus disease-19 (COVID-19) has affected millions of individuals around all corners of the globe. Symptoms and severities of infection with this highly contagious virus vary among individuals and there is disparity in the number of COVID-19-related casualties across different ethnic groups. The primary receptor for SARS-CoV-2 entry into the host cells is angiotensin-converting enzyme 2 (ACE2). Certain variants of ACE2 are known to be associated with COVID-19 comorbidities such as hypertension, cardiovascular complications, diabetes, chronic lung disease, etc. In this study, we looked into the geographic distribution of disease-associated variants of ACE2 as well as closely located PIR gene to explore any possible correlation with the disparities in COVID-19 severities and casualties across ethnic groups. Frequencies of the ACE2 variants associated with COVID-19 comorbidities are higher in the European and the admixed American populations. These variants are also present with stronger pairwise linkage disequilibrium (LD) in the European and the admixed American populations. On the other hand, the variants with protective role are more prevalent in the East and the South Asian populations. Strong pairwise LD exists among the activity modifying (modifier) variants of the PIR and ACE2 genes only in the European super-population. Absence of these PIR variants in the South Asian population may contribute to the overall lower COVID-19 case fatality rates (CFR) despite the dense population in this region.
Keywords: COVID-19, SARS-CoV-2, ACE2, PIR, Genetic variants, Linkage disequilibrium, Haplotype, Geographic distribution
1. Introduction
SARS-CoV-2 mediated COVID-19 has emerged as one of the most widespread and frightful pandemics in human history (Devaux et al., 2020; Esakandari et al., 2020). Symptoms and severities of infection with SARS-CoV-2 vary among individuals. While some of the infected individuals may be asymptomatic carriers or exhibit mild flu-like symptoms, the others may develop severe symptoms and progress to pneumonia, respiratory failure as well as death (Alimohamadi et al., 2020; Feng et al., 2020b; Wu and McGoogan, 2020). Till April 12, 2021 COVID-19 has spread in 215 countries and regions on earth with over 135,646,500 confirmed cases of infection and more than 2,930,500 deaths. There is disparity in the number of COVID-19 caused mortalities across ethnic groups in different geographic regions around the globe (World Health Organization, 2020; Yamamoto and Bauer, 2020). In addition to environmental, socioeconomic and regulatory factors, genetic variants may be responsible for the observed interpopulation and interethnic variability.
The primary receptor for SARS-CoV-2 entry into the host cell is the angiotensin-converting enzyme 2 (ACE2) (Davidson Anne et al., 2020; Lan et al., 2020). As SARS-CoV-2 occupies the ACE2 molecules on cell surface, it prevents ACE2-mediated degradation of angiotensin II (Ang II), which consequently raises Ang II level (Hirano and Murakami, 2020). Increased levels of tissue and circulating Ang II exert a number of direct pro-atherosclerotic effects and promotes monocyte and endothelial cell activation causing progression to atherosclerotic plaque formation (Thomas Merlin et al., 2010). Ang II induces production of proinflammatory cytokines and chemokines via Toll-like receptor 4 (TLR4) (Feng et al., 2020a). Alongside the lungs, SARS-CoV-2 invades other organs that express high level of ACE2. SARS-CoV-2 mediated damage occurs directly by viral propagation and indirectly by inflammatory and immune factors as well as imbalance between the renin angiotensin system (RAS) and ACE2/angiotensin-(1–7) axis caused by ACE2 deficiency (Ni et al., 2020). Genetic factors significantly affect the circulating level of ACE2 in individuals (Rice et al., 2006). The expression level of ACE2 correlates positively with COVID-19 susceptibility, whereas down-regulation of ACE2 activity exacerbate Ang II mediated inflammatory events (Behl et al., 2020).
The highly prevalent comorbidities in COVID-19 patients are hypertension, diabetes, cardiovascular complication and chronic lung disease, among which hypertension is the most common (Sanyaolu et al., 2020). COVID-19 patients with diabetes and hypertension often exhibit severe symptoms, and percentage of non-survivors is high in this group of patients (Richardson et al., 2020; Singh et al., 2020; Zhou et al., 2020). Hypertension per se is strongly associated with diabetes and other complications (Adler et al., 2000; de Boer et al., 2017). On the other hand, ACE2 variants are associated with hypertension, hypertension-related atrial fibrillation and left atrial remodeling (Lozano-Gonzalez et al., 2020; Luo et al., 2019; Patel et al., 2012).
This study explored the geographic distribution of disease associated variants of ACE2 as well as other genes in its vicinity to understand the interethnic variability in the susceptibility and severity of SARS-CoV-2 mediated COVID-19.
2. Materials and methods
2.1. Retrieval of disease-associated variants
ACE2 gene variants, that are associated with any disease, were retrieved from the DisGeNET database (version 7) (Piñero et al., 2020). The search was conducted using “ACE2” gene (Entrez Id: 59272) as the keyword. Redundant variants were removed from the list. Regulatory roles of the variants were retrieved from rSNPBase 3.1 (Guo and Wang, 2018). All filtering options (i.e. TF binding regions, chromatin interactive regions, topologically associated domains, lncRNAs coding regions, mature miRNAs regions, target sites of miRNAs and circRNA regions) were selected to identify any type of regulatory element.
2.2. Haplotype and linkage disequilibrium analysis
Frequencies of disease-associated ACE2 gene variants in 26 populations (Supplementary Table 1) across 5 super-populations (African, admixed American, East Asian, European and South Asian) were collected from the 1000 Genomes Database (1000 Genomes Project Consortium et al., 2015) using the Ensembl Allele Frequency Calculator based on the phase 3 data (Yates et al., 2020). Pairwise linkage disequilibrium (LD) among these variants and frequencies of haplotypes comprising these variants in the five super-populations were calculated using LDmatrix and LDhap modules of LDlink (version 5.0), respectively (Machiela and Chanock, 2015).
2.3. Identification of other modifier variants
LDproxy module of LDlink (Machiela and Chanock, 2015) was used to identify the genetic variants in absolute LD (r2 = 1) with any of the disease-associated variants in ACE2 within ±0.5 Mb (total 1mb) region in any of the super-populations. These SNPs were mapped to several genes. SNPs that are supposed to modify the activity (modifier) of the corresponding gene products were identified and selected using Ensembl Variant Effect Predictor (VEP) (McLaren et al., 2016). Frequencies of these modifier variants were retrieved from the 1000 Genomes Database using the Ensembl Allele Frequency Calculator based on the phase 3 data.
2.4. Identification of candidate genes associated with inflammation
UniProt database (Pundir et al., 2017) was searched to identify whether any of these genes is directly or indirectly associated with inflammation. Among the identified genes, only PIR was found to be associated with inflammation. LDmatrix was used to identify the LD patterns among the modifier variants of ACE2 and PIR genes in 5 super-populations.
2.5. Statistical analyses
The statistical tool available at Metaboanalyst (version 5) (Xia et al., 2009) was used for multivariate principal component analysis (PCA) based on the variant allele frequencies (VAFs) of the above mentioned disease-associated and modifier variants. In total five principal components were considered.
3. Result
3.1. Disease associated variants of ACE2
ACE2 harbors 15 variants that are associated with various diseases (Table 1 ). Except one (rs1514280), the other variants are associated with at least one of the following conditions- hypertension, dyslipidemia, type 2 diabetes mellitus, atrial fibrillation, and/or left ventricular hypertrophy. These are the major comorbidities of COVID-19 (Apicella et al., 2020; Das et al., 2020; Gawałko et al., 2020; Ma et al., 2020; Sanyaolu et al., 2020; Shibata et al., 2020; Stone et al., 2020). Among the identified variants, thirteen are intron variants, one (rs4830542) is a downstream gene variant, and one (rs2285666) is a splice region modifier. Intron variants can influence expression of the genes with introns through altering transcriptional activity or affecting splicing efficiency (Cooper, 2010; Lin et al., 2019). Splice site variants that cause creation of a cryptic splice site, incorrect recognition of splice site or disruption in activities of regulatory elements involved in splicing, can lead to generation of aberrant transcript or non-functional proteins from the corresponding gene (Moles-Fernández et al., 2018; Morrison et al., 2013). Gene expression can also be influenced by downstream gene variants as gene expression regulatory elements, such as enhancers, can be located downstream of the target gene (Benson et al., 2019; Pennacchio et al., 2013).
Table 1.
Disease associated variants of ACE2.
| Variant ID | Consequence | Alleles | Disease |
|---|---|---|---|
| rs4830542 | Downstream gene variant | C/G,T | Essential hypertension, atrial fibrillation, dyslipidemias |
| rs2074192 | Intron variant | C/T | Essential hypertension, Atrial fibrillation, dyslipidemias, left ventricular hypertrophy, hypertensive disease, pregnancy associated hypertension, metabolic syndrome X, cardiovascular diseases, carotid atherosclerosis |
| rs233575 | Intron variant | G/A | Dyslipidemias |
| rs1514280 | Intron variant | A/G | Negative affectivity, temporal pain, psychological symptom |
| rs4240157 | Intron variant | C/T | Essential hypertension, atrial fibrillation, dyslipidemias, hypertensive disease |
| rs879922 | Intron variant | C/G | Essential hypertension, dyslipidemias, carotid atherosclerosis, Non-insulin-dependent diabetes mellitus |
| rs4646156 | Intron variant | A/T | Angina pectoris, dyslipidemias |
| rs4646155 | Intron variant | C/T | Atrial fibrillation, essential hypertension, dyslipidemias |
| rs4646188 | Intron variant | A/G | Non-insulin-dependent diabetes mellitus, dyslipidemias, essential hypertension, hypertensive disease |
| rs4646142 | Intron variant | G/A,C | Dyslipidemias |
| rs2048683 | Intron variant | T/G | Essential hypertension |
| rs2285666 | Splice region variant | C/T | Dyslipidemias, non-insulin-dependent diabetes mellitus, hypertensive disease, orthostatic hypertension |
| rs6632677 | Intron variant | G/C | Atrial fibrillation, essential hypertension, ischemic stroke, hypertrophic cardiomyopathy |
| rs2106809 | Intron variant | A/G | Essential hypertension, hypertensive disease, cardiac event, hypertrophic cardiomyopathy, dyslipidemias, left ventricular hypertrophy, lone atrial fibrillation, ischemic stroke |
| rs1978124 | Intron variant | T/A,C | Dyslipidemias, hypertensive disease |
3.2. ACE2 variant allele frequencies in different populations
Variant allele frequencies (VAFs) of the retrieved disease associated variants of ACE2 in different populations are given in Table 2 . Variant alleles with frequencies ≥ 0.25 are shown as bold italic. The variant alleles at rs4646155, rs4646188 and rs6632677 loci are present with low frequencies in all populations. At the other loci, the distribution patterns of the variant alleles are apparently different among the super-populations. The variant alleles at rs4830542, rs2074192, rs233575, rs1514280, rs4240157, rs879922, rs4646156, rs2048683, and rs1978124 are relatively more prevalent in the European (EUR) populations. A similar VAF pattern is followed by two admixed American (AMR) populations- (Puerto Rican (PUR) and Colombian (CLM)). VAFs at majority of these loci are higher in these two populations compared to the other two admixed American populations (Mexican (MXL) and Peruvians (PEL)). The global minor (variant) alleles at rs4646142, rs2285666, and rs2106809 loci are present as the major alleles in the East Asian (EAS) populations as well as three South Asian (SAS) populations (Bangladeshi (BEB), Indian Telegu (ITU) and Sri Lankan Tamil (STU)), but are relatively less prevalent (frequency < 0.3) in the European populations. The variant allele at rs2074192 is present with high frequencies (> 0.3) in all, except the South Asian populations. In general, the variant alleles are present at much lower frequencies in the East Asian populations followed by the African (AFR) populations.
Table 2.
Frequencies of disease associated variants of ACE2 in world populations.
| Variant ID | Reference allele | Variant allele | Frequency of variant allele |
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR |
AMR |
EAS |
EUR |
SAS |
|||||||||||||||||||||||||
| ALLa | YRI | LWK | GWD | MSL | ESN | ASW | ACB | MXL | PUR | CLM | PEL | CHB | JPT | CHS | CDX | KHV | CEU | TSI | FIN | GBR | IBS | GIH | PJL | BEB | STU | ITU | |||
| rs4830542 | T | C | 0.32 | 0.63 | 0.48 | 0.59 | 0.56 | 0.55 | 0.42 | 0.52 | 0.21 | 0.36 | 0.37 | 0.15 | 0.04 | 0.03 | 0.03 | 0.06 | 0.03 | 0.38 | 0.40 | 0.34 | 0.28 | 0.35 | 0.31 | 0.33 | 0.22 | 0.26 | 0.29 |
| rs2074192 | C | T | 0.36 | 0.3 | 0.42 | 0.29 | 0.36 | 0.32 | 0.41 | 0.35 | 0.42 | 0.34 | 0.34 | 0.53 | 0.46 | 0.43 | 0.41 | 0.37 | 0.47 | 0.35 | 0.42 | 0.46 | 0.5 | 0.41 | 0.24 | 0.28 | 0.25 | 0.18 | 0.18 |
| rs233575 | A | G | 0.14 | 0.00 | 0.02 | 0.01 | 0.00 | 0.00 | 0.07 | 0.01 | 0.18 | 0.21 | 0.32 | 0.12 | 0.01 | 0.00 | 0.00 | 0.01 | 0.00 | 0.36 | 0.4 | 0.29 | 0.27 | 0.34 | 0.19 | 0.26 | 0.15 | 0.14 | 0.17 |
| rs1514280 | G | A | 0.20 | 0.20 | 0.15 | 0.30 | 0.23 | 0.25 | 0.18 | 0.14 | 0.20 | 0.29 | 0.34 | 0.12 | 0.01 | 0.00 | 0.00 | 0.01 | 0.00 | 0.38 | 0.39 | 0.33 | 0.27 | 0.35 | 0.22 | 0.26 | 0.17 | 0.15 | 0.18 |
| rs4240157 | T | C | 0.32 | 0.63 | 0.48 | 0.59 | 0.55 | 0.56 | 0.42 | 0.52 | 0.21 | 0.36 | 0.37 | 0.15 | 0.04 | 0.03 | 0.03 | 0.06 | 0.03 | 0.38 | 0.4 | 0.36 | 0.29 | 0.35 | 0.31 | 0.33 | 0.22 | 0.26 | 0.29 |
| rs879922 | G | C | 0.32 | 0.63 | 0.49 | 0.6 | 0.55 | 0.56 | 0.42 | 0.52 | 0.21 | 0.36 | 0.37 | 0.14 | 0.04 | 0.03 | 0.03 | 0.06 | 0.03 | 0.39 | 0.4 | 0.34 | 0.28 | 0.36 | 0.32 | 0.33 | 0.22 | 0.27 | 0.29 |
| rs4646156 | T | A | 0.20 | 0.22 | 0.24 | 0.19 | 0.17 | 0.17 | 0.18 | 0.20 | 0.17 | 0.36 | 0.30 | 0.12 | 0.01 | 0.00 | 0.00 | 0.02 | 0.01 | 0.40 | 0.38 | 0.33 | 0.29 | 0.34 | 0.23 | 0.28 | 0.16 | 0.17 | 0.17 |
| rs4646155 | C | T | 0.06 | 0.19 | 0.09 | 0.16 | 0.11 | 0.17 | 0.10 | 0.10 | 0.01 | 0.04 | 0.01 | 0.01 | 0.04 | 0.03 | 0.03 | 0.05 | 0.03 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.09 | 0.06 | 0.05 | 0.11 | 0.11 |
| rs4646188 | A | G | 0.04 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.03 | 0.05 | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.09 | 0.10 | 0.13 | 0.21 | 0.14 | 0.07 | 0.09 | 0.05 | 0.04 | 0.06 |
| rs4646142 | G | C | 0.36 | 0.19 | 0.17 | 0.31 | 0.24 | 0.28 | 0.26 | 0.24 | 0.36 | 0.27 | 0.37 | 0.36 | 0.51 | 0.55 | 0.56 | 0.55 | 0.53 | 0.26 | 0.19 | 0.26 | 0.24 | 0.25 | 0.4 | 0.4 | 0.55 | 0.53 | 0.52 |
| rs2048683 | G | T | 0.20 | 0.21 | 0.23 | 0.19 | 0.17 | 0.17 | 0.17 | 0.20 | 0.17 | 0.36 | 0.3 | 0.12 | 0.01 | 0.00 | 0.00 | 0.02 | 0.01 | 0.41 | 0.39 | 0.34 | 0.29 | 0.34 | 0.23 | 0.26 | 0.16 | 0.17 | 0.16 |
| rs2285666 | C | T | 0.35 | 0.17 | 0.16 | 0.3 | 0.21 | 0.19 | 0.24 | 0.21 | 0.36 | 0.27 | 0.37 | 0.36 | 0.51 | 0.55 | 0.56 | 0.55 | 0.52 | 0.26 | 0.19 | 0.26 | 0.22 | 0.25 | 0.40 | 0.40 | 0.55 | 0.53 | 0.52 |
| rs6632677 | G | C | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.04 | 0.01 | 0.03 | 0.06 | 0.09 | 0.10 | 0.08 | 0.04 | 0.03 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.02 | 0.01 | 0.00 |
| rs2106809 | A | G | 0.32 | 0.07 | 0.08 | 0.09 | 0.10 | 0.04 | 0.12 | 0.12 | 0.36 | 0.26 | 0.34 | 0.36 | 0.52 | 0.51 | 0.56 | 0.51 | 0.51 | 0.26 | 0.20 | 0.24 | 0.26 | 0.28 | 0.39 | 0.40 | 0.53 | 0.56 | 0.54 |
| rs1978124 | C | T | 0.21 | 0.13 | 0.10 | 0.08 | 0.07 | 0.11 | 0.16 | 0.06 | 0.21 | 0.40 | 0.38 | 0.12 | 0.01 | 0.00 | 0.00 | 0.02 | 0.01 | 0.52 | 0.52 | 0.48 | 0.41 | 0.43 | 0.27 | 0.28 | 0.18 | 0.19 | 0.17 |
All populations (ALL); African super-population (AFR)- ((Yoruba in Ibadan, Nigeria (YRI); Luhya in Webuye, Kenya (LWK); Gambian in Western Divisions in the Gambia (GWD); Mende in Sierra Leone (MSL); Esan in Nigeria (ESN); Americans of African Ancestry in SW USA (ASW); African Caribbeans in Barbados (ACB)); Ad Mixed American (AMR)- ((Mexican Ancestry from Los Angeles USA (MXL); Puerto Ricans from Puerto Rico (PUR); Colombians from Medellin, Colombia (CLM); Peruvians from Lima, Peru (PEL)); East Asian (EAS)- ((Han Chinese in Beijing, China (CHB); Japanese in Tokyo, Japan (JPT); Southern Han Chinese (CHS); Chinese Dai in Xishuangbanna, China (CDX); Kinh in Ho Chi Minh City, Vietnam (KHV)); European (EUR)- ((Utah Residents (CEPH) with Northern and Western European Ancestry (CEU); Toscani in Italia (TSI); Finnish in Finland (FIN); British in England and Scotland (GBR); Iberian Population in Spain (IBS)); South Asian (SAS)- ((Gujarati Indian from Houston, Texas (GIH); Punjabi from Lahore, Pakistan (PJL); Bengali from Bangladesh (BEB); Sri Lankan Tamil from the UK (STU); Indian Telugu from the UK (ITU)).
Drastic interpopulation variation exists for the VAFs at three loci- rs4830542, rs4240157 and rs879922 (Table 2). In the African populations, the VAFs at these loci range from 0.42 to 0.63. At the same loci, the VAFs in the East Asian populations range from 0.0 to 0.15. The other populations (in AMR, EUR and SAS) have intermediate VAFs at these loci. In the East Asian populations, the VAFs at rs1514280, rs4240157, rs879922, rs4646156, and rs4830542 are very low compared to the other populations.
3.3. LD among the ACE2 variants in different super-populations
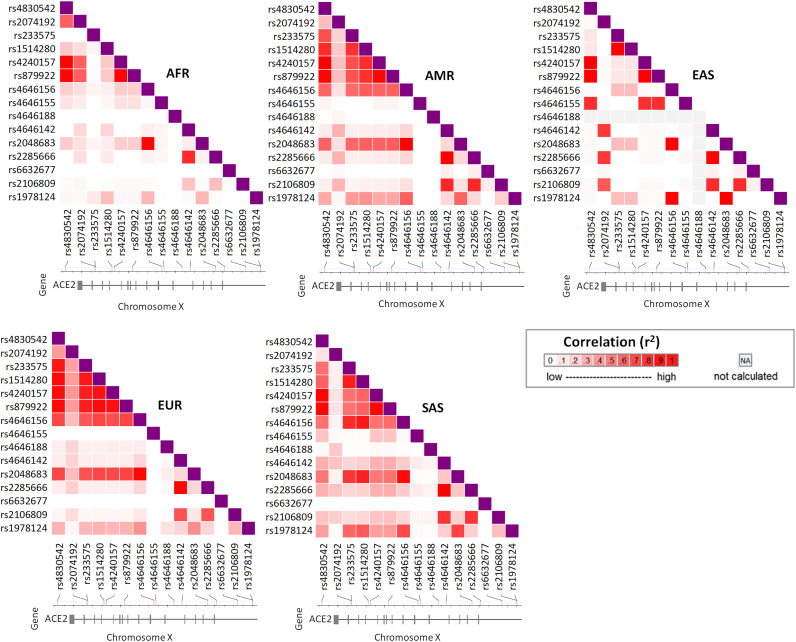
LD among the 15 disease associated variants in five super-populations is shown in Fig. 1 . In the EUR super-population, there is strong LD among rs4830542, rs233575, rs1514280, rs4240157, rs879922, rs4646156 and rs2048683. Although similar LD patterns for these variants are present in the admixed American and the South Asian (SAS) populations, these associations are not as strong as those observed in the European populations. The patterns of VAFs at these loci are similar in the EUR populations and the two admixed American populations (PUR and CLM) (Table 2), but quite different from the East Asian (EAS) populations. These variants, except rs1514280 and rs2048683, are associated with dyslipidemia (Table 1). Contrary to the other populations, relatively strong LD exists among rs2074192, rs4830542, rs4240157 and rs879922 in the African populations. rs4830542 has high pairwise LD with rs4240157 and rs879922 in all 5 super-populations.
Fig. 1.
Pairwise LD among the disease associated variants of ACE2. LD measures are given as r2. For the missing alleles in any population LD could not be calculated (NA). AFR = African, AMR = Admixed American, EAS = East Asian, EUR = European, SAS = South Asian.
Among all the super-populations, the East Asians appear quite different based on the LD patterns (Fig. 1). There is strong LD among rs4646155, rs4830542, rs4240157 and rs879922 in the East Asian super-population. Strong LD also exists among rs2074192, rs4646142, rs2285666 and rs2106809. In fact, majority of the LDs that are present in the East Asians are quite strong and distinct. rs4646188 variant allele is non-existent among the East Asians. Apparently, LD among the disease associated SNPs is the lowest in AFR compared to the other super-populations.
In all super-populations, rs4646142 is in high LD with rs2285666. Except in AFR, these two variants are in high LD with rs2106809. rs2048683 has high pairwise LD with rs4646156 in all super-populations. The pairwise LD of rs2048683 with the other SNPs vary among super-populations.
3.4. Haplotype frequencies in different super-populations
The haplotypes with frequencies ≥ 0.01 in different super-populations are shown in Table 3 (variant alleles are shown as bold italic). Among these, the C_C_G_A_C_C_A_C_A_G_T_C_G_A_T haplotype (#8 in Table 3) with variant alleles at rs4830542, rs233575, rs1514280, rs4240157, rs879922, rs4646156, rs2048683, and rs1978124 is present at ≥0.01 frequencies in EUR (0.277), AMR (0.179), SAS (0.164) and AFR (0.011) super-populations. The C_C_G_A_C_C_A_C_A_G_T_C_G_G_C haplotype (#9 in Table 3) harboring variant alleles at rs4830542, rs233575, rs1514280, rs4240157, rs879922, rs4646156, rs2048683 and rs2106809 is present only in the EUR (0.020) super-population at a frequency ≥ 0.01. Another haplotype C_C_G_A_C_C_T_C_A_C_G_T_G_G_C (#10 in Table 3) with variants at rs4830542, rs233575, rs1514280, rs4240157, rs879922, rs4646142, rs2285666, rs2106809 is present in EUR (0.022) and AMR (0.017) super-populations. The T_C_A_G_T_G_T_C_A_C_G_T_G_G_C (#14 in Table 3) and T_T_A_G_T_G_T_C_A_G_G_C_G_A_C (#17 in Table 3) haplotypes comprise 42.9% and 40.5% of all the haplotypes present in the EAS super-population. The latter haplotype harbors all the wild type alleles, except for rs2074192. The T_C_A_G_T_G_T_C_A_C_G_T_G_G_C (#14 in Table 3) haplotype is also highly prevalent in the SAS populations (0.429). The haplotype with none of the variant alleles (#15 in Table 3) is more frequent in the South Asian super-population (0.022).
Table 3.
Haplotypes with frequencies ≥ 0.01 in different super-populations.
| # | Haplotypesa,b | Frequencies in super-populations |
|---|---|---|
| 1. | C_C_A_A_C_C_A_C_A_G_T_C_G_A_T | AFR (0.072), AMR (0.021), EUR (0.014), SAS (0.013) |
| 2. | C_C_A_A_C_C_T_C_A_C_G_C_G_A_C | AFR (0.026) |
| 3. | C_C_A_A_C_C_T_C_A_C_G_T_G_A_C | AFR (0.091) |
| 4. | C_C_A_G_C_C_A_C_A_G_T_C_G_A_C | AFR (0.109), AMR (0.012) |
| 5. | C_C_A_G_C_C_T_C_A_C_G_T_G_G_C | AFR (0.016) |
| 6. | C_C_A_G_C_C_T_C_A_G_G_C_G_A_C | AFR (0.069) |
| 7. | C_C_A_G_C_C_T_T_A_G_G_C_G_A_C | AFR (0.129), AMR (0.015), EAS (0.030), SAS (0.075) |
| 8. | C_C_G_A_C_C_A_C_A_G_T_C_G_A_T | AFR (0.011), AMR (0.179), EUR (0.277), SAS (0.164) |
| 9. | C_C_G_A_C_C_A_C_A_G_T_C_G_G_C | EUR (0.020) |
| 10. | C_C_G_A_C_C_T_C_A_C_G_T_G_G_C | AMR (0.017), EUR (0.022) |
| 11. | T_C_A_G_T_G_T_C_A_C_G_T_C_G_C | AMR (0.036), EAS (0.064) |
| 12. | T_C_A_G_T_G_T_C_A_C_G_T_G_A_C | AFR (0.032), EAS (0.029), SAS (0.011) |
| 13. | T_C_A_G_T_G_T_C_A_C_G_T_G_A_T | AMR (0.013), EUR (0.022), SAS (0.014) |
| 14. | T_C_A_G_T_G_T_C_A_C_G_T_G_G_C | AFR (0.059), AMR (0.246), EAS (0.429), EUR (0.165), SAS (0.429) |
| 15. | T_C_A_G_T_G_T_C_A_G_G_C_G_A_C | AFR (0.012), AMR (0.019), EUR (0.016), SAS (0.022) |
| 16. | T_T_A_G_T_G_A_C_A_G_T_C_G_A_T | AMR (0.025), EUR (0.026) |
| 17. | T_T_A_G_T_G_T_C_A_G_G_C_G_A_C | AFR (0.325), AMR (0.288), EAS (0.405), EUR (0.144), SAS (0.124) |
| 18. | T_T_A_G_T_G_T_C_A_G_G_C_G_A_T | EUR (0.119) |
| 19. | T_T_A_G_T_G_T_C_A_G_G_C_G_G_C | EAS (0.012) |
| 20. | T_T_A_G_T_G_T_C_A_G_T_C_G_A_T | AFR (0.012), AMR (0.048), SAS (0.020) |
| 21. | T_T_A_G_T_G_T_C_G_G_G_C_G_A_C | AMR (0.032), EUR (0.107), SAS (0.061) |
| 22. | T_T_A_G_T_G_T_C_G_G_G_C_G_G_C | EUR (0.012) |
order of the variants in the haplotype: rs4830542_rs2074192_rs233575_rs1514280_rs4240157_rs879922_rs4646156_ rs4646155_rs4646188_rs4646142_rs2048683_rs2285666_rs6632677_rs2106809_rs1978124.
All variant haplotype: C_T_G_A_C_C_A_T_G_C_T_T_C_G_T.
3.5. Variants in the regulatory elements and their haplotype frequencies
Among the disease associated ACE2 variants, five (rs1514280, rs4240157, rs4646155, rs4646156, and rs6632677) were identified to be associated with regulatory elements as shown in Table 4 . rs1514280 and rs4240157 are associated with circular RNA (circRNA) coding region. circRNAs are produced from both protein-coding and non-coding regions of the genome and lack poly(A) tails and 5′ caps (Szabo and Salzman, 2016). These RNAs generated by backsplicing play key role in controlling the levels of endogenous miRNAs and act as protective factor against development of cardiac hypertrophy and atherosclerosis (Holdt et al., 2016; Stępień et al., 2018; Tan et al., 2017; Wang et al., 2016). rs4646155, rs4646156, and rs6632677 are associated with at least one gene targeting chromatin interactive region. Through formation of chromatin loop, distal regulatory elements are connected to their target genes (Peng et al., 2019; Sobhy et al., 2019). The frequencies of haplotypes containing these 5 regulatory SNPs in different super-populations are shown in Table 5 . The frequencies of the haplotypes with variant alleles at rs1514280, rs4240157 and rs4646156 (A_C_A_C_G) (#2 in Table 5) are > 0.2 in the AMR and EUR super-populations, whereas in the EAS super-population the frequency is 0.0026. The G_C_A_C_G (#5 in Table 5) haplotype with variant alleles at rs4240157 and rs4646156 is absent in EAS and SAS populations, but prevalent with > 0.1 frequencies in the AFR populations.
Table 4.
List of regulatory variants of ACE2.
| rs ID | Chromosome | Allele | Related regulatory elements | Target genes | rSNPa or LD-proxies | eQTL |
|---|---|---|---|---|---|---|
| rs1514280 | chrX:15586448 | C/T | circRNA region | N/A | rSNP | N |
| rs4240157 | chrX:15586964 | A/G | circRNA region | N/A | rSNP | N |
| rs4646155 | chrX:15597509 | G/A | Chromatin interactive region | 1 gene | rSNP | N |
| rs4646156 | chrX:15597043 | A/T | Chromatin interactive region | 1 gene | rSNP | Y |
| rs6632677 | chrX:15614872 | C/G | Chromatin interactive region | 2 genes | rSNP | N |
Regulatory SNP.
Table 5.
Haplotype frequencies of ACE2 regulatory variants in different super-populations.
| # | Haplotype a,b | ALL | AFR | AMR | EAS | EUR | SAS |
|---|---|---|---|---|---|---|---|
| 1 | G_T_T_C_G | 0.6477 | 0.4526 | 0.6565 | 0.8901 | 0.6084 | 0.6978 |
| 2 | A_C_A_C_G | 0.1528 | 0.0847 | 0.2099 | 0.0026 | 0.3146 | 0.1936 |
| 3 | G_C_T_T_G | 0.0607 | 0.1326 | 0.0191 | 0.0314 | 0.0013 | 0.085 |
| 4 | A_C_T_C_G | 0.045 | 0.1236 | 0.0363 | 0.0013 | 0.03 | 0.0042 |
| 5 | G_C_A_C_G | 0.0315 | 0.1107 | 0.0115 | – | 0.0026 | – |
| 6 | G_C_T_C_G | 0.0278 | 0.0917 | 0.0038 | 0.0013 | 0.0104 | 0.0028 |
| 7 | G_T_T_C_C | 0.0207 | 0.001 | 0.0363 | 0.0681 | 0.0013 | 0.007 |
| 8 | G_T_A_C_G | 0.013 | 0.001 | 0.0267 | 0.0039 | 0.0313 | 0.0097 |
| 9 | G_T_T_T_G | – | – | – | 0.0013 | – | – |
Order of variants: rs1514280_rs4240157_rs4646156_rs4646155_rs6632677.
All variant haplotype: A_C_A_T_C.
3.6. Modifier variants in genes adjacent to ACE2 and their LD patterns
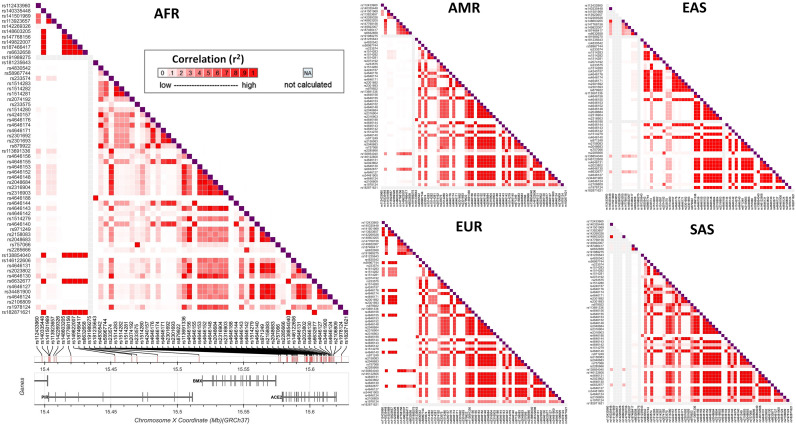
There are several genes on the X chromosome within ±0.5 Mbps of the disease associated variants of ACE2. Among these PIR is the only gene which is associated with inflammation. Frequencies of the modifier variants of ACE2 and PIR genes are given in supplementary Table 2 and pairwise LD between these variants is shown in Fig. 2. Modifier variants of PIR in the European super-population appear to have significantly stronger LD and form multiple small blocks compared to the other super-populations. Variants of PIR in the European super-population also have relatively more LDs with the variants of ACE2. PIR intronic modifier variant rs191989275 is present only in the European super-population (Fig. 2).
Fig. 2.
Pairwise LD among the modifier variants of ACE2 and PIR. LD measures are given as r2. For the missing alleles in any population LD could not be calculated (NA). AFR = African, AMR = Admixed American, EAS = East Asian, EUR = European, SAS = South Asian.
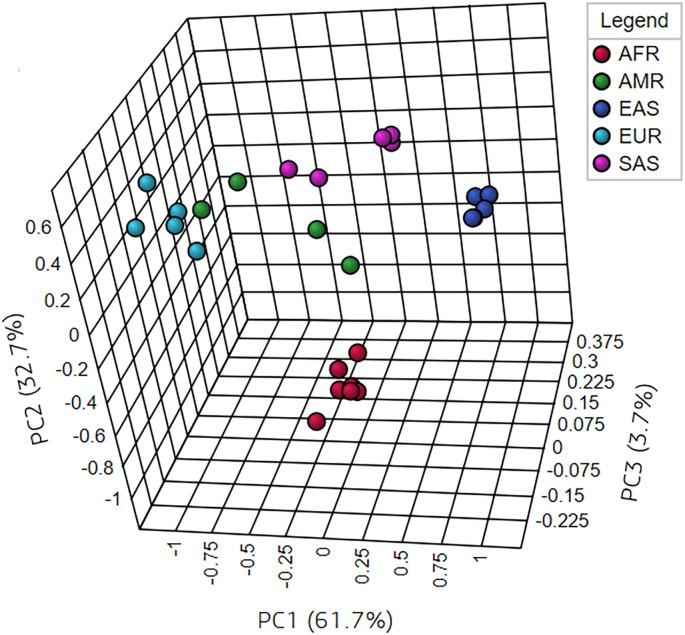
There is a striking difference in the frequencies of PIR modifier variants between the South Asian and other super-populations (Supplementary Table 2). PIR modifier variants at rs112433960, rs113923657, rs141501969, rs142269326, rs147768156, rs149822007, and rs191989275 loci are non-existent in the SAS super-population, although these are present in the AFR, AMR, EAS and/or EUR super-populations. In all super-populations, except SAS, the rs138854040 and rs6632677 variants of ACE2 form strong pairwise LD with several variants of the PIR gene. These differences in distribution of the above mentioned disease-associated and modifier variants are reflected in the un-supervised multivariate clustering of the populations based on VAFs (Fig. 3). As shown, populations of similar ethnic origins cluster together in the PCA plot.
Fig. 3.
Multivariate clustering of populations based on disease-associated and modifier VAFs of ACE2 and PIR. Clustering was performed using unsupervised principal component analysis (PCA) method. Only the first three principal components (PCs) are shown in this fig. (5 PCs were considered). As shown in the Fig. 3, the first three components could explain most part of the variance (cumulative variance for the PC1, PC2 and PC3 is 98.1%).
4. Discussion
COVID-19 patients with history of chronic illnesses such as hypertension, diabetes, cardiovascular disease, chronic lung disease and obesity are at higher risk of developing severe illness and deteriorating outcomes like acute respiratory distress syndrome (ARDS) and pneumonia (Chen et al., 2020b; Sanyaolu et al., 2020; Singh et al., 2020). ACE2 is a strong hypertensive quantitative trait locus (QTL) (Crackower et al., 2002). Several variants of ACE2 are also associated with dyslipidemia, type 2 diabetes mellitus, atrial fibrillation and left ventricular hypertrophy, among others. Based on the global distribution of COVID-19, the cases and mortality rates have been higher in the European and the American populations (World Health Organization, 2020; Yamamoto and Bauer, 2020). In this study, we have explored the interethnic and interpopulation variability in the distribution of ACE2 genetic variants that are associated with COVID-19 comorbidities.
Generally, ACE2 confers a protective effect on the epithelial cells of the lungs through the ACE2-Angiotensin (1–7)-MAS receptor axis that reduces inflammation, dilates blood vessels and reduces thrombosis. When ACE2 level is reduced on cell surface, the protective effect through ACE2-Angiotensin (1–7)-MAS receptor axis is minimized and amplification of the ACE-Angiotensin II-AT1 receptor axis occurs. ACE-Angiotensin II-AT1 receptor axis is responsible for various effects (such as vasoconstriction, increased inflammation, fibrosis and pulmonary damage) mediated by the Ang II (Verdecchia et al., 2020b). Individuals with higher ACE activity (thus, a more active ACE-Angiotensin II-AT1 receptor axis) in conjunction with reduced ACE2 activity (thus, a less active ACE2-Angiotensin (1–7)-MAS receptor axis to counter-regulate the ACE-Angiotensin II-AT1 receptor axis effects) are more susceptible to hypertension in association with classical cardiovascular risk factors such as old age, dyslipidemia, and diabetes (Ghafouri-Fard et al., 2020; Pinheiro et al., 2019).
When SARS-CoV-2 enters the lung epithelial cells, the ACE2 receptor is internalized and, thus, removed from the functional site, compromising the balance between ACE and ACE2 activity. This, therefore, remove the protective effect of the ACE2-Angiotensin (1–7)-MAS receptor axis. Decreased level of ACE2 contributes to vasoconstriction, increased inflammation, fibrosis and pulmonary damage and ultimately severe consequences of SARS-CoV-2 infection (Samavati and Uhal, 2020; Verdecchia et al., 2020a). In addition, ACE2 deficiency leads to Ang II accumulation. This elevated level of Ang II induces the production of pro-inflammatory cytokines and soluble IL-6 receptor, followed by STAT3 and NF-κB pathway-mediated activation of IL-6, which in turn causes overproduction of pro-inflammatory cytokines and chemokines including IL-6, and thus sets off a positive feedback loop (Hirano and Murakami, 2020). It is also known that ACE2 deficiency comes with old age, more in male than in females, and also diabetes mellitus (possibly due to excessive glycosylation) (Verdecchia et al., 2020b). ACE2 deficiency is also associated with Ang II induced exacerbation of hypertension and cardiac hypertrophy and maladaptive left ventricular remodeling after myocardial infarction (Kassiri et al., 2009; Pal and Bhansali, 2020; Xudong et al., 2006). Thus, people with existing ACE2 deficiency may be affected more severely by the loss of ACE2 following SARS-CoV-2 infection (Dijkman et al., 2012; Verdecchia et al., 2020a). The opposite may be true as well.
In the Indian population, a strong correlation exists between the variant allele at rs2285666 locus and lower COVID-19 infection rate as well as lower case fatality rates (CFR) (Srivastava et al., 2020). The variant allele at rs2285666 is present with > 0.5 frequencies in the EAS populations and three South Asian populations (BEB, STU, and ITU) (Table 2). The homozygous variant genotype at rs2285666 increases expression of ACE2 by 50% compared to the homozygous wild-type genotype (Asselta et al., 2020; Srivastava et al., 2020). Using the Human Splicing Finder (Desmet et al., 2009), Asselta et al. predicted the presence of variant allele at this splice region variant to increase the splice site strength by about 9.2% (Asselta et al., 2020). But in other studies, no significant splicing alteration by the variant allele at rs2285666 was found using the same tool (Novelli et al., 2020; Strafella et al., 2020). This calls for further analysis to have a clear understanding of the potential role of rs2285666 variant allele in the enhancement of ACE2 protein production.
In Han Chinese females, the heterozygous genotype at rs2285666 locus serves a protective role against essential hypertension (Zhang et al., 2018). Increased circulating Ang-(1–7) level was observed in females with the variant allele at the rs2106809 locus in Chinese Han population (Liu et al., 2016). The haplotype T_C_A_G_T_G_T_C_A_C_G_T_G_G_C with the variant alleles at rs4646142, rs2285666 and rs2106809 is highly prevalent in the EAS (0.429) and the SAS (0.429), but much lower in the EUR (0.165) populations.
rs4646188 variant allele is associated with hypertension, left ventricular hypertrophy and dyslipidemia (Luo et al., 2019; Pan et al., 2018; Patel et al., 2012). This variant allele is absent in the EAS populations (Table 2 and Fig. 1), which might be another candidate variant associated with protective role against severity of COVID-19.
Variant alleles at rs4240157, rs1978124, rs2048683, rs4646188, rs4646156 and rs879922 are associated with type 2 diabetes mellitus (Liu et al., 2018). Variant alleles at rs4830542, rs2106809, rs4240157, rs4646188, and rs879922 loci are associated with essential hypertension as well as RAS activation (Luo et al., 2019; Pan et al., 2018). rs4646155, rs4830542 and rs4240157 variants are associated with hypertension-related atrial fibrillation, and left atrial remodeling (Luo et al., 2019; Zhang et al., 2018). rs4646156 is a dyslipidemia associated chromatin interactive region related regulatory variant (Table 4). rs2074192 variant allele increases susceptibility to left ventricular hypertrophy (Fan et al., 2019). The variant allele at rs233575 is associated with higher left ventricular mass index (Lieb et al., 2006) and progression to type 2 diabetes mellitus (Liu et al., 2018). A strong LD block is seen in the EUR super-population among rs4830542, rs4240157, rs2074192, rs233575, rs1514280, rs879922 and rs4646156 loci (Fig. 1), all of which have modifier effect (Supplementary Table 2). A similar but weaker LD block is observed in the AMR populations. The VAFs at this LD block is >0.2 in two AMR (PUR and CLM) populations and the EUR populations (Table 2) and the haplotype with variant alleles at all these positions, except rs2074192, (C_C_G_A_C_C_A_C_A_G_T_C_G_A_T) has >0.01 frequencies in the EUR (0.277) and the AMR (0.179) populations. This might indicate that the people in these regions are at higher risk of pathological phenotypic effects associated with the variant alleles at rs4830542, rs4240157, rs2074192, rs233575, rs1514280, rs879922, and rs4646156 loci.
Admixed Latin American populations- CLM (Colombian), MXL (Mexican), PEL (Peruvians), and PUR (Puerto Ricans)- have distinct patterns of continental genetic admixture (Norris et al., 2018). The Puerto Ricans and the Colombians inherited more genetic content from the European ancestry than the Peruvians and the Mexicans (Homburger et al., 2015; Norris et al., 2018; Ruiz-Linares et al., 2014; Salzano and Sans, 2014). It is not unlikely to expect similar phenotypic effects in the European, Puerto Ricans and Colombians by the variants of ACE2 and PIR. In a previous study on drug response-related genetic variants, CLM and PUR formed a closer cluster with the Europeans in a population dendrogram (Ahsan et al., 2020). Fig. 3 shows a similar clustering of the populations based on the VAFs of the disease-associated and modifier variants of ACE2 and PIR.
The T_G_G_C haplotype consisting of the variant alleles at rs4646156, rs879922, rs4240157, and rs233575 may be associated with higher values for left ventricular mass index and septal wall thickness with a higher odds ratio for left ventricular hypertrophy in men (Lieb et al., 2006). Interestingly, the haplotypes #8 and #9 in Table 3 contain this specific combination of the variant alleles. #9 is found only in the European super-population (0.020) and #8 is most frequent in the European (0.277) followed by the admixed Americans (0.179).
Among the disease associated variants of ACE2, rs1514280 appears as an outlier at the first look based on its association with conditions like negative affectivity, temporal pain and psychological symptoms (Table 1). Long-lasting headache in COVID-19 patients is a frequent event, which is often severe and resistant to analgesics, in conjunction with anosmia (loss of the sense of smell) and/or ageusia (loss of taste functions of the tongue) (Rocha-Filho and Magalhães, 2020; Uygun et al., 2020). The haplotype with 3 regulatory variants at rs1514280, rs4240157 and rs4646156 loci (A_C_A_C_G) has a high frequency in the EUR (0.3146) and very low frequency in the EAS (0.0026) populations (Table 5). rs1514280 and rs4240157 loci are associated with circRNA formation. circRNA dysregulation is observed in multiple viral infections (Nahand et al., 2020). rs4646156 is an eQTL locus and this locus can modulate the expression of PIR gene in multiple tissues including esophagus, thyroid, adipose and artery (GTEx Consortium, 2013).
Strong LD and high VAFs at rs4830542, rs4240157, rs2074192, rs233575 and rs879922 loci in the EUR and two AMR (PUR and CLM) populations (Table 2) may be associated with more critical illness and higher death rates in the COVID-19 patients in these regions. On the other hand, very low frequencies of these alleles in the EAS populations might play protective role against progression to severe state by the comorbidities in SARS-CoV-2 infection. Although the VAFs at rs4830542, rs2074192, rs4240157 and rs879922 are > 0.3 in the AFR populations, no significant LD block is present among these variants in AFR populations. Another variant with high frequencies (> 0.35) in the EUR populations as well as PUR and CLM is rs1978124. People with rs1978124 variant have a high risk of developing type 2 diabetes (Liu et al., 2018). This variant allele is also associated with left ventricular remodeling in women (Patel et al., 2012).
COVID-19 is often associated with cytokine release syndrome (CRS) and its severity is associated with an elevated level of inflammatory cytokines and chemokines such as interleukin (IL)-2, IL-6, IL-1β, IL-7, IL-10, tumor necrosis factor (TNF), granulocyte colony-stimulating factor (G-CSF), monocyte chemoattractant protein-1 (MCP-1), and macrophage inflammatory protein 1 alpha (MIP1α) upon SARS-CoV-2 infection (Hojyo et al., 2020; Liu et al., 2020; Tay et al., 2020). The cytokine storm induces ARDS following infection with SARS-CoV-2 (Hirano and Murakami, 2020). IL-6 level elevates with progressive deterioration of illness and is higher in non-survivors compared to the survivors (Chen et al., 2020a; Zhou et al., 2020).
As a consequence of ACE2 internalization following SARS-CoV-2 infection, homeostatic responses try to restore ACE2 level by up-regulating ACE2 expression along with the other genes (including PIR) residing in a co-regulated cluster (Shovlin and Vizcaychipi, 2020). Double elite associations exist between two ACE2 enhancers (GH0XJ015596 and GH0XJ015579) and the promoter of PIR, which encodes the Pirin protein (Shovlin and Vizcaychipi, 2020). Pirin substantially facilitates binding of NF-κB p65 to DNA (Liu et al., 2013). Increased activation of NF-κB p65 pathway is associated with multiple diseases involving chronic inflammation (Giridharan and Srinivasan, 2018). Hyperactivation of NF-κB pathways leads to cytokine storm syndrome and increases COVID-19 severity (Hariharan et al., 2020). Furthermore, Pirin protein can bind to NF-κB, and modulate its DNA binding properties (Liu et al., 2013). Additionally, Pirin is overexpressed in response to oxidative stress (Brzóska et al., 2011). Oxidative stress can play a role in COVID-19 pathogenesis, perpetuate the cytokine storm cycle, blood clotting mechanism, and exacerbate hypoxia (Cecchini and Cecchini, 2020). In addition, cigarette smoke up-regulates PIR gene expression and the overexpressed Pirin protein may direct NF-κB towards a pro-apoptotic response causing death of airway epithelial cells (Gelbman et al., 2007). Active smoking is associated with increased COVID-19 severity (Gülsen et al., 2020). Hence, Pirin protein may be related to the severity of COVID-19.
Strong pairwise LD among variants in the PIR and ACE2 genes exists only in the European super-population. Because of the modifying nature of the variants, the presence of a certain combination of these SNPs, even when the individual variants have a low frequency, might bring about a significant change. The complete absence of the PIR variants in the South Asian population may contribute to an overall lower COVID-19 case to mortality rate despite the dense population in this region. More studies are needed to elucidate the contribution of this particular LD pattern to COVID-19-related mortality and severity.
5. Conclusion
In this study, we retrieved data on the disease associated genetic variants of ACE2 from databases and explored the VAFs and haplotype frequencies in 26 populations of five super-populations. We also explored the extents and levels of LDs among these variants in worldwide populations. Activity modifying (modifier) variants within ACE2 as well as closely located PIR genes show different and distinct distribution in populations of different ethnic origins. However, to confirm the population dependent associations with COVID-19 related severity, further studies with more representative samples from different populations are required.
List of the populations used in the study (from the 1000 Genomes Project database).
Frequencies of the modifier variants of ACE2 and PIR.
Funding
This study was conducted with a research grant from the University Grants Commission, Bangladesh.
Authors' contributions
TA, AAS- study design, TA, SSS, KF- data analysis, SSS, TA, KF- manuscript preparation, AAS- review of the manuscript.
Declaration of Competing Interest
There is no known conflict of interest.
Acknowledgments
This study was supported by a grant a research grant from the University Grants Commission, Bangladesh. The authors are thankful for the grant.
References
- Adler A.I., Stratton I.M., Neil H.A., Yudkin J.S., Matthews D.R., Cull C.A., Wright A.D., Turner R.C., Holman R.R. Association of systolic blood pressure with macrovascular and microvascular complications of type 2 diabetes (UKPDS 36): prospective observational study. BMJ (Clinical Research ed.) 2000;321:412–419. doi: 10.1136/bmj.321.7258.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan T., Urmi N.J., Sajib A.A. Heterogeneity in the distribution of 159 drug-response related SNPs in world populations and their genetic relatedness. PLoS One. 2020;15 doi: 10.1371/journal.pone.0228000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimohamadi Y., Sepandi M., Taghdir M., Hosamirudsari H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. Journal of Preventive Medicine and Hygiene. 2020;61:E304–E312. doi: 10.15167/2421-4248/jpmh2020.61.3.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apicella M., Campopiano M.C., Mantuano M., Mazoni L., Coppelli A., Del Prato S. COVID-19 in people with diabetes: understanding the reasons for worse outcomes. The Lancet Diabetes & Endocrinology. 2020;8:782–792. doi: 10.1016/S2213-8587(20)30238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging. 2020;12:10087–10098. doi: 10.18632/aging.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behl T., Kaur I., Bungau S., Kumar A., Uddin M.S., Kumar C., Pal G., Sahil Shrivastava K., Zengin G., Arora S. The dual impact of ACE2 in COVID-19 and ironical actions in geriatrics and pediatrics with possible therapeutic solutions. Life Sci. 2020;257:118075. doi: 10.1016/j.lfs.2020.118075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson K.K., Hu W., Weller A.H., Bennett A.H., Chen E.R., Khetarpal S.A., Yoshino S., Bone W.P., Wang L., Rabinowitz J.D., Voight B.F., Soccio R.E. Natural human genetic variation determines basal and inducible expression of PM20D1, an obesity-associated gene. Proc. Natl. Acad. Sci. U. S. A. 2019;116:23232–23242. doi: 10.1073/pnas.1913199116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzóska K., Stępkowski T.M., Kruszewski M. Putative proto-oncogene Pir expression is significantly up-regulated in the spleen and kidney of cytosolic superoxide dismutase-deficient mice. Redox Rep. 2011;16:129–133. doi: 10.1179/1351000211Y.0000000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini R., Cecchini A. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med. Hypotheses. 2020;143:110102. doi: 10.1016/j.mehy.2020.110102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J.A., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D.N. Functional intronic polymorphisms: buried treasure awaiting discovery within our genes. Hum Genomics. 2010;4:284–288. doi: 10.1186/1479-7364-4-5-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y., Scholey J., Ferrario C.M., Manoukian A.S., Chappell M.C., Backx P.H., Yagil Y., Penninger J.M. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- Das S., K R A., Birangal S.R., Nikam A.N., Pandey A., Mutalik S., Joseph A. Role of comorbidities like diabetes on severe acute respiratory syndrome coronavirus-2: a review. Life Sci. 2020;258:118202. doi: 10.1016/j.lfs.2020.118202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson Anne M., Wysocki J., Batlle D. Interaction of SARS-CoV-2 and other coronavirus with ACE (angiotensin-converting enzyme)-2 as their Main receptor. Hypertension. 2020;76:1339–1349. doi: 10.1161/HYPERTENSIONAHA.120.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer I.H., Bangalore S., Benetos A., Davis A.M., Michos E.D., Muntner P., Rossing P., Zoungas S., Bakris G. Diabetes and hypertension: a position statement by the American Diabetes Association. Diabetes Care. 2017;40:1273. doi: 10.2337/dci17-0026. [DOI] [PubMed] [Google Scholar]
- Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.-M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. Journal of Microbiology, Immunology, and Infection = Wei mian yu gan ran za zhi. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Deijs M., Milewska A., Pyrc K., Buelow E., van der Bijl A., van der Hoek L. Replication-dependent downregulation of cellular angiotensin-converting enzyme 2 protein expression by human coronavirus NL63. J. Gen. Virol. 2012;93:1924–1929. doi: 10.1099/vir.0.043919-0. [DOI] [PubMed] [Google Scholar]
- Esakandari H., Nabi-Afjadi M., Fakkari-Afjadi J., Farahmandian N., Miresmaeili S.-M., Bahreini E. A comprehensive review of COVID-19 characteristics. Biological Procedures Online. 2020;22:19. doi: 10.1186/s12575-020-00128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Wu G., Yue M., Ye J., Chen Y., Xu B., Shu Z., Zhu J., Lu N., Tan X. Hypertension and hypertensive left ventricular hypertrophy are associated with ACE2 genetic polymorphism. Life Sci. 2019;225:39–45. doi: 10.1016/j.lfs.2019.03.059. [DOI] [PubMed] [Google Scholar]
- Feng Q., Liu D., Lu Y., Liu Z. The interplay of renin-angiotensin system and toll-like receptor 4 in the inflammation of diabetic nephropathy. J Immunol Res. 2020;2020:6193407. doi: 10.1155/2020/6193407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Yu Q., Yao S., Luo L., Zhou W., Mao X., Li J., Duan J., Yan Z., Yang M., Tan H., Ma M., Li T., Yi D., Mi Z., Zhao H., Jiang Y., He Z., Li H., Nie W., Liu Y., Zhao J., Luo M., Liu X., Rong P., Wang W. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nat. Commun. 2020;11:4968. doi: 10.1038/s41467-020-18786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawałko M., Kapłon-Cieślicka A., Hohl M., Dobrev D., Linz D. COVID-19 associated atrial fibrillation: incidence, putative mechanisms and potential clinical implications. IJC Heart Vasc. 2020;30:100631. doi: 10.1016/j.ijcha.2020.100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbman B.D., Heguy A., O’Connor T.P., Zabner J., Crystal R.G. Upregulation of pirin expression by chronic cigarette smoking is associated with bronchial epithelial cell apoptosis. Respir. Res. 2007;8:10. doi: 10.1186/1465-9921-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium, Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafouri-Fard S., Noroozi R., Omrani M.D., Branicki W., Pośpiech E., Sayad A., Pyrc K., Łabaj P.P., Vafaee R., Taheri M., Sanak M. Angiotensin converting enzyme: a review on expression profile and its association with human disorders with special focus on SARS-CoV-2 infection. Vasc. Pharmacol. 2020;130:106680. doi: 10.1016/j.vph.2020.106680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giridharan S., Srinivasan M. Mechanisms of NF-κB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018;11:407–419. doi: 10.2147/JIR.S140188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gülsen A., Yigitbas B.A., Uslu B., Drömann D., Kilinc O. The effect of smoking on COVID-19 symptom severity: systematic review and meta-analysis. Pulm Med. 2020;2020:7590207. doi: 10.1155/2020/7590207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wang J. rSNPBase 3.0: an updated database of SNP-related regulatory elements, element-gene pairs and SNP-based gene regulatory networks. Nucleic Acids Res. 2018;46:D1111–D1116. doi: 10.1093/nar/gkx1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan A., Hakeem A.R., Radhakrishnan S., Reddy M.S., Rela M. The role and therapeutic potential of NF-kappa-B pathway in severe COVID-19 patients. Inflammopharmacology. 2020:1–10. doi: 10.1007/s10787-020-00773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52:731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojyo S., Uchida M., Tanaka K., Hasebe R., Tanaka Y., Murakami M., Hirano T. How COVID-19 induces cytokine storm with high mortality. Inflammation and Regeneration. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A., Gäbel G., Beutner F., Scholz M., Thiery J., Musunuru K., Krohn K., Mann M., Teupser D. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homburger J.R., Moreno-Estrada A., Gignoux C.R., Nelson D., Sanchez E., Ortiz-Tello P., Pons-Estel B.A., Acevedo-Vasquez E., Miranda P., Langefeld C.D., Gravel S., Alarcón-Riquelme M.E., Bustamante C.D. Genomic insights into the ancestry and demographic history of South America. PLoS Genet. 2015;11:e1005602. doi: 10.1371/journal.pgen.1005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiri Z., Zhong J., Guo D., Basu R., Wang X., Liu Peter P., Scholey James W., Penninger Josef M., Oudit Gavin Y. Loss of angiotensin-converting enzyme 2 accelerates maladaptive left ventricular remodeling in response to myocardial infarction. Circ. Heart Fail. 2009;2:446–455. doi: 10.1161/CIRCHEARTFAILURE.108.840124. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lieb W., Graf J., Götz A., König I.R., Mayer B., Fischer M., Stritzke J., Hengstenberg C., Holmer S.R., Döring A., Löwel H., Schunkert H., Erdmann J. Association of angiotensin-converting enzyme 2 (ACE2) gene polymorphisms with parameters of left ventricular hypertrophy in men. J. Mol. Med. 2006;84:88–96. doi: 10.1007/s00109-005-0718-5. [DOI] [PubMed] [Google Scholar]
- Lin H., Hargreaves K.A., Li R., Reiter J.L., Wang Y., Mort M., Cooper D.N., Zhou Y., Zhang C., Eadon M.T., Dolan M.E., Ipe J., Skaar T.C., Liu Y. RegSNPs-intron: a computational framework for predicting pathogenic impact of intronic single nucleotide variants. Genome Biol. 2019;20:254. doi: 10.1186/s13059-019-1847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Rehmani I., Esaki S., Fu R., Chen L., de Serrano V., Liu A. Pirin is an iron-dependent redox regulator of NF-κB. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9722–9727. doi: 10.1073/pnas.1221743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Chen Y., Zhang P., Zhong J., Jin L., Zhang C., Lin S., Wu S., Yu H. Association between circulating levels of ACE2-Ang-(1-7)-MAS axis and ACE2 gene polymorphisms in hypertensive patients. Medicine. 2016;95 doi: 10.1097/MD.0000000000003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li Y., Guan T., Lai Y., Shen Y., Zeyaweiding A., Zhao H., Li F., Maimaiti T. ACE2 polymorphisms associated with cardiovascular risk in Uygurs with type 2 diabetes mellitus. Cardiovasc. Diabetol. 2018;17:127. doi: 10.1186/s12933-018-0771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020;111:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Gonzalez K., Padilla-Rodríguez E., Texis T., Gutiérrez M.N., Rodríguez-Dorantes M., Cuevas-Córdoba B., Ramírez-García E., Mino-León D., Sánchez-García S., Gonzalez-Covarrubias V. Allele frequency of ACE2 intron variants and its association with blood pressure. DNA Cell Biol. 2020;39:2095–2101. doi: 10.1089/dna.2020.5804. [DOI] [PubMed] [Google Scholar]
- Luo Y., Liu C., Guan T., Li Y., Lai Y., Li F., Zhao H., Maimaiti T., Zeyaweiding A. Association of ACE2 genetic polymorphisms with hypertension-related target organ damages in South Xinjiang. Hypertens. Res. 2019;42:681–689. doi: 10.1038/s41440-018-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Song K., Huang Y. Coronavirus Disease-2019 (COVID-19) and cardiovascular complications. J. Cardiothorac. Vasc. Anesth. 2020;S1053-0770(1020):30400–30406. doi: 10.1053/j.jvca.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics (Oxford, England) 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles-Fernández A., Duran-Lozano L., Montalban G., Bonache S., López-Perolio I., Menéndez M., Santamariña M., Behar R., Blanco A., Carrasco E., López-Fernández A., Stjepanovic N., Balmaña J., Capellá G., Pineda M., Vega A., Lázaro C., de la Hoya M., Diez O., Gutiérrez-Enríquez S. Computational tools for splicing defect prediction in breast/ovarian cancer genes: how efficient are they at predicting RNA alterations? Front. Genet. 2018;9:366. doi: 10.3389/fgene.2018.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison F.S., Locke J.M., Wood A.R., Tuke M., Pasko D., Murray A., Frayling T., Harries L.W. The splice site variant rs11078928 may be associated with a genotype-dependent alteration in expression of GSDMB transcripts. BMC Genomics. 2013;14:627. doi: 10.1186/1471-2164-14-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahand J.S., Jamshidi S., Hamblin M.R., Mahjoubin-Tehran M., Vosough M., Jamali M., Khatami A., Moghoofei M., Baghi H.B., Mirzaei H. Circular RNAs: new epigenetic signatures in viral infections. Front. Microbiol. 2020;11:1853. doi: 10.3389/fmicb.2020.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., Hou C., Wang H., Liu J., Yang D., Xu Y., Cao Z., Gao Z. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24:422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris E.T., Wang L., Conley A.B., Rishishwar L., Mariño-Ramírez L., Valderrama-Aguirre A., Jordan I.K. Genetic ancestry, admixture and health determinants in Latin America. BMC Genomics. 2018;19:861. doi: 10.1186/s12864-018-5195-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli A., Biancolella M., Borgiani P., Cocciadiferro D., Colona V.L., D’Apice M.R., Rogliani P., Zaffina S., Leonardis F., Campana A., Raponi M., Andreoni M., Grelli S., Novelli G. Analysis of ACE2 genetic variants in 131 Italian SARS-CoV-2-positive patients. Hum Genomics. 2020;14:29. doi: 10.1186/s40246-020-00279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R., Bhansali A. COVID-19, diabetes mellitus and ACE2: the conundrum. Diabetes Res. Clin. Pract. 2020;162 doi: 10.1016/j.diabres.2020.108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Wang T., Li Y., Guan T., Lai Y., Shen Y., Zeyaweiding A., Maimaiti T., Li F., Zhao H., Liu C. Association of ACE2 polymorphisms with susceptibility to essential hypertension and dyslipidemia in Xinjiang, China. Lipids Health Dis. 2018;17:241. doi: 10.1186/s12944-018-0890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.K., Wai B., Ord M., MacIsaac R.J., Grant S., Velkoska E., Panagiotopoulos S., Jerums G., Srivastava P.M., Burrell L.M. Association of ACE2 genetic variants with blood pressure, left ventricular mass, and cardiac function in Caucasians with type 2 diabetes. Am. J. Hypertens. 2012;25:216–222. doi: 10.1038/ajh.2011.188. [DOI] [PubMed] [Google Scholar]
- Peng Y., Xiong D., Zhao L., Ouyang W., Wang S., Sun J., Zhang Q., Guan P., Xie L., Li W., Li G., Yan J., Li X. Chromatin interaction maps reveal genetic regulation for quantitative traits in maize. Nat. Commun. 2019;10:2632. doi: 10.1038/s41467-019-10602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Bickmore W., Dean A., Nobrega M.A., Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro D.S., Santos R.S., Jardim P.C.B.V., Silva E.G., Reis A.A.S., Pedrino G.R., Ulhoa C.J. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: a genetic association study in Brazilian patients. PLoS One. 2019;14 doi: 10.1371/journal.pone.0221248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pundir S., Martin M.J., O'Donovan C. UniProt Protein Knowledgebase. Methods Mol. Biol. 2017;1558:41–55. doi: 10.1007/978-1-4939-6783-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Jones A.L., Grant P.J., Carter A.M., Turner A.J., Hooper N.M. Circulating activities of angiotensin-converting enzyme, its homolog, angiotensin-converting enzyme 2, and neprilysin in a family study. Hypertension. 2006;48:914–920. doi: 10.1161/01.HYP.0000244543.91937.79. [DOI] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Northwell C.-R.C., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Filho P.A.S., Magalhães J.E. Headache associated with COVID-19: frequency, characteristics and association with anosmia and ageusia. Cephalalgia: an International Journal of Headache. 2020;40:1443–1451. doi: 10.1177/0333102420966770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Linares A., Adhikari K., Acuña-Alonzo V., Quinto-Sanchez M., Jaramillo C., Arias W., Fuentes M., Pizarro M., Everardo P., de Avila F., Gómez-Valdés J., León-Mimila P., Hunemeier T., Ramallo V., Silva de Cerqueira C.C., Burley M.-W., Konca E., de Oliveira M.Z., Veronez M.R., Rubio-Codina M., Attanasio O., Gibbon S., Ray N., Gallo C., Poletti G., Rosique J., Schuler-Faccini L., Salzano F.M., Bortolini M.-C., Canizales-Quinteros S., Rothhammer F., Bedoya G., Balding D., Gonzalez-José R. Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzano F.M., Sans M. Interethnic admixture and the evolution of Latin American populations. Genet. Mol. Biol. 2014;37:151–170. doi: 10.1590/s1415-47572014000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavati L., Uhal B.D. ACE2, much more than just a receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020;10:317. doi: 10.3389/fcimb.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., Hosein Z., Padda I., Mangat J., Altaf M. SN Comprehensive Clinical Medicine. 2020. Comorbidity and its impact on patients with COVID-19; pp. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S., Arima H., Asayama K., Hoshide S., Ichihara A., Ishimitsu T., Kario K., Kishi T., Mogi M., Nishiyama A., Ohishi M., Ohkubo T., Tamura K., Tanaka M., Yamamoto E., Yamamoto K., Itoh H. Hypertension and related diseases in the era of COVID-19: a report from the Japanese Society of Hypertension Task Force on COVID-19. Hypertens. Res. 2020;43:1028–1046. doi: 10.1038/s41440-020-0515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shovlin C.L., Vizcaychipi M.P. Vascular inflammation and endothelial injury in SARS-CoV-2 infection: the overlooked regulatory cascades implicated by the ACE2 gene cluster. QJM. 2020 doi: 10.1093/qjmed/hcaa241. QJM, hcaa241, (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Gupta R., Ghosh A., Misra A. Diabetes in COVID-19: prevalence, pathophysiology, prognosis and practical considerations. Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 2020;14:303–310. doi: 10.1016/j.dsx.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhy H., Kumar R., Lewerentz J., Lizana L., Stenberg P. Highly interacting regions of the human genome are enriched with enhancers and bound by DNA repair proteins. Sci. Rep. 2019;9:4577. doi: 10.1038/s41598-019-40770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Bandopadhyay A., Das D., Pandey R.K., Singh V., Khanam N., Srivastava N., Singh P.P., Dubey P.K., Pathak A., Gupta P., Rai N., Sultana G.N.N., Chaubey G. Genetic association of ACE2 rs2285666 polymorphism with COVID-19 spatial distribution in India. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.564741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stępień E., Costa M.C., Kurc S., Drożdż A., Cortez-Dias N., Enguita F.J. The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases. Acta Pharmacol. Sin. 2018;39:1085–1099. doi: 10.1038/aps.2018.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E., Kiat H., McLachlan C.S. Atrial fibrillation in COVID-19: a review of possible mechanisms. FASEB J. 2020;34:11347–11354. doi: 10.1096/fj.202001613. [DOI] [PubMed] [Google Scholar]
- Strafella C., Caputo V., Termine A., Barati S., Gambardella S., Borgiani P., Caltagirone C., Novelli G., Giardina E., Cascella R. Analysis of ACE2 genetic variability among populations highlights a possible link with COVID-19-related neurological complications. Genes (Basel) 2020;11:741. doi: 10.3390/genes11070741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L., Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W.L.W., Lim B.T.S., Anene-Nzelu C.G.O., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D.A., Li P.Y.Q., Richards A.M., Foo R.S.Y. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas Merlin C., Pickering Raelene J., Tsorotes D., Koitka A., Sheehy K., Bernardi S., Toffoli B., Nguyen-Huu T.-P., Head Geoffrey A., Fu Y., Chin-Dusting J., Cooper Mark E., Tikellis C. Genetic Ace2 deficiency accentuates vascular inflammation and atherosclerosis in the ApoE knockout mouse. Circ. Res. 2010;107:888–897. doi: 10.1161/CIRCRESAHA.110.219279. [DOI] [PubMed] [Google Scholar]
- Uygun Ö., Ertaş M., Ekizoğlu E., Bolay H., Özge A., Kocasoy Orhan E., Çağatay A.A., Baykan B. Headache characteristics in COVID-19 pandemic-a survey study. The Journal of Headache and Pain. 2020;21:121. doi: 10.1186/s10194-020-01188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. European Journal of Internal Medicine. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Long B., Liu F., Wang J.-X., Liu C.-Y., Zhao B., Zhou L.-Y., Sun T., Wang M., Yu T., Gong Y., Liu J., Dong Y.-H., Li N., Li P.-F. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2020. WHO Coronavirus Disease (COVID-19) Dashboard. [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xudong X., Junzhu C., Xingxiang W., Furong Z., Yanrong L. Age- and gender-related difference of ACE2 expression in rat lung. Life Sci. 2006;78:2166–2171. doi: 10.1016/j.lfs.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Bauer G. Apparent di ff erence in fatalities between Central Europe and East Asia due to SARS-COV-2 and COVID-19: four hypotheses for possible explanation. Med. Hypotheses. 2020;144:110160. doi: 10.1016/j.mehy.2020.110160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., Billis K., Boddu S., Marugán J.C., Cummins C., Davidson C., Dodiya K., Fatima R., Gall A., Giron C.G., Gil L., Grego T., Haggerty L., Haskell E., Hourlier T., Izuogu O.G., Janacek S.H., Juettemann T., Kay M., Lavidas I., Le T., Lemos D., Martinez J.G., Maurel T., McDowall M., McMahon A., Mohanan S., Moore B., Nuhn M., Oheh D.N., Parker A., Parton A., Patricio M., Sakthivel M.P., Abdul Salam A.I., Schmitt B.M., Schuilenburg H., Sheppard D., Sycheva M., Szuba M., Taylor K., Thormann A., Threadgold G., Vullo A., Walts B., Winterbottom A., Zadissa A., Chakiachvili M., Flint B., Frankish A., Hunt S.E., Iisley G., Kostadima M., Langridge N., Loveland J.E., Martin F.J., Morales J., Mudge J.M., Muffato M., Perry E., Ruffier M., Trevanion S.J., Cunningham F., Howe K.L., Zerbino D.R., Flicek P. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cong M., Wang N., Li X., Zhang H., Zhang K., Jin M., Wu N., Qiu C., Li J. Association of angiotensin-converting enzyme 2 gene polymorphism and enzymatic activity with essential hypertension in different gender: a case-control study. Medicine. 2018;97:e12917. doi: 10.1097/MD.0000000000012917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the populations used in the study (from the 1000 Genomes Project database).
Frequencies of the modifier variants of ACE2 and PIR.