Graphical abstract
Review constructs molecular underpinning of innate immune responses generated against SARS-CoV-2. Model depicts mechanism driving proinflammatory and interferon responses against SARS-CoV-2 in vivo. SARS-CoV-2 surface proteins and RNA genome are sensed by TLRs and RIG1 like receptors triggering proinflammatory and interferon responses. TLR3 mediated TBK1 and IRF3 activation have been reported. Likewise, SARS-CoV-2 RNA genome sensing by MDA5 and subsequent IFN production have also been shown. The de-ISGylation of MDA5 allows escaping of SARS-CoV-2 sensing and antiviral responses.
Keywords: SARS-CoV-2, Interferon regulatory factors, Alveolar macrophages, Toll like receptors
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections have triggered global pandemic that continue to impact adversely human health. New understanding has emerged about the innate and adaptive immune responses elicited in SARS-CoV-2 infection. The understanding of innate immune responses generated in hosts early in SARS-CoV-2 infection is vital for treatment efforts. Antiviral cytokines are released by innate immune cells in response to viral infections that play a pivotal role in limiting viral replication, pathology and generating optimal adaptive immune responses alongside the long-term memory responses against reinfections. One aspect of innate immune response generated against SARS-CoV-2 in vivo and which has received much attention has been high proinflammatory cytokine release in COVID-19 patients. Another vital discovery has been that the antiviral cytokine type I Interferon (IFN) family IFN-α mediates upregulation of angiotensin converting enzyme 2 (ACE2) membrane protein in airway epithelial cells. ACE2 is a receptor that SARS-CoV-2 binds to infect host cells. New understanding has emerged about the mechanism of SARS-CoV-2 induced exaggerated proinflammatory cytokine release as well as transcriptional regulation of ACE2. This review discusses various mechanisms underlying SARS-CoV-2 induced exaggerated proinflammatory cytokine response as well as transcriptional regulation of ACE2 receptor. We further elaborate on adaptive and memory responses generated against SARS-CoV-2.
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is an ongoing global pandemic that has imposed huge burden on human health (Zhang and Holmes, 2020; Wu et al., 2020; Coronaviridae Study Group of the International Committee on Taxonomy of V, 2020). Intense efforts are being invested to develop new therapeutics and more effective vaccines to control the disease. SARS-CoV-2 infection causes pneumonia like symptoms including bilateral lung involvement followed by acute respiratory distress, and lung damage in advanced stages (Lai et al., 2020). SARS-CoV-2 has also been reported to cause cardiovascular complications (Tavazzi et al., 2020; Li et al., 2020). A significant number of SARS-CoV-2 infected patients develop protective immunity even though the nature and duration of protective immunity is yet to be fully determined. Excessive levels of proinflammatory cytokines have been detected in COVID-19 patients that function as mediators of acute respiratory distress and cardiovascular complications such as acute myocarditis and endothelitis (Huang et al., 2020; Ong et al., 2020; Zhang et al., 2020a). New knowledge and mechanisms leading to proinflammatory cytokine release in SARS-CoV-2 and inflammation related damage to lungs and cardiovascular system has become available (Tavazzi et al., 2020; Li et al., 2020; Tersalvi et al., 2020). Involvement of the vascular system in COVID-19 patients could be via direct infection of endothelium and driven by inflammatory responses causing endotheliitis and hypercoagulable state. The ACE2 is a cellular receptor for SARS-CoV-2 entry into host. SARS-CoV-2 binds ACE2 with high affinity through receptor binding domain (RBD) of S-protein (Wrapp et al., 2020; Wan et al., 2020). ACE2 is expressed in gastrointestinal tissues, airway epithelium, and on endothelial cells (Bao et al., 2020). ACE2 expression in airway epithelial tissues and endothelium represents a major source of SARS-CoV-2 entry into lung epithelial tissues and possibly of endothelium potentially leading to cardiovascular complications (Imai et al., 2005; Ferrario et al., 2005; Varga et al., 2020). This notion is supported by the in vitro data that suggested SARS-CoV-2 could infect engineered human blood vessel organoids (Monteil et al., 2020). Patient data further supported virus presence within endothelial cells and accumulation of inflammatory cells in the heart that could potentially explain the mechanism underlying impairment of cardiovascular functions in COVID-19 patients (Varga et al., 2020). SARS-CoV-2 entry require S-protein cleavage into S1 and S2 by host transmembrane protease, the serine 2 (TMPRSS2) protease. S1 binds to the ACE2 receptor whereas S2 activity is required to form membrane fusion with the host cells (Ziegler et al., 2020). The TMPRSS2 protease has also been associated with gender related mortality differences observed between males and females infected with SARS-CoV-2 with reports indicating increased prevalence of COVID-19 cases in males compared with females (Mjaess et al., 2020; Jin et al., 2020). This effect could be related to coregulation of androgen hormone receptor and TMPRSS2 gene in what is described as TMPRSS2-androgens axis. The TMPRSS2 gene expression is regulated by androgen receptors (Lin et al., 1999). Androgen receptor gene is located in close proximity with TMPRSS2 locus (Shen et al., 2017).
Model organisms could help investigate the mechanisms related to cardiac and lung injury associated with SARS-CoV-2. The K18-hACE2 transgenic mouse model generated by Jackson Laboratories could be one such useful resource for studying SARS-CoV-2 pathogenesis. K18-hACE2 transgenic mice expresses human ACE2-coding sequences under transcriptional regulation of human cytokeratin K18 promoter driving ACE2 expression predominantly in epithelial cells. Thus, allowing SARS-CoV-2 to bind lung epithelium and potentially generating respiratory distress resembling human COVID-19 severe acute respiratory syndrome, which is responsible for majority of hospitalization and disease related deaths. Boa et al; created a new hACE2 transgenic mouse model (Bao et al., 2020). This model was tested for SARS-CoV-2 pathogenicity. SARS-CoV-2 was delivered intranasally and following the disease progression histopathological examination was performed that revealed interstitial pneumonia and lung damage. The lung infiltrates appear to have accumulation of lymphocytes and alveolar macrophages suggesting a possible lung damage due to excessive proinflammatory cytokine response and macrophage activation (Bao et al., 2020). The study does not examine in detail the involvement of TLRs and cytokine release from innate immune cells. One drawback with hACE2 transgenic mice model is that it cannot reliably recapitulate human innate and adaptive responses elicited against SARS-CoV-2. Therefore, a humanized mouse model expressing human airway epithelial tissues and human immune system may be a reliable model. The Lung only Mice (LoM) model may be one such model. The LoM model was created by implanting human lung tissues on an immunodeficient background. However, this mouse model lacks an immune system. Alternatively, the BLT-L (bone marrow, liver, thymus and lung tissue) which is a humanized mouse model that can generate hematopoietic system from transplanted bone marrow. The BLT-L humanized mouse model was created by the same group that developed the Lung-only Mice. In BLT-L model the autologous bone marrow, liver, thymus tissue, and lung tissue are engrafted on an immunodeficient background. The benefit of BLT-L model over LoM is that this model can generate innate and adaptive responses against viral infections (Wahl et al., 2019), thus, could serve as a model to investigate human immune responses against SARS-CoV-2 infection.
A crosstalk between innate and adaptive immune responses is essential for eradicating SARS-CoV-2 infection as well as for generating long term memory responses against reinfections. There is little understanding about the innate and adaptive immune responses elicited in SARS-CoV-2 infections in vivo in humans and in animal models. This review in below sections construct an innate and adaptive immune response model based on the recently published data on SARS-CoV-2 pathogenesis and immune responses measured in COVID-19 patients.
2. Cellular receptors sensing SARS-CoV-2
2.1. Viral associated molecular patterns
Innate immune responses against viral infections are elicited after host innate immune cells through their pattern recognition receptors (PRRs) recognize virus associated molecular patterns. Viruses utilize coat/membrane proteins to enter host cells by binding specifically on receptors/coreceptors. Host cells respond by internalizing viruses through receptor mediated endocytosis (Mercer et al., 2010). A second mechanism involves viruses attaching and aggregating non-specifically on host cell membranes e.g., cell membrane polysaccharides triggering clathrin mediated endocytosis or other less characterized endocytosis mechanisms such as micropinocytosis or caveolar dependent endocytosis (Mercer et al., 2010; Gruenberg, 2001). Once inside the host cells, internalized viruses are delivered to endosomes. Endosomes have low pH and are rich in proteases that among others include the cathepsins, their activity leads to uncoating of viruses and subsequent penetration into cytoplasm (Chandran et al., 2005). No direct evidence exists yet of SARS-CoV-2 engaging endosomal pathway. However, based on other CoV families such as SARS-CoV and Middle Eastern Respiratory Syndrome (MERS-CoV), which have been shown to engage the endocytic pathways (Yang and Shen, 2020; Wang et al., 2008; Qing et al., 2020; Simmons et al., 2005). It is most likely, therefore, that SASR-CoV-2 may be also engaging receptor mediated endocytosis, which is being explored as a potential therapeutic target to control SARS-CoV-2 infection and COVID-19 (Yang and Shen, 2020).
Endosomes are rich in endosomal toll-like receptors (TLRs) which sense viral genomes. The retinoic acid-inducible gene-I (RIG1) like receptors (RLRs) are second class of intracellular receptors that sense viral genomes after their penetration into cytoplasm. Innate immune cells as well as epithelial cell surfaces form the first line of defense against pathogens, they express both TLRs and RLRs (Akira et al., 2006). The endosomal TLR3 detects viral double-stranded RNA (ds-RNA) genome, which is transient replication intermediate of single stranded RNA (ss-RNA) viruses (Alexopoulou et al., 2001). Likewise, endosomal TLR7 and TLR8 detect viral ss-RNA (Diebold et al., 2004). Currently, it is not clear if endosomal TLRs or RLRs or both sense SARS-CoV-2 genome. SARS-CoV-2 contain RNA genome therefore involvement of both classes of receptors in sensing SARS-CoV-2 genome cannot be ruled out.
2.1.1. Mechanism of SARS-CoV-2 recognition by host pattern recognition receptors
The mechanism driving exaggerated proinflammatory cytokine response and deleterious T cell activity in COVID-19 patients is presently unclear (Ong et al., 2020; Zhou et al., 2020). The data discussed above suggest a potential involvement of Interferon regulatory factors that become activated through TLRs or RLRs mediated signaling pathways. No direct evidence exists yet that demonstrates SARS-CoV-2 structural proteins binding to any of the transmembrane TLRs, however, the likely candidates may be TLR2 or TLR4 as these have already been shown to engage viral coat proteins (Fig. 1 ). The herpes simplex virus 1 (HSV-1) and respiratory syncytial virus (RSV) envelope proteins have been shown to bind TLR2 and TLR4 triggering proinflammatory cytokine release (Kurt-Jones et al., 2004, 2000). A recent study demonstrated increased TLR2 mRNA fold expression in mild/moderate COVID-19 patients compared with healthy controls (Hadjadj et al., 2020). Similarly, the viral ds-RNA genomes bind to endosomal TLR3 whereas viral ss-RNA genomes bind to TLR7 and TLR8 (Akira et al., 2006; Alexopoulou et al., 2001; Diebold et al., 2004; Heil et al., 2004). Therefore, it may be anticipated that TLR7/8 could sense the SARS-CoV-2 genome (Fig. 1). However, no direct evidence exists showing SARS-CoV-2 genome binding to TLR7/8. Even the data related to TLRs sensing RNA genomes of SARS family viruses e.g., SARS-CoV and MERS-CoV is not clear with the exception that TLR adaptor MyD88 and TRIF were shown to be important for innate antiviral immunity in mouse models (Totura et al., 2015). Hadjadj et al., in recently published article demonstrated an increased expression of TLR7 in mild/moderate COVID-19 patients compared with healthy controls, whereas TLR8 expression was also elevated in severely ill COVID-19 patients compared with mild/moderate cases (Hadjadj et al., 2020). Interestingly, in the paper the interferon gene expression heatmap analysis revealed that IRF5 and IRF7 expression to be low in healthy controls as expected whereas their expression increased strongly in mild/moderate and severely ill COVID-19 patients followed by abrupt decrease in critically ill patients (Hadjadj et al., 2020). The kinetics of IRF5, IRF7 expression data could potentially explain the mechanism underlying reduced levels of type I and III interferon detection in severely ill COVID-19 patients (Trouillet-Assant et al., 2020; Gardinassi et al., 2020).
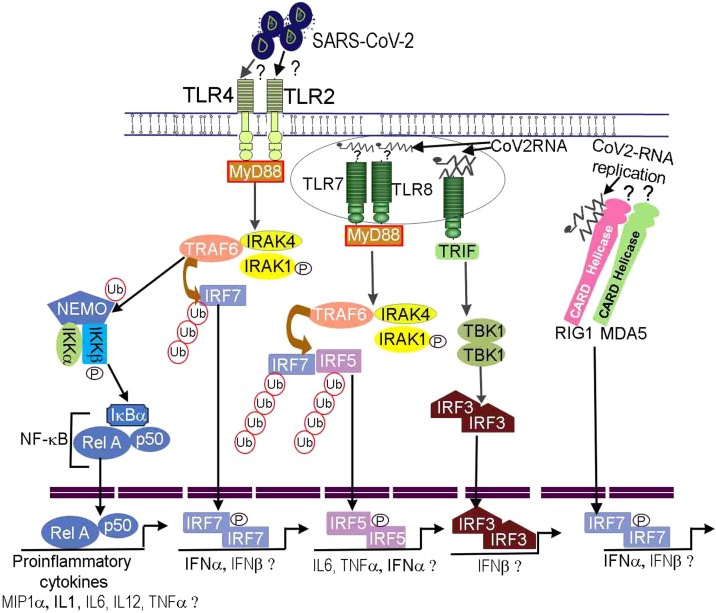
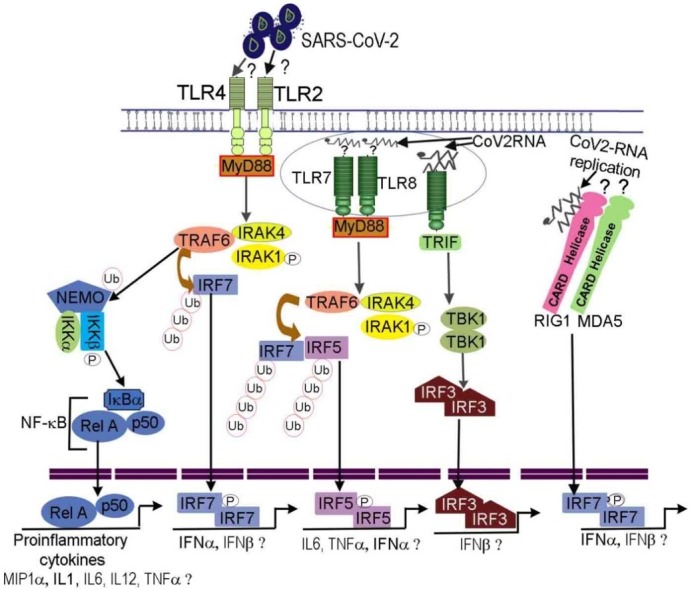
Fig. 1.
Predicted model depicting innate immune responses against SARS-CoV-2. SARS-CoV-2 enters epithelial/endothelial cells utilizing ACE2 receptor. The viral genome and surface proteins are sensed by TLRs and RIG like receptor family present in tissue resident and migratory dendritic cells triggering inflammatory and chemokine responses. Proinflammatory cytokines have been detected in COVID-19 patients suggesting SARS-CoV-2 surface proteins may be potentially recognized by TLR4 or TLR2 activating MyD88→TRAF6-IRAK. This assumption is based on studies showing HIV, HSV viral structural proteins binding to TLR2 (Henrick et al., 2015; Leoni et al., 2012) as well as recent RNA-sequencing data in COVID-19 patients (Hadjadj et al., 2020). The TRAF6 functions as E3 ligase to ubiquitinate IRF7 as well as NEMO of IKK complex. Ubiquitinated IRF7 also becomes phosphorylated, dimerize, and translocate into the nucleus to initiate transcription of antiviral cytokines. The IKKβ kinase of IKK complex phosphorylate IκBα of NF-κB releasing p65/p50 that enter the nucleus initiating proinflammatory cytokine gene transcription program. Likewise, SARS-CoV-2 genome binding to TLR7 or TLR8 activates MyD88→TRAF6-IRAK complex. The TLR7 and TLR8 have been found to be expressed in mild/moderate and severely ill COVID-19 patients respectively (Hadjadj et al., 2020). The TRAF6 ligase ubiquitinates IRF5. The IRF5 undergo phosphorylation, dimerize, and translocate into nucleus to transcribe proinflammatory cytokines, type I IFN e.g., IFNα/β. SARS-CoV-2 could bind to TLR3, MDA5 or RIG1 helicases during its replication wherein it generates a transient double stranded RNA structure. The dsRNA binding to TLR3 recruits TRIF adaptor to activate IRF3. Likewise, the dsRNA binding to RIG1 could activate IFN gene transcription via IRF7. The IFN-α/β has been shown to upregulate ACE2 receptor in airway epithelial cells. The IFN-α/β bind to IFNA receptor. The binding leads to IFNA receptor dimerization and ITAM phosphorylation to serve as a docking site for JAK receptor tyrosine kinases that become activated through autophosphorylation within tyrosine residues. The activated JAKs phosphorylate STATs leading to their dimerization and nuclear translocation. STAT dimers activate antiviral gene transcription program some of which are involved in cleaving viral proteins and viral RNAs in infected cells. STAT dimers also potentially bind to the ACE2 upregulating its expression. Interferon regulatory factors IRF7, IRF5, IRF3 and NF-κB including elements of interferon and STAT signaling pathways have been detected in peripheral lymphocytes (Wilk et al., 2020) and immune cells present in bronchoalveolar lavage of COVID-19 patients (Liao et al., 2020).
2.1.2. Involvement of TLR mediated signaling pathways in SARS-CoV-2 infection
The sensing of SARS-CoV-2 ss-RNA genome is anticipated to activate TLR7/8-MyD88-TRAF6-IRF5/IRF7 or TLR7/8-MyD88-IKK-NF-κB signaling pathways (Fig. 1 (Akira et al., 2006; Diebold et al., 2004; Honda et al., 2004)). Hadjadj et al., publication predicted the involvement of NF-κB signaling pathway especially in relation with its transcriptional regulation of IL6 and TNFα (Hadjadj et al., 2020). The role of intracellular RLRs such as MDA5, RIG1 sensing SARS-CoV-2 viral genome is actively being investigated. It may be anticipated that after sensing viral genome by MDA5 or RIG1 that signal via caspase activation and recruitment domain (CARD) bind to mitochondrial MAVS protein. MAVS recruit TRAF3-TRADD-RIP1 activating IKK complex that in turn activate NF-κB, IRF3 and IRF7 transcription factors (Fig. 1 (Yoneyama et al., 2004; Honda et al., 2005)). Therefore, we expect the above described signaling pathways to be activated in innate immune cells sensing SARS-CoV-2 thereby leading to synthesis of proinflammatory cytokines and IFNs. IFNs subsequently mediate antiviral protein synthesis in infected host epithelial cells. The activated innate immune cells release type I IFN that bind to IFNA-R activating JAK-STAT signaling pathway inducing transcription of antiviral proteins as well as ACE2 receptor upregulation as predicted in the model (Ziegler et al., 2020). Earlier studies on SARS-CoV have indicated that SARS-CoV accessory open reading frame 6 (ORF6) could antagonize interferon responses by inhibiting STAT1 nuclear localization (Frieman et al., 2007; Kopecky-Bromberg et al., 2007). Similarly, SARS-CoV-2 ORF6 (which shares ∼66 % sequence homology with SARS-CoV ORF6) have been reported to be involved in suppressing IFN production potentially through inhibition of IRF3 and STAT1 nuclear localization in a mechanism that seem to be independent of STAT1 phosphorylation (Lei et al., 2020). Coronavirus families use proteases to cleave replicase proteins. Novel SARS-CoV gene 1 product encodes replicase polypeptide that is processed into nonstructural proteins (NSPs) through activity of two proteases among which include papain-like protease (PLpro) (Harcourt et al., 2004). PLpro is an essential component of protease machinery in SARS-CoV-2. Its activity is involved in evasion of innate antiviral responses through cleavage of ubiquitin-like interferon-stimulated gene 15 (ISG15) protein linked with IRF3 as well as partial deubiquitination especially K48-Ub2 of other substrates in vitro. Cleavage of ISG15 posttranslational modification of IRF3 contributed to reduced type I IFN production whereas inhibiting PLpro promote TBK-IRF3 pathway demonstrating utility of targeting negative regulators of IFN pathway for control of SARS-CoV-2 and COVID-19 (Shin et al., 2020). Liu et al., recently demonstrated that SARS-CoV-2 papain-like protease mediate de-ISGylation of melanoma differentiation antigen (MDA5), which senses coronavirus RNA genome (Menachery et al., 2014; Liu et al., 2021). The de-ISGylation of MDA5 allows escape of SARS-CoV-2 sensing and antiviral responses (Liu et al., 2021).
The antiviral transcription factor IRF5 has been recognized to be involved in transcriptional upregulation of proinflammatory cytokines, chemokines, and type I IFNs especially IFNα4, IFN-β in response to TLR7/8 recognizing single stranded RNA viruses in vivo e.g., influenza A virus, murine norovirus, West Nile Virus, Newcastle disease virus (Graham et al., 2007; Forbester et al., 2020; Barnes et al., 2004). IRF5 is also associated with systemic lupus erythematosus (SLE), an autoimmune disease characterized by highly elevated type I IFN in patient serum (Graham et al., 2007; Eames et al., 2016). Both human and mouse IRF5 undergo Lys-63 ubiquitination in vitro in response to sensing TLR7/8 agonist R848 (resiquimod), a potent innate immune cell activator (Balkhi et al., 2008). K63-ubiquitination of IRF5 has been reported to be vital for its stability, nuclear translocation, recruitment to virus response elements present in proinflammatory and type I IFN genes (Balkhi et al., 2008; Paun et al., 2008). IRF5 becomes activated via the MyD88-TRAF6-IRAK pathway. TRAF6 is the E3-Ligase that catalyses K63-ubiquitination of IRF5 (Balkhi et al., 2008). Correspondingly, the serum collected from mice infected with NDV virus or plasmacytoid dendritic cells deficient in IRF5 were shown to produce reduced amounts of proinflammatory cytokines and type I IFNs (Paun et al., 2008; Takaoka et al., 2005). IRF7 is another transcription factor which is vital for antiviral innate and adaptive immunity. IRF7 becomes K63-ubiquitinated in MyD88-TRAF6 pathway in response to TLR7/8 sensing viral single strand RNA genomes. Abrogating K63-ubiquitination inhibits type I IFN transcription (Honda et al., 2005; Kawai et al., 2004). Interestingly, the early response IFNα4 which is regulated by IRF5 in positive feedback mechanisms activates delayed IFN genes as well as IRF7 (Marie et al., 1998). The role of IRF5 and IRF7 and the mechanism of their activation through ubiquitination or phosphorylation during SARS-CoV-2 infection is unknown. Furthermore, it is also unknown if these two vital anti-viral regulatory factors are involved in regulating transcription of excessive proinflammatory cytokines in response to SARS-CoV-2 infection (Fig. 1). Recent reports using single cell RNA sequencing performed on peripheral blood mononuclear cells of COVID-19 patients compared to healthy controls revealed elements of interferon signaling pathways, IRF7, IRF5, IRF3 to be highly elevated suggesting their potential role in generating interferon responses (Wilk et al., 2020). Likewise, the bronchoalveolar lavage immune cell profiling using single cell RNA sequencing also revealed an enrichment of expression clusters consistent with activation of innate immune cells and interferon-STAT signaling pathways as elaborated in above sections (Liao et al., 2020). In a study performed by Zhang et al., loss of function variant alleles was detected in COVID-19 patients presenting life threatening pneumonia. The loss of functional allele variants correspond to TLR and IFN genes including autosomal-recessive deficiencies of IRF7 and IFNAR1 and autosomal-dominant deficiencies of TLR3, UNC93B1, TICAM1, TBK1, IRF3, IRF7, IFNAR1, and IFNAR2 (Zhang et al., 2020b). In the same study, Zhang et al., further demonstrated that IRF7 deficient plasmacytoid dendritic cells were refractory to IFN production upon SASR-CoV-2 infection lending credibility to the model presented in Fig. 1. Taken together, elucidating the role of IRF3, IRF5, IRF7, and mechanisms of their activation can prove vital for our understanding of innate immune responses elicited against SARS-CoV-2 infection. As additional evidence begins to emerge, their role is also becoming more clearer especially related to excessive proinflammatory cytokine release and perturbation of type I interferon production during progression of SARS-CoV-2 infection from moderate to severely ill stages (Lei et al., 2020). Correspondingly, whether NF-κB, IRF5 could serve as drug targets in controlling excessive IL6 release in COVID-19 patients’ needed to be investigated. There is also a potential for enhancing activity of IRF7, IRF3 to promote type I IFN production (Shin et al., 2020; Zhang et al., 2020b). Even though the clinical relevance of these factors in controlling SARS-CoV-2 and for generating protective immunity in COVID-19 remains unknown. Synagren, the respiratory drug discovery and development company in June 2020 released a report on a phase II trial performed with its experimental drug SNG001 in COVID-19 patients (NCT04385095). SNG001 is an IFN-β inhalation drug with reports of benefit in COVID-19 patients (https://www.synairgen.com).
2.1.3. Interferon and inflammatory responses in SARS-CoV-2 infection
Some of the earlier studies performed on COVID-19 patients utilized plasma samples or RNA extracted from whole blood to examine cytokine profile and gene expression. These studies revealed elevated levels of proinflammatory cytokines in patient serum such as IL-6, IL8, IL1α, IL1β, TNFα, as well as type I interferon, IFN-α/β (Huang et al., 2020; Ong et al., 2020). However, the data presented in these two publications lacked quantitation, it was not clear how high the IFN-α/β levels were present in COVID-19 patients. It is interesting to note that excessive proinflammatory cytokines have also been reported in SARS-CoV and MERS-CoV infections (Peiris et al., 2003; Drosten et al., 2003). Despite IFN response in lung tissues there is no definitive proof yet of systemic IFN response in COVID-19 patients (Trouillet-Assant et al., 2020; Gardinassi et al., 2020; Zhang et al., 2020c; Song et al., 2020). A study by Blanco-Melo D et al., detected only low levels of type I and III interferon whereas higher levels of chemokines and IL6 were detected in primary human bronchial epithelial cells in vitro, ferret animal model, and in serum of SARS-CoV-2 infected patients (Blanco-Melo et al., 2020). In another study by Hadjadj J, et al., no IFN-α/β were detected in severely ill COVID-19 patients whereas elevated levels of IL6 and TNFα could be detected (Hadjadj et al., 2020). Several other publications have also reported of detecting only low levels of type I and III IFNs in peripheral lymphocytes and in plasma of COVID-19 patients whereas IL6, C-reactive protein (CRP), and chemokine IFNγ–induced protein 10 (IP-10) could be detected at higher levels (Trouillet-Assant et al., 2020; Gardinassi et al., 2020). The serum cytokine levels in COVID-19 patients can show age dependent variation. For examples, in children and young adults <24 years of age Interleukin 17A and IFN-γ were detected to be high however their levels dropped significantly in older patients (>24 years) (Tang et al., 2008). The persistently elevated levels of serum IL1 and IL6 in COVID-19 patients resemble macrophage activation syndrome (MAS)/hemophagocytic lymphohistiocytosis (HLH) (Tang et al., 2008). Two types of HLH have been described, the primary and secondary. A rare autosomal recessive trait causes primary HLH in children whereas secondary HLH/MAS may be driven by infections or autoimmune diseases (Risma and Jordan, 2012; Barrett et al., 2014). HLH/MAS is characterized by histiocytes infiltration of vital organs and abnormal immune activation as well excessive proinflammatory cytokine presence in serum (Barrett et al., 2014; Grupp et al., 2013). COVID-19 patients who present with acute respiratory distress syndrome (ARDS) have been shown to respond to anti-IL-6R blockade with tocilizumab (Yao et al., 2020; Giamarellos-Bourboulis et al., 2020). Tocilizumab is standard therapy applied to reverse clinical symptoms associated with Cytokine storm syndrome (CRS) in patients who receive autologous chimeric antigen receptor (CAR) modified T cells (Grupp et al., 2013; Davila et al., 2014).
Multisystem Inflammatory syndrome in children. Some children with COVID-19 present multiorgan inflammation referred to as Multisystem Inflammatory syndrome in children (MIS-C) associated with SARS-CoV-2 (Harwood et al., 2020; Godfred-Cato et al., 2020). The disorder presents clinical features characterized by shock, macrophage activation, and Kawasaki like disease. Kawasaki is a rare acute disease characterized by vasculitis affecting small and medium size arteries, some present with long-term effect on coronary arteries leading to coronary artery dilation or aneurysms (Kato et al., 1996; Verdoni et al., 2020). The median age of children with MIS-C is ∼8 years. Such children present with multiorgan inflammation in gastrointestinal, cardiovascular, respiratory, hematologic and mucocutaneous tissues (Feldstein et al., 2020; Jiang et al., 2020; Dufort et al., 2020). The elevated serum cytokines especially IL1 and IL6 and other proinflammatory cytokines detected in COVID-19 patients suggest an early innate immune response elicited against SARS-CoV-2 infection potentially involves TLR mediated recognition of viral PAMPs (Zhang et al., 2020b). The TLRs recognizing SARS-CoV-2 PAMPs can be present on alveolar macrophages, tissue resident dendritic cells (DC’s) and DC’s in blood (Ong et al., 2020; Yoshikawa et al., 2009). The data further imply that nasal epithelial cells, innate immune cells especially conventional dendritic cells (cDCs), and alveolar macrophages may shape the early adaptive immune responses in COVID-19 patients. It also appears from the study that attenuated T cell response leads to prolong survival of COVID-19 patients whereas strong T cell response induced lung damage (Ong et al., 2020). The deleterious T cell and proinflammatory cytokine responses in some COVID-19 patients could be triggered by exaggerated proinflammatory cytokine production involving activity of IRF3, IRF5, IRF7, and others (Ong et al., 2020). IRF3 and IRF7 being the key regulatory factors involved in type I IFN gene transcription. IFN-α/β promotes upregulation of MHC Class I molecules which is important for CD8 mediated adaptive immune responses (Matsuyama et al., 1993; Taniguchi et al., 2001). It is possible that interferon regulatory factors may also be involved in regulating SARS-CoV-2 viral antigen presentation and, thus, shaping adaptive immune responses.
3. Adaptive immune responses in SARS-CoV-2
3.1. Autoantibodies and B cell responses in COVID-19 patients
Autoantibodies against interferons have been detected in severely ill COVID-19 patients presenting pneumonia. The IgG autoantibodies were detected against IFN-ω and IFN-α2. Correspondingly, in the in vitro studies these autoantibodies neutralized IFN-ω, IFN-α2. These data support the protective role of IFNs against SARS-CoV-2 pathogenesis in a subset of patients (Bastard et al., 2020). The exact mechanism underlying B cell responses against SARS-CoV-2 in vivo is presently unclear. The professional dendritic cells and helper T cells orchestrate B cell responses. Soluble viral antigens or antigens dragged into lymph nodes by migratory APCs can crosslink BCRs to activate antibody mediated responses, the fixation of complemet pathways, and memory B cell development (Wykes et al., 1998). For B cell responses CD4 T cell help is vital. The CD4 T cell responses in SARS-CoV-2 were correlated with protective IgG and IgA antibody titers confirming T cell dependent B cell responses in COVID-19 patients (Grifoni et al., 2020a). Since the CD4 T cell responses in SARS-CoV-2 appeared to be TH1, it is essential to examine cytokine profiles to confirm the role of other T helper subtypes. The predicted T cell responses generated against SARS-CoV-2 infections in vivo has been represented as a model (Fig. 2 ). Several elements of the model have been revealed recently. The nature of CD4 and CD8 T cells responding to SARS-CoV-2 infection appeared to be conventional effector T cells capable of generating robust T helper and memory responses against reinfections (Dan et al., 2021; Grifoni et al., 2020b; Suthar et al., 2020) (Fig. 2). Dan et al., demonstrated memory B cells, antigen specific CD4 and CD8 T cells persist ∼8 months post SARS-CoV-2 infection. SARS-CoV-2 S-protein, RBD specific anti-IgG titers that correlated with neutralizing responses (Suthar et al., 2020), and anti-IgA antibodies could be detected ∼6 to 8 months post infection. Likewise, memory B cells expressing IgG isotype showed similar pattern of persistence. SARS-CoV-2 protein specific CD4 and CD8 T memory cells were also detected, however, in contrast with memory B cells, their number declined ∼50 % within 6–8 months (Dan et al., 2021), which is not unprecedented. A study by Gaebler et al., also demonstrated RBD specific IgG and IgM presence in SARS-CoV-2 infected individuals that decreased significantly, however, the neutralizing antibody decreased only by ∼5 fold whereas RBD specific memory B cells persist around ∼6-months (Gaebler et al., 2021). Interestingly, apart from humans, the IgG antibody adoptive transfer from convalescent SARS-CoV-2 rhesus macaques provided protection to naive rhesus macaques upon challenge with SARS-CoV-2 (McMahan et al., 2021). These studies demonstrated a robust elicitation of both humoral and T cells mediated effector and memory responses against SARS-CoV-2 that persist and, thus, can provide long-term protection against reinfections.
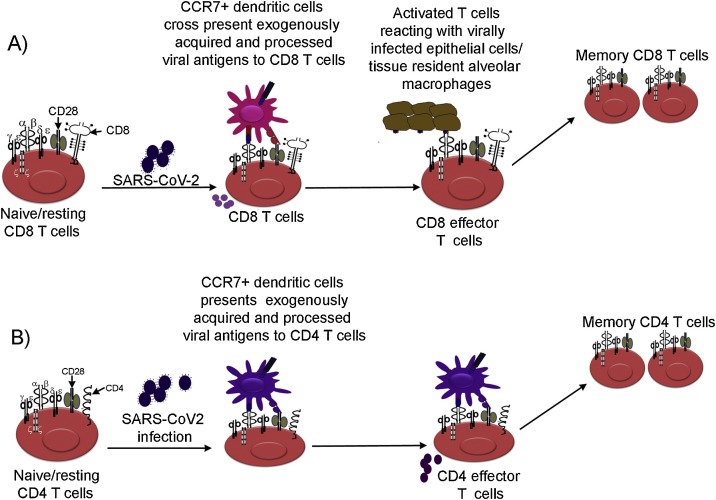
Fig. 2.
A predicted model depicting T cell responses againstSARS-CoV-2 infection. The model depicts CD8 T cells (A) and CD4 T cells (B) responses in SARS-CoV-2 infection. The CCR7+ migratory dendritic cells present exogenously acquired and processed SARS-CoV-2 antigens to CD8 T cells via cross presentation or to CD4 T cells via canonical presentation in tissues and lymph nodes. Tissue resident alveolar macrophages (TRAMs) also become infected with SARS-CoV-2, thus, can present endogenously processed viral antigens to CD8 T cells. Antigen reactive CD4 and CD8 T cells differentiate into effector T cells. With the elimination of infections effector T cells differentiate into terminal effector T cells followed by their contraction generating alongside long lived memory T cells. The phenotype of memory precursors and their ability to generate recall responses against SARS-CoV-2 reinfection is presently unclear. Activated CD4 T cells release effector cytokines that help B cells responses. CD8 T cells appear to follow a linear differentiation program in COVID-19 patients (Wilk et al., 2020).
3.1.1. Innate and adaptive immune system interact in SARS-CoV-2 infection
The mechanism of antigen specific T cell differentiation into effector and memory precursors in SARS-CoV-2 infection is poorly understood. In acute viral infections, the naïve/resting CD8 T cells become activated after interacting with professional antigen presenting cells that present viral antigens via canonical endogenous pathway or non-canonical cross presentation in tissues and draining lymph nodes. The activated T cells react and eliminate virally infected tissues (Gerlach et al., 2013; Kakaradov et al., 2017). With the elimination of acute infections, majority of antigen reactive effector T cells become programmed to terminally differentiate and undergo apoptosis where as a small percentage of cells survive that differentiate along memory T cell lineage (Balkhi, 2020). CD4 helper T cells via cytokine secretion play a vital role in B cell survival and antibody class switch recombination as well as CD8 memory T cell generation (Ahmed and Gray, 1996; Sun and Bevan, 2003). The interaction between innate and adaptive immune system in SARS-CoV-2 infection has recently been revealed. A landmark study by Grant et al., examined cross talk between innate and adaptive T cells in SARS-CoV-2 infected severely ill patients who have developed pneumonia. The RNA-seq data demonstrated enrichment of CD4 T cells, CD8 T cells, monocytes, CCR7+ migratory dendritic cells present in alveolar spaces, and TRAMs (tissue resident alveolar macrophages). Through single cell RNA-seq analysis, Grant et al., further demonstrated SARS-CoV-2 infection of alveolar macrophages (Grant et al., 2021). Additional evidence obtained through RNA sequencing analysis performed on peripheral lymphocytes and bronchoalveolar infiltrates indicated robust generation of SARS-CoV-2 specific effector CD8 T cell responses in moderately ill COVID-19 patients. The effector CD8 T cell response seem to decline in severely ill patients (Liao et al., 2020). Despite some of these recent findings, lack of suitable animal model remains a major bottleneck that hampers complete understanding of T cells responses generated against SARS-CoV-2 in vivo. Nevertheless, several pieces of data have been published on human MERS-CoV, SARS-CoV infections in humans and non-human primates that revealed robust T cell responses. The MERS-CoV specific CD4 and CD8 T cells were detected in patients exposed to MERS-CoV as well as patients that recovered from the disease. Using MERS-CoV specific 20-mer peptide corresponding to viral structural components i.e., surface, spike (S), membrane (M), and envelope (E) proteins, both CD4 and CD8 T cells could be detected in MERS-CoV infected patients indicating generation of both T helper and antiviral cytotoxic T cell responses (Alshukairi et al., 2018; Zhao et al., 2017). Remarkably, apart from effector T cells, both CD8 and CD4 T cell effector memory (CD45RA−) population were detected in MERS-CoV survivors indicating generation of effector memory T cell responses (Zhao et al., 2017). An excellent study published recently demonstrated SARS-CoV-2 specific T cell responses in COVID-19 convalescent patients. The study utilized an In silico predicted SARS-CoV-2 HLA I and II peptides designed to be recognized by antigen activated CD4 and CD8 TCRs (Grifoni et al., 2020a; Havenar-Daughton et al., 2016). In the same study, CD4 T cell recognition of SARS-CoV-2 S-protein specific peptides were detected to be more robust. CD4 T cell responses are vital for vaccine efforts and the results are important because S-protein is explored as the dominant epitope for vaccine -development against SARS-CoV-2 (Grifoni et al., 2020a; Gray et al., 2018). Activated CD4 T cells release cytokines especially IL4, IL5, IL2, IFNγ, TNFα that play vital role in antibody class switch recombination, CD8 T cells activation, memory generation, and maturations of professional dendritic cells (Fig. 2, (Sun and Bevan, 2003; Banchereau and Steinman, 1998; Bennett et al., 1998; Leist et al., 1989)).
4. Concluding remarks
Several new resources have been developed recently that can help investigate the immune responses elicited against SARS-CoV-2 in vivo in the mouse and cell line models. The heat-inactivated SARS-CoV-2 (ATCC VR-1986HK), total SARS-CoV-2 RNA genome (ATCC VR-740D), and SARS-CoV-2 expression vectors (Addgene) can be utilized to identify cellular pattern recognition receptors sensing SARS-CoV-2 associated molecular patterns as well as elucidating signaling pathways involved in synthesis of proinflammatory cytokines, type I interferons, and antiviral proteins. Furthermore, these resources can help explore the additional mechanisms underlying regulation of ACE2 in airway epithelial cells as well as transcriptional regulation of protease TMPRSS2.
Availability of data and material
I agree to sharing all the data and content of this review.
Author contributions
M.Y.B. conceived, planned, and wrote the review article.
Ethics approval and consent to participate
Not Applicable.
Consent for publication
I consent to publication.
Declaration of Competing Interest
The authors report no declarations of interest.
References
- Ahmed R., Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Alshukairi A.N., Zheng J., Zhao J., Nehdi A., Baharoon S.A., Layqah L., et al. High prevalence of MERS-CoV infection in camel workers in Saudi Arabia. mBio. 2018:9. doi: 10.1128/mBio.01985-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhi M.Y. Receptor signaling, transcriptional, and metabolic regulation of T cell exhaustion. Oncoimmunology. 2020;9 doi: 10.1080/2162402X.2020.1747349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhi M.Y., Fitzgerald K.A., Pitha P.M. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol. Cell. Biol. 2008;28:7296–7308. doi: 10.1128/MCB.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barnes B.J., Richards J., Mancl M., Hanash S., Beretta L., Pitha P.M. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J. Biol. Chem. 2004;279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- Barrett D.M., Teachey D.T., Grupp S.A. Toxicity management for patients receiving novel T-cell engaging therapies. Curr. Opin. Pediatr. 2014;26:43–49. doi: 10.1097/MOP.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020:370. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S.R., Carbone F.R., Karamalis F., Flavell R.A., Miller J.F., Heath W.R. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021:371. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila M.L., Riviere I., Wang X., Bartido S., Park J., Curran K., et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008226. 224ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Dufort E.M., Koumans E.H., Chow E.J., Rosenthal E.M., Muse A., Rowlands J., et al. Multisystem inflammatory syndrome in children in New York State. N. Engl. J. Med. 2020;383:347–358. doi: 10.1056/NEJMoa2021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames H.L., Corbin A.L., Udalova I.A. Interferon regulatory factor 5 in human autoimmunity and murine models of autoimmune disease. Transl. Res. 2016;167:167–182. doi: 10.1016/j.trsl.2015.06.018. [DOI] [PubMed] [Google Scholar]
- Feldstein L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., Son M.B.F., et al. Multisystem inflammatory syndrome in U.S. Children and adolescents. N. Engl. J. Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C.M., Jessup J., Chappell M.C., Averill D.B., Brosnihan K.B., Tallant E.A., et al. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111:2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- Forbester J.L., Clement M., Wellington D., Yeung A., Dimonte S., Marsden M., et al. IRF5 promotes influenza virus-induced inflammatory responses in human induced pluripotent stem cell-derived myeloid cells and murine models. J. Virol. 2020:94. doi: 10.1128/JVI.00121-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardinassi L.G., Souza C.O.S., Sales-Campos H., Fonseca S.G. Immune and metabolic signatures of COVID-19 revealed by transcriptomics data reuse. Front. Immunol. 2020;11:1636. doi: 10.3389/fimmu.2020.01636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C., Rohr J.C., Perie L., van Rooij N., van Heijst J.W., Velds A., et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635–639. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfred-Cato S., Bryant B., Leung J., Oster M.E., Conklin L., Abrams J., et al. COVID-19-Associated Multisystem Inflammatory Syndrome in Children - United States, March-July 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1074–1080. doi: 10.15585/mmwr.mm6932e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.R., Kyogoku C., Sigurdsson S., Vlasova I.A., Davies L.R., Baechler E.C., et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.I., Westerhof L.M., MacLeod M.K.L. The roles of resident, central and effector memory CD4 T-cells in protective immunity following infection or vaccination. Immunology. 2018 doi: 10.1111/imm.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of t cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of t cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Biol. 2001;2:721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- Grupp S.A., Kalos M., Barrett D., Aplenc R., Porter D.L., Rheingold S.R., et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood R., Allin B., Jones C.E., Whittaker E., Ramnarayan P., Ramanan A.V., et al. A national consensus management pathway for paediatric inflammatory multisystem syndrome temporally associated with COVID-19 (PIMS-TS): results of a national Delphi process. Lancet Child Adolesc Health. 2020 doi: 10.1016/S2352-4642(20)30304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Reiss S.M., Carnathan D.G., Wu J.E., Kendric K., Torrents de la Pena A., et al. Cytokine-independent detection of antigen-specific germinal center t follicular helper cells in immunized nonhuman Primates Using a live cell activation-induced marker technique. J. Immunol. 2016;197:994–1002. doi: 10.4049/jimmunol.1600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Henrick B.M., Yao X.D., Rosenthal K.L., team Is HIV-1 structural proteins serve as PAMPs for TLR2 heterodimers significantly increasing infection and innate immune activation. Front. Immunol. 2015;6:426. doi: 10.3389/fimmu.2015.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Yanai H., Mizutani T., Negishi H., Shimada N., Suzuki N., et al. Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15416–15421. doi: 10.1073/pnas.0406933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Tang K., Levin M., Irfan O., Morris S.K., Wilson K., et al. COVID-19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.M., Bai P., He W., Wu F., Liu X.F., Han D.M., et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front. Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakaradov B., Arsenio J., Widjaja C.E., He Z., Aigner S., Metz P.J., et al. Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 2017;18:422–432. doi: 10.1038/ni.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Sugimura T., Akagi T., Sato N., Hashino K., Maeno Y., et al. Long-term consequences of Kawasaki disease. A 10- to 21-year follow-up study of 594 patients. Circulation. 1996;94:1379–1385. doi: 10.1161/01.cir.94.6.1379. [DOI] [PubMed] [Google Scholar]
- Kawai T., Sato S., Ishii K.J., Coban C., Hemmi H., Yamamoto M., et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt-Jones E.A., Popova L., Kwinn L., Haynes L.M., Jones L.P., Tripp R.A., et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones E.A., Chan M., Zhou S., Wang J., Reed G., Bronson R., et al. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist T.P., Kohler M., Eppler M., Zinkernagel R.M. Effects of treatment with IL-2 receptor specific monoclonal antibody in mice. Inhibition of cytotoxic T cell responses but not of T help. J. Immunol. 1989;143:628–632. [PubMed] [Google Scholar]
- Leoni V., Gianni T., Salvioli S., Campadelli-Fiume G. Herpes simplex virus glycoproteins gH/gL and gB bind Toll-like receptor 2, and soluble gH/gL is sufficient to activate NF-kappaB. J. Virol. 2012;86:6555–6562. doi: 10.1128/JVI.00295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Y., Guan Z., Li H., Ye M., Chen X., et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lin B., Ferguson C., White J.T., Wang S., Vessella R., True L.D., et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- Liu G., Lee J.H., Parker Z.M., Acharya D., Chiang J.J., van Gent M., et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021;6:467–478. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie I., Durbin J.E., Levy D.E. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T., Kimura T., Kitagawa M., Pfeffer K., Kawakami T., Watanabe N., et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S., et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-o-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J., Schelhaas M., Helenius A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- Mjaess G., Karam A., Aoun F., Albisinni S., Roumeguere T. COVID-19 and the male susceptibility: the role of ACE2, TMPRSS2 and the androgen receptor. Prog. Urol. 2020;30:484–487. doi: 10.1016/j.purol.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Haja Mohideen S.M., et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27:879–882. doi: 10.1016/j.chom.2020.03.021. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun A., Reinert J.T., Jiang Z., Medin C., Balkhi M.Y., Fitzgerald K.A., et al. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. J. Biol. Chem. 2008;283:14295–14308. doi: 10.1074/jbc.M800501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E., Hantak M.P., Galpalli G.G., Gallagher T. Evaluating MERS-CoV entry pathways. Methods Mol. Biol. 2020;2099:9–20. doi: 10.1007/978-1-0716-0211-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risma K., Jordan M.B. Hemophagocytic lymphohistiocytosis: updates and evolving concepts. Curr. Opin. Pediatr. 2012;24:9–15. doi: 10.1097/MOP.0b013e32834ec9c1. [DOI] [PubMed] [Google Scholar]
- Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.W., Zhang C., Fan X., Meng F.P., Xu Z., Xia P., et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Bevan M.J. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- Tang Y., Xu X., Song H., Yang S., Shi S., Wei J., et al. Early diagnostic and prognostic significance of a specific Th1/Th2 cytokine pattern in children with haemophagocytic syndrome. Br. J. Haematol. 2008;143:84–91. doi: 10.1111/j.1365-2141.2008.07298.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Tavazzi G., Pellegrini C., Maurelli M., Belliato M., Sciutti F., Bottazzi A., et al. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur. J. Heart Fail. 2020;22:911–915. doi: 10.1002/ejhf.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersalvi G., Vicenzi M., Calabretta D., Biasco L., Pedrazzini G., Winterton D. Elevated troponin in patients with coronavirus disease 2019: possible mechanisms. J. Card. Fail. 2020;26:470–475. doi: 10.1016/j.cardfail.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Whitmore A., Agnihothram S., Schafer A., Katze M.G., Heise M.T., et al. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio. 2015;6:e00638–15. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillet-Assant S., Viel S., Gaymard A., Pons S., Richard J.C., Perret M., et al. Type I IFN immunoprofiling in COVID-19 patients. J. Allergy Clin. Immunol. 2020;146:206–208. doi: 10.1016/j.jaci.2020.04.029. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A., De C., Abad Fernandez M., Lenarcic E.M., Xu Y., Cockrell A.S., et al. Precision mouse models with expanded tropism for human pathogens. Nat. Biotechnol. 2019;37:1163–1173. doi: 10.1038/s41587-019-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020:94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykes M., Pombo A., Jenkins C., MacPherson G.G. Dendritic cells interact directly with naive B lymphocytes to transfer antigen and initiate class switching in a primary T-dependent response. J. Immunol. 1998;161:1313–1319. [PubMed] [Google Scholar]
- Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.H., Li T.Y., He Z.C., Ping Y.F., Liu H.W., Yu S.C., et al. [A pathological report of three COVID-19 cases by minimal invasive autopsies] Zhonghua Bing Li Xue Za Zhi. 2020;49:411–417. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T., Hill T., Li K., Peters C.J., Tseng C.T. Severe acute respiratory syndrome (SARS) coronavirus-induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte-derived macrophages and dendritic cells. J. Virol. 2009;83:3039–3048. doi: 10.1128/JVI.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang X., Ni L., Di X., Ma B., Niu S., et al. COVID-19: melatonin as a potential adjuvant treatment. Life Sci. 2020;250 doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020:370. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Y., Wang X.M., Xing X., Xu Z., Zhang C., Song J.W., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017:2. doi: 10.1126/sciimmunol.aan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890. doi: 10.1016/j.chom.2020.04.017. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
I agree to sharing all the data and content of this review.