Abstract
Coronavirus disease 2019 (COVID-19) is an ongoing global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Type I and III interferon (IFN) responses act as the first line of defense against viral infection and are activated by the recognition of viruses by infected cells and innate immune cells. Dysregulation of host IFN responses has been known to be associated with severe disease progression in COVID-19 patients. However, the reported results are controversial and the roles of IFN responses in COVID-19 need to be investigated further. In the absence of a highly efficacious antiviral drug, clinical studies have evaluated recombinant type I and III IFNs, as they have been successfully used for the treatment of infections caused by two other epidemic coronaviruses, SARS-CoV-1 and Middle East respiratory syndrome (MERS)-CoV. In this review, we describe the strategies by which SARS-CoV-2 evades IFN responses and the dysregulation of host IFN responses in COVID-19 patients. In addition, we discuss the therapeutic potential of type I and III IFNs in COVID-19.
Keywords: COVID-19, SARS-CoV-2, interferon, interferon-stimulated gene, therapeutics
INTRODUCTION
Coronavirus disease 2019 (COVID-19) was first identified in Wuhan, China at the end of 2019, and it rapidly spread across the globe.1 On March 11, 2020, the World Health Organization declared the COVID-19 outbreak a pandemic. COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a β-coronavirus with high sequence homology to bat coronaviruses (CoVs). SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) receptor for viral entry into host cells.2,3 Human CoVs include two other highly pathogenic viruses, SARS-CoV-1 and Middle East respiratory syndrome (MERS)-CoV, which caused epidemics in 2003 and 2012, respectively, as well as endemic common-cold CoVs, such as OC43, HKU1, 229E, and NL63.4 Although SARS-CoV-2 is not as lethal as SARS-CoV-1 or MERS-CoV,5 its extensive spread during the current pandemic has caused tremendous pressure and disastrous consequences for public health and the medical system worldwide. No highly effective antiviral drug is currently available for the treatment of COVID-19.
Type I and III interferons (IFNs) act as major first-line defenses against viruses. Virus-infected cells and innate immune cells recognize viral infections through pattern recognition receptors (PRRs) and produce type I and III IFNs. Type I IFNs comprise IFN-α, IFN-β, IFN-ε, IFN-κ, and IFN-ω in humans,6 and all of them bind to the ubiquitously expressed IFNα/β receptor, which is composed of the IFNAR1 and IFNAR2 subunits. Although type I IFNs can be secreted by many types of cells, plasmacytoid dendritic cells (pDCs) are the main source of type I IFNs during viral infection.7 When type I IFNs bind to IFNα/β receptor, the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway is activated, and the expression of hundreds of IFN-stimulated genes (ISGs) is upregulated.8,9
In humans, type III IFNs include four different IFN-λs, known as IFN-λ1/IL-29, IFN-λ2/IL-28A, IFN-λ3/IL-28B, and IFN-λ4. IFN-λs bind to the IFNλ receptor, a heterodimeric receptor formed by IFNLR1/IL28Rα and IL10Rβ that is exclusively expressed on epithelial cells and certain types of myeloid cells.10 Due to this specific expression pattern, the antiviral effects of IFN-λs are especially prominent at epithelial barriers, such as those in the gastrointestinal, respiratory, and reproductive tracts.11,12,13
Although type I and III IFNs are genetically distinct and use different receptors, the downstream signaling pathways and the transcriptional responses activated by type I and III IFNs exhibit substantial overlap. The major difference is that type I IFN signaling results in a rapid, systemic induction and decline in ISG expression, whereas type III IFN signaling induces a sustained upregulation of ISGs in epithelial cells mediated by unphosphorylated STATs.14 In this manner, type III IFNs provide antiviral protection at epithelial surfaces as a front-line defense that confers less collateral damage than the more potent type I IFN response.15
As type I and III IFNs are involved in host protection against viruses,16,17,18 many viruses have developed mechanisms to evade and suppress the antiviral functions of IFNs and ISGs.19,20 In this review, we describe how host cells sense CoV infection and how SARS-CoV-2 evades the type I and III IFN responses. Furthermore, we describe the dysregulated IFN responses in COVID-19 patients and discuss the therapeutic potential of type I and III IFNs in COVID-19.
EVASION OF IFN RESPONSES BY SARS-COV-2
Recognition of CoVs by the innate immune system
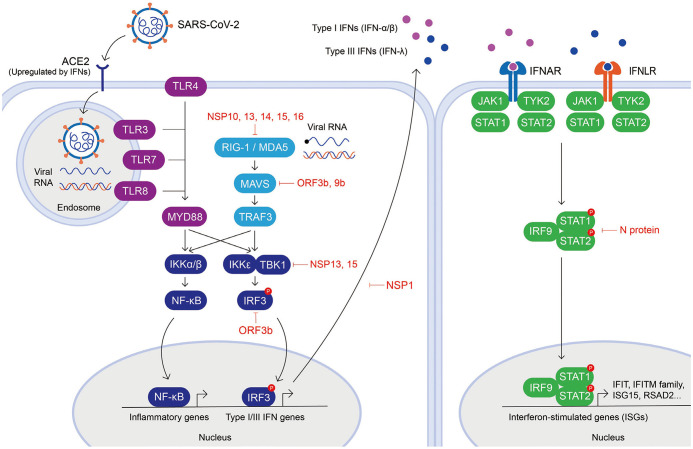
The innate immune system detects viral pathogens by recognizing their pathogen-associated molecular patterns (PAMPs) through various PRRs. Viral PAMPs are distinct molecular patterns that do not exist in host cells, including viral single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA).21 Although our current understanding of the specific innate immune sensing of SARS-CoV-2 is limited, the virus-host interactions of SARS-CoV-2 are predicted to resemble those of other CoVs due to their shared sequence homology. Host cells recognize viral RNA mainly through two different classes of PRRs, Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) (Fig. 1). RLRs are widely expressed by the majority of cell types and localized in the cytosol, whereas TLRs are usually expressed by innate immune system cells and localized on the cell membrane and cellular compartments like endosomes. Downstream signaling of TLRs and RLRs upon ligand binding activates transcription factors, such as IRF3, to produce type I and III IFNs and nuclear factor-κB (NF-κB) to express pro-inflammatory cytokines, which together induce antiviral programs in host cells.18,22,23
Fig. 1. Innate immune recognition of viral infection and evasion mechanisms by SARS-CoV-2. Viral infection is sensed by various innate immune receptors, including cytoplasmic RNA sensors (RIG-I and MDA5) and TLRs (TLR3, TLR4, TLR7, and TLR8). Upon recognition, proinflammatory genes and IFNs are upregulated by transcription factors, NF-κB, and IRF3. The secreted type I (IFN-α and -β) and III (IFN-λ) IFNs bind to IFNα/β receptor and IFNλ receptor, respectively, which activate the JAK-STAT signaling pathway to upregulate ISG expression. SARS-CoV-2 proteins that have been reported to interfere with IFN responses are indicated. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TLR, Toll-like receptor; IFN, interferon; NF-κB, nuclear factor-κB; IRF3, interferon regulatory factor 3; JAK-STAT, Janus kinase-signal transducer and activator of transcription; ISG, IFN-stimulated genes.
In the endosome, TLR3 detects dsRNA, while TLR7 and TLR8 detect ssRNA. CoVs are positive-sense ssRNA viruses that form dsRNA intermediates during their replication, which can be detected by TLR3 in the endosome, and by RIG-I, MDA5, and PKR in the cytosol. The ssRNA can also be detected by TLR7 or TLR8 in the endosome and potentially by RIG-I and PKR in the cytosol.24 The TLR located on the surface of innate immune cells, TLR4, recognizes viral glycoproteins, such as the respiratory syncytial virus fusion protein.25 Differences in the location of PAMP engagement can determine the type of IFN produced. For example, TLR4 engagement in the endosome results in the production of type I IFNs,26 whereas TLR4 signaling at the plasma membrane induces type III IFNs,27 which could explain the protective activity of type III IFNs at epithelial barriers that continually encounter PAMPs. TLR7 is crucial for sensing various CoVs, and is required for IFN-α production by pDCs in CoV infection.28,29 The cytosolic RLRs, RIG-I and MDA5, sense viral RNAs by detecting uncapped RNA bearing a 5′ triphosphate terminus, RNA with a non-methylated or incompletely methylated cap structure, and replicative intermediates consisting of dsRNA.30
Evasion of innate immune sensing by SARS-CoV-2
CoVs, including SARS-CoV-1 and MERS-CoV, suppress PRR activation by either evading recognition or antagonizing PRR signaling (Fig. 1).31,32,33,34,35,36 To evade innate recognition, dsRNA is processed in ER-derived double membrane vesicles that are formed during viral replication.36,37 Viral RNA evades RLR recognition by generating a guanosine cap and methylation at the 5′ end by non-structural proteins (NSPs) 10, 13, 14, and 16.31,32,35 CoVs also evade dsRNA sensors, especially MDA5, by encoding an endoribonuclease, NSP15, which cleaves 5′ polyuridines from the negative-sense viral RNA formed during viral replication.33,34
Recent studies have emphasized the possibility that SARS-CoV-2 is more efficient than other CoVs in inhibiting IFN signaling and activity.38,39,40,41 SARS-CoV-2 proteins have high amino acid sequence homology along with those of SARS-CoV-1, including NSP14, NSP15, NSP16, and N protein,41 suggesting that the evasion mechanisms of SARS-CoV-1 are likely preserved in SARS-CoV-2. In addition, NSP1, NSP3, NSP12, NSP13, NSP14, ORF3, ORF6, and M protein inhibit virus-induced IFN-β promoter activation; and ORF6 inhibits type I IFN production and its downstream signaling.42 A SARS-CoV-2 protein interaction map obtained from the analysis of 26 SARS-CoV-2 proteins expressed in human cells identified host proteins that physically interact with SARS-CoV-2 proteins.43 SARS-CoV-1 ORF3b was found to inhibit the induction of type I IFNs by inhibiting IRF3.44,45 Even though SARS-CoV-2 ORF3b protein is shorter than SARS-CoV-1 ORF3b, it was recently found to inhibit IFN induction more efficiently.46 Moreover, a natural variant encoding a longer ORF3b reading frame exhibits enhanced suppression of IFN induction.46 In addition, SARS-CoV-2 ORF9b, similar to SARS-CoV-1 ORF9b, has been found to localize on mitochondria and suppress IFN responses through association with TOM70.47,48 Furthermore, SARS-CoV-2 NSP13 and NSP15 have been found to interact with TBK1 and the TBK1 activator ring finger protein 41 (RNF41)/Nrdp1.47 SARS-CoV-2 NSP1 was also recently found to bind 40S and 80S ribosomes, shutting down capped mRNA translation and obstruction of the mRNA entry tunnel, thereby blocking RIG-I-dependent innate immune responses. This feature was previously demonstrated for NSP1 encoded by other CoVs, including SARS-CoV-1.49,50,51 When cells are stimulated by IFNs, SARS-CoV-2 N protein antagonizes IFN signaling by inhibiting phosphorylation of STAT1 and STAT2.52
As described above, SARS-CoV-2 has diverse mechanisms for evading IFN responses. However, these IFN signaling evasion mechanisms are able to work only in SARS-CoV-2-infected cells in which viral proteins exist, but not in other non-infected innate immune cells. This could explain how the innate immune cells can participate in delayed but exacerbated IFN responses in COVID-19 patients.
DYSREGULATION OF IFN RESPONSES IN COVID-19
Impaired IFN responses in COVID-19
Impaired IFN responses have been reported in COVID-19 patients, particularly in patients with severe disease (Table 1).38,39,53,54,55,56 SARS-CoV-1 infection has been shown to induce the production of pro-inflammatory cytokines and chemokines but suppress the induction of IFNs.57,58 Accordingly, negligible amounts of IFN-β and IFN-λ have been detected in the sera of COVID-19 patients, whereas moderate levels of ISGs and strong expression of chemokines have been found consistently across in vitro, ex vivo, and in vivo models of SARS-CoV-2 infection.38 Another study has reported that patients with severe and critical COVID-19 exhibit a highly impaired type I IFN response, characterized by low levels of IFN-α and IFN-β and low levels of ISG expression.39 In addition, the majority of COVID-19 patients with acute respiratory failure have profound suppression of type I and II IFN responses compared to patients with acute influenza.54 The impaired IFN production in COVID-19 patients can be explained by pDC depletion, as pDCs are the main producers of type I IFNs. In severe cases of COVID-19, the number of pDCs is significantly decreased in the peripheral blood39,59 and bronchoalveolar lavage (BAL) fluid.60
Table 1. Summary of Published Studies Regarding IFN Production and ISG Response in COVID-19 Patients.
| Cohort | Specimen | ISG response | Production of IFN | Refs |
|---|---|---|---|---|
| 24 COVID-19, 24 healthy | PBMC | Moderate ISG response, strong chemokine expression | Low IFN-I and IFN-III level | 38 |
| 50 COVID-19 (15 mild-to-moderate, 17 severe, 18 critical), 18 healthy | PBMC | Impaired ISG response in severe and critical patients | No IFN-β low IFN-α production and activity in severe and critical patientes | 39 |
| 8 COVID-19, 5 severe influenza, 4 healthy | PBMC | Strong type I IFN response co-existing with TNF-IL-1β-driven inflammation in classical monocytes of severe patients | nd | 75 |
| 113 COVID-19 | PBMC | nd | Increased IFN-α production in severe patients | 74 |
| 26 COVID-19 (critical) | PBMC | Low ISG expression, ISG correlated with IFN-α2 measurement | Low or no IFN-α production, no IFN-β and IFN-λ production | 57 |
| 76 COVID-19, 69 healthy | PBMC | Increased ISG expression in T cells and monocytes which correlated with IFN-α concentration in plasma | Low IFN-α production, lack of type I IFN gene expression | 56 |
| 8 COVID-19, 20 healthy | BALF | Increased ISG expression and chemokine-dominant hypercytokinemia | nd | 72 |
| 7 COVID-19 (4 ARDS), 6 healthy | PBMC | Positive correlation between ISG of CD14+ monocytes and age, and negative correlation with time from fever onset | nd | 60 |
| 10 COVID-19, 5 healthy | BALF, naso-oropharyngeal swab | nd | Increased IFN-α, IFN-β, IFN-λ mRNA in BALF | 67 |
| 19 COVID-19, 5 healthy | Nasopharyngeal/pharyngeal swab | Overexpression of cytokine/chemokine genes in non-resident macrophages of the airway epithelium in critical patients | nd | 71 |
| 9 COVID-19 (3 moderate, 6 severe/critical), 4 healthy | BALF | Type I IFN response mainly expressed by neutrophils and FCN+ classical monocytes | nd | 76 |
| 5 COVID-19 (4 moderate, 1 severe), 2 IAV, 3 healthy | PBMC | Increased ISG expression, and severe patients show stronger response to IFN and virus infection | nd | 65 |
| 10 COVID-19, 5 healthy | PBMC | Increased ISG expression in CD14++ inflammatory monocytes | nd | 66 |
| 16 COVID-19, 6 normal | Post-mortem lung samples | Two distinct pattern: ISGhigh, high cytokine production, high viral loads, limited pulmonary damage/ISGlow, low viral loads, high infiltrating activated CD8+ T cells and macrophages | nd | 68 |
| 79 COVID-19 (35 ARDS), 26 influenza (7 ARDS), 15 healthy | PBMC | Lower expression of IFN-α response genes compared to influenza | nd | 55 |
IFN, interferon; COVID-19, coronavirus disease 2019; ISG, IFN-stimulated genes; TNF, tumor necrosis factor; ARDS; acute respiratory distress syndrome; PBMC, peripheral blood mononuclear cell; BALF, bronchoalveolar lavage fluid.
Other studies have suggested that IFN induction may be delayed rather than completely impaired. Analysis of SARS-CoV-1-infected bronchial epithelial cells revealed that the production of IFNs is delayed compared to the production of pro-inflammatory cytokines.61 Furthermore, the induction of IFN-α, IFN-λ, and ISGs in SARS-CoV-1- and MERS-CoV-infected cells is delayed compared to that in influenza A virus (IAV)-infected cells.62 SARS-CoV-1-infected mice with severe symptoms exhibit robust viral replication and delayed type I IFN signaling. Type I IFNs induce an influx of inflammatory monocytes/macrophages and vascular leakage, and the pathology is diminished in the absence of IFN signaling.63
Enhanced IFN responses in severe COVID-19
Paradoxically, elevated IFN production and ISG expression are correlated with worse disease outcomes in CoV infection, including COVID-19 (Table 1).64,65,66,67 Clinically well-described SARS patients with poor outcomes have high levels of IFN-α and ISG expression, which could be associated with atypical innate and adaptive immune responses.68 In addition, IFN-α production is significantly correlated with the severity of MERS-CoV, and no apparent IFN-α response has been detected in patients with mild symptoms.69
BAL fluid samples from COVID-19 patients exhibit increased transcriptional levels of IFNA2, IFNB1, IFNL2, and IFNL3,70 as well as robust innate immune responses with notable hypercytokinemia and increased expression of ISGs, particularly ISG15, RSAD2/viperin, IFIT, and IFITM family members.71 High levels of IFN-α levels in sera 5–10 days from symptom onset have been associated with the severity of COVID-19.72 In a longitudinal study, patients with severe COVID-19 exhibited increased IFN-α production over time, whereas patients with moderate COVID-19 had decreased IFN-α levels.73 Single-cell RNA sequencing (scRNA-seq) analysis of peripheral blood mononuclear cells of COVID-19 patients showed hyper-inflammatory signatures across all types of immune cells.74 Specifically, classical monocytes from severe patients exhibited a type I IFN response in combination with TNF/IL-1β-driven inflammation, whereas those from mild patients exhibited only features of TNF/IL-1β-driven inflammation, suggesting a pivotal role of the type I IFN response in exacerbating inflammation in the progression to severe COVID-19.74 Other scRNA-seq studies of peripheral blood mononuclear cells have observed heterogeneous ISG signatures in CD14+ monocytes, with higher ISG scores showing a positive correlation with patient's age,59 and a broad type I IFN response genes expressed mainly by neutrophils and, to a lesser extent, by FCN1+ classical monocytes.75
The pro-inflammatory roles of IFNs have been well described in a mouse model of SARS-CoV-1, which demonstrated that delayed but considerable type I IFN responses in SARS-CoV-1-infected BALB/c mice trigger the accumulation of monocytes and macrophages as well as the production of pro-inflammatory cytokines, resulting in lethal pneumonia, vascular leakage, and insufficient T cell responses.63 Pro-inflammatory roles of type I IFNs have also been shown in human ACE2 expressing mice infected with SARS-CoV-2.76 Using Ifnar-/- mice or Irf3-/- Irf7-/- mice, this study proved that type I IFN responses are necessary for the recruitment of pro-inflammatory monocytes and macrophages to the infected lungs. In addition, type I IFNs have been found to reprogram the macrophage epigenome to promote inflammatory activation.77 TNF is a classical pro-inflammatory cytokine, but it also has a paradoxical anti-inflammatory function to limit inflammation-associated toxicity.78 This effect is mediated by tolerizing genes encoding inflammatory molecules, causing hyporesponsiveness to additional TLR signals in monocytes and macrophages.77 Type I IFNs were found to abolish the tolerizing effect of TNF and potentiate monocytes and macrophages responsive to additional TLR signals by priming chromatin to prevent the silencing of target genes of NF-κB.77 Park, et al.77 identified a gene module that was previously unresponsive to TLR signals due to TNF-induced tolerance but became responsive to TLR signals with type I IFNs pretreatment. This gene module was found to be significantly upregulated in the transcriptome of classical monocytes from patients with severe COVID-19, indicating a feedforward mechanism of type I IFN-induced hyperinflammation in severe COVID-19 cases.74 These results demonstrate that IFN responses are not impaired in COVID-19 patients and highlight their possible role in exacerbating inflammation, particularly in cases of severe COVID-19.
Recent studies have shown that SARS-CoV-2 receptor ACE2 in human airway epithelial cells is an ISG upregulated by type I and type II IFNs.79,80,81 These studies imply that exacerbated IFN responses could contribute to the cellular entry of SARS-CoV-2 and expand its cellular tropism, thereby promoting SARS-CoV-2 replication. Nevertheless, the antiviral action of IFNs against SARS-CoV-2 was shown to counterbalance the pro-viral effects of IFN-induced ACE2 upregulation.82
THERAPEUTIC POTENTIAL OF IFNS IN COVID-19
Therapeutic potential of type I IFNs in COVID-19
Numerous in vitro and in vivo studies have demonstrated the therapeutic efficacy of type I IFNs in SARS and MERS.83 Treatment with IFNs in cell culture and organoids has been shown to efficiently inhibit the replication of CoVs, including SARS-CoV-1, SARS-CoV-2, and MERS-CoV.40,41,84,85,86,87,88 Recent in vitro studies have highlighted that SARS-CoV-2 is highly sensitive to both IFN-α and IFN-β.40,41 In these studies, viral titers were remarkably reduced when IFN-α and IFN-β was administered prior to infection and reduced to a lesser extent when treatment was administered after infection, indicating that type I IFNs may be effective as either prophylactic or early treatment for COVID-19 patients. In China, guidelines for the treatment of COVID-19 recommend vapor inhalation of IFN-α twice a day in conjunction with ribavirin administration,89 which offers the advantage of delivering IFN-α specifically to the respiratory tract.
Several clinical trials have been registered to evaluate the efficacy of type I IFNs as a single or combination therapy for COVID-19 (Table 2). The multicenter, adaptive, randomized, open clinical trial DisCoVeRy is currently evaluating the efficacy of IFN-β1a as a treatment for COVID-19 in hospitalized adults in Europe (NCT04315948). A recent phase 2 trial of COVID-19 patients in Hong Kong has shown that the triple combination of IFN-β1b, lopinavir-ritonavir, and ribavirin is safe and superior to lopinavir-ritonavir alone in alleviating symptoms and shortening the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19.90 In a study of 77 adults hospitalized with COVID-19 in Wuhan, China who were treated with nebulized IFN-α2b, arbidol, or a combination of the two, IFN-α2b treatment with or without arbidol significantly reduced the duration of detectable virus and inflammatory markers IL-6 and C-reactive protein.91 Inhalation of nebulized IFN-β1a has also been reported to be safe and efficient in another study of COVID-19 patients in the UK.92 Another study conducted in Hubei Province showed that treatment with recombinant IFN-α nasal drops could prevent COVID-19 incidence without adverse effects, as the incidence among the 2944 healthcare workers treated with daily IFN-α for 28 days was zero.93
Table 2. Ongoing Clinical Trials Evaluating Efficacy of IFNs in COVID-19.
| Phase | IFN | Form | Drug combination | Status | NCT number |
|---|---|---|---|---|---|
| 4 | IFN-β-1a | Recombinant | Hydroxychloroquine, lopinavir/ritonavir | Enrolling by invitation | NCT04350671 |
| 3 | IFN-α-1b | Recombinant | Thymosine alpha 1 | Recruiting | NCT04320238 |
| 3 | IFN-β-1a | Recombinant | Remdesivir, lopinavir/ritonavir, hydroxychloroquine | Recruiting | NCT04315948 |
| 3 | IFN-β-1a | Pegylated | Recruiting | NCT04552379 | |
| 3 | IFN-β-1a | Recombinant | Not recruiting yet | NCT04647669 | |
| 3 | IFN-β-1a | Recombinant | Remdesivir | Active, not recruiting | NCT04492475 |
| 3 | IFN-β | Recombinant | Recruiting | NCT04324463 | |
| 2 | IFN-α-2b | Pegylated | Recruiting | NCT04480138 | |
| 2 | IFN-β-1a | Recombinant | Not recruiting yet | NCT04449380 | |
| 2 | IFN-β-1a (inhalation) | Recombinant | Recruiting | NCT04385095 | |
| 2 | IFN-β-1a, IFN-β-1b | Recombinant | Hydroxychloroquine, lopinavir/ritonavir | Completed (April 27, 2020) | NCT04343768 |
| 2 | IFN-β-1b | Recombinant | Clofazimine | Recruiting | NCT04465695 |
| 2 | IFN-β-1b | Recombinant | Hydroxychloroquine | Completed (July 7, 2020) | NCT04350281 |
| 2 | IFN-β-1b | Recombinant | Ribavirin | Recruiting | NCT04494399 |
| 2 | IFN-β-1b | Recombinant | Lopinavir/ritonavir, ribavirin | Completed (March 31, 2020) | NCT04276688 |
| 2 | IFN-β-1b | Recombinant | Lopinavir/ritonavir | Not recruiting yet | NCT04521400 |
| 2 | IFN-β-1b | Recombinant | Remdesivir | Recruiting | NCT04330690 |
| 2 | IFN-β-1b | Recombinant | Remdesivir | Recruiting | NCT04647695 |
| 2 | IFN-β-1b (inhalation) | Recombinant | Suspended | NCT04469491 | |
| 2 | IFN-λ-1a | Pegylated | Recruiting | NCT04354259 | |
| 2 | IFN-λ-1a | Pegylated | Recruiting | NCT04344600 | |
| 2 | IFN-λ-1a | Pegylated | Not recruiting yet | NCT04388709 | |
| 2 | IFN-λ | Pegylated | Recruiting | NCT04534673 | |
| 2 | IFN-λ | Pegylated | Enrolling by invitation | NCT04343976 | |
| 1,2 | IFN-α-2b | Recombinant | Rintatolimod | Recruiting | NCT04379518 |
| 1 | IFN-α-1b | Recombinant | Not recruiting yet | NCT04293887 |
IFN, interferon; COVID-19, coronavirus disease 2019.
Therapeutic potential of type III IFNs in COVID-19
Type III IFNs also trigger signals through the JAK-STAT pathway, inducing the upregulation of a pane of ISGs that substantially overlap with those induced by type I IFNs but demonstrate different context-specific functions. For example, IFN-λs are the predominant IFNs produced in the early phase of viral infection, as shown in IAV infection.94 IFN-λs act on IFNλ receptors, which are preferentially expressed by epithelial cells to control viral replication without causing hyper-inflammation.94 There is growing evidence that IFN-λs provide an important first line of defense against viral infection in the respiratory and gastrointestinal tracts. In mice, IFN-λs have been shown to protect respiratory epithelial cells from infection by respiratory viruses, including SARS-CoV-1.95 A recent study using human colon-derived cell lines and primary non-transformed human colon organoids revealed that SARS-CoV-2 infection can be controlled by both type I and III IFNs, although type III IFNs are more efficient at controlling viral replication.88 Furthermore, in a newly developed mouse model of SARS-CoV-2 infection, both prophylactic and therapeutic administration of pegylated IFN-λ1a diminished viral replication.96 However, recent studies have demonstrated that IFN-λs produced by lung dendritic cells in response to viral RNA lead to barrier damage, causing susceptibility to lethal bacterial superinfections.70,97 In addition, prolonged IFN-λ responses cause p53 activation, which reduces epithelial proliferation and differentiation, increasing susceptibility to bacterial superinfections and their severity.97 Therefore, although the therapeutic potential of type III IFNs is promising, the clinical safety of type III IFNs in COVID-19 patients still needs thorough investigation. Currently, four clinical trials (NCT04343976, NCT04354259, NCT04388709, and NCT04344600) using pegylated IFN-λs are ongoing, all of them currently in phase 2 (Table 2).
CONCLUDING REMARKS
Type I and III IFNs are key players in the control of viral replication, but their roles in hyper-inflammation need to be further elucidated. Contradictory results regarding impaired or enhanced IFN responses in severe COVID-19 patients may be explained by differences in the definition of disease severity, sampling time points, and/or type of experimental readout (e.g., IFN itself or cellular responses to IFNs) among studies.98 Although there are some discrepancies in the roles of IFNs in COVID-19, recent clinical trials conducted with type I and III IFNs have shown promising results when treated in the early phase. A retrospective cohort study of 446 COVID-19 patients revealed that early administration of IFN-α2b is associated with reduced in-hospital days, whereas late IFN therapy increases mortality and delayed recovery.99 Other in vitro and in vivo studies support prophylactic treatment with IFNs as an ideal option. Therefore, in order to use type I or III IFNs as therapeutics for COVID-19 patients with minimal side effects, early treatment or prophylactic treatment before symptom onset would be optimal. Nevertheless, recent studies suggest that caution is needed when using IFN therapies, as prolonged IFN responses may cause lung epithelial barrier damage and lead to susceptibility to lethal bacterial superinfections.70,97 Nevertheless, further clinical studies are needed to determine the efficacy and safety of recombinant type I and III IFNs for the treatment of patients with COVID-19.
ACKNOWLEDGEMENTS
This research was supported by the 2020 Joint Research Project of Institutes of Science and Technology.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Hojun Choi and Eui-Cheol Shin.
- Data curation: Hojun Choi.
- Formal analysis: Hojun Choi and Eui-Cheol Shin.
- Funding acquisition: Eui-Cheol Shin.
- Project administration: Eui-Cheol Shin.
- Software: Hojun Choi.
- Supervision: Eui-Cheol Shin.
- Validation: Hojun Choi and Eui-Cheol Shin.
- Visualization: Hojun Choi.
- Writing—original draft: Hojun Choi.
- Writing—review & editing: Hojun Choi and Eui-Cheol Shin.
- Approval of final manuscript: all authors.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fung TS, Liu DX. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 5.Fauci AS, Lane HC, Redfield RR. COVID-19-navigating the uncharted. N Engl J Med. 2020;382:1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 8.Schoggins JW. Interferon-stimulated genes: what do they all do? Annu Rev Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 9.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotenko SV, Rivera A, Parker D, Durbin JE. Type III IFNs: beyond antiviral protection. Semin Immunol. 2019;43:101303. doi: 10.1016/j.smim.2019.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 12.Wells AI, Coyne CB. Type III interferons in antiviral defenses at barrier surfaces. Trends Immunol. 2018;39:848–858. doi: 10.1016/j.it.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazear HM, Nice TJ, Diamond MS. Interferon-λ: immune functions at barrier surfaces and beyond. Immunity. 2015;43:15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung PS, Cheon H, Cho CH, Hong SH, Park DY, Seo HI, et al. Roles of unphosphorylated ISGF3 in HCV infection and interferon responsiveness. Proc Natl Acad Sci U S A. 2015;112:10443–10448. doi: 10.1073/pnas.1513341112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazear HM, Schoggins JW, Diamond MS. Shared and distinct functions of Type I and Type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mesev EV, LeDesma RA, Ploss A. Decoding type I and III interferon signalling during viral infection. Nat Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park A, Iwasaki A. Type I and Type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sa Ribero M, Jouvenet N, Dreux M, Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma DY, Suthar MS. Mechanisms of innate immune evasion in reemerging RNA viruses. Curr Opin Virol. 2015;12:26–37. doi: 10.1016/j.coviro.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelemans T, Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11:961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 26.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odendall C, Voak AA, Kagan JC. Type III IFNs are commonly induced by bacteria-sensing TLRs and reinforce epithelial barriers during infection. J Immunol. 2017;199:3270–3279. doi: 10.4049/jimmunol.1700250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cervantes-Barragan L, Züst R, Weber F, Spiegel M, Lang KS, Akira S, et al. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheuplein VA, Seifried J, Malczyk AH, Miller L, Höcker L, Vergara-Alert J, et al. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol. 2015;89:3859–3869. doi: 10.1128/JVI.03607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kindler E, Thiel V. To sense or not to sense viral RNA--essentials of coronavirus innate immune evasion. Curr Opin Microbiol. 2014;20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with Severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoops K, Kikkert M, van den Worm SHE, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stertz S, Reichelt M, Spiegel M, Kuri T, Martínez-Sobrido L, García-Sastre A, et al. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J Virol. 2020;94:e01410-20. doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freundt EC, Yu L, Park E, Lenardo MJ, Xu XN. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang HW, Zhang HN, Meng QF, Xie J, Li Y, Chen H, et al. SARS-CoV-2 ORF9B suppresses type I interferon responses by targeting TOM70. Cell Mol Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, et al. Severe acute respiratory syndrome coronavirus NSP1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lokugamage KG, Narayanan K, Huang C, Makino S. Severe acute respiratory syndrome coronavirus protein NSP1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural basis for translational shutdown and immune evasion by the NSP1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mu J, Fang Y, Yang Q, Shu T, Wang A, Huang M, et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:65. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mudd PA, Crawford JC, Turner JS, Souquette A, Reynolds D, Bender D, et al. Targeted immunosuppression distinguishes COVID-19 from influenza in moderate and severe disease. medRxiv. 2020 Jan 01; [Preprint] [Google Scholar]
- 55.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trouillet-Assant S, Viel S, Gaymard A, Pons S, Richard JC, Perret M, et al. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol. 2020;146:206–208.e2. doi: 10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Subbarao K. The immunobiology of SARS. Annu Rev Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 58.Reghunathan R, Jayapal M, Hsu LY, Chng HH, Tai D, Leung BP, et al. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 61.Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menachery VD, Eisfeld AJ, Schafer A, Josset L, Sims AC, Proll S, et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. Mbio. 2014;5:e01174-14. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu L, Yang P, Zhao Y, Zhuang Z, Wang Z, Song R, et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53:685–696.e3. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 67.Nienhold R, Ciani Y, Koelzer VH, Tzankov A, Haslbauer JD, Menter T, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11:5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim ES, Choe PG, Park WB, Oh HS, Kim EJ, Nam EY, et al. Clinical progression and cytokine profiles of Middle East respiratory syndrome coronavirus infection. J Korean Med Sci. 2016;31:1717–1725. doi: 10.3346/jkms.2016.31.11.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Lo Cascio A, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwon JS, Kim JY, Kim MC, Park SY, Kim BN, Bae S, et al. Factors of severity in patients with COVID-19: cytokine/chemokine concentrations, viral load, and antibody responses. Am J Trop Med Hyg. 2020;103:2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee JS, Park S, Jeong HW, Ahn JY, Choi SJ, Lee H, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park SH, Kang K, Giannopoulou E, Qiao Y, Kang K, Kim G, et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat Immunol. 2017;18:1104–1116. doi: 10.1038/ni.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park SH, Park-Min KH, Chen J, Hu X, Ivashkiv LB. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat Immunol. 2011;12:607–615. doi: 10.1038/ni.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith JC, Sausville EL, Girish V, Yuan ML, Vasudevan A, John KM, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev Cell. 2020;53:514–529.e3. doi: 10.1016/j.devcel.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, 3rd, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Busnadiego I, Fernbach S, Pohl MO, Karakus U, Huber M, Trkola A, et al. Antiviral activity of type I, II, and III interferons counterbalances ACE2 inducibility and restricts SARS-CoV-2. Mbio. 2020;11:e01928-20. doi: 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hensley LE, Fritz EA, Jahrling PB, Karp CL, Huggins JW, Geisbert TW. Interferon-β1a and SARS coronavirus replication. Emerg Infect Dis. 2004;10:317–319. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ströher U, DiCaro A, Li Y, Strong JE, Aoki F, Plummer F, et al. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon-α. J Infect Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RWAL, et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J Gen Virol. 2013;94(Pt 8):1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stanifer ML, Kee C, Cortese M, Zumaran CM, Triana S, Mukenhirn M, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sallard E, Lescure FX, Yazdanpanah Y, Mentre F, Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Q, Chen V, Shannon CP, Wei XS, Xiang X, Wang X, et al. Interferon-α2b treatment for COVID-19. Front Immunol. 2020;11:1061. doi: 10.3389/fimmu.2020.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Monk PD, Marsden RJ, Tear VJ, Brookes J, Batten TN, Mankowski M, et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meng Z, Wan T, Chen L, Chen X, Li L, Qin X, et al. An experimental trial of recombinant human interferon alpha nasal drops to prevent COVID-19 in medical staff in an epidemic area. medRxiv. 2020 Jan 01; doi: 10.2174/1568026621666210429083050. [Preprint] [DOI] [PubMed] [Google Scholar]
- 94.Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, et al. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890.e6. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 95.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Major J, Crotta S, Llorian M, McCabe TM, Gad HH, Priestnall SL, et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee JS, Shin EC. The type I interferon response in COVID-19: implications for treatment. Nat Rev Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang N, Zhan Y, Zhu L, Hou Z, Liu F, Song P, et al. Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID-19 patients. Cell Host Microbe. 2020;28:455–464. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]