Abstract
Sickle cell disease (SCD) and APOL1 G1/G2 variants increase chronic kidney disease (CKD) risk in African Americans by poorly understood mechanisms. We applied bioinformatics to identify new candidate genes associated with SCD-related CKD. An interaction network demonstrated APOA1 connecting HBB and APOL1 with 36 other candidate genes. Gene expression revealed upregulation of ELMO1 and down-regulation of APOA1 in the kidney cortex of SCD vs. non-SCD mice. Analysis of candidate genes identified ELMO1 rs10951509 to be associated with albuminuria and APOA1 rs11216132 with hemoglobinuria in SCD patients. A bioinformatic approach highlights ELMO1 and APOA1 as potentially associated with SCD nephropathy.
Keywords: Kidney disease, Sickle cell disease, APOL1, ELMO1, APOA1
When the hemoglobin S mutation, an E6V variant in the HBB gene, is inherited in the homozygous state (sickle cell anemia, HbSS), up to 44% of adults have chronic kidney disease (CKD),(1) which is approximately a two-fold higher prevalence than the general African-American adult population.(2) Homozygosity or compound heterozygosity for G1 and G2 variants of APOL1, encoding apolipoprotien L1, are commonly observed in people of African descent and account for up to 70% of the risk for kidney disease in non-diabetic African Americans.(3) The molecular mechanisms for how the HbS mutation or the APOL1 risk variants lead to CKD are not well understood.
A bioinformatic approach may help to identify candidate genes and pathophysiologic pathways for the development of kidney disease. This approach was previously used by Chasman and colleagues to identify candidate genes based on biological connections to 24 seed genes associated with kidney function in genome-wide association studies in European populations.(4) For example, LRP2, which encodes megalin and functions to reabsorb low molecular weight proteins from the urine, was selected as a candidate gene based on its connection to the seed gene, DAB2. The LRP2 rs10490130 variant was then found to be associated with estimated glomerular filtration rate in both European and African-American cohorts.
The purpose of this study was to identify genes associated with kidney disease in patients with sickle cell disease (SCD) using 1) gene interaction networks and pathway analysis, 2) gene expression studies in the kidney cortex of transgenic sickle cell mice, and 3) testing the association of candidate gene variants with kidney disease in SCD patients.
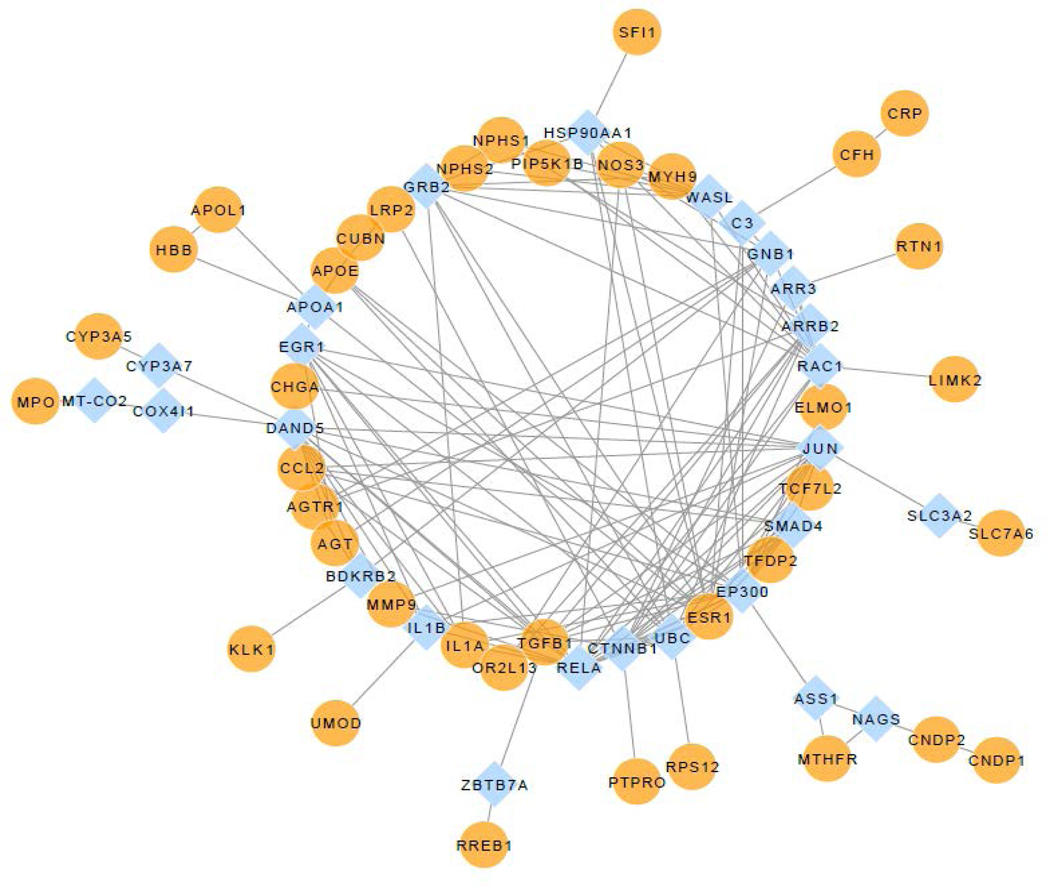
We performed a literature search for genetic variants associated with CKD in African Americans using the search terms, “gene” and “kidney” and “African”. We found 961 articles published prior to 12/31/2019 and selected 148 additional studies cited by these articles. Forty-six candidate genes with replicated variants associated with CKD in African Americans were identified (Supplementary Table 1–4). An interaction network was developed based on these 46 genes plus APOL1 and HBB using Cytoscape for the analysis.(5) This approach identified a network that was dependent on 36 of the candidate genes plus HBB, APOL1, and 26 additional genes not yet implicated in CKD (Figure 1). APOA1, encoding apo-lipoprotein A1, a gene not previously associated with CKD in African Americans, provided a functional connection among HBB, APOL1 and the other candidate genes in the network.
Figure 1.
Interaction network of candidate genes associated with kidney disease in African Americans. Orange circles (n = 38) represent the candidate genes, HBB, and APOL1; cyan diamonds (n = 26) represent the linker genes that facilitate connection of the candidate genes.
The DAVID bioinformatics resources (6) were used to develop gene enrichment pathway analysis using the 64 genes (36 candidate genes, 26 additional genes, HBB, and APOL1) included in this interaction network. Pathways with a Benjamini-adjusted P-value ≤ 0.01 were considered statistically significant. A gene enrichment pathway analysis using all 64 genes in the gene interaction network revealed 11 Kegg pathways that were enriched (Supplementary Table 5). One of these pathways, chemokine signaling, consists of six genes including ELMO1 and has a 6.2-fold enrichment (P = 0.006).
We tested whether the 46 candidate genes with replicated variants associated with CKD in African Americans (Supplementary Tables 1 – 4) were differentially expressed in the kidney cortex of transgenic sickle cell (HbSS) versus non-sickle cell (HbAA) mice. Renal cortical tissue from transgenic sickle cell mice (Townes model, Jackson laboratory; Bar Harbor, U.S.A.) were dissected. The RNA was extracted and processed by the University of Illinois at Chicago (UIC), Core Genomics Facility from ten samples (5 HbSS and 5 HbAA; all female and 9 months of age). The Affymetrix Mouse Gene Array 2.0 (Thermo Fisher Scientific, Waltham, MA) was employed for the study. Raw and FDR corrected P-values following the Benjamini-Hochberg procedure of differential expression for the candidate genes were calculated. Twenty-one of the 46 genes were differentially expressed (18 upregulated, 3 down-regulated) (FDR < 0.01) (Table 1). Elmo1 was the gene with the strongest differential expression in the renal cortex (FDR = 5.5 × 10−5). Due to the proximal position of APOA1 in the interaction of APOL1 with other candidate genes in the interaction network (Figure 1), we tested the differential expression of APOA1 and found it to be down-regulated in HbSS versus HbAA mice (fold change −0.7, P = 3.7 × 10−5, FDR = 0.001).
Table 1:
Candidate gene expression changes in the kidney cortex of sickle cell versus non-sickle cell transgenic mice
| Gene | Fold-Change* | FDR Value |
|---|---|---|
| Elmo1 | 1.21 | 0.00006 |
| Tfdp2 | 1.19 | 0.0005 |
| Frmd3 | 0.93 | 0.0036 |
| Cyp3a5 | 0.82 | 0.0051 |
| Fbxl20 | 1.16 | 0.0051 |
| Nphs1 | 1.28 | 0.0054 |
| Auh | 0.86 | 0.0054 |
| Lrp2 | 1.55 | 0.0068 |
| Umod | 1.21 | 0.0069 |
| Cubn | 1.61 | 0.0071 |
| Tgfb1 | 1.20 | 0.0072 |
| Ptpro | 1.37 | 0.0072 |
| Myh9 | 1.50 | 0.0072 |
| Tcf7l2 | 1.24 | 0.0075 |
| Nphs2 | 1.22 | 0.0075 |
| Nos3 | 1.12 | 0.0075 |
| Pip5k1B | 1.12 | 0.0075 |
| Sash1 | 1.20 | 0.0079 |
| Rreb1 | 1.32 | 0.0079 |
| Plekha1 | 1.28 | 0.0079 |
| Agtr1 | 1.25 | 0.0085 |
Fold-change represents the average change in gene expression in HbSS versus Hb AA mice; FDR, false discovery rate
We focused on polymorphisms of the 21 candidate genes associated with CKD in African Americans that were differentially expressed in the sickle mouse kidney cortex. We examined the association of 48 SNPs in these 21 genes with kidney disease in 299 SCD patients enrolled in a prospective registry at UIC. The study was approved by the institutional review board and all subjects provided written informed consent. The median age of this cohort was 32 years, 41% were female, and 48% were on hydroxyurea. Chronic kidney disease was present in 53% of SCD patients. Other baseline characteristics of the cohort are provided in Table 2. Genotyping was carried out using Affymetrix Axiom genome-wide Pan-African GeneChip array at the Core Genomics Facility at UIC, as previously described.(7) Allele dosages were associated with hemoglobinuria and markers of kidney disease adjusting for age, sex, SCD genotype, and population structure. Chronic kidney disease was defined as urine albumin concentration ≥ 30 mg/g creatinine or estimated glomerular filtration rate (eGFR), calculated by the chronic kidney disease epidemiology (CKD-EPI) equation,(8) as ≤ 60 mL/min/1.73m2 on two consecutive outpatient visits.(9) Eighty tag-SNPs for APOA1 were identified based on the phased genotypes using a greedy algorithm with a linkage disequilibrium threshold set at r2=0.5.(10) Bonferroni corrected P-values are provided for the association between the APOA1 tag-SNPs with kidney phenotypes. A polymorphism of ELMO1, rs10951509 (minor allele frequency 0.35), correlated with urine albumin concentration (β −0.39, P = 0.048) and CKD (OR 0.71, P = 0.089). A tag-SNP of APOA1, rs11216132, correlated with hemoglobinuria (minor allele frequency 0.31; OR 0.39, P = 0.055).
Table 2.
Baseline variables of 299 patients with sickle cell disease from the University of Illinois at Chicago.
| Patient Characteristics | Value |
|---|---|
| Age (years) | 32 (23 – 43) |
| Sex (male : female) | 59% : 41% |
| Genotype | |
| Hemoglobin SS or Sβ0-thalassemia | 245 (82%) |
| Hemoglobin SC | 39 (13%) |
| Hemoglobin Sβ+-thalassemia | 15 (5%) |
| Hydroyxurea therapy (%) | 145 (48%) |
| Chronic kidney disease (%) | 157 (53%) |
| eGFR (ml/min/1.73m2) | 131 (100 – 149) |
| Albuminuria (mg/g creatinine) | 39 (13 – 192) |
| Hemoglobinuria | 62 (21%) |
Median (interquartile) values provided; eGFR, estimated glomerular filtration rate
ELMO1 encodes a member of the engulfment and cell motility protein family involved in promoting phagocytosis and cell migration that has been implicated in diabetic nephropathy in African Americans.(11–13) Increased Elmo1 expression is observed in the kidney cortex of diabetic versus nondiabetic mice. Overexpression of ELMO1 in COS cells leads to increased expression of genes (TGFB1, FN1, COL1A1) which cause an over-accumulation of extracellular matrix proteins.(12) In Akita type I diabetic mice genetically altered to have a graded expression of Elmo1, a direct association between increased Elmo1 expression and albuminuria, glomerulosclerosis, and TGFB1 is observed,(13) while decreased expression is associated with protection from these changes. The ELMO1 rs10951509 variant has been associated with a reduced risk of diabetic nephropathy in two independent cohorts of African Americans.(11) Consistent with these findings, we observed that Elmo1 expression is increased in the kidney cortex of the transgenic sickle cell mice and that ELMO1 rs10951509 was associated with lower urine albumin concentrations in SCD patients highlighting the potential role of ELMO1 in SCD-related glomerulopathy.
The gene interaction analysis pointed to direct functional links among HBB, APOL1 and APOA1. APOL1 is a component of the trypanosome lytic factor complex that scavenges cell-free hemoglobin.(14) APOL1 G1/G2 risk variants are associated with hemoglobinuria in SCD patients providing clinical evidence for this functional link.(15) APOA1 encodes the major structural protein of HDL particles, including the trypanosome lytic factor.(16) In two multi-ethnic populations from the Atherosclerosis Risk in Communities (ARIC) cohort and from the Third National Health and Nutrition and Examination Survey (NHANES III), lower plasma APOA1 concentrations have been associated with a higher prevalence of CKD.(17, 18) Lower plasma APOA1 concentrations have been associated with endothelial dysfunction and elevated systolic pulmonary artery pressure, as estimated by echocardiography(19) and by right heart catheter-defined pulmonary hypertension,(20) in SCD patients. Furthermore, treatment of arterioles from SCD mice with an APOA1 mimetic, L-4F, protects vascular endothelial function by reducing endothelial xanthine oxidase and improving vasodilation.(21) Our observation for the association of APOA1 rs11216132 with hemoglobinuria may indicate a potential role for this apolipoprotein in cell-free hemoglobin scavenging, either in circulation or in the kidney cortex.
In summary, we selected APOL1, HBB, and 46 other genes with validated variants associated with CKD in African Americans and applied a bioinformatic approach to identify additional candidate genes for SCD-related CKD. Gene expression analysis identified that 21 of the 46 candidate genes were differentially expressed in the kidney cortex of SCD versus non-SCD mice, including the increased expression of ELMO1 and decreased expression of APOA1. This approach points to ELMO1 as a potential candidate gene for SCD-related nephropathy and suggests a role for a new candidate gene, APOA1. Future studies investigating these candidate genes may improve our understanding of the molecular pathways and serve as targets for future research in African American and SCD-related CKD.
Supplementary Material
Acknowledgements:
The project described was supported by the National Institutes of Health through grants K23-HL125984, R03-HL146788, and R01-HL153161 (S.L.S); K24-DK092290 (J.P.L.); R01- DK117445, R01- MD012765, and R21- HL140385 (N.F.)
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References:
- 1.Drawz P, Ayyappan S, Nouraie M, Saraf S, Gordeuk V, Hostetter T, et al. Kidney Disease among Patients with Sickle Cell Disease, Hemoglobin SS and SC. Clinical journal of the American Society of Nephrology : CJASN. 2016;11(2):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flessner MF, Wyatt SB, Akylbekova EL, Coady S, Fulop T, Lee F, et al. Prevalence and awareness of CKD among African Americans: the Jackson Heart Study. American journal of kidney diseases : the official journal of the National Kidney Foundation. 2009;53(2):238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedman BI, Kopp JB, Langefeld CD, Genovese G, Friedman DJ, Nelson GW, et al. The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. Journal of the American Society of Nephrology : JASN. 2010;21(9):1422–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chasman DI, Fuchsberger C, Pattaro C, Teumer A, Boger CA, Endlich K, et al. Integration of genome-wide association studies with biological knowledge identifies six novel genes related to kidney function. Human molecular genetics. 2012;21(24):5329–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Zhang W, Ma SF, Desai AA, Saraf S, Miasniakova G, et al. Hypoxic response contributes to altered gene expression and precapillary pulmonary hypertension in patients with sickle cell disease. Circulation. 2014;129(16):1650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, et al. A new equation to estimate glomerular filtration rate. Annals of internal medicine. 2009;150(9):604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.K/DOQI. National Kidney Foundation. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. American journal of kidney diseases : the official journal of the National Kidney Foundation. 2002;39(2 Suppl 1):S1–266. [PubMed] [Google Scholar]
- 10.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. American journal of human genetics. 2004;74(1):106–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leak TS, Perlegas PS, Smith SG, Keene KL, Hicks PJ, Langefeld CD, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Annals of human genetics. 2009;73(2):152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54(4):1171–8. [DOI] [PubMed] [Google Scholar]
- 13.Hathaway CK, Chang AS, Grant R, Kim HS, Madden VJ, Bagnell CR Jr., et al. High Elmo1 expression aggravates and low Elmo1 expression prevents diabetic nephropathy. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(8):2218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widener J, Nielsen MJ, Shiflett A, Moestrup SK, Hajduk S. Hemoglobin is a co-factor of human trypanosome lytic factor. PLoS pathogens. 2007;3(9):1250–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saraf SL, Zhang X, Shah B, Kanias T, Gudehithlu KP, Kittles R, et al. Genetic Variants and Cell-Free Hemoglobin Processing in Sickle Cell Nephropathy. Haematologica. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Perez-Morga D. The molecular arms race between African trypanosomes and humans. Nature reviews Microbiology. 2014;12(8):575–84. [DOI] [PubMed] [Google Scholar]
- 17.Lamprea-Montealegre JA, Sharrett AR, Matsushita K, Selvin E, Szklo M, Astor BC. Chronic kidney disease, lipids and apolipoproteins, and coronary heart disease: the ARIC study. Atherosclerosis. 2014;234(1):42–6. [DOI] [PubMed] [Google Scholar]
- 18.Goek ON, Kottgen A, Hoogeveen RC, Ballantyne CM, Coresh J, Astor BC. Association of apolipoprotein A1 and B with kidney function and chronic kidney disease in two multiethnic population samples. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2012;27(7):2839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuditskaya S, Tumblin A, Hoehn GT, Wang G, Drake SK, Xu X, et al. Proteomic identification of altered apolipoprotein patterns in pulmonary hypertension and vasculopathy of sickle cell disease. Blood. 2009;113(5):1122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anjum F, Lazar J, Soh J, Albitar M, Gowda S, Hussain MM, et al. Dysregulation of ubiquitin-proteasome pathway and apolipoprotein A metabolism in sickle cell disease-related pulmonary arterial hypertension. Pulmonary circulation. 2013;3(4):851–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou J, Ou Z, Jones DW, Holzhauer S, Hatoum OA, Ackerman AW, et al. L-4F, an apolipoprotein A-1 mimetic, dramatically improves vasodilation in hypercholesterolemia and sickle cell disease. Circulation. 2003;107(18):2337–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.