Abstract
Background
SARS-CoV-2 infection in children does not seem to follow the same pattern as in adults. Limited information is published on the level of antibody production and the duration of antibody response in children with COVID-19. Moreover, it is unknown if all children have a similar immune response to the infection, or if there are age dependent differences. In these data, we look at the IgM and IgG levels and duration of two age groups infected by the SARS-CoV-2 virus.
Methods
Residual laboratory specimens from pediatric patients positive for SARS-CoV-2 infection were tested for IgM and IgG against SARS-CoV-2 using an automated Abbott ARCHITECT i1000. We tested 181 specimens from 41 patients with a positive molecular result. Data was grouped either as time after nucleic acid amplification test (NAAT) or time after symptom onset. Patient samples were divided into 2 age groups: 0 to 11 years old and 12 to 19 years old. The assays detect IgM against the spike protein and IgG against the nucleocapsid protein.
Keywords: COVID-19, SARS-CoV-2, IgG, IgM, paediatrics, serology, Spike protein, Nucleocapsid protein
Specifications Table
| Subject | Health and medical sciences, Laboratory Medicine |
| Specific subject area | COVID-19 serology in pediatrics |
| Type of data | Figures |
| How data were acquired | Methods: Abbott SARS-CovV-2 IgG, AdviseDx SARS-CoV-2 IgM Make and model and of the instruments used: Abbott Architect i1000 |
| Data format | Analyzed |
| Parameters for data collection | Residual pediatric specimens from patients that tested positive for SARS-CoV-2 were collected longitudinally |
| Description of data collection | Residual pediatric specimens from patients that tested positive for SARS-CoV-2 were collected longitudinally and assayed with the Abbott IgG and IgM methods |
| Data source location | Institution: Children's Healthcare of Atlanta City/Town/Region: Atlanta, Georgia Country: USA Primary data sources: Core Laboratory at Children's Healthcare of Atlanta Egleston Campus, and Mendeley Data (see Data accessibility below) |
| Data accessibility | https://data.mendeley.com/datasets/gjyj5g9k9n/1 |
| Related research article | Co-submission: Interiano C, Muze S, Turner B, Gonzalez M, Rogers B, Jerris R, Weinzierl E, Elkhalifa M and Leung-Pineda V. Longitudinal Evaluation of the Abbott Architect SARS-CoV-2 IgM and IgG Assays in a Pediatric Population. Submitted to Practical Laboratory Medicine. Manuscript Number: PLM-D-21-00011 https://doi.org/10.1016/j.plabm.2021.e00208 |
Value of the Data
-
•
These data shows that different age groups in children have distinct immune response to SARS-COV-2 infection
-
•
Investigators interested in the immune response in pediatric patients to SARS-CoV-2 infection would benefit from this dataset
-
•
Our data can be used by others for statistical power in producing meta-analysis to study the immune response of children to SARS-CoV-2 infection
1. Data Description
The data shows the IgM and IgG results from residual COVID-19 positive patient samples. Fig. 1 are the serology results based on time after positive molecular test results for SARS-CoV-2 infection. Fig. 2 shows IgM and IgG results based on time after start of COVID-19 symptoms. Each figure divides the patient into two groups based on their age. The original study that produced these data did not separate results by age [1].
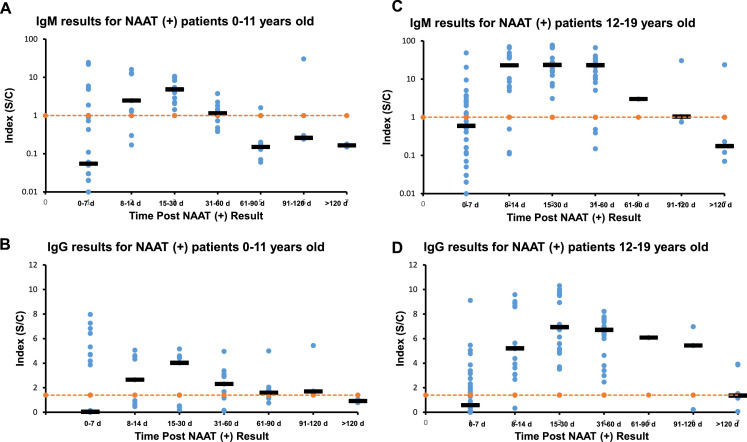
Fig. 1.
Longitudinal Results of Abbott SARS-Co-V2 IgM and IgG in COVID-19 Positive Patients Post NAAT results Divided by Age. IgM data for patients 0–11 years old (A) and patients 12–19 years old (C). IgG data for patients 0–11 years old (B) and patients 12–19 years old (D). Orange dotted line represents assay cutoffs, IgM (1.0) and IgG (1.4). Dark horizontal bars represent the median value for that time period. For the 0–11 years old group 71 specimens from 16 patients were tested. For the 12–19 years old group 109 samples from 25 patients were assayed.
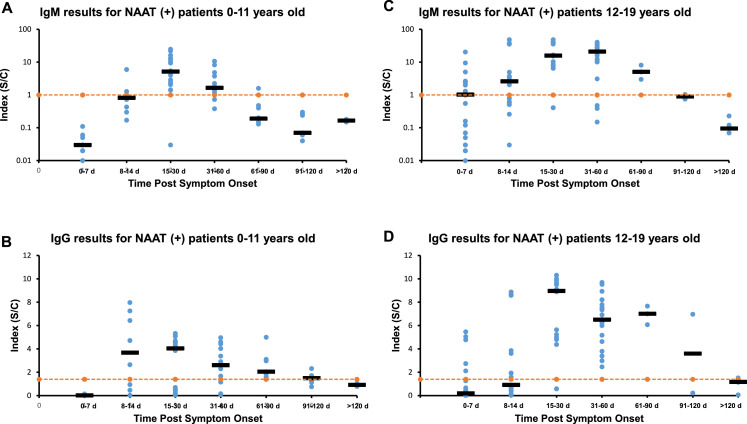
Fig. 2.
Longitudinal Results of Abbott SARS-Co-V2 IgM and IgG in COVID-19 Positive Patients Post Symptom Onset Divided by Age. IgM data for patients age 0–11 years old (A) and patients 12–19 years old (C). IgG data for patients 0–11 years old (B) and patients 12–19 years old (D). Orange dotted line represents cutoffs, IgM (1.0) and IgG (1.4). Dark horizontal bars represent the median value for that time period. For the 0–11 years old group 70 specimens from 15 patients were tested. For the 12–19 years group 86 samples from 22 patients were assayed.
In Fig. 1A, the results for IgM tests (y-axis) are plotted against the time (x-axis) when the specimens were collected after a positive molecular SARS-CoV-2 test for samples from patients 0 to 11 years old. The y-axis is the numerical result from the automated instrument and is in log-scale to encompass the range of results. The orange line represents the cutoff of the assay (1.00). The x-axis is divided in the following categories: 0–7 days, 8–14 days, 15–30 days, 31–60 days, 61–90 days, 91–120 days and >120 days after positive molecular results. The black horizontal bars for each time group represent the median value of IgM results for that particular time group. In Fig. 1B, the results for IgG tests (y-axis) are plotted against the time (x-axis) when the specimens were collected after a positive molecular SARS-CoV-2 test for samples from patients 0 to 11 years old. The y-axis is in normal scale and the orange line represents the cutoff of the IgG assay (1.4). The x-axis time groups are the same as Fig. 1A. The black horizontal bars represent the median value of IgG results for that particular time group. Fig. 1C is similar to Fig. 1A, except that the data comes from patients 12 to 19 years old. Fig. 1D is similar to Fig. 1B but data comes from patients 12 to 19 years old. For Fig. 1, the 0–11 years old group was comprised of 16 patients with 71 samples and the 12-19 year old group was composed of 25 patients with 109 samples
For Fig. 2A, IgM results (y-axis) are plotted against time (x-axis) after the patients reported start of COVID-19 symptoms for the 0–11 year old age group. The y-axis is in log scale and represents the signal/calibrator result that the instrument reports. The orange line represents the IgM cutoff. The x-axis time groups are the same as in Fig. 1, but they represent time after onset of COVID-19 symptoms. The black horizontal bars again represent the median IgM result for that specific time group. Fig. 2B shows the IgG results for the 0–11 year old group with the orange line representing the IgG cutoff (1.4) and the black horizontal bars representing the median IgG values for each time group. Fig. 2C shows the post symptom IgM data for the 12–19 year old group. Fig. 2D displays the IgG data for the 12–19 year old group. Fig. 2 represents data from 70 specimens from 15 patients for the 0–11 year old group and 86 specimens from 22 pediatric individuals for the 12–19 year old group.
2. Experimental Design, Materials and Methods
Specimens tested were leftover clinical blood samples from pediatric patients that tested positive for SARS-CoV-2 by a molecular method. Serial samples from patients were saved, when available, from April to November of 2020. Leftover specimens were stored at –20 °C within 5 days from collection. Leftover specimens were stored at 4 °C before transferring to frozen storage. Specimens were thawed to room temperature once to run IgM and IgG tests. Date of positive molecular result and date of start of symptoms was obtained from medical notes. The institutional review board (IRB) at Children's Healthcare of Atlanta approved this investigation (study #00000621) with a waiver of informed consent. The results were split into two age groups: 0–11 years old and 12–19 years old. Age of the patients was based on their age when they first presented for care. Specimens from the same patient were divided into different time periods (0–7 days, 8–14 days, 15–30 days, 31–60 days, 61–90 days, 91–120 days, >120 days after) relative to the date of positive molecular test or date of symptom onset. Data analysis, sorting and statistics was performed using Microsoft Excel.
Tests used to detect IgM and IgG against SARS-CoV-2 were manufactured by Abbott Laboratories using the Abbott ARCHITECT i1000 (Abbott Park, IL) and used according to the manufacturer's instructions. Specimens were thawed at room temperature, aliquoted and centrifuged for 5 minutes at 3,000 X g before loading on to the instrument. The IgM assay detects immunoglobulins against the spike protein and the IgG assay detects antibodies against the N protein of the virus. IgM results ≥ 1.00 and IgG results ≥ 1.4 are positive. Both assays are automated chemiluminescent microparticle immunoassays and required a minimum volume of 75µL for each test.
Ethics Statement
This study used residual clinical samples in the clinical laboratory. This study was approved by the institutional review board (IRB) at Children's Healthcare of Atlanta with a waiver of informed consent (study #00000621).
CRediT Author Statement
Cristina Interiano: Conceptualization, Investigation, Methodology, Data curation, Writing Reviewing & Editing; Sheicho Muze: Investigation, Methodology; Brian Turner: Methodology; Mark Gonzalez: Conceptualization, Writing - Reviewing & Editing; Beverly Rogers: Supervision, Writing - Reviewing & Editing; Robert Jerris: Conceptualization, Writing - Reviewing & Editing; Elizabeth Weinzierl: Writing - Reviewing & Editing; Van Leung-Pineda: Conceptualization, Investigation, Supervision, Data curation, Visualization, Writing - Original draft preparation, Reviewing & Editing.
Declaration of Competing Interest
This study was supported by a grant from Abbott Laboratories.
Acknowledgments
The authors wish to thank Kevin Pannell and Wilfred Morales for collecting residual specimens and Randal Schneider for coordinating reagent procurement.
Reference
- 1.Interiano C., Muze S., Turner B., Gonzalez M., Rogers B., Jerris R., Weinzierl E., Elkhalifa M., Leung-Pineda V. Longitudinal evaluation of the abbott architect SARS-CoV-2 IgM and IgG assays in a pediatric population. Pract. Lab. Med. 2021 doi: 10.1016/j.plabm.2021.e00208. Accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]