Abstract
Exosomes, the nanoscale phospholipid bilayer vesicles, enriched in selected proteins, nucleic acids and lipids, which they participated in a variety of biological processes in the body, including physiology and pathology. CircRNAs (circular RNAs) are a class of single-stranded closed molecules with tissue development specific expression patterns that have crucial regulatory functions in various diseases. Non-coding RNAs (such as microRNAs and long non‑coding RNAs) in exosomes have also been shown to play an important regulatory role in humans. However, little research has focused on exosomal circRNAs. Recently, CircRNAs have been identified to be enriched and stably expressed in exosomes. In this review, we summarize the biogenesis and biological functions of exosomes and circRNA, and further revealed the potential role of exosome-derived circRNA in different diseases. Besides, we propose its use as a diagnostic marker and therapeutic punctuation for diseases, especially in cancer.
Keywords: Exosomes, circRNAs, Exosomal circRNAs, Function, Liquid biomarker
Introduction
Exosomes, 30–150 nm in size, originate from the membrane vesicles of the endosomes and are secreted by almost all cells from different organisms, and are considered to be key roles in biological processes under normal and pathological conditions [1, 2]. Exosomes contain a variety of functional molecules, including various growth factors, proteins, metabolites, DNA, lipids, transfer RNA (tRNA), messenger RNA (mRNA), ribosomal RNA (rRNA), microRNA (miRNA), long non‑coding RNA (lncRNA), circRNA and so on [3]. Numerous studies have demonstrated that exosomes are crucial mediators of intercellular communication. After exosomes are released into the tissue fluid, through a series of movements, exosomes arrive at the recipient cells and begin to deliver their internal active substances (such as circRNAs), thus initiating functional responses and inducing subsequent phenotypic changes [4–6].
Exosomes not only target the target cells to trigger downstream signals but also transfer genetic material to target cells, exerting anti-inflammatory, antiapoptotic and immune-suppressive effects, promoting tissue repair and improving cytokine levels [7, 8]. In addition, exosomes are involved in the development of many pathological processes such as in cancer, diseases of the nervous system and endocrine system [9–11]. Exosomes accumulate and circulate freely in different body fluids, and also contain a variety of functional molecular carriers. Therefore, exosomes may become a very promising non-invasive liquid biomarker, which will play an important role in future clinical diagnosis and treatment [12, 13]. CircRNAs are new endogenous non-coding RNAs, which are produced by splicing events or back-splicing events via exons or (and) introns circularization of the original transcription [14]. With the classical linear RNA with 5′caps and 3′poly(A) tails, circRNAs have a covalent closed-loop structure to avoid being degraded by RNA external enzymes or RNase R, so the number of circRNAs is more stable than linear mRNA, which makes them more rich than the typical linear transcription products of the same gene [15]. Due to the application of RNA sequencing (RNA-seq) technology coupled with novel bioinformatics approaches, circRNA has been found to occur widely and stably in human, animal and plant cells, even in mammalian tissues [16]. CircRNAs could regulate gene expression at transcriptional, posttranscriptional and translational levels. They participate in many pathological processes such as Alzheimer’s disease, diabetes, atherosclerosis and myogenesis through regulating alternative splicing, sponging miRNAs, sequestering functional proteins or even encoding proteins [17–20]. Particularly, circRNAs play an important role in cancer growth, metastasis, recurrence and therapy resistance [21]. Surprisingly, recent studies have shown that large and stable expression of circRNA can be detected in exosomes extracted from human circulation and urine [22]. For instance, it has been reported that tumor-derived exosomes are involved in the development, invasion and metastasis of gastric cancer, and circRNA can be transmitted to recipient cells through exosomes to promote the malignant process of gastric cancer [23, 24]. Moreover, exosomal circRNAs were also found in pancreatic ductal adenocarcinoma (PDAR) [25], hepatocellular carcinoma [26], platelet-derived extracellular vesicles [27]. As can be seen from the above studies, exosome circRNAs may participate in a variety of life activities and pathological processes in the human body, and the combination of the two may further increase the potential clinical applications of these molecules as diagnostic and prognostic markers. In the past, exosomal lncRNAs and miRNAs have been widely reported, but relatively little attention has been devoted to exosomal circRNAs. In this review, we briefly address the biogenesis and biological functions of exosome and circRNAs, the significance and role of exosomal circRNAs in various diseases. Besides, we propose exosomal circRNAs can as a promising diagnostic marker and therapeutic punctuation for diseases, especially in cancer.
Biogenesis and function of exosome
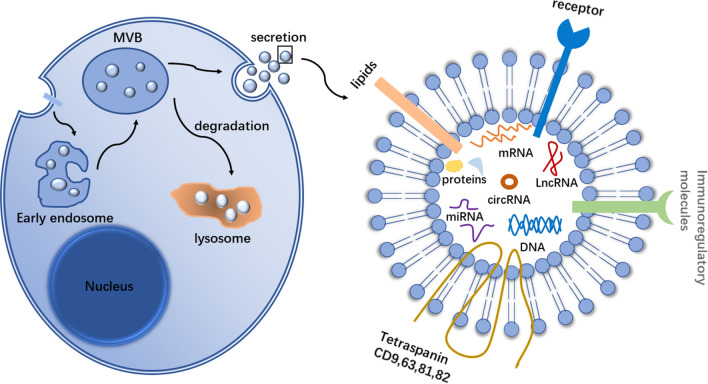
Exosomes are a type of extracellular vesicle consisting of a phospholipid bimolecular cell membrane, which are found in various body fluids such as blood, saliva, urine, breast milk and so on [28]. Early endosomes were formed by intracavity vesicles on the plasma membrane, which were further folded by the phospholipid bilayer to produce multiple intracavity vesicles, thus forming multi-vesicular body (MVB). It could not only fuse with the plasma membrane to release its contents, namely exosomes, but also transport to lysosomes to degrade the contents of vesicles [29, 30]. In addition, the membranes of exosomes are rich in a variety of proteins and lipids, such as cholesterol and sphingolipids. There are other proteins on the surface of exosomes, including four major transmembrane proteins (CD9, CD63, CD81 and CD82), which are enriched on their membranes and considered as ideal markers for exosome characterization [31], as shown in Fig. 1. It is noteworthy that exosomes play an important role in regulating normal physiological processes, such as repair ischemic injury [32], healthy pregnancy [33] and tissue repair [34]. In addition, exosomes also participate in the development of various diseases in the human body, playing a role that cannot be ignored. Studies have shown that CD317 protein and epidermal growth factor receptor (EGFR) are highly concentrated on the surface of exosomes in serum of non-small cell lung cancer (NSCLC), which is therefore considered as an important marker for the diagnosis of NSCLC [35]. Exosomes released by cancer cells can mediate phenotypical changes in the cells of tumor microenvironment (TME) to promote tumor growth and therapy resistance, for example, fibroblast- and macrophages-induced differentiation [36]. Therefore, exosomes are biomarkers with diagnostic and prognostic significance, and we will further study their potential in clinical application.
Fig. 1.
Biogenesis and structure of exosome
Early endosomes were formed by intracavity vesicles on the plasma membrane, which were further folded by the phospholipid bilayer to produce multiple intracavity vesicles, thus forming MVB. It can fuse with the plasma membrane to release its contents, namely exosomes, also transport to lysosomes to degrade the contents of vesicles.
The structure of exosomes include, tetraspanins (CD9, CD63, CD81, CD82). Lipid rafts, such as sphingolipids, cholesterol. Immunoregulatory molecules include major histocompatibility complex I, II (MHC I, MHC II), CD86. The receptors on the exosome membrane are different depending on the host cell, eg. EGFR. Exosome contents have proteins, nucleic acids such as DNA, tRNA, mRNA, rRNA, miRNA, lncRNA, circRNA and so on.
Isolation and detection of exosomes
Exosomes have great potential in clinical diagnosis, biomarker detection, target recognition of molecular therapy and personalized medical application. However, there are still some obstacles such as super-sensitive detection technology, efficient isolation method and quantitative analysis. In the clinical environment, a fast and reliable test standard is urgently needed. A large number of approaches have been used for exosome and extracellular vesicle (EV) isolation, such as ultracentrifugation, density-gradient centrifugation, size exclusion chromatography, microfiltration, immunomagnetic beads, and several polymer-based precipitation techniques [37–41]. These separation techniques rely either on the size and density of the EVs or the existing specific surface proteins on the exosomes such as, CD9, CD63, CD81, TSG101 and HSP70 [41]. Fortunately, with the in-depth research, different approaches with recognition element were reported, including nanoparticles tracking analysis (NTA), tunable resistive pulse sensing (RPS), dynamic light scattering (DLS), flow cytometry, cryo-EM, a platform combining surface plasmon resonance (SPR) with atomic Force Microscope (AFM), or by other techniques with similar capabilities have been developed [42–44]. Besides, Western Blotting can be utilized to verify the presence of biomarker proteins in EVs. Traditional flow cytometry is suitable for identification of large vesicles, but not nanoscale ones. High-sensitivity flow cytometer (HSFCM) has been found that can increase the detection limit from about 500 nm to as low as 40 nm, making it possible to detect other large and small vesicle populations, such as exosomes [45]. Although these techniques and methods are fast-changing, there are still many limitations and shortcomings in their clinical application. First, many of methods require state-of-the-art instruments. Second, impure recoveries with regard to remnant matrix species (e.g., proteins, genetic material) and are performed on clinically irrelevant time and volume scales. Third, the selectivity for some specific exosomes is deficient. Fourth, the accuracy is often influenced by unreliable nanoparticle counting, which is from large proteins and lipoprotein particles, and even some exosomes less than a certain size being omitted [46]. We expect that in the future a technology will be developed to break down these barriers and translate exon detection of disease sources into clinical applications for early screening in primary care settings.
Origins, biogenesis and functions of circRNA
Origins of circRNA
CircRNA was discovered as early as the 1970s, in 1976, Sanger et al. obtained a source of viroids from tomatoes and purified the viroid RNA. Hydrodynamic and thermo-dynamic studies proved that circRNA exists in the viroid [47], which is an earlier research study we discovered that proved the existence of circRNA. But CircRNAs were thought to be a result of splicing errors for several decades, so scientists have paid little attention to how the molecule works in humans [15]. Subsequently, circRNAs have since been found in humans, mice, rats, fungi and other organisms [48–50]. However, with the development of modern technology, many advanced methods for circRNA detection and identification have been developed, such as bioinformatics and high-throughput sequencing technologies, which provide further convenience for us to study the biological functions of circRNA. Since then, circRNA-related research has grown rapidly and has gradually become a new star in the non-coding endogenous RNA field.
Biogenesis of circRNA
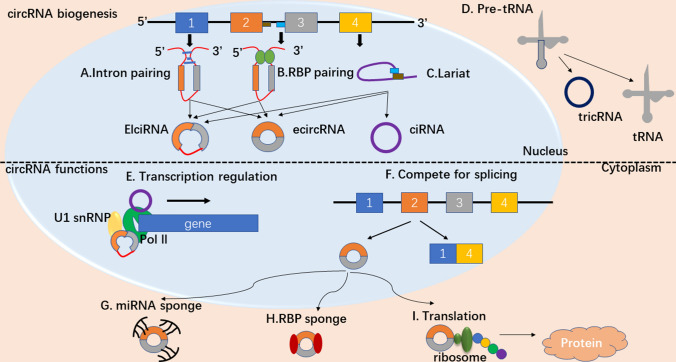
circRNAs differ from other RNAs in their remarkable continuous closed-loop structure, which is covalently linked by free 3′and 5′ends [51]. This closed-loop structure, which is also called a “back-splicing” structure, which require the spliceosome machinery to join a 5′ splice site (donor) of an exon to an 3′ splice site (acceptor) [52]. The biogenesis of circRNAs as shown in Fig. 2, which can be divided into three types: ciRNAs (circular intron RNAs), Ecircnas (circular exon RNAs), and EIciRNAs (exon–intron circular RNAs) [53]. The formation mechanism is as follows: the cycle driven by intron pairing is realized by direct base pairing of introns on both sides of complementary sequences or reverse repeats [54, 55]. In the RBP (RNA-associated binding protein) associated pairing drive, the RBPs are embedded into the precursor RNA during transcription and bind to the lateral intron sequence, causing the precursor RNA to bend and fold to form a circular cross region [56]. RBP pairing and intron pairing drive circularization in the direct back-splicing pathway of circRNA formation. In lariat-driven models, it takes place in exon jump events or the removal of introns from the precursor mRNAs [57]. In addition, another conserved mode of circRNA biogenesis is a small part of intron-derived circRNAs are from pre-tRNA. During pre-tRNA maturation, the tRNA splicing endonuclease complex cuts an intron-containing pre-tRNA at a canonical bulge-helix bulge (BHB) motif; and the intron termini are also joined by RtcB ligase to form a stable circRNA, named tricRNA [58, 59], as shown in Fig. 2.
Fig. 2.
Biogenesis and function of circRNAs. a Intron pairing driven circularization. The cycle driven by intron pairing is realized by direct base pairing of introns on both sides of complementary sequences or reverse repeats. The introns are removed or retained to form ecircRNA or EIciRNA. b RBP pairing driven circularization. The RBPs are embedded into the precursor RNA during transcription and bind to the lateral intron sequence, causing the precursor RNA to bend and fold to form a circular cross region. c Lariat-driven circularization. It takes place in exon jump events or the removal of introns from the precursor mRNAs. d TricRNAs are synthesized from introns spliced from pre-tRNA. The tRNA splicing endonuclease complex cuts an intron-containing pre-tRNA at a canonical bulge-helix bulge (BHB) motif; and the intron termini are also joined by RtcB ligase to form a stable circRNA, named tricRNA. e EIciRNAs or ciRNAs can interact with transcription complexes in the promoter region of their host gene to induce gene transcription by interacting with U1 snRNP or RNA pol II. f The back-splicing and linear splicing can compete with each other during splicing. As a result, a linear RNA or an ecircRNA is generated. g circRNAs can act as miRNA sponges to inhibit miRNA activity. h circRNAs can interact with RBPs and affect their functions and translocations. i circRNAs have protein-coding capacity and can encode proteins
Functions of circRNA
CircRNAs are new molecules in the field of ncRNA research, and their biological functions have been extensively studied. Their functions can be divided into four parts: as miRNA sponges regulating their functions, as transcription or translation regulators, interacting with proteins to regulate gene expression, and having the potential to encode proteins. These functions are shown in Fig. 2.
miRNA sponge
CircRNAs have been shown to act as miRNA sponges that can isolate miRNAs from their target mRNAs, thereby alleviating miRNA-mediated gene suppression or expression. The most typical example is Cdr1as, abundant circRNAs in mammalian brain, which contains more than 70 conserved miR7 binding sites and miR-671 sites that are completely complementary to the entire mature miR-671 sequence and can directly regulate the function of miRNAs and their collaborative gene [60]. CircHIPK3 has been shown to promote cancer cell growth by binding to multiple miRNAs, including the tumor suppressor miR-124 [61]. The existing results significantly demonstrate that the circRNA-miRNA interaction is critical for the organism.
Regulation of transcription and alternative splicing
Although circRNAs are mostly exons and distributed in the cytoplasm, ciRNA and EIciRNA are abundant in the nucleus and play a role as transcriptional regulatory cis-elements [62]. Some circRNAs regulate the transcription of their parent genes by promoting the extended activity of RNA polymerase II (Pol II) [62, 63]. EIciRNA with Pol II and its coding gene U1 small nuclear ribonucleoprotein (U1 snRNP) promoter interaction, enhance its parent gene transcription [64]. Therefore, although ciRNA and EIciRNA cannot function as miRNA sponges, they can regulate gene expression and transcription through other pathways, which has been confirmed in studies. Back-splicing can also compete with linear mRNA splicing, so disruption of the balance between circular and linear RNA may promote abnormal gene expression, which may be further involved in pathological activity [56].
Translation
Some circRNAs have an open reading frame (ORF) and an internal ribosomal binding site (IRES), showing potential for protein translation [65–67]. CircLINC-PINT can encode the peptide chain and inhibit the transcription of oncogenes, thus playing an anticancer role in glioblastoma [68]. Another study showed that in the presence of translation initiation factors EIF4G2 and YTHDF3, modification of N6-methyladenosine (m6A) was sufficient to promote the initiation of circRNAs protein translation in human cells [69].
Interactions with RBPs
CircRNAs may have the potential to regulate protein function by binding, storing, sequencing and isolating proteins to specific subcellular locations, and act as dynamic scaffold molecules regulating protein–protein interactions [70]. Recent studies have shown that eight RNA-binding proteins interact with circRNAs in liver cancer cells, and SCD-circRNA 2 can regulate RNA-binding sequence protein 3 (RBM3) and play an important role in liver cancer [71]. On the other hand, circRNAs can influence the translation process by competitively binding RBP to their mRNAs,and the extensive binding of circPABPN1 to HuR prevented the interaction between HuR and PABPN1, thus inhibiting the translation of PABPN1 [72]. All the evidence suggests that circRNAs are more likely to regulate protein function by acting as scaffolds for protein complexes rather than simply binding to individual proteins.
Origin and mechanism of exosomal circRNAs
Numerous studies have shown that exosomes contain various nucleic acid substances, such as mRNA, miRNA, lncRNA, rRNA, circRNA, etc., when they transferred to the recipient cells, causing a series of biological reactions and play an important role in regulating the physiological process of cells [73–75]. Moreover, the rich and stable expression of circRNAs in extracellular vesicles has been identified, providing new clues and support for the functional research of circRNAs. It was shown that compared with parental cells, circRNAs in exosomes were more abundant, larger in number and more stable in expression than linear mRNA subtypes [22, 76]. Possible mechanisms for circRNAs to be more abundant in exosomes than in its producing cells include, first, the relatively long half-life of circRNAs, as they are covalently closed-loop structures without a poly A tail or a 5′–3′ end, thereby avoiding degradation by endonuclease [62]. Studies have shown that most circRNAs have a half-life of more than 48 h, while linear RNAs have a half-life of less than 20 h. Therefore, compared with linear transcripts in cells, circRNAs are more stable and enriched [15]. Second, the extracellular vesicles release circRNAs, which are then exported to the extracellular microenvironment to form a mechanism for circRNA clearance, so circRNAs are more enriched in the extracellular space [77]. Finally, due to the protective effect or certain characteristics of the exosome envelope, it can safely prevent further removal of contents or degradation by RNA enzyme, so exosomal circRNAs are highly stable and abundant [78]. Exosomes contain different types of active substances whose sources reflect the state and type of cells; however, exosomes from different or identical sources may contain the same or different molecules, indicating that the molecular subsets in exosomes are tissue-specific and uniquely species-specific. Reverse transcriptional quantitative PCR and high-throughput analysis were used to detect exosome-derived circRNA, and the results revealed the difference of circRNA types in serum and exosomes [22], which also indicated that circRNA entered exosomes actively, including selectivity in the sorting process [79]. In addition, studies have shown that RNA-binding complexes endosoma-sorting complex required for transport II (ESCRT‑II) play an important role in the development of MVB and may help EV to selectively incorporate RNA [1]. Other studies have shown that intracellular miRNA may regulate the process of circRNA entering exosomes [22].
In summary, we need to further investigate and elucidate the complex mechanisms by which RNAs selectively and specifically enters exosomes, and we speculate that multiple types of sorting mechanisms may be at work simultaneously. These findings will greatly expand the potential of circRNAs as a prognostic and diagnostic biomarker.
Exosomal circRNAs in cancers
Increasing evidences show that exosomal circRNAs play a crucial role in a variety of cancers and participate in many important biological processes, promoting or inhibiting cancer. It could provide us with further insights into the functional mechanism of exosomal circRNAs. Table 1 illustrates the role of some exosomal circRNAs in different cancers.
Table 1.
Dysregulation of exosomal circRNAs in cancer biology
| circRNA | Cancer type | Expression | Biological function | Mechanism | Citation |
|---|---|---|---|---|---|
| circ-0051443 | HCC | Downregulated | Promote cell apoptosis and arrest the cell cycle | circ-0051443/miR-331-3p/BAK1 | [80] |
| circ-DB | HCC | Upregulated | Promote growth and reduce DNA damage | circ-DB/miR-34a /USP7 | [81] |
| Cdr1as | HCC | Upregulated | Promote proliferation and migration | Cdr1as/miR-1270/AFP | [82] |
| circNRIP1 | GC | Upregulated | Promote tumour metastasis | circNRIP1/miR-149-5p/AKT1 | [83] |
| ciRS-133 | GC | Upregulated | Promote cancer-associated cachexia | ciRS-133/miR-133/PRDM16 | [5] |
| Circ-RanGAP1 | GC | Upregulated | Promote invasion and metastasis | circ-RanGAP1/miR-877-3p/VEGFA | [24] |
| circLONP2 | CRC | Upregulated | Promote invasion and metastasis | circLONP2 /miR-17/DGCR8 | [84] |
| ciRS-122 | CRC | Upregulated | Promote glycolysis and drug resistance | ciRS-122/miR-122/PKM2 | [85] |
| circ-ABCC1 | CRC | Upregulated | Promote cell stemness, sphere formation and metastasis | circ-ABCC1/Wnt /β-catenin | [86] |
| circIFT80 | CRC | Upregulated | Promote the proliferation, migration and invasion | circIFT80 /miR-1236-3p /HOXB7 | [87] |
| circFMN2 | CRC | Upregulated | Promote proliferation and migration | circFMN2/miR-1182 /hTERT | [88] |
| circ-PDE8A | PDAC | Upregulated | Promote proliferation and invasion | Circ-PDE8A /miR-338/MACC1 | [25] |
| Circ-IARS | PDAC | Upregulated | Promoting tumor invasion and metastasis | circ-IARS/miR-122/RhoA | [89] |
| CircPUM1 | ovarian cancer | Upregulated | Promote proliferation, migration and invasion | CircPUM1 /miR-615-5p/miR-6753-5p/NF-Κb/MMP2 | [90] |
| CircWHSC1 | ovarian cancer | Upregulated | Promote proliferation and metastasis | CircWHSC1/miR-145/miR-1182 | [91] |
| Cdr1as | ovarian cancer | Downregulated | Inhibit proliferation and promot cisplatin-induced cell apoptosis | Cdr1as/miR-1270/SCAI | [92] |
| CircSATB2 | NSCLC | Upregulated | Promotes proliferation, migration and invasion | circSATB2/miR-326/FSCN1 | [93] |
| Circ_0044516 | prostate cancer | Upregulated | Promote growth and metastasis | Circ_0044516/miR‐29a‐3p | [94] |
| CircPRMT5 | UCB | Upregulated | Promote growth and metastasis | circPRMT5/miR-30c/SNAIL1/E-cadherin | [95] |
| circRASSF2 | LSCC | Upregulated | Promote proliferation | circRASSF2/miR-302b-3p/IGF-1R | [96] |
| mc-COX2 | CLL | Upregulated | Promote proliferation and inhibit apoptosis | – | [97] |
| CircRASSF2 | PTC | Upregulated | Promote proliferation and inhibit apoptosis | CircRASSF2/miR-1178/TLR4 | [98] |
HCC hepatocellular carcinoma, GC gastric cancer, CRC colorectal cancer, PDAC pancreatic ductal adenocarcinoma, NSCLC non-small cell lung cancer, UCB urothelial carcinoma of the bladder, PTC papillary thyroid carcinoma, LSCC laryngeal squamous cell carcinoma, CLL chronic lymphocytic leukemia
Hepatocellular carcinoma (HCC)
Circ-0051443 is significantly lower in the plasma exosomes and tissues from patients with HCC than healthy controls. Circ-0051443 is transmitted from normal cells to HCC cells via exosomes and suppresses the malignant biological behaviors by promoting cell apoptosis and arresting the cell cycle. Therefore, exosomal circ-0051443 can serve as a predictor and potential therapeutic target for HCC [80].
exosome circ-deubiquitination (circ-DB) is up-regulated in HCC patients with higher body fat ratios. Moreover, exo-circ-DB promotes HCC growth and reduces DNA damage via the suppression of miR-34a and the activation of deubiquitination-related USP7 [81]. Exosomes extracted from HCC cells overexpressing circRNA Cdr1as accelerated the proliferative and migratory abilities of surrounding normal cells. In all, circRNA Cdr1as serves as a ceRNA to promote the progression of HCC. Meanwhile, it is directly transferred from HCC cells to surrounding normal cells exosomes to further mediate the biological functions of surrounding cells [82]. The study aimed to identify differentially-expressed circRNAs (DECs) in human blood exosomes from patients with HCC and to investigate their diagnostic value. Additionally, compared with the normal samples, hsa_circ_0004001, hsa_circ_0004123, hsa_circ_0075792, and a combination of the three were utilized as valuable diagnostic biomarkers in HCC [99].
Gastric cancer (GC)
It was proven that circNRIP1 can be transmitted by exosomal communication between GC cells, and exosomal circNRIP1 promoted tumor metastasis in vivo. In addition, circNRIP1 sponges miR-149-5p to affect the expression level of AKT1 and eventually acts as a tumor promotor in GC [83]. Exosomes derived from GC cells deliver ciRS-133 into preadipocytes, promoting the differentiation of preadipocytes into brown-like cells by activating PRDM16 and suppressing miR-133. Thus, exosome-delivered circRNAs are involved in white adipose tissue (WAT) browning and play a key role in cancer-associated cachexia [5]. Circ-RanGAP1 was significantly up-regulated in both GC tissues and exosomes from the plasma of GC patients. High circ-RanGAP1 expression was closely associated with an advanced TNM stage, lymph node metastases, and worse survival. The circ-RanGAP1-mediated miR-877-3p/VEGFA axis promotes GC progression [24]. Low hsa_circ_0065149 expression levels in GC tissues were significantly associated with tumor diameter, perineural invasion and longer overall survival. More important, hsa_circ_0065149 levels were significantly decreased in plasma exosomes of early GC patients. As a screening biomarker for early GC, hsa_circ_0065149 in plasma exosomes has higher sensitivity and specificity than traditional clinical biomarkers [100].
Colorectal cancer (CRC)
Studies indicate that circLONP2 acts as key metastasis-initiating molecule during CRC progression through modulating the intracellular maturation and intercellular transfer of miR-17, resulting in dissemination of metastasis-initiating ability in primary site and acceleration of metastasis formation in foreign organs. circLONP2 could serve as an effective prognostic predictor and/or novel anti-metastasis therapeutic target in CRC treatment [84]. Furthermore, exosomes from oxaliplatin-resistant cells delivered ciRS-122 to sensitive cells, thereby promoting glycolysis and drug resistance through miR-122 sponging and PKM2 upregulation [85]. Exosomes from CD133 cells carrying circ-ABCC1 can promote cell stemness, sphere formation and metastasis in CRC, unveiling that circ-ABCC1 serves as a novel candidate target for CRC treatment [86]. Studies have shown that hsa_circ_0067835 (circIFT80) and hsa-circ-0005100 (circFMN2) were significantly up-regulated in exosomes from CRC patients’ serum. They could promote the proliferation, migration and invasion of cancer cells. These results showed that these exosomal levels of circRNAs were tumor-derived and they can serve as a diagnostic or prognostic biomarker for CRC [87, 88]. Cancer-derived exosomal circPACRGL plays an oncogenic role in CRC proliferation and metastasis. CircPACRGL serves as a sponge for miR-142-3p/miR-506-3p to facilitate the transforming growth factor-β1 (TGF-β1) expression, providing mechanistic insights into the roles of circRNAs in CRC progression and a valuable marker for CRC treatment [101].
Pancreatic cancer
Exosomal circ-PDE8A was associated with progression and prognosis in PDAC patients. Circ-PDE8A acts as a ceRNA for miR-338 to regulate MACC1 and stimulates invasive growth via the MACC/MET/ERK or AKT pathways [25]. Circ-IARS was up-regulated in plasma exosomes in pancreatic cancer tissues and patients with metastatic disease, and was found to enter HUVECs through exosomes, promoting tumor invasion and metastasis. In addition, circ-IARS expression was positively correlated with liver metastasis, vascular infiltration and tumor-node-metastasis (TNM) stage, and negatively correlated with postoperative survival time, suggesting that circ-IARS may be a potential prognostic marker for pancreatic cancer [89].
Ovarian cancer
CircPUM1 up-regulated the expression of nuclear factor kappa B (NF-κB) and MMP2 by sponging miR-615-5p and miR-6753-5p to promote ovarian cancer proliferation, migration and invasion, which also acts on the peritoneum and contributes to metastasis of cancer in the form of cancer-derived exosomes [90]. CircWHSC1, which is highly expressed in ovarian cancer, acts on the peritoneal mesothelium in the form of exosomes to induce tumor metastasis. It is also found to promote tumor development through sponging miR-145 and miR-1182 [91]. Cdr1as was down-regulated in serum exosomes from cisplatin-resistant patients. Overexpression of Cdr1as inhibited cell proliferation and promoted the cisplatin-induced cell apoptosis in ovarian cancer cells by regulating the miR-1270/SCAI signaling pathway [92].
Breast cancer
Both of hsa-circRNA-00005795 and hsa-circRNA-0088088 may function as competing endogenous RNAs and may play vital roles in BCa, which hold significant value for the prediction of BCa by examining the expression of the circRNAs in serum exosomes from BCa patients compared with that in healthy donors. The study provides a theoretical basis for using stable exosomal circRNAs as new biomarkers for predicting the tumorigenesis, development and metastasis of BCa [102].
Lung cancer
The CXCR4-related circular RNA exo-hsa_circRNA_0056616 in exosomes was more expressed in lung adenocarcinoma tissues with lymph node metastasis. This circRNA represents a potential biomarker for predicting lymph node metastasis in lung adenocarcinoma [103]. CircSATB2 is highly expressed in serum exosomes of lung cancer patients, with high sensitivity and specificity in clinical detection, and is associated with lung cancer metastasis. In addition, circSATB2 promotes proliferation, migration and invasion of NSCLC cells through exosome. The results revealed that circSATB2 may be a promising diagnostic marker in lung cancer [93]. CircRNA-002178 could be delivered into CD8 T cells to induce PD1 expression via exosomes. It could be detected in exosomes of plasma from lung adenocarcinoma (LUAD) patients and could serve as biomarkers for LUAD early diagnosis [104].
Urinary system cancer
Circ_0044516 was verified to be significantly up-regulated in the exosomes from prostate cancer patients and the cell lines. Inhibition of circ_0044516 expression in prostate cancer cells decreased prostate cancer cell growth and metastasis, which regulated prostate cancer cell biological functions by sponging miR‐29a‐3p [94]. CircPRMT5 was up-regulated in serum and urine exosomes from patients with UCB, and significantly correlated with tumor metastasis. Clinical analysis from two independent UCB cohorts showed that the circPRMT5/miR-30c/SNAIL1/E-cadherin pathway was essential in supporting UCB progression [95].
Other tumors
There is a circRASSF2/miR-302b-3p/insulin-like growth factor 1 receptor (IGF-1R) axis in LSCC progression. Importantly, study demonstrated that circRASSF2 was up-regulated in serum exosomes from LSCC patients, indicating the significance of exosomal circRNAs in tumor cell proliferation [96]. Mitochondrial genome-derived (mt)-circRNAs were found to be highly expressed in the plasma exosomes of CLL patients. The endogenous reduction of mc-COX2 can affect mitochondrial functions, suppress cell proliferation, and induce cell apoptosis. The upregulation of mc-COX2 was positively associated with leukemogenesis and worsening survival of CLL patients. Our findings prove that mc-COX2, a critical mt-circRNA highly expressed in plasma, derived from CLL cells and delivered by exosomes, is associated with the progression and prognosis of CLL [97]. CircRASSF2 was up-regulated in serum exosomes from PTC patients. The study demonstrates that circRASSF2 modulates PTC progression through the miR-1178/TLR4 pathway, which indicate that circRASSF2 may serve as a promising therapeutic target for the treatment of PTC patients [98]. In terms of clinical diagnostic biomarkers, studies have demonstrated the profiles of differentially-expressed circRNAs in extracellular vesicles in serum of patients with thyroid papillary carcinoma [105] and endometrial carcinoma [106]. These results suggest that circRNAs derived from EV has diagnostic potential.
In summary, a large number of studies have shown the great potential of exosomes associated circRNAs as potential biomarkers in liquid biopsies, as illustrated in the Table 2. We will further explore the mechanisms involved in the disease process in the future.
Table 2.
Dysregulation of exosomal circRNAs in other diseases
| circRNA | Diseases | Expression | Biological function | Mechanism | Citation |
|---|---|---|---|---|---|
| CircHIPK3 | Cardiomyocytes | Upregulated | Decrease in oxidative stress-induced CMVECs dysfunction | CircHIPK3/miR-29a/IGF-1 | [107] |
| cZFP609 | Ischemic diseases | Upregulated | Inhibit endothelial angiogenic activity induced by hypoxia | cZFP609 /HIF1α/VEGFA | [108] |
| circHIPK3 | Ischemic injury of the skeletal muscle | Downregulated | Improve blood perfusion, inhibit ischemia induced pyroptosis | circHIPK3/miR-421 /FOXO3a | [32] |
| circ-Rtn4 | Osteoporosis | Upregulated | Increase cell activity and inhibit apoptosis | circ-Rtn4/miR-146a/TNF-α | [109] |
| circRNA-0077930 | Vascular smooth muscle aging | Upregulated | Promote cellular senescence of VSMCs | circRNA-0077930/miR-622/Kras | [110] |
CMVECs cardiac microvascular endothelial cells
Exosomal circRNAs in other disease
Not only do exosomal circRNAs play a vital role in tumors, but also play an important role in diseases other than tumors, such as nervous system diseases and cardiovascular diseases, as summarized in Table 3.
Table 3.
Exosomal circRNAs in clinical applications
| CircRNA | Disease type | Specimen source | Specimen type | Expression | AUC | Citation |
|---|---|---|---|---|---|---|
| hsacirc_020135 | PTC | PTC/benign thyroid goiter | Serum | Downregulated | – | [105] |
| hsa circ 0002577 | Endometrial cancer | Endometrial cancer/healthy | Serum | Upregulated | – | [106] |
| hsa_circ_0065149 | GC | GC/healthy/gastritis | Plasma | Downregulated | 0.769 | [100] |
| three-circRNA signature | HCC | HCC/healthy | Serum | Upregulated | 0.89 | [99] |
| hsa-circRNA-0088088 | Breast cancer | Breast cancer/healthy | Serum | Upregulated | – | [102] |
| hsa-circRNA-00005795 | Breast cancer | Breast cancer/healthy | Serum | Downregulated | – | [102] |
| CircRNA-002178 | LUAD | LUAD/healthy | Serum | Upregulated | 0.9956 | [104] |
| hsa_circRNA_0056616 | LUAD | With lymph node metastasis/without | Plasma | Upregulated | 0.812 | [103] |
| Circ-0051443 | HCC | HCC/healthy | Plasma | Downregulated | 0.8089 | [80] |
| circ-0004771 | CRC | CRC/healthy | Serum | Upregulated | 0.88 | [114] |
| hsa_circ_0087862 | Immune-mediated demyelinating | Immune-mediated demyelinating/healthy | Cerebrospinal fluid | Upregulated | 1 | [115] |
| circ-KIAA1244 | GC | GC/healthy | Plasma | Downregulated | 0.7481 | [116] |
PTC papillary thyroid carcinoma, GC gastric cancer, HCC hepatocellular carcinoma, LUAD lung adenocarcinoma
Human platelets are particularly rich in circRNA compared to other hematopoietic cell types, and since platelets are essential for central physiological processes such as hemostasis, wound healing, inflammation, and cancer metastasis, these findings could greatly expand the potential of circRNA as a prognostic and diagnostic biomarker [27, 111, 112]. Exosomes from the extracellular space of the brain, where differentially-expressed circRNAs might be related to the growth and repair of neurons, the development of the nervous system, and the transmission of nerve signals. The most highly correlated pathways that identified were involved primarily with glutamatergic synapse and the cyclic guanosine monophosphate-protein kinase G signaling pathway [113]. CircHIPK3 in hypoxic exosomes (HPC-exos) plays a role in cardiac microvascular endothelial cells (CMVECs) under oxidative conditions through miR-29a-mediated IGF-1 expression, leading to a decrease in oxidative stress-induced CMVECs dysfunction. These data suggest that the exosomal circRNA in CMs is a potential target to control CMVECs dysfunction under oxidative conditions [107].
To explored the role of smooth muscle SIRT1 in endothelial angiogenesis after ischemia and the underlying mechanisms, we performed a femoral artery ligation model using VSMC specific human SIRT1 transgenic (−Tg) and knockout (KO) mice. −Tg VSMCs inhibited endothelial angiogenic activity induced by hypoxia via the exosome cZFP609. The cZFP609 was delivered into ECs, and detained HIF1α in the cytoplasm via its interaction with HIF1α, thereby inhibiting VEGFA expression and endothelial angiogenic functions [108].
Conclusion
As an important carrier of exosomes, circRNAs play an important regulatory role in various physiological and pathological biological processes, such as inflammation, tissue repair, cell proliferation, migration, invasion, metastasis, apoptosis, and chemotherapy & radiotherapy resistance. Due to their unique features and high specificity, their ability to mediate cell communication, what is more, exosomal circRNAs is abundant, stable, and present in different extracellular fluids, including saliva, blood, and urine [28], making it an ideal diagnostic marker and therapeutic target as well as potential vectors for drug delivery. However, compared with other ncRNAs, such as miRNA and lncRNA, the studies of circRNAs in exosomes are still incomplete. Therefore, we need to further explore its functional mechanism.
At present, the study of the structure of endogenous circRNAs sponges may contribute to the design and development of effective artificial sponges to regulate disease progression. As a stable and effective miRNA inhibitor, the artificial miRNA "sponge" technique may be a new strategy for RNA gene therapy in the future. It can simultaneously inhibit the expression of other miRNAs and produce a more long-lasting inhibiting effect. Therefore, the use of nanoparticles to deliver circRNAs to generate specific types of exosomes provides an option for selective targeted cancer cell elimination [117–120].
However, the specificity of exosomal circRNAs structure and the insufficiency of technology limit the discussion of its function. There are still many challenges and difficulties in clinical application. First, a standardized method for collecting, processing and isolating exosome samples has not yet been established [28, 121]. Ultracentrifugation method is the gold standard for exosome separation, but it is time and labor-consuming, requires a large amount of raw materials and expensive equipment, and is inefficient for high-throughput measurement [122], which cannot be effectively applied to clinical work. Second, in the clinical collection of patient samples, blood or other body fluids contain many impurities, such as protein complexes, nucleic acid lysates, etc., which are challenging to extract correct exosomes. Third, circRNA and its corresponding linear mRNA sequences are partially overlapped, so circRNA expression and function cannot be accurately assessed. Fourth, due to the low abundance of circRNAs, it is difficult to detect them in exosomes using accurate methods. Therefore, it is important to develop and use appropriate methods and techniques to elucidate the molecular mechanisms and regulatory networks of exosomal circRNAs.
Abbreviations
- AFM
Atomic force microscope
- CD
Cluster of differentiation
- CircRNAs
Circular RNAs
- COX
Cyclooxygenase
- EM
Electron microscope
- ESCRT
Endosoma-sorting complex required for transport
- EV
Extracellular vesicle
- MHC
Major histocompatibility complex
- MMP
Matrix metalloprotease
- HSP
Heat shock protein
- HUVECS
Human umbilical vein endothelial cells
- lncRNA
Long non-coding RNA
- miRNA/miR
Micro-RNA
- mRNA
Messenger RNA
- MVB
Multi-vesicular body
- TEM
Transmission electron microscopy
Author contributions
Writing—original draft preparation: GX; writing—review and editing: GX, TW and WC. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 81774261, Project of Guangdong Bureau of Traditional Chinese Medicine, grant number 20191001 and 20171001.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Informed consent
For this type of study formal consent is not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
X. Guo, Email: guoxuan85111@163.com
W. Tan, Email: tanweioy@163.com
C. Wang, Email: gzwchj@126.com
References
- 1.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 2.Marbán E. The secret life of exosomes what bees can teach us about next-generation therapeutics. J Am Coll Cardiol. 2018;71(2):193–200. doi: 10.1016/j.jacc.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22:34–45. doi: 10.1038/cdd.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, Yang H, Sun W, Wang X, Zhu K, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144:2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, Li X, Chen J, Liu K, Li C, Zhu G. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.CAN-15-1625. [DOI] [PubMed] [Google Scholar]
- 7.Burrello J, Monticone S, Gai C, et al. Stem cell-derived extracellular vesicles and immune-modulation. Front Cell Dev Biol. 2016;4:83. doi: 10.3389/fcell.2016.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorge IM, Kim SY, Mano JF, Kalionis B, Chrzanowski W. Extracellular vesicles, exosomes and shedding vesicles in regenerative medicine—a new paradigm for tissue repair. Biomater Sci. 2017;6:60–78. doi: 10.1039/c7bm00479f. [DOI] [PubMed] [Google Scholar]
- 9.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian T, Zhang HX, He CP, Fan S, Zhu YL, Qi C, Huang NP, Xiao ZD, Lu ZH, Tannous BA, Gao J. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137–149. doi: 10.1016/j.biomaterials.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Pan Y, Hui X, Hoo RLC, Ye D, Chan CYC, Feng T, Wang Y, Lam KSL, Xu A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J Clin Invest. 2019;129:834–849. doi: 10.1172/JCI123069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiteside TL. The potential of tumor-derived exosomes for noninvasive cancer monitoring: an update. Expert Rev Mol Diagn. 2018;18:1029–1040. doi: 10.1080/14737159.2018.1544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whiteside TL. The emerging role of plasma exosomes in diagnosis, prognosis and therapies of patients with cancer. Contemp Oncol (Poznan, Poland) 2018;22:38–40. doi: 10.5114/wo.2018.73882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7) Genes. 2016;7(12):116. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5(1):12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lococo F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y, Fu G, Ye Y, Ming L. Exosomes participate in the carcinogenesis and the malignant behavior of gastric cancer. Scand J Gastroenterol. 2017;52:499–504. doi: 10.1080/00365521.2016.1278458. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, Zhang Y, Li H, Zhang Q, Yang Y, Chen G. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–445. doi: 10.1016/j.ebiom.2018.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preusser C, Hung LH, Schneider T, Schreiner S, Hardt M, Moebus A, Santoso S, Bindereif A. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. 2018;7:1424473. doi: 10.1080/20013078.2018.1424473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boriachek K, Islam MN, Moller A, Salomon C, Nguyen NT, Hossain MSA, Yamauchi Y, Shiddiky MJA. Biological functions and current advances in isolation and detection strategies for exosome Nanovesicles. Small. 2017;1702153:1–21. doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- 29.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 31.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113(8):E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bing Y, Zhang Yu, Chun L, et al. Stem cell-derived exosomes prevent pyroptosis and repair ischemic muscle injury through a novel exosome/circHIPK3/FOXO3a pathway. Theranostics. 2020;10:6728–6742. doi: 10.7150/thno.42259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czernek L, Düchler M. Exosomes as messengers between mother and fetus in pregnancy. Int J Mol Sci. 2020;21:4264. doi: 10.3390/ijms21124264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin S, Ji C, Wu P, Jin C, Qian H. Human umbilical cord mesenchymal stem cells and exosomes: bioactive ways of tissue injury repair. Am J Transl Res. 2019;11:1230–1240. [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;2015(4):1–10. doi: 10.3402/jev.v4.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos P, Almeida F. Role of exosomal miRNAs and the tumor microenvironment in drug resistance. Cells. 2020;9:1450. doi: 10.3390/cells9061450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szatanek R, Baj-Krzyworzeka M, Zimoch J, Lekka M, Siedlar M, Baran J. The methods of choice for extracellular vesicals (EVs) characterization. Int J Mol Sci. 2017;18(6):1153. doi: 10.3390/ijms18061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of extracellular v esicles: general methodologies and latest trends. Biomed Res Int. 2018;2018:8545347. doi: 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobb R, Möller A. Size exclusion chromatography: a simple and reliable method for exosome purification. In: Methods in molecular biology, Clifton. 2017. [DOI] [PubMed]
- 41.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, et al. Comparison of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Koritzinsky EH, Street JM, Star RA, Yuen PST. Quantification of exosomes. J Cell Physiol. 2017;232:1587–1590. doi: 10.1002/jcp.25387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(2018):1–43. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao J, Yang M, Duan Y. Chemistry, biology, and medicine of fluorescent nanomaterials and related systems: new insights into biosensing, bioimaging, genomics, diagnostics, and therapy. Chem Rev. 2014;114(12):6130–6178. doi: 10.1021/cr200359p. [DOI] [PubMed] [Google Scholar]
- 45.Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12(1):671–680. doi: 10.1021/acsnano.7b07782. [DOI] [PubMed] [Google Scholar]
- 46.He F, Liu H, Guo X, Yin BC, Ye BC. Direct exosome quantification via bivalent-cholesterol-labeled DNA anchor for signal amplification. Anal Chem. 2017;89:12968–12975. doi: 10.1021/acs.analchem.7b03919. [DOI] [PubMed] [Google Scholar]
- 47.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Lovell Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 49.Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA. 1996;93:6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Meng XW, Li X, Zhang PJ, et al. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18(4):547–557. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 54.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Kadener S, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 57.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017;14:978–984. doi: 10.1080/15476286.2017.1317911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt CA, et al. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res. 2019;47:6452–6465. doi: 10.1093/nar/gkz311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:6357. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 61.Zheng QP, Bao CY, Guo WJ, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Zhang X, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 64.Li ZY, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 65.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9(1):4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Y, Fan XJ, Mao MW, et al. Extensive translation of circular RNAs driven by N-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du WW, Zhang C, Yang WN, et al. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong W, Dai ZH, Liu FC, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155–167. doi: 10.1016/j.ebiom.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J, Wang Z, Chen X, Zhang W, Yokoyama S, et al. Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res. 2014;24:1164–1180. doi: 10.1038/cr.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shao N, Xue L, Wang R, Luo K, Zhi F, Lan Q. MiR-454-3p is an exosomal biomarker and functions as a tumor suppressor in glioma. Mol Cancer Ther. 2019;18:459–469. doi: 10.1158/1535-7163.MCT-18-0725. [DOI] [PubMed] [Google Scholar]
- 75.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS ONE. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, Kim JH, Choi DS, Kim YK, Hwang D, Gho YS. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genom. 2009;10:556. doi: 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei C, Yingyao Q, Shaoyi F, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 81.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Yang Su, Xiurui Lv, Wei Y, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) 2019;11:8182–8203. doi: 10.18632/aging.102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(20):2501–2515. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kai H, Feng-Wei W, Chen-Hui C, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60. doi: 10.1186/s12943-020-01184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, Liu R, Fan Q, Zhu K, Li J, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hong Z, Chen S, Fu Q. Exosomes from CD133 cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J Cell Biochem. 2020;121:3286–3297. doi: 10.1002/jcb.29600. [DOI] [PubMed] [Google Scholar]
- 87.Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang Y, Cui G. CircIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol Ther Nucleic Acids. 2019;18:375–387. doi: 10.1016/j.omtn.2019.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci. 2019;133:2463–2479. doi: 10.1042/CS20190715. [DOI] [PubMed] [Google Scholar]
- 89.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xue G, Zhi-Hong Z, Yao L, et al. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–892. doi: 10.1016/j.omtn.2019.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zong Zhi-Hong Du, Yu-Ping GX, et al. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38:437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao Z, Mei Ji, Qianqing W, et al. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids. 2019;18:24–33. doi: 10.1016/j.omtn.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Nan Z, Aruo N, Lijian C, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian Li, Xiangzhou S, Liheng C. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121:2118–2126. doi: 10.1002/jcb.28239. [DOI] [PubMed] [Google Scholar]
- 95.Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 96.Tian L, Cao J, Jiao H, Zhang J, Ren X, Liu X, Liu M, Sun Y. CircRASSF2 promotes laryngeal squamous cell carcinoma progression by regulating the miR-302b-3p/IGF-1R axis. Clin Sci (Lond) 2019;133:1053–1066. doi: 10.1042/CS20190110. [DOI] [PubMed] [Google Scholar]
- 97.Zijuan Wu, Handong S, Chunling W, et al. Mitochondrial genome-derived circRNA mc-COX2 functions as an oncogene in chronic lymphocytic leukemia. Mol Ther Nucleic Acids. 2020;20:801–811. doi: 10.1016/j.omtn.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guojun Wu, Wenhong Z, Xiaona L, et al. circRASSF2 acts as ceRNA and promotes papillary thyroid carcinoma progression through miR-1178/TLR4 signaling pathway. Mol Ther Nucleic Acids. 2020;19:1153–1163. doi: 10.1016/j.omtn.2019.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Xiang-Hong S, Yu-Tong W, Guo-Fu Li, et al. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:226. doi: 10.1186/s12935-020-01302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yongfu S, Xueping X, Lu R, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. 2020;26:1475–1482. doi: 10.1007/s12253-019-00716-y. [DOI] [PubMed] [Google Scholar]
- 101.Shang Anquan Gu, Chenzheng WW, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Su-Jin Y, Dan-Dan W, Si-Ying Z, et al. Identification of circRNA-miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics. 2020;12:101–125. doi: 10.2217/epi-2019-0058. [DOI] [PubMed] [Google Scholar]
- 103.Fei He, Xuejing Z, Zheng L, et al. Plasma exo-hsa_circRNA_0056616: a potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer. 2020;11:4037–4046. doi: 10.7150/jca.30360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.JunFeng W, XuHai Z, YanBo W, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang C, Wei Y, Yu L, Xiao Y. Identification of altered circular RNA expression in serum exosomes from patients with papillary thyroid carcinoma by high-throughput sequencing. Med Sci Monit. 2019;25:2785–2791. doi: 10.12659/MSM.915658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu H, Gong Z, Shen Y, Fang Y, Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10:187–197. doi: 10.2217/epi-2017-0109. [DOI] [PubMed] [Google Scholar]
- 107.Yan W, Ranzun Z, Weiwei L, et al. Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid Med Cell Longev. 2019;2019:7954657. doi: 10.1155/2019/7954657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yong-Qing D, Peng K, Chang-Lin Li, et al. Smooth muscle SIRT1 reprograms endothelial cells to suppress angiogenesis after ischemia. Theranostics. 2020;10:1197–1212. doi: 10.7150/thno.39320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cao G, Meng X, Han X, et al. Exosomes derived from circRNA Rtn4-modified BMSCs attenuate TNF-α-induced cytotoxicity and apoptosis in murine MC3T3-E1 cells by sponging miR-146a. Biosci Rep. 2020;40. [DOI] [PMC free article] [PubMed]
- 110.Wang S, Zhan J, Lin X, et al. CircRNA-0077930 from hyperglycaemia-stimulated vascular endothelial cell exosomes regulates senescence in vascular smooth muscle cells. Cell Biochem Funct. 2020. [DOI] [PubMed]
- 111.Smyth SS, McEver RP, Weyrich AS, Morrell CN, Hoffman MR, Arepally GM, French PA, Dauerman HL, Becker RC, Platelet CP. Platelet functions beyond hemostasis. J Thromb Haemost. 2009;7:1759–1766. doi: 10.1111/j.1538-7836.2009.03586.x. [DOI] [PubMed] [Google Scholar]
- 112.French PA. Platelet functions beyond hemostasis, antiplatelet therapies in high-risk patient subgroups: the fourth annual Platelet Colloquium. J Thromb Thrombolysis. 2009;28:252–254. doi: 10.1007/s11239-009-0334-2. [DOI] [PubMed] [Google Scholar]
- 113.Zhao RT, Zhou J, Dong XL, Bi CW, Jiang RC, Dong JF, Tian Y, Yuan HJ, Zhang JN. Circular ribonucleic acid expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice. J Neurotrauma. 2018;35:2056–2066. doi: 10.1089/neu.2017.5502. [DOI] [PubMed] [Google Scholar]
- 114.Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jinting He, Ming R, Haiqi Li, et al. Exosomal circular RNA as a biomarker platform for the early diagnosis of immune-mediated demyelinating disease. Front Genet. 2019;10:860. doi: 10.3389/fgene.2019.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tang Weiwei Fu, Kai SH, et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17:137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Usman WM, Pham TC, Kwok YY, Vu LT, Ma V, Peng B, Chan YS, Wei L, Chin SM, Azad A, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9:2359. doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Roma-Rodrigues C, Raposo LR, Cabral R, Paradinha F, Baptista PV, Fernandes AR. Tumor microenvironment modulation via gold nanoparticles targeting malicious exosomes: implications for cancer diagnostics and therapy. Int J Mol Sci. 2017;18:E162. doi: 10.3390/ijms18010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim SM, Yang Y, Oh SJ, Hong Y, Seo M, Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release. 2017;266:8–16. doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 120.Qi H, Liu C, Long L, Ren Y, Zhang S, Chang X, Qian X, Jia H, Zhao J, Sun J, et al. Blood exosomes endowed with magnetic and targeting properties for cancer therapy. ACS Nano. 2016;10:3323–3333. doi: 10.1021/acsnano.5b06939. [DOI] [PubMed] [Google Scholar]
- 121.Maas SL, de Vrij J, van der Vlist EJ, Geragousian B, van Bloois L, Mastrobattista E, Schiffelers RM, Wauben MH, Broekman ML, Nolte-‘t Hoen EN. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J Control Release. 2015;200:87–96. doi: 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Minciacchi V, Zijlstra A, Rubin MA, Di Vizio D. Extracellular vesicles for liquid biopsy in prostate cancer: where are we and where are we headed? Prostate Cancer Prostatic Dis. 2017;20:251–258. doi: 10.1038/pcan.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]