Abstract
Tumor microenvironment denotes the non-cancerous cells and components presented in the tumor, including molecules produced and released by them. The constant interactions between tumor cells and the tumor microenvironment play decisive roles in tumor initiation, progression, metastasis, and response to therapies. The tumor microenvironment as a therapeutic target in cancer has attracted great research and clinical interest. Here we summarize the current progress in targeting the tumor microenvironment in both drug development and clinical trials; highlight challenges in targeting the tumor microenvironment to achieve therapeutic efficacy; explore new technologies and approaches to better decipher the tumor microenvironment; and discuss strategies to intervene in the pro-tumor microenvironment and maximize therapeutic benefits.
Keywords: Tumor microenvironment, target, therapy, drug, resistance
1. Introduction
Cancer is a genetic disease driven by the accumulation of mutations in cancer cells. Even though non-cancerous cells in tumors, such as stromal cells and immune cells, have been known for quite some time to play critical roles in tumor progression and therapeutic responses, attention has been predominantly focused on cancer cells. However, only 5–10% of cancer cases can be attributed to genetic defects, with 90–95% of all cancer cases having their roots in the environment and lifestyle1. Over time, cancer has been recognized as an evolutionary and ecological process2, involving constant, dynamic, and reciprocal interactions between cancer cells and the tumor microenvironment (TME). The TME comprises all the non-cancerous host cells in the tumor, including fibroblasts, endothelial cells, neurons, adipocytes, adaptive, and innate immune cells, as well as its non-cellular components, including the extracellular matrix (ECM) and soluble products such as chemokines, cytokines, growth factors, and extracellular vesicles.
The acquisition and maintenance of the hallmarks of cancer—such as sustaining proliferative signaling, resisting cell death, inducing angiogenesis, activating invasion and metastasis, triggering tumor-promoting inflammation, and avoiding immune destruction—depend, to various degrees, on the contributions from the TME. This reliance on the TME offers an opportunity for therapeutic intervention by targeting TME elements or its signaling pathways. Given the increased understanding of the crucial roles of the TME in tumor development and therapeutic resistance, many efforts have been devoted to targeting components of the TME to achieve therapeutic benefits in cancer patients. There is a significant therapeutic advantage in targeting the TME compared with directly targeting cancer cells, because cancer cells are prone to drug resistance due to their genomic instability, whereas non-tumor cells in the TME have a genetically more stable nature and are more vulnerable. On the other hand, therapies targeting the TME must be specifically directed to cancer-related phenotypic changes in the non-tumor cells to avoid potential adverse effects caused by targeting normal healthy cells in other tissues. To achieve this requires an in-depth understanding of the differences between the pro-tumor host cells in the TME and normal host cells at the molecular and cellular levels. However, many non-tumor host cells present in the TME have a broad spectrum functional status, which brings challenges to deciphering the mechanisms and identifying targets for intervention in the pro-tumor TME.
In this review, we summarize current progress in targeting the TME in both drug development and clinical trials, highlight the challenges and opportunities, and discuss new technologies and strategies that can be adapted to exploring the roles of TME and developing drugs for therapeutic targeting of the TME.
2. Targeting tumor microenvironment: from bench to bedside
2.1. Targeting tumor-infiltrating T-cells
Because targeting immune checkpoints on tumor-infiltrating T-cells to booster anti-tumor immunity and the functions of T-cells have been discussed extensively in many reviews3–8, we will not reassess these here.
2.2. Targeting cancer-associated fibroblasts and the extracellular matrix
Among the stromal cells present in the TME, cancer-associated fibroblasts (CAFs) are the most abundant9. CAFs are critically involved in tumorigenesis, metastasis, angiogenesis, immune evasion, and drug resistance via cell-cell contact, secretion of abundant regulatory molecules and extracellular vesicles, and particularly, synthesizing and remodeling the extracellular matrix (ECM)10. Due to their abundance and well-established pro-tumor roles in many tumor types, CAFs have been promising therapeutic targets for cancer intervention, and numerous drugs targeting CAFs have been developed and tested in preclinical studies.
2.2.1. The roles of CAFs and ECM in tumor progression and therapeutic resistance
CAFs constitute a complex population of cells with wide varieties of cells-of-origin, heterogeneous phenotypes, and diverse functions10, all of which are also shared by many other non-cancerous host cells in the TME. Pro-tumor activities of CAFs have been reported in prostate cancer11, breast cancer12,13, pancreatic cancer14, and colorectal cancer15. As an example, CAF-derived CXC-chemokine ligand 12 (CXCL12; also known as stromal cell-derived factor 1, SDF-1) promoted breast cancer cell proliferation and rapid tumor growth via binding to CXC-chemokine receptor 4 (CXCR4) expressed on breast cancer cells12. A recent study found that activated pancreatic stellate cells (PSCs, resident pancreatic fibroblasts) released a key paracrine factor, leukemia inhibitory factor (LIF), which facilitated pancreatic cancer progression by inhibition of cancer cell differentiation16. In addition to secreted factors, CAFs also enhance cancer cell invasion and metastasis through remodeling ECM or cell adhesions. Many ECM components, such as certain types of collagens17 and matrix metalloproteinases (MMPs)18, stimulate invasion and metastasis by promoting cancer cell epithelia-mesenchymal transition (EMT)19 or collective invasion of cancer cells20. Mechanically, active adhesion between N-cadherin on CAFs and E-cadherin on cancer cells drives cooperative tumor invasion21. Recent in-depth studies of the heterogeneous subsets of CAFs revealed some previous unappreciated functions of CAFs, including antigen presenting22, depletion of tumor antigen-specific cytotoxic T lymphocytes (CTLs)23, and tumor suppression9, suggesting the multiplex functions of CAFs.
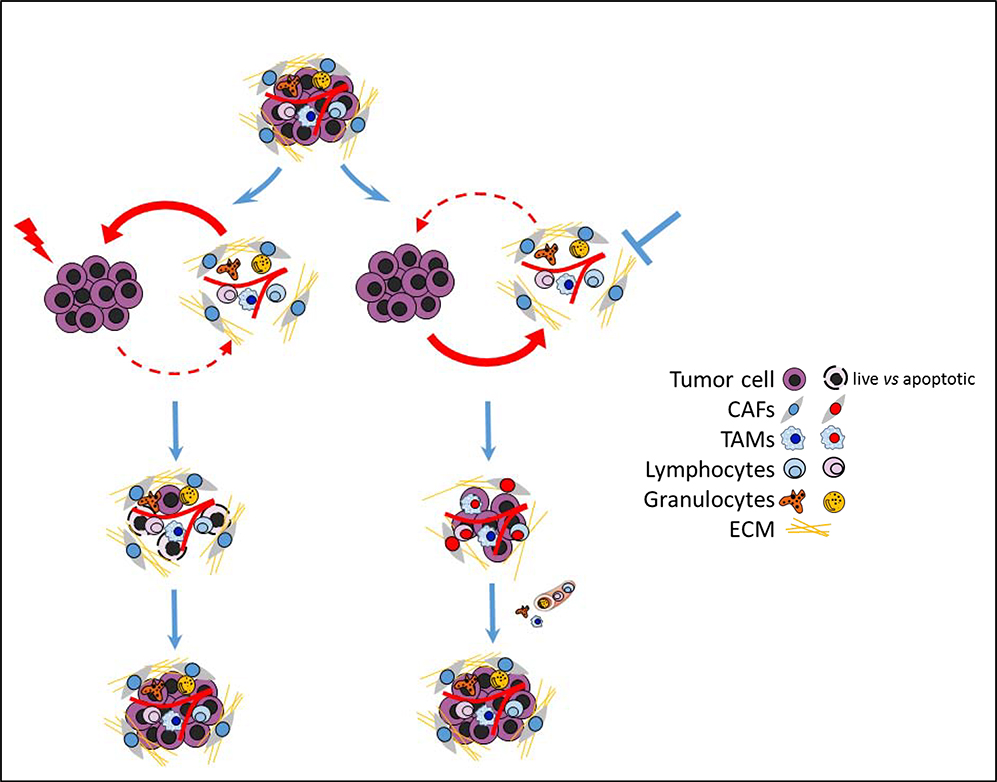
Numerous studies have also shown that CAFs, in addition to their extensive influences on tumor development, confer cancer chemotherapy resistance24 (Fig.1, left). One of the most well-known examples is pancreatic ductal adenocarcinoma (PDAC), which manifests a unique fibrotic TME and is quite resistant to chemotherapy25. Some of the CAF-mediated resistant mechanisms include delivery of exosomes stimulating cancer cell survival26; promoting cancer cell EMT, which decreases expression of transporters responsible for drug uptake27; and scavenging chemo drug to reduce the amount of intra-tumoral chemotherapy drug28. CAFs also contribute to targeted therapy resistance. For example, CAF-derived hepatocyte growth factor (HGF) elicits innate resistance of BRAF-mutant cancer cells to RAF inhibitors29. Evidence also indicates that CAFs are involved in immune evasion and resistance to immunotherapies. For example, fibroblast activation protein (FAP)-positive CAFs secrete CXCL12, which protected PDAC cancer cells from anti-tumor T-cells and caused unresponsiveness to immune checkpoint blockade (ICB) therapies in mouse PDAC models30. Interestingly, a recent study also found that a distinct ECM gene signature is correlated with immune evasion and immunotherapy failure31, suggesting critical roles of CAFs in responses to immunotherapy.
Fig.1.
Schematic diagram of targeting tumor cell or TME only and their potential resistant mechanisms. Left: Targeting tumor cell only (such as chemotherapies) kills majority tumor cells. However, the residue tumor cells may survive with the help from TME, leading to tumor relapse. Right: Targeting TME can inhibit recruitment and activation of pro-tumor cells (stromal cells and immune cells) and enhance anti-tumor responses. However, the TME will be reconstituted by tumor cells via active recruitment and programming of bone marrow derived cells or local resident stromal/immune cells.
2.2.2. Developing drugs targeting CAFs and ECM
Various drugs targeting CAFs or ECMs are under preclinical and/or clinical studies (Table 1). Among the potential targets identified in CAFs, FAP is the most extensively studied. FAP is a serine protease expressed on activated CAFs in greater than 90% of human cancers32. FAP-targeting immunotoxin successfully depleted FAP+ CAFs in vivo and showed potent tumor inhibitory activities in multiple cancer models33,34. Other approaches to target FAP also have been developed, including DNA vaccine35 and chimeric antigen receptor (CAR) T cells36. However, contradictory results generated from different groups raised cautions in the targeting of FAP. For example, the multipotent bone marrow stromal cells also express FAP and can be killed by immunotoxin, leading to potential toxicity and cachexia37. In addition, FAP-targeting immunotoxin failed in an early phase II trial (NCT02198274) in patients with advanced colorectal cancer due to limited therapeutic response38. Another potential surface target of CAF is GPR77, which is specifically expressed in a subset CAF population with pro-tumor and chemo-resistance activities39. Similar to FAP, GPR77+ CAFs can be selectively targeted by specific antibodies for deletion, which led to a significantly improved response to chemotherapy39. Other surface targets of CAF include vitamin D receptor (VDR)40 and platelet-derived growth factor receptor (PDGFR)41, both of which can be targeted by small molecules to enhance responses to chemotherapy. VDR has been identified as a crucial suppressor of CAF activation and vitamin D analogs induced notable stromal remodeling in preclinical PDAC models40. Several Phase I/II clinical trials are conducted to investigate if paricalcitol, an analog of vitamin D2, will improve response to chemotherapies or immunotherapies in PDAC patients (Table 1) and most results are still pending. Platelet-derived growth factors (PDGF) facilitate the formation of a prominent tumor stroma via recruiting/ activating CAFs and blockade of stromal PDGFR may benefit cancer treatment42. However, PDGFR inhibitor Imatinib failed to show therapeutic benefit in patients with advanced stage PDAC in a multi-institutional Phase 2 clinical trial (NCT00161213)43. In addition to surface targets, CAF-derived cytokines (IL644), chemokines (CXCL1212), and growth factors (LIF16) that support tumor cells or the pro-tumor TME also can be blocked to achieve therapeutic benefit. For example, CXCL12 derived from CAFs plays a critical role in local immunosuppression by excluding T-cells from infiltration. The CXCR4 inhibitor AMD3100, which blocks CXCL12 and CXCR4 interaction, restored anti-tumor immunity by enhancing infiltration of CD8+ T-cells into tumor30. A recent phase IIa COMBAT clinical trial (NCT02826486) indicated that the combination of pembrolizumab (anti-PD1) and BL-8040 (motixafortide), a CXCR4 antagonist, expanded the benefit of chemotherapy in PDAC45. Importantly, BL-8040 treatment effectively reprogramed TME, characterized by increased CD8+ effector T-cell infiltration and decreased immunosuppressive cell populations, including myeloid-derived suppressor cells (MDSCs) and circulating regulatory T-cells45. Given their critical roles in promoting tumor progression, ECM molecules such as tenascin C46, hyaluronan47, and MMPs48 have also attracted much attention in drug development. In several phase I and II clinical trials47,49,50, recombinant human hyaluronidase (PEGPH20), which depleted hyaluronan in tumors, showed promising clinical activities in pancreatic cancer and non-small cell lung cancer, especially hyaluronan high tumors. However, a recent phase III HALO-301 trial of PEGPH20 (NCT02715804) missed it’s primary endpoint in patients with metastatic pancreas cancer and further development of PEHPH20 was halted51. This failure suggests that targeting desmoplasia alone in PDAC treatment may not be sufficient. Other clinical trials targeting LOXL252 and MMPs53 are also mostly disappointing. Nevertheless, although many potential targets of CAFs have been identified and drugs targeting them have shown some promising results in preclinical tests, only a few such drugs have moved into the clinic in the end (Table 1)32.
Table 1.
Selected drugs potentially targeting cancer-associated fibroblasts (CAFs) or the extracellular matrix (ECM) in preclinical and clinical studies
| Drugs | Target and mechanism | Cancer types | National Clinical Trial number | Status | Ref. |
|---|---|---|---|---|---|
| Sibrotuzumab | 131I-labeled anti-FAP mAb | Colorectal, non-small cell lung, breast, or head and neck cancers |
NCT02198274
NCT02209727 |
Phase I | 183 |
| Calcipotriol Paricalcitol | Vitamin D analogue | Early-stage skin cancer, breast cancer, pancreatic cancer |
NCT03596073
NCT04617067 NCT02030860 NCT03138720 NCT04054362 |
Phase I/II | 40 |
| Pamrevlumab (FG-3019) | Anti-CTGF mAb | Pancreatic cancer | NCT03941093 | Phase III | 184 |
| Plerixafor (AMD3100) BL-8040 (motixafortide) | CXCR4 receptor antagonist | Pancreatic cancer |
NCT04177810
NCT02179970 NCT02826486 NCT03193190 |
Phase I/II | 30,45 |
| IPI-926 | Smoothened inhibitor | Pancreatic cancer | NCT01130142 | Phase I | 185 |
| S-3304 | MMP inhibitor | Advanced solid tumors |
NCT00078390
NCT00033215 |
Phase I | 48 |
| 131I-m81C6 | 131I-labeled anti-tenascin mAb | Brain tumors |
NCT00002752
NCT00003461 |
Phase II | 186 |
| Imatinib | PDGFR inhibitor | Advanced solid tumors |
NCT00161213
NCT00281996 NCT01048320 NCT00485485 |
Phase I/II | 187 |
| GS-6624 (simtuzumab) | LOXL2 mAb | Pancreatic cancer |
NCT01472198
NCT01479465 |
Phase II | 52 |
| Tetrathiomolybdate | Copper chelator, target LOX |
Breast cancer, prostate cancer |
NCT00195091
NCT00150995 NCT00405574 |
Phase II | 188 |
| pegvorhyaluronidase alfa (PEGPH20; PVHA) | Recombinant Human Hyaluronidase | Lung cancer, Pancreatic cancer, |
NCT01453153
NCT02563548 NCT01839487 NCT02715804 |
Phase I/II/III |
2.3. Targeting tumor-associated macrophages
Tumor-associated macrophages (TAMs), which are abundant in the TME of most cancer types, are generally associated with poor clinical outcome in cancer patients54,55. The established pro-tumoral roles of TAMs include stimulating pro-tumor inflammation, facilitating cancer cell immune evasion, promoting angiogenic switch, and accelerating tumor cell invasion/metastasis. Because TAMs are one of the dominant innate immune cell populations in tumors, the reciprocal interactions between cancer cells and TAMs shape the tumor immune landscape. TAMs are emerging as a key target in immunotherapy.
2.3.1. The roles of TAMs in tumor progression and therapeutic resistance
As an important source of cytokines/chemokines, TAMs are critically involved in the initiation and maintenance of chronic inflammation, which is associated with tumorigenesis and tumor progression56. For example, TAM-derived pro-inflammatory cytokines such as IL-1β57, IL-658, and IL-2359,60 promoted tumor growth and progression in colorectal cancer and other cancer types. The chronic inflammation is induced and maintained by the feed-forward networks formed between cancer cells and TAMs, which may be also involved with mucosal barrier defects and complex interactions with the microbiome54,61. In addition to pro-tumor inflammation, TAMs also play a central role in suppression of adaptive antitumor immunity55. For example, TAMs express elevated levels of immune checkpoint ligands, such as PD-L1, PD-L262, and VISTA63, which can suppress T-cell activities directly. Furthermore, TAMs can recruit regulatory T-cells (T-reg) and facilitate expansion of T-reg in the TME, ultimately resulting in suppression of T-cell immunity64. Angiogenic switch, which refers to the induction of a tumor vasculature, is required for tumor growth and expansion65. TAMs promote angiogenic switch by delivering critical pro-angiogenic factors66, including vascular endothelial growth factors (VEGF)67, IL-868, and MMP9 69, which reinforce the recruitment and activation of endothelial cells and other cells supporting the generation of the vascular networks. In addition, a subset of TAMs, which express the angiopoietin-1 receptor TIE2 (TIE2+) and cluster around tumor blood vessels, regulates angiogenesis in various mouse tumor models and is correlated with micro-vessel density in human tumors70. Given the essential roles of TAMs in stimulating the pro-tumor TME, it is anticipated that metastatic cancer cells will attract bone marrow-derived monocytes, which differentiate into TAMs once extravasated from blood vessels. Indeed, highly invasive cancer cells secrete elevated levels of CCL2 and/or CSF1 to recruit and expand TAMs, which then engage in vicious cycles with cancer cells and promote cancer cell migration71, intravasation72, and outgrowth in the metastatic sites73. Our team has found that PTEN loss in brain metastases leads to an increased secretion of CCL2, which attracted CCR2+ (CCL2 receptor) microglial cells into the brain; CCL2-knowkdown MDA-MB-231 cells significantly prolonged survival of mice bearing brain metastases74.
Accumulation of TAMs has been frequently observed in tumors that have received various therapeutic treatments75, including radiotherapy76, chemotherapy77, and anti-VEGF therapy78. The protective role of TAMs in response to therapeutic treatment was confirmed by enhanced therapeutic efficacy when recruitment or polarization of TAMs was blocked79. Distinct mechanisms may be involved in TAM-mediated therapeutic resistance, including but not limited to restoration of vascularization and suppression of cytotoxic T-lymphocytes. A recent study found that the metabolites released by TAMs in PDAC, particularly the pyrimidines, inhibited gemcitabine through molecular competition and resulted in chemotherapy resistance80. TAMs also contribute to adaptive resistance to targeted therapy in cancer. As an example, treatment of melanoma patients with MAPK pathway inhibitor increased TAMs in the tumor; consequently, TAMs-derived TNFα as a crucial melanoma growth factor caused resistance to MAPK pathway inhibitors81. As one of the dominant immunosuppressive cell populations in the TME, TAMs are an important determinant of response to immunotherapy55. TAMs prevented cytotoxic CD8+ T-cells from reaching the tumors and led to unresponsiveness to anti-PD1 immunotherapy82.
2.3.2. Developing drugs targeting TAMs
Drugs targeting monocytes/macrophages with ongoing clinical trials are summarized in Table 2. CSF1R signaling has attracted the most attention so far because CSF1/CSF1R plays a central role in the production, differentiation, and function of macrophages. Inhibitors (PLX3397, JNJ-40346527, PLX7486, and ARRY-382) and neutralizing antibodies (RG7155, IMC-CS4, and FPA008) targeting CSF1R successfully depleted TAMs and microglia in preclinical animal models and cancer patients54,83. Some of the inhibitors and antibodies under clinical evaluation induced anti-tumor responses in patients with glioma, lymphoma, or other advanced-stage solid tumors. Among the CSF1R inhibitors, PLX3397 is currently under investigation in multiple clinical trials. PLX3397 was well tolerated but showed no efficacy in glioblastomas (NCT01349036) 84. Another clinical study of PLX3397 and Pembrolizumab (anti-PD1) (NCT02452424) to treat advanced melanoma and other solid tumors also terminated early due to lack of clinical efficacy (https://clinicaltrials.gov/). Similarly, CSF1R neutralizing antibody FPA008 did not meet its primary end point of progression-free survival in a phase II clinical trial in patients with advanced pancreatic cancer (NCT03336216, https://bwnews.pr/391gi3X). In addition, the potential risk and toxicity caused by depletion of all the macrophages, including normal resident macrophages, need to be carefully evaluated54. The CCL2-CCR2 axis is another focus of developing drugs targeting TAMs. CCL2 has been identified as a potent chemoattractant for monocytes/macrophages. Despite the fact that anti-CCL2 monoclonal antibody (mAb) was able to significantly reduce TAMs in the tumor during treatment, withdrawal of the antibody accelerated lung metastasis in several breast cancer mouse models due to fast rebound of monocyte recruitment82. Additionally, compensatory and redundant mechanisms leading to the failure of CCL2-CCR2 axis blockade were uncovered in preclinical models54, suggesting that a better understanding of monocyte recruitment/retention is necessary before performing clinical trials. A phase 1b study of PF-04136309, a small molecule antagonist of human CCR2, in combination with chemotherapies (NCT02732938) raised concern for synergistic pulmonary toxicity and did not show extra benefit over chemotherapies in treatment of metastatic PDAC85. On the other hand, an anti-CCR2 antibody (MLN1202) tested in a phase II clinical trial (NCT01015560) for metastatic cancer showed therapeutic effects in 14 out of 43 patients with bone metastases85. In addition to depletion of TAMs or blocking recruitment of monocytes, there have also been attempts to develop drugs that can reprogram TAMs, such as agonistic anti-CD40 antibodies86, histone deacetylase inhibitors87, and PI3Kγ inhibitors88,89. The goal is to convert TAMs from a pro-tumor phenotype to an anti-tumor phenotype, which includes releasing TAM-mediated immunosuppression of adaptive immunity, abolishing pro-angiogenic function, and interrupting the bad cycle between TAMs and tumor cells. Despite that the antitumor effects of targeting CD40 for monotherapy or combination therapy have been observed in some tumors, the therapeutic efficacy has been moderate and heterogeneous90. Macrophage PI3Kγ controls an important switch between immune stimulation and suppression during inflammation and cancer89. Initial clinical results from IPI-549, a PI3Kγ inhibitors, combined with nivolumab (anti-PD1) in advanced solid tumors (NCT02637531) demonstrated favorable tolerability, evidence of immune stimulation, and early signs of clinical activity91. CD47-mediated “do not eat me” signaling is another potential target to invigorate anti-tumor activities of macrophages. CD47, which is highly expressed on tumor cells, interacts with thrombospondin 1 and signal regulatory protein-α (SIRPα) expressed by macrophages, activating the “do not eat me” signal and protecting tumor cells from phagocytosis by macrophages92. Blockade of the CD47-SIRPα axis by anti-CD47 antibodies93 or recombinant SIRPα-crystallizable fragment (Fc) fusion protein94 increased tumoricidal activity of macrophages in preclinical studies and early clinical trials. Gilead’s Magrolimab (Hu5F9-G4), a first-in-class investigational anti-CD47 mAb, is tested in multiple clinical trials. A phase Ib/II clinical study of magrolimab in combination with cetuximab (anti-EGFR) showed encouraging responses in colorectal cancer patients95.
Table 2.
Drugs targeting monocyte/macrophage populations in preclinical and clinical studies
| Drugs | Target and mechanism | Cancer types | National Clinical Trial number | Status | Ref. |
|---|---|---|---|---|---|
| FPA008, PLX3397, IMC-CS4, AMG 820, PLX7486, | CSF1R blocking antibodies or inhibitors | Advanced tumors, pancreatic cancer |
NCT01346358
NCT02452424 NCT03336216 NCT01349036 |
Phase I/II | 83,173 |
| MLN1202, PF-04136309 | CCR2 blocking antibodies or antagonists | Bone metastasis, pancreatic cancer |
NCT01015560
NCT01413022 NCT02732938 |
Phase II | 83,189 |
| CP-870,893, APX005M | CD40 agonist antibodies | Melanoma, breast cancer, non-small cell lung cancer, renal cell cancer, esophageal cancer, brain tumor |
NCT02157831
NCT01456585 NCT03165994 NCT03123783 NCT03719430 NCT04130854 NCT02482168 |
Phase I/II | 90,190 |
| IPI-549 | PI3Kγ inhibitor | Advanced solid tumors |
NCT02637531
NCT03961698 |
Phase I/II | 91 |
| Hu5F9-G4, CC-95251, SRF231 | SIRPα or CD47 blocking antibodies, target CD47/SIRPα axis | Lymphoma, leukemia, colorectal cancer |
NCT03783403
NCT02953782 NCT03558139 NCT02216409 |
Phase I/II | 191 |
| AZD9150 | Stat3 inhibitors | Advanced pancreatic, non-small cell lung cancer, colorectal cancer |
NCT03421353
NCT03394144 NCT01839604 NCT03334617 NCT02499328 |
Phase II | 192 |
| Reparixin, AZD5069, SX-682 | CXCR1/2 antagonist | Metastatic breast cancer, Colon cancer, Prostate cancer |
NCT02001974
NCT02370238 NCT02499328 NCT04599140 |
Phase I/II | 193 |
2.4. Targeting tumor-associated neutrophils
Neutrophils, which originate from myeloid precursors, are the most abundant population of white blood cells in peripheral blood and the first responders of innate immunity96,97. Tumor-associated neutrophils (TANs) constitute an important part of the TME and are actively involved in tumor progression and metastasis. Many preclinical studies have demonstrated the pro-tumor roles of TANs, which are achieved partially by stimulating the ECM and inflammation of the TME. Neutrophil-released granules consist of various proteases, including matrix metalloprotease 9 (MMP-9)98, neutrophil elastase99, and cathepsin G100, which can remodel ECM and promote tumor invasion101. Besides proinflammatory cytokines/chemokines, TANs also produce immunosuppressive factors, such as arginase 1 and TGFβ, which suppress adaptive immunity102, and release growth factors, including HGF103, which support tumor progression. Recent studies found that not only TANs in the TME but also neutrophils in the peripheral blood facilitate tumor progression and metastasis104. Interactions between circulating tumor cells (CTC) and neutrophils in the bloodstream drive tumor cell cycle progression104, stimulate tumor cell extravasation105, and significantly enhance metastatic potential of CTC. Consistent with these findings, a high circulating neutrophil-to-lymphocyte ratio (NLR) in patients represented a poor prognostic factor in breast, colon, liver, and many other types of cancer96,102. Targeting of neutrophils has emerged as a potential therapeutic approach for cancer patients106. One of the most promising targets is CXCR2, a critical regulator for neutrophil mobilization107. Small inhibitors or antibodies blocking interactions between CXCR2 and its ligand (CXCL8), which showed anti-tumor activities and enhanced responses to chemotherapies in preclinical studies, are under clinical evaluation106. Several clinical trials of SX-682, a potent inhibitor of CXCR1/2, are proposed to test whether SX-682 can block cancers from attracting MDSCs and enhance therapeutic efficacy in combination with immunotherapies108. Recently, our team revealed that EZH2 can be phosphorylated at tyrosine (Y) 696 by Src, and p-696-EZH2 works as a transcription factor to upregulate G-CSF, which recruits immunosuppressive neutrophils into brain metastases lesions; G-CSF antibodies, and combinatorial immune checkpoint blockade plus Src inhibitors can block neutrophil infiltration to inhibit brain metastasis109.
2.5. Targeting tumor-promoting chronic inflammation
Inflammation is a fundamental innate immune response to disturbed tissue homeostasis110. Chronic inflammation is recognized as one of the hallmarks of cancer111, which fuels tumorigenesis and promotes cancer progression. Anti-inflammatory drugs, including aspirin, significantly reduced cancer risks in large population studies112,113, suggesting that inflammation is a promising target for cancer therapy. Macrophages, which are the prime source of inflammation, and tumor cells both can produce proinflammatory cytokines and inflammatory mediators, which sustain tumor cell proliferation and survival58, immune evasion114, angiogenesis115, ECM remodeling116, metastasis56, chemoresistance117, and radioresistance118. Targeting either the key mediators involved in proinflammatory pathways, such as primary inflammatory cytokines (e.g., IL-1, TNF, IL-6), or the master regulators of the inflammatory response, such as transcription factors NF-κB and STAT3, may inhibit cancer-promoting inflammation119. TGFβ is also an attractive target since it is a master regulator of chronic inflammation. Different classes of TGFβ inhibitors, including neutralizing, and bifunctional antibodies, receptor kinase inhibitors, antisense oligonucleotides, and TGFβ-related vaccines have been developed and tested in multiple clinical trials.120 However, despite extensive efforts in > 15 years since the first clinical trial was engaged, the translation of TGFβ inhibitors from bench to bedside has been slow and challenging. For example, the antitumor activity of PF-03446962, a fully human IgG2 monoclonal antibody that blocks TβRI, has been investigated and failed in hepatocellular carcinoma121, urothelial cancer122, malignant pleural mesothelioma123, and metastatic colorectal cancer124. Galunisertib (LY2157299), a first-in-class small inhibitor of TGFβRI, didn’t show clinical benefit in patients with malignant glioma in two separate studies125,126. With ongoing clinical trials of next generation TGβRI inhibitors and bifunctional antibodies combining TGFβ and immune checkpoint inhibition120, TGFβ is now considered an appealing therapeutic target in the era of immunotherapy. Another option is targeting macrophages, because they are the major source of inflammatory factors127. Currently, some antibodies/inhibitors have shown anti-tumor activities in preclinical studies and a few of them have been explored in early-stage clinical trials54. A challenge of targeting inflammation is how to achieve selective inhibition of pro-tumor chronic inflammation without ruining anti-tumor immunity.
2.6. Targeting tumor angiogenesis
Angiogenesis is considered essential for tumor development and progression. Many antiangiogenic drugs have been developed and tested in the clinic to treat various human malignancies. However, antiangiogenic therapy provides only transitory improvements in a small subset of cancer patients128. One major reason for failure of the therapy is that the hypoxia TME, which is prompted by disruption of tumor vessels, drives tumor neovascularization and regrowth. On the other hand, more in-depth studies found that antiangiogenic therapies, such as inhibition of VEGF and the VEGF receptor (VEGFR), create a more mature and functional vasculature, which will enhance drug delivery and efficacy129. Therefore, targeting tumor angiogenesis may have potential in combinatory therapies.
2.7. Targeting other components of the tumor microenvironment
Other components in the TME, including B-lymphocytes130,131, Treg132, adipocytes133–135, mesenchymal stem cells136, and exosomes137,138, also influence tumor progression and therapeutic responses. For example, adipocytes can promote ovarian cancer metastasis and chemoresistance via adipokine release and lipid transfer133,134. However, the functions and mechanisms of these components underlying their relationships with cancer cells are much less appreciated compared with those discussed above, and how to maneuver them to achieve therapeutic effects remains unclear.
3. Challenges of targeting the tumor microenvironment
3.1. Paradox: friends or foes?
To develop effective therapies targeting the TME, it is critical to determine the roles of non-cancerous host cells and non-cellular components in the TME. Host cells or non-cellular components with pro-tumor roles need to be restrained by antagonists to block their activities or reduce their amounts, while cells or components with anti-tumor functions could be targeted by agonists to enhance their activities or increase their amounts. One of the greatest challenges to targeting the TME is that various host cells or non-cellular components within the TME may have contradictory relationships with cancer cells. In addition to the conventional pro-tumor roles of CAFs and TANs, recent studies found some populations of CAFs and TANs have tumor-suppressive activities. Depletion of alpha smooth muscle actin positive (α-SMA+) CAFs in PDAC mouse models increased cancer stem cells (CSC) and induced more aggressive tumors that ultimately diminished survival139. Similarly, targeting tumor-derived sonic hedgehog signaling, which drove the desmoplasia in PDAC, accelerated tumor progression despite reduced fibrotic contents140. These data suggest that at least some CAFs may restrain PDAC growth. Studies on TANs also resulted in controversy regarding the role of neutrophils in cancer141–143. In fact, TANs with a pro-tumor phenotype (N2) would switch to an anti-tumor phenotype (N1) upon blockade of TGFβ signaling, suggesting multifaceted and dynamic roles of TANs144. Although it is still not clear whether certain subtypes of macrophages can restrain tumor growth, the anti-tumor efficacy of anti-CD47 in preclinical studies indicates potential tumoricidal activity of TAMs94. In sum, the paradoxical roles of TME components discovered in recent years indicate important knowledge gaps in our understanding of the fundamental components of the TME. This may be a reason why attempts to eliminate CAFs or TAMs largely failed to show efficacy in cancer clinical trials as monotherapies. How to distinguish the contradictory functions of TME components and precisely target subsets with desired functionality remains a huge challenge.
3.2. Complexity: heterogeneous and dynamic phenotypes
One major barrier to deciphering and targeting the TME is its complexity. The TME comprises numerous cell types, each of which consists of heterogeneous subsets with various phenotypes and diverse functions. Recent studies have found that the immune cells in the TME show significantly increased heterogeneity of cell states and marked phenotypic expansions compared with those in blood and normal tissues145. For example, 14 unique myeloid and 17 unique T-cell clusters were found in human breast tumor tissues, double the number of observed clusters in normal breast tissue145. The heterogeneity of non-cancerous cells likely derives from a wide variety of cells-of-origin and the exposure of cells to diverse local microenvironments, which differ in the extent of hypoxia, inflammation, nutrient supply, growth factors, and other inputs.
Conventional classification, which separates non-cancerous cells into a few mutually exclusive categories marked by discrete activation or differentiation status (polarization), has been widely adopted to analyze CD4+ T helper cells (Th1 vs. Th2), macrophages (M1 vs. M2), and neutrophils (N1 vs. N2). However, the latest evidence from CyTOF and single-cell sequencing indicated that the conventional classification, based mostly on markers generated in in vitro experiments, may utterly oversimplify the phenotypic complexity of non-cancerous cell populations in the TME146,147. Recent analysis of T-cells in the TME revealed that T-cells reside along a broad continuum of activation status145. The phenotypic heterogeneity of T-cells in the TME is likely shaped by a combination of antigen TCR stimulation and local microenvironmental stimuli, which together may lead to more discrete states of T-cells despite the fact that many of the responses individually (e.g., inflammation, hypoxia) represent phenotypic continuums. The myeloid cells in the TME are even more complicated in their heterogeneity148, and their effect on tumor progression remains poorly characterized. Analysis of TAMs in human breast cancer at single-cell resolution challenged not only the traditional macrophage polarization model (M1 vs. M2) but also the refined model in which TAMs reside along a spectrum between the M1 and M2 states145. Instead, this analysis suggested the co-existence of M1 and M2 states in TAMs145.
The TME is not static but undergoes massive dynamic changes during tumor development. However, little is known about these changes of TME components in human cancer. Additionally, cancer therapies, including radiotherapy, chemotherapy, targeted therapy, and immunotherapy, have enormous impact on the TME. For example, chemotherapies can eliminate most neutrophils in the TME149. On the other hand, chemotherapeutic agents can influence multiple aspects of TAMs150, such as recruitment of monocytes/macrophages to the tumor sites, depletion of monocyte/macrophage linages, and regulation of macrophage phenotypes. Immunotherapy also has broad impacts on the TME, including myeloid cells151. Whether and how these impacts on TME will affect patient responses to treatment is still largely unknown.
The complexity of the TME imposes vast challenges to a full understanding of the TME components and structure, and to development of effective drugs to target them. However, recent technological advances, including single-cell omics and artificial intelligence (AI), provide unprecedented opportunities to unbiasedly decipher the complexities of the TME at the single-cell level on a genome scale.
4. Profiling and deciphering the tumor microenvironment
4.1. Profiling the tumor microenvironment
Attempts to dissect the complexity of the TME have motivated development of computational approaches, such as deconvolution methods, to integrate the estimation of type-specific expression profiles of tumor cells, immune cells, and the tumor stroma. However, small subsets of TME components are either invisible or only partially characterized when interrogated using standard analyses that average data across a bulk population of cells. The ongoing revolution in single-cell omics, with single-cell RNA sequencing (scRNA-seq) leading the way, addresses the problem and provides novel and important insights into highly complex and dynamic biological systems, such as the TME.
Characterization of single-cell programs at the genome scale allows more precisely the deciphering of heterogeneous cellular subsets in the TME and uncovers previously unappreciated functions. As an example, scRNA-seq identified a subset of CAFs characterized by LRRC15 expression (LRRC15+) and TGF-β signaling activation as a determinant of resistance to immunotherapy152. Similarly, antigen-presenting CAFs (apCAF), which express MHC II and are capable of activating CD4+ T cells in an antigen-specific fashion, were found in both a mouse PADC model and human PDACs for the first time22. These findings highlight the need to revise the cell subset structures based on unbiased analyses of scRNA-seq data instead of limited protein marker expression defined by previous studies.
Single-cell sequencing allows profiling of small quantities of tumor cells and TME cellular constituents simultaneously, which makes it valuable to study co-evolution of tumor cells and the TME during tumor development. Analysis of pancreatic cancer precursors at single-cell resolution revealed increased proinflammatory immune components in the TME at an early stage, which was progressively depleted and taken over by stromal myofibroblast populations during neoplastic progression153. Profiling of patients’ melanoma at single-cell resolution uncovered two distinct tumor cell states, Melanocyte inducing transcription factor (MITF) dominant and AXL dominant, which corresponded to distinct tumor microenvironmental patterns154, including specific interactions between cancer cells and their TME. Single-cell sequencing can also be applied to track the dynamic changes of the TME during therapeutic treatment. Using paired scRNA-seq and T-cell receptor sequencing of cells from patients with basal or squamous cell carcinoma before and after anti-PD-1 therapy, T-cells responding to checkpoint blockade were found to mainly derived from a distinct repertoire of T-cell clones that had not been observed before treatment, instead of pre-existing tumor-infiltrating T lymphocytes155. Taken together, single-cell sequencing of tumor cells and the TME cells will provide immense insights on tumor evolution, patients’ tumor classification, and guidance for cancer therapies.
The phenotype and function of cells in the TME may be highly dependent on a cell’s exact spatial location and interaction with adjacent cells. Coupling scRNA-seq with spatial information offers in-depth mechanistic insights into cell-cell interactions and relationships among different cellular components. Direct quantification of individual RNA in cells can be achieved in intact tissue by fluorescence in situ hybridization (FISH)156,157. Advances in spatially resolved technologies for tumor profiling, including multiplex tissue imagine158, as well as the integration of data from disparate technologies, will significantly improve our understanding of cell-cell interactions and spatial effects in the TME159,160.
Along with scRNA-seq, new methods and technologies to profile genetic, epigenetic, proteomic, spatial, and lineage information in individual cells have been invented and are advancing rapidly148,161. These single-cell multiomics technologies can reveal cellular heterogeneity at multiple molecular layers. Integrative analysis of multiomics data will provide profound novel insights into the fundamental mechanisms driving cellular heterogeneity and help to identify targetable cellular subsets/signaling essential for cancer cell adaptation to the TME162.
4.2. Organ chips for dissection of local TME
Animal models are precious for elucidating functions and mechanisms of the TME, but they do not always faithfully recapitulate human-relevant tissue and organ microenvironment. As a result, preclinical animal models generally perform poorly in predicting therapeutic responses in human clinical trials163. Even when orthotopic models effectively display some human cancer behaviors, it is difficult to decipher cellular interactions and physiological processes due to the lack of temporal resolution and sensitivity of human tissues in most animal models. To overcome these challenges, human organs-on-chips (also known as organ chips) based on microfluidic cell culture technology provide opportunities to model cancer cell behaviour within the human-relevant TME in vitro. Organ chips are microfluidic cell culture devices containing perfused hollow microchannels made from optically clear plastic, glass, or flexible polymers164. The microchannels are inhabited by tumor cells, normal epithelial cells, stromal cells, endothelial cells, and immune cells under conditions mimicking in vivo organ-level physiology and pathophysiology by recreating tissue-level and organ-level structures and functions in vitro165. Organ chips enable dissection of the local TME and study of the effects of a single microenvironmental component on cancer cells, cultured either alone or in various combinations with different cell types, numerous mechanical cues, and even microbiota, while visualizing key steps of the human cancer cascade, such as angiogenesis and invasion/metastasis, and responses to therapy in real time and a controlled manner at high resolution. As an example, a study using a lung cancer chip found that healthy alveolar epithelium promoted human non-small-cell lung cancer cell proliferation, while secreted factors from endothelial cells partially suppressed tumor growth166. In a breast cancer chip, a complex HER2+ breast cancer ecosystem, composed of cancer cells, endothelial cells, CAFs, and immune cells, was reconstituted and the therapeutic response to trastuzumab was visualized ex vivo in real time167. Along with more and more scRNA-seq data generated from human tissues/organs and patients’ cancer samples, the emerging single cell expression atlas of human organs/tissues will guide the research and development of more human-relevant next-generation organ chips. The incorporation of human organ chips into drug development pipelines will significantly accelerate discovery of new drugs targeting the human TME, which could not have been realized using traditional in vitro systems and available preclinical animal models.
4.3. Artificial intelligence (AI) empowerment of TME research
Artificial intelligence (AI) has shown immense potential for recognizing patterns in large amounts of high dimensional data, deciphering relationships among complex features in big data (including images), and identifying characteristics in big data, all of which are critical for understanding the complex TME. It is conceivable that AI technology will become a key component of TME research to manage and analyze big data describing the highly heterogeneous and convoluted TME. AI-based analysis will be especially useful for extracting quantitative information about the TME from using whole-slide histopathologic images168,169, flow and mass cytometry, and bulk and single-cell transcriptomic approaches. Integration of AI technology and multiplex biomarker analysis will enable analysts to gain deep insights into the TME170. Using deep learning (DL), a subset of machine learning (ML) in AI technology that applies artificial neural networks to learn from data without supervision, Joel et al. analyzed tumor-infiltrating lymphocytes (TILs) based on H&E images from 13 TCGA tumor types171. DL helped to uncover relationships between TIL patterns and tumor genetic features, immune landscape, cancer type, and patient outcome. Another study applied an ML approach to analyze immune-related gene expression in patients with non-small cell lung cancer and identified an immune-related gene signature that successfully predicted patients’ responses to immune checkpoint blockade therapies172. New paradigms arising from the incorporation of ML, organ chip technology, time-lapse microscopy analysis, and single-cell transcriptomic approaches may achieve reconstitution of human TME ex vivo and potentially diminish the gaps between in vivo and similar ex vivo scenarios. Furthermore, DL may even help to recapitulate and predict patients’ therapeutic responses via in silico experiments.
5. Strategies to target the pro-tumor microenvironment
5.1. Elimination
Elimination of pro-tumor elements in the TME has been considered an effective way to target the TME and has been widely investigated in preclinical and clinical studies. For example, treatment with anti-CCL2 antibody or anti-CSF1R antibody can significantly reduce the amount of TAMs in the TME, whereas anti-FAP antibody is able to specifically deplete FAP+ CAFs. However, some concerns have been raised recently in regards to its potential toxicity, off-target effects, and long term effects. Anti-FAP antibody targeted not only CAFs but potentially also muscle cells and bone marrow cells37. Similarly, anti-CSF1R antibody treatment depleted tissue resident macrophages, such as microglia in brain and Kupffer cells in liver173, in addition to TAMs in the tumor. Elimination of these normal cells may induce toxicity during treatment. Moreover, depletion of an entire population of cells in the TME may eliminate potential subsets that have anti-tumor activities or are critical for therapeutic response. One example is eradication of SMA+ CAFs in pancreatic cancer, which led to more aggressive cancer139, suggesting that some subsets of SMA+ CAFs might suppress PDAC cells174. Depletion of TAMs in the TME may also remove some macrophages with tumoricidal activities. Furthermore, emerging evidence indicates that macrophages in the TME play critical roles for anti-tumor immunity and immunotherapy response, suggesting that depletion of all the macrophages in the TME may not be totally beneficial55. Lastly, the long-term effects of the elimination procedure, including the compensatory response, may lead to the failure of the treatment. To achieve long-term therapeutic efficacy, cellular homeostasis will be a great challenge for monotherapies using elimination procedure (Fig 1, right).
5.2. Normalization
Other than direct elimination of pro-tumor elements, it is appealing to target the TME by blocking the pro-tumor activities of the TME or even reprogram the pro-tumor phenotype/function of the TME to an anti-tumor phenotype/function. The idea of “normalization” is to revert the overall tumor-favoring TME to a normal tissue environment, which generally restrains early tumor development and improves the delivery and efficacy of anticancer therapeutics175. To reach this goal requires mechanistic insights into the pro-tumor phenotype/function in the TME. Some molecules/pathways contributing to the pro-tumor phenotypes/functions of TME have been identified and targeted to “normalize” TME. Blockade of GPR77-mediated signaling abolished pro-tumor activities of CAFs and made cancer cells vulnerable to chemotherapy39. Another example is PI3Kγ, which is highly expressed in myeloid cells in the TME, and inhibition of PI3Kγ signaling switched the immunosuppressive phenotype of TAMs to an immunostimulatory transcriptional program that restored CD8+ T cell anti-tumor immunity89. PD1 is also considered a target to achieve “immune normalization” for negating tumor-induced immune deficiency selectively in the tumor microenvironment176. Compared with elimination, normalization strategy intervenes to tip the balance toward anti-tumor activities by selective targeting of pro-tumor signaling in the TME, which reduces overall potential toxicity and off-target effects. However, whether “normalization” of one pro-tumor signaling pathway will be sufficient to convert the TME and achieve therapeutic efficacy is not clear. Meanwhile, given the high complexity and heterogeneity of the TME, different normalization approaches may be needed to target various subtypes of the TME. The recent discovery of six different immune subtypes of cancer across multiple cancer types has shed some light on the inter-tumor heterogeneity of the tumor immune microenvironment177. Biomarker-guided normalization approaches may be necessary to achieve precise targeting of the TME and improve therapeutic efficacy.
5.3. Targeting tumor cells to intervene in the pro-tumor microenvironment
Despite the advantages of targeting the TME directly, such an approach might achieve only transient and limited efficacy as monotherapy due to cellular homeostasis of the TME (Fig. 1, right). In the presence of cancer cells, which can recruit and manipulate non-cancerous cells, the TME will be replenished after withdrawal of the treatment. Without targeting cancer cells, it is difficult to achieve broad and sustainable effects on TME.
Cancer cells are the major driving force behind co-evolution of cancer cells and the TME. Many studies have indicated that cancer cells facilitate cancer progression by manipulating a cancer-favoring microenvironment. Compared with targeting TME directly, targeting cancer cells may treat the root cause of the pro-tumor TME instead of the symptoms178. Some oncogenic/suppressive signaling may modulate the pro-tumor TME, in addition to their well-known cell autonomous functions. For example, KRAS mutation in pancreatic cancer cells activates the Sonic Hedgehog (SHh) signaling, which drives CAF activation and promotes the synthesis of ECM components and MMPs179. Tp53 loss or mutation in cancer cells is associated with reduced CTLs and NK cells in TME180, which can accelerate cancer cell immune evasion. Recent study indicated that loss of Tp53 in head and neck cancer also drives adrenergic transdifferentiation of tumor-associated sensory nerves, which in turn stimulate tumor progression181. NF1 loss in glioblastoma cells correlates with increased TAMs and altered immune landscape in glioma182. Despite these emerging findings, intrinsic drivers dictating stromal and immune contexture of the tumors are largely unknown.
The latest advances in technology offer unparalleled opportunities to investigate the biomarkers and intrinsic determinants of the TME by delineating the multi-omics profiles of tumor cells and the TME. Analysis of genetic alterations in cancer cells with matched tumor stromal/immune landscape may yield in-depth mechanistic insights and enable rational decision-making in the clinical use of personalized intervention strategies for cancer patients. Instead of normalizing one signaling pathway in the TME, targeting the intrinsic determinants of tumor stromal and immune landscape in cancer cells may rewire the over-all entire communication networks between cancer cells and the TME, which may switch TME from pro-tumor to antitumor activities.
6. Summary and perspectives
As the key element of the evolutionary and ecological process in cancer development and cancer therapy, the TME has been attracting more and more attention in research and drug development. Recent advances in cutting-edge technologies, such as single-cell multi-omics and AI, enable deciphering the TME via multi-omics profiling and will significantly reduce our current knowledge gaps. Emerging evidence suggests that in many cases, targeting elements of the TME alone may be inadequate for executing broad and sustainable therapeutic efficacy in cancer patients.
Despite important progress made in the past decade, many clinical trials targeting the TME have failed to show promising efficacy in cancer patients. The only exception is immunotherapy, including immune checkpoint blockade therapies. The success of developing immunotherapies from bench to clinic highlights several key points. Perhaps the first is the importance of basic research. The breakthrough of immunotherapies emerged from a careful deciphering of basic biology that spanned many years, including fundamental mechanisms of T-cell activation and inhibition. Our current understanding of fibroblasts, macrophages, and other TME elements has not reached a level similar to that for T-cells. Lack of an in-depth understanding of the fundamental mechanisms of these TME elements impedes the discovery and development of novel drugs targeting TME. The second point concerns biomarker-guided therapy. Immunotherapies, such as anti-PD1/PDL1 treatment, show significantly more efficacy in the treatment of tumors with pre-existing anti-tumor immunity, such as more T-cell infiltration and higher levels of IFNγ and PDL1 expression in tumors. Similarly, given the high heterogeneity of the TME, development of reliable biomarkers to guide TME-targeted therapies will be essential to achieve clinical efficacy. The third clue is the value of the combinatory approach. Despite remarkable and durable efficacy in some cancer patients who received immunotherapies, currently a majority of cancer patients do not benefit from immunotherapies. It has been recognized that combinatory therapy is promising for improving response to immunotherapy. TME-targeted therapies may have to be combined with other therapies to maximize efficacy and benefit more patients. With all these insights from success in immunotherapies, we expect that TME-targeted therapy will not take long to achieve a breakthrough and reach its first milestone.
Acknowledgements
We thank Michael Worley of Scientific Publications of MD Anderson Cancer Center (MDACC) for manuscript editing. This work was supported by National Institutes of Health (NIH) grants R01-CA112567-06 (D.Y.), R01CA184836 (D.Y.), R01CA208213 (D.Y.), NIH Cancer Center Support Grant P30CA016672 to MDACC, Duncan Family Institute Seed Funding Research Program, and METAvivor Research Award #56675. Dr. Yu is the Hubert L. & Olive Stringer Distinguished Chair in Basic Science, MDACC.
Abbreviations
- α-SMA
alpha smooth muscle actin
- AI
artificial intelligence
- CAF
cancer-associated fibroblast
- CAR
chimeric antigen receptor
- CSC
cancer stem cell
- CTC
circulating tumor cell
- CTL
cytotoxic T lymphocytes
- CXCL12
CXC-chemokine ligand 12
- CXCR4
CXC-chemokine receptor 4
- DL
deep learning
- ECM
extracellular matrix
- EMT
epithelia-mesenchymal transition
- FAP
fibroblast activation protein
- FISH
fluorescence in situ hybridization
- HGF
hepatocyte growth factor
- ICB
immune checkpoint blockade
- LIF
leukemia inhibitory factor
- mAb
monoclonal antibody
- MDSC
myeloid-derived suppressor cell
- MITF
melanocyte inducing transcription factor
- ML
machine learning
- MMP
matrix metalloproteinase
- NLR
neutrophil-to-lymphocyte ratio
- PDAC
pancreatic ductal adenocarcinoma
- PDGF
platelet-derived growth factor
- PDGFR
platelet-derived growth factor receptor
- PSC
pancreatic stellate cell
- scRNA-seq
single-cell RNA sequencing
- SHh
Sonic Hedgehog
- SIRP
signal regulatory protein
- TAM
tumor associated macrophage
- TAN
Tumor-associated neutrophil
- TIL
tumor-infiltrating lymphocytes
- TME
tumor microenvironment
- VDR
vitamin D receptor
- VEGF
vascular endothelial growth factor
Footnotes
Conflict of Interest Statement:
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anand P et al. Cancer is a Preventable Disease that Requires Major Lifestyle Changes. Pharm Res-Dordr 25, 2097–2116, doi: 10.1007/s11095-008-9661-9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merlo LMF, Pepper JW, Reid BJ & Maley CC Cancer as an evolutionary and ecological process. Nature Reviews Cancer 6, 924–935, doi: 10.1038/nrc2013 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Savas P et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol 13, 228–241, doi: 10.1038/nrclinonc.2015.215 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Pardoll DM The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews Cancer 12, 252–264, doi: 10.1038/nrc3239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentles AJ et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Cancer Research 75, doi: 10.1158/1538-7445.Transcagen-Pr09 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joyce JA & Fearon DT T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80, doi: 10.1126/science.aaa6204348/6230/74 [pii] (2015). [DOI] [PubMed] [Google Scholar]
- 7.Postow MA, Callahan MK & Wolchok JD Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol 33, 1974–U1161, doi: 10.1200/Jco.2014.59.4358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribas A & Wolchok JD Cancer immunotherapy using checkpoint blockade. Science 359, 1350-+, doi: 10.1126/science.aar4060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R The biology and function of fibroblasts in cancer. Nature Reviews Cancer 16, 582–598, doi: 10.1038/nrc.2016.73 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Chen XM & Song EW Turning foes to friends: targeting cancer-associated fibroblasts. Nature Reviews Drug Discovery 18, 99–115, doi: 10.1038/s41573-018-0004-1 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Olumi AF et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Research 59, 5002–5011 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orimo A et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121, 335–348, doi: 10.1016/j.cell.2005.02.034 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Shekhar MPV, Werdell J, Santner SJ, Pauley RJ & Tait L Breast stroma plays a dominant regulatory role in breast epithelial growth and differentiation: Implications for tumor development and progression. Cancer Research 61, 1320–1326 (2001). [PubMed] [Google Scholar]
- 14.Hwang RF et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res 68, 918–926, doi: 10.1158/0008-5472.CAN-07-5714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henriksson ML et al. Colorectal cancer cells activate adjacent fibroblasts resulting in FGF1/FGFR3 signaling and increased invasion. Am J Pathol 178, 1387–1394, doi: 10.1016/j.ajpath.2010.12.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature 569, 131–135, doi: 10.1038/s41586-019-1130-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Provenzano PP et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. Bmc Med 4, doi:Artn 3810.1186/1741-7015-4-38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaggioli C et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol 9, 1392–U1392, doi: 10.1038/ncb1658 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Zhang K et al. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol 15, 677-+, doi: 10.1038/ncb2743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedl P & Wolf K Tube travel: The role of proteases in individual and collective a cancer cell invasion. Cancer Research 68, 7247–7249, doi: 10.1158/0008-5472.Can-08-0784 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Labernadie A et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol 19, 224–237, doi: 10.1038/ncb3478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elyada E et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov 9, 1102–1123, doi: 10.1158/2159-8290.Cd-19-0094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakins MA, Ghorani E, Munir H, Martins CP & Shields JD Cancer-associated fibroblasts induce antigen-specific deletion of CD8(+) T Cells to protect tumour cells. Nature Communications 9, doi:ARTN 94810.1038/s41467-018-03347-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennen WN, Isaacs JT & Denmeade SR Rationale Behind Targeting Fibroblast Activation Protein-Expressing Carcinoma-Associated Fibroblasts as a Novel Chemotherapeutic Strategy. Mol Cancer Ther 11, 257–266, doi: 10.1158/1535-7163.Mct-11-0340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erkan M et al. The role of stroma in pancreatic cancer: diagnostic and therapeutic implications. Nat Rev Gastro Hepat 9, 454–467, doi: 10.1038/nrgastro.2012.115 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Richards KE et al. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 36, 1770–1778, doi: 10.1038/onc.2016.353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530, doi: 10.1038/nature16064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hessmann E et al. Fibroblast drug scavenging increases intratumoural gemcitabine accumulation in murine pancreas cancer. Gut 67, 497–507, doi: 10.1136/gutjnl-2016-311954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straussman R et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504, doi: 10.1038/nature11183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feig C et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A 110, 20212–20217, doi: 10.1073/pnas.1320318110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakravarthy A, Khan L, Bensler NP, Bose P & De Carvalho DD TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun 9, 4692, doi: 10.1038/s41467-018-06654-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X & Song E Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 18, 99–115, doi: 10.1038/s41573-018-0004-110.1038/s41573-018-0004-1 [pii] (2019). [DOI] [PubMed] [Google Scholar]
- 33.Ostermann E et al. Effective immunoconjugate therapy in cancer models targeting a serine protease of tumor fibroblasts. Clin Cancer Res 14, 4584–4592, doi: 10.1158/1078-0432.CCR-07-5211 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Fang JX et al. A potent immunotoxin targeting fibroblast activation protein for treatment of breast cancer in mice. Int J Cancer 138, 1013–1023, doi: 10.1002/ijc.29831 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duperret EK et al. Alteration of the Tumor Stroma Using a Consensus DNA Vaccine Targeting Fibroblast Activation Protein (FAP) Synergizes with Antitumor Vaccine Therapy in Mice. Clinical Cancer Research 24, 1190–1201, doi: 10.1158/1078-0432.Ccr-17-2033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang LCS et al. Targeting Fibroblast Activation Protein in Tumor Stroma with Chimeric Antigen Receptor T Cells Can Inhibit Tumor Growth and Augment Host Immunity without Severe Toxicity. Cancer Immunol Res 2, 154–166, doi: 10.1158/2326-6066.Cir-13-0027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts EW et al. Depletion of stromal cells expressing fibroblast activation protein-alpha from skeletal muscle and bone marrow results in cachexia and anemia. J Exp Med 210, 1137–1151, doi: 10.1084/jem.20122344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofheinz RD et al. Stromal antigen targeting by a humanised monoclonal antibody: an early phase II trial of sibrotuzumab in patients with metastatic colorectal cancer. Onkologie 26, 44–48, doi: 10.1159/000069863 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Su S et al. CD10(+)GPR77(+) Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 172, 841–856 e816, doi: 10.1016/j.cell.2018.01.009 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Sherman MH et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 159, 80–93, doi: 10.1016/j.cell.2014.08.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pietras K, Pahler J, Bergers G & Hanahan D Functions of paracrine PDGF signaling in the proangiogenic tumor stroma revealed by pharmacological targeting. PLoS Med 5, e19, doi: 10.1371/journal.pmed.0050019 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderberg C et al. Paracrine signaling by platelet-derived growth factor-CC promotes tumor growth by recruitment of cancer-associated fibroblasts. Cancer Res 69, 369–378, doi: 10.1158/0008-5472.CAN-08-2724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss RA et al. A Multi-institutional Phase 2 Study of Imatinib Mesylate and Gemcitabine for First-Line Treatment of Advanced Pancreatic Cancer. Gastrointest Cancer Res 5, 77–83 (2012). [PMC free article] [PubMed] [Google Scholar]
- 44.Studebaker AW et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res 68, 9087–9095, doi: 10.1158/0008-5472.CAN-08-0400 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Bockorny B et al. BL-8040, a CXCR4 antagonist, in combination with pembrolizumab and chemotherapy for pancreatic cancer: the COMBAT trial. Nat Med 26, 878-+, doi: 10.1038/s41591-020-0880-x (2020). [DOI] [PubMed] [Google Scholar]
- 46.O’Connell JT et al. VEGF-A and Tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proc Natl Acad Sci U S A 108, 16002–16007, doi: 10.1073/pnas.1109493108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hingorani SR et al. Phase Ib Study of PEGylated Recombinant Human Hyaluronidase and Gemcitabine in Patients with Advanced Pancreatic Cancer. Clin Cancer Res 22, 2848–2854, doi: 10.1158/1078-0432.CCR-15-2010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiappori AA et al. A phase I pharmacokinetic and pharmacodynamic study of s-3304, a novel matrix metalloproteinase inhibitor, in patients with advanced and refractory solid tumors. Clin Cancer Res 13, 2091–2099, doi: 10.1158/1078-0432.CCR-06-1586 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Hingorani SR et al. HALO 202: Randomized Phase II Study of PEGPH20 Plus Nab-Paclitaxel/Gemcitabine Versus Nab-Paclitaxel/Gemcitabine in Patients With Untreated, Metastatic Pancreatic Ductal Adenocarcinoma. J Clin Oncol 36, 359–366, doi: 10.1200/JCO.2017.74.9564 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Ward JP et al. Efficacy and safety of pegvorhyaluronidase alfa (PEGPH20; PVHA) and pembrolizumab (pembro) combination therapy in patients (Pts) with stage III/IV non-small cell lung cancer (NSCLC). Ann Oncol 30 (2019). [Google Scholar]
- 51.Hakim N, Patel R, Devoe C & Saif MW Why HALO 301 Failed and Implications for Treatment of Pancreatic Cancer. Pancreas (Fairfax) 3, e1–e4, doi: 10.17140/POJ-3-e010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benson AB 3rd et al. A Phase II Randomized, Double-Blind, Placebo-Controlled Study of Simtuzumab or Placebo in Combination with Gemcitabine for the First-Line Treatment of Pancreatic Adenocarcinoma. Oncologist 22, 241–e215, doi: 10.1634/theoncologist.2017-0024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winer A, Adams S & Mignatti P Matrix Metalloproteinase Inhibitors in Cancer Therapy: Turning Past Failures Into Future Successes. Mol Cancer Ther 17, 1147–1155, doi: 10.1158/1535-7163.Mct-17-0646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cassetta L & Pollard JW Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov 17, 887–904, doi: 10.1038/nrd.2018.169 (2018). [DOI] [PubMed] [Google Scholar]
- 55.DeNardo DG & Ruffell B Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 19, 369–382, doi: 10.1038/s41577-019-0127-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grivennikov SI, Greten FR & Karin M Immunity, inflammation, and cancer. Cell 140, 883–899, doi: 10.1016/j.cell.2010.01.025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greten FR et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118, 285–296, doi: 10.1016/j.cell.2004.07.013 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Grivennikov S et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113, doi: 10.1016/j.ccr.2009.01.001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langowski JL et al. IL-23 promotes tumour incidence and growth. Nature 442, 461–465, doi: 10.1038/nature04808 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Kortylewski M et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 15, 114–123, doi: 10.1016/j.ccr.2008.12.018 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grivennikov SI et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258, doi: 10.1038/nature11465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umezu D et al. Inhibitory functions of PD-L1 and PD-L2 in the regulation of anti-tumor immunity in murine tumor microenvironment. Cancer Immunol Immunother 68, 201–211, doi: 10.1007/s00262-018-2263-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blando J et al. Comparison of immune infiltrates in melanoma and pancreatic cancer highlights VISTA as a potential target in pancreatic cancer. P Natl Acad Sci USA 116, 1692–1697, doi: 10.1073/pnas.1811067116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noy R & Pollard JW Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 41, 49–61, doi: 10.1016/j.immuni.2014.06.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bergers G & Benjamin LE Tumorigenesis and the angiogenic switch. Nat Rev Cancer 3, 401–410, doi: 10.1038/nrc1093 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Lin EY et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res 66, 11238–11246, doi:0008–5472.CAN-06–1278 [pii] 10.1158/0008-5472.CAN-06-1278 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Cursiefen C et al. VEGF-A stimulates lymphangiogenesis and hemangiogenesis in inflammatory neovascularization via macrophage recruitment. J Clin Invest 113, 1040–1050, doi: 10.1172/JCI20465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koch AE et al. Interleukin-8 as a Macrophage-Derived Mediator of Angiogenesis. Science 258, 1798–1801, doi:DOI 10.1126/science.1281554 (1992). [DOI] [PubMed] [Google Scholar]
- 69.Huang SY et al. Contributions of stromal metalloproteinase-9 to angiogenesis and growth of human ovarian carcinoma in mice. J Natl Cancer I 94, 1134–1142 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Lewis CE, Harney AS & Pollard JW The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell 30, 18–25, doi: 10.1016/j.ccell.2016.05.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goswami S et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Research 65, 5278–5283, doi:Doi 10.1158/0008-5472.Can-04-1853 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Harney AS et al. Real-Time Imaging Reveals Local, Transient Vascular Permeability, and Tumor Cell Intravasation Stimulated by TIE2hi Macrophage-Derived VEGFA. Cancer Discov 5, 932–943, doi: 10.1158/2159-8290.CD-15-0012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Q, Zhang XH & Massague J Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell 20, 538–549, doi: 10.1016/j.ccr.2011.08.025S1535-6108(11)00357-6 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang L et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104, doi: 10.1038/nature15376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruffell B & Coussens LM Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472, doi: 10.1016/j.ccell.2015.02.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen FH et al. Radiotherapy decreases vascular density and causes hypoxia with macrophage aggregation in TRAMP-C1 prostate tumors. Clin Cancer Res 15, 1721–1729, doi: 10.1158/1078-0432.CCR-08-1471 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hughes R et al. Perivascular M2 Macrophages Stimulate Tumor Relapse after Chemotherapy. Cancer Res 75, 3479–3491, doi: 10.1158/0008-5472.CAN-14-3587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ferrara N Role of myeloid cells in vascular endothelial growth factor-independent tumor angiogenesis. Curr Opin Hematol 17, 219–224, doi: 10.1097/MOH.0b013e3283386660 (2010). [DOI] [PubMed] [Google Scholar]
- 79.Brown JM, Recht L & Strober S The Promise of Targeting Macrophages in Cancer Therapy. Clin Cancer Res 23, 3241–3250, doi: 10.1158/1078-0432.CCR-16-3122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Halbrook CJ et al. Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab 29, 1390–1399 e1396, doi: 10.1016/j.cmet.2019.02.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith MP et al. The Immune Microenvironment Confers Resistance to MAPK Pathway Inhibitors through Macrophage-Derived TNF alpha. Cancer Discov 4, 1214–1229, doi: 10.1158/2159-8290.Cd-13-1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peranzoni E et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc Natl Acad Sci U S A 115, E4041–E4050, doi: 10.1073/pnas.1720948115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poh AR & Ernst M Targeting Macrophages in Cancer: From Bench to Bedside. Front Oncol 8, 49, doi: 10.3389/fonc.2018.00049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Butowski N et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro Oncol 18, 557–564, doi: 10.1093/neuonc/nov245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noel M et al. Phase 1b study of a small molecule antagonist of human chemokine (C-C motif) receptor 2 (PF-04136309) in combination with nab-paclitaxel/gemcitabine in first-line treatment of metastatic pancreatic ductal adenocarcinoma. Invest New Drugs 38, 800–811, doi: 10.1007/s10637-019-00830-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khalil M & Vonderheide RH Anti-CD40 agonist antibodies: preclinical and clinical experience. Update Cancer Ther 2, 61–65, doi: 10.1016/j.uct.2007.06.001 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guerriero JL et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature 543, 428–432, doi: 10.1038/nature21409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaneda MM et al. Macrophage PI3K gamma Drives Pancreatic Ductal Adenocarcinoma Progression. Cancer Discov 6, 870–885, doi: 10.1158/2159-8290.Cd-15-1346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaneda MM et al. PI3K gamma is a molecular switch that controls immune suppression. Nature 539, 437–442, doi: 10.1038/nature19834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li DK & Wang W Characteristics and clinical trial results of agonistic anti-CD40 antibodies in the treatment of malignancies. Oncol Lett 20, 176, doi: 10.3892/ol.2020.12037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sullivan RJ et al. Initial results from first-in-human study of IPI-549, a tumor macrophage-targeting agent, combined with nivolumab in advanced solid tumors. J Clin Oncol 36, doi:DOI 10.1200/JCO.2018.36.15_suppl.3013 (2018). [DOI] [Google Scholar]
- 92.Barclay AN & van den Berg TK The Interaction Between Signal Regulatory Protein Alpha (SIRP alpha) and CD47: Structure, Function, and Therapeutic Target. Annu Rev Immunol 32, 25–50, doi: 10.1146/annurev-immunol-032713-120142 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Chao MP et al. Anti-CD47 Antibody Synergizes with Rituximab to Promote Phagocytosis and Eradicate Non-Hodgkin Lymphoma. Cell 142, 699–713, doi: 10.1016/j.cell.2010.07.044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petrova PS et al. TTI-621 (SIRP alpha Fc): A CD47-Blocking Innate Immune Checkpoint Inhibitor with Broad Antitumor Activity and Minimal Erythrocyte Binding. Clinical Cancer Research 23, 1068–1079, doi: 10.1158/1078-0432.Ccr-16-1700 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Fisher GA et al. A phase Ib/II study of the anti-CD47 antibody magrolimab with cetuximab in solid tumor and colorectal cancer patients. J Clin Oncol 38 (2020). [Google Scholar]
- 96.Shaul ME & Fridlender ZG Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol 16, 601–620, doi: 10.1038/s41571-019-0222-4 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Coffelt SB, Wellenstein MD & de Visser KE Neutrophils in cancer: neutral no more. Nature Reviews Cancer 16, 431–446, doi: 10.1038/nrc.2016.52 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Coussens LM, Tinkle CL, Hanahan D & Werb Z MMP-9 supplied by bone marrow-derived cells contributes to skin carcinogenesis. Cell 103, 481–490, doi: 10.1016/s0092-8674(00)00139-2 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tohme S et al. Neutrophil Extracellular Traps Promote the Development and Progression of Liver Metastases after Surgical Stress. Cancer Res 76, 1367–1380, doi: 10.1158/0008-5472.CAN-15-1591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Erpenbeck L & Schon MP Neutrophil extracellular traps: protagonists of cancer progression? Oncogene 36, 2483–2490, doi: 10.1038/onc.2016.406 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Dumitru CA, Lang S & Brandau S Modulation of neutrophil granulocytes in the tumor microenvironment: mechanisms and consequences for tumor progression. Semin Cancer Biol 23, 141–148, doi: 10.1016/j.semcancer.2013.02.005 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Grecian R, Whyte MKB & Walmsley SR The role of neutrophils in cancer. Brit Med Bull 128, 5–14, doi: 10.1093/bmb/ldy029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wislez M et al. Hepatocyte growth factor production by neutrophils infiltrating bronchioloalveolar subtype pulmonary adenocarcinoma: role in tumor progression and death. Cancer Res 63, 1405–1412 (2003). [PubMed] [Google Scholar]
- 104.Szczerba BM et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 566, 553–557, doi: 10.1038/s41586-019-0915-y (2019). [DOI] [PubMed] [Google Scholar]
- 105.Huh SJ, Liang S, Sharma A, Dong C & Robertson GP Transiently entrapped circulating tumor cells interact with neutrophils to facilitate lung metastasis development. Cancer Res 70, 6071–6082, doi: 10.1158/0008-5472.CAN-09-4442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Granot Z Neutrophils as a Therapeutic Target in Cancer. Frontiers in Immunology 10, doi:ARTN 171010.3389/fimmu.2019.01710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Powell DR & Huttenlocher A Neutrophils in the Tumor Microenvironment. Trends Immunol 37, 41–52, doi: 10.1016/j.it.2015.11.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Greene S et al. Inhibition of MDSC Trafficking with SX-682, a CXCR1/2 Inhibitor, Enhances NK-Cell Immunotherapy in Head and Neck Cancer Models. Clin Cancer Res 26, 1420–1431, doi: 10.1158/1078-0432.Ccr-19-2625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang L, W. Y., Zou ZF, Yao J, Li P, Qu JK, Badu-Nkansah A, Yuan XL, Chang WC, Saunus J, Lakhani S, Hung MC, Yu D. Blocking immunosuppressive neutrophils deters pY696-EZH2-driven brain metastases. In press. Science Translational Medicine (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Elinav E et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nature Reviews Cancer 13, 759–771, doi: 10.1038/nrc3611 (2013). [DOI] [PubMed] [Google Scholar]
- 111.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674, doi: 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- 112.Rothwell PM et al. Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death: analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet 379, 1602–1612, doi: 10.1016/S0140-6736(11)61720-0 (2012). [DOI] [PubMed] [Google Scholar]
- 113.Rothwell PM et al. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet 379, 1591–1601, doi: 10.1016/S0140-6736(12)60209-8 (2012). [DOI] [PubMed] [Google Scholar]
- 114.Zelenay S et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell 162, 1257–1270, doi: 10.1016/j.cell.2015.08.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ghosh N, Chaki R, Mandal V & Mandal SC COX-2 as a target for cancer chemotherapy. Pharmacol Rep 62, 233–244, doi:Doi 10.1016/S1734-1140(10)70262-0 (2010). [DOI] [PubMed] [Google Scholar]
- 116.Hojilla CV, Wood GA & Khokha R Metalloproteinases as common effectors of inflammation and extracellular matrix breakdown in breast cancer. Breast Cancer Research 10, doi:ARTN 20510.1186/bcr1980 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kelly MG et al. TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. Cancer Research 66, 3859–3868, doi: 10.1158/0008-5472.Can-05-3948 (2006). [DOI] [PubMed] [Google Scholar]
- 118.McKelvey KJ, Hudson AL, Back M, Eade T & Diakos CI Radiation, inflammation and the immune response in cancer. Mammalian Genome 29, 843–865, doi: 10.1007/s00335-018-9777-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Coussens LM, Zitvogel L & Palucka AK Neutralizing Tumor-Promoting Chronic Inflammation: A Magic Bullet? Science 339, 286–291, doi: 10.1126/science.1232227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ciardiello D, Elez E, Tabernero J & Seoane J Clinical development of therapies targeting TGFbeta: current knowledge and future perspectives. Ann Oncol 31, 1336–1349, doi: 10.1016/j.annonc.2020.07.009 (2020). [DOI] [PubMed] [Google Scholar]
- 121.Simonelli M et al. Phase I study of PF-03446962, a fully human monoclonal antibody against activin receptor-like kinase-1, in patients with hepatocellular carcinoma. Ann Oncol 27, 1782–1787, doi: 10.1093/annonc/mdw240 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Necchi A et al. PF-03446962, a fully-human monoclonal antibody against transforming growth-factor beta (TGFbeta) receptor ALK1, in pre-treated patients with urothelial cancer: an open label, single-group, phase 2 trial. Invest New Drugs 32, 555–560, doi: 10.1007/s10637-014-0074-9 (2014). [DOI] [PubMed] [Google Scholar]
- 123.Wheatley-Price P et al. A Phase II Study of PF-03446962 in Patients with Advanced Malignant Pleural Mesothelioma. CCTG Trial IND.207. J Thorac Oncol 11, 2018–2021, doi: 10.1016/j.jtho.2016.06.024 (2016). [DOI] [PubMed] [Google Scholar]
- 124.Clarke JM et al. A phase Ib study of the combination regorafenib with PF-03446962 in patients with refractory metastatic colorectal cancer (REGAL-1 trial). Cancer Chemother Pharmacol 84, 909–917, doi: 10.1007/s00280-019-03916-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brandes AA et al. A Phase II randomized study of galunisertib monotherapy or galunisertib plus lomustine compared with lomustine monotherapy in patients with recurrent glioblastoma. Neuro Oncol 18, 1146–1156, doi: 10.1093/neuonc/now009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wick A et al. Phase 1b/2a study of galunisertib, a small molecule inhibitor of transforming growth factor-beta receptor I, in combination with standard temozolomide-based radiochemotherapy in patients with newly diagnosed malignant glioma. Invest New Drugs 38, 1570–1579, doi: 10.1007/s10637-020-00910-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leon-Cabrera S, Schwertfeger KL & Terrazas LI Inflammation as a Target in Cancer Therapy. Mediat Inflamm, doi:Artn 197169810.1155/2019/1971698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rivera LB & Bergers G Tumor angiogenesis, from foe to friend. Science 349, 694–695, doi: 10.1126/science.aad0862 (2015). [DOI] [PubMed] [Google Scholar]
- 129.Jain RK Normalizing tumor vasculature with anti-angiogenic therapy: A new paradigm for combination therapy. Nature Medicine 7, 987–989, doi:DOI 10.1038/nm0901-987 (2001). [DOI] [PubMed] [Google Scholar]
- 130.Tsou P, Katayama H, Ostrin EJ & Hanash SM The Emerging Role of B Cells in Tumor Immunity. Cancer Res 76, 5597–5601, doi: 10.1158/0008-5472.CAN-16-0431 (2016). [DOI] [PubMed] [Google Scholar]
- 131.Yuen GJ, Demissie E & Pillai S B lymphocytes and cancer: a love-hate relationship. Trends Cancer 2, 747–757, doi: 10.1016/j.trecan.2016.10.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tanaka A & Sakaguchi S Regulatory T cells in cancer immunotherapy. Cell Res 27, 109–118, doi: 10.1038/cr.2016.151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nieman KM et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 17, 1498–1503, doi: 10.1038/nm.2492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang J et al. Adipocytes promote ovarian cancer chemoresistance. Sci Rep 9, 13316, doi: 10.1038/s41598-019-49649-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Quail DF & Dannenberg AJ The obese adipose tissue microenvironment in cancer development and progression. Nat Rev Endocrinol 15, 139–154, doi: 10.1038/s41574-018-0126-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ridge SM, Sullivan FJ & Glynn SA Mesenchymal stem cells: key players in cancer progression. Molecular Cancer 16, doi:ARTN 3110.1186/s12943-017-0597-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang TT et al. Functions of Exosomes in the Triangular Relationship between the Tumor, Inflammation, and Immunity in the Tumor Microenvironment. Journal of Immunology Research 2019, doi:Artn 419782910.1155/2019/4197829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhao H et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 5, e10250, doi: 10.7554/eLife.10250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ozdemir BC et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival (vol 25, pg 719, 2014). Cancer Cell 28, 831–833, doi: 10.1016/j.ccell.2015.11.002 (2015). [DOI] [PubMed] [Google Scholar]
- 140.Rhim AD et al. Stromal Elements Act to Restrain, Rather Than Support, Pancreatic Ductal Adenocarcinoma. Cancer Cell 25, 735–747, doi: 10.1016/j.ccr.2014.04.021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Finisguerra V et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature 522, 349-+, doi: 10.1038/nature14407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gershkovitz M et al. TRPM2 Mediates Neutrophil Killing of Disseminated Tumor Cells. Cancer Res 78, 2680–2690, doi: 10.1158/0008-5472.CAN-17-3614 (2018). [DOI] [PubMed] [Google Scholar]
- 143.Mensurado S et al. Tumor-associated neutrophils suppress pro-tumoral IL-17+ gammadelta T cells through induction of oxidative stress. PLoS Biol 16, e2004990, doi: 10.1371/journal.pbio.2004990 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mantovani A The yin-yang of tumor-associated neutrophils. Cancer Cell 16, 173–174, doi: 10.1016/j.ccr.2009.08.014 (2009). [DOI] [PubMed] [Google Scholar]
- 145.Azizi E et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236, doi: 10.1016/j.cell.2018.05.060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gaublomme JT et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell 163, 1400–1412, doi: 10.1016/j.cell.2015.11.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC & Schultze JL The Myeloid Cell Compartment-Cell by Cell. Annual Review of Immunology, Vol 37, 2019 37, 269–293, doi: 10.1146/annurev-immunol-042718-041728 (2019). [DOI] [PubMed] [Google Scholar]
- 148.Wagner J et al. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 177, 1330–1345 e1318, doi: 10.1016/j.cell.2019.03.005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Z, Till B & Gao Q Chemotherapeutic agent-mediated elimination of myeloid-derived suppressor cells. Oncoimmunology 6, e1331807, doi: 10.1080/2162402X.2017.1331807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Larionova I et al. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology 8, doi: 10.1080/2162402x.2019.1596004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Strauss L, Weaver J, Freeman GJ & Boussiotis VA PD-1 blockade alters fate commitment of myeloid progenitors during tumor-mediated “emergency” myelopoiesis. J Immunol 198 (2017). [Google Scholar]
- 152.Dominguez CX et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov, doi: 10.1158/2159-8290.CD-19-0644 (2019). [DOI] [PubMed] [Google Scholar]
- 153.Bernard V et al. Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clinical Cancer Research 25, 2194–2205, doi: 10.1158/1078-0432.Ccr-18-1955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196, doi: 10.1126/science.aad0501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yost KE et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nature Medicine 25, 1251-+, doi: 10.1038/s41591-019-0522-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee JH et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 10, 442–458, doi: 10.1038/nprot.2014.191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090, doi: 10.1126/science.aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Keren L et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387 e1319, doi: 10.1016/j.cell.2018.08.039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Levitin HM, Yuan J & Sims PA Single-Cell Transcriptomic Analysis of Tumor Heterogeneity. Trends Cancer 4, 264–268, doi: 10.1016/j.trecan.2018.02.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Suva ML & Tirosh I Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges. Mol Cell 75, 7–12, doi: 10.1016/j.molcel.2019.05.003 (2019). [DOI] [PubMed] [Google Scholar]
- 161.Chappell L, Russell AJC & Voet T Single-Cell (Multi)omics Technologies. Annu Rev Genomics Hum Genet 19, 15–41, doi: 10.1146/annurev-genom-091416-035324 (2018). [DOI] [PubMed] [Google Scholar]
- 162.Stuart T & Satija R Integrative single-cell analysis. Nat Rev Genet 20, 257–272, doi: 10.1038/s41576-019-0093-7 (2019). [DOI] [PubMed] [Google Scholar]
- 163.Day CP, Merlino G & Van Dyke T Preclinical mouse cancer models: a maze of opportunities and challenges. Cell 163, 39–53, doi: 10.1016/j.cell.2015.08.068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sontheimer-Phelps A, Hassell BA & Ingber DE Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer 19, 65–81, doi: 10.1038/s41568-018-0104-6 (2019). [DOI] [PubMed] [Google Scholar]
- 165.Biselli E et al. Organs on chip approach: a tool to evaluate cancer -immune cells interactions. Sci Rep 7, 12737, doi: 10.1038/s41598-017-13070-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hassell BA et al. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep 23, 3698, doi: 10.1016/j.celrep.2018.06.028 (2018). [DOI] [PubMed] [Google Scholar]
- 167.Nguyen M et al. Dissecting Effects of Anti-cancer Drugs and Cancer-Associated Fibroblasts by On-Chip Reconstitution of Immunocompetent Tumor Microenvironments. Cell Rep 25, 3884–3893 e3883, doi: 10.1016/j.celrep.2018.12.015 (2018). [DOI] [PubMed] [Google Scholar]
- 168.Nagy ML et al. Quantitative image profiling of the tumor microenvironment on double stained immunohistochemistry images using deep learning. J Clin Oncol 37, e14619–e14619, doi: 10.1200/JCO.2019.37.15_suppl.e14619 (2019). [DOI] [Google Scholar]
- 169.Yoo SY et al. Whole-Slide Image Analysis Reveals Quantitative Landscape of Tumor-Immune Microenvironment in Colorectal Cancers. Clin Cancer Res, doi: 10.1158/1078-0432.CCR-19-1159 (2019). [DOI] [PubMed] [Google Scholar]
- 170.Koelzer VH, Sirinukunwattana K, Rittscher J & Mertz KD Precision immunoprofiling by image analysis and artificial intelligence. Virchows Arch 474, 511–522, doi: 10.1007/s00428-018-2485-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Saltz J et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep 23, 181–193 e187, doi: 10.1016/j.celrep.2018.03.086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wiesweg M et al. Machine learning-based predictors for immune checkpoint inhibitor therapy of non-small-cell lung cancer. Ann Oncol 30, 655–657, doi: 10.1093/annonc/mdz049 (2019). [DOI] [PubMed] [Google Scholar]
- 173.Cannarile MA et al. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer 5, 53, doi: 10.1186/s40425-017-0257-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mizutani Y et al. Meflin-Positive Cancer-Associated Fibroblasts Inhibit Pancreatic Carcinogenesis. Cancer Res 79, 5367–5381, doi: 10.1158/0008-5472.CAN-19-0454 (2019). [DOI] [PubMed] [Google Scholar]
- 175.Jain RK Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J Clin Oncol 31, 2205–2218, doi: 10.1200/JCO.2012.46.3653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sanmamed MF & Chen L A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 175, 313–326, doi: 10.1016/j.cell.2018.09.035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Thorsson V et al. The Immune Landscape of Cancer. Immunity 48, 812–830 e814, doi: 10.1016/j.immuni.2018.03.023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wellenstein MD & de Visser KE Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity 48, 399–416, doi: 10.1016/j.immuni.2018.03.004 (2018). [DOI] [PubMed] [Google Scholar]
- 179.Dias Carvalho P et al. KRAS Oncogenic Signaling Extends beyond Cancer Cells to Orchestrate the Microenvironment. Cancer Res 78, 7–14, doi: 10.1158/0008-5472.CAN-17-2084 (2018). [DOI] [PubMed] [Google Scholar]
- 180.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61, doi: 10.1016/j.cell.2014.12.033S0092-8674(14)01639-0 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Amit M et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 578, 449–454, doi: 10.1038/s41586-020-1996-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Q et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 32, 42–56 e46, doi: 10.1016/j.ccell.2017.06.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Siveke JT Fibroblast-Activating Protein: Targeting the Roots of the Tumor Microenvironment. J Nucl Med 59, 1412–1414, doi: 10.2967/jnumed.118.214361 (2018). [DOI] [PubMed] [Google Scholar]
- 184.Neesse A et al. CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proc Natl Acad Sci U S A 110, 12325–12330, doi: 10.1073/pnas.1300415110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jimeno A et al. Phase I study of the Hedgehog pathway inhibitor IPI-926 in adult patients with solid tumors. Clin Cancer Res 19, 2766–2774, doi: 10.1158/1078-0432.CCR-12-3654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Reardon DA et al. Salvage radioimmunotherapy with murine iodine-131-labeled antitenascin monoclonal antibody 81C6 for patients with recurrent primary and metastatic malignant brain tumors: phase II study results. J Clin Oncol 24, 115–122, doi: 10.1200/JCO.2005.03.4082 (2006). [DOI] [PubMed] [Google Scholar]
- 187.Reilley MJ et al. Phase I clinical trial of combination imatinib and ipilimumab in patients with advanced malignancies. J Immunother Cancer 5, 35, doi: 10.1186/s40425-017-0238-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chan N et al. Influencing the Tumor Microenvironment: A Phase II Study of Copper Depletion Using Tetrathiomolybdate in Patients with Breast Cancer at High Risk for Recurrence and in Preclinical Models of Lung Metastases. Clin Cancer Res 23, 666–676, doi: 10.1158/1078-0432.CCR-16-13261078-0432.CCR-16-1326 [pii] (2017). [DOI] [PubMed] [Google Scholar]
- 189.Lim SY, Yuzhalin AE, Gordon-Weeks AN & Muschel RJ Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget 7, 28697–28710, doi: 10.18632/oncotarget.7376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vonderheide RH CD40 Agonist Antibodies in Cancer Immunotherapy. Annu Rev Med 71, 47–58, doi: 10.1146/annurev-med-062518-045435 (2020). [DOI] [PubMed] [Google Scholar]
- 191.Weiskopf K Cancer immunotherapy targeting the CD47/SIRPalpha axis. Eur J Cancer 76, 100–109, doi: 10.1016/j.ejca.2017.02.013 (2017). [DOI] [PubMed] [Google Scholar]
- 192.Hong D et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med 7, 314ra185, doi: 10.1126/scitranslmed.aac5272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schott AF et al. Phase Ib Pilot Study to Evaluate Reparixin in Combination with Weekly Paclitaxel in Patients with HER-2-Negative Metastatic Breast Cancer. Clin Cancer Res 23, 5358–5365, doi: 10.1158/1078-0432.CCR-16-2748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]