Abstract
Purpose
To train and validate a predictive model of mortality for hospitalized COVID-19 patients based on lung densitometry.
Methods
Two-hundred-fifty-one patients with respiratory symptoms underwent CT few days after hospitalization. “Aerated” (AV), “consolidated” (CV) and “intermediate” (IV) lung sub-volumes were quantified by an operator-independent method based on individual HU maximum gradient recognition. AV, CV, IV, CV/AV, IV/AV, and HU of the first peak position were extracted. Relevant clinical parameters were prospectively collected. The population was composed by training (n = 166) and validation (n = 85) consecutive cohorts, and backward multi-variate logistic regression was applied on the training group to build a CT_model. Similarly, models including only clinical parameters (CLIN_model) and both CT/clinical parameters (COMB_model) were developed. Model’s performances were assessed by goodness-of-fit (H&L-test), calibration and discrimination. Model’s performances were tested in the validation group.
Results
Forty-three patients died (25/18 in training/validation). CT_model included AVmax (i.e. maximum AV between lungs), CV and CV/AE, while CLIN_model included random glycemia, C-reactive protein and biological drugs (protective). Goodness-of-fit and discrimination were similar (H&L:0.70 vs 0.80; AUC:0.80 vs 0.80). COMB_model including AVmax, CV, CV/AE, random glycemia, biological drugs and active cancer, outperformed both models (H&L:0.91; AUC:0.89, 95%CI:0.82–0.93). All models showed good calibration (R2:0.77–0.97). Despite several patient's characteristics were different between training and validation cohorts, performances in the validation cohort confirmed good calibration (R2:0–70-0.81) and discrimination for CT_model/COMB_model (AUC:0.72/0.76), while CLIN_model performed worse (AUC:0.64).
Conclusions
Few automatically extracted densitometry parameters with clear functional meaning predicted mortality of COVID-19 patients. Combined with clinical features, the resulting predictive model showed higher discrimination/calibration.
Keywords: COVID-19, CT, Lung densitometry, Respiratory distress syndrome
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 2, was identified in China and very rapidly spread around the world [1], [2], resulting in the current coronavirus disease 2019 (COVID-19) pandemic with tens of millions of confirmed cases worldwide.
In a relevant number of patients, the virus can cause severe interstitial pneumonia with subsequent acute respiratory distress syndrome (ARDS), responsible for dramatic respiratory failure including fatal outcome.
Chest Computed Tomography (CT) plays a fundamental role in diagnosing and characterizing lung involvement in COVID-19 patients, recognizing different imaging patterns based on the duration of the tissue inflammation [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14]. The disease has a wide variety of CT findings, which depend on both clinical severity and time elapsed since the symptoms onset [6], [7]. Radiological hallmarks of COVID-19 pneumonia are: bilateral ground glass opacities, crazy paving pattern and/or consolidations predominantly in subpleural locations in the lower lobes [11], [12], [15], [16], [17].
CT changes in the lungs were reported to be associated with more severe symptoms, longer time to recovery as well as an increased risk of death [12], [16], [18], [19], [20], [21], [22], [23]. However, only a limited number of predictive models of disease severity and/or mortality were based on quantitative features [16], [18], [20], [22], [23], the majority lacking of sufficient validation and then usability, claiming for an urgent need of robust and validated models [21].
Our Institution was largely involved in the first wave of the pandemic in northern Italy. Several hundred patients were hospitalized and a large number of them underwent chest CT scan shortly after hospitalization. Several clinical predictors were primarily found to predict short-term mortality and time to recovery [24], [25].
In this rapidly changing scenario, focusing on objective CT-based outcome predictors, we first aimed to develop and implement an automated, operator-independent quantitative method to characterize lungs of COVID-19 patients based on individually optimized Hunsfield Unit (HU) thresholds [26]. The proposed method was based on an interpretable and intuitive phenomenological characterization of lungs with the explicit aim to be easily implemented independently of software availability and/or post-processing tools.
It permits to individually assess HU thresholds able to automatically divide the lungs into three regions, namely the aerated, intermediate and consolidation volumes and to extract parameters characterizing lungs appearance based on this classification. This is an important step aiming to make the interpretation of the images simultaneously operator-independent and interpretable, differently from most AI based approaches [29], [30].
The aim of current study was to train and validate a CT-based model able to predict mortality using this previously developed operator-independent extraction method. We compared the discriminative performances of this model with that of an exclusively clinical-based model and finally combining CT and clinical features to derive a third model and test its accuracy in early mortality prediction.
2. Materials and methods
2.1. Patients and clinical data collection
This study is a secondary analysis within our COVID-19 Institutional study (the COVID-BioB, Clinical trials govNCT04318366). All patients aged > 18 years, hospitalized for COVID-19 during the period February-April 2020, who underwent at least one CT scan during hospitalization were considered for the present study. COVID-19 diagnosis was made based on positive SARS-CoV-2 real-time reverse-transcriptase polymerase chain reaction (RT-PCR) and/or radiological findings suggestive of COVID-19 pneumonia. Details on patient management during hospitalization, clinical predictors of adverse outcome in our population, time to recovery or death, and data collection procedures were reported elsewhere [24], [25].
All patients signed an informed consent. The study was approved by the Institutional Review Board (protocol number 34/INT/2020) and conforms to the Declaration of Helsinki.
2.2. CT scanning
The first CT scan after hospitalization was considered for the current investigation. Patients were scanned on three different scanners: Incisive (64sl)-Philips, Brilliance (64sl)-Philips and Lightspeed VCT (64sl)-GE Medical System. All patients were scanned with the following parameters: X-ray tube voltage of 120 kV and automatic current modulation ([149–549] mA), slice thickness 1–1.25 mm, matrix 512 × 512. The raw data were reconstructed using standard kernels with filtered back projection as well as adaptive statistical iterative reconstruction. CT images were retrieved from the hospital Picture Archiving and Communication System (PACS). The inter-scanner variability on the assessment of HU values was previously investigated by phantom measurements and found to be negligible [26].
2.3. Lung segmentation and HU-based sub-segmentation
CT images were exported from the Institutional PACS and then uploaded in the Eclipse system (v13.7, Varian Inc.) for segmentation purposes. Five well trained operators with>5 years’ experience in contouring for radiotherapy planning segmented both lungs using automated tools (such as maximum gradient search and thresholding selection) combined with manual delineation/correction. Only one observer contoured the lungs for each patient (i.e.: contouring was not repeated among the five operators). Two expert radiologists (>5 year experience) independently reviewed a sample set (n = 30 patients) for consensus against the contours delineated by the five observers, finding the delineation acceptable in all cases.
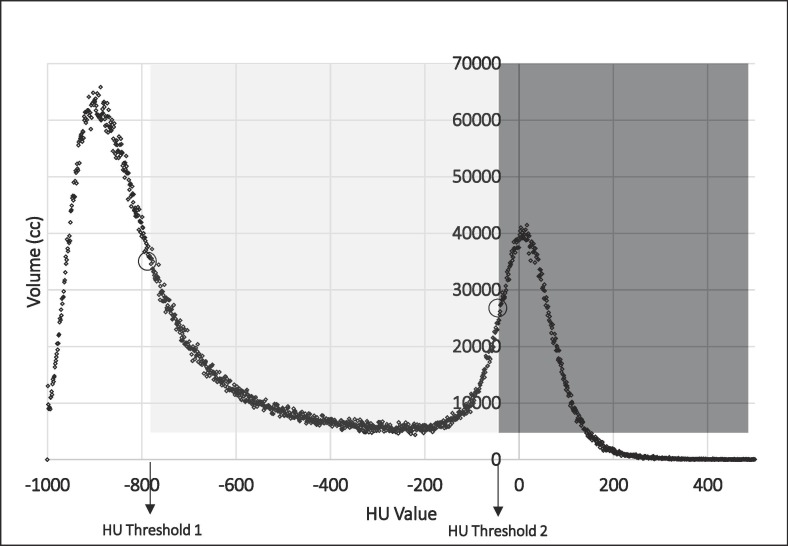
After the lungs segmentation was accomplished, contours and images were transferred to the MIM 6 v 6.9.6 software platform. In order to reduce the impact of the different CT discretization and voxel/pixel dimension used, CT images were resampled with an isotropic 1.5 × 1.5 × 1.5 mm3 voxel size. The original lung contours were recorded on the resampled images. Histograms of HU of the lungs were then extracted and used to define three different sub-volumes named as “Aerated” (AV), “Consolidated” (CV) and “Intermediate” (IV). Briefly, two typical peak values, one in the low-density region (typically −1000 HU, −700 HU) and one next to the water HU value (−34 HU, 0 HU) were present, as shown in the example of Fig. 1 . A Matlab script was developed to find individually the inflexion points and the corresponding HU thresholds (th1 1 and th2) of the HU density histogram according to a maximum gradient computation. They corresponded to the descending portion of the curve after the first peak and to the ascending portion of the curve before the second peak, as described by Mazzilli et al. [26] and previously suggested for lung densitometry characterization of idiopathic pulmonary fibrosis [27]. Then, the resulting operator-independent thresholds individually identified the three regions AV, IV and CV, reflecting their expected functional meaning. Despite inter-observer variability in contouring lungs was not quantified, “little” inter-observer variations in lung contouring cannot not expected to significantly influence the assessment of the sub-volumes, given the largely different densitometry patterns compared to normal lung.
Fig. 1.
Graphical representation of the threshold values found by searching inflexion points of the HU-density histograms. The Aerated Volume (AV) in white ranges between −1000 HU and HU Threshold 1; the Intermediate Volume (IV) in light grey ranges between HU Threshold 1 and HU Threshold 2; the Consolidated Volumes (CV) in dark grey ranges from HU Threshold 2 until higher HU values.
2.4. Quantitative CT parameters
As previously described [26], HU histograms data were interpolated with an integral smooth function f(HU) and AV, CV, IV, the ratios CV/AV, IV/AV, the HU value corresponding to the peak positions (MaxPeakAerated, MaxPeakConsolidated), the width and height of IV (Width_Intermediate, Height_Intermediate) in terms of HU range and the mean HU value of IV were extracted. They were considered both as single lung and as paired organ, considering both lungs; as single organ, maximum and minimum values between the two lungs were considered. The formulae of the mentioned parameters were defined as (referred to single lung):
where and are the HU values corresponding to the thresholds th1 and th2.
2.5. Analyses: training predictive models
According to the TRIPOD 2 level of models generalizability [31], the population was composed by a training (n = 166) and a validation (n = 85) cohorts; models were trained on the training cohort data and tested onto the validation cohort.
The two cohorts were consecutive (not randomized), due to the variable availability of the operators for lung delineation. Due to the rapid change of patient characteristics at hospital admittance, the variable availability of intensive care admittance and the changes in the applied therapies, the two populations could be expected to be different. This was considered to be an additional value for our validation purposes and we decided to deliberately keep these two populations as they were, without any additional merging.
The differences between patients characteristics (both clinical and densitometry) of the two cohorts were tested by two-tailed t-tests and chi-square tests, where appropriate.
The end-point was early death, defined as death occurring during hospitalization as a consequence of respiratory and/or other COVID-19-related manifestation.
All the previously extracted quantitative CT parameters were considered and tested on the training group as potential predictors through Univariate Logistic Regression (ULR). First, ULR was carried out and only variables with p < 0.05 were selected for further analysis; then, a Multivariate Logistic Regression (MLR) backward analysis was conducted on the previous selected variables by retaining in the final model variables with p < 0.20; this choice was arbitrarily followed aiming to retain in the resulting models potentially relevant features with “large” odds ratios.
The resulting model including only CT parameters was named CT_model. The individual resulting probabilities computed by MLR were considered and named CT_index. Similarly, the same procedure was followed to assess the best clinical predictors, deriving a model including only clinical variables (CLIN_model) and the corresponding CLIN_index. The following clinical parameters including demographics data, comorbidities and laboratory data were considered: sex, age, race, arterial hypertension, coronary artery disease, diabetes mellitus, chronic obstructive pulmonary disease, chronic kidney disease, active malignances, peripheral oxygen saturation (SpO 2), the ratio of arterial oxygen partial pressure, (PaO2) in mmHg to fractional inspired oxygen (FiO2) expressed as a fraction (SatO2/FiO2), the ratio of SpO2 to FiO2 (SpOP2 /FiO2), body temperature, hemoglobin, absolute lymphocytes, random glycemia, aspartate transaminase, alanine transaminase, lactate transaminase, C-reactive protein, and creatinine levels at hospitals admission and the use of biological drugs.
Finally, the same procedure was followed by considering both CT and clinical parameters to assess a combined model (COMB_model) and in the same way the corresponding COMB_index. The goodness of fit of the three models was quantified by the Hosmer and Lemeshow (H&L) test and calibration plots. The discriminative power of the models was quantified by their AUCs, sensitivity and specificity, based on the maximization of the Youden index and AUCs were compared by the De Long method [28]. Positive and negative predictive values (PPV, NPV) were also calculated, relative to the same best cut-off values identified by the Youden index. Analyses were performed using Medcalc v 19.5.3 and R-software.
2.6. Validating models
The performances of the models developed on the training cohort were tested in the validation group. In particular, CLIN_index, CT_index and COMB_index were derived for all patients of the validation group using MLR coefficients of models developed in the training; then indexes were tested using ROC analysis. Significance (p-value) in stratifying the events was first verified and calibration plots for each model were generated for the validation cohort.
3. Results
Demographics, clinical, laboratory and respiratory function features of patients are summarized in Table 1 : they are split between training and validation cohorts with their p-value of the t-test (or chi-square for dichotomic variables) for distribution difference. Similarly, a summary of the densitometry parameters was shown in Table 2 , reporting the differences between the two cohorts. A number of clinical characteristics were different between the two groups; in general, the validation group included patients with better lung functionality and HU-based parameters (higher AV and lower IV and CV) compared to the training group. On the other hand, age, weight, BMI and incidence of obstructive pulmonary disease were slightly higher in the validation group. The therapy received by most patients during hospitalization was the association of hydroxychloroquine with lopinavir/ritonavir, which was the standard of care for COVID-19 at our Institution at the time of patient enrolment in the COVID-BioB study. The severity of the clinical picture guided the administration of further specific treatments in selected patients. Specifically, biological drugs were used in 57/251 patients with a significant unbalance between training and validation cohorts. The median time (interquartile range, IQR) from hospital access to CT was 1 day (0–4).
Table 1.
patients’ clinical characteristics of the training and validation groups.
| Training Group | Validation Group | p –value | |
|---|---|---|---|
| Demographic Characteristics | |||
| age, years (mean; median; range) | 61; 61; 20–86 | 65; 66; 18–95 | 0.0004 |
| sex (Male; Female) | 123; 43 | 57; 28 | 0.8434 |
| weight (mean; median; range) | 79; 80; 45–124 | 75; 75; 39–120 | 0.0099 |
| height (mean; median; range) | 170; 170; 150–190 | 169; 170; 142–187 | 0.0170 |
| BMI (mean; median; range) | 27; 27, 18–43 | 26; 26; 18–47 | 0.0097 |
| race (Caucasian; Hispanic; Asiatic; Afro-american) | 138; 12; 2; 1 | 81; 2; 1; 1 | 0.9990 |
| Comorbidities | |||
| Arterial hypertension (y; n, missing) | 67; 82; 17 | 40; 42; 3 | 0.0350 |
| Coronary disease (y; n, missing) | 12; 137; 17 | 15; 67; 3 | 0.2666 |
| Diabetes mellitus (y; n, missing) | 43; 126; 17 | 15; 67; 3 | 0.0980 |
| Obstructive pulmonary disease (y; n, missing) | 4; 166; 17 | 10; 73; 3 | 0.0021 |
| Chronic renal disease (y; n, missing) | 12; 137; 17 | 11; 71; 3 | 0.4080 |
| Active Cancer (y; n, missing) | 10; 140; 16 | 9; 74; 2 | 0.3489 |
| ICU (y; n, missing) | 37; 99; 30 | 12; 71; 2 | 0.4260 |
| Biological drugs (y; n; missing) | 55; 97; 14 | 78; 7; 0; 0 | 0.0415 |
| satO2 (mean; median; range) | 91; 93; 50–100 | 93; 95; 63–100 | 0.0025 |
| FiO2 (mean; median; range) | 1; 1; 1–1 | 0.27; 0.21; 0.21–1 | 0.1654 |
| satO2/FiO2 (mean; median; range) | 408; 438; 70–476 | 409; 447; 93–476 | 0.0126 |
| EGAPaO2 (mean; median; range) | 66; 63; 28–251 | 68; 66; 37–127 | 0.2512 |
| EGAFiO2 (mean; median; range) | 0.32; 0.21; 0.21–1.00 | 0.3; 0.21; 0.21–1 | 0.0065 |
| PaO2/FiO2 (mean; median; range) | 262; 281; 47–667 | 283; 300; 58–586 | 0.1301 |
| Body temperature (mean; median; range) | 38; 38; 36–41 | 38; 38; 36–41 | 0.0222 |
| Laboratory results | |||
| Hemoglobin (mean; median; range) | 14; 14; 7–51 | 13; 14; 8–18 | 0.1067 |
| Absolute lymphoncytes (mean; median; range) | 1.27; 0.90; 0.30–42.00 | 1.14; 1.10; 0.10–5.70 | 0.8592 |
| Glycemia (mean; median; range) | 131; 109; 58–500 | 117; 104; 71–305 | 0.5807 |
| Aspartate transaminase (mean; median; range) | 58; 46; 13–378 | 54; 39; 13–225 | 0.5626 |
| Alanine transaminase (mean; median; range) | 52; 37; 8–578 | 48; 28; 11–275 | 0.7346 |
| Lactate deidrogenase (mean; median; range) | 427;409; 115–1101 | 392; 320; 128–2017 | 0.3303 |
| C-reactive protein (mean; median; range) | 113; 91; 3–410 | 82; 66; 0–313 | 0.0925 |
| Creatinine (mean; median; range) | 1.08; 1.03; 0.44–5.71 | 1.18; 0.98; 0.56–7.57 | 0.8038 |
| Endpoints | |||
| Deaths (y; n) | 25; 141 | 18; 85 | 0.9900 |
Table 2.
Patients’ densitometry parameters of the training and validation groups.
| TRAINING |
VALIDATION |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| min | max | mean | median | min | max | mean | median | p-value | |
| Aerated_Volume_Max | 46.67 | 2908.43 | 1025.13 | 942.83 | 149.05 | 2764.59 | 1141.12 | 1004.00 | 0.399 |
| Intermediate_Volume_Max | 402.33 | 1955.32 | 1041.30 | 1005.37 | 250.11 | 1890.81 | 964.92 | 888.98 | <0.001 |
| Consolidated_Volumed_Max | 24.76 | 589.99 | 165.23 | 135.51 | 39.24 | 1102.60 | 176.69 | 127.16 | <0.001 |
| ConsolidatedVolume/AeratedVolume_Max | 0.03 | 15.15 | 0.42 | 0.18 | 0.03 | 5.86 | 0.32 | 0.11 | 0.074 |
| IntermediateVolume/AeratedVolume_Max | 0.48 | 34.65 | 2.02 | 1.39 | 0.36 | 6.48 | 1.08 | 0.90 | <0.001 |
| Width_Intermediate_Max | 543.00 | 846.00 | 754.80 | 780.00 | 508.00 | 874.00 | 761.51 | 781.00 | <0.001 |
| Height_Intermediate_Max | 178.73 | 742.76 | 408.31 | 396.58 | 145.88 | 793.61 | 377.34 | 359.19 | <0.001 |
| Aerated_Volume_Min | 35.19 | 2074.58 | 752.28 | 657.89 | 36.66 | 2684.95 | 911.36 | 801.59 | <0.001 |
| Intermediate_Volume_Min | 256.35 | 1747.75 | 860.98 | 851.55 | 166.29 | 1819.25 | 789.50 | 733.96 | <0.001 |
| Consolidated_Volumed_Min | 17.85 | 442.05 | 118.59 | 96.94 | 25.18 | 686.78 | 119.68 | 82.85 | <0.001 |
| ConsolidatedVolume/AeratedVolume_Min | 0.02 | 5.49 | 0.21 | 0.11 | 0.02 | 11.65 | 0.42 | 0.10 | 0.220 |
| IntermediateVolume/AeratedVolume_Min | 0.34 | 17.37 | 1.31 | 0.95 | 0.38 | 17.50 | 1.32 | 0.89 | 0.005 |
| Width_Intermediate_Min | 396.00 | 846.00 | 743.34 | 780.00 | 394.00 | 839.00 | 730.35 | 760.00 | <0.001 |
| Height_Intermediate_Min | 125.75 | 612.12 | 340.09 | 341.56 | 125.06 | 727.99 | 316.08 | 306.85 | <0.001 |
| Aerated_Volume_Tot | 81.86 | 4983.01 | 1777.41 | 1546.59 | 214.03 | 5445.38 | 2052.48 | 1788.28 | <0.001 |
| Intermediate_Volume_Tot | 791.38 | 3697.63 | 1902.28 | 1885.08 | 416.40 | 3654.62 | 1754.41 | 1671.30 | <0.001 |
| Consolidated_Volumed_Tot | 48.35 | 964.85 | 283.82 | 235.35 | 69.44 | 1597.17 | 296.37 | 206.31 | <0.001 |
| ConsolidatedVolume/AeratedVolume_Tot | 0.03 | 9.50 | 0.28 | 0.14 | 0.03 | 6.64 | 0.34 | 0.10 | 0.108 |
| IntermediateVolume/AeratedVolume_Tot | 0.43 | 24.54 | 1.58 | 1.16 | 0.37 | 8.37 | 1.14 | 0.89 | <0.001 |
In total, 43/251 (17%) patients died during hospitalization, 25 and 18 in the training and validation group respectively.
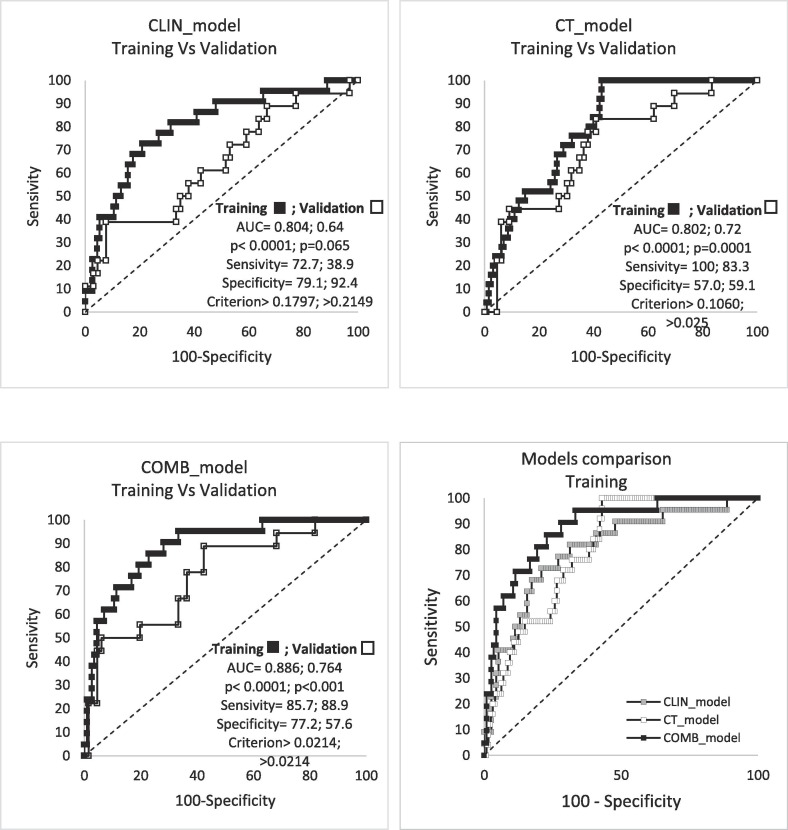
Results of ULR (training cohort) are reported in Table S1 of Supplementary Materials; Table 3 summarizes the results of MLR; Table 4 , the performances of the three models in the training cohort in terms of AUC, significance p-value, sensitivity, PPV and NPV. In short, the combination of three CT parameters predicts the risk of early death with discrimination equal to 80%, similarly to the model obtained using only clinical variables. Combining CT and clinical parameters significantly improved the performance of the resulting COMB_model, with an increase of AUC from 0.80 to 0.89, as also shown in Table 3 and Fig. 2 .
Table 3.
Multivariable Regression Logistic analysis; only the variables with p < 0.05 in the URL analysis were selected for MRL.
| Clinical model | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Coefficient | P | OR | 95%CL | AUC | 95%Cl | Variable | Coefficient | P | OR | 95%CL | Hosmer | AUC | 95%CL |
| Glycemia | 0.0076028 | 0.018 | 1.0076 | 1,0013 to 1,0140 | 0.803 | 0,727 to 0,866 | Clinical_index | 6.13842 | 0.0001 | 463.3207 | 22,6987 to 9457,1944 | P = 0,8387 | 0.804 | 0,727 to 0,866 |
| Biological drugs | −1.76113 | 0.0243 | 0.1719 | 0,0371 to 0,7953 | Constant | −2.8879 | <0,0001 | |||||||
| C-reactive protein | 0.0054004 | 0.047 | 1.0054 | 1,0001 to 1,0108 | ||||||||||
| Constant | −3.0147 | <0,0001 | ||||||||||||
| CT model | ||||||||||||||
| Variable | Coefficient | P | OR | 95%CL | AUC | 95%Cl | Variable | Coefficient | P | OR | 95%CL | Hosmer | AUC | 95%CL |
| Aerated_Volume_Max | −0.0037859 | 0.0049 | 0.9962 | 0,9936 to 0,9989 | 0.802 | 0,730 to 0,862 | CT_index | 6.3065 | <0,0001 | 548.1232 | 26,6008 to 11294,3689 | P = 0,2899 | 0.802 | 0,730 to 0,862 |
| Consolidated_Volume_Tot | 0.0062398 | 0.005 | 1.0063 | 1,0019 to 1,0107 | Constant | −2.93635 | <0,0001 | |||||||
| Consolidated/AeratedVolume_Tot | −3.17537 | 0.1268 | 0.0418 | 0,0007 to 2,4623 | ||||||||||
| Constant | 0.42004 | 0.7001 | ||||||||||||
| Combined model | ||||||||||||||
| Variable | Coefficient | P | OR | 95%CL | AUC | 95%Cl | Variable | Coefficient | P | OR | 95%CL | Hosmer | AUC | 95%CL |
| Aerated_Volume_Max | −0.0038748 | 0.0186 | 0.9961 | 0,9929 to 0,9994 | 0.886 | 0,820 to 0,934 | Combined_index | 6.69175 | <0,0001 | 805.7315 | 57,6530 to 11260,5234 | P = 0,6060 | 0.886 | 0,819 to 0,934 |
| Consolidated_Volume_Tot | 0.0067809 | 0.007 | 1.0068 | 1,0019 to 1,0118 | Constant | −3.24624 | <0,0001 | |||||||
| Consolidated/AeratedVolume_Tot | −3.06428 | 0.1483 | 0.0467 | 0,0007 to 2,9745 | ||||||||||
| Glycemia | 0.0057383 | 0.0707 | 1.0058 | 0,9995 to 1,0120 | ||||||||||
| Biological drugs | −1.79185 | 0.0315 | 0.1667 | 0,0325 to 0,8535 | ||||||||||
| Active Cancer | 1.56007 | 0.109 | 4.7592 | 0,7064 to 32,0645 | ||||||||||
| Constant | −0.4444 | 0.7475 | ||||||||||||
Table 4.
ROC analysis results on the Training group (values of sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) refer to the best cut-off value assessed by the maximization of the Youden index).
| Variable | AUC | 95% CL | Significance level P | Youden index J | Associated criterion | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|---|
| Clinical index | 0.804 | 0.727 to 0.866 | <0.0001 | 0.519 | >0.179 | 72.73 | 79.13 | 40.00 | 93.80 |
| CT index | 0.802 | 0.730 to 0.862 | <0.0001 | 0.570 | >0.106 | 100 | 57.03 | 31.2 | 100.00 |
| Combined index | 0.886 | 0.819 to 0.934 | <0.0001 | 0.629 | >0.153 | 85.71 | 77.19 | 40.90 | 96.70 |
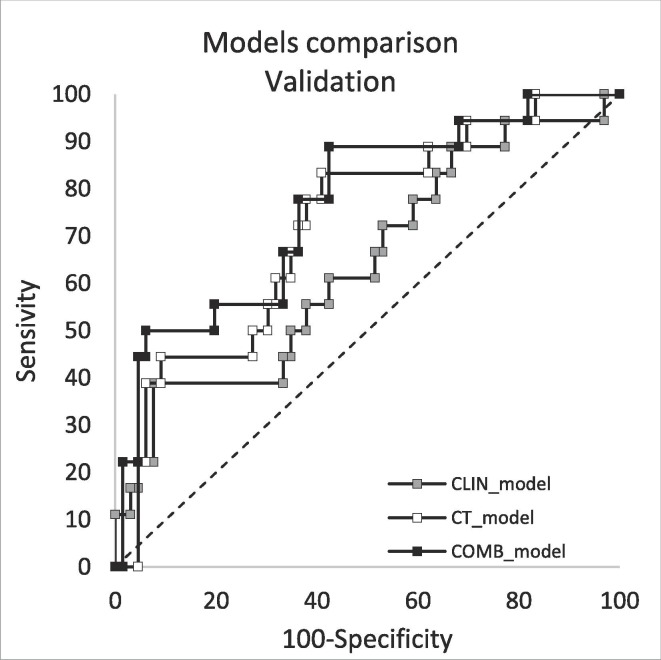
Fig. 2.
ROC curves of the predictive indexes of the three models and their comparison in the training and validation group.
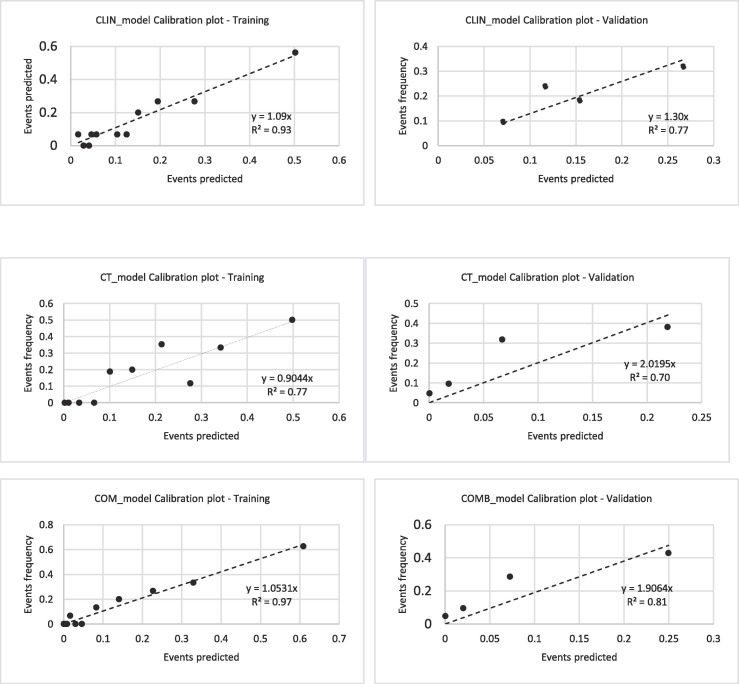
The calibration plots of the three models are shown in Fig. 3 : slope and R2 ranged between 0.89 and 0.93 and 0.77–0.97 respectively.
Fig. 3.
Calibration plots of the predictive indexes in the training and validation group.
The results regarding the validation of the three models are reported in Table 5 : they confirmed the training cohort results, although CLIN_Index was found to be of borderline significance (AUC = 0.64, p = 0.065). On the other hand, both CT_model and COMB_model showed much better performances (AUC = 0.72, p = 0.001 and AUC = 0.76, p < 0.001 respectively) confirming the ability of CT parameters to predict the risk of death. The calibration plots showed slightly worse performances compared to the training cohort, although R2 remained satisfactorily high, ranging between 0.70 and 0.81. Very importantly, NPV was very similar (and high) in both the training and the validation cohorts.
Table 5.
ROC analysis results on the Validation group values of sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) refer to the best cut-off value assessed by the maximization of the Youden index).
| Variable | AUC | 95% CL | Significance level P | Youden index J | Associated criterion | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|---|
| Clinical index | 0.641 | 0.53 to 0.74 | 0.0650 | 0.313 | >0.215 | 38.94 | 92.44 | 58.33 | 84.73 |
| CT index | 0.722 | 0.614 to 0.814 | 0.0007 | 0.424 | >0.025 | 83.34 | 59.13 | 35.71 | 92.94 |
| Combined index | 0.764 | 0.659 to 0.850 | <0.0001 | 0.465 | >0.021 | 88.94 | 57.63 | 36.42 | 95.10 |
4. Discussion
The literature regarding the diagnostic performances of CT in COVID-19 patients is large and includes several reviews and meta-analyses; however, despite recent efforts [12], [16], [18], [23], [32], the availability of quantitative models predicting clinical outcome based on CT biomarkers remains limited. The current study trained and validated models to predict early death in a cohort of COVID-19 patients from a single center during the first wave of the pandemic. The investigation tested whether quantitative, operator-independent (and interpretable) CT features could capture the majority of the clinical and prognostic picture. A phenomenological approach for sub-segmenting the lungs in three main regions was implemented, adapting a maximum-gradient method previously suggested as optimal in characterizing lungs of patients with idiopathic pulmonary fibrosis [27].
The combination of only three features was able to predict mortality with classification performances near to 80%, showing very high sensitivity and relatively low specificity, translating in a very high negative predictive power. The same model considering only the two most robust parameters (CV and maximum AV value between the two lungs) showed similar performance with an AUC of 0.79.
Of course, clinical features including patient characteristics as well as the different individual response to different therapeutic actions (i.e. external ventilation and/or antiviral drugs) are expected to explain at least part of the residual lack of discrimination of the CT_model. Indeed, the addition of few clinical parameters such as random glycemia at hospital admission and the use of biological drugs in the resulting COMB_model was able to improve discrimination up to AUC equal to 0.89, outperforming the CLIN_model. Importantly, the performances of the models were replicated successfully in a validation group. As partly expected, due to the lower numbers and to the choice of keeping a relatively large p-value threshold during backward selection of variables (with the aim of accounting for most of the potential predictors), the performances of the models were worse in the validation group, and this was especially true for CLIN_model.
Very importantly, the worse results for CLIN_model can also be explained by the different clinical characteristics of the two cohorts only partly overlapping in terms of hospital admission day.
On the other hand, results show the strength of the extracted densitometry features in correctly predicting the risk of mortality also in a cohort of patients significantly different from the point of view of several clinical characteristics.
Results regarding the predictive value of quantitative CT parameters are consistent with few previous studies: Colombi et al [20] first showed IV < 73% assessed at admission CT as able to predict the patients’ mortality in a cohort of 236 patients; the corresponding predictive model combining this feature with several clinical parameters slightly, but significantly, outperformed discrimination compared to a model including only clinical information (AUC: 0.86 vs 0.83). A limitation of this study was the unreported performance of any model including only CT parameters and the lack of any validation cohort. On the other hand, this was the first large and clear demonstration of the potentials of using quantitative HU-based features to predict mortality.
Regarding validation, to our knowledge, up to now no studies reported independent validation nor following the actual TRIPOD-2 like approach (i.e. splitting a single center cohort into training and validation groups [31]) neither with external validation studies (TRIPOD-3 and 4). More in general, the need for improving reliability of diagnostic and predictive models of COVID-19 cohorts based on imaging biomarkers was underlined by a recent review [21].
Others authors reported quantitative CT biomarkers for predicting severity of symptoms, recovery and mortality [12], [16], [18], [20], [22], [23]. As an example, Leonardi et al [22] combined CV derived by semi-automatic segmentation of lungs (with large manual intervention) showing very high AUC (0.96) in assessing critically ill patients. Similarly, CV obtained semi-automatically with the intervention of a radiologists and combined with other clinical parameters was found to correctly classify 106 COVID-19 patients based on adverse outcome (defined as death or need of mechanical ventilation) with an AUC = 0.92 using support vector machine [23]. Major limitations of this study was the risk of overfitting and the operator-dependent segmentation, although their result is again consistent with our findings. Others used macroscopic quantitative CT parameters as well as AI-based solutions with discriminative power typically ranging between 0.70 and 0.90 [16], [18], [33], [34].
In general, most studies showed good to excellent performance in predicting outcome. However, as previously underlined they were often affected by a high risk of bias, due to poor reporting and poor methodologic aspects [21]. Moreover, in most of them, machine learning and AI algorithms found predictors in a complex way, which makes challenging their interpretation. These considerations suggest that their predictive performance when trying to apply on new patients can be expected to be significantly lower than that reported. This is also why we choose death as (objective) outcome and an approach focused on trying to capture few, interpretable features explaining the larger part of the events.
Our study has several limitations: a major one is the need of delineating the lungs, which is a cumbersome procedure, subject to inter-observer variability. In general, the inter-observer agreement in manually delineating “normal” lungs for patients with thoracic cancer is assumed to be very small, due to the good visibility of lungs; recently, an acceptably low inter-observer variability for lung delineation was also reported for COVID-19 patients with pneumonia, with an average Dice index equal to 0.79 [35]. This suggests that the accuracy of our manual-based segmentation approach should be expected to be sufficiently robust.
Instead, in order to overcome the problem of the long time necessary for manual delineation, an atlas based on the available manually segmented lungs is actually under development and validation; preliminary results promise to drastically reduce the time for segmentation in the future. Another limitation concerns the still limited number of patients, not yet able to depict the whole picture.
In conclusion, we demonstrated that few CT-based quantitative features extracted with an operator-independent approach based on lung densitometry of COVID-19 patients can be combined to build a model with moderately high discrimination in classifying patients based on their risk of death. The model can be significantly improved when combining them with few clinical parameters such as random glycemia at hospital admission, use of biological drugs and presence of active cancer.
Although mortality rate is hopefully expected to decrease also in patients with compromised lungs (i.e.: having a predicted high risk of mortality) during the next waves, the prediction of the risk of death from the first wave should remain as a clinically relevant, objective score for predicting illness severity in the future. External validations on other cohorts are warranted.
Of note, the Matlab scripts to extract the three lung components from the HU histogram and an excel form to calculate the risk of mortality are available upon request to the authors.
Acknowledgments
Acknowledgments
Dr M Mori is funded by an AIRC grant ( IG 23015 ).
The authors thank Davide Raspanti (TEMA Sinergie) for the precious support in the development and implementation of MIM workflows.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmp.2021.04.022.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agricola E., Beneduce A., Esposito A., Ingallina G., Palumbo D., Palmisano A. Heart and Lung Multimodality Imaging in COVID-19. JACC Cardiovasc Imaging. 2020;13:1792–1808. doi: 10.1016/j.jcmg.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X et al., CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 2020;295:202–207, DOI:10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed]
- 5.Ding X, Xu J, Zhou J, Long Q. Chest CT findings of COVID-19 pneumonia by duration of symptoms, Eur. J. Radiol. 2020;127:109009. DOI:10.1016/j.ejrad.2020.109009. [DOI] [PMC free article] [PubMed]
- 6.Pan F., Ye T., Sun P., Gui S., Liang B., Lingli L. novel coronavirus (COVID-19) pneumonia. Radiology. 2019;2020 doi: 10.1148/radiol.2020200370. [DOI] [Google Scholar]
- 7.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J. Radiological findings from 81 patients with COVID- 19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojha V., Mani A., Pandey N.N., Sharma S., Kumar S. CT in coronavirus disease 2019 (COVID-19): a systematic review of chest CT findings in 4410 adult patients. Eur Radiol. 2020;30(11):6129–6138. doi: 10.1007/s00330-020-06975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Z., Zhang Y., Wang Y.i., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol. 2020;30(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z., Guo D., Li C., Fang Z., Chen L., Yang R. initial chest CT findings. Eur Radiol. 2020;30(8):4398–4406. doi: 10.1007/s00330-020-06816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hani C., Trieu N.H., Saab I., Dangeard S., Bennani S., Chassagnon G. COVID-19 pneumonia: a review of typical CT findings and differential diagnosis. Diagn Interv Imaging. 2020;101(5):263–268. doi: 10.1016/j.diii.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (covid-19) pneumonia: a multicenter study. AJR Am J Roentgenol. 2020;214(5):1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 13.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. (COVID-19) in China: a report of 1014 cases. Radiology. 2019;2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Hong H, Yoon SH, Diagnostic performance of CT and reverse transcriptase-polymerase chain reaction for coronavirus disease 2019: a meta- analysis, Radiology 2020: 201343, DOI:10.1148/radiol.2020201343. [DOI] [PMC free article] [PubMed]
- 15.Hamer Okka Wilkea, Salzberger Bernd, Gebauer Johannes, Stroszczynski Christian, Pfeifer Michael. CT morphology of COVID-19: Case report and review of literature. RoFo Fortschritte Auf Dem Gebiet Der Rontgenstrahlen Und Der Bildgeb Verfahren. 2020;192(05):386–392. doi: 10.1055/a-1142-4094. [DOI] [PubMed] [Google Scholar]
- 16.Li Kunhua, Wu Jiong, Wu Faqi, Guo Dajing, Chen Linli, Fang Zheng. The Clinical and Chest CT Features Associated With Severe and Critical COVID-19 Pneumonia. Invest Radiol. 2020;55(6):327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansell David M., Bankier Alexander A., MacMahon Heber, McLoud Theresa C., Müller Nestor L., Remy Jacques. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246(3):697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]
- 18.Liu F., Zhang Q., Huang C., Shi C., Wang L., Shi N. CT quantification of pneumonia lesions in early days predicts progression to severe illness in a cohort of COVID-19 patients. Theranostics. 2020;10(12):5613–5622. doi: 10.7150/thno.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meiler Stefanie, Schaible Jan, Poschenrieder Florian, Scharf Gregor, Zeman Florian, Rennert Janine. 109256. Can CT performed in the early disease phase predict outcome of patients with COVID 19 pneumonia? Analysis of a cohort of 64 patientsfrom Germany. Eur J Radiol. 2020;131:109256. doi: 10.1016/j.ejrad.2020.109256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombi Davide, Bodini Flavio C., Petrini Marcello, Maffi Gabriele, Morelli Nicola, Milanese Gianluca. Well-aerated lung on admitting chest CT to predict adverse outcome in COVID-19 pneumonia. Radiology. 2020;296(2):E86–E96. doi: 10.1148/radiol.2020201433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ,2020:369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed]
- 22.Leonardi Andrea, Scipione Roberto, Alfieri Giulia, Petrillo Roberta, Dolciami Miriam, Ciccarelli Fabio. Role of computed tomography in predicting critical disease in patients with covid-19 pneumonia: a retrospective study using a semiautomatic quantitative method. Eur J Radiol. 2020;130:109202. doi: 10.1016/j.ejrad.2020.109202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matos J., Paparo Francesco, Mussetto Ilaria, Bacigalupo Lorenzo, Veneziano Alessio, Perugin Bernardi Silvia. Evaluation of novel coronavirus disease (COVID-19) using quantitative lung CT and clinical data: prediction of short-term outcome. Eur Radiol Exp. 2020;4(1) doi: 10.1186/s41747-020-00167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zangrillo A., Beretta L., Scandroglio Monti G, Fominskiy E., Colombo S., Morselli F. Characteristics, treatment, outcomes and cause of death of invasively ventilated patients with COVID-19 ARDS in Milan, Italy. Crit Care Resusc. 2020;3:200–211. doi: 10.1016/S1441-2772(23)00387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciceri F, Castagna A, Rovere-Querini P, Decobelli F, Ruggeri A, Galli A et al. Early predictors of clinical outcomes of COVID-19 outbreak in Milan, Italy, Clinical Immunology 2020:217,17:108509. doi:10.1016/j.clim.2020.108509.Epub 2020 Jun 12. [DOI] [PMC free article] [PubMed]
- 26.Mazzilli Aldo, Fiorino Claudio, Loria Alessandro, Mori Martina, Esposito Pier Giorgio, Palumbo Diego. An automatic approach for individual HU-based characterization of lungs in COVID-19 patients. Appl Sciences. 2021;11(3):1238. doi: 10.3390/app11031238. [DOI] [Google Scholar]
- 27.Loeh Benjamin, Brylski Lukas T., von der Beck Daniel, Seeger Werner, Krauss Ekaterina, Bonniaud Philippe. Lung CT densitometry in idiopathic pulmonary fibrosis for the prediction of natural course, severity and mortality. Chest. 2019;155(5):972–981. doi: 10.1016/j.chest.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 28.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 29.Li Lin, Qin Lixin, Xu Zeguo, Yin Youbing, Wang Xin, Kong Bin. Using Artificial Intelligence to Detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaganti Shikha, Grenier Philippe, Balachandran Abishek, Chabin Guillaume, Cohen Stuart, Flohr Thomas. Automated Quantification of CT Patterns Associated with COVID-19 from Chest CT. Radiol Artif Intell. 2020;2(4):e200048. doi: 10.1148/ryai.2020200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins G S, Reitsma J B, Altman D G, Moons K G M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Br J Cancer. 2015;112(2):251–259. doi: 10.1038/bjc.2014.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jajodia A, Ebner L, Heidinger B, Chaturvedi A, Prosch H. Imaging in corona virus disease 2019 (COVID-19)—A Scoping review. Eur Jour Radiol Open, 2020 7, art. no. 100237. DOI:10.1016/j.ejro.2020.100237. [DOI] [PMC free article] [PubMed]
- 33.Yip S, Klanecek Z, Naganawa S, Kim J, Studen A, Rivetti L et al. Performance and Robustness of Machine Learning-based Radiomic COVID-19 Severity Prediction. Medrxiv, 2020: DOI:10.1101/2020.09.07.20189977. (white paper on web).
- 34.Mushtaq J, Pennella R, Lavalle S, Colarieti A, Steidler S, Martinenghi CMA et al. Initial chest radiographs and artificial intelligence (AI) predict clinical outcomes in COVID-19 patients: analysis of 697 Italian patients. Eur Radiol 2020. In press. [DOI] [PMC free article] [PubMed]
- 35.Li Zhang, Zhong Zheng, Li Yang, Zhang Tianyu, Gao Liangxin, Jin Dakai. From community-acquired pneumonia to COVID-19: a deep learning-based method for quantitative analysis of COVID-19 on thick-section CT scans. Eur Radiol. 2020;30(12):6828–6837. doi: 10.1007/s00330-020-07042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.