Abstract
Recently, the incidence of hypertension has significantly increased among young adults. While aerobic exercise intervention (AEI) has long been recognized as an effective treatment, individual differences in response to AEI can seriously influence clinicians' decisions. In particular, only a few studies have been conducted to predict the efficacy of AEI on lowering blood pressure (BP) in young hypertensive patients. As such, this paper aims to explore the implications of various cardiopulmonary metabolic indicators in the field by mining patients' cardiopulmonary exercise testing (CPET) data before making treatment plans. CPET data are collected “breath by breath” by using an oxygenation analyzer attached to a mask and then divided into four phases: resting, warm-up, exercise, and recovery. To mitigate the effects of redundant information and noise in the CPET data, a sparse representation classifier based on analytic dictionary learning was designed to accurately predict the individual responsiveness to AEI. Importantly, the experimental results showed that the model presented herein performed better than the baseline method based on BP change and traditional machine learning models. Furthermore, the data from the exercise phase were found to produce the best predictions compared with the data from other phases. This study paves the way towards the customization of personalized aerobic exercise programs for young hypertensive patients.
1. Introduction
As a prevalent chronic disease, hypertension has been widely considered as a major risk factor for cardio-cerebrovascular events [1]. Strikingly, hypertension incidence is increasing most dramatically in young adults [2, 3]. As an alternative to antihypertensive drugs, lifestyle adjustments, including body weight control, diet, and exercise, can also be used to lower blood pressure (BP) [4, 5]. In particular, aerobic exercise not only directly reduces BP but also indirectly achieves similar effects by controlling body weight, reducing stress, and improving vascular endothelial function, along with other mechanisms [6–8]. Therefore, aerobic exercise intervention (AEI) has been widely recommended for the treatment of hypertension [9, 10]. Unfortunately, specific guidelines for effectively administering aerobic exercise aimed at antihypertension have not been widely accepted as there is significant individual variation in BP lowering achieved by the same exercise program, with the same exercise type, time, frequency, and duration [11–13]. Understanding the individual responsiveness to AEI before formulating comprehensive hypertension management plans will help to improve both effectiveness and efficiency of BP management. To our knowledge, research in this field is still very limited, thus motivating us to perform the work conducted in this paper.
For the clinical feasibility and practicality, this work provided an investigation on the feasibility of utilizing machine learning techniques to predict the efficacy of AEI on young hypertensive patients. Taking into account the prognostic ability of key cardiopulmonary variables, data mining was performed based on the data generated by cardiopulmonary exercise testing (CPET) before treatment. CPET provides a comprehensive physiological assessment of multiorgan system function, including not only cardiovascular and pulmonary but also musculoskeletal and hematopoietic systems [14]. It can help clinicians identify the severity of the disease and evaluate the response to treatments, thus playing an important role in formulating aerobic exercise training prescription and cardiac rehabilitation [15, 16]. In this paper, CPET being used is an electric bicycle with many sensors (see Figure 1) as the main ergometer to measure the changes of various cardiopulmonary metabolic indicators over time. To provide the best measure of the response to exercise, these data were collected “breath by breath” by an oxygenation analyzer attached to a mask. The specific test scheme guided by clinicians included four phases: (1) resting for 1 minute to relieve the patient's tension; (2) load-free cycling (no resistance on the pedals) for 3 minutes to warm up; (3) exercise for 5–12 minutes with increasing resistance on the pedals (20–35 watt/min increment) until maximal exertion; and (4) recovery for 6 minutes with the first 3 minutes of load-free cycling and the second 3 minutes of sitting still.
Figure 1.

Illustration of cardiopulmonary exercise testing. To protect privacy, the patient's eyes were partially blurred.
Based on the professional advice of clinicians, this paper first utilized a simple method as the baseline to predict the BP-lowering effect of AEI for young hypertensive patients. Just to be clear, BP in this paper was equal to the sum of systolic blood pressure (SBP) and diastolic blood pressure (DBP). This method compared BP at the 6th minute of recovery (R6BP) with BP at the pre-exercise resting (PEBP) in a single CPET before AEI. Patients with R6BP ≤ PEBP were predicted to be strong responders to AEI. If the converse was true, they were predicted to be weak responders. Subsequent experiments showed that the accuracy of this method was typically 50%–60%, closely approximating a random guess, and far beneath the requirement for making effective and accurate clinical exercise prescriptions. To meet this challenge, machine learning techniques were utilized to fully capitalize on the information present within several cardiopulmonary metabolic indicators provided by CPET. As such, this work provides useful insights into the formulation of personalized AEI prescriptions for young hypertensive patients. The main contributions of this paper are as follows:
A sparse representation classifier based on analytic dictionary learning was designed to accurately predict the efficacy of AEI on BP lowering. This model can not only alleviate the interference of redundant information and noise brought by breath-by-breath collection but also overcome the deficiency of the existing sparse representation-based classifier which needs a large number of training samples.
The significance of various cardiopulmonary metabolic indicators at different phases of CPET for this task was discussed through comparative experiments. The results showed that the data from the exercise phase can produce the best predictions compared with the data from other phases. Among various metabolic indicators, oxygen pulse (i.e., oxygen intake per heartbeat) was recommended as a powerful indicator for predicting the individual responsiveness to AEI.
The remainder of the paper is structured as follows. Section 2 introduces various metabolic indicators of CPET used in this paper. Section 3 briefly introduces the related works, including the development of application scenarios and research methods. Section 4 describes the designed model in detail based on the shortcomings of the existing model. Section 5 reports the experimental results along with analyses. Finally, conclusions and future works are summarized in Section 6.
2. Main Metabolic Indicators of CPET
CPET provides time-varying information regarding multiple indicators related to circulation, respiration, and gas metabolism at different levels of exercise intensity [17]. The nine indicators recommended by professional clinicians for this work are briefly described in the following:
Heart rate (HR): the number of heartbeats per minute. Normally, HR is 60–100 beats per minute at rest. HR varies individually according to age, sex, and other physiological factors.
Stroke volume (SV): the volume of blood ejected from either ventricle of the heart in a single beat. The main affecting factors of SV are myocardial contractility, venous return blood volume (preload), arterial BP (afterload), and so forth.
Cardiac output (CO): the volume of blood that flows out of the heart in a given period, usually denoted as liters per minute. It can be obtained by multiplying the average SV per beat by HR, varying with metabolism and activity. For example, it increases with muscle movement, emotional agitation, pregnancy, and so forth.
Oxygen pulse (VO2/HR): the volume of the oxygen intake per heartbeat. Hence, it is the amount of oxygen that the tissues of the body extract from oxygen carried by each SV. A higher oxygen pulse suggests better cardiopulmonary function. This can be used as a comprehensive index to determine the cardiopulmonary function.
Oxygen consumption/kilogram (VO2/kg): the volume of oxygen consumed by the metabolic processes of the body over a period of time, usually denoted as milliliters of oxygen per kilogram of body weight per minute. It reflects the body's ability to use oxygen and is usually determined by the maximum cardiac output, arterial oxygen content, cardiac output to the distribution index of the exercise muscle, and muscle oxygen capacity.
Tidal volume (VT): the volume of air inhaled or exhaled during a normal breath. It is related to age, sex, volume and surface, breathing habits, body metabolism, and so forth.
Ventilation volume/minute (VE): the volume of air inhaled or exhaled from the lungs in a minute, which can be obtained by multiplying VT by the respiratory rate.
Respiratory exchange ratio (R): the ratio of the carbon dioxide (CO2) output to the oxygen (O2) uptake (i.e., VCO2/VO2) during the same period. It reflects not only the exchange of tissue metabolism of gas but also the influence of transient change in gas storage.
Carbon dioxide ventilation equivalent (VE/VCO2): the ability of the body to discharge carbon dioxide, calculated as the ratio between the required ventilation volume and carbon dioxide output.
To illustrate the characteristics of these indicators more vividly, Figure 2 shows a visualization of the above nine indicators for a patient during the exercise phase of a CPET before AEI. Since each breath represents a sampling point, the information of each metabolic indicator collected by the breath-by-breath technique can be stored as a time series [18].
Figure 2.
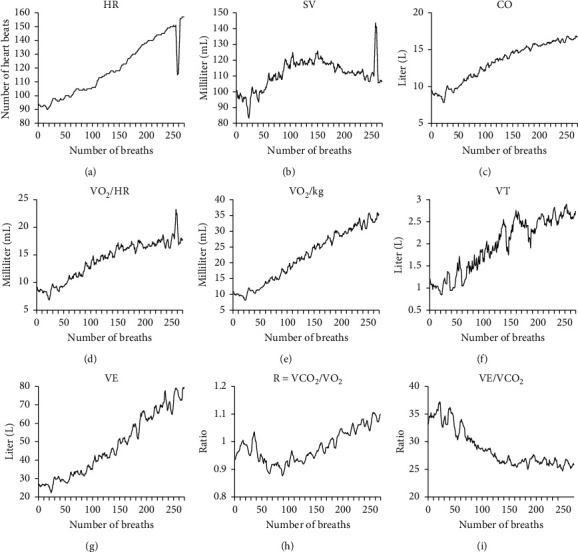
Visualization of several indicators for a patient during the exercise phase of a CPET. (a) Heart rate. (b) Stroke volume. (c) Cardiac output. (d) Oxygen pulse. (e) Oxygen consumption/kilogram. (f) Tidal volume. (g) Ventilation volume/minute. (h) Respiratory exchange ratio. (i) Carbon dioxide ventilation equivalent.
3. Related Works
CPET is a dynamic, noninvasive diagnostic method to evaluate cardiopulmonary function during increasing load exercise. Recently, the application of CPET in clinical decision-making for various diseases has been significantly developed. For example, CPET is playing a growing role in cardiology, including heart failure, valve diseases, and ischemic heart disease [19]. Buys et al. evaluated the predictive value of CPET for the incidence of hypertension in patients undergoing aortic coarctation surgery and determined the high-risk boundary as VE/VCO2 slope ≥27 and peak SBP ≥220 mmHg through Cox regression analysis [20]. Keller et al. suggested that BP overresponse in CPET might be a diagnostic tool for identifying high-risk groups of hypertension [21]. Besides, CPET can be used as a tool for preoperative risk stratification of patients (not limited to cardiopulmonary surgery) to predict postoperative adverse outcomes [22, 23]. Currently, one of the most impressive advances is that the integration of CPET and other tests has been introduced to diagnose several diseases [24]. Exercise stress echocardiography and CPET have been successfully combined in the dynamic assessment of heart failure for hypertensive patients [25]. Similarly, CPET combined with echocardiography of the right ventricle was applied to predict the prognosis of patients with pulmonary arterial hypertension [26].
From the perspective of research methods, in addition to traditional statistical analysis, data mining of CPET using machine learning techniques is gradually becoming a research hotspot. Leopold et al. developed a greedy heuristic algorithm based on feature clustering to study the ability of CPET to predict the anaerobic mechanical power outputs [27]. Braccioni et al. used a random forest algorithm to analyze the relationship between symptoms and cardiopulmonary parameters of lung transplant recipients based on incremental CPET [28]. Sakr et al. evaluated the performance of six machine learning techniques in predicting the individuals at risk of hypertension through treadmill stress tests on a massive crowd [29]. Unfortunately, the above work only selected some special values of cardiopulmonary metabolic indicators (such as peaks or slope) as features for analysis, without taking into account their time-varying characteristics. Our previous work has proved that time-varying data of some metabolic indicators obtained through CPET could be used to predict the efficacy of AEI [30], but how to further improve the predictive accuracy is still a challenge, especially in the case of insufficient training samples. This encourages us to perform the research conducted in this paper.
In fact, the prediction of the BP-lowering effect of AEI by using a certain metabolic indicator can be transformed into time series classification (TSC) for data mining. To date, researchers have proposed hundreds of approaches for TSC in different application scenarios. TSC algorithms can be roughly divided into seven categories: (1) the whole-series-based method, (2) the interval-based method, (3) the shaped method, (4) the word-frequency-based method, (5) the model-based method, (6) the integration-based method, and (7) the deep learning-based method. Bagnall et al. evaluated the latest progress of TSC algorithms on 85 datasets in the University of California, Riverside (UCR) archive [31]. They recommended 1-nearest neighbor with dynamic time warping (1NN-DTW) and random forest (RF) as the baseline classifiers for comparison with other classifiers. Besides, they also concluded that the integration-based method can achieve high accuracy by utilizing multiple classifiers on one or more feature spaces. For example, Bagnall et al. integrated 35 classifiers on the time, frequency, change, and shapelet transformation domains [32]. On this basis, Lines et al. added two new classifiers, two additional transformation domains, and a hierarchical structure of probability voting to further improve the performance [33]. Recently, the method based on deep learning has gradually become a research hotspot [34]. Deep learning is characterized by learning hidden and more abstract representations of data from the original time series to achieve better classification performance. This method is widely used for end-to-end learning including methods such as convolutional neural networks (CNNs) [35] and echo state network (ESN) [36]. The common disadvantage of these methods is that they require a large amount of data and computational cost for model training. As this work represents the first stage in a larger experiment, the relatively small number of samples means that the above approach is not appropriate. Moreover, the robustness of the method to signal-to-noise ratio also needs to be considered because the process involved in collecting CPET data is usually very noisy. For the above reasons, a classifier based on sparse representation is recommended for the task in this paper.
4. Sparse Representation-Based Classifier
In this section, a sparse representation classifier based on dictionary learning was designed to accurately predict the efficacy of AEI on BP lowering. This method firstly eliminated redundant information and reduced noise by feature extraction based on the sparse representation. At the same time, it took advantage of learning of an analytic dictionary without requiring as many training samples as the existing sparse representation-based classifier.
4.1. Brief Introduction for Sparse Representation
Recently, sparse representation has received increasing attention in many fields. While initially developed for use in image analysis and signal processing, sparse representation has been successfully utilized for dealing with more general tasks in the machine learning field [37]. Specifically, given a signal x ∈ Rm of m observations and an overcomplete dictionary D ∈ Rm×n (n ≫ m) in which the column vector di (1 ≤ i ≤ n) is known as an atom, the main goal of the sparse representation is the reconstruction of a signal perfectly with the least possible number of atoms. Its objective function is as follows:
| (1) |
where α ∈ Rn is the sparse representation (or sparse solution) of x and ‖·‖0 refers to the number of nonzero elements in α. Due to the noise in the real signal, the solution of equation (1) can be approximated by either of the following two equations:
| (2) |
| (3) |
where δ can be considered as noise or a reconstruction residual; the sparse factor k is a predefined integer not less than 1. Besides, based on the Lagrange multiplier theorem, solving sparse representation can be equivalently transformed into an unconstrained minimization problem:
| (4) |
where λ is a positive constant used to achieve a tradeoff between the reconstruction residual and the sparse solution.
It should be noted that since obtaining the optimal solution with l0-norm minimization is an NP-hard problem, many algorithms have been proposed to deal with it. The strategies commonly used in these algorithms mainly include greedy pursuit strategy and convex relaxation strategy [38, 39]. The greedy pursuit strategy represented by the orthogonal matching pursuit (OMP) algorithm is to gradually approach the optimal solution through the sequential selection of column vectors (atoms) until the end of iteration [40]. For the convex relaxation strategy, the main idea is to replace the l0-norm minimization term with the l1-norm minimization term. Taking equation (3) as an example, it can be approximately equivalent to the lasso problem:
| (5) |
where ‖·‖1 represents the sum of the absolute values of nonzero elements in α and ε is a positive constant given beforehand. The advantage of this strategy is that the l1-norm minimization problem has an analytical solution and can be effectively solved by several methods, such as least angle regression (LAR) [41], coordinate descent algorithm (CDA) [42], iterative shrinkage-thresholding algorithm (ISTA) [43], and many variations of them.
4.2. The Existing Sparse Representation-Based Classifier
Proposed by Wright et al., a sparse representation-based classifier (SRC) was first applied in the field of face recognition and then successfully extended to TSC [44, 45]. Specifically, the sparse representation of an unlabeled sample is first solved based on the dictionary composed of all labeled samples. Then, the reconstruction residuals of each class are calculated by using the samples of each class and the corresponding elements in the sparse representation. Finally, the classification is performed by examining which class leads to the minimum residual of the unlabeled sample. The steps to implement SRC are as follows:
-
(1)
The l2-norm normalization is preprocessed for each sample of the whole dataset with a class number of c.
-
(2)
A dictionary D = [D1, ⋯, Dj, ⋯, Dc] is generated, where Dj (1 ≤ j ≤ c) is a subdictionary composed of jth-class normalized samples in the training set as column vectors (atoms).
-
(3)
The sparse representation α of the unlabeled sample y is obtained by using the algorithm described above.
-
(4)The unlabeled sample y is reconstructed, respectively, using each Dj and corresponding αj, where αj (1 ≤ j ≤ c) is a subvector consisting of the elements in α corresponding to all atoms in Dj. The label is determined based on the minimum residual, as shown in the following equation:
(6)
Figure 3 shows the SRC schematic for a two-class problem. The success of the SRC depends on the hypothesis that the unlabeled sample can be best reconstructed by a linear representation of samples within the same class. However, once the samples of different classes look similar to each other, the performance of SRC is very unstable [46]. Besides, the dictionary cannot satisfy the overcompleteness if the number of labeled samples is less than the dimension of samples, which will also affect the performance of the SRC [47]. To overcome the shortcoming of the SRC, a sparse representation classifier based on an analytic dictionary was designed, and then its accuracy was improved by using dictionary learning. For the sake of simplicity, the model was called SRC-AL for short. The principle is described in the following.
Figure 3.
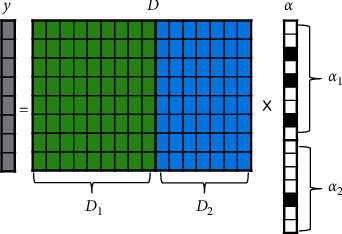
The SRC schematic for a two-class problem, where y is the unlabeled sample. D1 and D2 represent subdictionaries composed of all normalized samples belonging to class 1 and class 2, respectively. The sparse representation α can be divided into two subvectors (α1 and α2) according to the number of columns in D1 and D2. The black-filled blocks in α represent nonzero elements.
4.3. The Designed Sparse Representation-Based Classifier
In the application domain of sparse representation, an overcomplete dictionary can be usually generated using data implementation or analytic approach [48]. The approach based on data implementation is to construct an explicit dictionary directly by using the raw data. This is exactly the way adopted by the SRC, intending to obtain the residuals of the unlabeled sample reconstructed by the samples of different classes. Unlike SRC, SRC-AL generates an implicit dictionary based on the analytic approach as the initial dictionary. This approach generally utilizes some fixed transformations, such as discrete Fourier transform (DFT), discrete cosine transform (DCT), and discrete wavelet transform (DWT) [49]. Compared to the data implementation, the analytic approach has the advantage of allowing an overcomplete dictionary of any size without being limited by the number of labeled samples. However, due to the poor adaptability, the analytic dictionary often requires further optimization through dictionary learning. K-singular value decomposition (K-SVD) is a popular algorithm for dictionary learning, which updates the used atoms one by one in an iterative manner to train the overcomplete dictionary most suitable for the training set [50].
Inspired by the sparse representation predictor for time series proposed by our previous work [51], the workflow of SRC-AL consists of the following six steps:
-
(1)
Generate an initial dictionary D ∈ R(m+c)×n by utilizing the analytic approach, where m is the dimension of the sample, c is the number of classes, and n is an arbitrary integer much larger than (m + c). The upper and lower parts of the dictionary are represented by Dup∈Rm×n and Dlw∈Rc×n, respectively.
-
(2)
Normalize each sample of the training dataset with l2-norm, and convert its label into one-hot encoding. Combine the above two parts into the new training sample x∈R(m+c).
-
(3)According to the training set composed of new samples, update the initial dictionary through dictionary learning, with the purpose of better reconstructing the samples. The objective function of dictionary learning can be described as
(7) where r is the number of samples in the training set and αi is a sparse representation of sample xi.
-
(4)
Normalize the unlabeled sample y∈Rm with l2-norm, and then obtain its sparse representation αy ∈ Rn based on the upper part of the learned dictionary (Dup' ∈ Rm×n).
-
(5)
Multiply the lower part of the learned dictionary (Dlw' ∈ Rc×n) by the sparse representation αy ∈ Rn to obtain the label vector Ly∈Rc.
-
(6)Determine the label of y according to the index of the element with the largest absolute value in Ly, as shown in the following equation:
(8) where Ly(i) represents the ith element in vector Ly.
Figure 4 shows the SRC-AL schematic for a two-class problem. Assuming that sample x1 belongs to class 1, the green-filled blocks represent the normalized sample, and the following “10” represents the one-hot encoding of the label. Similarly, the blue-filled blocks represent the normalized sample of class 2, and the following “01” represents the one-hot encoding of its label. The dictionary filled with orange is generated by the analytic approach. To better reconstruct all training samples, a dictionary-learning algorithm (such as K-SVD) should be applied to constantly update the dictionary. Based on the upper part of the learned dictionary (Dup'), the sparse representation αy of the unlabeled sample y (grey-filled blocks) is solved, and then Dlw' × αy is used to obtain the label vector Ly. Finally, the element with the largest absolute value in Ly is set to 1, and the other elements are set to 0. This one-hot encoding is used to replace the question mark in Figure 4 to achieve the classification of y.
Figure 4.
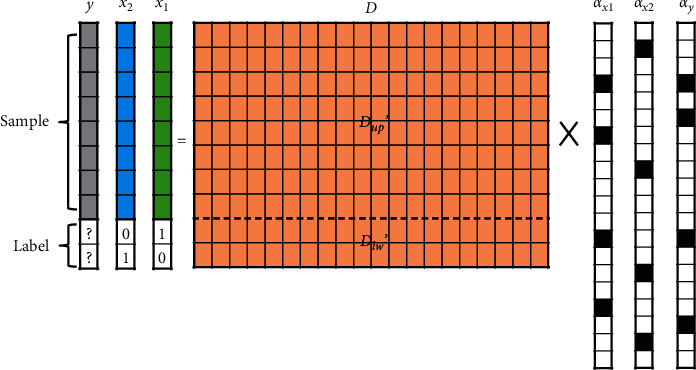
The SRC-AL schematic for a two-class problem, where x1 and x2 represent the samples of class 1 and class 2, respectively, and y is the unlabeled sample. Due to the limited space, only two labeled samples are drawn in the figure. In fact, all labeled samples are used for dictionary learning. Dup' and Dlw' represent the upper and lower parts of the learned dictionary. αx1, αx2, and αy are the sparse representations of x1, x2, and y where the black-filled blocks represent nonzero elements.
5. Experiments and Results
CPET data from 24 young patients with stage I hypertension before AEI treatment were used for the experiments. The dataset was provided by the Department of Cardiology, First Affiliated Hospital of Sun Yat-sen University, China. The whole exercise process of all the people was completed under the supervision of professional medical staff in the hospital. Blood pressure before and after exercise was assessed using both dynamic and exercise blood pressure results. Although the cost of each sample is very large, the data are highly comparable and reliable due to the guaranteed amount of exercise and more comprehensive monitoring indicators.
The performance of various machine learning models based on the data from the exercise phase was compared with the baseline method given by the clinician. Note that the baseline method only focused on BP change between pre-exercise and postexercise within a single CPET, while the machine learning model took into account the time series of metabolic indicators during CPET. After verifying the effectiveness of the designed model, the significance of the data from different phases in CPET for predicting the efficacy of AEI on BP lowering was further evaluated.
5.1. Description of the Dataset
-
(1)
Inclusion criteria: between the ages of 18 and 45; stage I hypertension (SBP: 140–160 mmHg; DBP: 90–100 mmHg) either without medication or with discontinuation of antihypertensive drugs for more than two weeks and still presenting stage I hypertension; no regular exercise for four months prior to admission; willingness to participate in follow-ups for more than 6 months.
-
(2)
Treatment prescription: patients underwent aerobic exercise with an Italian COSMED K4 electric bicycle. Training intensity corresponded to the metabolic equivalent of task (MET) of 70% of maximal oxygen consumption (VO2max). Get aerobic exercise 5 times per week, each time 45 minutes (exercise intensity equivalent to 2,000–3,000 kcal per week), lasting 12 weeks.
-
(3)Classification standard: patients were categorized as strong or weak responders of AEI treatment according to the therapeutic effect. The classification process is as follows:
- All patients received 24-hour dynamic BP monitoring before and after AEI to obtain their daily mean BP.
- The rate of BP change before and after treatment was calculated for each patient: ri=‖MBPB − MBPA‖/MBPB, where MBPB and MBPA indicated the mean BP of 24 hours before and after treatment, respectively.
- Z-score standardization was performed for ri as follows: zi=(ri − μ)/σ, where μ and σ were the mean and standard deviation, respectively. The role of zi was to determine whether the antihypertensive efficacy of the ith patient was above average.
- Classify according to zi. Patients with zi >0 (14 individuals in total) were identified as the strong antihypertensive responders of AEI, while patients with zi <0 (10 individuals in total) were classified as weak responders. The real labels of 24 patients are detailed in Table 1.
Table 1.
Real labels of 24 young patients with stage I hypertension.
| Sample | MBPB (mmHg) | MBPA (mmHg) | r i (%) | z i | Label |
|---|---|---|---|---|---|
| 1 | 242 | 202 | 16.529 | 2.209 | Strong |
| 2 | 229 | 195 | 14.847 | 1.792 | Strong |
| 3 | 221 | 192 | 13.122 | 1.365 | Strong |
| 4 | 241 | 212 | 12.033 | 1.094 | Strong |
| 5 | 235 | 213 | 9.362 | 0.432 | Strong |
| 6 | 253 | 230 | 9.091 | 0.365 | Strong |
| 7 | 223 | 203 | 8.969 | 0.334 | Strong |
| 8 | 249 | 227 | 8.835 | 0.301 | Strong |
| 9 | 209 | 191 | 8.612 | 0.246 | Strong |
| 10 | 244 | 223 | 8.607 | 0.245 | Strong |
| 11 | 214 | 196 | 8.411 | 0.196 | Strong |
| 12 | 246 | 226 | 8.130 | 0.127 | Strong |
| 13 | 204 | 188 | 7.843 | 0.055 | Strong |
| 14 | 244 | 225 | 7.787 | 0.041 | Strong |
| 15 | 244 | 226 | 7.377 | −0.060 | Weak |
| 16 | 231 | 214 | 7.359 | −0.065 | Weak |
| 17 | 240 | 223 | 7.083 | −0.133 | Weak |
| 18 | 231 | 215 | 6.926 | −0.172 | Weak |
| 19 | 214 | 207 | 3.271 | −1.079 | Weak |
| 20 | 221 | 214 | 3.167 | −1.104 | Weak |
| 21 | 222 | 216 | 2.703 | −1.220 | Weak |
| 22 | 211 | 208 | 1.422 | −1.537 | Weak |
| 23 | 207 | 205 | 0.966 | −1.650 | Weak |
| 24 | 207 | 208 | 0.483 | −1.770 | Weak |
As can be seen from Table 1, all patients except the last one exhibited certain antihypertensive effects following 12 weeks of AEI treatment. The average antihypertensive change rate was 7.582%. The individual showing the best antihypertensive effect exhibited a 40 mmHg (or 16.529%) BP decrease after AEI. However, the absence of obvious changes in BP of some individuals also proved that the efficacy of AEI is significantly different in hypertensive patients.
5.2. Experimental Results
In this paper, accuracy and F1-score (the harmonic average of precision and recall) obtained by the confusion matrix (see Figure 5) were used to evaluate the performance of the model. For them, higher values indicate positive benefits.
Figure 5.
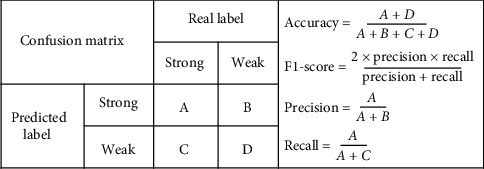
Illustration of the confusion matrix.
5.2.1. The Performance of the Baseline Method Based on BP Change
An intuitive way to predict the BP-lowering effect of AEI is to determine whether the BP of patients after exercise is lower than that before exercising in CPET. Specifically, the pre-exercise resting BP (PEBP) was subtracted from BP at the 6th minute of the recovery phase (R6BP) to obtain BP change (△BP) for each patient. A patient with △BP less than 0 was considered to be unable to benefit from AEI, meaning the predicted label was weak. Conversely, a patient would exhibit a strong, beneficial antihypertensive response to AEI. The predicted labels of the baseline method based on BP change are shown in Table 2. Using the confusion matrix, the accuracy of the baseline method was 0.542, and F1-score was 0.56. This meant that the baseline method was only slightly superior to the random guess (accuracy = 0.5), far less than the requirement for clinical applications.
Table 2.
Predicted labels of the baseline method based on BP change, where △BP = PEBP − R6BP.
| Sample | PEBP (mmHg) | R6BP (mmHg) | △BP (mmHg) | Predicted label | Real label |
|---|---|---|---|---|---|
| 1 | 242 | 267 | −25 | Weak | Strong |
| 2 | 233 | 260 | −27 | Weak | Strong |
| 3 | 199 | 170 | 29 | Strong | Strong |
| 4 | 236 | 216 | 20 | Strong | Strong |
| 5 | 190 | 223 | −33 | Weak | Strong |
| 6 | 238 | 246 | −8 | Weak | Strong |
| 7 | 209 | 205 | 4 | Strong | Strong |
| 8 | 274 | 256 | 18 | Strong | Strong |
| 9 | 201 | 203 | −2 | Weak | Strong |
| 10 | 224 | 214 | 10 | Strong | Strong |
| 11 | 204 | 211 | −7 | Weak | Strong |
| 12 | 262 | 255 | 7 | Strong | Strong |
| 13 | 216 | 218 | −2 | Weak | Strong |
| 14 | 219 | 194 | 25 | Strong | Strong |
| 15 | 222 | 232 | −10 | Weak | Weak |
| 16 | 245 | 229 | 16 | Strong | Weak |
| 17 | 247 | 250 | −3 | Weak | Weak |
| 18 | 176 | 173 | 3 | Strong | Weak |
| 19 | 233 | 225 | 8 | Strong | Weak |
| 20 | 219 | 227 | −8 | Weak | Weak |
| 21 | 205 | 206 | −1 | Weak | Weak |
| 22 | 223 | 235 | −12 | Weak | Weak |
| 23 | 216 | 242 | −26 | Weak | Weak |
| 24 | 224 | 185 | 39 | Strong | Weak |
5.2.2. The Performance of Machine Learning Models Based on the Metabolic Indicators
Time series of the nine metabolic indicators described above during the exercise phase were selected for analysis using machine learning models. Of note, patients had distinct exercise durations based on different physical conditions, resulting in different numbers of sampling points for individuals (ranging from 85 to 270). As most machine learning models required samples to have the same dimension, linear interpolation was first applied to unify the sampling numbers of all patients to 270 points. Afterward, SRC-AL presented herein was compared with SRC and some popular models of TSC, including 1NN-DTW, random forest (RF), and support vector machine (SVM). Due to the limited samples, the leave-one-out cross-validation was adopted to carry out the experiments [52]. All the above models were implemented by MATLAB. For SRC and SRC-AL, OMP and K-SVD algorithms in the SPAMS toolbox were used to solve the sparse representation and dictionary learning, respectively. Besides, the optimal sparse factor was obtained by grid search in a specific interval. Finally, for SRC-AL, the size of the initial dictionary was defined as a matrix where the number of columns was twice the number of rows, which was realized by the discrete cosine transform. The experimental results of each model are shown in Tables 3 and 4, where the last column of each table shows the average performance of each metabolic indicator based on different machine learning models.
Table 3.
Accuracy of various machine learning models based on the data from the exercise phase of CPET.
| Indicator | SRC-AL | SRC | 1NN-DTW | RF | SVM | Mean |
|---|---|---|---|---|---|---|
| VO2/HR | 1.000 | 0.792 | 0.625 | 0.583 | 0.667 | 0.733 |
| VE | 0.875 | 0.708 | 0.625 | 0.458 | 0.542 | 0.642 |
| VO2/kg | 0.917 | 0.667 | 0.500 | 0.500 | 0.583 | 0.633 |
| CO | 0.917 | 0.667 | 0.500 | 0.583 | 0.792 | 0.692 |
| HR | 0.833 | 0.625 | 0.500 | 0.500 | 0.417 | 0.575 |
| SV | 0.667 | 0.542 | 0.458 | 0.542 | 0.542 | 0.550 |
| VE/VCO2 | 0.708 | 0.542 | 0.500 | 0.458 | 0.458 | 0.533 |
| VT | 1.000 | 0.458 | 0.583 | 0.500 | 0.333 | 0.575 |
| R | 0.958 | 0.417 | 0.375 | 0.500 | 0.583 | 0.567 |
Table 4.
F1-score of various machine learning models based on the data from the exercise phase of CPET.
| Indicator | SRC-AL | SRC | 1NN-DTW | RF | SVM | Mean |
|---|---|---|---|---|---|---|
| VO2/HR | 1.000 | 0.815 | 0.667 | 0.667 | 0.714 | 0.773 |
| VE | 0.903 | 0.741 | 0.640 | 0.581 | 0.593 | 0.692 |
| VO2/kg | 0.933 | 0.692 | 0.539 | 0.600 | 0.643 | 0.681 |
| CO | 0.923 | 0.692 | 0.571 | 0.667 | 0.828 | 0.736 |
| HR | 0.846 | 0.640 | 0.539 | 0.600 | 0.462 | 0.617 |
| SV | 0.667 | 0.667 | 0.552 | 0.645 | 0.621 | 0.630 |
| VE/VCO2 | 0.778 | 0.667 | 0.600 | 0.606 | 0.581 | 0.646 |
| VT | 1.000 | 0.381 | 0.615 | 0.571 | 0.429 | 0.599 |
| R | 0.966 | 0.462 | 0.516 | 0.571 | 0.643 | 0.632 |
5.2.3. The Performance of SRC-AL Based on the Data from Different Phases of CPET
Since SRC-AL performed best in the above model, it was directly used to evaluate the significance of the data generated in the three important phases of CPET for predicting the individual responsiveness to AEI. These three phases included warm-up, exercise, and recovery. Similar to the exercise phase, the data dimensions of different patients in the other two phases were also inconsistent. For the warm-up phase, the shortest time series of metabolic indicators had only 38 sample values, while the longest had 81 sample values. For the recovery phase, the shortest one had only 113 sample values, while the longest one had 195 sample values. Therefore, linear interpolation should be used first to unify the data dimensions of different patients into the same. Besides, the dictionary learned in the exercise phase cannot be applied to the other two phases due to different data dimensions. The experimental results of SRC-AL based on the data of the above three phases of CPET are shown in Table 5.
Table 5.
The performance of SRC-AL based on the data from the three phases of CPET.
| Indicator | Warm-up accuracy | Exercise accuracy | Recovery accuracy | Warm-up F1-score | Exercise F1-score | Recovery F1-score |
|---|---|---|---|---|---|---|
| VO2/HR | 0.583 | 1.000 | 0.625 | 0.737 | 1.000 | 0.743 |
| VE | 0.625 | 0.875 | 0.833 | 0.757 | 0.903 | 0.875 |
| VO2/kg | 0.750 | 0.917 | 0.750 | 0.824 | 0.933 | 0.824 |
| CO | 0.917 | 0.917 | 0.708 | 0.923 | 0.923 | 0.720 |
| HR | 0.833 | 0.833 | 0.792 | 0.857 | 0.846 | 0.828 |
| SV | 0.958 | 0.667 | 0.625 | 0.963 | 0.667 | 0.757 |
| VE/VCO2 | 0.750 | 0.708 | 0.708 | 0.800 | 0.778 | 0.788 |
| VT | 0.583 | 1.000 | 0.792 | 0.737 | 1.000 | 0.828 |
| R | 0.917 | 0.958 | 0.708 | 0.923 | 0.966 | 0.759 |
5.3. Analyses of Experimental Results
This work investigated the ability of metabolic indicators to discriminate between strong and weak responses to AEI in patients. Through the analysis of the above experimental results, the following insights can be obtained to help clinicians predict the efficacy of AEI on young hypertensive patients based on CPET.
From Tables 3 and 4, SRC-AL and SRC were superior to other traditional classifiers in predicting the individual responsiveness to AEI based on the time series of metabolic indicators. This is mainly because the process of collecting these metabolic indicator data is prone to generate many interference signals, while the sparse representation can effectively extract the main features of time series and enhance the robustness to noise to the maximum extent.
The performance of SRC-AL was significantly better than that of SRC regardless of the time series based on any indicator, although both were based on sparse representation. This indicates that SRC needs an adequate set of training samples to form an overcomplete dictionary for better performance. On the contrary, SRC-AL can always guarantee the overcompleteness because it generates dictionaries employing the analytic approach. Through dictionary learning, the initial dictionary can be gradually updated to better fit the training samples and their labels.
According to the last column of Table 3, except for the indicator VE/VCO2, the average accuracy of all the other metabolic indicators based on the five machine learning models was higher than that of the baseline method based on BP change (accuracy = 0.542). However, if evaluated by the average F1-score, all metabolic indicators were superior to BP change alone (F1-score = 0.56), as shown in the last column of Table 4. This interesting finding suggests that the multipoint characteristics of cardiopulmonary metabolic indicators formed by collecting breath data can more accurately reflect the individual responsiveness to AEI. Figure 6 visualizes the comparison between the predictive performance of each indicator obtained by machine learning models and that of BP change obtained by the baseline method, where Figure 6(a) shows the average/optimal accuracy and Figure 6(b) shows the average/optimal F1-score. Note that the optimal performance of all metabolic indicators was obtained by SRC-AL designed herein.
Table 5 illustrates the significance of data from different phases in CPET for predicting the BP-lowering effect of AEI. VO2/HR, VE, VO2/kg, VT, and R had the best predictive effect by using the time series of the exercise phase, while HR, SV, and VE/VCO2 performed better according to the time series of the warm-up phase. The performance of CO was consistent in both the exercise and the warm-up phases. Finally, the data in the recovery phase were less important than in the previous two phases. The reason may be that the patient is only active for the first three minutes during the recovery phase and remains inactive for the next three minutes. In other words, the metabolic data of patients in the active state are more significant for predicting the individual responsiveness to AEI.
Figure 6.
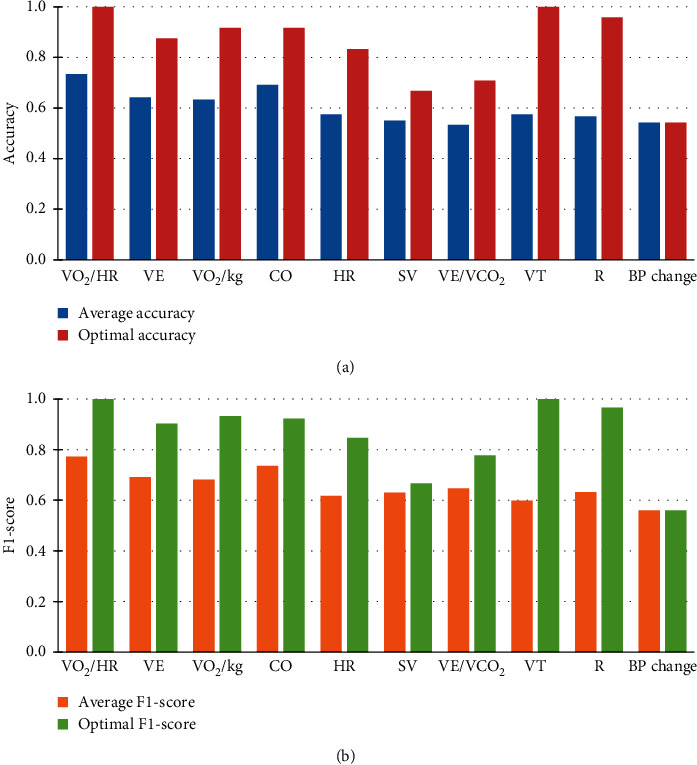
Comparison between the predictive performance of each indicator obtained by machine learning models and that of BP change obtained by the baseline method. (a, b) The average and optimal performance of metabolic indicators, respectively.
5.4. Additional Experiments
Considering that the sample size of the aforementioned experiments is rather limited, six datasets from the UCR Time Series Classification Archive were selected for additional experiments to further verify the effectiveness of our proposed model [53]. The common characteristics of these datasets include the following: (1) the number of classes is two and (2) the number of training samples is less than or close to the length of the sample. This results in the dictionary based on data implementation not being overcomplete, which may reduce the classification accuracy of SRC. Nevertheless, the proposed model based on analytic dictionary learning (SRC-AL) should not be affected. The detailed description of the datasets is shown in Table 6. According to the results demonstrated in Table 7, SRC-AL achieved the best classification in all the datasets, indicating that SRC-AL is particularly suitable for datasets with fewer training samples than the sample length.
Table 6.
Brief description of six datasets from the UCR Time Series Classification Archive.
| Type | Dataset | Classes | Length | Training set | Testing set |
|---|---|---|---|---|---|
| ECG | ECGFiveDays | 2 | 136 | 23 | 861 |
| ECG | ECG2000 | 2 | 96 | 100 | 100 |
| Sensor | SonyAIBORobotSurface1 | 2 | 70 | 20 | 601 |
| Spectro | Ham | 2 | 431 | 109 | 105 |
| Image | Herring | 2 | 512 | 64 | 64 |
| Image | BeetleFly | 2 | 512 | 20 | 20 |
Table 7.
Classification accuracy of various machine learning models on UCR datasets.
| Dataset | SRC-AL | SRC | 1NN-DTW | RF | SVM |
|---|---|---|---|---|---|
| ECGFiveDays | 0.974 | 0.971 | 0.768 | 0.787 | 0.974 |
| ECG2000 | 0.920 | 0.900 | 0.770 | 0.819 | 0.770 |
| SonyAIBORobotSurface1 | 0.890 | 0.757 | 0.725 | 0.733 | 0.677 |
| Ham | 0.762 | 0.619 | 0.467 | 0.722 | 0.619 |
| Herring | 0.672 | 0.609 | 0.531 | 0.572 | 0.625 |
| BeetleFly | 0.900 | 0.650 | 0.700 | 0.825 | 0.900 |
In addition, considering that SRC-AL is an extended sparse representation classifier, an interesting question is whether or not other machine learning models can be modified to handle the problem addressed in this paper with better performance. To answer this question, the improved versions of some machine learning models were used to be compared with SRC-AL. For example, in order to reduce the huge feature space of the random forest, time series forest (TSF) was used to divide a time series into random intervals (m is the length of the time series), and then the mean, standard deviation, and slope of each interval were all taken as features for classification [54]. Similarly, in order to improve the classification accuracy of 1NN-DTW, 1NN-sharpDTW was first adopted to convert the time series into a sequence of shape descriptors, and then the locally similar structures were paired [55]. Aiming to extract different characteristics of the domain data, three description functions were utilized to encode local shape information in this paper: raw subsequence (RAWS), discrete wavelet transform (DWT), and slope. Specifically, RAWS was applied to directly take a subsequence of the data around a sampling point of a time series as its shape descriptor. On this basis, DWT was used to decompose each subsequence into three levels, and then all the coefficients were serialized into a shape descriptor. Alternatively, the slope function was first adopted to divide each subsequence into several intervals, and then the slopes of the fitting lines of all the intervals were concatenated into a shape descriptor. According to the results shown in Table 8, SRC-AL performed best in all the improved versions. This fully demonstrates the significance of sparse representation in feature extraction and noise reduction of CPET data.
Table 8.
Accuracy of improved versions of various machine learning models based on the data from the exercise phase of CPET.
| Indicator | SRC-AL | 1NN-sharpDTW (RAWS) | 1NN-sharpDTW (DWT) | 1NN-sharpDTW (slope) | TSF |
|---|---|---|---|---|---|
| VO2/HR | 1.000 | 0.625 | 0.583 | 0.542 | 0.675 |
| VE | 0.875 | 0.583 | 0.583 | 0.667 | 0.392 |
| VO2/kg | 0.917 | 0.458 | 0.500 | 0.500 | 0.450 |
| CO | 0.917 | 0.833 | 0.833 | 0.667 | 0.558 |
| HR | 0.833 | 0.500 | 0.542 | 0.542 | 0.517 |
| SV | 0.667 | 0.583 | 0.625 | 0.542 | 0.592 |
| VE/VCO2 | 0.708 | 0.625 | 0.583 | 0.542 | 0.542 |
| VT | 1.000 | 0.542 | 0.542 | 0.625 | 0.392 |
| R | 0.958 | 0.583 | 0.583 | 0.625 | 0.517 |
6. Conclusions and Future Works
In recent years, the incidence of hypertension has shown a clear trend towards presenting in younger patients. Note that AEI has been recognized as an effective treatment among young hypertensive patients. Unfortunately, research regarding how to predict the individual responsiveness to AEI for young hypertensive patients is still lacking. As such, a sparse representation classifier based on analytic dictionary learning, a.k.a. SRC-AL, was designed to mine the time series of multiple cardiopulmonary metabolic indicators from CPET data to accurately estimate the effectiveness of AEI on patients' BP management.
In summary, the experimental results first showed that the machine learning model, especially SRC-AL, which is based on the time series of metabolic indicators, can better predict the individual responsiveness to AEI than the baseline method that is based on scalar values of BP change alone. Secondly, data from the exercise phase in CPET are the first choice for data mining, with the second choice being data from the warm-up phase. Thirdly, VO2/HR is strongly recommended as a powerful, new prognostic indicator for predicting aerobic exercise efficacy as an antihypertensive, with an average accuracy of about 75% and up to 100%. Besides, CO is also a good choice not only because its average performance is second only to VO2/HR but also due to the fact that its performance is very stable in both warm-up and exercise phases. As such, this will likely prove to be useful to clinicians for more accurately selecting comprehensive antihypertensive treatment measures without requiring extra clinical testing.
Note that the predictive model in this study is a qualitative prediction that predicts whether or not an individual hypertensive patient's response to aerobic exercise intervention is ideal. In future work, the quantitative prediction model of BP reduction caused by AEI is planned to be studied. Besides, BP defined in the current model is the sum of SBP and DBP. It may make more sense to analyze SBP and DBP separately in the subsequent work. Finally, the work presented here includes data generated from 24 young patients with stage I hypertension. Due to the limited sample size of this dataset, more samples should be collected in the future to prove the robustness of the proposed method. At the same time, further optimization can be attempted through the data augmentation technologies.
Acknowledgments
The authors wish to thank the research team of the Department of Cardiology, First Affiliated Hospital of Sun Yat-sen University, for providing the experimental data. This work was supported by the National Natural Science Foundation of China (Grant nos. 61772136 and 61672159) and the Natural Science Foundation of Fujian Province (Grant no. 2018J07005).
Data Availability
The data used to support the findings of this study cannot be made freely available in order to protect patient privacy. Requests for access to these data should be made to the corresponding author.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Carey R. M., Whelton P. K. Prevention, detection, evaluation, and management of high blood pressure in adults: synopsis of the 2017 American college of cardiology/American heart association hypertension guideline. Annals of Internal Medicine. 2018;168(5):351–358. doi: 10.7326/m17-3203. [DOI] [PubMed] [Google Scholar]
- 2.Yano Y., Reis J. P., Colangelo L. A., et al. Association of blood pressure classification in young adults using the 2017 American College of Cardiology/American Heart Association blood pressure guideline with cardiovascular events later in life. Journal of the American Medical Association. 2018;320(17):1774–1782. doi: 10.1001/jama.2018.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu S., Song Y., Chen S., et al. Blood pressure classification of 2017 associated with cardiovascular disease and mortality in young Chinese adults. Hypertension. 2020;76(1):251–258. doi: 10.1161/hypertensionaha.119.14239. [DOI] [PubMed] [Google Scholar]
- 4.Brook R. D., Appel L. J., Rubenfire M., et al. Beyond medications and diet: alternative approaches to lowering blood pressure. Hypertension. 2013;61(6):1360–1383. doi: 10.1161/hyp.0b013e318293645f. [DOI] [PubMed] [Google Scholar]
- 5.Wen H., Wang L. Reducing effect of aerobic exercise on blood pressure of essential hypertensive patients: a meta-analysis. Medicine. 2017;96(11):p. e6150. doi: 10.1097/md.0000000000006150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao L., Li X., Yan P., et al. The effectiveness of aerobic exercise for hypertensive population: a systematic review and meta‐analysis. The Journal of Clinical Hypertension. 2019;21(7):868–876. doi: 10.1111/jch.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedralli M. L., Eibel B., Waclawovsky G., et al. Effects of exercise training on endothelial function in individuals with hypertension: a systematic review with meta-analysis. Journal of the American Society of Hypertension. 2018;12(12):e65–e75. doi: 10.1016/j.jash.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Gorostegi-Anduaga I., Corres P., MartinezAguirre-Betolaza A., et al. Effects of different aerobic exercise programmes with nutritional intervention in sedentary adults with overweight/obesity and hypertension: EXERDIET-HTA study. European Journal of Preventive Cardiology. 2018;25(4):343–353. doi: 10.1177/2047487317749956. [DOI] [PubMed] [Google Scholar]
- 9.Pedersen B. K., Saltin B. Exercise as medicine-evidence for prescribing exercise as therapy in 26 different chronic diseases. Scandinavian Journal of Medicine & Science in Sports. 2015;25:1–72. doi: 10.1111/sms.12581. [DOI] [PubMed] [Google Scholar]
- 10.Lopes S., Mesquita-Bastos J., Alves A. J., Ribeiro F. Exercise as a tool for hypertension and resistant hypertension management: current insights. Integrated Blood Pressure Control. 2018;11:65–71. doi: 10.2147/ibpc.s136028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacke C., Nunan D., Weisser B. Do exercise trials for hypertension adequately report interventions? A reporting quality study. International Journal of Sports Medicine. 2018;39(12):902–908. doi: 10.1055/a-0649-1040. [DOI] [PubMed] [Google Scholar]
- 12.Ozemek C., Arena R. Precision in promoting physical activity and exercise with the overarching goal of moving more. Progress in Cardiovascular Diseases. 2019;62(1):3–8. doi: 10.1016/j.pcad.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Ross R., Goodpaster B. H., Koch L. G., et al. Precision exercise medicine: understanding exercise response variability. British Journal of Sports Medicine. 2019;53(18):1141–1153. doi: 10.1136/bjsports-2018-100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albouaini K., Egred M., Alahmar A., Wright D. J. Cardiopulmonary exercise testing and its application. Postgraduate Medical Journal. 2007;83(985):675–682. doi: 10.1136/hrt.2007.121558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balady G. J., Arena R., Sietsema K., et al. Clinician’s guide to cardiopulmonary exercise testing in adults. Circulation. 2010;122(2):191–225. doi: 10.1161/cir.0b013e3181e52e69. [DOI] [PubMed] [Google Scholar]
- 16.Youn J.-C., Kang S.-M. Cardiopulmonary exercise test in patients with hypertension: focused on hypertensive response to exercise. Pulse. 2015;3(2):114–117. doi: 10.1159/000431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mezzani A. Cardiopulmonary exercise testing: basics of methodology and measurements. Annals of the American Thoracic Society. 2017;14(Supplement_1):S3–S11. doi: 10.1513/annalsats.201612-997fr. [DOI] [PubMed] [Google Scholar]
- 18.Drescher U., Koschate J., Hoffmann U. Oxygen uptake and heart rate kinetics during dynamic upper and lower body exercise: an investigation by time-series analysis. European Journal of Applied Physiology. 2015;115(8):1665–1672. doi: 10.1007/s00421-015-3146-4. [DOI] [PubMed] [Google Scholar]
- 19.Guazzi M., Bandera F., Ozemek C., Systrom D., Arena R. Cardiopulmonary exercise testing. Journal of the American College of Cardiology. 2017;70(13):1618–1636. doi: 10.1016/j.jacc.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Buys R., Van De Bruaene A., Müller J., et al. Usefulness of cardiopulmonary exercise testing to predict the development of arterial hypertension in adult patients with repaired isolated coarctation of the aorta. International Journal of Cardiology. 2013;168(3):2037–2041. doi: 10.1016/j.ijcard.2013.01.171. [DOI] [PubMed] [Google Scholar]
- 21.Keller K., Stelzer K., Ostad M. A., Post F. Impact of exaggerated blood pressure response in normotensive individuals on future hypertension and prognosis: systematic review according to PRISMA guideline. Advances in Medical Sciences. 2017;62(2):317–329. doi: 10.1016/j.advms.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Older P. O., Levett D. Z. H. Cardiopulmonary exercise testing and surgery. Annals of the American Thoracic Society. 2017;14(Supplement_1):S74–S83. doi: 10.1513/annalsats.201610-780fr. [DOI] [PubMed] [Google Scholar]
- 23.Stubbs D. J., Grimes L. A., Ercole A. Performance of cardiopulmonary exercise testing for the prediction of post-operative complications in non-cardiopulmonary surgery: a systematic review. PLoS One. 2020;15(2):p. e0226480. doi: 10.1371/journal.pone.0226480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santoro C., Sorrentino R., Esposito R., et al. Cardiopulmonary exercise testing and echocardiographic exam: an useful interaction. Cardiovascular Ultrasound. 2019;17(1):p. 29. doi: 10.1186/s12947-019-0180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nedeljkovic I., Banovic M., Stepanovic J., et al. The combined exercise stress echocardiography and cardiopulmonary exercise test for identification of masked heart failure with preserved ejection fraction in patients with hypertension. European Journal of Preventive Cardiology. 2016;23(1):71–77. doi: 10.1177/2047487315604836. [DOI] [PubMed] [Google Scholar]
- 26.Badagliacca R., Papa S., Valli G., et al. Echocardiography combined with cardiopulmonary exercise testing for the prediction of outcome in idiopathic pulmonary arterial hypertension. Chest. 2016;150(6):1313–1322. doi: 10.1016/j.chest.2016.07.036. [DOI] [PubMed] [Google Scholar]
- 27.Leopold E., Navot-Mintzer D., Shargal E., et al. Prediction of the Wingate anaerobic mechanical power outputs from a maximal incremental cardiopulmonary exercise stress test using machine-learning approach. PLoS One. 2019;14(3):p. e0212199. doi: 10.1371/journal.pone.0212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braccioni F., Bottigliengo D., Ermolao A., et al. Dyspnea, effort and muscle pain during exercise in lung transplant recipients: an analysis of their association with cardiopulmonary function parameters using machine learning. Respiratory Research. 2020;21(1):1–11. doi: 10.1186/s12931-020-01535-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakr S., Elshawi R., Ahmed A., et al. Using machine learning on cardiorespiratory fitness data for predicting hypertension: the Henry Ford ExercIse Testing (FIT) Project. PLoS One. 2018;13(4):p. e0195344. doi: 10.1371/journal.pone.0195344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang G., Leng X., Huang F., et al. Use CPET data to predict the intervention effect of aerobic exercise on young hypertensive patients. Proceedings of 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); November 2019; San Diego, CA, USA. IEEE; pp. 1699–1702. [Google Scholar]
- 31.Bagnall A., Lines J., Bostrom A., Large J., Keogh E. The great time series classification bake off: a review and experimental evaluation of recent algorithmic advances. Data Mining and Knowledge Discovery. 2017;31(3):606–660. doi: 10.1007/s10618-016-0483-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagnall A., Lines J., Hills J., Bostrom A. Time-series classification with COTE: the collective of transformation-based ensembles. IEEE Transactions on Knowledge and Data Engineering. 2015;27(9):2522–2535. doi: 10.1109/tkde.2015.2416723. [DOI] [Google Scholar]
- 33.Lines J., Taylor S., Bagnall A. Time series classification with HIVE-COTE: the hierarchical vote collective of transformation-based ensembles. ACM Transactions on Knowledge Discovery from Data. 2018;12(5):p. 52. [Google Scholar]
- 34.Fawaz H. I., Forestier G., Weber J., et al. Deep learning for time series classification: a review. Data Mining and Knowledge Discovery. 2019;33(4):917–963. [Google Scholar]
- 35.Zhao B., Lu H., Chen H., Liu J., Wu D. Convolutional neural networks for time series classification. Journal of Systems Engineering and Electronics. 2017;28(1):162–169. doi: 10.21629/jsee.2017.01.18. [DOI] [Google Scholar]
- 36.Ma Q., Shen L., Chen W., Wang J., Wei J., Yu Z. Functional echo state network for time series classification. Information Sciences. 2016;373:1–20. doi: 10.1016/j.ins.2016.08.081. [DOI] [Google Scholar]
- 37.Zhang Z., Xu Y., Yang J., Li X., Zhang D. A survey of sparse representation: algorithms and applications. IEEE Access. 2015;3:490–530. doi: 10.1109/access.2015.2430359. [DOI] [Google Scholar]
- 38.Tropp J. A., Gilbert A. C., Strauss M. J. Algorithms for simultaneous sparse approximation. Part I: greedy pursuit. Signal Processing. 2006;86(3):572–588. doi: 10.1016/j.sigpro.2005.05.030. [DOI] [Google Scholar]
- 39.Tropp J. A. Algorithms for simultaneous sparse approximation. Part II: convex relaxation. Signal Processing. 2006;86(3):589–602. doi: 10.1016/j.sigpro.2005.05.031. [DOI] [Google Scholar]
- 40.Tropp J. A., Gilbert A. C. Signal recovery from random measurements via orthogonal matching pursuit. IEEE Transactions on Information Theory. 2007;53(12):4655–4666. doi: 10.1109/tit.2007.909108. [DOI] [Google Scholar]
- 41.Efron B., Hastie T., Johnstone I., et al. Least angle regression. The Annals of Statistics. 2004;32(2):407–499. doi: 10.1214/009053604000000067. [DOI] [Google Scholar]
- 42.Wu T. T., Lange K. Coordinate descent algorithms for lasso penalized regression. The Annals of Applied Statistics. 2008;2(1):224–244. doi: 10.1214/07-aoas147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck A., Teboulle M. A fast iterative shrinkage-thresholding algorithm for linear inverse problems. SIAM Journal on Imaging Sciences. 2009;2(1):183–202. doi: 10.1137/080716542. [DOI] [Google Scholar]
- 44.Wright J., Yang A. Y., Ganesh A., et al. Robust face recognition via sparse representation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2008;31(2):210–227. doi: 10.1109/TPAMI.2008.79. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z., Zuo W., Hu Q., Lin L. Kernel sparse representation for time series classification. Information Sciences. 2015;292:15–26. doi: 10.1016/j.ins.2014.08.066. [DOI] [Google Scholar]
- 46.Zhang L., Yang M., Feng X. Sparse representation or collaborative representation: which helps face recognition. Proceedings of International Conference on Computer Vision; November 2011; Barcelona, Spain. IEEE; pp. 471–478. [Google Scholar]
- 47.Deng W., Hu J., Guo J. Extended SRC: undersampled face recognition via intraclass variant dictionary. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012;34(9):1864–1870. doi: 10.1109/TPAMI.2012.30. [DOI] [PubMed] [Google Scholar]
- 48.Rubinstein R., Bruckstein A. M., Elad M. Dictionaries for sparse representation modeling. Proceedings of the IEEE. 2010;98(6):1045–1057. doi: 10.1109/jproc.2010.2040551. [DOI] [Google Scholar]
- 49.Wang P., Kong L., Du T., Wang L. Orthogonal sparse dictionary based on Chirp echo for ultrasound imaging. Applied Acoustics. 2019;156:359–366. doi: 10.1016/j.apacoust.2019.07.031. [DOI] [Google Scholar]
- 50.Aharon M., Elad M., Bruckstein A. K-SVD: an algorithm for designing overcomplete dictionaries for sparse representation. IEEE Transactions on Signal Processing. 2006;54(11):4311–4322. [Google Scholar]
- 51.Yu Z., Zheng X., Huang F., et al. A framework based on sparse representation model for time series prediction in smart city. Frontiers of Computer Science. 2020;15(1):1–13. doi: 10.1007/s11704-019-8395-7. [DOI] [Google Scholar]
- 52.Alkhodari M., Islayem D. K., Alskafi F. A., Khandoker A. H. Predicting hypertensive patients with higher risk of developing vascular events using heart rate variability and machine learning. IEEE Access. 2020;8:192727–192739. doi: 10.1109/access.2020.3033004. [DOI] [Google Scholar]
- 53.Dau H. A., Bagnall A., Kamgar K., et al. The UCR time series archive. IEEE/CAA Journal of Automatica Sinica. 2019;6(6):1293–1305. doi: 10.1109/jas.2019.1911747. [DOI] [Google Scholar]
- 54.Deng H., Runger G., Tuv E., Vladimir M. A time series forest for classification and feature extraction. Information Sciences. 2013;239:142–153. doi: 10.1016/j.ins.2013.02.030. [DOI] [Google Scholar]
- 55.Zhao J., Itti L. Shapedtw: shape dynamic time warping. Pattern Recognition. 2018;74:171–184. doi: 10.1016/j.patcog.2017.09.020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study cannot be made freely available in order to protect patient privacy. Requests for access to these data should be made to the corresponding author.


