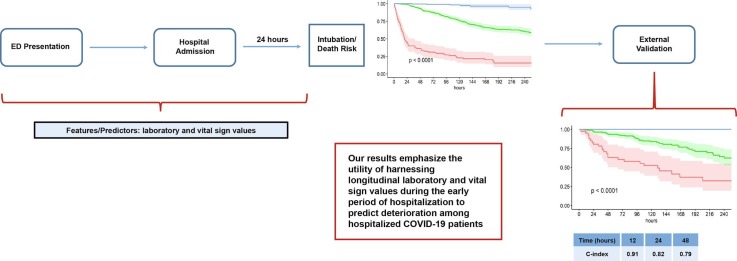
Graphical abstract
Keywords: COVID-19, Prediction, Machine learning, EMR, Deterioration, Intubation
Abstract
From early March through mid-May 2020, the COVID-19 pandemic overwhelmed hospitals in New York City. In anticipation of ventilator shortages and limited ICU bed capacity, hospital operations prioritized the development of prognostic tools to predict clinical deterioration. However, early experience from frontline physicians observed that some patients developed unanticipated deterioration after having relatively stable periods, attesting to the uncertainty of clinical trajectories among hospitalized patients with COVID-19. Prediction tools that incorporate clinical variables at one time-point, usually on hospital presentation, are suboptimal for patients with dynamic changes and evolving clinical trajectories. Therefore, our study team developed a machine-learning algorithm to predict clinical deterioration among hospitalized COVID-19 patients by extracting clinically meaningful features from complex longitudinal laboratory and vital sign values during the early period of hospitalization with an emphasis on informative missing-ness. To incorporate the evolution of the disease and clinical practice over the course of the pandemic, we utilized a time-dependent cross-validation strategy for model development. Finally, we validated our prediction model on an external validation cohort of COVID-19 patients served in a demographically distinct population from the training cohort. The main finding of our study is the identification of risk profiles of early, late and no clinical deterioration during the course of hospitalization. While risk prediction models that include simple predictors at ED presentation and clinical judgement are able to identify any deterioration vs. no deterioration, our methodology is able to isolate a particular risk group that remain stable initially but deteriorate at a later stage of the course of hospitalization. We demonstrate the superior predictive performance with the utilization of laboratory and vital sign data during the early period of hospitalization compared to the utilization of data at presentation alone. Our results will allow efficient hospital resource allocation and will motivate research in understanding the late deterioration risk group.
1. Introduction
The COVID-19 pandemic has disrupted the United States (US) healthcare system in unprecedented ways. As of October 7, 2020, more than seven million confirmed cases of COVID-19 and over two hundred thousand deaths were recorded in the US alone [16]. New York City (NYC) was the epicenter during the initial surge of the pandemic in the US from early March to mid-May 2020. It served as an early example for hospital systems nationwide preparing for their own surge of cases. COVID-19 patients overwhelmed NYC hospital systems with shortages in supply of intensive care unit (ICU) beds, ventilators, inpatient floor beds, and personal protective equipment (PPE). Adequate surge staffing necessitated redeployment of medical professionals to unfamiliar roles, including physicians who were confronted by a novel disease and challenged to triage patients with unpredictable clinical courses [17]. Care management and telemedicine protocols had to adapt rapidly to unknown disease progression.
In anticipation of ventilator shortages and limited ICU bed capacity, hospital operations prioritized the development of prognostic tools to predict clinical deterioration. As a result, several prognostic tools for clinical deterioration have been published, a systematic review of which, though not comprehensive, is reported here [11]. This systematic review validated the findings of 17 studies in an external cohort of 411 patients admitted to a London hospital and found areas under the receiver operating characteristic (AUROC) curve ranged from 0.56 to 0.78 across the models. They concluded that no prognostic model demonstrated consistently higher net benefit compared to admission oxygen saturation on room air and age.
Most of these models utilize as predictors the clinical characteristics of patients at the time of ED presentation or hospital admission and ignore the longitudinal clinical trajectory in the course of hospitalization. Prognostic models that do consider longitudinal clinical trajectories often do so by reducing the longitudinal information into discrete instances e.g., few recorded values [10] or minimum/maximum value prior to prediction [21] or by over-simplifying the information into a few summary measures like linear trends [8]. Moreover, these features are sensitive to missing data which are typically imputed with single or multiple imputations [10], [21]. Imputing missing data assumes the underlying missing data mechanism is missing completely at random (MCAR) or missing at random (MAR) and is arguably only reliable when the missing rate is 30% or less. Not only are missing rates amongst many clinical laboratory (lab) markers high (because lab tests are often only drawn when necessitated), but by extension MCAR and MAR assumptions are violated because missingness is informative of clinical course. In this paper, we develop novel features from complex longitudinal clinical trajectories that account for informative missingness and also preserve clinical meaningfulness.
Additionally, most published prognostic models focus on a binary outcome in order to identify patients who develop severe disease at some point (whether needing ICU resources, intubation, or experiencing in-hospital mortality) or on the contrary, to identify patients who will not need intensive resources and can be safely discharged. Such models are unable to distinguish when deterioration occurs in the course of hospitalization due to the binary nature of the outcome. In their early experience, frontline physicians observed that some patients developed unanticipated deterioration after having relatively stable periods, attesting to the uncertainty of clinical trajectories among hospitalized patients with COVID-19 [19], [23]. Therefore, we develop prognostic tools operationalizing outcome as a time to event, or survival outcome, in order to distinguish stable patients who do not need ventilation from patients who might deteriorate at a later stage in their hospital course and from patients who deteriorate early in the hospital course. This will allow efficient hospital resource allocation (e.g. ICU beds, ventilators, staffing) as well as inform transfer decisions in healthcare settings without intensive care capabilities.
Uniquely positioned within the New York-Presbyterian (NYP) 10-hospital healthcare system, we developed predictive models of clinical deterioration among hospitalized COVID-19 patients by using machine-learning algorithms and harnessing longitudinal lab and vital sign values during hospitalization. We present novel methodology for extracting linear and complex non-linear trend features from longitudinal lab and vital sign values while considering informative missingness. The methodology of feature extraction from longitudinal laboratory values and vital signs is innovative in the following ways: 1) taking into account informative missingness of features extracted from longitudinal records (recorded with different frequency and time period for each patient); 2) eliminates the need to delete or impute informative missing records in the calculation of linear trends of recorded values, their frequency, their variability and whether they belong within a clinically normal range; 3) non-linear trends of records are captured by constructing clusters of longitudinal trajectories that take into account informative missingness and the fact that records are captured over different time-periods and frequency for each patient; and finally, 4) all defined features are simple, categorical and clinically interpretable. In order to account for timing of deterioration, we modelled deterioration as a survival outcome, or time to event outcome, in which time to event represented time to intubation/death. A key feature of a prognostic model is the definition of the index time at which the predictive model is supposed be applied in clinical practice. The index time determines which patients are included in the model because predictors are recorded prior to the index time and outcome necessarily occurs after the index time. In this paper, we considered 1) an early index time (i.e., 24 h since hospitalization) that has the advantage of predicting clinical deterioration early on in the course of hospitalization and 2) an index time during any point during the hospitalization (e.g. a randomly selected time-point) that ensures more accuracy due to the longer period of observation. Both definitions of index time have the potential to inform efficient hospital resource allocation by predicting time of clinical deterioration. Further, to incorporate the evolution of clinical practice over the course of the pandemic, we utilized a time-dependent cross-validation strategy for model development. Finally, we validated our final prediction model on an external cohort of COVID-19 patients being served in a different hospital in NYC from the training cohort.
2. Methods
2.1. Study population
The study included a prospective cohort of COVID-19 adult patients admitted to two New York Presbyterian hospitals: Weill Cornell Medicine (NYP-WCM) and Lower Manhattan Hospital (NYP-LMH), from March 3 (date of first positive case) to May 15, 2020. Inclusion required (1) hospital admission through the Emergency Department (ED) with at least 24 h free of invasive mechanical ventilation, (2) a positive reverse transcription polymerase chain reaction assay for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus upon hospital admission, (3) age at least 18 years, and (4) at least one lab or vital sign value recorded before 24 h of hospitalization. The WCM Institutional Review Board approved the study with waiver of informed consent.
2.2. Primary outcome
The primary outcome was intubation, defined as the requirement of invasive mechanical ventilation at any point during the hospital stay, or in-hospital mortality with no preceding intubation. The outcome for some patients was not observed due to hospital transfer, discharge, or last chart review for patients still in the hospital as of the date of data-extraction. For that reason, a time-to-event or survival analysis was performed that takes censoring of outcome into account. The index time started at 24 h post hospital admission for each patient, which we will refer to as the 24-hour index time. A subsequent analysis considered index time as a randomly selected time between this 24-hour index time and the patient-specific end time (i.e. intubation, death, discharge, transfer, or last chart review), which we will refer to as the random index time. A timeline diagram depicting 24-hour and random index time is included in Supplemental Fig. 1.
2.3. Predictor variables
Predictor variables collected at ED presentation included demographic characteristics, comorbidities, presenting symptoms, ED supplemental oxygen requirements, and initial chest X-ray results. Additional predictor variables included standardized features of longitudinal lab and vital sign values extracted from the EMR from patient-specific date of ED admission through index time. The following lab values were included in the analysis: white blood cell count, neutrophil percent, hemoglobin, platelet count, sodium level, blood urea nitrogen (BUN), potassium, glucose, albumin, total bilirubin, troponin-I, international normalization ratio, procalcitonin, pH (arterial and venous), pO2 (arterial), pCO2 (arterial and venous), carbon dioxide, creatinine, D-dimer, ferritin level, erythrocyte sedimentation rate, C-reactive protein, lactate W/B (arterial), total Sequential Organ Failure Assessment (SOFA) aggregate score, FiO2, SpO2, PaO2, SF ratio and PF ratio. Vital signs included temperature, heart rate, blood pressure (systolic and diastolic), respiratory rate, and SpO2. Level of supplemental oxygen support was also monitored from ED presentation to index time. There were six levels of supplemental oxygen support: room air (i.e. no support), oxygen mask or nasal cannula, high-flow nasal cannula, non-rebreather mask, non-invasive mechanical ventilation, and invasive mechanical ventilation.
2.4. Feature extraction of laboratory values and vital signs
The lab and vital sign measurements among the predictors were longitudinal in nature as they could have been measured repeatedly for a patient’s hospital stay. The longitudinal trajectories of lab and vital signs are likely to represent the complexities of a patient’s response to the disease and its treatment in the hospital. Therefore, it is necessary to extract meaningful and informative information from the labs and vital signs. Existing methods for extracting features from longitudinal measurements of lab values and vital signs are often limited to simple linear trends that are sensitive to missing data or utilize the first or last available values which ignore the time point at which they are measured [10], [21]. We present a methodology to extract features from longitudinal trajectories of lab values and vital signs that is novel in the way it captures both linear and non-linear trends and in the way these trends take into account an informative missing data mechanism. Missing EMR data is typically imputed either by single or multiple imputation methods [2], [5], [25] that assume the underlying missing data mechanism to be missing at random. Moreover, missing values should not be imputed when the missing rate is >30%, which is often the case for certain labs. While utilizing feature extraction algorithms (e.g. neural networks) can result in superior predictive performance, they have limited clinical relevance. Our feature extraction method retains the clinical relevance and interpretability of the features.
The mechanisms of recording data for labs and vitals are different. Vital signs come from machine-enabled automatic extraction, and thus are frequently recorded. Meanwhile, lab values are recorded following a physician’s order that depends on the patient’s clinical course, and thus are sparse and missing lab values are likely to be informative. Therefore, we conceptualize our feature extraction pipeline into three domains below. Definition of features may differ depending on the index time (24-hour or random) and types of measurements (lab values or vital signs) (See Table 1 ).
Table 1.
Features of labs and vitals for 24-hour and random index time.
| Feature | Labs |
Vitals |
|||
|---|---|---|---|---|---|
| 24-hr | random | 24-hr | random | ||
| Missing indicator | Y | Y | N | N | |
| Trend | 1) Originally recorded values | Y | Y | Y | Y |
| 2) Number of values recorded per calendar day | N* | Y | N | N | |
| 3) Variance of values recorded per calendar day | N* | Y | N | N | |
| 4) (Lower/Upper) abnormal values | Y | Y | Y | Y | |
| Clustering | LGMM | Y | Y | Y | N |
| DTW + Hierarchical clustering | N | N | N | Y | |
‘Y’ if the feature was calculated, ‘N’ if not.
Except SOFA score and variables used for defining SOFA score.
2.4.1. Handling of missing lab values
It is often the case that a patient has no value for an otherwise important lab marker because their clinical course did not necessitate it. On the other hand, a patient having a value for this marker may inform of a perhaps declining clinical course. During the early stages of the pandemic when there were shortages of personal protective equipment, lab tests were administered more judiciously. Therefore, the mechanism of missingness of labs is likely informative or missing not at random. Out of 38 measurements (32 labs and 6 vital signs), 9 (23.7%) measurements had a >50% missing rate, which makes imputing missing values inappropriate. To capture the informative missingness, a missing indicator of lab values was included as a feature. The same definition of missing indicator was used for both 24-hour and random index time. Vital signs are automatically extracted with much higher frequencies than lab values and missing indicator is either irrelevant or uninformative.
2.4.2. Trend features
To capture the longitudinal trends of lab values and vital signs, we defined a collection of trend features that represent the patient’s clinical course. Linear trends are typically defined as regression coefficients of labs or vitals with time. In the presence of missing values, traditional trend features would delete otherwise usable lab markers that have a high degree of missingness. We used a novel principle of defining trend features so that missing values are handled without deleting observations; i.e. we coded missing values as ‘zero’ in a derived feature such that the non-missing ‘zero’ of the derived feature and the missing value code ‘zero’ reflect similar information. The trend features were defined as correlations with time so that they remain unit-free and allows us to avoid the exclusion of missing data. The correlation coefficient was further discretized to three levels (−1, 0, +1); a strong positive correlation between +0.3 and +1 was coded as +1, a strong negative correlation between −1 and −0.3 was coded as −1, and a weak/no correlation between −0.3 an d 0.3 was coded as 0. Defining trend features in this fashion allowed us to code a missed lab data point as ‘zero’ to indicate a zero trend or no change over time.
In addition to the modification of trend features described above to handle missing data, we constructed four new sets of trend features of lab values and vital signs that capture a more nuanced picture of clinical deterioration using the same principles to deal with missing data. Specifically, the trend in daily frequency by which a certain lab is ordered, or the trend in daily variability of a lab marker or vital sign, or the trend in frequency by which a lab marker or vital sign is above or below the normal range, lend insights into clinical deterioration. For that reason, we defined trend by calculating the correlation coefficient with time, in the manner described above, of the following features: 1) originally recorded values, 2) number of values recorded per calendar day, 3) variance of values recorded within a calendar day, and 4) (lower/upper) abnormal values. The second and third sets were calculated to monitor the change in daily frequency and variability of lab values. A strong positive trend in daily frequency and variability may imply clinical deterioration while a strong negative trend may indicate improvement or stabilization of the condition therefore requiring less clinical attention. Vital signs were excluded for these two sets because the frequency and variability of automatic extraction of vital signs are not informative. Due to small number of calendar days from ED presentation to 24-hour post hospitalization, these two sets were calculated only for random index time. Using the principle of defining unit-free features in order avoid exclusion of missing data, lab values or vital signs outside their clinical normal range were labeled as lower- or upper- abnormal and the trend of the occurrence of each was tracked over time. Lower- and upper- abnormalities were considered separately due to their different clinical implication. A strong positive trend indicates a deteriorating condition in that labs or vitals are more likely to be outside their clinical normal range over time. Meanwhile, a strong negative trend indicates a clinical improvement with less chance of lower- or upper- abnormality in labs and vitals. As noted before, a missing lab value will be coded as a ‘zero’ trend in their lower- or upper- abnormal equating them to consistent normal lab values.
2.4.3. Clustering of trajectories
Although trend features capture linear trends over time, certain lab or vital signs might present non-linear trends (e.g. initial improvement with a rapid decline). To capture the non-linearity of a large number of features in an automated data-driven fashion, we performed unsupervised cluster analysis of trajectories of lab values and vital signs in order to identify sub-groups of individuals who have similar trajectories of lab values and vital signs. We adopted two different methods for clustering the trajectories of lab and vital values – namely latent growth mixture models (LGMM) [20] and dynamic time warping (DTW) [3], [4].
LGMM was used in the analysis of lab values with both 24-hour and random index time. LGMM assumes the existence of a finite number of latent clusters within a sample of longitudinal data and identifies the distinct pattern of trajectories within each latent cluster. We varied the number of clusters and determined the number of clusters based on the Bayesian Information Criterion (BIC) [22]. As the LGMM with two clusters was chosen for majority of lab values based on BIC, we fitted the LGMM with two clusters for all lab values for consistency. Patients with missing lab values were classified into a separate cluster, which resulted in the total of 3 clusters of trajectories.
For vital signs, two different strategies were employed for identifying clusters of trajectories. In the 24-hour index time analysis, the LGMM with two clusters was fitted. As vital signs were more densely observed than lab values, the average in a window of 2 h with a rolling overlap of 1 h was used for LGMM. Meanwhile, the random index time analysis of vital signs involved aligning temporal sequences of vital signs because each patient has a different length and frequency of recorded vital signs. We assumed that patients may exhibit a similar pattern of vital signs over time but the timing of certain state of vital signs may be different. DTW takes two sets of longitudinal data with different lengths and calculates the distance required for obtaining the optimal match of each. We implemented DTW with a constraint that the first [last] record of the one longitudinal data is matched with the first [last] record of the other longitudinal data. Pairwise distance between temporal sequences of vital signs via DTW was then calculated and clusters were determined using hierarchical clustering [13]. Compared to LGMM, the clustering of trajectories using DTW accounts for heterogeneity in timing of state of vital signs across patients and captures the changes in vital signs regardless of different length of observations.
2.5. Model development and cross validation (NYP-WCM Cohort)
We used a subset of the final cohort (patients last located at NYP-WCM) in order to train models for predicting time to intubation or death from index time. Predictor features for modelling included features recorded at the time of ED presentation and our novel lab and vital sign features derived up to index time (24-hour or random index time). The primary goal of our prediction modelling is to aid clinical decision making by providing a time-dependent predictive risk e.g. risk of intubation/death after 24 or 48 h post index time. For that reason, we utilized a time-dependent concordance index (Harrell’s C-index) [12] for assessing predictive accuracy at these various time points. In pre-processing, missing values at presentation were imputed by K-nearest neighbors.
To test the utility of our developed lab and vital sign features, we ran a sequence of models testing different combinations of the 3 domain groups of features (missing labs, trend features, and clustering of trajectories). These models included: 1) just baseline features recorded at the time of ED presentation, 2) baseline features and missing lab indicators, 3) baseline features and trend features, 4) baseline features and cluster features, 5) baseline features, missing lab indicators, and trend features, 6) baseline features, missing lab indicators, and cluster features, 7) baseline features, trend features, and cluster features, and 8) baseline features, missing lab indicators, trend features, and cluster features. All features besides baseline features were derived up to the index time of evaluation (24-hour or random index time).
2.5.1. Time-dependent cross validation
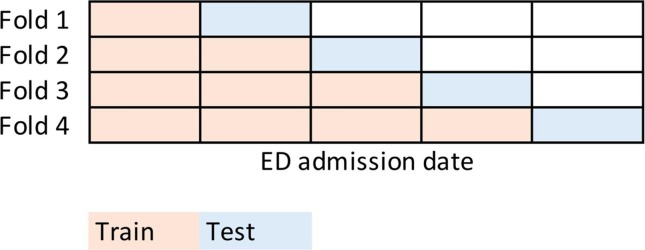
Unique to our prediction problem was the rapidly evolving care landscape at the surge of the COVID-19 pandemic in NYC. Medical professionals were redeployed to unfamiliar roles and challenged to triage patients with unpredictable clinical courses. The introduction of a novel disease required adaptation of care management and telemedicine protocols over time. In order to mimic this real-world phenomenon, we implemented a cross-validation strategy accounting for admission date. The training data was divided into five equally sized folds in the order of the admission date (see Fig. 1 ). Models were trained on the first fifth of the data (patients admitted earliest at the start of the pandemic) and C-indices calculated on the next fifth. In sequential fashion, methods learned on the data by adding the next fifth of patients and testing on the succeeding fifth.
Fig. 1.
Time-dependent cross-validation scheme.
Within each training step, cross-validation tuned specified hyper-parameters. Pre-processing steps were carried out within each fold that included eliminating predictors with near-zero variance and highly correlated predictors based on a 0.70 correlation coefficient cutoff.
2.5.2. Black-box method
We chose random survival forests (RSF) for right-censored time-to-event [14] outcome for our black-box method for two reasons – first, random forests is based on decision trees which naturally model higher order interactions between predictors and second, it has well validated open-source code that implements time-dependent c-indices. RSF is an extension of Breiman’s random forest (RF) method [6]. As in RF, RSF consists of individual trees grown from multiple bootstrapped samples of the data to create an ensemble of individual trees (forests) that reduces generalization error due to the introduction of randomness [6], [18].
In our case, optimal node splitting was determined by the log-rank test. Ensemble C-indices were obtained for each fold of cross-validation and averaged for estimated predictive performance. Forests were built with 1000 trees and the number of variables randomly selected at node splits was set as the square root of the total number of predictor variables.
2.5.3. Interpretable methods
We further implemented two interpretable methods to compare their performance with RSF in order assess the need of a black-box method. The first of which was classification and regression trees (CART) for survival outcome as outlined by Breiman et al. [7]. A simplified version of random forests, CART produces one learner tree, lending to interpretation by allowing a user to follow decision rules at node splits down the tree. To prevent overfitting, we tuned the cost complexity (cp) parameter within each fold via 5-fold cross validation. In CART, the cp parameter controls the growth of the tree by regularizing the size of the tree. C-indices were averaged across the outer cross-validation folds to estimate the predictive accuracy.
The second interpretable method we implemented was Cox proportional hazards regression. Because we had over 300 predictors for modelling, within each cross validation fold we performed a Cox proportional hazards elastic net (Cox-Elastic Net) regression with a shrinkage penalty, tuned through inner 10-fold cross-validation. From the fitted elastic net, we arbitrarily selected the top predictors with non-zero coefficients, up to a maximum of 10. These predictors were used to build a final Cox proportional hazards model within each fold. C-indices were averaged across outer cross-validation folds for estimated predictive performance.
2.5.4. Interpretation and risk stratification
As RSF is an ensemble of a large number of trees, it is not readily interpretable. We provided a ranking of the important predictors using a measure of variable importance (VIMP) that is based on Breiman-Cutler (BC) [6] and Ishwaran-Kogalur (IK) [14], [15] VIMP. In addition, we stratified patients into three risk profiles - early, late, and no deterioration - based on the bottom 10%, middle 10–70%, and top 30% of the ensemble predicted survival probability distribution at 24 h post index time (probability distribution of being alive and remaining intubation-free at 24 h post index time). Kaplan-Meier estimates of survival probabilities and 95% confidence intervals were then calculated and plotted for each risk profile and time point of evaluation.
2.6. External validation cohort (NYP-LMH)
Lastly, we applied the final RSF model on the cohort of patients last located at NYP-LMH in order to test performance. We calculated C-indices at 12, 24, and 48 h post index time. Cutoff values of the ensemble predicted survival probabilities at 24 h post index time were determined from the stratification of risk profiles in the training cohort and used to stratify the patients in the validation cohort into risk profiles. Kaplan-Meier estimates of survival probabilities and 95% confidence intervals were calculated and plotted for each risk profile and time point of interest.
3. Results
There were 1,045 patients in the NYP-WCM training cohort and 292 patients included in the NYP-LMH validation cohort. Median ages were 66 and 70, respectively. The NYP-WCM cohort had more male patients and fewer patients with age 65 years or more compared to the validation NYP-LMH cohort. Majority of the NYP-WCM patients were of white race whereas majority of NYP-LMH cohort were of Asian race (Table 2 ). The NYP-WCM patients had a higher frequency of requiring supplemental oxygen within 3 h of ED arrival, End State Renal Disease (ESRD), any cancer, any immunosuppression, symptoms of fever, symptoms of dyspnea and a lower rate of COPD compared to NYP-LMH cohort (see Table 2).
Table 2.
Patient characteristics at ED presentation: training (NYP-WCM) and validation (NYP-LMH) Cohorts.
| NYP-WCM |
NYP-LMH |
Overall |
|
|---|---|---|---|
| (N = 1045) | (N = 292) | (N = 1337) | |
| Demographics | |||
| Age | |||
| >=65 | 551 (52.7%) | 186 (63.7%) | 737 (55.1%) |
| Race | |||
| White | 400 (38.3%) | 61 (20.9%) | 461 (34.5%) |
| Black | 144 (13.8%) | 39 (13.4%) | 183 (13.7%) |
| Asian | 109 (10.4%) | 115 (39.4%) | 224 (16.8%) |
| Other | 220 (21.1%) | 47 (16.1%) | 267 (20.0%) |
| Not Specified | 172 (16.5%) | 30 (10.3%) | 202 (15.1%) |
| Sex | |||
| Male | 622 (59.5%) | 157 (53.8%) | 779 (58.3%) |
| BMI (kg/m^2) | |||
| <25 | 351 (33.6%) | 137 (46.9%) | 488 (36.5%) |
| 25 to <30 | 347 (33.2%) | 78 (26.7%) | 425 (31.8%) |
| >=30 | 332 (31.8%) | 69 (23.6%) | 401 (30.0%) |
| Missing | 15 (1.4%) | 8 (2.7%) | 23 (1.7%) |
| Active and/or former smoker/vaper | 295 (28.2%) | 86 (29.5%) | 381 (28.5%) |
| ED Supplemental Oxygen | |||
| Required supplemental oxygen within the first 3 h of arrival | 576 (55.1%) | 142 (48.6%) | 718 (53.7%) |
| Comorbidities | |||
| Diabetes Mellitus (DMI, DMII) | 321 (30.7%) | 96 (32.9%) | 417 (31.2%) |
| Hypertension (HTN) | 594 (56.8%) | 170 (58.2%) | 764 (57.1%) |
| Chronic Obstructive Pulmonary Disease (COPD) | 51 (4.9%) | 26 (8.9%) | 77 (5.8%) |
| Chronic Kidney Disease (CKD) | 51 (4.9%) | 16 (5.5%) | 67 (5.0%) |
| End Stage Renal Disease (ESRD) | 73 (7.0%) | 16 (5.5%) | 89 (6.7%) |
| Coronary Artery Disease (CAD) | 157 (15.0%) | 47 (16.1%) | 204 (15.3%) |
| Any Cancer | 85 (8.1%) | 13 (4.5%) | 98 (7.3%) |
| Any Immunosuppression | 35 (3.3%) | 0 (0%) | 35 (2.6%) |
| Symptoms | |||
| Fever | 734 (70.2%) | 168 (57.5%) | 902 (67.5%) |
| Cough | 724 (69.3%) | 191 (65.4%) | 915 (68.4%) |
| Diarrhea | 279 (26.7%) | 81 (27.7%) | 360 (26.9%) |
| Nausea or vomitingNausea or vomiting | 202 (19.3%) | 55 (18.8%) | 257 (19.2%) |
| Myalgias | 218 (20.9%) | 51 (17.5%) | 269 (20.1%) |
| Dyspnea | 683 (65.4%) | 157 (53.8%) | 840 (62.8%) |
| Initial Chest X-ray Findings | |||
| Unilateral Infiltrate | 110 (10.5%) | 43 (14.7%) | 153 (11.4%) |
| Bilateral Infiltrates | 764 (73.1%) | 193 (66.1%) | 957 (71.6%) |
| Pleural Effusion | 57 (5.5%) | 14 (4.8%) | 71 (5.3%) |
| Other | 58 (5.6%) | 21 (7.2%) | 79 (5.9%) |
| Outcome | |||
| intubation and/or in-hospital mortality | 295 (28.2%) | 69 (23.6%) | 364 (27.2%) |
Almost 30% (N = 295, 28%) of NYP-WCM patients experienced invasive ventilation or in-hospital mortality compared with 24% (N = 69) of NYP-LMH patients. There were 1339 patients included in analyses involving random index time (1047 from NYP-WCM and 292 from NYP-LMH), determined as having any lab or vital sign value recorded by this random time point.
3.1. Feature extraction
Our feature extraction pipeline was applied to thirty-two lab values and six vital signs that are reported in section 2.4. When using 24-hour post hospitalization as the index time, 5 features were calculated for each lab value including missing indicator, trend of originally recorded values, trend of lower- and upper- abnormality, and clusters of trajectories using LGMM. Accounting for two additional trend features for SOFA score (frequency and variance of values recorded per calendar day), a total of 160 features were calculated for labs. The same set of features with the exception of the missing data indicator was calculated for each vital sign resulting in a total of 24 vital sign features.
In the NYP-WCM training cohort, <1% of patients underwent an arterial blood gas test between ED presentation and 24-hour post hospitalization and had recorded pH, pCO2, and lactate W/B. On the other hand, lab values with the lowest rate of missing (≤1%) were white blood cell, hemoglobin, platelet count, and total SOFA aggregate score. Measurements with the strongest trend in originally recorded values (either +1 or −1) included glucose level (75.7%), white blood cell count (74.0%), BUN (73.6%), and platelet count (73.3%).
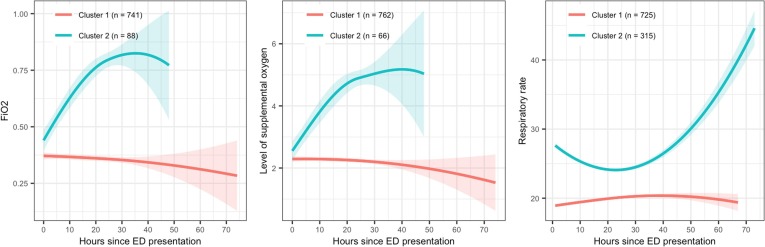
Clusters of trajectories for both lab values and vital signs were determined using LGMM for the 24-hour index time. Three clusters from LGMM were coded as 1, 2, and 9 where patients in Cluster 1 presented low/decreasing values of labs or vitals over time compared to those in Cluster 2 (high/increasing) on average. Cluster 9 indicates those with missing labs and plays an important role in comparing groups with certain trajectories of lab values to those with no lab tests.
Panels in Fig. 2 illustrate the clusters of trajectories of three lab/vitals in the NYP-WCM training cohort: FiO2 (left), level of supplementary oxygen (center), and respiratory rate (right). A solid line is the locally weighted scatterplot smoothing (LOESS) curve of individual trajectories of patients in one cluster. The shades are 95% confidence interval for the LOESS curve. The lines and shades are color-coded for LCMM clusters. Cluster 1 in these three panels represents patients with more stable condition, but the interpretation of clusters may be different for other labs and vitals.
Fig. 2.
Examples of LGMM clusters of trajectories. FiO2 (left), level of supplemental oxygen (center), respiratory rate (right) Red is for Cluster 1 and blue is for Cluster 2. Solid line: locally weighted scatterplot smoothing (LOESS) curve Shades: 95% confidence interval.
3.2. Predictive performance
The predictive performance, determined by cross-validated c-indices in the training cohort (NYP-WCM), of the proposed methods are presented in Table 3 .
Table 3.
Cross-validated C-indices: Training Cohort (NYP-WCM).
| Model 1 | Model 2 | Model 3 | |
|---|---|---|---|
| 12 hrs | |||
| RSF | 0.698 | 0.933 | 0.943 |
| CART | 0.725 | 0.813 | 0.850 |
| Cox-Elastic Net | 0.724 | 0.839 | 0.894 |
| 24 hrs | |||
| RSF | 0.690 | 0.920 | 0.927 |
| CART | 0.688 | 0.689 | 0.831 |
| Cox-Elastic Net | 0.687 | 0.792 | 0.868 |
| 48 hrs | |||
| RSF | 0.648 | 0.852 | 0.891 |
| CART | 0.661 | 0.565 | 0.821 |
| Cox-Elastic Net | 0.660 | 0.708 | 0.845 |
‘hrs’=hours since index time where index time contingent on model scenario.
‘CART’=Classification and Regression Tree for survival.
‘Cox-Elastic Net’=Cox proportional hazards regression with predictors chosen through elastic net regression.
‘Model 1’=predictors at ED presentation alone; index time defined at 24 h of hospitalization.
‘Model 2’=predictors at ED presentation including longitudinal laboratory and vital sign features extracted up to 24 h; index time defined at 24 h of hospitalization.
‘Model 3’=predictors at ED presentation including longitudinal laboratory and vital sign features extracted up to random index time; index time defined at random time as discussed in text.
Specifically, the c-indices at 12, 24 and 48 h post index time were obtained for three predictive methods (RSF, CART and Cox-Elastic Net) and three model scenarios: Model 1 – prediction made at 24-hour index time and predictors restricted to those obtained at ED presentation only, Model 2 – prediction made at 24-hour index time with predictors at ED presentation along with all derived lab and vital sign predictor features for 24-hour index time, and Model 3 – prediction made at random index time with predictors at ED presentation along with all derived lab and vital sign predictor features for random index time. The predictive performance of the multiple model scenarios presented in Table 3 suggests that predictors restricted to those at ED presentation show weak predictive performance of the outcome at 12, 24 and 48 h from the index time. Use of longitudinal features for the 24 h and random index time improves predictive performance substantially especially for the risk of intubation/death evaluated at 12 h and 24 h post-index time. The ability of all models to predict the outcome at 48 h post-index time is lower than that at 12 and 24 h post index time. A comparison of the predictive performance (model 2 of Table 3) using time-dependent cross-validation versus using conventional cross-validation showed that the time-dependent CV gave slightly more conservative estimates of predictive performance compared to conventional CV (Supplemental Table 1).
3.3. Comparative performance of features
We compared the predictive performance (cross-validated c-index) of each set of features extracted from lab values and vital signs i.e. missing lab indicators, trend features and clusters of trajectories (non-linear features) with the best performing ML algorithm – random survival forests and 24-hour index time. Specifically, we progressively added each group of features in 8 separate models as shown in Table 4 . The cluster features have the highest gain in predictive performance (model 4 vs 1), followed by the missing lab indicators (Model 2 vs 1) and trend features (Model 3 vs 1). In the presence of cluster features and missing lab features (Model 6), trend features do not improve the predictive performance (Model 8).
Table 4.
Comparative predictive performance of features (c-index).
| Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | Model 6 | Model 7 | Model 8 | |
|---|---|---|---|---|---|---|---|---|
| 12 hrs | ||||||||
| RSF | 0.698 | 0.892 | 0.807 | 0.924 | 0.892 | 0.941 | 0.911 | 0.933 |
| 24 hrs | ||||||||
| RSF | 0.690 | 0.850 | 0.793 | 0.894 | 0.869 | 0.924 | 0.895 | 0.920 |
| 48 hrs | ||||||||
| RSF | 0.648 | 0.776 | 0.745 | 0.827 | 0.803 | 0.839 | 0.835 | 0.852 |
All models employed 24-hour index time.
‘RSF’=Random Survival Forest.
‘Model 1’=baseline features.
‘Model 2’=baseline features, missing lab indicators.
‘Model 3’=baseline features, trend features.
‘Model 4’=baseline features, cluster features.
‘Model 5’=baseline features, missing lab indicators, trend features.
‘Model 6’=baseline features, missing lab indicators, cluster features.
‘Model 7’=baseline features, trend features, cluster features.
‘Model 8’=baseline features, missing lab indicators, trend features, cluster features.
3.4. Predictor importance
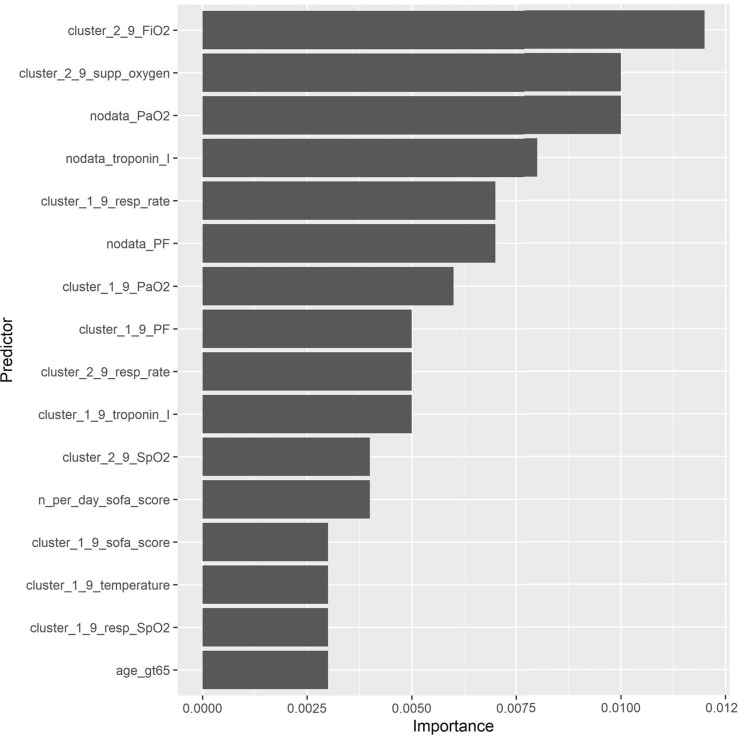
The predictive performance of RSF, a black box method, outperforms the interpretable methods such as Cox-Elastic Net and CART in terms of predictive accuracy. We chose the RSF model at 24-hour index time utilizing baseline features and all domain features (i.e. scenario 2 for RSF of Table 3 or analogously scenario 8 of Table 4) as our final model for validation in the NYP-LMH cohort. The top most important predictors determined as those that explain 70% of the total cumulative importance are shown in Fig. 3 .
Fig. 3.
Predictor importance from RSF on training cohort (NYP-WCM). The top most important predictors determined as those that explain 70% of the total cumulative importance are shown. ‘age_gt65’=≥65 years of age. For labs and vitals, predictors are labeled as ‘<feature>_<lab/vital>’. Features are labeled as below: ‘cluster_1_9’=Low/decreasing (Cluster 1) compared to high/increasing (Cluster 2) or no data/missing value (Cluster 9). Clusters were identified using LGMM. ‘cluster_2_9’=High/increasing (Cluster 2) compared to low/decreasing (Cluster 1) or no data/missing value (Cluster 9). Clusters were identified using LGMM. ‘nodata’=No available data or missing lab or vital sign ‘n_per_day_trend’=Trend in the number of values recorded per calendar day Labs and vital signs are labeled as below: ‘supp_oxygen’=Level of supplemental oxygen. ‘resp_rate’=Respiratory rate.
We found that three of the top five strongest predictors were LGMM cluster features for FiO2, level of supplemental oxygen, and respiratory rate. Specifically, the identification of cluster 2 for FiO2 (patients with increasing levels over time) was the strongest predictor (compared to stable/decreasing or no data), followed by the identification of cluster 2 for level of supplemental oxygen (patients with increasing levels over time compared to stable/decreasing or no data), and the identification of cluster 1 for respiratory rate (patients with lower initial respiratory rates which remain stable over time compared to higher initial respiratory rates which increase over time or no data). Fig. 2 represents the cluster-specific average trajectories for each of these measures. Patients not belonging to either cluster 1 or cluster 2 for each measure had no data available and were identified as cluster 9 (not shown). Besides these three cluster features, indicators of no available data for PaO2 and troponin I were also among the top five. Having no data for these measures is informative as physicians most likely did not suspect deterioration.
3.5. Risk stratification
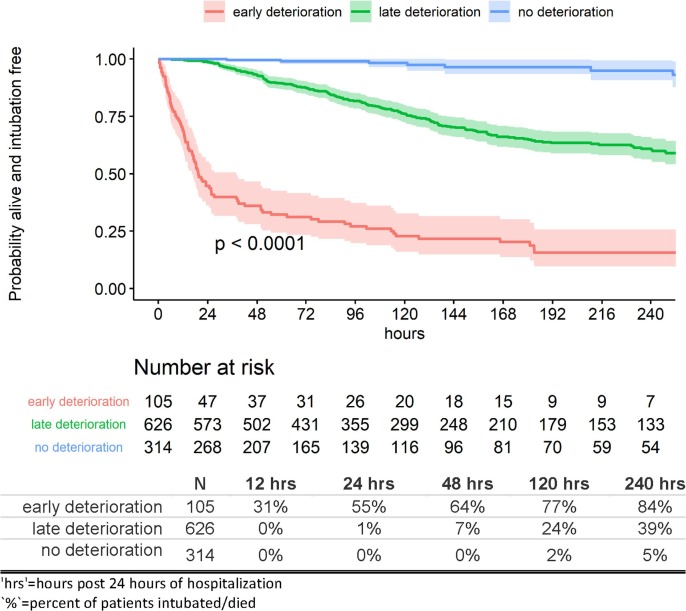
Three risk profiles (early, late and no deterioration) were created from the ensemble predicted survival probabilities at 24 h post index time. The probability of being alive and remaining intubation-free through 24 h after a stable initial 24 h of hospitalization ranged from 0% to 85.6% for the early deterioration risk profile, >85.6% to 98.6% for the late deterioration risk profile, and >98.6% for the no deterioration risk profile.
Notably, these results demonstrate that patients in the late deterioration risk profile have lower probability of deteriorating at the beginning of their hospital stay and the probability of deterioration increases during the later phase of their hospitalization (Fig. 4 ).
Fig. 4.
Kaplan-Meier estimates by risk profile: training cohort (NYP-WCM).
Specifically, 55% of patients deteriorate by 24 h after an initial stable condition within 24 h of hospitalization in the early deterioration risk profile compared to 1% in the late deterioration risk profile and 0% in the no deterioration risk profile. By 5 days after index time, 77%, 24%, and 2% deteriorate and by 10 days after index time, 84%, 39%, and 5% of patients deteriorate, respectively (Fig. 4).
Table 5 shows the differences in patient characteristics at ED presentation between these risk profiles. The frequency of a number of patient characteristics decreased with decreasing risk profile, including the proportion of patients ≥ 65 years of age, patients reporting a history of smoking/vaping, requiring supplemental oxygen within the first 3 h of ED arrival, presenting with dyspnea, and initial chest radiograph infiltrates. In addition to these characteristics, male sex and the presence of several comorbid conditions, particularly diabetes mellitus, hypertension, end stage renal disease, and coronary artery disease, further distinguished between the late deterioration and no deterioration groups. Of note, the no deterioration group had a higher proportion of patients present with GI symptoms (diarrhea, nausea or vomiting).
Table 5.
Patient Characteristics at ED Presentation by Risk Profile: Training Cohort (NYP-WCM).
| Early deterioration |
Late deterioration |
No deterioration |
|
|---|---|---|---|
| (N = 105) | (N = 626) | (N = 314) | |
| Demographics | |||
| Age | |||
| >=65 | 68 (64.8%) | 369 (58.9%) | 114 (36.3%) |
| Race | |||
| White | 39 (37.1%) | 241 (38.5%) | 120 (38.2%) |
| Black | 12 (11.4%) | 89 (14.2%) | 43 (13.7%) |
| Asian | 7 (6.7%) | 72 (11.5%) | 30 (9.6%) |
| Other | 26 (24.8%) | 121 (19.3%) | 73 (23.2%) |
| Not Specified | 21 (20.0%) | 103 (16.5%) | 48 (15.3%) |
| Sex | |||
| Male | 63 (60.0%) | 395 (63.1%) | 164 (52.2%) |
| BMI (kg/m^2) | |||
| <25 | 34 (32.4%) | 228 (36.4%) | 89 (28.3%) |
| 25 to <30 | 29 (27.6%) | 202 (32.3%) | 116 (36.9%) |
| >=30 | 40 (38.1%) | 187 (29.9%) | 105 (33.4%) |
| Missing | 2 (1.9%) | 9 (1.4%) | 4 (1.3%) |
| Active and/or former smoker/vaper | 37 (35.2%) | 184 (29.4%) | 74 (23.6%) |
| ED Supplemental Oxygen | |||
| Required supplemental oxygen within the first 3 h of arrival | 88 (83.8%) | 365 (58.3%) | 123 (39.2%) |
| Comorbidities | |||
| Diabetes Mellitus (DMI, DMII) | 38 (36.2%) | 206 (32.9%) | 77 (24.5%) |
| Hypertension (HTN) | 67 (63.8%) | 390 (62.3%) | 137 (43.6%) |
| Chronic Obstructive Pulmonary Disease (COPD) | 8 (7.6%) | 36 (5.8%) | 7 (2.2%) |
| Chronic Kidney Disease (CKD) | 6 (5.7%) | 38 (6.1%) | 7 (2.2%) |
| End Stage Renal Disease (ESRD) | 4 (3.8%) | 55 (8.8%) | 14 (4.5%) |
| Coronary Artery Disease (CAD) | 24 (22.9%) | 108 (17.3%) | 25 (8.0%) |
| Any Cancer | 8 (7.6%) | 59 (9.4%) | 18 (5.7%) |
| Any Immunosuppression | 3 (2.9%) | 24 (3.8%) | 8 (2.5%) |
| Symptoms | |||
| Fever | 77 (73.3%) | 433 (69.2%) | 224 (71.3%) |
| Cough | 72 (68.6%) | 432 (69.0%) | 220 (70.1%) |
| Diarrhea | 29 (27.6%) | 161 (25.7%) | 89 (28.3%) |
| Nausea or vomiting | 16 (15.2%) | 103 (16.5%) | 83 (26.4%) |
| Myalgias | 22 (21.0%) | 123 (19.6%) | 73 (23.2%) |
| Dyspnea | 80 (76.2%) | 415 (66.3%) | 188 (59.9%) |
| Initial Chest X-ray Findings | |||
| Unilateral Infiltrate | 9 (8.6%) | 80 (12.8%) | 21 (6.7%) |
| Bilateral Infiltrates | 90 (85.7%) | 454 (72.5%) | 220 (70.1%) |
| Pleural Effusion | 10 (9.5%) | 38 (6.1%) | 9 (2.9%) |
| Other | 5 (4.8%) | 36 (5.8%) | 17 (5.4%) |
| Outcome | |||
| Intubation and/or in-hospital mortality | 86 (81.9%) | 195 (31.2%) | 14 (4.5%) |
Supplemental Table 2 shows the differences in distribution of the top predictors between the risk profiles. As noted earlier, three of the top five strongest predictors were LGMM cluster features for FiO2, level of supplemental oxygen, and respiratory rate. These findings further support the previously discussed trends in respiratory status characteristics across risk profiles. There was decreasing representation in the following predictor clusters with decreasing risk profile: FiO2, cluster 2 and level of supplemental oxygen, cluster 2. Cluster 2 for both of these predictors represents increasing oxygenation requirements over time. (Fig. 2) In terms of respiratory rate, 46% (N = 48/105) of patients in the early deterioration group fell into cluster 1 compared to 62% (N = 389/626) of patients in the late deterioration group and 92% (N = 288/314) of patients in the no deterioration group. Cluster 1 represents low and stable values over time (Fig. 2).
3.6. Validation cohort
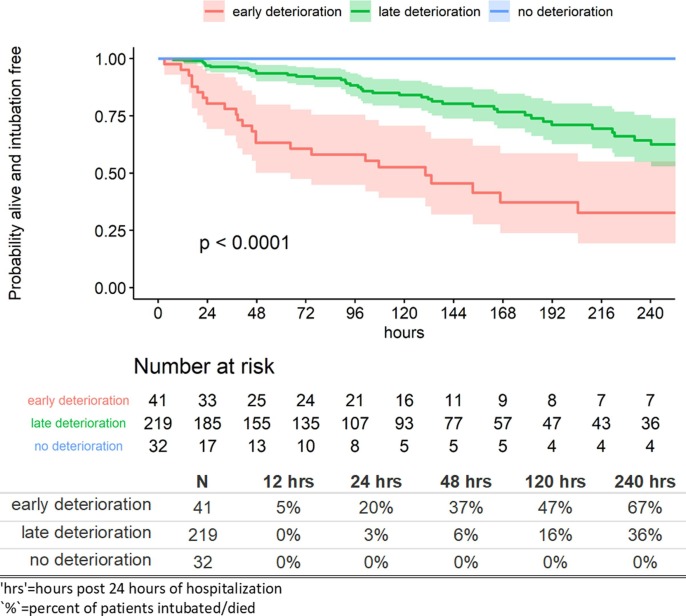
The RSF model developed in the training NYP-WCM cohort was applied to the NYP-LMH validation cohort which yielded c-indices of 0.913, 0.824, and 0.790 at 12, 24, and 48 h post 24-hour index time, respectively. Applying risk stratification to the validation cohort based on ensemble predicted survival probabilities at 24 h post index time produced similar trajectories to the training cohort (Fig. 5 ).
Fig. 5.
Kaplan-Meier estimates by risk profile: validation Cohort (NYP-LMH).
Specifically, 20% of patients deteriorate by 24 h after a stable initial 24 h of hospitalization in the early deterioration risk profile compared to 3% in the late deterioration risk profile and 0% in the no deterioration risk profile. By 5 days after index time, 47%, 16%, and 0% deteriorate, and by 10 days after index time, 67%, 36%, and 0% of patients deteriorate, respectively (Fig. 5). Compared to the training NYP-WCM cohort, the early deterioration risk profile deteriorated less rapidly in the validation NYP-LMH cohort. Only 5% of patients in this group from NYP-LMH were intubated or died within 12 h post 24-hour index time compared to 31% from NYP-WCM. At 24 h, 20% deteriorated compared to 55% and at 48 h 37% deteriorated compared to 64%. The other two risk profiles had similar trends of deterioration between the two cohorts. Importantly, patients classified into the no deterioration risk profile within the validation NYP-LMH cohort had no deterioration. For those in the early deterioration risk profile, 61% went on to deteriorate at some point during hospitalization compared to 20% in the late deterioration risk profile.
4. Discussion
We developed predictive models of clinical deterioration using machine-learning algorithms among hospitalized COVID-19 patients by harnessing longitudinal lab and vital sign values during the early period of hospitalization. Our main innovation was to develop a pipeline to extract clinically meaningful features of longitudinal trajectories of lab values and vital signs paying particular attention to informative missing values. Our features are defined simply and categorically so that they retain clinical interpretation while capturing subtle nuances of in-hospital clinical trajectories. Specifically, we conceptualized three sets of features for lab values and vital signs - linear trends, missing value indicators and clusters of trajectories (non-linear trends) and found that trajectory clusters provide the highest gain in predictive performance followed by missing indicators and trend features. We also found superior predictive performance of random survival forests with the utilization of lab and vital sign data during the early period of hospitalization compared to the utilization of data at ED presentation alone. We demonstrated through external validation that our model accurately predicts clinical trajectories of hospitalized COVID-19 patients.
The main finding of this study is that we identified a late deterioration risk profile in which the probability of being alive and remaining intubation free in the next 24 h after a stable initial 24 h of hospitalization was high, despite deterioration later during hospitalization. Key features of the late deterioration risk profile compared to no deterioration included older age (≥65), a history of smoking/vaping, and initial presence of dyspnea, supplemental oxygen requirement, and chest radiograph infiltrates. The strongest predictors were high/increasing FiO2, level of supplemental oxygen, and respiratory rate. These results suggest that our algorithm can distinguish between late and no deterioration despite the appearance of a stable clinical trajectory at the 24-hour mark of hospitalization. Although prediction of early or no deterioration is supported by clinical judgement and can be achieved with prognostic models that include simpler predictors at ED presentation [11], our results indicate that prognostic models with predictors at ED presentation may not be sufficient to identify this third risk group of patients showing late deterioration with high degree of accuracy. The ability to predict in-hospital clinical deterioration from the early period of hospitalization would allow efficient hospital resource allocation (e.g. ICU beds, ventilators, staffing) as well as inform early clinical interventions, such as transfer decisions in healthcare settings without intensive care capabilities. Thus, the major strength of our study is the development of an informatics tool that extracts clinically meaningful features from the complex longitudinal information of a patient’s clinical course in order to accurately predict late deterioration of hospitalized COVID + patients. Such a set of standardized features defined in this way is useful in a variety of operational and research endeavors beyond that observed for deterioration of COVID + patients that require predictive modeling using Electronic Medical Records (EMR).
Clinical data during pandemics presents many complexities. For example, within the NYP hospital system in March 2020 there was no default ED lab order set for suspected COVID-19 patients; however, in early April as clinical experience matured a COVID-19 clinical order set was implemented with incomplete penetrance. In this case, the presence of lab values in the EMR from an ED admission could reflect clinical necessity or more simply the date of ED admission. As another example, care teams had to don PPE when in close contact with COVID-19 patients, reducing the frequency of lab studies performed in order to preserve PPE supply and mitigate the spread of infection. These circumstances inflated the frequency of lab values for patients with suspected deterioration compared to patients with suspected stability. These examples illustrate the complexities in the utilization of EMR data to predict clinical deterioration. To this end, our pipeline of extracting predictors from the EMR with a particular focus on informative missing-ness is essential in reducing selection bias in prognostic model development and ensures higher accuracy and greater generalizability.
Adding to the strength of our methods was the utilization of a time-dependent cross-validation scheme to account for the rapid changes in the clinical knowledge of the disease and its care. The COVID-19 pandemic overwhelmed NYC hospitals from early March to mid-May 2020 [24]. Hospitals responded to a saturation of intensive care resources with the conversion of alternative spaces to ICUs, redeployment of clinical staff to unfamiliar environments and the rapid development and dissemination of management protocols of a novel disease. It is important during a pandemic to recognize that these evolving circumstances can introduce biases into model development and produce lack of generalizability. Our time-dependent cross-validation strategy estimated predictive performance conservatively compared to conventional cross-validation.
It is important to emphasize the methodological importance of our study while simultaneously acknowledging its limitations. While we have demonstrated the need and advantage of including longitudinal clinical trajectories to predict late deterioration, such predictions cannot be obtained with a simple risk calculator that requires as input only a few easily available risk factors by the bedside; rather the predictive model has to be embedded within the EMR system with an automated feature extraction tool. Such implementation would have to overcome challenges of big data management platforms that include but are not limited to existence of multiple data standards, structures, types and format; rapid growth in data necessitating re-training the model on a periodic basis as new variants of the virus emerge; unavailability of open-source tools (such as R) to execute the predictive models and high costs. Although it is feasible to overcome these barriers, implementing our risk prediction model in the EMR system has additional challenges due to complexities of feature extraction. For that reason, we prioritized a feature extraction pipeline that is easy to implement in code, has clinical interpretation and does not need computation intensive imputation strategies to handle missing data. An exception to this principle are the clusters of lab and vital trajectories, which can be computationally intensive to implement in real-time. A solution to this computational burden would be to define these trajectories once sufficient data is collected and assign a new patient to one of these clusters based on the distance of the new patient’s trajectory to the average trajectory of a cluster. Moreover, additional research is needed in order isolate currently unidentified clinical characteristics that predict late deterioration and that could be used for simplified risk prediction tools used for clinical care.
Another challenge of predictive model implementation in the real world setting is the mechanism and timing of alerting the clinician regarding the concern about deterioration, the expected response to the alert, and the effectiveness of the response. Alert fatigue [1] is a well-recognized, but unintended consequence of decision support where accuracy and actionability are poor. As ML algorithms are trained to optimize accuracy, they need to be made sensitive to clinician behavior by explicitly adding additional optimization criteria. Even if such short-term issues are overcome, there could be additional medium and longer-term issues with implementation such as automation bias and reinforcement of outmoded practice [9]. Specifically, clinicians may become complacent about the monitoring of a patient classified in the “no deterioration” group (automation bias) although a few in that group may still deteriorate. This issue is compounded for a black-box model. On the other hand, as members of the “late deterioration” group are successfully identified and intervened on, the models would need to be retrained to avoid the risk of reinforcing outmoded practice [9]. Retraining the models would need the costly endeavor of a continuously learning system which would reinforce a feedback loop between prognosis and practice could result in self-fulfilling predictions. Such challenges and barriers of implementing a predictive model is not particular to our case. The uniqueness of the pandemic where surge in cases created inpatient bed capacity issues, we believe our predictive model is still useful in making critical triage decision for patient disposition.
Another limitation of our study is that it was developed using patient data within one hospital at the surge of the pandemic in NYC. It is unknown how our model generalizes to other healthcare systems and other geographical regions or to other time-periods of the COVID-19 pandemic. As with any prognostic tool, the underlying model must be continually developed and validated for generalizability. However, NYP-WCM included one of the largest COVID-19 patient populations in NYC at the surge of the pandemic from early March through mid-May and included a diverse patient population. Moreover, we validated our final model on patients at NYP-LMH, which represents a demographically distinct patient population. These strengths help compensate for this limitation.
Lastly, while we demonstrated substantial improvement in predictive performance with Random Survival Forests compared to interpretable methods, we did not consider more novel machine learning algorithms such as deep learning for survival outcomes that has the potential for improvement in prediction accuracy.
5. Conclusion
In summary, we developed an informatics pipeline that harnesses complex longitudinal clinical characteristics (laboratory values and vital signs) of hospitalized COVID+ patients and accurately predicts clinical deterioration with high accuracy and identifies a risk profile of patients who are initially stable but deteriorate later in their hospitalization course. Prediction models that include simple risk factors of patients recorded at ED presentation or clinical judgement are unable to isolate this group of patients showing late deterioration. The ability to detect clinical deterioration at different stages of hospitalization would allow efficient hospital resource allocation (e.g., ICU beds, ventilators, staffing) as well as inform early clinical interventions, such as transfer decisions in healthcare settings without intensive care capabilities. The major strength of our methodology is our pipeline to extract clinically meaningful and easily implementable features from longitudinal trajectories with a particular emphasis on informative missing-ness and our time-dependent cross-validation strategy that reflects the changing course of this novel disease.
Funding Sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [Monika Safford received salary support for investigator-initiated research on CVD risk reduction strategies using large databases.].
Acknowledgments
Acknowledgements
SB, EM, JL, HZ are supported partially by National Institute of Mental Health P50 MH113838 and 2UL1 TR000457 and IE and KH were supported by 2UL1 TR000457.
JC, IE, RK, SB, and MW are funded by PCORI grant contract # HSD-1604-35187.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2021.103794.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Ash J.S., Sittig D.F., Poon E.G., Guappone K., Campbell E., Dykstra R.H. The extent and importance of unintended consequences related to computerized provider order entry. J. Am. Med. Inform. Assoc. 2007;14(4):415–423. doi: 10.1197/jamia.M2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaulieu-Jones B.K., Lavage D.R., Snyder J.W., Moore J.H., Pendergrass S.A., Bauer C.R. Characterizing and managing missing structured data in electronic health records: data analysis. JMIR Med. Informat. 2018;6(1):e11. doi: 10.2196/medinform.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D. Berndt, C. J., Using dynamic time warping to find patterns in time series, in: AAAIWS'94: Proceedings of the 3rd International Conference on Knowledge Discovery and Data Mining, 1994.
- 4.Berndt D.J., Clifford J. Paper Presented at the KDD Workshop. 1994. Using dynamic time warping to find patterns in time series. [Google Scholar]
- 5.Bounthavong M., Watanabe J.H., Sullivan K.M. Approach to addressing missing data for electronic medical records and pharmacy claims data research. Pharmacotherapy: J. Human Pharmacol. Drug Therapy. 2015;35(4):380–387. doi: 10.1002/phar.1569. [DOI] [PubMed] [Google Scholar]
- 6.Breiman L. Random forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- 7.Breiman L., Friedman J., Stone C.J., Olshen R.A. CRC Press; 1984. Classification and Regression Trees. [Google Scholar]
- 8.Burdick H., Lam C., Mataraso S., Siefkas A., Braden G., Dellinger R.P., et al. Prediction of respiratory decompensation in Covid-19 patients using machine learning: The READY trial. Comput. Biol. Med. 2020;124:103949. doi: 10.1016/j.compbiomed.2020.103949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challen R., Denny J., Pitt M., Gompels L., Edwards T., Tsaneva-Atanasova K. Artificial intelligence, bias and clinical safety. BMJ Quality Saf. 2019;28(3):231–237. doi: 10.1136/bmjqs-2018-008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng F.-Y., Joshi H., Tandon P., Freeman R., Reich D.L., Mazumdar M., et al. Using machine learning to predict ICU transfer in hospitalized COVID-19 patients. J. Clin. Med. 2020;9(6):1668. doi: 10.3390/jcm9061668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R.K., Marks M., Samuels T.H., Luintel A., Rampling T., Chowdhury H., et al. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: An observational cohort study. Eur. Respir. J. 2020 doi: 10.1183/13993003.03498-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrell F.E., Califf R.M., Pryor D.B., Lee K.L., Rosati R.A. Evaluating the yield of medical tests. JAMA. 1982;247(18):2543–2546. [PubMed] [Google Scholar]
- 13.Hartigan J.A. John Wiley & Sons Inc.; 1975. Clustering Algorithms. [Google Scholar]
- 14.Ishwaran H., Kogalur U.B., Blackstone E.H., Lauer M.S. Random survival forests. Ann. Appl. Stat. 2008;2(3):841–860. [Google Scholar]
- 15.H. Ishwaran, K. U., E. Blackstone, M. Lauer, Random survival forests, Ann. Appl. Stat. (2008).
- 16.Johns Hopkins University & Medicine Coronavirus Resource Center. (2020). Retrieved from https://coronavirus.jhu.edu/.
- 17.Kumaraiah D., Yip N., Ivascu N., Hill L. Innovative ICU physician care models: Covid-19 pandemic at NewYork-Presbyterian. NEJM Catalyst Innovat. Care Deliv. 2020;1(2) [Google Scholar]
- 18.B. L, Random forests. Machine Learn. (2001).
- 19.O'Carroll O., MacCann R., O'Reilly A., Dunican E.M., Feeney E.R., Ryan S., et al. Remote monitoring of oxygen saturation in individuals with COVID-19 pneumonia. Eur. Respir. J. 2020;56(2) doi: 10.1183/13993003.01492-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proust-Lima C., Philipps V., Liquet B. Estimation of extended mixed models using latent classes and latent processes: The R package lcmm. J. Stat. Softw. 2017;78(2):1–56. [Google Scholar]
- 21.Razavian N., Major V.J., Sudarshan M., Burk-Rafel J., Stella P., Randhawa H., et al. A validated, real-time prediction model for favorable outcomes in hospitalized COVID-19 patients. npj Digital Med. 2020;3(1):1–13. doi: 10.1038/s41746-020-00343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz G. Estimating the dimension of a model. Ann. Stat. 1978;6(2):461–464. [Google Scholar]
- 23.Shah S., Majmudar K., Stein A., Gupta N., Suppes S., Karamanis M., et al. Novel use of home pulse oximetry monitoring in COVID-19 patients discharged from the emergency department identifies need for hospitalization. Acad. Emerg. Med. 2020;27(8):681–692. doi: 10.1111/acem.14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.US News & World Report. (2020, March 23). Gov. Andrew Cuomo Orders Hospitals to Increase Capacity by 50% [Press release]. Retrieved from https://www.usnews.com/news/health-news/articles/2020-03-23/new-york-gov-andrew-cuomo-orders-hospitals-to-increase-capacity-by-50.
- 25.Wells B.J., Chagin K.M., Nowacki A.S., Kattan M.W. Strategies for handling missing data in electronic health record derived data. Egems. 2013;1(3) doi: 10.13063/2327-9214.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.