Abstract
The nuclear pore complex (NPC) is a massive nuclear envelope-embedded protein complex, the canonical function of which is to mediate selective nucleocytoplasmic transport. In addition to its transport function, the NPC has been shown to interact with the underlying chromatin and to influence both activating and repressive gene regulatory processes, contributing to the establishment and the epigenetic maintenance of cell identity. In this review, we discuss diverse gene regulatory functions of NPC components and emerging mechanisms underlying these functions, including roles in genome architecture, transcription complex assembly, chromatin remodeling, and coordination of transcription and mRNA export. These functional roles highlight the importance of the NPC as a nuclear scaffold directing genome organization and function.
Keywords: nuclear pore complex, nucleoporin, chromatin, genome organization, genome architecture
Introduction
Interactions between the genome and nuclear scaffolds are known to facilitate genome organization, transcriptional regulation, and epigenetic maintenance, thus playing a key role in defining cellular gene expression programs [1]. One of the most prominent nuclear scaffolds is the Nuclear Pore Complex (NPC), which forms a transport channel through the Nuclear Envelope (NE) to allow selective trafficking of macromolecules [2]. The NPC consists of multiples of 30 different subunits, called Nucleoporins (Nups), which comprise its various sub-complexes and sub-structures. The NE-embedded core of the NPC is shaped as a ring with an eight-fold rotational symmetry and is built by Nups of the outer and inner ring sub-complexes [3]. Once assembled, Nups of these sub-complexes remain associated with the NPC core during interphase and are thus termed stable [4,5]. The NPC core anchors other important sub-structures (Figure 1), such as the nuclear basket and the channel Nups that set up the selective transport barrier [6]. In contrast to stable Nups, some of these Nups have been shown to associate with the NPC core dynamically [7], and are often characterized by extended intrinsically disordered domains of Phenylalanine-Glycine (FG) repeats. Although the majority of Nups are present ubiquitously across cell types, many Nups, both stable and dynamic, exhibit varying expression in specific cell lineages and tissues [8].
Figure 1. Current models for mechanisms and functions of Nup-genome interactions.
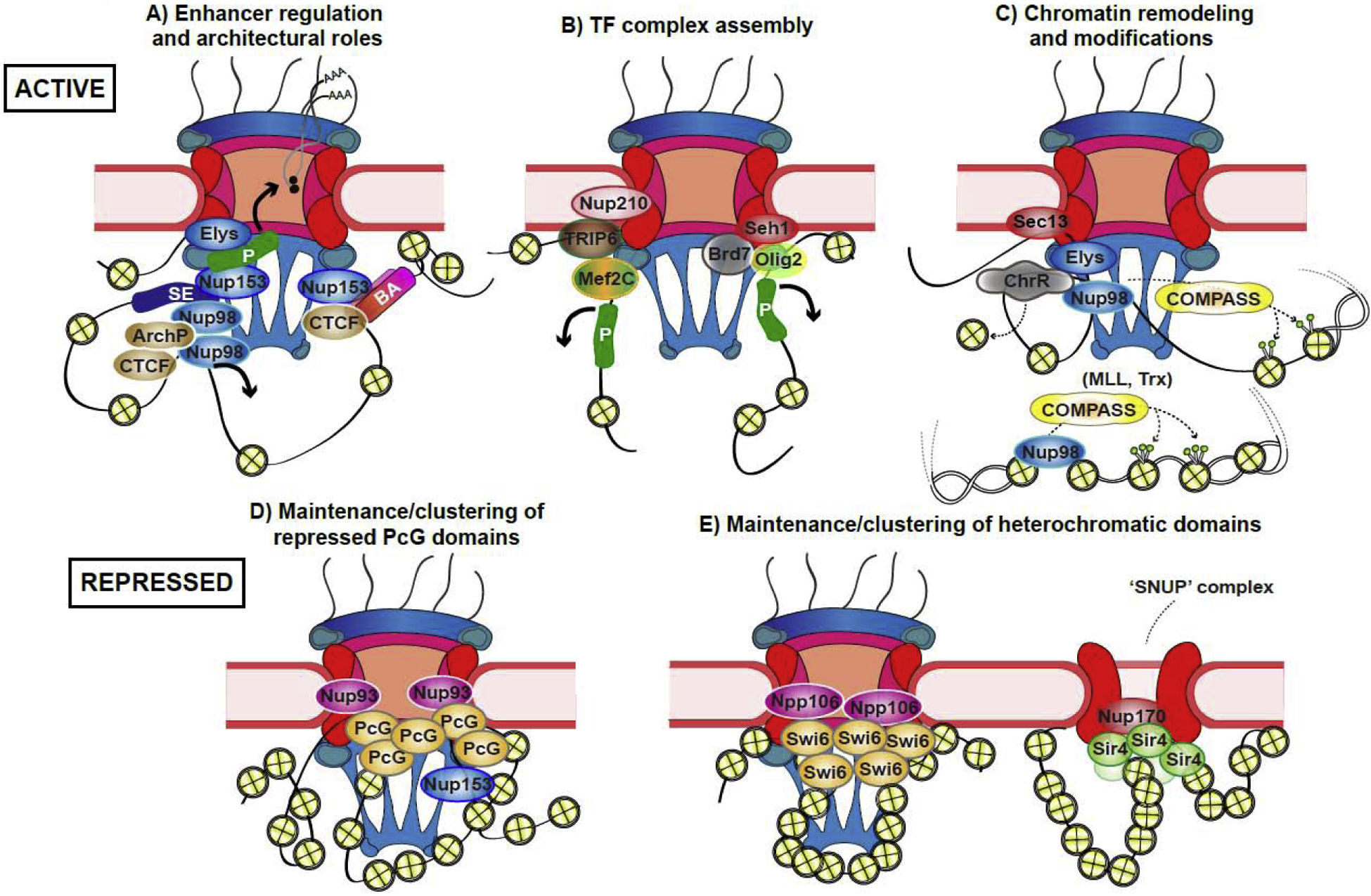
A) Depiction of proposed architectural roles of Nups, such as Nup153, Nup98, and Elys, at enhancers or super-enhancers (SEs), promoters (P), and elements with boundary activity (BA) in scaffolding genomic loops, strengthening CTCF-bound boundaries, and coupling to mRNA export. ArchP (brown ovals) stands for architectural proteins. B) Model of proposed roles of Nups such as Nup210 and Seh1 in assembly of transcription factor (TF) complexes. Specific TFs are discussed in text. C) Model of proposed functions of Nups Sec13, Elys, and Nup98, in chromatin remodeling and H3K4 di- and tri-methylation. ChR (grey ovals) stands for chromatin remodelers. COMPASS (which can include MLL or Trx) is an H3K4 HMTase complex. H3K4 di- or tri-methylation can occur on or off-pore. D) and E) Depiction of identified roles of Nup93 (metazoan) or its homologue Npp106 (S. pombe) in maintenance and clustering of repressed Polycomb (PcG) or Swi6-targeted heterochromatic regions, respectively. Nup153 has also been shown to target PcG regions in mouse ES cells. Right part of E) depicts an NPC-like structure Sir4-associated Nup (SNUP) complex, containing Nup170, which has been proposed to maintain Sir4-targeted heterochromatin, such as subtelomeric regions, in S. cerevisiae. Stable NPC core is shown in red (inner and outer ring sub-complexes), auxiliary structures such as the nuclear basket in blue. Yellow circles represent nucleosomes in various states of compaction. References for these models are discussed in text.
In recent years, NPC components have been implicated in a variety of physiological processes and pathologies, including cancer and neurodegenerative disorders [9], stirring interest in the molecular mechanisms behind these roles. In addition to their classical function in nucleocytoplasmic transport, Nups are also known to contribute to genome regulation via physical interactions with chromatin. Although the nuclear periphery is generally regarded as a repressive environment [10], early cytological observations revealed the condensed heterochromatin at the NE punctuated by less condensed chromatin in close associated with NPCs [11]. Genome-wide and locus-specific studies in multiple organisms have now demonstrated binding of Nups to subsets of genes and regulatory elements, and have revealed functional roles of NPC components in gene regulation [12–14]. Interestingly, Nups have been found to play a role in both activating and repressive processes, with certain NPC components or sub-complexes preferentially involved in gene activation or silencing. Despite this variety of functions, several mechanistic themes have emerged from recent studies on the gene regulatory roles of Nups, and these themes are the focus of this review. Below we discuss current knowledge on the proposed molecular roles of NPC components in genome architecture, transcriptional complex assembly, regulation of chromatin structure, and in coupling of transcription and transport. Investigation of these mechanisms is important for understanding physiological and disease-related functions of Nups, and for fully unraveling principles of genome architecture.
Genomic binding and functions of Nups at active regions
The majority of NPC-related chromatin binding and functional studies in recent years have highlighted the roles of Nups at active genes. Multiple investigations, including some of the earliest, in S. cerevisiae and C. elegans found that inducible genes, such as the nutrient-induced INO1, GAL10, HXK1 and the heat shock-induced hsp16, are recruited to nuclear pores upon activation [15–20]. Genome-wide studies in Drosophila and mammalian cells have subsequently demonstrated that certain Nups, particularly dynamic ones such as Nup98 and Nup153, associate with actively transcribing regions and target a subset of active promoters [21–25]. In fly and mammalian cells, many of these binding events were found to take place in the nucleoplasm, away from the NE-embedded nuclear pores [21–23,25–27], demonstrating that dynamic Nups can associate with chromatin throughout nuclear space and that such Nup-genome contacts primarily involve active genes.
A recent development in the field has been the discovery by multiple groups that in addition to promoters, metazoan NPC components frequently target enhancers and in particular, super-enhancers (SEs) [28–31] (Figure 1A). SEs are enhancers that are characterized by particularly high levels of H3K27 acetylation and by clusters of binding sites for multiple transcription factors (TFs), and are thought to be particularly important for cell identity [32]. In two different human cell types, Nup153 was identified at a large fraction of classified SEs [31], and multiple Nups, such as Nup153, Nup133 and Elys, were recently found to be enriched at highly connected enhancers and oncogenic super-enhancers (OSEs) in colon cancer cells [28]. The targeting of Nups to enhancers was also identified by a dCas9-based proteomic approach, which characterized protein composition of specific clustered enhancers in erythroid cells and in addition to expected regulators of enhancer function such as GATA1 and chromatin remodelers, detected high abundance of FG Nups Nup98, Nup153, and Nup214 [30]. In Drosophila, Nup98 was similarly identified at a significant subset of enhancers genome-wide, including at genes regulated by a steroid hormone ecdysone and at complex enhancers of Hox gene clusters [29]. In addition to enhancers, Nups have also been identified at architectural features such as TAD boundaries - in mouse ES cells, as many as 67% of TAD boundaries were reported to be targeted by Nup153 [33]. Nup98 was similarly identified at a subset of TAD boundaries, characterized by high insulator strength, in the fly genome [34].
Together, these studies revealed that Nups target enhancers, as well as architectural elements, with key roles in cell differentiation. Correspondingly, depletion of specific Nups has been shown to result in down- or mis-regulation of their gene targets in multiple differentiation systems (Figure 1A–C). For example, knockdown of Nup153 in rodent neural progenitor cells (NeuPCs) leads to down-regulation of genes, promoters of which are targeted by Nup153, and to disruption of NeuPC maintenance [35]. Depletion of Nup153 in human U2OS cells or IMR90 fibroblasts similarly causes a widespread disruption of transcriptional programs, with a preferential effect on genes associated with the Nup153-targeted SEs [31]. Likewise, loss of Nup210 in mammalian myoblasts results in compromised expression of muscle genes, many of which are binding targets of Nup210, and in inhibition of muscle maturation [36]. In Drosophila, depletion of Nup98 has been shown to lead to reduced expression of Hox genes in developing larval tissues [24] and of genes involved in anti-viral response in embryonic S2 cells [37]. Several Nups have also been reported to regulate parent-specific expression of the imprinted Kcnq1ot1 locus, where expression of a noncoding RNA and the associated silencing of paternal alleles were found to depend on Nup107, Nup153, and Nup62 [38].
A distinct gene regulatory function linked to Nup-genome interactions is their role in transcriptional memory. Transcriptional memory describes a process via which previously activated genes respond more robustly upon subsequent activation, after a period of repression [39]. Nup98 homologues are required for transcriptional memory and the heightened re-activation dynamics of a number of inducible genes across species, including inositol-inducible INO1 gene in yeast and interferon-gamma-inducible genes in human cells [27], and ecdysone-inducible genes in fly cells [29]. In addition to Nup98, the nuclear basket component Mlp1 and an FG Nup Nup42 have been similarly implicated in transcriptional memory of galactose-induced HXK1 and salt stress-responsive genes in yeast, respectively [40,41]. As transcriptional memory can be maintained over cell generations, i.e. epigenetically, this role of Nups may be particularly important for maintenance of specific transcription programs and cell identity.
Genomic binding and functions of Nups at repressed regions
Original chromatin binding studies of NPC-genome interactions in yeast revealed that not all Nups target transcriptionally active chromatin [42]. Several subsequent reports supported the idea that certain NPC components associate with and regulate silent or heterochromatic regions. ChIP-chip characterization of Nup93 in HeLa cells uncovered preferential association of Nup93 with regions marked by repressive histone modifications [43]. Nup93 is a component of the inner ring Nup93-Nup155 sub-complex, multiple components of which have been implicated in gene silencing. The yeast Nup155 homologue Nup170 was found to target sub-telomeric heterochromatic regions and to be functionally required for their silencing [44,45], while Nup155 in rat cardiomyocytes has been shown to interact with a histone deacetylase HDAC4 [46]. These studies have suggested that unlike other reported roles of Nups, the inner ring sub-complex of the NPC contributes to gene repression.
Several recent studies have provided evidence for this role and highlighted the unique and highly conserved function of the Nup93 sub-complex in maintenance of silent chromatin. In Drosophila cells, high-resolution chromatin binding studies compared genome-wide distribution of Nup107 and Nup93, two representative components of the outer and inner ring sub-complexes, respectively, and found that Nup93 specifically targets silent Polycomb domains [47] (Figure 1D). Polycomb or Polycomb group (PcG) proteins are conserved gene silencing regulators, which deposit the H3K27 tri-methylation mark over genomic domains as part of their repressive mechanism. Nup93 was identified at approximately one third of Polycomb domains in the fly genome, and was found to be required for efficient silencing of these regions [47]. In contrast, Nup107 was detected primarily at transcriptionally active regions, in line with the known relationship between NPCs and active genes. The link between Nup93 and Polycomb repression has also been suggested by studies in a colorectal cancer cell line, where loss of Nup93 was found to result in de-repression and reduction of H3K27 tri-methylation at genes within the HoxA cluster [48]. Interestingly, Nup153, despite its connections to gene activation described above, has also been shown to target a subset of Polycomb regions and promote their repression in mES cells [49].
The relationship between the Nup93 sub-complex and repressive chromatin has been independently identified in S. pombe, where proteomic analysis of isolated native heterochromatin identified inner ring Nups as part of the heterochromatin-specific proteome [50]. The S.pombe heterochromatin includes pericentromeric and subtelomeric repetitive regions, and relies on Swi6, homologue of the metazoan HP1, and H3K9 methylation for repression. Deletion of the Nup93 homologue Npp106 was found to disrupt epigenetic maintenance of heterochromatic silencing, impair distribution of Swi6 along heterochromatic domains and reduce levels of H3K9 methylation [50]. Furthermore, deletions of non-inner ring Nups did not produce these phenotypes, demonstrating that the Nup93/Npp106 inner ring sub-complex is specifically involved in the maintenance of Swi6-associated heterochromatin (Figure 1E).
Since the Nup93 sub-complex represents a stable part of the NPC core, these repressive interactions are expected to take place at NE-embedded NPCs, but whether all such contacts occur at canonical NPCs remains an open question. Biochemical and imaging studies have shown that the S. cerevisiae Nup170, in association with several other stable Nups and the silencing factor Sir4, may exist as a distinct NE-associated structure, termed the Sir4-associated Nup (Snup) complex [44]. The Snup complex, which is thought to be separate from canonical NPCs (Figure 1E, right), has been proposed to mediate the NE tethering and maintenance of sub-telomeric heterochromatin [44]. If such complexes exist in animal cells, it is conceivable that Polycomb domains are predominantly associated with distinct NPC-like complexes containing Nup93. Interestingly, a separate study has identified another NE-associated factor Amo1 that plays a key role in heterochromatin maintenance and perinuclear tethering in S. pombe [51]. Amo1 appears to partially co-localize with NPCs, raising the possibility that it may also be part of a unique NPC-like complex or of a special subset of nuclear pores. Further work will be required to determine whether specialized NPCs or membrane-associated NPC-like complexes function as unique platforms for silent chromatin.
Given the variety of functional roles for Nups described above, one of the main outstanding questions in the field has been the molecular mechanisms, by which nuclear pore proteins carry out these gene regulatory functions. Below, we discuss four non-mutually exclusive mechanisms via which Nups have been proposed to regulate active and repressed regions, and the current evidence supporting these ideas.
Mechanisms - roles of Nups in genome architecture
The reported presence of Nups such as Nup98 and Nup153 at subsets of enhancers, promoters, and TAD boundaries suggested that Nups may function as regulators of genome architecture and long-range genomic contacts (Figure 1A). In agreement with this role, studies in the yeast system have found that the nuclear basket Mlp1 is required for the formation of a transcriptional 5’–3’ loop [41], and for interchromosomal clustering of GAL1–10 alleles [52]. Another nuclear basket component Nup2 was demonstrated to be involved in long-range clustering of genes, controlled by specialized nuclear positioning elements termed DNA zip codes [53]. Depletion of Drosophila Nup98 was shown to impair formation of enhancer-promoter loops at ecdysone-activated genes, in a manner that can be separated from transcriptional activity itself [29,54]. Namely, while RNAi depletion of Nup98 led to the loss of ecdysone-induced increase in enhancer-promoter contacts, it did not compromise the on-going transcriptional output of such promoters during initial exposure to ecdysone. Instead, loss of these contacts correlated with reduced transcriptional re-activation during subsequent ecdysone exposure, suggesting that Nup98 may stabilize enhancer-promoter contacts as part of the transcriptional memory system [29,54]. This concept is similar to the proposed function of the yeast Mlp1 in forming “memory gene loops” [41], but in both cases, the causative role of loop formation, whether enhancer-promoter or 5’–3’, in driving epigenetic memory remains to be determined. Interestingly, the detected interactions between Nups and enhancers in fly or mammalian cells were found to predominantly occur at actual NPCs [28,29,31], suggesting that NE-embedded NPCs may function as scaffolds for spatial folding of complex enhancers.
In further support of architectural functions of Nups, Drosophila Nup98 was reported to interact with several architectural factors, including the well-known CTCF [29]. This interaction between Nups and architectural machinery appears to be conserved, as recently, murine Nup153 was shown to physically interact with CTCF and cohesion complex subunits in mES cells [33]. Importantly, depletion of Nup153 was found to compromise chromatin targeting of CTCF and of cohesin subunit SMC3 genome-wide, with a substantial fraction of CTCF sites exhibiting decreased occupancy in the absence of Nup153. This Nup153-dependent loss of CTCF from cis-regulatory elements of a subset of EGF-induced genes was also associated with compromised transcriptional response and with reduced presence of RNAP II before and shortly after induction [33]. Together, these findings illustrated that Nup153 can control chromatin binding of architectural machinery, with functional outcomes for inducible gene expression.
The role of Nups in genome architecture appears to also be a part of their function in gene repression. Depletion of Nup93 was found to disrupt nuclear clustering of distant Nup93-bound Polycomb domains in fly cells [47], and such clustering has been proposed to contribute to stabilization of Polycomb silencing [55]. Interestingly, comparison to Hi-C maps revealed that Nup93-targeted Polycomb domains preferentially interact with each other, further supporting the notion that Nup93 promotes long-range interactions of repressed chromatin regions [47]. Similarly, in S. pombe, loss of the Nup93 homologue Npp106 lead to de-clustering of heterochromatic foci, particularly at telomeres and the mating type locus, demonstrating the normal function of Npp106 and likely other inner ring sub-complex components in bringing together distant heterochromatic regions [50].
Mechanisms - roles of Nups in assembly and stabilization of transcriptional complexes
Another mechanism highlighted by recent research is the role of Nups in the assembly of regulatory complexes at key target genes or in the stabilization of such complexes on chromatin (Figure 1B). Recent work identified the transmembrane NPC component Nup210 to be required for myoblast differentiation into mature myofibers in mammalian and zebrafish systems [36,56]. This function was found to converge on the ability of Nup210 to recruit the muscle transcription factor (TF) Mef2C and a transcriptional co-regulator Trip6 to the NPC, where Nup210 is involved in their targeting to key muscle genes [36]. Nup210 physically associates with such genes and promotes recruitment of Mef2C, and together, these findings put forth a model for the function of Nup210 in fostering the assembly of a competent Mef2C transcriptional complex, which in turn drives muscle maturation [36]. Likewise, Nup210 was found to play a functional role in the immune system via promoting activation of key target genes in CD4+ T lymphocytes [57]. Nup210 knockout mice were found to display a specific decrease in the number of circulating CD4+ T cells, accompanied by the inability of naïve CD4+ T cells to sustain or transmit T cell receptor (TCR) signaling. Mechanistically, Nup210 was found to modulate TCR signaling through activation of the Cav2 gene, which was positioned at the nuclear periphery and depended on Nup210 for rapid activation dynamics in response to TCR stimulation [57], supporting the notion that NPCs can function as scaffolds for the assembly of specific transcriptional complexes.
In addition to muscle and immune systems, several Nups have been found to regulate the development and functions of the nervous system [35,58]. Recently, a component of the stable outer ring Nup107 sub-complex, Seh1 was discovered to play a key role in differentiation of mammalian oligodendrocytes, which are responsible for myelination of axons in the CNS [59]. Seh1 knockout was found to inhibit oligodendrocyte maturation, and this effect was independent from defects in cell proliferation, nuclear pore assembly, or transport. Instead, Seh1 was found to bind multiple key target genes and to physically interact with Olig2, a central TF in oligodendrocyte differentiation, and a chromatin remodeling co-factor Brd7. Paralleling findings on Nup210, the interaction of Seh1 with Olgi2 and Brd7 occurs at the NE, and Seh1 is required for efficient targeting of Olig2 and Brd7 to key target genes. The authors propose that the Seh1-scaffolded complex of Olig2 and Brd7 promotes expression of the oligodendrocyte differentiation program [59], underscoring another example of the NPC functioning in the assembly of a cell type specific transcriptional complex.
How widespread such mechanisms may be in gene regulation? Interestingly, recent work in yeast has highlighted the capacity of many TFs to interact with the NPC [60]. A large screen of DNA binding factors for their ability to re-position a genetic locus to the NPC discovered that as many as 65% of these can target the URA3 locus to the nuclear periphery in a manner that depends on the NPC component Nup2. A portion of such TFs were also found to require Nup100, a homologue of Nup98, for gene re-positioning to the NPC. If extended to animal systems, these findings suggest that many developmental gene expression programs may utilize such NPC-TF interactions. Furthermore, given the reported roles of Nup98 in transcriptional memory, Nup98 homologues may help maintain transcriptional programs by stabilization of a given chromatin-bound transcriptional complex through cell divisions. In support of this notion, Drosophila Nup98 is able to maintain its interaction with ecdysone receptor (EcR), a central TF of the ecdysone response, during the period of memory maintenance in the absence of ecdysone [29]. Similarly, Nup98 in mammalian cells is necessary to maintain poised RNAP II at interferon-gamma-inducible genes during the repression period [27], further illustrating the role of NPC components in stabilization as well as assembly of competent transcriptional complexes.
Mechanisms - roles of Nups in modulating chromatin structure
Early cytological images of mammalian nuclei revealed the close physical association between decondensed chromatin and nuclear pores, prompting the idea of a functional connection between open chromatin and NPCs [11]. Recent studies have provided evidence for this connection and demonstrated another mechanism by which Nups can influence gene expression - via regulation of chromatin structure (Figure 1C). Targeting Drosophila Nups such as Sec13 and Nup62 to ectopic locations in the genome via the LacI-lacO tethering system was found to result in visible decompaction of condensed chromatin, providing gain-of-function evidence to the ability of Nups to induce chromatin opening [61]. Mechanistically, this chromatin-opening activity was found to rely on another Nup Elys and to require the ATP-dependent chromatin remodeling BRM complex, with which both Sec13 and Elys associated physically. In support of these findings, previous studies in C. elegans identified genetic and biochemical interactions between the homologue of Elys, MEL-28, and the SWI/SNF chromatin remodeling complex components [62,63]. Seh1, which is structurally related to Sec13, was similarly found to associate with several chromatin remodelers in oligodendrocytes, most notably Brd7 [59]. Similarly to Elys [61], loss of Seh1 affected chromatin compaction, leading to decreased chromatin accessibility at a subset of target genes that normally become accessible during oligodendrocyte differentiation [59]. Importantly, in cases of either Seh1 or Sec13/Elys, the observed changes in chromatin compaction do not appear to be simply a consequence of changes in transcription [59,61]. For instance, Seh1 appears to interact with the TF Olig2 and the chromatin remodeler Brd7 independently, such that knockdown of one does not compromise the interaction of Seh1 with the other (Liu), suggesting that assembly of transcription and chromatin remodeling machinery may be two separate but coordinated functions of Nups.
Deposition of histone modifications is another pathway that regulates chromatin compaction and function, and it has also been shown to interface with Nups, most notably Nup98. Several studies have identified an evolutionary conserved interaction between Nup98 and the histone methyl transferases (HMTs) that modify H3K4, also known as the COMPASS/MLL complexes [24,27,64,65]. Deposition of both H3K4 di-methylation and trimethylation at promoters has been shown to depend on mammalian Nup98 in different context [27,64]. Drosophila Nup98 has been found to physically and genetically interact with the fly version of MLL, Trithorax [24], while murine Nup98 was demonstrated to associate with Wdr82, a COMPASS/Set1A complex component, in hematopoietic cells [64]. It is particularly telling that a study that looked at recurring protein domains of Nups across eukaryotic evolution reported a repeated occurrence of Nup98 homologs containing a SET domain, normally found in MLL/Trx/Set1A HMTs [66], although the enzymatic activity of SET-containing Nup98 homologues has not been explored. Together, these findings highlight Nup98 as a component of the H3K4 methylation pathway and point to another key function of Nups in chromatin structure.
Mechanisms - roles of Nups in connecting transcription and mRNA export
The initial proposal for the function of NPC-genome contacts, as outlined by the gene gating hypothesis [11], envisioned that decondensed regions found in proximity to nuclear pores are active genes, transcription of which is coupled to efficient mRNA export via physical linkage to NPCs. This model has received substantial support from studies in yeast, which identified physical and functional connections between nuclear basket Nups, transcriptional complexes SAGA and Mediator, and the Transcription and Export complex 2 (TREX-2) [67,68]. The role of NPC components in transcription-export coordination has also been recently demonstrated in human colorectal cancer DLD-1 cells, where auxin-induced degradation of the nuclear basket component TPR was found to disrupt both nascent transcription and mRNA export of a subset of genes [69]. Moreover, RNA-seq changes resulting from acute TPR depletion closely paralleled expression changes caused by acute depletion of TREX-2 complex components [69]. These findings are further supported by the previously identified role of Mlp1, the yeast homologue of TPR, in attenuating transcription in response to mRNA export defects, thus coordinating the two processes [70].
Interestingly, a recent paper demonstrated that the reported targeting of SEs to NPCs can facilitate mRNA export of associated genes [28]. In this case, NPC targeting of the Myc gene and its associated OSE was found to result in aberrantly high expression of Myc in colon cancer cells, yet no changes in nascent transcription of Myc were detected. Instead, increased expression stemmed from elevated mRNA export and the resulting increase in stability of mRNA once exported to the cytoplasm. This regulation and NPC anchoring of the Myc-driving OSE were found to depend on Nup Elys, which in turn physically associated with OSE-bound beta-catenin, revealing integration of Wnt signaling and NPC tethering in enhancer-driven regulation of Myc [28]. These results present a particularly novel paradigm that certain SEs can drive gene expression through boosting rates of mRNA export by utilizing NPC association (Figure 1A). Although compelling, it is unclear how general this paradigm is, since other studies of gene-NPC associations, such as the association of Nup210 with muscle genes described above, did not identify any effects on mRNA export [36]. It remains to be determined whether other non-oncogenic NPC-targeted enhancers similarly influence not just transcription, but also export of target mRNAs.
Conclusions and perspectives
As discussed above, the NPC has emerged as a complex nuclear scaffold capable of influencing both activating and silencing events via a wide range of potential mechanisms. This complexity may not be surprising given the number of individual NPC components and their reported involvement in unique interactions with chromatin and transcriptional regulators. The specificity of these interactions can be further enforced by cell type-specific increases in expression of a given Nup, such as those reported for Nup210 and Seh1 [56,59]. Additionally, different nuclear pores or NPC-like assemblies may engage in unique chromatin interactions via the specific Nup or NPC sub-complex, aiding in separation of the different mechanisms used by Nups to influence gene expression. Unraveling these mechanisms further will entail determining whether different Nup-dependent mechanisms are interconnected or consequences of each other, and which ones are primary versus secondary, or direct versus indirect. Deciphering these questions is intimately tied to our general understanding of the precise relationship between regulation of chromatin, transcription, and genome architecture.
A possible broad model that can account for the diverse mechanisms of Nups involves their role in creating or stabilizing a particular micro-environment, consisting of chromatin-bound regulators and necessary genomic loops. Nup153 in particular appears to be widely utilized by different gene regulatory processes, and it is possible that a key role of Nup153 involves functioning as “molecular glue” for a particular macro-assembly. In this manner, Nup153 may be recruited by activating and repressive regulators to aid in stabilizing or connecting a chromatin-bound complex, suggesting that function of Nup153 may be highly locus and context-specific. Likewise, Nup98 has been implicated in interactions with architectural proteins, transcription factors, mRNA export machinery, and histone modifying enzymes, and it may similarly be used in sealing a particular expression or epigenetic state. It is of note that both Nup153 and Nup98 possess a high number of intrinsically disordered FG repeats, and it has been hypothesized that such FG-rich Nups may create phase-separated sub-compartments around target loci, functioning to concentrate, stabilize, and separate a given environment [54]. Such models remain to be tested, but it is becoming increasingly clear that NPC components carry the capacity to integrate a large number of diverse gene regulatory steps and exert a significant impact on cellular gene expression states.
Acknowledgements
We apologize to our colleagues whose work could not be cited due to space limitations. We thank members of the Capelson lab, particularly Jennifer Aleman, Terra Kuhn, and Ashley Duke, for helpful discussions. M.C. is supported by the NIH R01GM124143 grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare no conflict of interest.
References
*of special interest
**of outstanding interest
- 1.Buchwalter A, Kaneshiro JM, Hetzer MW: Coaching from the sidelines: the nuclear periphery in genome regulation. Nat Rev Genet 2019, 20:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wente SR, Rout MP: The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2010, 2:a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Angelo MA, Hetzer MW: Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 2008, 18:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabut G, Lenart P, Ellenberg J: Dynamics of nuclear pore complex organization through the cell cycle. Curr Opin Cell Biol 2004, 16:314–321. [DOI] [PubMed] [Google Scholar]
- 5.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, Hetzer MW: Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013, 154:971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knockenhauer KE, Schwartz TU: The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 2016, 164:1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabut G, Doye V, Ellenberg J: Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 2004, 6:1114–1121. [DOI] [PubMed] [Google Scholar]
- 8.Guglielmi A, Sakuma S, D’Angelo MA: Nuclear pore complexes in development and tissue homeostasis. Development 2020, 147:dev183442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakuma S, D’Angelo MA: The roles of the nuclear pore complex in cellular dysfunction, aging and disease. Semin Cell Dev Biol 2017, 68:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabianca DS, Gasser SM: Spatial segregation of heterochromatin: Uncovering functionality in a multicellular organism. Nucleus 2016, 7:301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blobel G: Gene gating: a hypothesis. Proc Natl Acad Sci U S A 1985, 82:8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ptak C, Wozniak RW: Nucleoporins and chromatin metabolism. Curr Opin Cell Biol 2016, 40:153–160. [DOI] [PubMed] [Google Scholar]
- 13.Sood V, Brickner JH: Nuclear pore interactions with the genome. Curr Opin Genet Dev 2014, 25:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Shi Y, Yildirim E: The Nuclear Pore Complex in Cell Type-Specific Chromatin Structure and Gene Regulation. Trends Genet 2019, 35:579–588. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH: DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 2010, 12:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH: H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 2007, 5:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieppois G, Iglesias N, Stutz F: Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 2006, 26:7858–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P: Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol 2013, 200:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK: Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell 2006, 21:379–391. [DOI] [PubMed] [Google Scholar]
- 20.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM: Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 2006, 441:774–778. [DOI] [PubMed] [Google Scholar]
- 21.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW: Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010, 140:372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalverda B, Pickersgill H, Shloma VV, Fornerod M: Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010, 140:360–371. [DOI] [PubMed] [Google Scholar]
- 23.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW: Dynamic association of NUP98 with the human genome. PLoS Genet 2013, 9:e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascual-Garcia P, Jeong J, Capelson M: Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep 2014, 9:433–442. [DOI] [PubMed] [Google Scholar]
- 25.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A: Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 2010, 6:e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franks TM, Benner C, Narvaiza I, Marchetto MC, Young JM, Malik HS, Gage FH, Hetzer MW: Evolution of a transcriptional regulator from a transmembrane nucleoporin. Genes Dev 2016, 30:1155–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH: A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 2013, 11:e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **28.Scholz BA, Sumida N, de Lima CDM, Chachoua I, Martino M, Tzelepis I, Nikoshkov A, Zhao H, Mehmood R, Sifakis EG, et al. : WNT signaling and AHCTF1 promote oncogenic MYC expression through super-enhancer-mediated gene gating. Nat Genet 2019, 51:1723–1731. [DOI] [PubMed] [Google Scholar]; This study reports targeting of the Myc gene and associated cancer cell line-specific oncogenic super-enhancer (OSE) to nuclear pores in human HCT-116 cells. Strikingly, the increased expression of Myc in HCT-116 cells is shown to result not from increased transcription, but from elevated export of Myc mRNA that leads to its increased stability in the cytoplasm. This regulation is dependent on Nup Elys/AHCTF1, which is found to associate with the beta-catenin/TCF4 complex at the OSE, demonstrating the interplay of Wnt signaling and the OSE-NPC interaction to promote aberrant mRNA export.
- *29.Pascual-Garcia P, Debo B, Aleman JR, Talamas JA, Lan Y, Nguyen NH, Won KJ, Capelson M: Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol Cell 2017, 66:63–76 e66. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript reports the targeting of specific Nups such as Nup98 to enhancers of the Drosophila genome and identifies a functional role of Nup98 in the formation and maintenance of enhancer-promoter loops of ecdysone-inducible genes. Importantly, the identified stabilization of enhancer-promoter loops by Nup98 is found to be independent from on-going transcription during initial exposure to ecdysone. Instead, such Nup98-stabilized enhancer-promoter loops are proposed to contribute to the epigenetic memory of activating events.
- 30.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, et al. : In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 2017, 170:1028–1043 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibarra A, Benner C, Tyagi S, Cool J, Hetzer MW: Nucleoporin-mediated regulation of cell identity genes. Genes Dev 2016, 30:2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA: Super-enhancers in the control of cell identity and disease. Cell 2013, 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Kadota S, Ou J, Shi Y, Lee JT, Sun J, Yildirim E: Nucleoporin 153 links nuclear pore complex to chromatin architecture by mediating CTCF and cohesin binding. Nat Commun 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate physical interactions between Nup153 and architectural proteins CTCF and cohesins in mouse ES cells, where loss of Nup153 leads to decreased chromatin binding of CTCF and cohesin SMC3 at a number of enhancers, promoters, and TAD boundaries. Depletion of Nup153 and of associated CTCF targeting result in compromised transcriptional response of EGF-induced immediate early genes, underscoring the role of Nup153 in genome regulation.
- 34.Ramirez F, Bhardwaj V, Arrigoni L, Lam KC, Gruning BA, Villaveces J, Habermann B, Akhtar A, Manke T: High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 2018, 9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toda T, Hsu JY, Linker SB, Hu L, Schafer ST, Mertens J, Jacinto FV, Hetzer MW, Gage FH: Nup153 Interacts with Sox2 to Enable Bimodal Gene Regulation and Maintenance of Neural Progenitor Cells. Cell Stem Cell 2017, 21:618–634 e617. [DOI] [PubMed] [Google Scholar]
- *36.Raices M, Bukata L, Sakuma S, Borlido J, Hernandez LS, Hart DO, D’Angelo MA: Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex. Dev Cell 2017, 41:540–554 e547. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work describes the molecular mechanism behind the tissue-specific function of Nup210 in verterbrate myoblast differentiation into mature muscle cells. The authors report the ability of Nup210 to target muscle differentiation genes, and identify a requirement of Nup210 in the NPC-coupled assembly of the Mef2C transcriptional complex at such targets. The Nup210-driven assembly of this key transcription complex is found to be necessary for the maintanence of the muscle-specific gene expression program.
- 37.Panda D, Pascual-Garcia P, Dunagin M, Tudor M, Hopkins KC, Xu J, Gold B, Raj A, Capelson M, Cherry S: Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila. Proc Natl Acad Sci U S A 2014, 111:E3890–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sachani SS, Landschoot LS, Zhang L, White CR, MacDonald WA, Golding MC, Mann MRW: Nucleoporin 107, 62 and 153 mediate Kcnq1ot1 imprinted domain regulation in extraembryonic endoderm stem cells. Nat Commun 2018, 9:2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Urso A, Brickner JH: Epigenetic transcriptional memory. Curr Genet 2017, 63:435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan Q, Haroon S, Bravo DG, Will JL, Gasch AP: Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics 2012, 192:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ: Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 2009, 23:2610–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA: Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004, 117:427–439. [DOI] [PubMed] [Google Scholar]
- 43.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA: Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev 2008, 22:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapetina DL, Ptak C, Roesner UK, Wozniak RW: Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes. J Cell Biol 2017, 216:3145–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW: A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 2013, 152:969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kehat I, Accornero F, Aronow BJ, Molkentin JD: Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol 2011, 193:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **47.Gozalo A, Duke A, Lan Y, Pascual-Garcia P, Talamas JA, Nguyen SC, Shah PP, Jain R, Joyce EF, Capelson M: Core Components of the Nuclear Pore Bind Distinct States of Chromatin and Contribute to Polycomb Repression. Mol Cell 2020, 77:67–81 e67. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigates genome-wide distribution of core nuclear pore proteins Nup93 and Nup107, which are representative components of the inner and outer ring sub-complexes, respectively, in Drosophila cells. While Nup107 is predominantly found at active genes, Nup93 is located at repressed chromatin regions, bound by Polycomb proteins. Depletion of Nup93 leads to de-repression of Polycomb targets, co-bound by Nup93, and to de-clustering of distant Polycomb regions. This work and the study by Iglesias et al., which were published back-to-back, reveal the specific function of the inner ring sub-complex Nups in silencing and nuclear organization of repressed chromatin domains.
- 48.Labade AS, Karmodiya K, Sengupta K: HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenetics Chromatin 2016, 9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacinto FV, Benner C, Hetzer MW: The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev 2015, 29:1224–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **50.Iglesias N, Paulo JA, Tatarakis A, Wang X, Edwards AL, Bhanu NV, Garcia BA, Haas W, Gygi SP, Moazed D: Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance. Mol Cell 2020, 77:51–66 e58. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this manuscript, the authors report biochemical isolation and proteomic characterization of native heterochromatin and euchromatin in S. pombe. In addition to known heterochromatic regulators and nuclear membrane proteins, the heterochromatin-specific proteome is found to contain Nups of the inner ring NPC sub-complex. Inner ring Nups, and particularly Npp106, the homologue of metazoan Nup93, are discovered to function in epigenetic maintenance of heterochromatic silencing and in nuclear clustering of heterochromatic domains, uncovering the unique function of the inner ring sub-complex in chromatin repression.
- 51.Holla S, Dhakshnamoorthy J, Folco HD, Balachandran V, Xiao H, Sun LL, Wheeler D, Zofall M, Grewal SIS: Positioning Heterochromatin at the Nuclear Periphery Suppresses Histone Turnover to Promote Epigenetic Inheritance. Cell 2020, 180:150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH: Subnuclear positioning and interchromosomal clustering of the GAL1–10 locus are controlled by separable, interdependent mechanisms. Mol Biol Cell 2016, 27:2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH: Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell 2012, 22:1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pascual-Garcia P, Capelson M: Nuclear pores in genome architecture and enhancer function. Curr Opin Cell Biol 2019, 58:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G: Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 2011, 144:214–226. [DOI] [PubMed] [Google Scholar]
- 56.D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW: A change in nuclear pore complex composition regulates cell differentiation. Dev Cell 2012, 22:446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *57.Borlido J, Sakuma S, Raices M, Carrette F, Tinoco R, Bradley LM, D’Angelo MA: Nuclear pore complex-mediated modulation of TCR signaling is required for naïve CD4 + T cell homeostasis. Nat Immunol 2018, 19:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript identifies a tissue-specific function of Nup210 in the maintenance and survival of mouse CD4+ T lymphocytes. Nup210-deficient T cells are found to have compromised ability to transmit TCR signaling and to exhibit elevated levels of Fas, leading to increased cell death. Nup210 is shown to be required for the expression of NPC-bound genes with roles in TCR signaling and cell death regulation, emphasizing the function of Nup210 as a scaffold for the assembly of tissue-specific transcriptional complexes.
- 58.Lupu F, Alves A, Anderson K, Doye V, Lacy E: Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell 2008, 14:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, Yan M, Liang Y, Liu M, Zhang K, Shao D, Jiang R, Li L, Wang C, Nussenzveig DR, et al. : Nucleoporin Seh1 Interacts with Olig2/Brd7 to Promote Oligodendrocyte Differentiation and Myelination. Neuron 2019, 102:587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **60.Brickner DG, Randise-Hinchliff C, Lebrun Corbin M, Liang JM, Kim S, Sump B, D’Urso A, Kim SH, Satomura A, Schmit H, et al. : The Role of Transcription Factors and Nuclear Pore Proteins in Controlling the Spatial Organization of the Yeast Genome. Dev Cell 2019, 49:936–947 e934. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, the authors determine factors that mediate NPC-chromatin interactions directly, and identify a positioning domain within transcriptional regulator Gcn4 capable of targeting genes to the nuclear pore. Importantly, using a large candidate screen, they discover that a substantial number of yeast transcription factors (TFs) are capable of gene targeting to the NPC, suggesting widespread utilization of NPC-TF contacts in gene regulation. Some of these TFs depend on Nup100, the homologue of metazoan Nup98, for NPC targeting, while others appear to utilize other NPC components.
- *61.Kuhn TM, Pascual-Garcia P, Gozalo A, Little SC, Capelson M: Chromatin targeting of nuclear pore proteins induces chromatin decondensation. J Cell Biol 2019, 218:2945–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports an intrinsic ability of certain Nups to promote chromatin decompaction. Targeting Sec13 and Nup62 to ectopic locations in the Drosophila genome via the LacI-lacO tethering system leads to decondensation of condensed chromatin regions on polytene chromosomes. This Nup-induced decondensation is shown to involve Elys, which interacts with the chromatin remodeling BRM complex and which is required for normal chromatin accessibility of endogenous gene targets of Nups.
- 62.Ertl I, Porta-de-la-Riva M, Gomez-Orte E, Rubio-Pena K, Aristizabal-Corrales D, Cornes E, Fontrodona L, Osteikoetxea X, Ayuso C, Askjaer P, et al. : Functional Interplay of Two Paralogs Encoding SWI/SNF Chromatin-Remodeling Accessory Subunits During Caenorhabditis elegans Development. Genetics 2016, 202:961–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernandez AG, Mis EK, Lai A, Mauro M, Quental A, Bock C, Piano F: Uncovering buffered pleiotropy: a genome-scale screen for mel-28 genetic interactors in Caenorhabditis elegans. G3 (Bethesda) 2014, 4:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franks TM, McCloskey A, Shokirev MN, Benner C, Rathore A, Hetzer MW: Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev 2017, 31:2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D’Urso A, Takahashi YH, Xiong B, Marone J, Coukos R, Randise-Hinchliff C, Wang JP, Shilatifard A, Brickner JH: Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katsani KR, Irimia M, Karapiperis C, Scouras ZG, Blencowe BJ, Promponas VJ, Ouzounis CA: Functional genomics evidence unearths new moonlighting roles of outer ring coat nucleoporins. Sci Rep 2014, 4:4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Kohler A: The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell 2015, 162:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E: Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 2004, 116:75–86. [DOI] [PubMed] [Google Scholar]
- 69.Aksenova V, Smith A, Lee H, Bhat P, Esnault C, Chen S, Iben J, Kaufhold R, Yau KC, Echeverria C, et al. : Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. Nat Commun 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F: Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J 2005, 24:813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]


