Abstract
Osteoporosis is a common skeletal disease, affecting ~200 million people around the world. As a complex disease, osteoporosis is influenced by many factors, including diet (e.g. calcium and protein intake), physical activity, endocrine status, coexisting diseases and genetic factors. In this review, we first summarize the discovery from genome-wide association studies (GWASs) in the bone field in the last 12 years. To date, GWASs and meta-analyses have discovered hundreds of loci that are associated with bone mineral density (BMD), osteoporosis, and osteoporotic fractures. However, the GWAS approach has sometimes been criticized because of the small effect size of the discovered variants and the mystery of missing heritability, these two questions could be partially explained by the newly raised conceptual models, such as omnigenic model and natural selection. Finally, we introduce the clinical use of GWAS findings in the bone field, such as the identification of causal clinical risk factors, the development of drug targets and disease prediction. Despite the fruitful GWAS discoveries in the bone field, most of these GWAS participants were of European descent, and more genetic studies should be carried out in other ethnic populations to benefit disease prediction in the corresponding population.
Subject terms: Calcium and phosphate metabolic disorders, Osteopetrosis
Introduction
Osteoporosis is a common skeletal disease affecting ~200 million people around the world; it is characterized by decreased bone density, bone microstructural damage and a consequent increase in bone fragility.1,2 Nearly 22 million women and 5.5 million men were estimated to have osteoporosis in Europe3 and 10 million in the United States, and this number continues to rise.4 In China, ~83.9 million people are estimated to suffer from osteoporosis, and this number, including osteopenia, should increase to ~212 million people by 2050.5 Bone fragility is a poor outcome of osteoporosis, where long-term therapy and medical management are needed, especially in elderly individuals.1 By 2050, it is estimated that ~51.1% of worldwide hip fracture cases will be from Asia.6 Accordingly, the burden of treatment for osteoporosis and osteoporotic fractures has been rising very rapidly, with an annual cost of $17 billion to treat fractures in the United States.4,7 In China, ~2.33 million osteoporotic fractures occurred in 2010, costing $9.45 billion, and the annual costs are estimated to double by 2035.8 Therefore, as aging-related diseases, osteoporosis and osteoporotic fracture inflict a substantial economic, social, and clinical burden.
Osteoporosis, as a complex disease, is influenced by many factors, including diet (calcium and protein intake), physical activity, endocrine status, coexisting diseases, and genetic factors.1 Osteoporosis is mainly characterized by low bone mineral density (BMD), which is highly heritable, with heritability ranging from 50% to 80%.2,9 To date, genome-wide association studies (GWASs) (Supplemental Note Box 1) and meta-analyses have discovered many loci that are associated with BMD, osteoporosis, and osteoporotic fractures.10–12 Furthermore, next-generation sequencing (NGS) of large-scale samples has also uncovered novel rare/low-frequency variants in susceptible genes/loci for BMD, osteoporosis and fracture.13,14 Recently, the approach of Mendelian randomization was widely used to identify the causative risk factors for osteoporosis by using GWAS results.15
In this article, we first reviewed the fruitful discovery achieved by GWASs and meta-analyses for osteoporosis and related traits in the last 12 years (Fig. 1). We introduced several newly raised conceptual models, such as omnigenic models and natural selection, which might explain the mystery of missing heritability of complex traits. We then summarized the clinical use of GWAS findings in the bone field, such as the identification of causal clinical risk factors, the development of drug targets, and disease prediction.
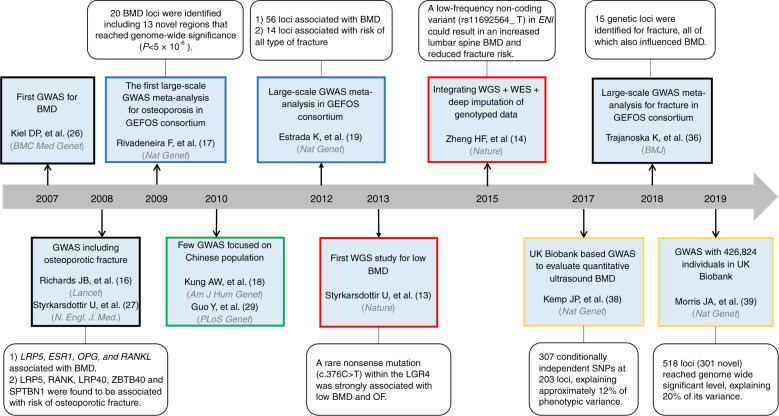
Fig. 1.
Timeline highlighting important milestones during the 12 years of GWAS discoveries for osteoporosis and related traits. Blue boxes indicate the studies from the GEFOS and GENOMOS consortia. The green box indicates the studies focused on the Chinese population. Red boxes indicate GWASs including rare variants. Yellow boxes indicate the UK Biobank-based GWAS. BMD bone mineral density, ESR1 estrogen receptor 1, GWAS genome-wide association study, LRP5 low-density lipoprotein receptor-related protein 5, LRP40 low-density lipoprotein receptor-related protein 4, OPG osteoprotegerin, RANK receptor activator of nuclear factor-kappa β, RANKL RANK ligand, SPTBN1 spectrin beta, nonerythrocytic 1, WES, whole-exome sequencing, WGS whole-genome sequencing, ZBTB40 zinc finger and BTB domain containing 40
GWAS in the bone field
Measurement of bone mass
Most studies have focused on areal BMD (aBMD) obtained from a 2-dimensional projection scan with dual energy X-ray absorptiometry (DXA).14,16–19 The T-score is measured in standard deviation (SD), a mathematical term that calculates how much one’s bone mass varies from the average. It defines an individual’s bone mass as normal (above −1 SD), osteopenia (between −1.0 and −2.5 SD) and osteoporosis (below −2.5 SD).20 This measurement could be influenced by several different skeletal parameters, such as periosteal expansion, trabecular volumetric BMD (vBMD), cortical BMD, cortical thickness, trabecular number and trabecular thickness.21
Bone mass can also be assessed with other radiological imaging tools, such as quantitative computed tomography (QCT), which has the advantage of revealing unique bone information. Paternoster et al.22 performed the first GWAS on cortical vBMD measured by QCT and found that the genetic variant rs1021188 near the RANKL gene was associated with the density of cortical bone, and rs9287237 on FMN2 was associated with the trabecular bone fraction, while the other three SNPs (rs271170 on LINC00326 near EYA4, rs7839059 on COLEC10 near OPG and rs6909279 on CCDC170 near ESR1) had previously been reported to be associated with aBMD. However, QCT was not applicable to the WHO definition of osteoporosis that was based on DXA measurement, and QCT was more expensive with a higher dosage of exposure to radiation but might not predict fractures better than DXA measurement.23
An alternative method of estimating bone mass is derived from quantitative ultrasound (QUS). This measurement is quick, safe, and relatively inexpensive and can therefore be assessed in very large sample sizes, such as ~500 thousand samples in the UK Biobank. The advantages over DXA make QUS a complementary (not replacement) approach to bone health assessment. QUS consists of the use of two separate ultrasound measurements, speed of sound (SOS) and broadband ultrasound attenuation (BUA), typically at the heel. Measures of estimated BMD derived from ultrasound were moderately correlated with DXA-derived BMD at the hip and spine.24 A meta-analysis of GWASs25 using heel ultrasound parameters identified a novel locus (rs597319 near TMEM135) and replicated 6 previously reported loci (ESR1, SPTBN1, RSPO3, WNT16, DKK1 and GPATCH1).
Early GWAS design
It has been established that the variation in BMD is the most important predictor for osteoporosis and fracture. Therefore, GWASs for osteoporosis mainly investigated the effect of genetic influence on BMD. In 2007, Kiel et al.26 published the first GWAS, including 1 141 Framingham Heart Study subjects, and they identified 40 SNPs that could potentially be associated with several bone phenotypes (Fig. 1). Unfortunately, owing to the small sample size, none of the P values exceeded the threshold of genome-wide significance (P < 5 × 10−8). In 2008, two GWASs were published and identified 4 loci associated with BMD (LRP5, ESR1, OPG, and RANKL); in addition, LRP5, RANK, LRP40, ZBTB40, and SPTBN1 were found to be associated with the risk of osteoporotic fracture (Fig. 1).16,27 Soon afterwards, a deluge of GWASs were conducted on osteoporosis and related traits (Fig. 2, Table 1 and Supplemental Table 1).
Fig. 2.
Genetic loci reported by the GWAS catalog for osteoporosis and related traits. # Fracture occurring at any site, except fingers, toes and skull, after age 18. BMC bone mineral content, BMD bone mineral density, BUA broadband ultrasound attenuation, FN femoral neck, LL lower limbs, OF osteoporotic fractures, SOS speed of sound
Table 1.
Genome-wide association studies conducted on osteoporosis and related traits
| Studies | Traits | Discovery sample number and ancestry | Replication sample number and ancestry | Design | Population | Measurement | |
|---|---|---|---|---|---|---|---|
| Kiel DP (2007)26 | BMD (spine, femoral neck), hip geometry | 1 117 European | – | GWAS | Adult | DXA | |
| Richards JB (2008)16 | BMD (spine, femoral neck), osteoporosis, osteoporotic fractures | 2 094 European women | 6 463 European | GWAS | 91.8% women | DXA | |
| Styrkarsdottir U (2008)27 |
BMD (spine, total hip or femoral neck), low-trauma fractures |
5 861 European | 7 925 European | GWAS | Elderly adults | DXA | |
| Liu YZ (2008)51 | Bone size (hip), low-trauma hip fractures | 1 000 European |
1 216 European; Chinese (266 cases, 177 controls) |
GWAS | Adult | DXA | |
| Styrkarsdottir U (2009)144 | BMD (spine, hip), low-trauma fractures | 6 865 European | 8 510 European | GWAS | Adult | DXA | |
| Xiong DH (2009)145 | BMD (spine, total hip, femoral neck) | 1 000 European | 4 925 European, 3 655 Chinese, 908 African | GWAS | Adult | DXA | |
| Timpson NJ (2009)44 | BMD (femoral neck, total body minus the head) | 1 518 European | 4 312 European | GWAS | Children | DXA | |
| Liu YZ (2009)146 | BMD (spine, femoral neck) | 1 000 European | 3 355 European | GWAS | Adult | DXA | |
| Rivadeneira F (2009)17 | BMD (spine, femoral neck) | 19 195 European | – | GWAS-meta | Adult | DXA | |
| Guo Y (2010)29 | Osteoporotic fractures (hip) |
Asian Chinese (350 cases and 350 controls) |
Asian Chinese (390 cases and 516 controls) BMD (9 962 European and Chinese) |
GWAS | Elderly adults | DXA | |
| Kung AW (2010)18 |
BMD (spine, femoral neck), osteoporotic fractures |
785 Asian Chinese | 13 913 European,1 584 Asian Chinese | GWAS | Adult | DXA | |
| Koller DL (2010)46 | BMD (spine, femoral neck) | 1 524 European | 669 European | GWAS | Premenopausal women | DXA | |
| Guo Y (2010)147 | BMD (femoral neck) | 983 European | 2 557 European | GWAS | Adult | DXA | |
| Hsu YH (2010)148 |
BMD (spine, femoral neck), geometric indices of the hip |
3 569 European | 7 633 European women | GWAS-meta | Elderly adults | DXA | |
| Tan L (2010)149 | BMD (forearm) | 1 000 European | 1 628 East Asian | GWAS | Adult | DXA | |
| Paternoster L (2010)22 | Cortical BMD | 1 934 European | 3 835 European | GWAS-Meta | Adolescent, adult | pQCT | |
| Kou I (2011)30 | Osteoporosis | Japanese (713 cases and 3 094 controls) | Japanese (1 566 cases and 1 577 controls) | GWAS | Elderly adults | DXA | |
| Duncan EL (2011)150 | BMD (spine, total hip, femoral neck) | 1 955 European | 20 898 European | GWAS | Postmenopausal women | DXA | |
| Lei SF (2012)52 | Bone size (spine) | 1 627 Chinses | 1 728 European | GWAS | Adult | DXA | |
| Estrada K (2012)19 |
BMD (spine, femoral neck), low-trauma fractures |
32 961 European, East Asian | 50 933 European | GWAS-meta | Elderly adults | DXA | |
| Zheng HF (2012)48 |
BMD (forearm), cortical thickness, forearm fracture |
5 672 European; Fracture: 2 023 cases, 3 740 controls, European |
GWAS-Meta | Adult | DXA | ||
| Medina-Gomez, C (2012)45 | BMD (total body minus the head) | 2 600 European | 11 052 European | GWAS | Children, adult | DXA | |
| Liu CT (2012)49 | BMD (spine, femoral neck) | 25 353 European | 24 763 European, East Asian and others | GWAS-Meta | Adult | DXA | |
| Guo YF (2013)53 | Bone size (hip) | 1 627 Chinese | 2 286 European | GWAS | Adult | DXA | |
| Koller DL (2013)47 | BMD (spine, femoral neck) | 4 061 European, Asian | 4 744 European, Asian | GWAS-meta | Premenopausal women | DXA | |
| Deng FY (2013)151 | Bone size (spine) | 2 286 European | 3 503 European; 1 627 Chinses | GWAS | Adult | DXA | |
| Hwang JY (2013)32 | Any low-trauma osteoporotic fractures | Korean (288 cases and 1 139 controls) | Asian (831 cases and 2 305 controls) | GWAS-meta | Elderly adults | Radiographs | |
| Zheng HF (2013)152 | BMD (forearm), forearm fracture | 5 866 European, 715 Mexican American; forearm fracture: 2 023 cases and 3 740 controls | GWAS-Meta | Adult | DXA | ||
| Oei L (2014)153 | Osteoporotic vertebral fractures | 329 cases and 2 666 controls, European | 26 217 European, 1 294 East Asian | GWAS | Elderly adults | Radiographs | |
| Zhang L (2014)35 | BMD (spine, total hip, femoral neck) | 8 472 European, 1 547 East Asian, 1 124 others | 10 732 European, 5 139 East Asian | GWAS-meta | Adult | DXA | |
| Kemp JP (2014)154 | BMD (forearm, lower limb, skull, total body minus the head) | 8 007 European, 1 409 others | – | GWAS-Meta | Children | DXA | |
| Chesi A (2015)43 | BMD (forearm), BMC (forearm) | 908 European; 163 others | 481 European | GWAS | Children | DXA | |
| Zheng HF (2015)14 |
BMD (spine, femoral neck, forearm), fractures# |
BMD (total = 53 236) Fracture (total = 508 253) |
– | WGS, GWAS-meta | Adults | DXA | |
| Tan LJ (2015)155 | BMD (spine, total hip) | 826 Chinese | 1 728 European;709 African American or Afro-Caribbean;408 Hispanic or Latin American | GWAS | Adult | DXA | |
| Styrkarsdottir U (2016)156 | BMD (spine, total hip or whole body), osteoporosis, osteoporotic fractures | European (2 894 cases, 206 485 controls) | – | WGS | Adult | DXA | |
| Styrkarsdottir U (2016)157 | BMD (spine, hip), osteoporotic fractures |
BMD (20 100 Icelanders, 10 091 European and East Asian) osteoporotic fractures (10 389 cases and 264 522 controls) |
GWAS | Adult | DXA | ||
| Mullin BH (2016)158 | BMD (spine, total hip, femoral neck) | 1 042 European | 5 654 European | GWAS | Adult | DXA | |
| Hwang JY (2016)33 | SOS | 1 895 Asian Korean | 7 263 Asian Korean | GWAS-Meta | Adult | Ultrasound | |
| Taylor KC (2016)159 | Fractures# | 10 305 African American or Afro-Caribbean | – | GWAS-meta | Only women | DXA, radiographs, self-report | |
| Choi HJ (2016)160 | BMD (spine, total hip, femoral neck) | 2 729 Korean | 1 547 Chinese; 3 237 European | GWAS | Adult | DXA | |
| Pei YF (2016)161 | BMD (hip Ward’s triangle) | 4 305 European; 1 579 Asian; 1 295 others | GWAS-Meta | Adult | DXA | ||
| Pei YF (2016)162 | BMD (hip trochanter and intertrochanter) | 6 912 European | 971 European, 1 291 others | GWAS | Adult | DXA | |
| Chesi A (2017)50 | BMD (spine, total hip, femoral neck, forearm) | 933 European American | 486 European American | GWAS | Children | DXA | |
| Mullin BH (2017)163 | SOS, BUA | 1 268 European | 1 610 and 13 749 European | GWAS-Meta | Adult | QUS | |
| Villalobos-Comparán M (2017)164 |
BMD (spine, femoral neck) Bone geometry (femoral neck) |
411 Hispanic or Latin American | 420 Hispanic or Latin American | GWAS | Postmenopausal women | DXA | |
| Kemp JP (2017)38 | BMD (heel) | 142 487 European | – | GWAS | Adult | QUS | |
| Peng C (2017)165 | BMD (spine, femoral neck) | 53 236 NR; 5 152 East Asian | – | GWAS-meta | Adult | DXA | |
| Lu S (2017)166 | BMD (spine, hip and total body) | 2 069 European | – | GWAS | Adult | DXA | |
| Alonso N (2018)167 | Osteoporotic vertebral fractures | European (1 553 cases, 4 340 controls) | European (1 028 cases, 3 762 controls) | GWAS | Only women | DXA | |
| Inaba H (2018)168 | BMD (spine) | 235 European; 48 African American or Afro-Caribbean; 13 NR | GWAS | Children | QCT | ||
| Pei YF (2018)169 | BMD (spine, femoral neck) | 37 657 European; 1 539 East Asian; 1 295 others | – | GWAS-meta | Adult | DXA | |
| Lin X(2018)170 | BMD (femoral neck) | 49 988 European | – | GWAS | Adult | DXA | |
| Kim SK (2018)171 | BMD (heel), osteoporosis and fracture |
394 929 European; osteoporosis: 455 cases, 28 819 controls Fracture: 59 378 cases, 348 296 controls |
GWAS | Adult | QUS | ||
| Qiu C (2018)172 | BMD (spine) |
1 547 Asian, 3 237 Caucasian, 712 African, 409 Hispanic |
– | GWAS meta | Adult | DXA | |
| Trajanoska K (2018)36 | Fractures# |
37 857 cases, 227 116 controls, European or East Asian |
147 200 cases, 150 085 controls, European | GWAS-meta | Adult | Diagnosis, self-report | |
| Gregson CL (2018)173 | BMD (spine, total hip) | 1 380 European | 30 970 NR | GWAS | Adult | DXA | |
| Naito T (2018)31 | BMD (spine, femoral neck) | Japanese (173 cases, 81 controls) | – | GWAS | Adult | DXA | |
| Liang X (2018)174 | BMD (spine, hip, femoral neck, forearm, total body) | 2 286 European; 3 404 Framingham Heart Study (FHS) subjects | GWAS | Adult, children | DXA | ||
| Morris JA (2019)39 | BMD (heel), fracture# | 426 824 individuals | – | GWAS-Meta | Adult | QUS, diagnosis, self-report | |
| Hsu YH (2019)175 | Hip geometry | 18 719, mostly European | Mostly European (N > 9 000) | GWAS-Meta | Adult | DXA | |
| Baird DA (2019)54 | Hip shape | 10 217 European, 5 717 NR | – | GWAS-Meta | Adult | DXA | |
| Styrkarsdottir U (2019)56 | Bone size (spine, hip, femoral neck, trochanter, intertrochanteric region) | European (N ≥ 28 954) |
European and East Asian (N = 13 608–21 277) |
GWAS | Adult | DXA | |
| Zhang H (2020)55 |
BMD (hip) Bone size (hip) |
3 267 European; 1 619 East Asian; 843 African American or Afro-Caribbean; 446 Hispanic or Latin American |
GWAS | Adult | DXA | ||
BMD bone mineral density, BMC bone mineral content, BUA broadband ultrasound attenuation, DXA dual-energy X-ray absorptiometry, GWAS genome-wide association study, NA not available, QCT quantitative computed tomography, pQCT peripheral QCT, QUS quantitative ultrasound, SOS speed of sound, WGS whole-genome sequencing; # fracture occurring at any site, except fingers, toes and skull, after age 18
GWASs in the East Asian population
It is worth noting that the success of GWASs mainly came from studies performed in Caucasian populations, while only a few GWASs focused on East Asian populations (Fig. 1 and Table 1). Yang et al.28 performed a case-control GWAS in 700 elderly Chinese Han subjects (350 hip fracture patients and 350 healthy matched controls) and found that UGT2B17 copy number variation was associated with hip fracture. The same GWAS dataset was reanalyzed by Guo et al.29 who found that the rs13182402 SNP in ALDH7A1 at 5q31 was strongly associated with hip fracture (Fig. 1).29 Kung et al.18 conducted a GWAS and meta-analysis of BMD and fragility fractures in Chinese women (Hong Kong population) and found that the intronic SNP rs2273061 in the JAG1 gene was strongly associated with the BMD of lumbar vertebrae (Fig. 1). The first GWAS of osteoporosis conducted in a Japanese population30 found that a common variant (rs7605378 on FONG) at 2q33.1 conferred the risk of osteoporosis in elderly individuals using a total of ~6 700 subjects.30 Recently, a GWAS with only 254 Japanese patients with inflammatory bowel disease (IBD) found that no SNPs reached genome-wide significance (P < 5 × 10−8) for femoral neck (FN) and lumbar spine (LS) BMD.31 Hwang et al.32 performed an association study in 1 119 fracture patients and 3 444 controls in Korean and Japanese populations and found a new MECOM locus associated with osteoporotic fracture [P = 3.59 × 10−8, odds ratio (OR) = 1.39]32. Another multistage GWAS meta-analysis identified a novel heel SOS locus (rs2446422 on GLDN) in the Korean population,33 and the allele-specific epigenetic modifications of the SNP were confirmed using ENCODE annotations.33
GWAS meta-analysis
The allelic architectures of BMD and osteoporosis are likely to be multifactorial, with each factor imparting a relatively small effect. The identification of these loci with weak effects required studies with comprehensive coverage of the genome and very large sample sizes. The Genetic Factors of Osteoporosis (GEFOS) consortium (www.gefos.org) and the Genetic Markers for Osteoporosis (GENOMOS) consortium (www.genomos.eu) were employed to maximize the samples available for large GWAS meta-analyses, with a consequent increase in statistical power and new locus discovery. In 2009, a meta-analysis (GEFOS-1) of femoral neck and lumbar spine BMD was performed within the GEFOS consortium with 19 195 subjects of northern European descent and identified 20 BMD loci, including 13 novel regions that reached genome-wide significance (P < 5 × 10−8)17 (Fig. 1 and Table 1). In 2012, the GEFOS consortium released their second-round GWAS meta-analysis results (GEFOS-2);19 compared to the first GEFOS meta-analysis, the sample size of the study population increased significantly, which led to the identification of 56 loci associated with BMD (Fig. 1 and Table 1). However, not all genome-wide significant results of the first GEFOS meta-analysis could be replicated in the second study because increasing the sample size could also lead to sample heterogeneity.34 GEFOS-2 also revealed 6 loci associated with the risk of all types of fractures (FAM210A, SLC25A13, LRP5, MEPE, SPTBN1, and DKK1);19 however, the definition of fracture in this study was quite heterogeneous, including hip, spine, wrist and other types of fractures. Therefore, these findings should be interpreted with caution before independent validation in other samples with homogeneous fracture types.34 Zhang et al.35 conducted a three-stage GWAS meta-analysis, and two novel loci were identified in the pooled sample of males and females (SMOC1) and in the female-specific sample (CLDN14); they also independently confirmed 13 previously reported loci (ZBTB40, GPR177, FGFRL1, MEPE, MEF2C, ESR1, SHFM1, WNT16, OPG, SOX6, LRP5, AKAP11, and FOXL1). Further gene expression analysis in osteogenic cells implied a potential functional association of the SMOC1 and CLDN14 genes in bone metabolism35. For fracture risk, the largest GWAS meta-analysis to date, including 25 cohorts from Europe, the United States, East Asia and Australia, identified 15 genetic loci for fracture, all of which also influenced BMD.36
GWASs including rare variants
Early GWAS design included only common variants [minor allele frequency (MAF) > = 5%] and poorly covered low-frequency (1% <=MAF < 5%) and rare variants (MAF < 1%). Next-generation sequencing technology provides an approach to capture rare and low-frequency variants, which might be identified to be associated with complex traits with large effects. Styrkarsdottir et al.13 performed the first whole-genome sequencing (WGS) study for BMD in an Icelandic population and found a rare nonsense mutation (c.376 C > T) within LGR4 that was strongly associated with low BMD and osteoporotic fracture. However, the mutation was not present in the public Exome Variant Server (EVS) database or in the Australian samples (Fig. 1)13. In 2015, Zheng et al.14 integrated WGS data (n = 2 882), whole-exome sequencing data (n = 3 549), deep imputation of genotyped data (n = 26 534)37, and de novo replication genotyping data (n = 20 271) and found that a low-frequency noncoding variant (rs11692564_T, near EN1, MAF = 1.6%) could result in an increased lumbar spine BMD (effect size = +0.20 standard deviation, P = 2 × 10−14) and reduced fracture risk (OR = 0.85, P = 1 × 10−11) (Fig. 1). Conditional loss of En1 in a cre/flox mouse model resulted in osteopenia and increased skull bone resorption via an indirect effect since En1 was not expressed in osteoclasts.14
Large-scale biobank based GWASs
The UK Biobank (www.ukbiobank.ac.uk) recruited 502 647 individuals aged between 37 and 76 years from all over the country in 2006–2010, and the heel bone quality of the participants was evaluated by quantitative ultrasound SOS and BUA. In 2017, Kemp et al.38 conducted a GWAS of 142 487 individuals from the UK Biobank using BMD as estimated by quantitative ultrasound of the heel. They demonstrated that 307 conditionally independent SNPs attained a genome-wide significance level at 203 loci, explaining ~12% of phenotypic variance (Fig. 1). Next, they investigated the underlying mechanism of these SNPs by four steps (including a. bioinformatic, functional genomic annotation and human osteoblast expression studies; b. gene function prediction; c. skeletal phenotyping of 120 knockout mice with deletions in genes adjacent to lead independent SNPs; d. the analysis of gene expression in mouse osteoblasts, osteocytes and osteoclasts) and suggested that the GPC6 gene was a novel determinant of BMD and the pathophysiology of osteoporosis.38
A new study by Morris et al. evaluated genetic determinants of BMD as estimated by heel quantitative ultrasound in 426 824 individuals, identifying 518 loci (301 novel) that reached a genome-wide significance level, explaining 20% of its variance (Fig. 1).39 They also undertook a meta-analysis of ~1.2 million individuals and identified 13 fracture loci (all associated with heel BMD), highlighting the importance of BMD as a determinant of fracture risk.39 They found that target genes were enriched in those known to influence bone density and strength from cell-specific features (maximum OR = 58, P = 1 × 10−75) and found an increased abnormal skeletal phenotype frequency through the phenotyping of 126 knockout mice with disruptions in predicted target genes.39 Finally, DAAM2 showed critical effects on bone strength, porosity, composition and mineralization.
While most BMD GWASs analyzed data derived from DXA, these two studies of UK Biobank data used estimated BMDs derived from calcaneus ultrasound. Though Pearson’s correlation coefficients between DXA and QUS parameters showed a moderate association (r = 0.42–0.61)24,40 and quantitative ultrasound had the ability to predict the occurrence of fractures in older women41 and men,42 there were some essential differences. The GWASs did not replicate 18 known loci from previous studies utilizing DXA-derived BMD measures, and 6 loci had opposite effects on heel BMD in the study of Kemp et al.38 compared to previous DXA BMD studies. Although these differences may be due to various reasons, differences in measurement by QUS and DXA were likely to be causes.
Age- and sex-specific BMD GWASs
Although most studies have focused on adults, GWASs have also performed in younger individuals, including children,43–45 teenagers,22 and premenopausal women.46,47 The first GWAS reported for BMD in children identified the SP7 locus, which encodes the transcription factor osterix, as being associated with whole-body BMD, and replication was subsequently achieved in adult lumbar spine BMD.44 Recently, Chesi et al.43 found that two loci achieved genome-wide significance: rs7797976 within CPED1 in girls and rs7035284 near MTAP in boys at the distal radius. Actually, signals at the CPED1-WNT16-FAM3C locus have been previously reported to be associated with BMD at other skeletal sites in adults17 and children (skull and total body aBMD) of European ancestry45. Interestingly, this locus was also associated with cortical bone thickness, bone strength, and the risk of forearm fracture in adults;48 peak bone mass in premenopausal women;47 and BMD and fracture in elderly individuals.19 The integration of functional studies in Wnt16 knockout mice revealed reductions in bone mineral content (BMC), bone area and bone strength.45,48 Both natural variation in humans and functional studies in Wnt16 knockout mice demonstrated that WNT16 was an important determinant of the bone mass at different body sites in children and adults and the risk of fracture, suggesting that this genetic effect acted over the whole lifetime.
To detect genetic variants influencing variability in peak BMD in premenopausal women, Koller et al. conducted a GWAS in 1 524 US Caucasian women (aged 20–45 years) and 669 African American women (aged 20–44 years). A novel gene, CATSPERB, was identified to be significant in femoral neck BMD.46 CATSPERB was not found to be significant in the meta-analyses of GEFOS-2, although the samples from the above study were included in GEFOS-2.19 Later, a meta-analysis was carried out restricting samples to premenopausal white women from 4 cohorts (n = 4 061, aged 20–45 years), and two loci (WNT16) and (ESR1/C6orf97) were identified to influence the peak bone mass at the lumbar spine and femoral neck.47 Only 4 out of the 56 GEFOS-2 loci19 were observed to have P values below 5 × 10−5 in this meta-analysis.
Although most of the published GWASs on skeletal phenotypes did not have adequate power to test sex-specific genetic effects,49 there was suggestive evidence for an interaction between sex and SNP rs1021188 (near RANKL) (P = 0.01), with a stronger association in males than females (at age 15, males −6.77 mg·cm3 per C allele, P = 2 × 10−6; females −2.79 mg·cm3 per C allele, P = 0.004).22 In the GEFOS-2 study,19 two loci (Xp22.31 in men and 8q13.3 in women) were discovered in the sex-stratified meta-analysis; however, only the locus in Xp22.31 (near FAM9B) showed significant heterogeneity (Phet = 1.62 × 10−8), and the imbalance in sample size between women and men and the conservative heterogeneity test limited the ability to identify sex-specific findings. In a study of European American children (n = 1 419),50 four novel loci (IZUMO3, RBFOX1, SPBT, and TBPL2) were identified to be associated with BMD at the 1/3 distal radius, spine, total hip and femoral neck, two of which were sex-specific loci (SPTB in females and IZUMO3 in males).
Bone size/geometry GWASs
Bone size (BS) is also an important factor that influences bone geometry and bone strength. To date, a limited number of GWASs for BS have been conducted compared to studies on BMD. In Table 1, we summarized the current GWASs and meta-analyses in bone size/geometry. In early studies, GWASs of the bone area of the hip or lumbar spine using DXA did not find significant loci,26,51–53 possibly due to the small sample size. Recently, a study of a hip shape model (HSM) derived from statistical shape modeling of DXA scans found 8 loci associated with hip shape,54 and another GWAS meta-analysis identified 22 significant loci (P < 5.0 × 10−8) for hip bone size.55 Styrkarsdottir et al. reported a large GWAS of bone size using a simple parameter from DXA scans, the bone area,56 they found that 8 loci for the lumbar spine area, 5 loci for the total hip area, 4 loci for the intertrochanteric area, 3 loci for the trochanter area, and 1 locus for the femoral neck area satisfied the criteria of genome-wide significance (Table 1).
GWAS findings cannot perfectly explain the variance in bone mass
GWASs on osteoporosis and related traits have made great achievements in the past 12 years and have highlighted many genes/loci and related biological pathways that contribute to the pathophysiology of osteoporosis and/or fracture, such as the RANK-RANKL-OPG and WNT signaling pathways. These pathways are functionally relevant to bone metabolism and endochondral ossification, and their contribution to osteoporosis has been well established.57 However, at the same time, similar to other complex traits, the variance in bone mass could not be fully explained by GWAS findings.
Missing heritability and beyond
The genetic architecture of osteoporosis and fracture involves both common and rare functional variants,58,59 and the effect sizes of low-frequency and rare variants by genetic burden are larger than those of common variants.14 BMD is also a highly heritable trait, and the genetic effect was estimated to account for as much as 75% of the variance in BMD at the site of the femoral neck;60 however, only a small portion of heritability was explained by loci identified by GWASs. For example, in the GEFOS-1 study,17 15 LS-SNPs combined explained ~2.9% of the variance in LS-BMD, and 10 FN-SNPs combined explained ~1.9% of the variance in FN-BMD with 19 195 subjects. In the GEFOS-2 study,19 63 SNPs explained 5.8% of the total genetic variance in FN-BMD among ~84 000 individuals. To date, the number of associated loci has increased to ~1 000, explaining 20% of the variance in eBMD among 426 824 subjects.39 GWASs rely on the “common disease-common variant” hypothesis and lead to the identification of multiple genetic variants that explain, in aggregate, only a small portion of the BMD variance. This has been referred to as the mystery of the “missing heritability”.61 Therefore, larger-scale, better-powered GWASs could identify more variants, but it seems that the variance explained by these common variants is likely to remain minor.
Unlike Mendelian diseases that are caused by mutations in coding regions, most of the associated SNPs for osteoporosis and related traits are found in noncoding intergenic and intronic regulatory regions.62 Therefore, the greatest challenge was to understand the functional consequences of these SNPs and to accurately elucidate the biological mechanism by which these genes and SNPs act. To date, only a small fraction of SNPs/genes and their functional mechanisms have been successfully characterized,63 and these variants or regions could be transcription factor binding sites that regulate or affect gene expression.62
Polygenicity and negative selection
GWASs of osteoporosis and related traits often identified a number of SNPs that had significant p values but showed very low disease odds ratios (ORs). For example, in the GEFOS-2 study,19 13 of 14 SNPs associated with any low-trauma osteoporotic fracture had ORs <1.10. A recent GWAS involving 53 184 fractures and 373 611 controls39 identified 14 association SNPs, all of which had ORs <1.10. In a GWAS of Chinese fractures, the highest OR of rs13182402 in the ALDH7A1 gene was 2.25.29 Despite their statistical significance, the ORs were small and explained little about the genetic contribution to fracture.
Over the last few years, a commonly accepted explanation for the small OR was that osteoporosis was caused by a large number of interacting genes, each with a small effect size and additive increment to disease risk, called “polygenic inheritance”. It is known that common diseases have a polygenic genetic architecture.64 Thus, perhaps in many cases, the so-called problem of missing heritability might be synonymous with high polygenicity (defined as the total number of genetic loci or alleles with nonzero effects contributing to a phenotype).61,65 The classic polygenic model consists of contributions to disease risk from both common and rare variants.61 In 2018, using UK Biobank data, Zeng et al. confirmed that negative selection played a predominant role in shaping the relationship between effect size and MAF for complex traits.66 They found that 23 out of the 28 studied complex traits (including heel BMD) showed significant signatures of natural selection, and the genetic variants associated with heel BMD were under negative selection, with a moderate estimate (Sˆ = −0.381), where Sˆ reflected the strength of selection on the trait-associated SNPs.66
More recently, O’Connor et al. redefined polygenicity as the effective number of independently associated SNPs (Me). For the 33 complex traits they studied, the “Me” estimates for common SNPs ranged from 500 to 20 000, with a ‘Me’ estimate for heel BMD of ~800.67 This implied that most common SNPs were associated with complex traits and that heritability was spread evenly across the genome.67 They found that functionally important regions in the genome had higher polygenicity and higher heritability, but low-frequency SNPs had lower polygenicity than common SNPs on average. The conclusion was that negative selection not only constrained the effect sizes of common variants on average but also flattened their distribution across the genome.67
Polygenicity and omnigenicity
Recently, Boyle et al.68 proposed the “omnigenic model” in which gene regulatory networks were fully interconnected; that is, all genes expressed in disease-related cells were considered to affect disease phenotype, but most heritability could be explained by effects on genes outside core pathways. This model tried to answer 2 questions: (1). Why do the lead hits from GWASs for any given trait contribute so little to heritability? (2). Why does so much of the genome contribute to heritability? The key feature of this model was the classification of genes as “core” (direct roles in disease) or “peripheral” (essentially all other expressed genes can transregulate core genes). In fact, the “omnigenicity model” is one scenario of the “polygenicity model”, in which the “polygenicity” is partitioned into different parts.
In the latest point of view, they defined the “core gene” as the only gene from which the gene product (protein or RNA for a noncoding gene) had a direct effect—not mediated through the regulation of another gene—on cellular and organismal processes, leading to a change in the expected value of a particular phenotype; “peripheral genes” were defined as those expressed in relevant cell types that could affect the phenotype only indirectly through regulatory effects on core genes.69 This model assumed that the relationship between each core gene and the expected phenotype value was a linear function of the gene expression level; moreover, each core gene was likely affected by large numbers of weak trans (peripheral) variants, and most trait heritability was mediated through trans effects.69
Based on this model, most variants that contributed to heritability tended to be spread across the whole genome, and genes with specific functions for osteoporosis or related traits could only explain little heritability. This might explain why some loci/genes identified by GWASs for BMD were considered to have no contribution to the pathophysiology of osteoporosis and/or fracture, while some genes that had known functional relevance to bone metabolism and endochondral ossification tended to be core genes. For example, LRP5, which encodes low-density lipoprotein receptor-related protein 5, could function as a coreceptor together with the seven-transmembrane-spanning Frizzled for Wnt proteins to regulate intracellular signal transduction by β-catenin,70,71 and the activation of the Wnt pathway results in cytoplasmic β-catenin accumulation. Consequently, β-catenin translocates to the nucleus and in turn regulates osteoblast proliferation and differentiation, thus determining bone mass.72 Osteoblasts produce RANKL following the binding of RANKL to RANK on the surface of osteoclastic precursors, and subsequently, NF-κB is activated and translocates into the nucleus and interacts with NFATc1 to trigger osteoclastogenic gene transcription.73 OPG, a member of the tumor necrosis factor (TNF) receptor superfamily (TNFRS), also known as TNFRS member 11B (TNFRS11B), can bind to RANKL to prevent its coupling with RANK and inhibit the maturation of osteoclasts as a result of reducing bone resorption. Notably, these genes related to the bone metabolism pathway, such as LRP5,16 RANKL,22 ESR1,25,38,39 BMP4 (bone morphogenetic protein 4),38,74 and WNT16, were identified in GWAS signals.38,45,48
Clinical relevance of GWAS findings
The ultimate goal of genetic study is to translate the discoveries into clinical practice. GWAS discoveries for osteoporosis and related traits in the past 12 years are undoubtedly more fruitful than previous linkage analyses and candidate gene association analyses, and hundreds of loci (thousands of SNPs) have been identified that are significantly and robustly associated with osteoporosis and related traits (Fig. 2, Table 1 and Supplemental Table 1). However, it is still too early to understand the function of novel proteins identified by GWASs. This review is not meant to describe novel discovered loci and their interactions. We assumed that there are three ways in which GWAS findings could provide important clinical insight for osteoporosis. First, GWAS results could be employed to investigate the causal risk factors for osteoporosis by using the Mendelian randomization approach. Second, new drug targets and anti-osteoporotic therapeutics should be investigated. Despite the small effect of common variants identified by GWASs, it should be noted that the effect size of the genetic variant on molecular phenotypes could be large, and the drug effect on targets could also be magnified (e.g., statins).75 Third, genetic information could be applied to “personalized” medicine, for example, disease prediction and risk stratification, leading to the overall improvement in disease prevention or intervention.
Mendelian randomization approach to link clinical risk factors to osteoporosis and fracture
The identification of causative risk factors is essential for the prevention and treatment of osteoporosis, and a better understanding of causality could be conducive to further prevention strategies and clinical trials and to providing targets for effective lifestyle and drug intervention.15,76 Observational studies have identified associations of potential risk factors (for example, smoking, low body mass index (BMI), low vitamin D level, earlier age at menopause and physical inactivity) with fracture risk. However, because of confounding factors and reverse causality, bias might be introduced into observational studies, thereby reducing their reliability. The gold standard for evidence for causal effects could come from well-conducted randomized control trials (RCTs), but RCTs are resource-intensive and examine mainly short-term exposures. In addition, not all risk factors can be investigated by RCTs.15
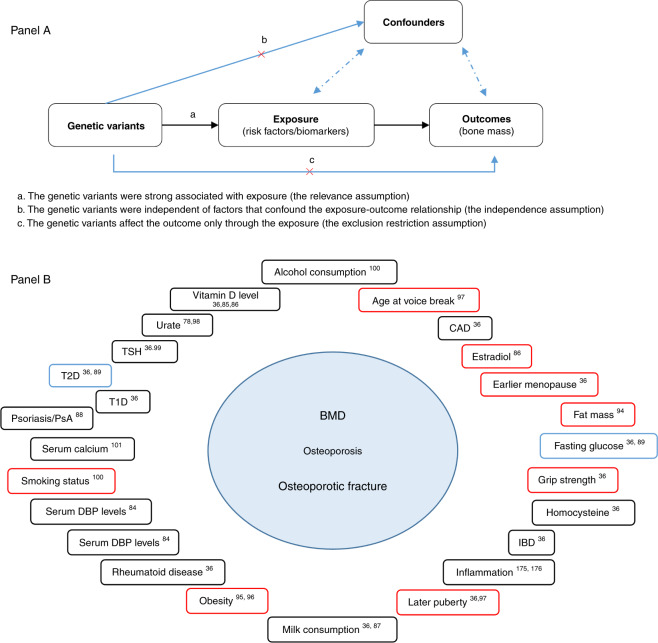
Recently, Mendelian randomization (MR) analyses have been widely used to illustrate the causal effect between exposures and outcomes using large-scale GWAS summary statistic data.77–79 MR is a type of analytical approach that takes genetic variants associated with a risk factor (e.g., calcium) as instrumental variables (IVs) to examine the causality between exposure and outcome (e.g., BMD).80,81 Since the genetic alleles are randomly assorted during conception, MR analyses are less susceptible to confounding factors; additionally, MR analyses are robust to reverse causation bias because genotypes are unlikely to be affected by disease. Further information can be found in Supplemental Note Box 2. Three main assumptions must be applied when conducting Mendelian randomization analyses.82 First, the genetic variants should be strongly associated with exposure (the relevance assumption); second, the genetic variants should be independent of factors that confounded the exposure-outcome relationship (the independence assumption); and third, the genetic variants affect the outcome only through the exposure (the exclusion restriction assumption) (Fig. 3, Panel A). This approach has advantages over traditional observational studies by minimizing confounding bias. To date, the MR approach in the bone field has been applied predominantly to assess causal relationships between different factors and BMD, osteoporosis and fracture (Fig. 3, Panel B and Table 2). Among these factors, vitamin D level, inflammatory disease, obesity and diabetes were frequently investigated.
Fig. 3.
Mendelian randomization in bone field. Panel A: Principal of Mendelian randomization. Panel B: The causality between the clinical risk factors and osteoporosis from the current literature. Red boxes indicate the causal relationship. Black boxes indicate the noncausal relationship. Blue boxes indicate controversial results. BMD bone mineral density, CAD coronary artery disease, IBD inflammatory bowel disease, DBP vitamin D binding protein, PsA psoriatic arthritis, T1D type 1 diabetes, T2D type 2 diabetes, TSH thyroid stimulating hormone
Table 2.
Mendelian randomization studies in the bone field
| Studies | Exposure | Outcome | IVs | Sample size; ethnicity | MR method | Unit | P value | Estimate (95% CI) | Interpretation |
|---|---|---|---|---|---|---|---|---|---|
|
Timpson NJ (2009)94 |
Obesity (fat mass) |
BMC | 2 SNPs (fat mass) |
Obesity (8 480) BMC (7 470), children; European |
IV regression with 2SLS | 1 g change in BMC per 1 kg change in fat mass | 0.000 2 | TB-BMC, 0.02 (−0.20, 0.15) | Fat mass is in the causal pathway for bone mass in children. |
| 0.03 | UL-BMC, 0.46 (0.31, 0.61) | ||||||||
| 0.002 | LL-BMC, 0.55 (0.41, 0.68) | ||||||||
| 2.30E-06 | LS-BMC, 0.48 (0.33, 0.63) | ||||||||
| Warodomwichit D (2013)96 | Obesity | BMD | 1 SNP |
Obesity (2 154) BMD (2 154), adults; Thai |
IV regression with 2SLS | 1 g.cm−2 change in BMD per 1 kg.m−2 change in BMI | 0.01 | TH-BMD, 0.02 (0.00, 0.03) | Obesity might be causally related to BMD at the femur but not at the spine. |
| 0.014 | FN-BMD, 0.01 (0.00, 0.03) | ||||||||
| NS | LS-BMD, 0.00 (−0.01, 0.01) | ||||||||
|
Oeil L (2014)176 |
Inflammation | Fracture | 29 SNPs (CRP) |
Inflammation (6 386) Fracture (1 561), adults; American |
Weighted genetic risk score | OR for fracture per 1 SD increase in CRP | 0.23 | Fracture, 1.00 (0.99, 1.00) | No causal association between CRP level and fracture. |
|
Leong A (2014)84 |
Serum DBP levels | BMD | 1 SNP |
Serum DBP levels (2 254) BMD (2 254), adults; Canadian |
IV regression with 2SLS | 1 g.cm−2 change in BMD per 1 SD change in DBP | 0.43 | FN-BMD, −0.005 (−0.02, 0.01) | No causal association between DBP level and BMD. |
|
Dalbeth N (2015)98 |
Urate | BMD | 5 SNPs |
Urate (2 501) BMD (2 501), adults; European |
IV regression with 2SLS | 1 g.cm−2 change in BMD per 1 mmol.L−1 change in urea levels | 0.06 | TF-BMD, −0.29 (−0.60, 0.01) | No causal association between urate and BMD. |
| 0.08 | FN-BMD, −0.27 (−0.58, 0.03) | ||||||||
| 0.68 | LS-BMD, 0.08 (−0.32, 0.48) | ||||||||
| Xiong A (2016)78 | Urate | BMD | 18 SNPs |
Urate (1 322) BMD (1 322), adults; Chinese |
IV regression with 2SLS | 1 g.cm−2 change in BMD per 1 mmol.L−1 change in urea levels | 0.5 | TH-BMD, 0.19 (−0.36, 0.74) | No causal association between urate and BMD. |
| 0.53 | FN-BMD, −0.19 (−0.42, 0.81) | ||||||||
| 0.26 | LS-BMD, 0.39 (−0.26, 0.98) | ||||||||
|
Kemp JP (2016)95 |
Obesity | BMD | 32 SNPs (BMI) |
Obesity (5 221) BMD (5 221), children; European |
MR Egger; Multivariable MR | SD change in BMD per SD increase in BMI | 0.78 | SK-BMD, −0.02 (−0.20, 0.15) | Obesity is causally related to increase in BMD at all sites except the skull. |
| <0.001 | UL-BMD, 0.46 (0.31, 0.61) | ||||||||
| <0.001 | LL-BMD, 0.55 (0.41, 0.68) | ||||||||
| <0.001 | LS-BMD, 0.48 (0.33, 0.63) | ||||||||
| <0.001 | PE-BMD, 0.39 (0.34, 0.64) | ||||||||
|
Li SS (2016)85 |
Vitamin D level | BMD | 4 SNPs |
Vitamin D level (1 824) BMD (1 824), postmenopausal women; Chinese |
IV regression with 2SLS | 1 g.cm−2 change in BMD per 1 log ng.mL−1 change in total 25OHD | 0.326 | TH-BMD, −0.04 (−0.13, 0.04) | No causal association between vitamin D and BMD. |
| 0.261 | FN-BMD, −0.04 (−0.13, 0.03) | ||||||||
| 0.384 | LS-BMD, 0.05 (−0.16, 0.06) | ||||||||
|
Ahmad OS (2017)89 |
T2D | BMD | 32 SNPs |
T2D (149 821) BMD (32 961), adults; European |
IVW approach | SD change in BMD per odds in log-odds of T2D | 0.044 | FN-BMD, 0.034 (0.001, 0.067) | Genetically increased T2D risk and genetically increased fasting glucose have weak positive effects on FN-BMD. |
| 0.148 | LS-BMD, 0.026 (−0.01, 0.061) | ||||||||
| Fasting glucose (FG) | BMD | 30 SNPs |
FG (133 010) BMD (32 961), adults; European |
IVW approach | SD change in BMD per 1 mmol.L−1 increase in GF | 0.034 | FN-BMD, 0.13 (0.01, 0.25) | ||
| 0.211 | LS-BMD, 0.082 (−0.045, 0.21) | ||||||||
| 2-h glucose | BMD | 6 SNPs |
2hGlu (133 010) BMD (32 961), adults; European |
IVW approach | SD change in BMD per 1 mmol.L−1 increase in 2hGlu | 0.134 | FN-BMD, 0.089 (−0.027, 0.20) | ||
| 0.354 | LS-BMD, 0.06 (−0.06, 0.18) | ||||||||
| Yang Q (2017)87 | Milk consumption | BMD | 1 SNP (lactose intolerance) |
Milk consumption (32 961) BMD (32 961), adults; European |
IVW approach | 1 SD change in BMD per 1 SD change of milk consumption | NA | FA-BMD, 0.049 (−0.128, 0.226) | No causal association between adult milk intake and BMD. |
| NA | FN-BMD, −0.015 (−0.089, 0.059) | ||||||||
| NA | LS-BMD, 0.015 (−0.073, 0.104) | ||||||||
|
Huang JV (2017)177 |
Inflammation | BMD | 16 SNPs (CRP) |
BMD (32 961); European |
MR-Egger IVW approach | 1 g.cm−2 change in BMD per 1 log mg.L−1 change in total hsCRP | 0.506 | FA-BMD, 0.054 (NA) | No causal association between hsCRP and BMD |
| 0.726 | FN-BMD, −0.014 (NA) | ||||||||
| 0.184 | LS-BMD, −0.074 (NA) | ||||||||
| Cousminer DL (2018)97 | Later puberty | BMD | 331 SNPs | 733 girls; European | Two-sample MR | 1 SD change in BMD per 1 year later onset of puberty | 0.004 6 | LS-BMD −0.179 | A causal association between later puberty and LS-BMD. |
| Age at voice break | BMD | 43 SNPs | 685 boys; European | Two-sample MR | 1 SD change in BMD per 1 year earlier onset of age at voice break | 0.000 3; 7.04E-05 | LS-BMD −0.119; FN-BMD: −0.113 | A causal association between later puberty and LS/FN-BMD. | |
|
Larsson SC (2018)86 |
Estradiol | BMD | 1 SNP |
Estradiol (2 767) eBMD (32 965), adults; European |
IVW approach | 1 SD change in BMD and g.cm−2 in eBMD per 10% increase in estradiol | 4.60E-06 | FN-BMD, 0.038 (NA) | A causal association between serum estradiol levels and increase BMD. |
| 0.001 | LS-BMD, 0.031 (NA) | ||||||||
| 6.00E-18 | eBMD, 0.030 (NA) | ||||||||
| Vitamin D level | BMD | 5 SNPs |
Vitamin D (42 274) BMD (32 961) eBMD (142 487), adults; European |
IVW approach | 1 SD change in BMD per 1 SD change in 25OHD (g.cm−2 eBMD) | 0.37 | FN-BMD, 0.02 (−0.03, 0.07) | No causal association between vitamin D and BMD. | |
| 0.49 | LS-BMD, 0.02 (−0.04, 0.08) | ||||||||
| 0.02 | eBMD, -0.03 (−0.05, −0.01) | ||||||||
| van Vliet NA (2018)99 | TSH | BMD | 20 SNPs |
TSH (26 420) BMD (32 735), adults; European (mostly) |
Two sample MR IVW approach | SD change in BMD per 1 SD decrease in serum TSH level | 0.92 | FN-BMD: 0.003 (−0.053, 0.048) | No causal association between serum TSH levels and BMD. |
| 0.73 | LS-BMD: 0.010 (−0.069, 0.049) | ||||||||
|
Guo R (2018)100 |
Smoking status | BMD | 139-142 SNPs |
Smoking status (32 735) BMD (445 921), adults; European (mostly) |
Two-sample MR IVW approach | NA | 0.053 | FN-BMD: −0.139 (NA) | A causal association between smoking and decreased heel BMD. |
| 0.976 | LS-BMD: −0.003 (NA) | ||||||||
| 0.077 | FA-BMD: −0.264 (NA) | ||||||||
| 0.003 | Heel BMD: −0.053 (NA) | ||||||||
| Alcohol consumption | BMD | 5–6 SNPs |
Alcohol consumption (32 735) BMD (445 921), adults; European (mostly) |
Two-sample MR IVW approach | NA | 0.964 | FN-BMD: −0.008 (NA) | No causal association between alcohol consumption and BMD. | |
| 0.742 | LS-BMD: 0.067 (NA) | ||||||||
| 0.593 | FA-BMD: 0.194 (NA) | ||||||||
| 0.822 | Heel BMD: 0.010 (NA) | ||||||||
| Trajanoska K (2018)36 | T2D | Fracture | 38 SNPs | T2D: 56 862 (12 171 cases) Fracture: 185 057 cases, 377 201 controls; mostly European | Two sample MR; MR-Egger | OR of fracture per doubling in odds of T2D susceptibility | 0.37 | Fracture: 0.99 (0.99, 1.01) | No causal association between T2D and fracture risk. |
| T1D | Fracture | 19 SNPs | T1D: 26 890 (9 934 cases) Fracture: 185 057 cases, 377 201 controls | Two sample MR; MR-Egger | OR of fracture per doubling in odds of T1D susceptibility | 0.57 | Fracture: 1.00 (1.00, 1.01) | No causal association between T1D and fracture risk. | |
| Fasting glucose | Fracture | 35 SNPs |
Fasting glucose (58 074) Fracture: 185 057 cases, 377 201 controls |
Two sample MR; MR-Egger | OR of fracture per 1 SD increase in fasting glucose level | 0.24 | Fracture: 1.04 (0.97, 1.12) | No causal association of fasting glucose levels with fracture risk. | |
| CAD | Fracture | 38 SNPs | CAD: 107 432 (41 513 cases) Fracture: 185 057 cases, 377 201 controls | Two sample MR; MR-Egger | OR of fracture per doubling in odds of CAD susceptibility | 0.76 | Fracture: 1.00 (0.99, 1.02) | No causal association between CAD and fracture risk. | |
| Rheumatoid disease | Fracture | 30 SNPs |
Rheumatoid disease: 58 284 (14 361 cases) Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR; MR-Egger | OR of fracture per doubling in odds of rheumatoid disease susceptibility | 0.14 | Fracture: 1.01 (1.10, 1.02) | No causal association between rheumatoid disease and fracture risk. | |
| Vitamin D | Fracture | 4 SNPs |
Vitamin D: 33 996 Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per 1 SD decrease in 25OHD | 0.07 | Fracture: 0.84 (0.70, 1.02) | No causal association of decreased 25OHD levels with increased fracture risk. | |
| Dairy calcium intake | Fracture | 1 SNP (lactose intolerance) | Dairy calcium intake: 171 213 Fracture: 185 057 cases, 377 201 controls; mostly European | Two sample MR MR-Egger | OR of fracture per 1 SD increase in milk consumption | 0.94 | Fracture: 1.01 (0.80, 1.23) | No causal association between milk consumption and fracture risk. | |
| FN-BMD | Fracture | 43 SNPs |
FN-BMD: 32 961 Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per 1 SD decrease in FN-BMD | <0.001 | Fracture: 1.55 (1.48, 1.63) | A causal association between decreased FN-BMD and increased fracture risk. | |
| LS-BMD | Fracture | 40 SNPs |
LS-BMD: 31 800 Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per 1 SD decrease in LS-BMD | <0.001 | Fracture: 1.43 (1.37, 1.50) | A causal association between decreased LS-BMD and increased fracture risk. | |
| Homocysteine | Fracture | 13 SNPs |
Homocysteine: 44 147 Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per 1 SD increase in homocysteine level | 0.78 | Fracture: 0.98 (0.92, 1.05) | No causal association between homocysteine level and fracture risk. | |
|
Inflammatory bowel disease (IBD) |
Fracture | 151 SNPs |
IBD: 34 652 (12 882 cases) Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per doubling in odds of inflammatory bowel disease susceptibility | 0.92 | Fracture: 1.00 (1.10, 1.01) | No causal association between inflammatory bowel disease and fracture risk. | |
| TSH | Fracture | 20 SNPs |
TSH: 26 523 Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per 1 SD decrease in serum TSH level | 0.78 | Fracture: 0.99 (0.94, 1.04) | No causal association between serum TSH levels and fracture risk. | |
| Grip strength | Fracture | 15 SNPs |
Grip strength: 142 035 Fracture: 185 057 cases, 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per 1 SD increase in grip strength | 0.01 | Fracture: 2.14 (1.13, 4.04) | A causal association between decreased grip strength and fracture risk. | |
| Age of puberty | Fracture | 54 SNPs | Age of puberty: 182 416 Fracture: 185 057 cases, 377 201 controls; mostly European | Two sample MR MR-Egger | OR of fracture per 1 SD change, i.e., 3.9 years earlier menopause | 0.05 | Fracture: 1.10 (1.00, 1.21) | No causal association between earlier menopause and fracture risk. | |
| Age at menopause | Fracture | 106 SNPs |
Age at menopause: 69 360 Fracture: 185 057 cases; 377 201 controls; mostly European |
Two sample MR MR-Egger | OR of fracture per 1 SD change, i.e., 1.4 years late puberty | 0.04 | Fracture: 1.06 (1.00, 1.13) | A causal association between late puberty and increased fracture risk. | |
|
Cerani A (2019)101 |
Serum calcium | BMD | 1 SNP |
Serum calcium: 61 079 BMD: 426 824; mostly European |
IVW approach | 1 SD change in BMD per 1 SD change in serum calcium concentration | 0.85 | heel BMD, 0.003 (−0.059–0.066) | No causal association between serum calcium consumption and heel BMD. |
| Serum calcium | Fracture | 1 SNP |
Serum calcium: 61 079 Fracture: 76 549 cases, 470 164 controls; mostly European |
IVW approach | OR of fracture per 1 SD increase in serum calcium concentration | 0.85 | Fracture, 1.01 (0.89–1.15) | No causal association between serum calcium consumption and fracture risk. | |
| Xia (2020)88 | Psoriasis | eBMD | 60 SNPs | 301 667, European | One-sample MR | SD change in BMD per odds in log-odds of psoriasis susceptibility | 0.24 | heel BMD, − 0.04 (−0.11–0.029) | No causal association between psoriasis and heel BMD. |
|
Psoriasis: 19 032 cases, 286 769 controls eBMD: 462 824; European |
Two sample MR | 0.28 | heel BMD, −0.002 (−0.009–0.002) | ||||||
| Psoriasis | Fracture | 60 SNPs |
Psoriasis: 19 032 cases, 286 769 controls Fracture: 45 087 cases, 317 775 controls; European |
Two sample MR | OR of fracture per doubling in odds of psoriasis susceptibility | 0.72 | Fracture, 1.00 (0.99–1.02) | No causal association between psoriasis and fracture. | |
| Psoriatic arthritis (PsA) | eBMD | 25 SNPs | 301 667; European | One-sample MR | SD change in BMD per odds in log-odds of psoriatic arthritis susceptibility | 0.88 | heel BMD, 0.002 (−0.025–0.030) | No causal association between psoriatic arthritis and heel BMD. | |
|
PsA: 3 061 cases, 13 670 controls eBMD: 462 824; European |
Two sample MR | 0.69 | heel BMD, −0.001 (−0.005–0.003) | ||||||
| Psoriatic arthritis | Fracture | 25 SNPs |
PsA: 3 061 cases, 13 670 controls Fracture: 45 087 cases, 317 775 controls; European |
Two sample MR | OR of fracture per doubling in odds of psoriatic arthritis susceptibility | 0.52 | Fracture, 0.99 (0.98–1.01) | No causal association between psoriatic arthritis and fracture. |
IVs instrument variables, 25OHD 25-hydroxyvitamin D, 2SLS two-stage least squares, BMC bone mineral content, BMD bone mineral density, BMI body mass, CAD Coronary Artery Disease, CI confidence interval, CRP C-reactive protein, DBP vitamin D binding protein, eBMD estimated bone mineral density from ultrasound, FA forearm, FHS Framingham Heart Study, FN femoral neck, IVW Inverse-variance weighted, IW Instrumental variable, LL lower limbs, LS lumbar spine, NA not available, OR odds ratio, PE pelvis, SD standard deviation, SK Skull, T1D type 1diabetes, T2D type 2 diabetes, TB total body, TSH Thyroid Stimulating Hormone, UL upper limbs
Vitamin D and BMD/fracture
Vitamin D, by improving intestinal calcium absorption, has pivotal roles in bone heath. Vitamin D insufficiency was reported as a risk factor for several common diseases and conditions, including osteoporosis and osteoporotic fracture.83 However, the influence of vitamin D on the etiology of low bone mass and osteoporosis is unclear due to inconsistent results from clinical studies.79 Leong et al.84 investigated the causal relationship between vitamin D-binding protein (DBP) levels and BMD in the Canadian Multicentre Osteoporosis Study (CaMos) using individual-level data, and the results demonstrated a strong causal relationship between serum DBP and 25OHD levels; however, serum DBP had no causal effect on femoral neck BMD or osteoporosis (Table 2). Furthermore, Li et al.85 found no evidence for a causal effect of vitamin D levels on BMD (total hip, FN and LS) in Chinese postmenopausal women using four SNPs, GC-rs2282679, NADSYN1-rs12785878, CYP2R1-rs10741657 and CYP24A1-rs6013897, as candidate instrumental variables in the MR analyses. Recently, using data from the GEFOS consortium and UK Biobank, Larsson et al.86 found that vitamin D levels had no effect on BMD (FN, LS, heel) (N = 32 965). Recently, a study also showed a lack of a causal relationship between vitamin D levels and fracture risk by using 37 857 cases and 227 116 controls from the GEFOS Consortium, UK Biobank, EPIC-Norfolk study and 23andMe36 (Fig. 3, Panel B and Table 2). Similarly, as a provider of protein, micronutrients and dairy calcium, milk was recommended by some dietary guidelines, particularly for bone health. However, MR studies using a SNP (rs4988235) located upstream of the lactase gene as an instrumental variable found that milk consumption had no causal effect on BMD87 or fracture36 (Fig. 3, Panel B and Table 2).
Diseases and BMD/fracture
To date, diseases such as type 2 diabetes (T2D) and inflammatory diseases have been studied for their effects on osteoporosis or fracture (Table 2). Trajanoska et al.36 found that IBD was not a causal factor for fracture risk in 185 057 cases and 377 191 controls. More recently, with 432 513 samples from the UK Biobank dataset, Xia et al. found that psoriatic arthritis might be a risk factor for low BMD, but the link was not genetically determined. Psoriasis without arthritis is not a risk factor for osteoporosis.88
By using SNPs as IVs [32 SNPs strongly associated with type 2 diabetes (T2D), 30 SNPs associated with fasting glucose and 4 SNPs associated with 2-h glucose (2hGlu)], Ahmad et al.89 found that a genetically increased risk of T2D and a genetically increased risk of fasting glucose both had weak effects on increasing femoral neck BMD, but no significant trends were observed for the effect of T2D and fasting glucose on lumbar spine BMD.89 Furthermore, Trajanoska et al.36 found that T2D and fasting glucose were not causal for fracture in 185 057 cases and 377 191 controls. The study also reported no causal effect of type 1 diabetes (T1D) and coronary artery disease (CAD) on fracture.36
Other factors and BMD/fracture
Fat mass might be a causal decisive factor of bone mass, but the evidence was contradictory.90–93 By using variants of two loci [FTO (fat mass and obesity-associated gene) and MC4R (melanocortin 4 receptor)] strongly associated with fat mass and obesity, Timpson et al. evaluated the relation between fat mass and bone outcomes in ~5 000 children at a mean age of 9.9 years from the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort and suggested that fat mass was the causal pathway for bone mass in children.94 In 2016, a study investigated whether adiposity was causal for BMD at the skull, upper limbs and lower limbs, pelvis and lumbar spine in 5 221 children from ALSPAC using 32 SNPs (strongly associated with BMI), and the results suggested that adiposity was causally related to increased BMD at all sites except the skull.95 The relationship between obesity and BMD was also investigated in adults, and it was found that obesity might be causally related to BMD at the femur but not at the spine.96 In addition, the MR approach has been used to show a positive causal association between serum estradiol concentrations and femoral neck BMD, lumbar spine BMD and heel BMD.86 Other studies demonstrated that earlier menopause and late puberty were causal factors for increasing fracture risk.36,97 However, urate,78,98 thyroid stimulating hormone (TSH),36,99 homocysteine,36 alcohol consumption100 and smoking status36,100 were not identified as causal factors for BMD or fracture by the MR approach (Fig. 3, Panel B). Notably, it was demonstrated that genetically decreased BMD was the only clinical risk factor with evidence for an effect on fracture risk among 15 clinically identified fracture factors.36 More recently, Cerani et al. undertook an MR study and found that a standard deviation increase in genetically derived serum calcium (0.13 mmol·L−1 or 0.51 mg·dL−1) was not associated with increased estimated BMD (426 824 subjects, P = 0.92) or a reduced risk of fractures (76 549 cases and 470 164 controls; P = 0.85).101
Therapeutic targets for osteoporosis
Despite the small effect size of common variants identified by GWASs, most of the osteoporosis agents in use (or undergoing trials) target pathways related to the GWAS-discovered BMD genes, and genetic information might significantly improve the search for drug targets and increase the success rate of preclinical and clinical trials.102 Moreover, it is well recognized that the effect size of association is not well correlated with clinical relevance, as many FDA-approved medications target proteins linked to common variants identified by GWASs.102–104 An example of success in the field was the use of GWAS data for drug repositioning studies. Sanseau et al.105 found that among the publicly relevant disease-related GWAS loci, 155 out of the 991 loci (15.6%) were related to drug development. Among them, the drug indications of 63 targeted proteins matched the corresponding GWAS traits, indicating that the pathogenic genes excavated by GWASs had a higher probability of being directly used as drug targets.105 For example, the IL12B (interleukin 12B) gene found in psoriasis GWASs encodes the target of ustekinumab, a newly proven drug for psoriasis. In addition, the gene was considered to be related to Crohn’s disease, and the development of related drugs was in a phase II clinical trial.105 Another example was denosumab, which is a drug marketed for the treatment of osteoporosis in postmenopausal women, targeting the gene TNFSF11 (tumor necrosis factor superfamily, member 11), also known as RANKL. Denosumab is a RANKL inhibitor that functions by preventing the development of osteoclasts. Recently, it was speculated that the drug might have a therapeutic effect on Crohn’s disease, as TNFSF11 was found to be significantly associated with Crohn’s disease in GWASs.106 The current drugs that are available for the treatment of osteoporosis and their most likely targets are listed in Table 3. Five anti-osteoporosis therapeutics currently approved or in advanced clinical trials were supported by GWAS data. It was reasonable to believe that the findings of GWASs could be potentially powerful in the identification of anti-osteoporosis drug targets and drug repositioning.
Table 3.
Present and potential near-term osteoporosis drug targets that have been linked to changes in BMD by GWAS. Table adapted from178
| Drug class | Drug target | Principle | Stage | Target locus identified through GWASs | Refs. |
|---|---|---|---|---|---|
| Denosumab | RANKL | Reduces bone resorption by selectively targeting RANKL | Approved for clinical use | RANKL | 179 |
|
Sclerostin inhibitors (Romosozumab) |
Sclerostin (SOST) | Improve the recruitment and activation of osteoblasts by targeting Wnt/β-catenin signaling pathways | Approved for clinical use in Japan, US and Europe | SOST | 107 |
| Selective estrogen receptor modulators | Estrogen receptor | Reduces bone resorption | Approved for clinical use | ESR1 | 180 |
| by targeting the OPG/RANK/RANKL pathway | |||||
| Parathyroid hormone analogs | Parathyroid hormone receptor | Majorly participate in the process of bone formation | Approved for clinical use | Not identified, but the pathway has been | 181,182 |
| by targeting the PKA pathway | highlighted through PTHLH (encodes PTHRP) | ||||
| Bisphosphonates | Farnesyl pyrophosphate | Inhibition of bone resorption | Approved for clinical use | Not identified | 183 |
| Estrogen ESR1 | Estrogen receptor | Reduces bone resorption | Approved for clinical use | ESR1 | 184 |
| by targeting the OPG/RANK/RANKL pathway | |||||
| Cathepsin K inhibitors | Cathepsin K | Inhibition of bone resorption | Terminated | Not identified | 185 |
| by targeting the OPG/RANK/RANKL pathway | |||||
| DKK1 inhibitors | DKK1 | Improve bone formation by targeting the Wnt/β-catenin signaling pathway | In the preclinical phase | DKK1 | 120 |
DKK1 dickkopf 1, ESR1 estrogen receptor 1, OPG osteoprotegerin, PKA protein kinase A, PTHLH parathyroid hormone-like hormone, PTHRP parathyroid hormone-related protein, RANK receptor activator of nuclear factor kβ, RANKL RANK ligand, SOST sclerostin, US United States
The SOST (sclerostin) gene was found to be strongly associated with BMD by GWASs;107 SOST produces sclerostin, which is a key Wnt pathway regulator that is preferentially expressed by osteocytes. Sclerostin acts by binding to the Wnt coreceptor LRP5/6 by competing with Wnt protein; as a consequence, sclerostin blocks the accumulation of β-catenin in the cytoplasm, inhibits the differentiation and proliferation of osteoblasts, enhances osteoclastogenesis and causes bone loss.108,109 Given the inhibitory effect of sclerostin on osteoblast function and bone formation, blocking the activity of sclerostin to activate this pathway seems to be a potential strategy in the treatment of osteoporosis. Romosozumab (AMG785/CDP-7851), a monoclonal humanized antibody to sclerostin, was evaluated for its efficacy. Compared with the traditional bone resorption inhibitor alendronate and the bone formation promoter teriparatide, the greatest feature of romosozumab was its ability to reverse postmenopausal osteoporosis in women with hormone deficiency.110–112 Saag et al. compared the effect between romosozumab (210 mg monthly administered subcutaneously) and alendronate (70 mg weekly) for 12 months, followed by open-label alendronate 70 mg weekly for another 12 months in postmenopausal women with osteoporosis and a fragility fracture113. After 24 months, a lower risk of fractures, including clinical fractures (27% lower), hip fractures (38%), new vertebral fractures (48% lower) and nonvertebral fractures (19%), was observed in the romosozumab-to-alendronate group than in the alendronate-to-alendronate group.113 A phase III clinical trial was conducted to estimate the effect of romosozumab (n = 206) versus teriparatide (n = 209) on osteoporosis in postmenopausal women who took oral bisphosphonate for at least 3 years, and it was found that romosozumab (210 mg once monthly) had a greater effect on hip BMD than subcutaneous teriparatide (20 μg once daily)114. Another trial recruited 7 180 postmenopausal women who had osteoporosis, and the subjects were randomly assigned to receive subcutaneous injections of romosozumab (at a dose of 210 mg) or placebo monthly for 12 months; thereafter, both groups received denosumab 60 mg every 6 months twice.115 At the end of the initial 12 months, romosozumab had decreased the incidence of new vertebral fractures and nonvertebral fractures by ~73% and 24%, respectively.115 At 24 months, a 75% lower risk of vertebral fractures was seen in the romosozumab group after the transition to denosumab.115
On 9 April 2019, the US Food and Drug Administration (FDA) approved romosozumab for the treatment of osteoporosis in postmenopausal women at high risk of fracture, with a boxed warning highlighting the risk of cardiovascular adverse events and a postmarketing requirement to assess the cardiovascular safety of romosozumab.116 On 28 June 2019, the European Medicines Agency (EMA) recommended the refusal of the marketing authorization for Evenity (romosozumab) because the results suggested that patients given Evenity had an increased risk of serious effects on the heart and circulatory system, such as heart attacks or strokes.117 In addition, there were more deaths in patients aged over 75 years who were given the medicine. As it was unclear why the medicine appeared to increase the risk of heart and circulatory problems, measures to reduce the risk could not readily be put in place.117 Dramatically, after re-examining initial opinions, the EMA noted that the medicine showed convincing evidence of benefit in women with severe osteoporosis, with better effect than alendronate, and it was suggested that only women who had no history of heart attack and stroke could take the medicine.117 On 17 October 2019, the EMA recommended that marketing authorization be granted but for a restricted indication in postmenopausal women with severe osteoporosis at high risk of fracture.117
Dickkopfs (DKKs) are secreted proteins composed of two cysteine-rich domains with four homologous forms (DKK-1~4) in vivo. DKK-1 inhibited the Wnt/β-catenin signaling pathway by directly binding to LRP5/6 and formed a complex with Kringen, a transmembrane protein containing a Kringle domain, which increased endocytosis and decreased LRP5/6 content, thus leading to the inactivation of the Wnt pathway.118,119 DDK1 is closely related to bone mass,120,121 and similar to sclerostin monoclonal antibodies, monoclonal antibodies to DKK-1 increase trabecular mass and density in mice122 and restore bone density in osteoporotic mice and rhesus monkeys.123 Monoclonal antibodies to DKK-1 included BHQ880124 and PF04840082,125 but both were in the preclinical phase.
Prediction of osteoporosis and fracture
One of the goals of genetic study is to improve the value of clinical application, for example, to predict osteoporosis or fracture risk from GWAS findings. Studies have shown that at least 150 loci with an OR value of 1.5 or 250 loci with an OR value of 1.25 were required for the prediction of disease risk.126 This suggested that any single locus could not be useful in clinical prediction, regardless of the size of the effect. However, theoretical and empirical studies have suggested that profiling multiple variants that are associated with bone phenotypes could improve the accuracy of fracture prediction and classification beyond that obtained by conventional clinical risk factors, such as the Fracture Risk Assessment Tool (FRAX).127
Polygenic risk scoring
Polygenic risk scoring was one primary approach for disease risk prediction. In a semisimulation study for fracture, it was shown that a profiling of up to 25 genes/variants (each with a relative risk of 1.10–1.35 and frequency ranging from 0.25 to 0.60) in the presence of clinical risk factors—with or without BMD—could achieve an AUC of 0.80.128 Ho-Le et al. took 62 BMD-associated SNPs to define the predictive value of genetic profiling for fracture prediction in 557 men and 902 women and found that individuals with a greater polygenic risk score (PRS) had a lower femoral neck BMD (P < 0.01); each unit increase in PRS was associated with a hazard ratio of 1.20 for fracture, and this association was independent of age, prior fractures and falls.129. However, polygenic risk scoring remained limited due to the linkage disequilibrium (LD) pruning of SNPs (prioritizing the most significant associations up to an empirically determined P value threshold, and pruning the SNPs based on LD).130 To remediate this issue, recent developments in machine learning may be a novel strategy.131,132 Machine learning approaches adapted a set of sophisticated statistical and computational algorithms to make predictions by mathematically mapping the complex associations between a set of risk SNPs to complex disease phenotypes.133 The optimal predictive ability for the target disease was obtained by mapping the pattern of selected features in the training genotype data, and at the end of the training stage, the model with the maximum predictive ability of the training dataset was selected for validation.131,134 Machine learning has been applied to the prediction of diseases or traits, such as inflammatory bowel disease,135 Alzheimer’s disease,136 cancers,137,138 heart failure139 and height.140
Machine learning methods
Through the analysis of 341 449 individuals from the UK Biobank, Forgetta et al. tested whether machine learning methods could provide a clinically relevant genomic prediction of quantitative ultrasound speed of sound (SOS)—a risk factor for osteoporotic fracture.141 In the Model Selection Set, age, sex and BMI explained 4.0% of the variance in SOS; the addition of the remaining FRAX clinical risk factors increased the variance explained to 4.8%, whereas when polygenic risk scores across different P value thresholds were added, the variance explained increased to at most 18.5%.141 Surprisingly, the machine learning algorithm improved the explained variance in SOS to a maximum of 25.0%. Then, they selected the top model (the machine learning algorithm selected 21 717 activated SNPs with a P value ≤ 10−4) from the Model Selection Set to test for its correlation with the SOS in the validation set and found that the model could explain 23.2% of the variance in the measured SOS. Subsequently, they evaluated the associations among SOS, genomically predicted SOS (gSOS), BMD and fracture and found that decreased SOS and fracture were both strongly associated with increased odds of incident fracture (gSOS had the highest risk per SD) in the univariate model. However, in multivariate models, gSOS was more strongly associated with major osteoporotic fracture than SOS or BMD.141 For fracture prediction, gSOS outperformed FRAX clinical risk factors alone. The machine learning algorithms provided better predictions than traditionally used polygenic risk scores. These findings suggested that genetic profiling of BMD-associated genetic variants could improve the accuracy of fracture prediction over and above that of clinical risk factors alone.
Perspective
Despite fruitful GWAS discoveries in the bone field, most of these GWAS participants were of European descent. In fact, if we extended to other complex traits, ~79% of GWASs were conducted in European populations according to the GWAS catalog. Martin et al.142 systematically evaluated the polygenic risk prediction accuracy in Japanese, British and African-descent individuals on the basis of using independent GWASs of equal sample sizes from BioBank Japan (BBJ) and UK Biobank, including 17 quantitative anthropometric and blood panel traits and five disease endpoints; they demonstrated that prediction accuracy was consistently higher with GWAS summary statistics from ancestry-matched summary statistics. The condition of genetic resources and analyses overwhelmingly centered on individuals of European ancestry would lead to imbalances in the subsequent translatability of findings. To realize the full and equitable potential of the polygenic risk score, it was encouraged that more GWASs and sequencing studies on osteoporosis, BMD and fracture should be carried out in additional ethnic populations, such as the Chinese population, which made up ~20% of the global population. Fortunately, the cost of whole-genome sequencing and genotyping has dramatically decreased, making the utility of genetic variants more affordable and practical. In addition, the prioritization of the recruitment and analysis of diverse cohorts would become smooth with an increasingly globalized and connected research community.143
In summary, the achievement of GWASs is unprecedented in the understanding of how genetic variants influence osteoporosis and fracture. In the future, by mining large databases with detailed characterization of relevant phenotypes, more causal genes/mutations will be identified. In addition, large-scale genetic data could provide a new way to identify new drug targets and could be translated into precision treatment options to prevent and treat osteoporosis and fracture.
Supplementary information
Box 1 Introduction of Genome-Wide Association Studies
Box 2 Introduction of Mendelian Randomization (MR)
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81871831 and 32061143019). The funding agencies had no role in the study design, data collection and analysis or the decision to publish or prepare the manuscript. We thank the peer reviewers for their thorough and helpful review of this manuscript. We also thank the High-Performance Computing Center at Westlake University for the facility support and technical assistance.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41413-021-00143-3.
References
- 1.Pouresmaeili F, Kamalidehghan B, Kamarehei M, Goh YM. A comprehensive overview on osteoporosis and its risk factors. Therapeutics Clin. Risk Manag. 2018;14:2029–2049. doi: 10.2147/TCRM.S138000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu X, Zheng H. Factors influencing peak bone mass gain. Front Med. 2021;15:53–69. doi: 10.1007/s11684-020-0748-y. [DOI] [PubMed] [Google Scholar]
- 3.Hernlund E, et al. Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA) Arch. Osteoporos. 2013;8:136. doi: 10.1007/s11657-013-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge R, et al. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005-2025. J. Bone Miner. Res. 2007;22:465–475. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 5.Liu ZH, Zhao YL, Ding GZ, Zhou Y. Epidemiology of primary osteoporosis in China. Osteoporos. Int. 1997;7(Suppl 3):S84–S87. doi: 10.1007/BF03194349. [DOI] [PubMed] [Google Scholar]
- 6.Cooper C, Campion G, Melton LJ., 3rd Hip fractures in the elderly: a world-wide projection. Osteoporos. Int. 1992;2:285–289. doi: 10.1007/BF01623184. [DOI] [PubMed] [Google Scholar]
- 7.Melton LJ., 3rd Adverse outcomes of osteoporotic fractures in the general population. J. Bone Miner. Res. 2003;18:1139–1141. doi: 10.1359/jbmr.2003.18.6.1139. [DOI] [PubMed] [Google Scholar]
- 8.Si L, Winzenberg TM, Jiang Q, Chen M, Palmer AJ. Projection of osteoporosis-related fractures and costs in China: 2010–2050. Osteoporos. Int. 2015;26:1929–1937. doi: 10.1007/s00198-015-3093-2. [DOI] [PubMed] [Google Scholar]
- 9.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr. Rev. 2002;23:303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 10.Zheng HF, Spector TD, Richards JB. Insights into the genetics of osteoporosis from recent genome-wide association studies. Expert Rev. Mol. Med. 2011;13:e28. doi: 10.1017/S1462399411001980. [DOI] [PubMed] [Google Scholar]
- 11.Trajanoska K, Rivadeneira F. The genetic architecture of osteoporosis and fracture risk. Bone. 2019;126:2–10. doi: 10.1016/j.bone.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat. Rev. Genet. 2012;13:576–588. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 13.Styrkarsdottir U, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497:517–520. doi: 10.1038/nature12124. [DOI] [PubMed] [Google Scholar]
- 14.Zheng HF, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson SC, Michaelsson K, Burgess S. Mendelian randomization in the bone field. Bone. 2019;126:51–58. doi: 10.1016/j.bone.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards JB, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivadeneira F, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kung AW, et al. Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am. J. Hum. Genet. 2010;86:229–239. doi: 10.1016/j.ajhg.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanis JA, Melton LJ, 3rd, Christiansen C, Johnston CC, Khaltaev N. The diagnosis of osteoporosis. J. Bone Miner. Res. 1994;9:1137–1141. doi: 10.1002/jbmr.5650090802. [DOI] [PubMed] [Google Scholar]
- 21.Bachrach LK. Acquisition of optimal bone mass in childhood and adolescence. Trends Endocrinol. Metab. 2001;12:22–28. doi: 10.1016/s1043-2760(00)00336-2. [DOI] [PubMed] [Google Scholar]
- 22.Paternoster L, et al. Genome-wide association meta-analysis of cortical bone mineral density unravels allelic heterogeneity at the RANKL locus and potential pleiotropic effects on bone. PLoS Genet. 2010;6:e1001217. doi: 10.1371/journal.pgen.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams JE. Quantitative computed tomography. Eur. J. Radiol. 2009;71:415–424. doi: 10.1016/j.ejrad.2009.04.074. [DOI] [PubMed] [Google Scholar]
- 24.Gonnelli S, et al. Quantitative ultrasound and dual-energy X-ray absorptiometry in the prediction of fragility fracture in men. Osteoporos. Int. 2005;16:963–968. doi: 10.1007/s00198-004-1771-6. [DOI] [PubMed] [Google Scholar]
- 25.Moayyeri A, et al. Genetic determinants of heel bone properties: genome-wide association meta-analysis and replication in the GEFOS/GENOMOS consortium. Hum. Mol. Genet. 2014;23:3054–3068. doi: 10.1093/hmg/ddt675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiel DP, et al. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Styrkarsdottir U, et al. Multiple genetic loci for bone mineral density and fractures. N. Engl. J. Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 28.Yang TL, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am. J. Hum. Genet. 2008;83:663–674. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Y, et al. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806. doi: 10.1371/journal.pgen.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kou I, et al. Common variants in a novel gene, FONG on chromosome 2q33.1 confer risk of osteoporosis in Japanese. PloS ONE. 2011;6:e19641. doi: 10.1371/journal.pone.0019641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naito T, et al. Clinical and genetic risk factors for decreased bone mineral density in Japanese patients with inflammatory bowel disease. J. Gastroenterol. Hepatol. 2018;33:1873–1881. doi: 10.1111/jgh.14149. [DOI] [PubMed] [Google Scholar]
- 32.Hwang JY, et al. Meta-analysis identifies a MECOM gene as a novel predisposing factor of osteoporotic fracture. J. Med. Genet. 2013;50:212–219. doi: 10.1136/jmedgenet-2012-101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang JY, Kim YJ, Choi BY, Kim BJ, Han BG. Meta analysis identifies a novel susceptibility locus associated with heel bone strength in the Korean population. Bone. 2016;84:47–51. doi: 10.1016/j.bone.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Liu YJ, Zhang L, Papasian CJ, Deng HW. Genome-wide Association Studies for Osteoporosis: A 2013 Update. J. Bone Metab. 2014;21:99–116. doi: 10.11005/jbm.2014.21.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum. Mol. Genet. 2014;23:1923–1933. doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trajanoska K, et al. Assessment of the genetic and clinical determinants of fracture risk: genome wide association and mendelian randomisation study. BMJ. 2018;362:k3225. doi: 10.1136/bmj.k3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai WY, et al. Genotype imputation and reference panel: a systematic evaluation on haplotype size and diversity. Brief Bioinform. 2019;21:1806–1817. doi: 10.1093/bib/bbz108. [DOI] [PubMed] [Google Scholar]
- 38.Kemp JP, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 2017;49:1468–1475. doi: 10.1038/ng.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris JA, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019;51:258–266. doi: 10.1038/s41588-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai WY, et al. Identification of PIEZO1 polymorphisms for human bone mineral density. Bone. 2020;133:115247. doi: 10.1016/j.bone.2020.115247. [DOI] [PubMed] [Google Scholar]
- 41.Bauer DC, et al. Broadband ultrasound attenuation predicts fractures strongly and independently of densitometry in older women. A prospective study. Study of Osteoporotic Fractures Research Group. Arch. Intern. Med. 1997;157:629–634. [PubMed] [Google Scholar]
- 42.Bauer DC, et al. Quantitative ultrasound predicts hip and non-spine fracture in men: the MrOS study. Osteoporos. Int. 2007;18:771–777. doi: 10.1007/s00198-006-0317-5. [DOI] [PubMed] [Google Scholar]
- 43.Chesi A, et al. A trans-ethnic genome-wide association study identifies gender-specific loci influencing pediatric aBMD and BMC at the distal radius. Hum. Mol. Genet. 2015;24:5053–5059. doi: 10.1093/hmg/ddv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timpson NJ, et al. Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Hum. Mol. Genet. 2009;18:1510–1517. doi: 10.1093/hmg/ddp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medina-Gomez C, et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012;8:e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koller DL, et al. Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. J. Clin. Endocrinol. Metab. 2010;95:1802–1809. doi: 10.1210/jc.2009-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koller DL, et al. Meta-analysis of genome-wide studies identifies WNT16 and ESR1 SNPs associated with bone mineral density in premenopausal women. J. Bone Miner. Res. 2013;28:547–558. doi: 10.1002/jbmr.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng HF, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. 2012;8:e1002745. doi: 10.1371/journal.pgen.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu CT, et al. Assessment of gene-by-sex interaction effect on bone mineral density. J. Bone Miner. Res. 2012;27:2051–2064. doi: 10.1002/jbmr.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chesi A, et al. A genomewide association study identifies two sex-specific loci, at SPTB and IZUMO3, influencing pediatric bone mineral density at multiple skeletal sites. J. Bone Miner. Res. 2017;32:1274–1281. doi: 10.1002/jbmr.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu YZ, et al. Identification of PLCL1 gene for hip bone size variation in females in a genome-wide association study. PloS ONE. 2008;3:e3160. doi: 10.1371/journal.pone.0003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lei SF, et al. Genome-wide association study identifies HMGN3 locus for spine bone size variation in Chinese. Hum. Genet. 2012;131:463–469. doi: 10.1007/s00439-011-1093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo YF, et al. Suggestion of GLYAT gene underlying variation of bone size and body lean mass as revealed by a bivariate genome-wide association study. Hum. Genet. 2013;132:189–199. doi: 10.1007/s00439-012-1236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baird DA, et al. Identification of novel loci associated with hip shape: a meta-analysis of genomewide association studies. J. Bone Miner. Res. 2019;34:241–251. doi: 10.1002/jbmr.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H, et al. Pleiotropic loci underlying bone mineral density and bone size identified by a bivariate genome-wide association analysis. Osteoporos. Int. 2020;31:1691–1701. doi: 10.1007/s00198-020-05389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Styrkarsdottir U, et al. GWAS of bone size yields twelve loci that also affect height, BMD, osteoarthritis or fractures. Nat. Commun. 2019;10:2054. doi: 10.1038/s41467-019-09860-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu XH, et al. Molecular genetic studies of gene identification for osteoporosis: the 2009 update. Endocr. Rev. 2010;31:447–505. doi: 10.1210/er.2009-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 60.Recker RR, Deng HW. Role of genetics in osteoporosis. Endocrine. 2002;17:55–66. doi: 10.1385/ENDO:17:1:55. [DOI] [PubMed] [Google Scholar]
- 61.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farber CR. Systems genetics: a novel approach to dissect the genetic basis of osteoporosis. Curr. Osteoporos. Rep. 2012;10:228–235. doi: 10.1007/s11914-012-0112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Q. Genetic study of complex diseases in the post-GWAS era. J. Genet. Genomics. 2015;42:87–98. doi: 10.1016/j.jgg.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 64.Shi H, Kichaev G, Pasaniuc B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am. J. Hum. Genet. 2016;99:139–153. doi: 10.1016/j.ajhg.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng J, et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 2018;50:746–753. doi: 10.1038/s41588-018-0101-4. [DOI] [PubMed] [Google Scholar]
- 67.O’Connor LJ, et al. Extreme polygenicity of complex traits is explained by negative selection. Am. J. Hum. Genet. 2019;105:456–476. doi: 10.1016/j.ajhg.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, Li YI, Pritchard JK. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177:1022–1034.e6. doi: 10.1016/j.cell.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tamai K, et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 71.Gong Y, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 72.Balemans W, Van Hul W. The genetics of low-density lipoprotein receptor-related protein 5 in bone: a story of extremes. Endocrinology. 2007;148:2622–2629. doi: 10.1210/en.2006-1352. [DOI] [PubMed] [Google Scholar]
- 73.Liu W, Zhang X. Receptor activator of nuclear factor-kappaB ligand (RANKL)/RANK/osteoprotegerin system in bone and other tissues (review) Mol. Med. Rep. 2015;11:3212–3218. doi: 10.3892/mmr.2015.3152. [DOI] [PubMed] [Google Scholar]
- 74.Kichaev G, et al. Leveraging polygenic functional enrichment to improve GWAS Power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Visscher PM, et al. 10 years of gwas discovery: biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao PP, et al. Relationship between alcohol use, blood pressure and hypertension: an association study and a Mendelian randomisation study. J. Epidemiol. Community Health. 2019;73:796–801. doi: 10.1136/jech-2018-211185. [DOI] [PubMed] [Google Scholar]
- 77.Sleiman PM, Grant SF. Mendelian randomization in the era of genomewide association studies. Clin. Chem. 2010;56:723–728. doi: 10.1373/clinchem.2009.141564. [DOI] [PubMed] [Google Scholar]
- 78.Xiong A, et al. No causal effect of serum urate on bone-related outcomes among a population of postmenopausal women and elderly men of Chinese Han ethnicity–a Mendelian randomization study. Osteoporos. Int. 2016;27:1031–1039. doi: 10.1007/s00198-015-3341-5. [DOI] [PubMed] [Google Scholar]
- 79.Trajanoska K, Rivadeneira F. Using mendelian randomization to decipher mechanisms of bone disease. Curr. Osteoporos. Rep. 2018;16:531–540. doi: 10.1007/s11914-018-0467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 81.Davey Smith G, Ebrahim S. What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ. 2005;330:1076–1079. doi: 10.1136/bmj.330.7499.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, Kraft P. Methodological challenges in mendelian randomization. Epidemiology. 2014;25:427–435. doi: 10.1097/EDE.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holick MF. Vitamin D deficiency. N. Engl. J. Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 84.Leong A, et al. The causal effect of vitamin D binding protein (DBP) levels on calcemic and cardiometabolic diseases: a Mendelian randomization study. PLoS Med. 2014;11:e1001751. doi: 10.1371/journal.pmed.1001751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li SS, et al. Genetically low vitamin d levels, bone mineral density, and bone metabolism markers: a mendelian randomisation study. Sci. Rep. 2016;6:33202. doi: 10.1038/srep33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Larsson SC, Melhus H, Michaelsson K. Circulating serum 25-hydroxyvitamin D levels and bone mineral density: mendelian randomization study. J. Bone Miner. Res. 2018;33:840–844. doi: 10.1002/jbmr.3389. [DOI] [PubMed] [Google Scholar]
- 87.Yang Q, et al. Genetically predicted milk consumption and bone health, ischemic heart disease and type 2 diabetes: a Mendelian randomization study. Eur. J. Clin. Nutr. 2017;71:1008–1012. doi: 10.1038/ejcn.2017.8. [DOI] [PubMed] [Google Scholar]
- 88.Xia J, et al. Systemic evaluation of the relationship between psoriasis, psoriatic arthritis and osteoporosis: observational and Mendelian randomisation study. Ann. Rheum. Dis. 2020;79:1460–1467. doi: 10.1136/annrheumdis-2020-217892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ahmad OS, et al. A mendelian randomization study of the effect of type-2 diabetes and glycemic traits on bone mineral density. J. Bone Miner. Res. 2017;32:1072–1081. doi: 10.1002/jbmr.3063. [DOI] [PubMed] [Google Scholar]
- 90.Wang MC, et al. The relative contributions of lean tissue mass and fat mass to bone density in young women. Bone. 2005;37:474–481. doi: 10.1016/j.bone.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 91.Clark EM, Ness AR, Tobias JH. Adipose tissue stimulates bone growth in prepubertal children. J. Clin. Endocrinol. Metab. 2006;91:2534–2541. doi: 10.1210/jc.2006-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Janicka A, et al. Fat mass is not beneficial to bone in adolescents and young adults. J. Clin. Endocrinol. Metab. 2007;92:143–147. doi: 10.1210/jc.2006-0794. [DOI] [PubMed] [Google Scholar]
- 93.Zhao LJ, et al. Correlation of obesity and osteoporosis: effect of fat mass on the determination of osteoporosis. J. Bone Miner. Res. 2008;23:17–29. doi: 10.1359/JBMR.070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Timpson NJ, Sayers A, Davey-Smith G, Tobias JH. How does body fat influence bone mass in childhood? A Mendelian randomization approach. J. Bone Miner. Res. 2009;24:522–533. doi: 10.1359/jbmr.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kemp JP, Sayers A, Smith GD, Tobias JH, Evans DM. Using Mendelian randomization to investigate a possible causal relationship between adiposity and increased bone mineral density at different skeletal sites in children. Int. J. Epidemiol. 2016;45:1560–1572. doi: 10.1093/ije/dyw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Warodomwichit D, et al. Causal inference of the effect of adiposity on bone mineral density in adults. Clin. Endocrinol. 2013;78:694–699. doi: 10.1111/cen.12061. [DOI] [PubMed] [Google Scholar]
- 97.Cousminer DL, et al. Genetically determined later puberty impacts lowered bone mineral density in childhood and adulthood. J. Bone Miner. Res. 2018;33:430–436. doi: 10.1002/jbmr.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dalbeth N, et al. Mendelian randomization analysis to examine for a causal effect of urate on bone mineral density. J. Bone Miner. Res. 2015;30:985–991. doi: 10.1002/jbmr.2434. [DOI] [PubMed] [Google Scholar]
- 99.van Vliet NA, et al. Thyroid stimulating hormone and bone mineral density: evidence from a two-sample mendelian randomization study and a candidate gene association study. J. Bone Miner. Res. 2018;33:1318–1325. doi: 10.1002/jbmr.3426. [DOI] [PubMed] [Google Scholar]
- 100.Guo R, Wu L, Fu Q. Is there causal relationship of smoking and alcohol consumption with bone mineral density? a mendelian randomization study. Calcif. tissue Int. 2018;103:546–553. doi: 10.1007/s00223-018-0452-y. [DOI] [PubMed] [Google Scholar]
- 101.Cerani A, et al. Genetic predisposition to increased serum calcium, bone mineral density, and fracture risk in individuals with normal calcium levels: mendelian randomisation study. BMJ. 2019;366:l4410. doi: 10.1136/bmj.l4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 103.Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nat. Neurosci. 2016;19:1397–1407. doi: 10.1038/nn.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wray NR, Wijmenga C, Sullivan PF, Yang J, Visscher PM. Common disease is more complex than implied by the core gene omnigenic model. Cell. 2018;173:1573–1580. doi: 10.1016/j.cell.2018.05.051. [DOI] [PubMed] [Google Scholar]
- 105.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat. Biotechnol. 2012;30:317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 106.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lewiecki EM. Sclerostin: a novel target for intervention in the treatment of osteoporosis. Discov. Med. 2011;12:263–273. [PubMed] [Google Scholar]
- 108.Winkler DG, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J. Biol. Chem. 2005;280:26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 110.Recker RR, et al. A randomized, double-blind phase 2 clinical trial of blosozumab, a sclerostin antibody, in postmenopausal women with low bone mineral density. J. Bone Miner. Res. 2015;30:216–224. doi: 10.1002/jbmr.2351. [DOI] [PubMed] [Google Scholar]
- 111.Geusens P. New insights into treatment of osteoporosis in postmenopausal women. RMD Open. 2015;1:e000051. doi: 10.1136/rmdopen-2015-000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Falk SS, Mittlmeier T, Gradl G. Results of geriatric distal radius fractures treated by intramedullary fixation. Injury. 2016;47(Suppl 7):S31–S35. doi: 10.1016/S0020-1383(16)30851-8. [DOI] [PubMed] [Google Scholar]
- 113.Saag KG, et al. Romosozumab or alendronate for fracture prevention in women with osteoporosis. N. Engl. J. Med. 2017;377:1417–1427. doi: 10.1056/NEJMoa1708322. [DOI] [PubMed] [Google Scholar]
- 114.Langdahl BL, et al. Romosozumab (sclerostin monoclonal antibody) versus teriparatide in postmenopausal women with osteoporosis transitioning from oral bisphosphonate therapy: a randomised, open-label, phase 3 trial. Lancet. 2017;390:1585–1594. doi: 10.1016/S0140-6736(17)31613-6. [DOI] [PubMed] [Google Scholar]
- 115.Cosman F, et al. Romosozumab treatment in postmenopausal women with osteoporosis. N. Engl. J. Med. 2016;375:1532–1543. doi: 10.1056/NEJMoa1607948. [DOI] [PubMed] [Google Scholar]
- 116.Amgen. FDA Approves EVENITYTM (romosozumab) for the treatment of osteoporosis in postmenopausal women at high risk for fracture. available at https://www.fda.gov/news-events/press-announcements/fda-approves-new-treatment-osteoporosis-postmenopausal-women-high-risk-fracture (2019).
- 117.Francisco, E. M. Approval of the marketing authorisation for Evenity (romosozumab). available at https://www.ema.europa.eu/en/documents/medicine-qa/questions-answers-approval-marketing-authorisation-evenity-romosozumab_en.pdf (2019).
- 118.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 119.Mao B, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 120.Canalis E. Update in new anabolic therapies for osteoporosis. J. Clin. Endocrinol. Metab. 2010;95:1496–1504. doi: 10.1210/jc.2009-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Korvala J, et al. Rare variations in WNT3A and DKK1 may predispose carriers to primary osteoporosis. Eur. J. Med. Genet. 2012;55:515–519. doi: 10.1016/j.ejmg.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 122.Glantschnig H, et al. Generation and selection of novel fully human monoclonal antibodies that neutralize Dickkopf-1 (DKK1) inhibitory function in vitro and increase bone mass in vivo. J. Biol. Chem. 2010;285:40135–40147. doi: 10.1074/jbc.M110.166892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Glantschnig H, et al. A rate-limiting role for Dickkopf-1 in bone formation and the remediation of bone loss in mouse and primate models of postmenopausal osteoporosis by an experimental therapeutic antibody. J. Pharmacol. Exp. Ther. 2011;338:568–578. doi: 10.1124/jpet.111.181404. [DOI] [PubMed] [Google Scholar]
- 124.Bodine PV, et al. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol. Endocrinol. 2004;18:1222–1237. doi: 10.1210/me.2003-0498. [DOI] [PubMed] [Google Scholar]
- 125.Betts AM, et al. The application of target information and preclinical pharmacokinetic/pharmacodynamic modeling in predicting clinical doses of a Dickkopf-1 antibody for osteoporosis. J. Pharmacol. Exp. Ther. 2010;333:2–13. doi: 10.1124/jpet.109.164129. [DOI] [PubMed] [Google Scholar]
- 126.Pepe MS, Gu JW, Morris DE. The potential of genes and other markers to inform about risk. Cancer Epidemiol., Biomark. Prev. 2010;19:655–665. doi: 10.1158/1055-9965.EPI-09-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kanis JA, Johnell O, Oden A, Johansson H, McCloskey E. FRAX and the assessment of fracture probability in men and women from the UK. Osteoporos. Int. 2008;19:385–397. doi: 10.1007/s00198-007-0543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tran BN, et al. Genetic profiling and individualized prognosis of fracture. J. Bone Miner. Res. 2011;26:414–419. doi: 10.1002/jbmr.219. [DOI] [PubMed] [Google Scholar]
- 129.Ho-Le TP, Center JR, Eisman JA, Nguyen HT, Nguyen TV. Prediction of bone mineral density and fragility fracture by genetic profiling. J. Bone Miner. Res. 2017;32:285–293. doi: 10.1002/jbmr.2998. [DOI] [PubMed] [Google Scholar]
- 130.Pare G, Mao S, Deng WQ. A machine-learning heuristic to improve gene score prediction of polygenic traits. Sci. Rep. 2017;7:12665. doi: 10.1038/s41598-017-13056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ho DSW, Schierding W, Wake M, Saffery R, O’Sullivan J. Machine learning SNP based prediction for precision medicine. Front. Genet. 2019;10:267. doi: 10.3389/fgene.2019.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bellot P, de Los Campos G, Perez-Enciso M. Can deep learning improve genomic prediction of complex human traits? Genetics. 2018;210:809–819. doi: 10.1534/genetics.118.301298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kruppa J, Ziegler A, Konig IR. Risk estimation and risk prediction using machine-learning methods. Hum. Genet. 2012;131:1639–1654. doi: 10.1007/s00439-012-1194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang J, et al. Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci. Rep. 2017;7:44048. doi: 10.1038/srep44048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Isakov O, Dotan I, Ben-Shachar S. Machine learning-based gene prioritization identifies novel candidate risk genes for inflammatory bowel disease. Inflamm. bowel Dis. 2017;23:1516–1523. doi: 10.1097/MIB.0000000000001222. [DOI] [PubMed] [Google Scholar]
- 136.Guo H, Zhang F, Chen J, Xu Y, Xiang J. Machine learning classification combining multiple features of a hyper-network of fMRI data in Alzheimer’s disease. Front. Neurosci. 2017;11:615. doi: 10.3389/fnins.2017.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lynch CM, et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int. J. Med. Inform. 2017;108:1–8. doi: 10.1016/j.ijmedinf.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jovic S, Miljkovic M, Ivanovic M, Saranovic M, Arsic M. Prostate cancer probability prediction by machine learning technique. Cancer Investig. 2017;35:647–651. doi: 10.1080/07357907.2017.1406496. [DOI] [PubMed] [Google Scholar]
- 139.Awan SE, Sohel F, Sanfilippo FM, Bennamoun M, Dwivedi G. Machine learning in heart failure: ready for prime time. Curr. Opin. Cardiol. 2018;33:190–195. doi: 10.1097/HCO.0000000000000491. [DOI] [PubMed] [Google Scholar]
- 140.Lello L, et al. Accurate genomic prediction of human height. Genetics. 2018;210:477–497. doi: 10.1534/genetics.118.301267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Forgetta, V. et al. Machine learning to predict osteoporotic fracture risk from genotypes. bioRxiv, 413716 (2018).
- 142.Martin AR, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Genetics for all. Nat. Genet. 51, 579-579 (2019). [DOI] [PubMed]
- 144.Styrkarsdottir U, et al. New sequence variants associated with bone mineral density. Nat. Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 145.Xiong DH, et al. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am. J. Hum. Genet. 2009;84:388–398. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu YZ, et al. Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PloS ONE. 2009;4:e6827. doi: 10.1371/journal.pone.0006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guo Y, et al. IL21R and PTH may underlie variation of femoral neck bone mineral density as revealed by a genome-wide association study. J. Bone Miner. Res. 2010;25:1042–1048. doi: 10.1359/jbmr.091040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hsu YH, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010;6:e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tan L, et al. A genome-wide association analysis implicates SOX6 as a candidate gene for wrist bone mass. Sci. China Life Sci. 2010;53:1065–1072. doi: 10.1007/s11427-010-4056-7. [DOI] [PubMed] [Google Scholar]
- 150.Duncan EL, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Deng FY, et al. Genome-wide association study identified UQCC locus for spine bone size in humans. Bone. 2013;53:129–133. doi: 10.1016/j.bone.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zheng HF, et al. Meta-analysis of genome-wide studies identifies MEF2C SNPs associated with bone mineral density at forearm. J. Med. Genet. 2013;50:473–478. doi: 10.1136/jmedgenet-2012-101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oei L, et al. Genome-wide association study for radiographic vertebral fractures: a potential role for the 16q24 BMD locus. Bone. 2014;59:20–27. [PMC free article] [PubMed] [Google Scholar]
- 154.Kemp JP, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014;10:e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tan LJ, et al. Bivariate genome-wide association study implicates ATP6V1G1 as a novel pleiotropic locus underlying osteoporosis and age at menarche. J. Clin. Endocrinol. Metab. 2015;100:E1457–E1466. doi: 10.1210/jc.2015-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Styrkarsdottir U, et al. Two rare mutations in the COL1A2 gene associate with low bone mineral density and fractures in Iceland. J. Bone Miner. Res. 2016;31:173–179. doi: 10.1002/jbmr.2604. [DOI] [PubMed] [Google Scholar]
- 157.Styrkarsdottir U, et al. Sequence variants in the PTCH1 gene associate with spine bone mineral density and osteoporotic fractures. Nat. Commun. 2016;7:10129. doi: 10.1038/ncomms10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mullin BH, et al. Genome-wide association study using family-based cohorts identifies the WLS and CCDC170/ESR1 loci as associated with bone mineral density. BMC Genomics. 2016;17:136. doi: 10.1186/s12864-016-2481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Taylor KC, et al. A genome-wide association study meta-analysis of clinical fracture in 10,012 African American women. Bone Rep. 2016;5:233–242. doi: 10.1016/j.bonr.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Choi HJ, et al. Genome-wide association study in East Asians suggests UHMK1 as a novel bone mineral density susceptibility gene. Bone. 2016;91:113–121. doi: 10.1016/j.bone.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 161.Pei YF, et al. Genome-wide association meta-analyses identified 1q43 and 2q32.2 for hip Ward’s triangle areal bone mineral density. Bone. 2016;91:1–10. doi: 10.1016/j.bone.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pei YF, et al. Association of 3q13.32 variants with hip trochanter and intertrochanter bone mineral density identified by a genome-wide association study. Osteoporos. Int. 2016;27:3343–3354. doi: 10.1007/s00198-016-3663-y. [DOI] [PubMed] [Google Scholar]
- 163.Mullin BH, et al. Genome-wide association study meta-analysis for quantitative ultrasound parameters of bone identifies five novel loci for broadband ultrasound attenuation. Hum. Mol. Genet. 2017;26:2791–2802. doi: 10.1093/hmg/ddx174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Villalobos-Comparan M, et al. A pilot genome-wide association study in postmenopausal Mexican-Mestizo women implicates the RMND1/CCDC170 locus is associated with bone mineral density. Int. J. Genomics. 2017;2017:5831020. doi: 10.1155/2017/5831020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Peng C, et al. Enhanced identification of potential pleiotropic genetic variants for bone mineral density and breast cancer. Calcif. Tissue Int. 2017;101:489–500. doi: 10.1007/s00223-017-0308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lu S, et al. Bivariate genome-wide association analyses identified genetic pleiotropic effects for bone mineral density and alcohol drinking in Caucasians. J. Bone Miner. Metab. 2017;35:649–658. doi: 10.1007/s00774-016-0802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Alonso N, et al. Identification of a novel locus on chromosome 2q13, which predisposes to clinical vertebral fractures independently of bone density. Ann. Rheum. Dis. 2018;77:378–385. doi: 10.1136/annrheumdis-2017-212469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Inaba H, et al. Bone mineral density in children with acute lymphoblastic leukemia. Cancer. 2018;124:1025–1035. doi: 10.1002/cncr.31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pei YF, et al. Joint study of two genome-wide association meta-analyses identified 20p12.1 and 20q13.33 for bone mineral density. Bone. 2018;110:378–385. doi: 10.1016/j.bone.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lin X, et al. Identifying potentially common genes between dyslipidemia and osteoporosis using novel analytical approaches. Mol. Genet. genomics. 2018;293:711–723. doi: 10.1007/s00438-017-1414-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim SK. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PloS ONE. 2018;13:e0200785. doi: 10.1371/journal.pone.0200785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Qiu C, Shen H, Fu X, Xu C, Deng H. Meta-analysis of genome-wide association studies identifies novel functional CpG-SNPs associated with bone mineral density at lumbar spine. Int. J. Genomics. 2018;2018:6407257. doi: 10.1155/2018/6407257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gregson CL, et al. Genome-wide association study of extreme high bone mass: Contribution of common genetic variation to extreme BMD phenotypes and potential novel BMD-associated genes. Bone. 2018;114:62–71. doi: 10.1016/j.bone.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liang X, et al. Assessing the genetic correlations between early growth parameters and bone mineral density: a polygenic risk score analysis. Bone. 2018;116:301–306. doi: 10.1016/j.bone.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hsu YH, et al. Meta-analysis of genomewide association studies reveals genetic variants for hip bone geometry. J. Bone Miner. Res. 2019;34:1284–1296. doi: 10.1002/jbmr.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Oei L, et al. Dissecting the relationship between high-sensitivity serum C-reactive protein and increased fracture risk: the Rotterdam Study. Osteoporos. Int. 2014;25:1247–1254. doi: 10.1007/s00198-013-2578-0. [DOI] [PubMed] [Google Scholar]
- 177.Huang JV, Schooling CM. Inflammation and bone mineral density: a Mendelian randomization study. Sci. Rep. 2017;7:8666. doi: 10.1038/s41598-017-09080-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat. Rev. Genet. 2012;13:576–588. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 179.Cummings SR, et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N. Engl. J. Med. 2009;361:756–765. doi: 10.1056/NEJMoa0809493. [DOI] [PubMed] [Google Scholar]
- 180.Ettinger B, et al. Reduction of vertebral fracture risk in postmenopausal women with osteoporosis treated with raloxifene: results from a 3-year randomized clinical trial. Multiple Outcomes of Raloxifene Evaluation (MORE) Investigators. JAMA. 1999;282:637–645. doi: 10.1001/jama.282.7.637. [DOI] [PubMed] [Google Scholar]
- 181.Neer RM, et al. Effect of parathyroid hormone (1-34) on fractures and bone mineral density in postmenopausal women with osteoporosis. N. Engl. J. Med. 2001;344:1434–1441. doi: 10.1056/NEJM200105103441904. [DOI] [PubMed] [Google Scholar]
- 182.Greenspan SL, et al. Effect of recombinant human parathyroid hormone (1-84) on vertebral fracture and bone mineral density in postmenopausal women with osteoporosis: a randomized trial. Ann. Intern. Med. 2007;146:326–339. doi: 10.7326/0003-4819-146-5-200703060-00005. [DOI] [PubMed] [Google Scholar]
- 183.Liberman UA, et al. Effect of oral alendronate on bone mineral density and the incidence of fractures in postmenopausal osteoporosis. The Alendronate Phase III Osteoporosis Treatment Study Group. N. Engl. J. Med. 1995;333:1437–1443. doi: 10.1056/NEJM199511303332201. [DOI] [PubMed] [Google Scholar]
- 184.Rossouw JE, et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results From the Women’s Health Initiative randomized controlled trial. JAMA. 2002;288:321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 185.Gauthier JY, et al. The discovery of odanacatib (MK-0822), a selective inhibitor of cathepsin K. Bioorg. Medicinal Chem. Lett. 2008;18:923–928. doi: 10.1016/j.bmcl.2007.12.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Box 1 Introduction of Genome-Wide Association Studies
Box 2 Introduction of Mendelian Randomization (MR)