Abstract
Splitting bioactive proteins into conditionally reconstituting fragments is a powerful strategy for building tools to study and control biological systems. However, split proteins often exhibit a high propensity to reconstitute even without the conditional trigger, limiting their utility. Current approaches for tuning reconstitution propensity are laborious, context-specific, or often ineffective. Here, we report a computational design strategy grounded in fundamental protein biophysics to guide experimental evaluation of a sparse set of mutants to identify an optimal functional window. We hypothesized that testing a limited set of mutants would direct subsequent mutagenesis efforts by predicting desirable mutant combinations from a vast mutational landscape. This strategy varies the degree of interfacial destabilization while preserving stability and catalytic activity. We validate our method by solving two distinct split protein design challenges, generating both design and mechanistic insights. This new technology will streamline the generation and use of split protein systems for diverse applications.
INTRODUCTION
Split proteins and conditional reconstitution systems are powerful tools for interrogating biology and controlling cell behavior.1-4 These systems work by splitting a protein into two fragments to disrupt the protein’s function. Each fragment is then fused to a partner domain such that the split protein is reconstituted, and its function is restored, only when the partner domains interact. This modular strategy may be applied to diverse functional proteins to control bioluminescence5,6, fluorescence7, proteolytic cleavage8-10 and transcription11,12. As a result, conditionally reconstituted split proteins have been employed in a variety of applications including probing and discovering new protein-protein interactions13-16, studying post-translational modifications17, establishing small molecule-regulated control over enzymatic activity18,19, and rewiring cellular signaling9,10.
Despite their utility in certain contexts, broader application of split protein systems is largely limited by the spontaneous reconstitution of fragments, resulting in high background activity (Fig. 1a). Past approaches to optimize split protein systems include the development of computational algorithms to computationally identify split sites that avoid extremes of spontaneous reconstitution or failure to reconstitute20, experimental screens to identify thermostabilizing split fragments as these tend to yield more functional split proteins21-23, and utilization of other computational methods to enable design-driven stabilization of existing proteins24-27. However, fundamental molecular rules on optimizing a split protein system remain poorly understood. Splitting a protein tends to expose its hydrophobic core, creating highly unfavorable interactions between the core and solvent. Reconstitution is driven by a strong inherent preference to desolvate by recombining the fragments. Evaluating alternative splitting sites can vary reconstitution propensity, but this approach often only partially ameliorates the problem because changing splitting sites may not substantially affect underlying hydrophobic forces. Therefore, it is necessary to identify variants with a reconstitution propensity that precludes spontaneous reconstitution but enables reconstitution under desired conditions. Variants with a range of reconstitution propensities can be generated by random mutagenesis and screened for the desired property.6,28 However, high-throughput screening is not readily available for all split protein systems, and low-throughput clonal testing of variants can be laborious and suffer from inefficient exploration of sequence space. Even when screens generate improved variants, it may be difficult to interpret why only certain mutants were successful, and as a result, generalizable rules cannot be transferred to guide the tuning of new split protein systems. Furthermore, split protein systems tuned by mutagenesis exhibit performance characteristics determined by (and limited to) the conditions used in the initial screens, again posing a barrier to general applications. This knowledge and capability deficit can be addressed by employing computational protein design tools.
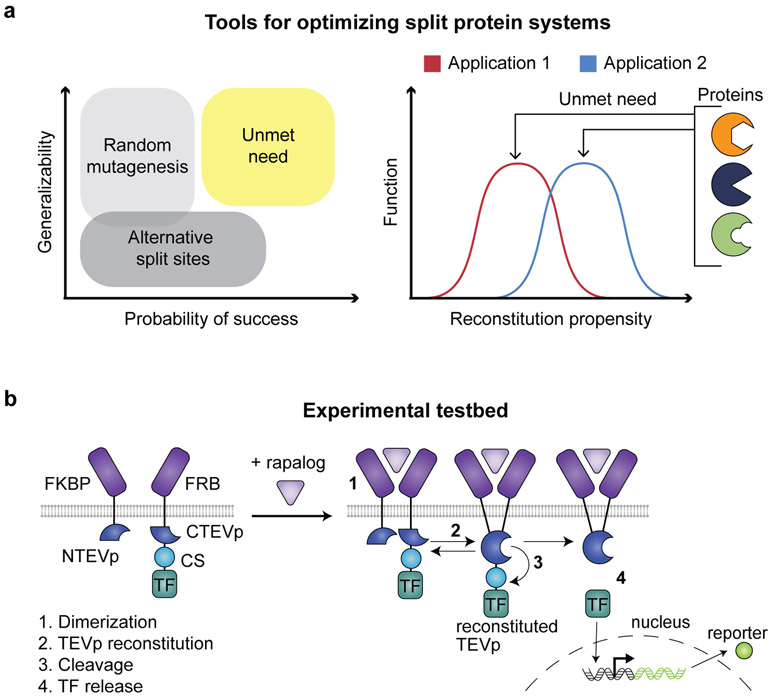
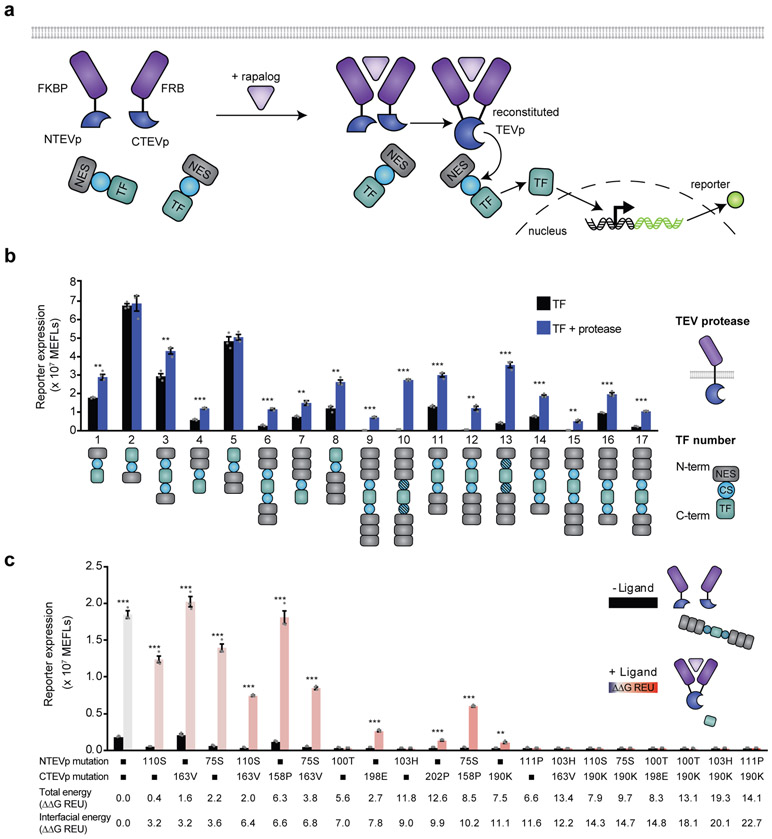
Fig. 1. Design-driven strategy for tuning split protein systems.
a, Current methods for optimizing split proteins are limited (left); an ideal tool would enable adapting split proteins for multiple applications, each of which may require distinct reconstitution propensities (right). b, This cartoon illustrates the experimental testbed used here. Ligand binding-induced chain dimerization results in split TEVp reconstitution, trans-cleavage, and release of a previously sequestered transcription factor to drive reporter expression.
Here, we report a general computational design strategy based on biophysical principles to guide experimental screening for optimizing split protein systems which we term Split Protein Optimization by Reconstitution Tuning, or SPORT. We demonstrate proof-of-concept by optimizing a split protease system for conditional reconstitution in two different contexts, and this exploration yields a straightforward design method that may be extended to tune other split protein systems and can be implemented by most research laboratories. This new method for efficiently engineering split protein systems will streamline the generation and use of split protein systems for diverse applications.
RESULTS
Formulation of the design challenge and strategy
As a first step toward developing a strategy that addresses the challenge of designing split proteins, we selected a model system based on the well-studied Tobacco Etch Virus protease (TEVp) and we sought to tune the reconstitution propensity of split TEVp. For this purpose, we modified a synthetic receptor system that we previously reported (Modular Extracellular Split Architecture, MESA)29 to report upon conditional split TEVp reconstitution. In this testbed, ligand binding-induced dimerization of a membrane receptor reconstitutes an intracellular split TEVp, which autolytically liberates a sequestered transcription factor to drive reporter gene expression (Fig. 1b). Initially, the canonical split TEVp (split between residues 118/119)30 showed high background (ligand-independent) signaling (Supplementary Fig. 1). As the original screens used to identify this split were performed in a soluble rather than membrane-bound context, these data suggest that tethering split TEVp to a membrane may promote reconstitution. This problem was not limited to the canonical split site, as other TEVp partitioning also yielded poor performance (Supplementary Fig. 2). Given these observations, we formulated a design goal: rationally mutate split TEVp to optimize two key MESA performance characteristics, minimal reporter expression in the absence of ligand and a substantial fold increase in reporter expression upon ligand addition.
Biophysical principles underlying SPORT
We developed SPORT, a computation-guided workflow to rationally design split protein interfaces to optimize reconstitution propensity. In this discussion, “reconstitution propensity” is a lumped term that describes the combined effects of the two reversible reactions: (1) fragment association and dissociation, governed by the equilibrium binding constant, and (2) fragment reconstitution, governed by the folding and unfolding equilibrium. SPORT employs Rosetta, a state-of-the-art software package for single-state protein design.31 Given a protein with a predetermined split site, we first identified key interfacial residues to target for mutagenesis. Residues with large differences in solvent-accessible surface area (SASA)—when comparing intact protein and split fragments—were classified as buried residues. These buried residues are ideal targets for mutagenesis as they likely contribute substantially to the driving force for spontaneous reconstitution. For each buried residue, we performed a comprehensive, in silico mutational scan to evaluate the energy perturbation of all possible single-point mutations on the interaction energy across the split protein interface (ΔΔGInterfacial) and total stability of the mutated protein (ΔΔGTotal) relative to the parent. The degree of disruption is a critical design consideration. Insufficient disruption may retain high background activity while excessive disruption may impair catalytic activity due to loss of overall protein stability. Therefore, the interface must be carefully tuned so that the driving force provided by ligand binding-induced dimerization promotes reconstitution. This “Goldilocks zone” likely differs for each individual protein and perhaps depends upon context, and this zone is difficult to define a priori. Therefore, our strategy was to identify the Goldilocks zone for a given protein by choosing mutations that span the range of ΔΔG values. We hypothesized that a limited test set would direct subsequent mutagenesis efforts by predicting desirable mutations from a vast amount of sequence space. Each of these propositions was tested using experimental case studies.
Tuning membrane-tethered split TEV protease
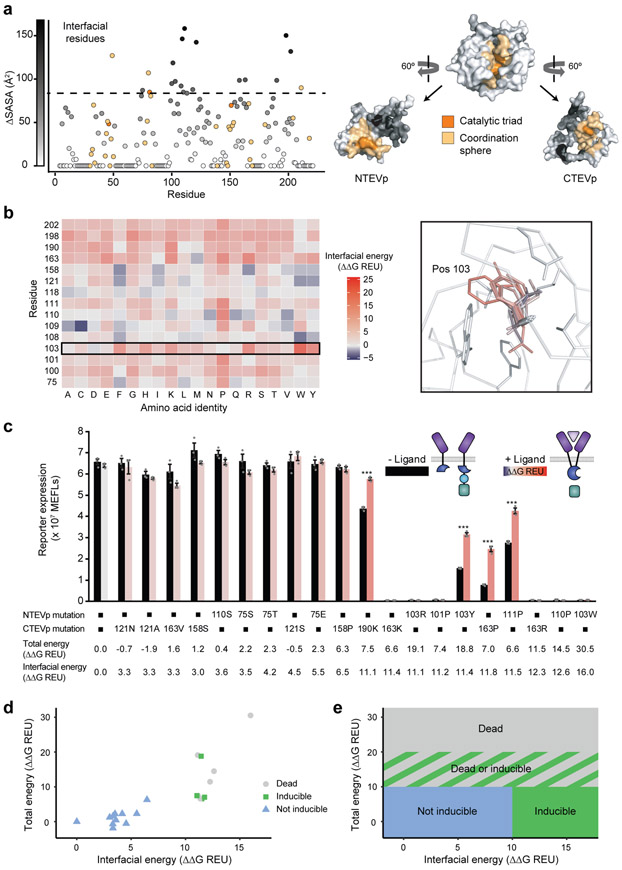
To investigate and validate SPORT, we applied our design workflow to the split TEVp MESA system. We first assessed the per-residue change in solvent accessible surface area (ΔSASA) between the intact form and split fragments (Fig. 2a). In total, 130 of the 218 residues showed increased SASA in the isolated fragments. We excluded from this set the catalytic triad and 27 residues lying within a 6-Å coordination sphere around the catalytic triad to avoid perturbing the catalytic function of reconstituted TEVp. Of the remaining 100 positions, we chose the 15 positions with the largest ΔSASA (9 in the N-terminal half (NTEVp) and 6 in the C-terminal half (CTEVp) of split TEVp) as candidates for mutagenesis. Next, we evaluated the energy perturbation of all possible single-point mutations (285 in total) at these positions using Rosetta. As expected, few mutations were predicted to increase stability, and a vast majority were destabilizing (Fig. 2b, Supplementary Figs. 3, 4). Many positions exhibited a variety of stabilizing, benign, and destabilizing point mutations that were consistent with structure. For instance, bulky sidechain substitutions (Trp, Phe, Tyr, Arg, Lys and His) at position 103 resulted in many steric clashes with neighboring residues (Fig. 2b, right panel) and subsequently conferred large decreases in predicted stability. To validate our selection of the 15 positions with the largest ΔSASA as optimal targets for mutagenesis, we evaluated the energy perturbation of all possible single-point mutations at 18 randomly selected positions not buried at the interface (ΔSASA = 0 Å2) and all positions moderately buried (50 ≤ ΔSASA ≤ 85 Å2) using Rosetta (Supplementary Fig. 4). Although the interface may be perturbed with mutations at moderately buried positions, the greatest diversity of energy perturbations was achieved at positions with the highest ΔSASA. Thus, we concluded that high ΔSASA positions exhibit the greatest potential for tuning reconstitution to various degrees.
Fig. 2. Computation guided method development and experimental analysis.
a, Left, characterization of the change in solvent-accessible surface area (SASA), between fragment and reconstituted forms, of each residue of 118/119 split TEVp. Right, 3D depiction of 118/119 split TEVp, showing the catalytic triad (orange), coordination sphere (yellow), and ΔSASA (greyscale). b, Mutational scanning of high ΔSASA residues (left) and example of all possible mutations of residue 103 (right), with change in interfacial energy indicated by color. Energetic perturbations to total protein stability are shown in Supplementary Fig. 3. c, Experimental analysis of TEVp mutations predicted to span a range of interfacial energies. We hypothesize that modest rapalog-induced toxicity or inhibition of protein expression could explain the slight decrease in signal observed upon rapalog addition for some mutants. Error bars depict S.E.M. Two-tailed Student’s t-test (*p ≤ 0.05, ***p ≤ 0.001). Experiments were performed in biological triplicate. Results are representative of two independent biological experiments. d, Experimental phenotypes observed in c were plotted on an energy landscape and annotated as indicated by color (reporter expression normalized to WT < 0.05 is “dead”, fold induction < 1.2 is “not inducible”, fold induction ≥ 1.2 is “inducible”). e, Proposed model for predicting zones of functional phenotypes based upon total and interfacial energy; the boundaries comprise hypotheses posed based upon observations using the initial 20 mutants tested in c.
Guided by these predictions, we next experimentally characterized 20 single-mutant split TEVp variants that span a wide range of ΔΔGInterfacial (3.1 to 16.1 Rosetta Energy Units, or REU) and ΔΔGTotal (−1.9 to 30 REU) energies (Fig. 2c). We observed high background signaling (i.e., reporter expression) for disruptions up to ΔΔGInterfacial ~6.6 REU, suggesting that destabilization was insufficient. However, four out of ten variants with ΔΔGInterfacial >10 REU exhibited reduced background signaling and substantial ligand-induced activation (Fig. 2c). The remaining six were completely inactive (or “dead”); they induced no signaling under any conditions. Energy-based partitioning across different phenotypes—inducible (fold induction ≥ 1.2), not inducible, and dead—became evident when comparing ΔΔGInterfacial and ΔΔGTotal of all 20 single mutant split TEVp variants (Fig. 2d). Variants with high background activity due to insufficient destabilization fell in the region where ΔΔGInterfacial < 10 and ΔΔGTotal < 10 REU. Variants with the dead phenotype had ΔΔGInterfacial > 10 and ΔΔGTotal > 10 REU; since these mutants were expressed, as confirmed by Western blot (Supplementary Fig. 5), the lack of signaling suggested that these mutations directly preclude reconstitution. Most of the inducible variants (three out of four) were observed in the energy window where ΔΔGInterfacial > 10 and ΔΔGTotal < 10 REU, which may represent the Goldilocks zone we hypothesized to exist. An additional region contained a mixture of inducible and dead phenotypes. By inspection of these results, we then proposed a model for broadly classifying experimental phenotypes based on energy partitions (Fig. 2e); this hypothesis was used to guide our selection of the subsequent round of mutants for experimental characterization.
SPORT predicts outcomes of combining mutations
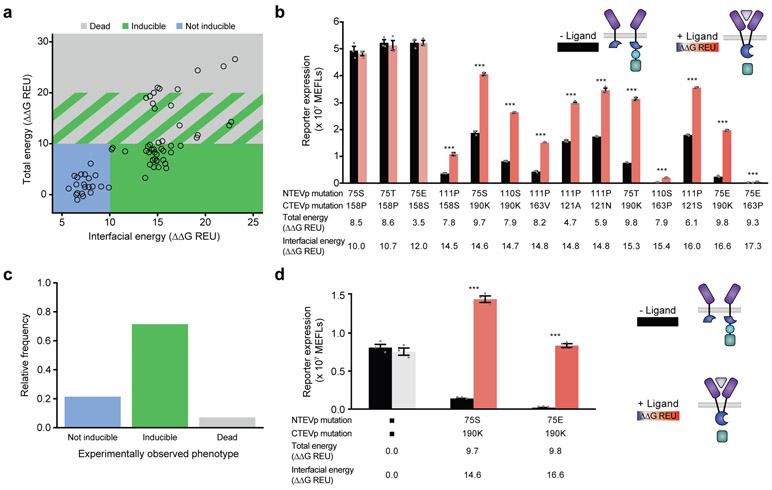
We next evaluated whether our proposed classifier model—a hypothesis proposed based upon observations with single mutants—could predict the phenotypes of combined NTEVp and CTEVp mutations, including both double (two mutations on one chain) and paired (one mutation on each chain) mutants derived by combining the initial 14 single, non-dead mutations tested, yielding 67 possible double and paired mutants. Of these 67 possible double and paired mutants, 28 mutants were predicted to be inducible (Fig. 3a). The region of predicted inducibility encompasses high ΔΔGInterfacial energy and low ΔΔGTotal energy; mutations that have a large effect on the interface and a small effect on overall folding are desired. We selected 14 mutants that span the interfacial energy range for experimental investigation. Ten mutants exhibited inducible signaling as predicted, one was dead, and three were not inducible (yielding an observed accuracy of 0.71 for inducible predictions) (Fig. 3b,c). Interestingly, three of the prediction failures fell at the low end of the range of predicted changes in interfacial energy, suggesting opportunities for refining our initial classification hypothesis. For the 67 mutants tested, the calculated ΔΔG for the double and paired mutants were nearly identical to the sums of the ΔΔG calculated for the associated single mutants (Supplementary Fig. 6), which is unsurprising given that our analysis focuses primarily on the local effects of each mutation. Thus, for subsequent analyses of combined mutants, we simply added the effects of single mutants in our calculations.
Fig. 3. Evaluation of model-predicted phenotypes for combined mutations.
a, Computed energies and computationally predicted phenotypes (open circle data points) based on the classifier model (shaded boxes)—proposed as a hypothesis in Fig. 2e—of all possible double and paired mutants constructed by combinatorial sampling of the 20 initial single mutants tested (omitting the 6 dead mutations) in Fig. 2c. b, Experimental evaluation of selected mutants predicted to be inducible. c, Experimentally observed phenotypes for the fourteen mutants predicted to be inducible (from b), showing that the model predicts inducibility at a fairly high rate (10/14). d, Normalizing protein expression levels improves performance (fold induction) of selected mutants (from b), whereas WT function is not changed. Fold inductions for 75S/190K and 75E/190K: 2.2 and 7.9 respectively (from b) and 10.3 and 43.3 respectively (from d). The normalization results in a statistically significant improvement of fold induction (between panels b and d) for each of the two mutant pairs analyzed (two-tailed Student’s t-test, p ≤ 0.001). Normalization was achieved using Western blot analysis (Supplementary Fig. 7) to adjust DNA doses transfected (per well, N-terminal chains: 0.4 ng WT, 1 ng 75S, 1.4 ng 75E; C-terminal chains: 5 ng WT, 12 ng 190K). Error bars depict S.E.M. (*p ≤ 0.05, ***p ≤ 0.001). Experiments were performed in biological triplicate. Results are representative of two independent biological experiments.
We next investigated how variations in expression level might impact inducibility. We used Western blot analysis to normalize and vary chain expression levels by adjusting DNA doses (Supplementary Fig. 7). Notably, multiple constructs remained inducible across the entire range of doses tested, suggesting that our SPORT-optimized mechanism is robust to variations in chain expression level and ratio. However, the performance of these constructs (i.e., fold induction of signaling upon ligand addition) could be substantially improved through tuning expression such that protein levels of each fragment are comparable (Fig. 3d, Supplementary Fig. 8). It is possible that some mutations altered both catalytic efficiency of the reconstituted TEVp and reconstitution propensity. For example, for mutants that exhibit decreased “ON” state compared to WT split TEVp, it is difficult to disentangle these effects. However, changes in catalytic efficiency alone cannot explain the behavior of many mutants exhibiting increased fold-induction upon ligand addition (Supplementary Fig. 8), because simply decreasing catalytic efficiency would decrease both background signaling and ligand-induced signaling29. Moreover, there exist several mutants for which the magnitude of the “ON” state is comparable to that of the WT, providing no evidence of altered catalytic efficiency. Taken together, these results suggest that a classifier calibrated with a limited set of experimental observations spanning the full range of ΔΔG can predict function of new mutants with high accuracy in a manner that is independent of expression level of the construct.
Mechanistic insights of SPORT-guided mutations
In order to generate insights into how the high-performing mutants identified above achieve improved function, we investigated overall protein stability, dimerization-induced stabilization, and reconstitution propensity. We investigated each of these phenomena in the cellular context to best enable comparisons with the functional evaluations used to identify these high-performing variants. We previously observed that each individual mutant fragment was expressed in the absence of ligand, many at levels comparable to those of the WT fragments (Supplementary Fig. 5); thus, these mutations did not act by diminishing global stability of the fragments. However, this does not rule out the possibility that some mutants exhibit some dimerization-induced stabilization or dimerization-induced stabilization plus altered reconstitution propensity, so we explored this possibility by quantifying levels of protein expression (accumulation) in cells as a contextually relevant measure of stability32. Chains were expressed in various pairings, and several mutant chain pairs showed some elevated accumulation in the presence of ligand (Supplementary Fig. 9a,b). Interestingly, the magnitude of this effect depended upon which chains were paired. However, no mutants exhibited enough destabilization in the absence of ligand to explain functional consequences such as diminished background protease activity (Supplementary Fig. 9c). Therefore, although some SPORT-guided mutations confer modest ligand-induced stabilization, this effect is insufficient to explain the functional ligand-induced signaling trends observed when comparing mutants to WT versions of split TEVp.
We next investigated how split TEVp domains interact using a single-cell resolution FRET-based flow cytometry assay we recently reported33. As illustrated in Supplementary Fig. 10a, adding FRET fluorophores mCerulean and mVenus onto the C-termini of the split TEVp chains enables the evaluation of association-dependent FRET signal in the absence and presence of ligand. Interestingly, mutant split TEVp variants did not exhibit substantially reduced FRET, compared to WT, in the absence of ligand (Supplementary Fig. 10b). This could indicate that FRET due to transient diffusive interaction between chains (that does not result in sufficient protease activity to yield reporter induction), is the dominant contributor to FRET in the absence of ligand (compared to FRET due to spontaneous reconstitution). Mutant pairs that were highly inducible in functional assays (e.g., all combinations including CTEVp 190K) generally exhibited an increase in FRET signal upon ligand addition, whereas WT split TEVp exhibited no such increase. There were also exceptions to this rule; for instance, all combinations including NTEVp 111P exhibited enhanced ligand-inducible signaling but no ligand-induced increase in FRET. Overall, these experiments provided additional mechanistic insights and demonstrated that SPORT-guided mutations may identify split protein variants that exhibit enhanced performance through subtly distinct mechanisms.
SPORT predicts phenotypes of novel variants
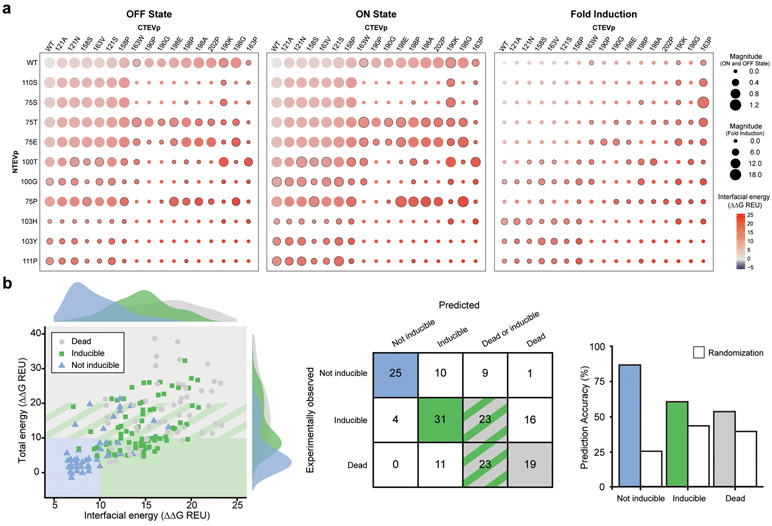
To investigate the accuracy of our classification scheme, we next tested a broad set of mutants (not included in our calibration set) including new single mutants and all combinations of paired mutations derived from both the original and this expanded mutation set (omitting dead constructs). Mutants were selected to explore the boundaries of the energy landscape classifier model and were expected to reflect a wide range of induced and uninduced reporter expression levels and ratios (Figs. 2e, 3a). We also sought to investigate whether the phenotypic partitioning demonstrated for inducibility (Fig. 3b,c) is extensible to the other phenotype classes (i.e., dead and not inducible). This expanded set paired 10 N-terminal mutations with 16 C-terminal mutations. In general, variants with larger ΔΔGInterfacial energies had lower reporter expression levels in both the background (OFF) and ligand-induced (ON) states (Fig. 4a left and middle panels; Supplementary Fig. 11a; Supplementary Fig.11b left and middle panels). Thus, the cost of lowering the OFF state is to also lower the ON state, but these reductions are not always proportional. This is evident by the diversity of calculated fold inductions (Fig. 4a, right panel, Supplementary Fig. 11b, right panel). Only one variant, 75P/198E, exhibited a substantial decrease in the OFF state and an increase in the ON state relative to wild type (WT). However, the variants with highest fold inductions, such as 75S/163P (17.3 fold induction) and 75T/163P (9.92 fold induction), exhibited substantially lower OFF and ON states than did the WT, reflecting a tradeoff between desirable performance characteristics.
Fig. 4. Evaluation of model-predicted phenotypes for novel mutations and combinations.
a, For each experimentally characterized construct, reporter output was quantified in the absence of ligand (OFF state) and following ligand addition (ON state), and the fold induction was calculated. Calculated interfacial energy for each construct is indicated by circle color, magnitude of reporter expression (or fold-induction) is indicated by circle size, and constructs with a fold induction ≥ 1.2 are denoted with a black border. Single mutants observed to be dead (Fig. 2c) were not carried forward to this analysis. b, Left, experimentally observed phenotypes (data point color) were mapped onto the proposed classifier model (shaded boxes) from Fig. 2e, with observed frequency distributions shown as histograms. Right, evaluation of model prediction accuracy compared to random assignment of phenotypes.
Overall, we observed moderate agreement between the experimental phenotypes (data point color) and predicted phenotypes (classification scheme proposed based upon initial observations—the shaded boxes) for these novel variants and combinations (Fig. 4b). The classifier model was most accurate for predicting the not-inducible phenotype (25 of 29, 86%). Many inducible phenotype predictions were confirmed (31 of 52, 60%). This success rate is impressive given that phenotypic classification boundaries were set roughly based upon the sparse calibration set (Fig. 2e). The distributions of total and interfacial energies indicate natural phenotype boundaries like those drawn from the calibration set (Fig. 4b). The “Goldilocks” zone of this extended data set appears to represent regions of sequence space where one can increase ΔΔGInterfacial as high as possible without incurring a substantial penalty in ΔΔGTotal. The “dead” zone may represent variants with high ΔΔGTotal and high ΔΔGTotal, which may indicate a disruption of proper protein folding.
In order to gain additional insight into how choice of calibration set and sample size may impact the accuracy of SPORT predictions, we performed retrospective bootstrapping analysis of the data presented in Figure 4 (see Supplementary Software 1 for full details). Experimental data were stratified by the energy landscape and partitioned randomly into calibration and prediction subsets. Linear discriminate analysis was applied to evaluate the accuracy of classification under various conditions (Supplementary Fig. 12). We observed robust prediction accuracy using multiple unique calibration sets and using sample sizes ranging from 4 to 28. This outcome suggests that our ability to generate a general classification model was not dependent upon the specific calibration data we used in our initial characterization experiment (Fig. 2c), and that a relatively small set of calibration data drawn from a distribution like that included in Figure 4 would be sufficient to generate a general classification model.
Extension of SPORT predictions to new design goals
A major limitation to current approaches for employing split proteins is that often a variant selected to perform well in one context fails in a different context. To investigate whether the SPORT design method is generalizable beyond our initial model system, we developed a distinct model system that employs split TEVp in a soluble form, where we hypothesized that a different reconstitution propensity would be required (compared to the membrane-bound model system). In this system (Fig. 5a), ligand binding domains were fused to split TEVp domains, and a separate soluble transcription factor flanked by protease cleavage sites and nuclear export signals (NES) was expressed; TEVp-mediated cleavage removes the NES from the transcription factor to enable nuclear localization and reporter expression. We first built and tested a panel of candidate constructs, varying the number of NES elements, their placement at the N and/or C termini of the transcription factor, and the P1’ residue of the TEVp cleavage sequence which governs cleavage kinetics34 (Fig. 5b). Several soluble transcription factor constructs exhibited the desired phenotype of low signaling in the absence of TEVp and high signaling when co-expressed with full TEVp; construct TF10 was selected for evaluating split TEVp variants.
Fig. 5. Model-guided design of a new split TEVp application in soluble context.
a, This cartoon illustrates the soluble split TEVp testbed. Ligand-binding-induced dimerization mediates reconstitution of split TEVp, which then cleaves one or more nuclear export sequence (NES) elements from a soluble transcription factor, leading to nuclear import and reporter expression. b, The testbed was developed by evaluating engineered transcription factors (TF) for consistency with the mechanism proposed in a; varying the number of NES elements, NES placement at the N and/or C terminus of the transcription factor, and the P1’ residue of the TEVp cleavage sequence, where shaded cleavage sequence (CS) domains indicate a Gly residue in the P1’ position, and unshaded CS domains indicate a Met residue in this position.34 c, Experimental analysis of single and paired mutants sampling a range of interfacial energies (indicated by color and labeled), employing TF10 from b. Error bars depict S.E.M. Two-tailed Student’s t-test (*p ≤ 0.05, ***p ≤ 0.001). Experiments were performed in biological triplicate. Results are representative of two independent biological experiments.
Using our soluble split TEVp test system, we evaluated a subset of mutants comprising 10 single TEVp mutants and 10 paired TEVp mutants that span a range of interfacial energies (Fig. 5c). The construct based upon WT split TEVp exhibited a substantial fold induction, which is consistent with the fact that this split protein was identified by screens performed in the soluble phase30. However, the WT construct also yielded high background signaling, indicating an opportunity to improve performance. For modest increases in ΔΔGInterfacial (~0–3.6 REU), background signaling persisted. For intermediate increases in ΔΔGInterfacial (~6-10 REU), a mixture of phenotypes was observed, including dead constructs and those with both substantially reduced background signaling and substantial fold inductions. For large increase in ΔΔGInterfacial > ~10 REU, constructs were generally weakly-inducible or dead. Thus, a focused evaluation of 20 design variants, guided by SPORT, yielded ~5 variants exhibiting improved performance compared to the WT construct. Altogether, these observations demonstrate that the SPORT method and associated energy landscape concept can be employed to efficiently solve distinct split protein optimization challenges.
DISCUSSION
In this study, we developed and validated what is to our knowledge the first computation-guided strategy for tuning split protein reconstitution propensity. Although the split TEVp MESA used as our first model system would have been deemed infeasible using standard evaluations of split proteins (Supplementary Figs. 1-2), application of SPORT to tune this system yielded multiple high-performing new synthetic receptor scaffolds (Fig. 3d). We show that unlike the classical MESA receptors we have characterized in prior work29,35,36, split TEVp MESA tuned by SPORT exhibit excellent performance characteristics (i.e., low-background and high fold-induction) in a manner that is robust to variations in both biosensor expression level and the ratio at which biosensor chains are expressed (Supplementary Fig. 8). This property is of great practical utility, as it precludes the need to carefully tune the implementation of each biosensor.
Several important insights emerged from this study. First, our approach demonstrated that testing a sparse set of mutants along the energy landscape is an effective strategy to choose optimal interfacial energies to promote conditional reconstitution. Second, multiple point mutations with similar energies exhibited similar performance, which suggests reconstitution propensity depends on the energy of destabilization but may be agnostic to specific mutations. Third, the concept of a Goldilocks zone is intuitively appealing and likely generalizable to different proteins and application contexts. Fourth, although the optimal energy window may have to be adjusted on a case-by-case basis, SPORT provides a simple design framework for engineering split protein systems. We find that membrane-bound split proteins must be destabilized to a greater degree than soluble split proteins in order to avoid spontaneous reconstitution. Altogether, these results show that split protein systems can be engineered based on fundamental principles of protein biophysics, which obviates the need for exhaustive screening and generates rules applicable to new candidate proteins.
There are several interesting opportunities for extending and improving SPORT in future work. First, it would be valuable to identify general principles as to when SPORT is a useful tool for tuning a given split protein of interest. For example, it could also be interesting to determine how properties such as intrinsic protein thermostability influence the opportunity for tuning reconstitution propensity through this method. In addition, although our analysis showed that SPORT can be used to identify mutations that confer specific energy changes, it does not yet enable a priori prediction of where the Goldilocks zone will fall for new applications. It is possible that subsequent analysis of many case studies could identify trends that enable such predictions and thus harness SPORT to further focus experimental investigations. Another opportunity is pairing SPORT with a multiparameteric optimization framework for exploring pareto-optimal tradeoffs between performance characteristics; for example, in our model system, there seems to be such a tradeoff between low background in the ligand-free state and high signaling in the ligand-induced state. Finally, the SPORT algorithm itself may be refined to better avoid false positives (e.g., dead mutants that share a region of the energy landscape with inducible variants). Altogether, our findings suggest many opportunities for expanding the utility of split proteins for many new applications and highlight the impact of SPORT-guided development of novel biochemical and synthetic biology tools.
METHODS
General DNA assembly
Plasmid cloning was performed using standard molecular biology techniques of PCR and restriction enzyme cloning with Phusion DNA Polymerase (NEB), restriction enzymes (NEB; Thermo Fisher), T4 DNA Ligase (NEB), and Antarctic Phosphatase (NEB). Development of the tTA-responsive YFP reporter plasmid was described previously29. Plasmids were transformed into chemically competent TOP10 E. coli (Thermo Fisher) and grown at 37°C.
Plasmid preparation
Plasmid DNA used for transfection was prepared using the PEG precipitation method, which was previously described in detail.37
Cell culture
HEK293FT cells (Life Technologies/Thermo) were maintained at 37°C incubator and 5% CO2. Cells were cultured in DMEM (Gibco #31600-091) with 4.5 g/L glucose (1 g/L, Gibco #31600-091; 3.5 g/L additional, Sigma #G7021), 3.7 g/L sodium bicarbonate (Fisher Scientific #S233), 10% FBS (Gibco #16140-071), 6 mM L-glutamine (2 mM from Gibco 31600-091 and 4 mM from additional Gibco 25030-081), penicillin (100 U/μL), and streptomycin (100 μg/mL) (Gibco 15140122). Cells were cultured in a 37°C incubator with 5% CO2.
Transfection
Transfections were performed in 24-well plates seeded at 1.5 x 105 cells in 0.5 mL of DMEM media (for functional experiments), or 6 well plates seeded at 7 x 105 cells in 2 mL DMEM media (for Western blot experiments). At 6-8 hours post-seeding, cells were transfected using calcium phosphate method with a total DNA content of 1-2 ug DNA per mL of media (see Supplementary Table 1 for plasmid doses used in each experiment), using DNA prepared by PEG precipitation. All experiments included blue fluorescent protein (BFP) as a control to assess transfection efficiency. Plasmid DNA was mixed in H2O, and 2 M CaCl2 was added to a final concentration of 0.3 M CaCl2. This mixture was added dropwise to an equal-volume solution of 2x HEPES-Buffered Saline (280 mM NaCl, 0.5 M HEPES, 1.5 mM Na2HPO4) and gently pipetted up and down four times. After 2.5 minutes, the solution was mixed vigorously by pipetting ten times. 100 μL of this mixture was added dropwise to each well of a 24-well plate, or 200 μL was added dropwise to each well of a 12-well plate, and the plates were swirled gently. For functional experiments, 12 hours post-transfection, media containing 0.1 μM rapamycin analog (Takara AP21967) or 0.1% ethanol as a control was added to cells. At 24-30 hours post-media change, cells were harvested for flow cytometry with Trypsin-EDTA (Gibco #25300-054), which was then quenched with medium, and the resulting cell solution was added to at least 2 volumes of FACS buffer (PBS pH 7.4 with 2–5 mM EDTA and 0.1% BSA). Cells were spun at 150 x g for 5 min, FACS buffer was decanted, and fresh FACS buffer was added. All experiments were performed in biological triplicate.
Flow Cytometry
Approximately 104 live cells from each transfected well of the 24-well plate were analyzed using a BD LSR Fortessa Special Order Research Product (Robert H. Lurie Cancer Center Flow Cytometry Core) running FACSDiva software. Flow cytometer lasers and filter sets used for data acquisition are listed in Supplementary Table 2 (for experiments involving reporter expression) and Supplementary Table 3 (for experiments involving FRET). Samples were analyzed using FlowJo v10 software (FlowJo, LLC). Fluorescence data were compensated for spectral bleed-through, and additional spectral bleed-through compensation for FRET experiments was performed to compensate fluorescence of either mCerulean or mVenus out of the AmCyan (FRET) channel. As shown in Supplementary Fig. 13, the HEK293FT cell population was identified by FSC-A vs. SSC-A gating, and singlets were identified by FSC-A vs. FSC-H gating. A control sample of cells—generated by transfecting cells with a mass of pcDNA (empty vector) equivalent to the mass of DNA used in other samples in the experiment—was used to distinguish transfected and non-transfected cells. For the single-cell subpopulation of the pcDNA-only sample, a gate was made to identify cells that were positive for the constitutively driven blue fluorescent protein (BFP) used as a transfection control in other samples such that the gate included no more than 1% of the non-fluorescent cells.
Quantification of reporter output
The mean fluorescence intensity (MFI) of the single-cell transfected population was calculated and exported for further analysis. To calculate reporter expression, the FITC channel MFI was averaged across three biological replicates and cell autofluorescence was subtracted. As shown in Supplementary Fig. 14, to convert MFI to Mean Equivalents of Fluorescein (MEFLs), UltraRainbow Calibration Particles (Spherotech URCP-100-2H) were run in each individual experiment. The bead population was identified by FSC-A vs. SSC-A gating, and 9 bead subpopulations were identified through two fluorescent channels, with MEFL values corresponding to each subpopulation supplied by the manufacturer. For each experiment, a calibration curve was generated for the experimentally determined MFI vs. the manufacturer MEFL values, a linear regression was performed, and the slope of the regression was used as a conversion factor. Standard error was propagated through all calculations.
Quantification of FRET by flow cytometry
The workflow used for quantification of FRET by flow cytometry was previously reported in detail33. Briefly, the mCerulean+/mVenus+ population was distinguished from samples transfected with the transfection control (miRFP670) and pcDNA only, mCerulean only, and mVenus only, by gating such that less than 1% of the single-color samples were included. The normalized FRET (NFRET) parameter38 was defined in the FlowJo workspace, as described by equation (1):
| (1) |
Where Comp AmCyan FI is the compensated fluorescence intensity (FI) of a cell in the AmCyan channel, Comp Pacific Blue FI is the compensated fluorescence intensity of that cell in the Pacific Blue channel, and Comp FITC FI is the compensated fluorescence intensity of that cell in the FITC channel. Average NFRET metrics were calculated for negative (cytosolic mCerulean and mVenus co-transfected) and positive (membrane-tethered mCerulean-mVenus fusion protein) controls included in each experiment. A calibrated NFRET parameter was defined with these controls such that the NFRET of all samples is scaled linearly between these controls, as described by equation (2):
| (2) |
The calibrated NFRET metrics were exported for further analysis. Standard error was propagated through all calculations.
Western blotting
Western blots were performed to evaluate protein expression and normalize total expression of each TEVp chain. A 3X-FLAG tagged NanoLuciferase was used as a normalization control. A detailed Western blot protocol was previously described.37 Antibodies used are Monoclonal ANTI-FLAG M2 antibody produced in mouse (Sigma Cat F#1804; RRID: AB_262044) ; Anti-mouse IgG, HRP-linked Antibody (Cell Signaling Technology Cat# 7076; RRID: AB_330924).
Quantification and expression normalization of split TEVp chains
Scanned Western blot images were imported into ImageJ and analyzed with the analyze gel feature. The intensity for each NTEVp or CTEVp chain band was quantified and reported as the percent of the total signal from all NTEVp or CTEVp chains on the blot. The intensity of the NanoLuciferase normalization control was quantified in the same way. For the expression normalization experiment, this analysis was repeated for multiple image captures at a range of exposure times. The calculated intensity was averaged across all exposure times, and then the NTEVp or CTEVp chain intensity (expression level) was divided by the NanoLuciferase intensity (expression level). This calculated value was compared to the intensity calculated for the WT CTEVp sample, which was included as an internal control for normalization and quantification experiments.
Solvent-accessible Surface Area
The structure of TEVp was obtained from the Research Crystallography for Structural Bioinformatics (RCSB) PDB (ID code: 1LVM). Per-residue solvent-accessible surface areas (SASA) were computed using GROMACS v2018.1, which utilizes the double cubic lattice method (DCLM) described by Eisenhaber et al.39 The change in solvent accessible area was computed as
where structures of the N and C-terminal fragments were isolated from the crystal structure.
Computational Interface Scanning
All modeling calculations were performed using the Rosetta molecular modeling suite v3.9. Single-point mutants were generated using the standard Relax application, which enables local conformational sampling to minimize energy (Supplementary Note 1 includes full details). The total energy (ΔGTotal) of each mutant was computed as the average of 100 relaxed models.40 The energy perturbation to total energy was computed as
The Rosetta Scripts application with the InterfaceAnalyzeMover was applied to each relaxed model to evaluate the average residue-residue interaction energies between the N- and C-terminal fragments (Supplementary Note 1 includes the full details). The interfacial energy was computed as the pair-wise sum of all short-range interaction energies as shown by
where i and j denote the sets of residues within each fragment. The energy perturbation of each mutation to the interfacial energy was then computed as
Phenotype Classifier
Experimentally characterized variants were assigned class labels (not-inducible, inducible and dead) based on reporter expression levels in the ligand-absent and ligand-induced states. Variants with ≥1.2 fold higher reporter expression in the ligand-induced state relative to the ligand-absent state were labeled as inducible. For variants with expression levels <5% of wild-type (WT) sequence in the ligand-induced state and <1.2 fold activation were classified as functionally dead. The remaining variants were assigned the not-inducible class label.
STATISTICAL ANALYSIS
Statistical details for each experiment are included in the figure legends. The data shown reflect the mean across these biological replicates of the mean fluorescence intensity (MFI) of approximately 2,000–3,000 single, transfected cells. Error bars represent the SEM (standard error of the mean). For statistical analyses, two-tailed Student’s t-tests were used to evaluate whether a significant difference exists between two groups of samples, and the reported comparisons meet the two requirements of this test: (1) the values compared are expected to be derived from a normal distribution, and (2) the variance of each group is expected to be comparable to that of the comparison group since the same transfection methodologies and data collection methods were used for all samples that were compared. A p value of ≤ 0.05 was considered to be statistically significant.
DATA AVAILABILITY
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request. Raw experimental data and computation generated data for main text figures are provided in Source Data. Raw experimental data for supplementary figures are provided in Supplementary Dataset 1. Plasmid maps are provided in Supplementary Dataset 2, and annotated descriptions of all plasmids are in Supplementary Dataset 3. The structure of TEVp was obtained from the Research Crystallography for Structural Bioinformatics (RCSB) PDB (ID code: 1LVM). A subset of plasmids used in this study will be made available on Addgene, including complete and annotated GenBank files, at https://www.addgene.org/Joshua_Leonard/.
CODE AVAILABILITY
Rosetta details and script are provided in Supplementary Note 1. Linear discriminate analysis details and script are provided in Supplementary Software 1. Jupyter notebook code used to make Figure 4a is provided in Supplementary Software 2.
Supplementary Material
Supplementary Data 1 Raw data for supplementary figures
Supplementary Software 1 Linear discriminate analysis script. Note that this is a text file with a file extension indicating that it is for use with R.
Supplementary Software 2 Jupyter notebook code for Figure 4a. Note that this is a text file with a file extension indicating that it is for use with Jupyter.
Supplementary Data 2 Archive of plasmid maps
Supplementary Data 3 List of all plasmids and annotation of key features
ACKNOWLEDGEMENTS
This work was supported in part by the National Institute of Biomedical Imaging and Bioengineering of the NIH under Award Number 1R01EB026510 (JNL); the Northwestern University Flow Cytometry Core Facility supported by Cancer Center Support Grant (NCI 5P30CA060553); T.B.D was supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG). J.D.B. and A.N.P. were supported by the National Science Foundation through Graduate Research Fellowships. J.D.B. and W.K.C. were supported in part by the National Institutes of Health Training Grant (T32GM008449) through Northwestern University’s Biotechnology Training Program. This work is also supported in part by the Great Lakes Bioenergy Research Center, U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0018409 (S.R and A.T.M). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, Department of Defense, Department of Energy or other federal agencies.
Footnotes
COMPETING INTERESTS
J.N.L is a co-inventor on a patent that covers the MESA technology used in this manuscript (US Patent 9,732,392 B2).
REFERENCES
- 1.Romei MG & Boxer SG Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Annual Review of Biophysics 48, 19–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shekhawat SS & Ghosh I Split-protein systems: beyond binary protein-protein interactions. Curr Opin Chem Biol 15, 789–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehr MC & Rossner MJ Split protein biosensor assays in molecular pharmacological studies. Drug Discovery Today 21, 415–429 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Muller J & Johnsson N Split-ubiquitin and the split-protein sensors: chessman for the endgame. Chembiochem 9, 2029–38 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Paulmurugan R & Gambhir SS Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem 75, 1584–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon AS et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem Biol 11, 400–8 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Ozawa T, Kaihara A, Sato M, Tachihara K & Umezawa Y Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Analytical Chemistry 73, 2516–2521 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Gray DC, Mahrus S & Wells JA Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell 142, 637–46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao XJ, Chong LS, Kim MS & Elowitz MB Programmable protein circuits in living cells. Science 361, 1252–1258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fink T et al. Design of fast proteolysis-based signaling and logic circuits in mammalian cells. Nature Chemical Biology 15, 115–122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B, Volz SE & Zhang F A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature biotechnology 33, 139–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nihongaki Y, Otabe T, Ueda Y & Sato M A split CRISPR–Cpf1 platform for inducible genome editing and gene activation. Nature Chemical Biology 15, 882–888 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Paulmurugan R, Umezawa Y & Gambhir SS Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci U S A 99, 15608–13 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fetchko M & Stagljar I Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods 32, 349–62 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Pandey N, Nobles CL, Zechiedrich L, Maresso AW & Silberg JJ Combining random gene fission and rational gene fusion to discover near-infrared fluorescent protein fragments that report on protein-protein interactions. ACS Synth Biol 4, 615–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones KA et al. Development of a Split Esterase for Protein–Protein Interaction-Dependent Small-Molecule Activation. ACS Central Science (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wehr MC, Reinecke L, Botvinnik A & Rossner MJ Analysis of transient phosphorylation-dependent protein-protein interactions in living mammalian cells using split-TEV. BMC Biotechnol 8, 55 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camacho-Soto K, Castillo-Montoya J, Tye B & Ghosh I Ligand-gated split-kinases. J Am Chem Soc 136, 3995–4002 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Camacho-Soto K, Castillo-Montoya J, Tye B, Ogunleye LO & Ghosh I Small molecule gated split-tyrosine phosphatases and orthogonal split-tyrosine kinases. J Am Chem Soc 136, 17078–86 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Dagliyan O et al. Computational design of chemogenetic and optogenetic split proteins. Nature Communications 9, 4042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silberg JJ, Endelman JB & Arnold FH SCHEMA-guided protein recombination. Methods Enzymol 388, 35–42 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Nguyen PQ, Liu S, Thompson JC & Silberg JJ Thermostability promotes the cooperative function of split adenylate kinases. Protein Engineering, Design and Selection 21, 303–310 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Lindman S, Hernandez-Garcia A, Szczepankiewicz O, Frohm B & Linse S In vivo protein stabilization based on fragment complementation and a split GFP system. Proceedings of the National Academy of Sciences 107, 19826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dantas G et al. High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J Mol Biol 366, 1209–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin S, Ding F & Dokholyan NV Eris: an automated estimator of protein stability. Nature Methods 4, 466–467 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Masso M & Vaisman II. Accurate prediction of stability changes in protein mutants by combining machine learning with structure based computational mutagenesis. Bioinformatics 24, 2002–9 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Lee T-S & York DM Computational Mutagenesis Studies of Hammerhead Ribozyme Catalysis. Journal of the American Chemical Society 132, 13505–13518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Y et al. Directed Evolution of Split APEX2 Peroxidase. ACS Chem Biol 14, 619–635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daringer NM, Dudek RM, Schwarz KA & Leonard JN Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth Biol 3, 892–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wehr MC et al. Monitoring regulated protein-protein interactions using split TEV. Nature Methods 3, 985–993 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Crescitelli R et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles 2(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yen H-CS, Xu Q, Chou DM, Zhao Z & Elledge SJ Global Protein Stability Profiling in Mammalian Cells. Science 322, 918–923 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Edelstein HI et al. Elucidation And Refinement Of Synthetic Receptor Mechanisms. Synthetic Biology (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapust RB, Tozser J, Copeland TD & Waugh DS The P1' specificity of tobacco etch virus protease. Biochem Biophys Res Commun 294, 949–55 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Hartfield RM, Schwarz KA, Muldoon JJ, Bagheri N & Leonard JN Multiplexing Engineered Receptors for Multiparametric Evaluation of Environmental Ligands. Acs Synthetic Biology 6, 2042–2055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz KA, Daringer NM, Dolberg TB & Leonard JN Rewiring human cellular input-output using modular extracellular sensors. Nat Chem Biol 13, 202–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES: METHODS-ONLY
- 37.Donahue PS et al. The COMET toolkit for composing customizable genetic programs in mammalian cells. Nature Communications 11, 779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z & Liu Y Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophysical journal 81, 2395–2402 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenhaber F, Lijnzaad P, Argos P, Sander C & Scharf M The double cubic lattice method: Efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. Journal of Computational Chemistry 16, 273–284 (1995). [Google Scholar]
- 40.Alford RF et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J Chem Theory Comput 13, 3031–3048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1 Raw data for supplementary figures
Supplementary Software 1 Linear discriminate analysis script. Note that this is a text file with a file extension indicating that it is for use with R.
Supplementary Software 2 Jupyter notebook code for Figure 4a. Note that this is a text file with a file extension indicating that it is for use with Jupyter.
Supplementary Data 2 Archive of plasmid maps
Supplementary Data 3 List of all plasmids and annotation of key features
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request. Raw experimental data and computation generated data for main text figures are provided in Source Data. Raw experimental data for supplementary figures are provided in Supplementary Dataset 1. Plasmid maps are provided in Supplementary Dataset 2, and annotated descriptions of all plasmids are in Supplementary Dataset 3. The structure of TEVp was obtained from the Research Crystallography for Structural Bioinformatics (RCSB) PDB (ID code: 1LVM). A subset of plasmids used in this study will be made available on Addgene, including complete and annotated GenBank files, at https://www.addgene.org/Joshua_Leonard/.