Abstract
O-GlcNAc is an essential and dynamic post-translational modification presented on thousands of nucleocytoplasmic proteins. Interrogating the role of O-GlcNAc on a single target protein is crucial yet challenging to perform in cells. Herein, we developed a nanobody-fused split O-GlcNAcase (OGA) as an O-GlcNAc eraser for selective deglycosylation of a target protein in cells. After systematic cellular optimization, we identified a split OGA with reduced inherent deglycosidase activity that selectively removed O-GlcNAc from the desired target protein when directed by a nanobody. We demonstrate the generality of the nanobody-fused split OGA using four nanobodies against five target proteins, and use the system to study functions for O-GlcNAc on the transcription factors c-Jun and c-Fos. The nanobody-directed O-GlcNAc eraser provides a new strategy for functional evaluation and engineering of O-GlcNAc via the selective removal of O-GlcNAc from individual proteins directly in cells.
Introduction
O-Linked N-acetyl glucosamine (O-GlcNAc) is a monosaccharide post-translational modification (PTM) installed on serine or threonine residues of numerous nucleocytoplasmic proteins across species1. O-GlcNAc is a reversible and dynamic modification regulated by a single pair of enzymes, the writer O-GlcNAc transferase2 (OGT) and the eraser O-GlcNAcase3 (OGA). Loss of O-GlcNAc homeostasis has been linked to many diseases, including neurodegeneration4, diabetes5, and cancer6. Understanding the functional contribution of O-GlcNAc will elucidate the essential roles this modification plays in maintaining nutrient homeostasis and cellular signaling7.
To uncover the functions of O-GlcNAc on a protein in cells, methods to either globally regulate O-GlcNAc or investigate specific glycosites have been developed8. Overexpression, genetic knockdown/knockout, or application of chemical inhibitors for OGT or OGA are common mechanisms to globally reduce or elevate O-GlcNAc, respectively9–11. However, these approaches produce wide-spread changes to O-GlcNAc levels that require additional studies to characterize the function of O-GlcNAc on a target protein. Furthermore, inhibitors of OGT and OGA have recently been shown to rapidly alter expression of OGT9 or OGA10. With the advent of glycoproteomic methods12–14, specific glycosites are more readily targeted by site-directed mutagenesis approaches to permanently add O-GlcNAc15 or block its installation. However, modification site mapping remains non-trivial and using site-directed mutagenesis to study O-GlcNAcylated proteins with multiple or unmapped glycosites is challenging. Additionally, O-GlcNAc has extensive cross-talk with other PTMs, including phosphorylation16 and ubiquitylation17, which may be disrupted by site-directed mutagenesis. An alternative method to selectively edit protein O-GlcNAcylation in cells will facilitate dissection of O-GlcNAc functions on the target protein.
Recently, we reported the selective installation of O-GlcNAc to a target protein in living cells using a nanobody fusion to OGT18. A nanobody is a small, single-domain protein binder that is capable of recognizing intracellular targets with high affinity and selectivity19. Building on the insights gained from engineering a nanobody-OGT and the recent crystal structures of human OGA20–22, we sought to develop an O-GlcNAc eraser to remove O-GlcNAc from a desired target protein in cells by leveraging the protein selectivity of the nanobody and the robust enzymatic activity of OGA after rational engineering (Fig. 1a). We developed a split OGA with minimal size and limited inherent activity by optimization and characterization of a series of OGA constructs, and demonstrated that fusion of a nanobody selectively restores deglycosidase activity on the target protein (Fig. 1b). The nanobody-fused split OGA is modular and generalizable for various nanobodies that recognize protein and peptide tags for the successful deglycosylation of a broad range of target proteins with excellent protein selectivity and minimal effects on global O-GlcNAc levels. We further demonstrated use of the O-GlcNAc eraser to distinguish global versus protein-specific functions for O-GlcNAc on transcription factors c-Jun and c-Fos activity in cells, two transcription factors that carry multiple or unmapped O-GlcNAc sites.
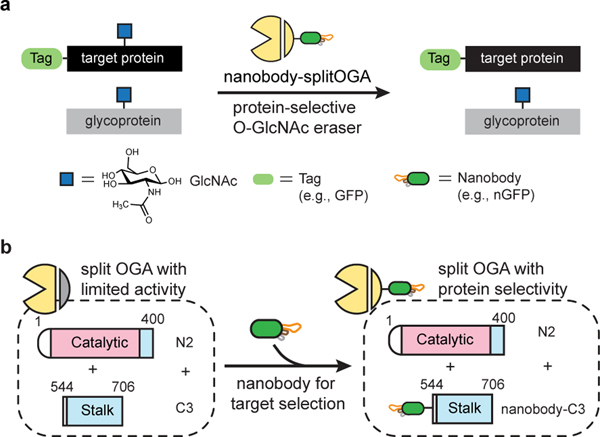
Fig. 1 |. Design and development of a nanobody-directed split OGA for O-GlcNAc removal in a protein-selective manner.
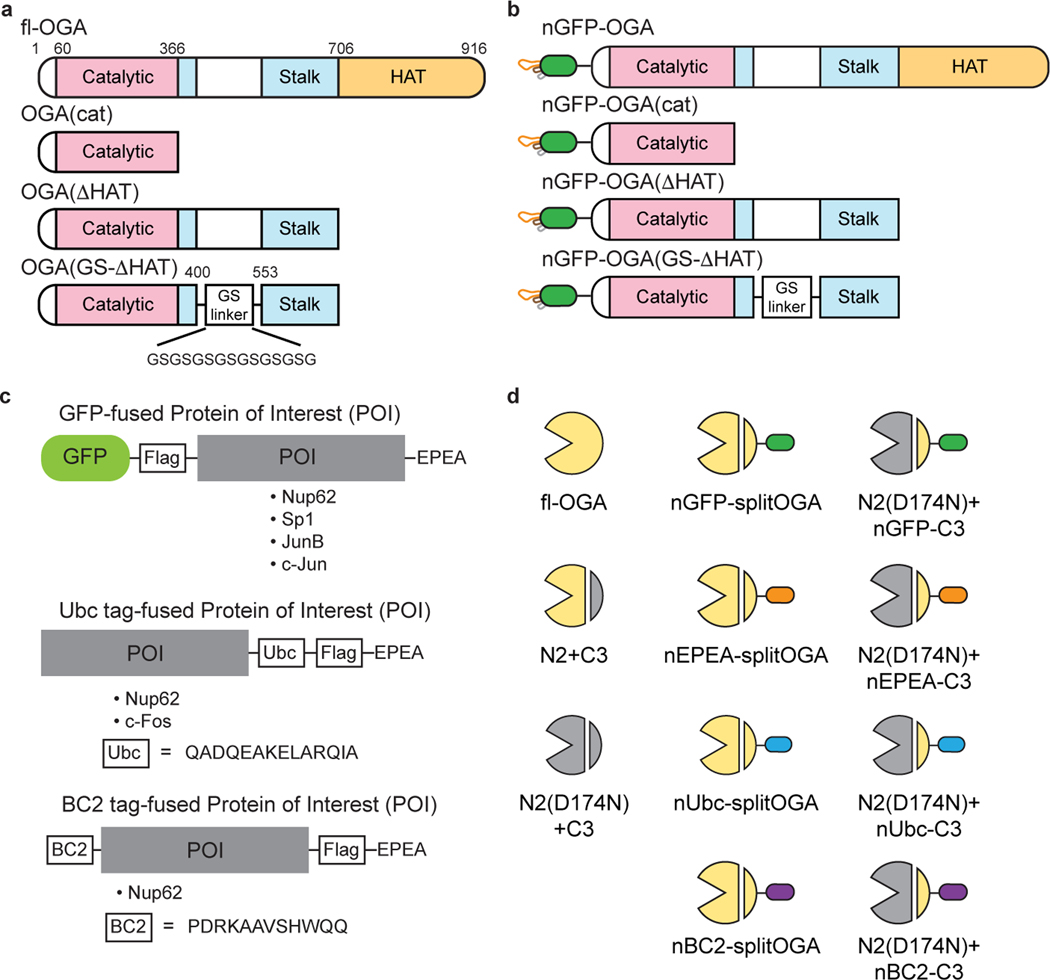
a, Overview of the approach for target protein deglycosylation using a nanobody-directed O-GlcNAc eraser. The nanobody is able to recognize a desired target and redirect the enzyme to remove the O-GlcNAc modification from the target protein. Tags used in this study are GFP, EPEA tag, BC2 tag, and Ubc tag. The corresponding nanobodies are nGFP, nEPEA, nBC2, and nUbc, respectively (see Extended Data Fig. 1c, d). b, Design of split OGA to achieve protein selectivity. OGA was engineered into a split and truncated form with limited inherent substrate activity. Introduction of a nanobody to the split OGA promoted localization to and deglycosylation of the desired target protein. The catalytic domain and stalk domain of split OGA are highlighted in pink and blue, respectively.
Results
Identifying the essential domains of active OGA.
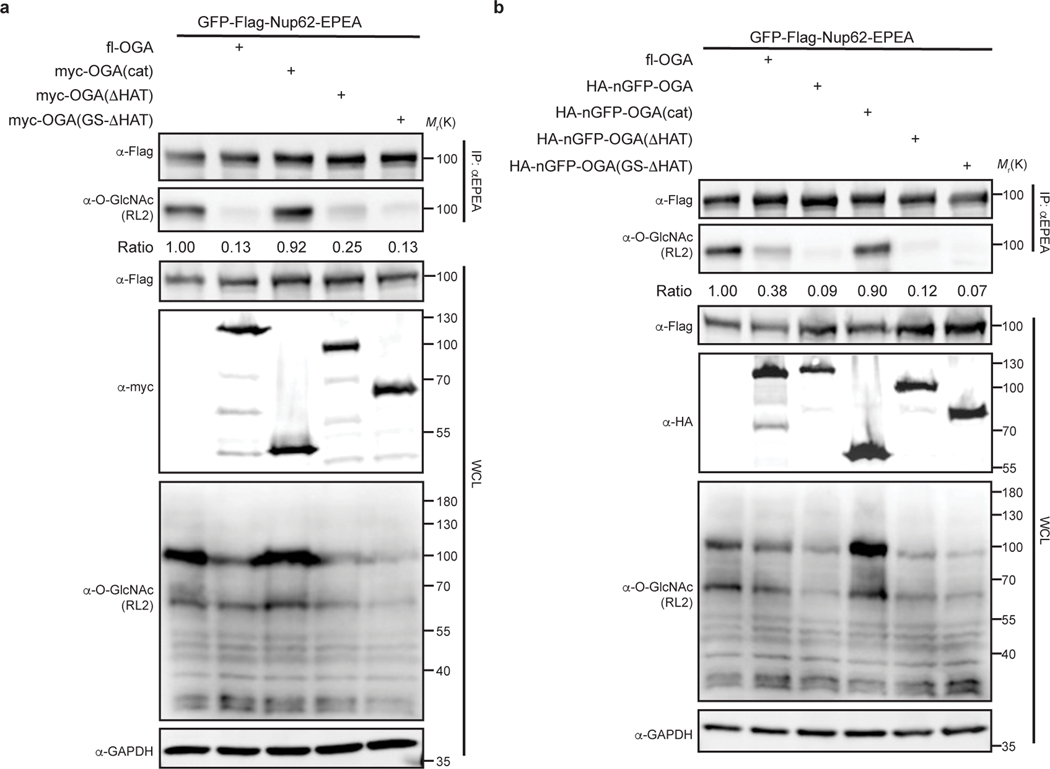
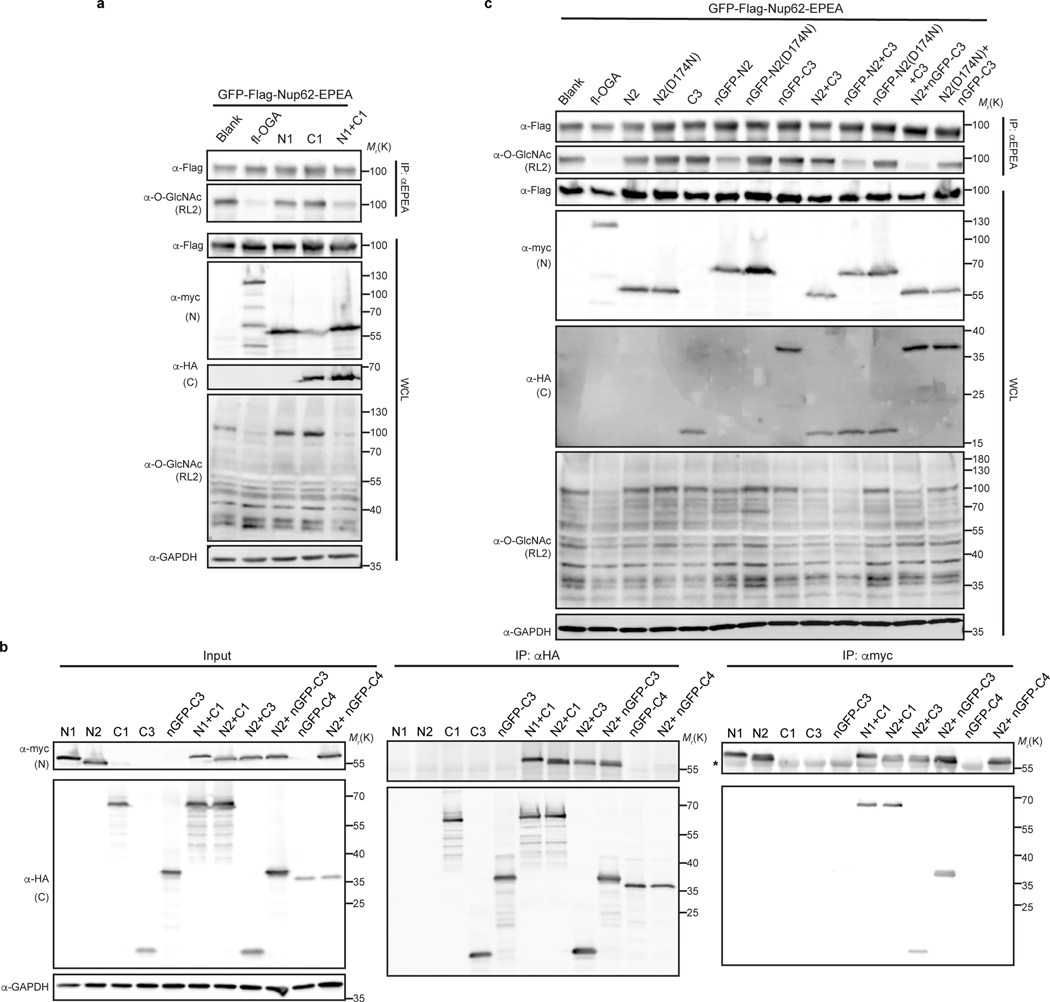
The long splice variant of human OGA is a 103-kDa hydrolase containing a catalytic domain, a stalk domain, and a pseudo-histone acetyltransferase (HAT) domain interspersed by several disordered regions23. Inspired by the recent structures of OGA20–22, we initiated efforts to identify a nominally functional OGA variant that targeted a substrate of interest in living cells using three constructs with or without the nanobody: (1) the catalytic domain alone, (2) a construct lacking the C-terminal HAT domain, and (3) a construct with a glycine-serine linker replacing a disordered region in OGA20 (Extended Data Fig. 1a,b). To evaluate the enzymatic activities of these constructs, we used the highly O-GlcNAcylated Nup62 tagged with GFP and a Flag-tag at the N-terminus for detection, and an EPEA-tag at the C-terminus for enrichment (GFP-Nup62, Extended Data Fig. 1c) as a target protein24. After co-expression with one of the three OGA constructs in HEK 293T cells, GFP-Nup62 was immunoprecipitated and the O-GlcNAc levels probed with the RL2 antibody. As expected20–22, we found that the catalytic domain and the stalk domain of OGA, but not the HAT domain, are required for deglycosylation of GFP-Nup62 within cells (Extended Data Fig. 2a). Deglycosylation of GFP-Nup62 was enhanced by recruitment of the active OGA constructs with an N-terminal nanobody against GFP (nGFP)25 (Extended Data Fig. 2b). However, further investigation revealed that these initial nanobody-OGA fusion proteins exhibited poor selectivity and affected the localization of the target protein (Extended Data Fig. 3). To improve the target protein selectivity, we pursued the further engineering of OGA to identify a construct capable of serving as a protein-selective O-GlcNAc eraser.
Optimizing a selective nanobody-directed split OGA.
With the aim of reducing the inherent activity of OGA and thereby improving selectivity for the target protein through the nanobody in living cells, we utilized a novel strategy to reduce the size and activity of OGA. Human OGA contains a caspase-3 cleavage site at Asp-413 that splits OGA during apoptosis into an N- and C-terminal fragment26, which can reconstitute enzymatic activity when co-expressed simultaneously in living cells (Extended Data Fig. 4a). Encouraged by this observation, we next sought to lower the association between the two fragments by first fine tuning the enzymatic activity of split OGA. We generated three N-fragments (N1–N3) and four C-fragments (C1–C4) by iteratively reducing the size of each fragment, starting from the original cleavage site Asp-413 (Fig. 2a). The deglycosidase activity of these fragments was screened against GFP-Nup62 beginning with the truncated N1–N3 fragments paired with the original C-fragment [C1, amino acid (aa) 414–916] (Fig. 2b and Supplementary Figure 1a). Following immunoprecipitation and probing for O-GlcNAc on GFP-Nup62, we found that N2 was the minimal N-fragment that possessed deglycosylation activity when paired with C1. No activity was seen with the shorter N3 fragment on GFP-Nup62 regardless of the C-fragment pairing (Supplementary Figure 2). We reasoned that part of the stalk domain (aa 367–400) forms critical contacts for the biochemical activity of OGA in cells, which is consistent with in vitro observations22.
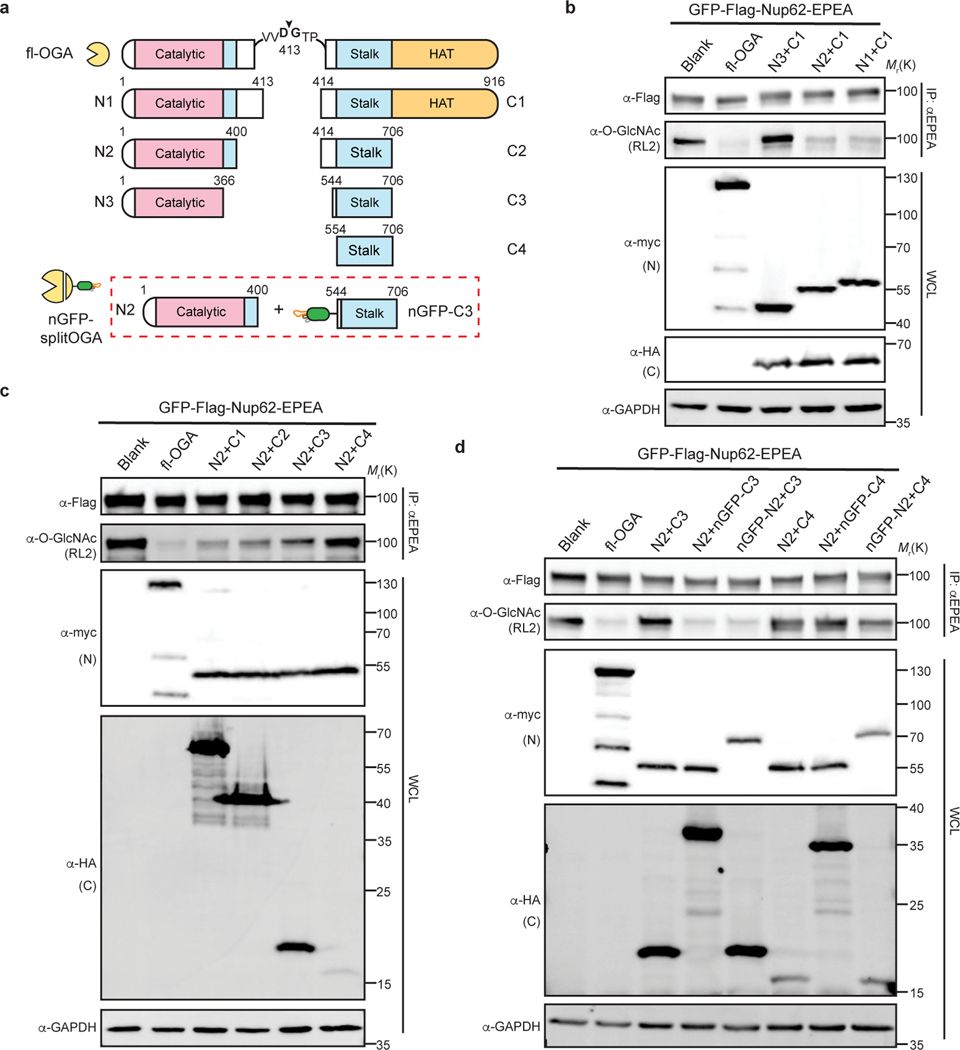
Fig. 2 |. Design and optimization of nanobody-fused split OGA for protein-selective deglycosylation on GFP-Nup62.
a, Schematic of the split site, N-fragments, and C-fragments tested in this study. Amino acid numbers appear on top. The optimal combination N2 with nGFP-fused C3 (shown in red rectangle) is termed nGFP-splitOGA. b, Optimization of the N-fragment with C1 on GFP-Nup62 to obtain a minimal N-fragment. c, Optimization of the C-fragment with N2 on GFP-Nup62 to obtain a minimal C-fragment with limited inherent substrate activity. d, nGFP fusion to split OGA regains activity on GFP-Nup62. The optimized split OGA (N2 + C3) shows limited substrate activity that is reinstated after fusion with nGFP on either fragment. GFP-Nup62 was co-expressed with the indicated constructs, enriched by anti-EPEA beads, and analyzed by immunoblotting to reveal the protein level and O-GlcNAc modification level respectively. WCL, whole cell lysate. The data in b-d are representative of two biological replicates.
Similarly, we screened the C-fragments in combination with N2 and found that shorter C-fragments corresponded to a decrease in split OGA activity on GFP-Nup62 (Fig. 2c and Supplementary Figure 1b). Among those combinations, the N2-C3 pair and the N2-C4 pair showed significantly reduced activity. Target protein deglycosylation was next evaluated by fusion of nGFP to the N-terminus of either the N-fragment (N2) or C-fragments (C3 or C4). Fusion of nGFP to either N2 or C3 selectively restored deglycosidase activity to GFP-Nup62. However, no activity on GFP-Nup62 was observed with the N2-C4 pair after fusion of nGFP (Fig. 2d and Supplementary Figure 1c). Likewise, only the N2 and nGFP-fused C3 pair were reciprocally co-immunoprecipitated (Extended Data Fig. 4b), which implies that amino acids 544–553 contribute to the association of the two fragments, likely via the formation of the OGA homodimer, and therefore reconstituting the active OGA in vivo22.
Finally, we confirmed the deglycosidase activity on the target protein derived from the nanobody-fused split OGA by mutation of D174, one of the two catalytic aspartate residues in OGA27, to asparagine (N) as an inactive negative control (Extended Data Fig. 4c). On comparison to the inactive control, we found that N2 with nGFP-fused C3 possessed the highest efficiency and selectivity at removal of O-GlcNAc on GFP-Nup62, and termed this combination nGFP-splitOGA in subsequent experiments (highlighted in the red rectangle, Fig. 2a).
Selective O-GlcNAc removal from target proteins.
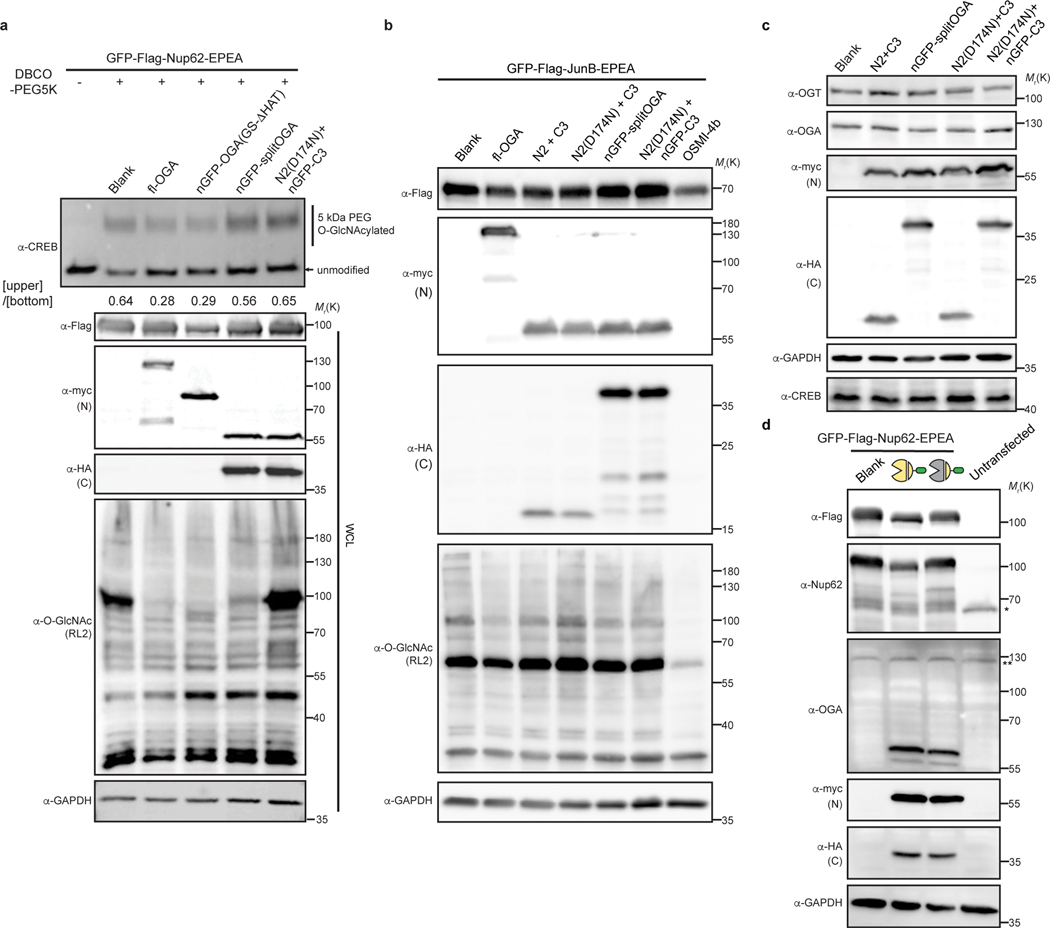
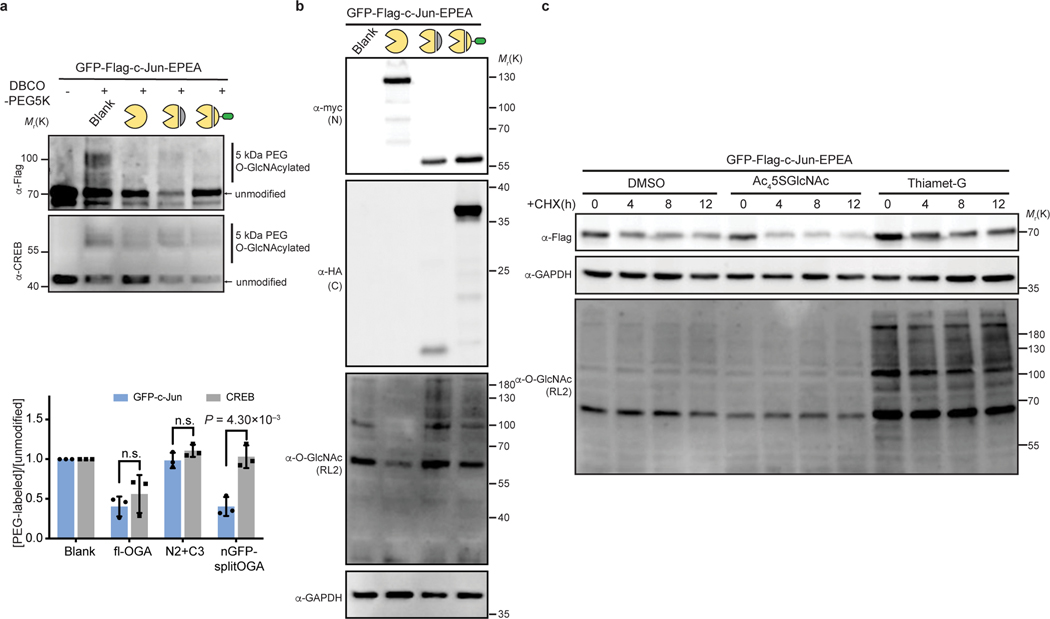
We next evaluated the generalizability of nGFP-splitOGA as a selective O-GlcNAc eraser against a range of target proteins. nGFP-splitOGA removed O-GlcNAc analogous to the full-length OGA (fl-OGA) from GFP-Nup62 in a nGFP-dependent manner, regardless of the GFP orientation on Nup62 (Fig. 3a, Supplementary Figure 3 & 4a). nGFP-splitOGA is similarly as effective as the fl-OGA at deglycosylation of the constitutively O-GlcNAc-modified protein, GFP-Sp124 following installation of the nanobody for protein targeting (Fig. 3b and Supplementary Figure 4b). A third target protein, GFP-JunB, was evaluated using a mass shift assay24 as this transcription factor possesses several glycosites at a relatively low occupancy28. Cell lysates co-transfected with nGFP-splitOGA and GFP-JunB were sequentially labeled with UDP-GalNAz and 5kDa DBCO-PEG to reveal the O-GlcNAcylation status by immunoblotting.
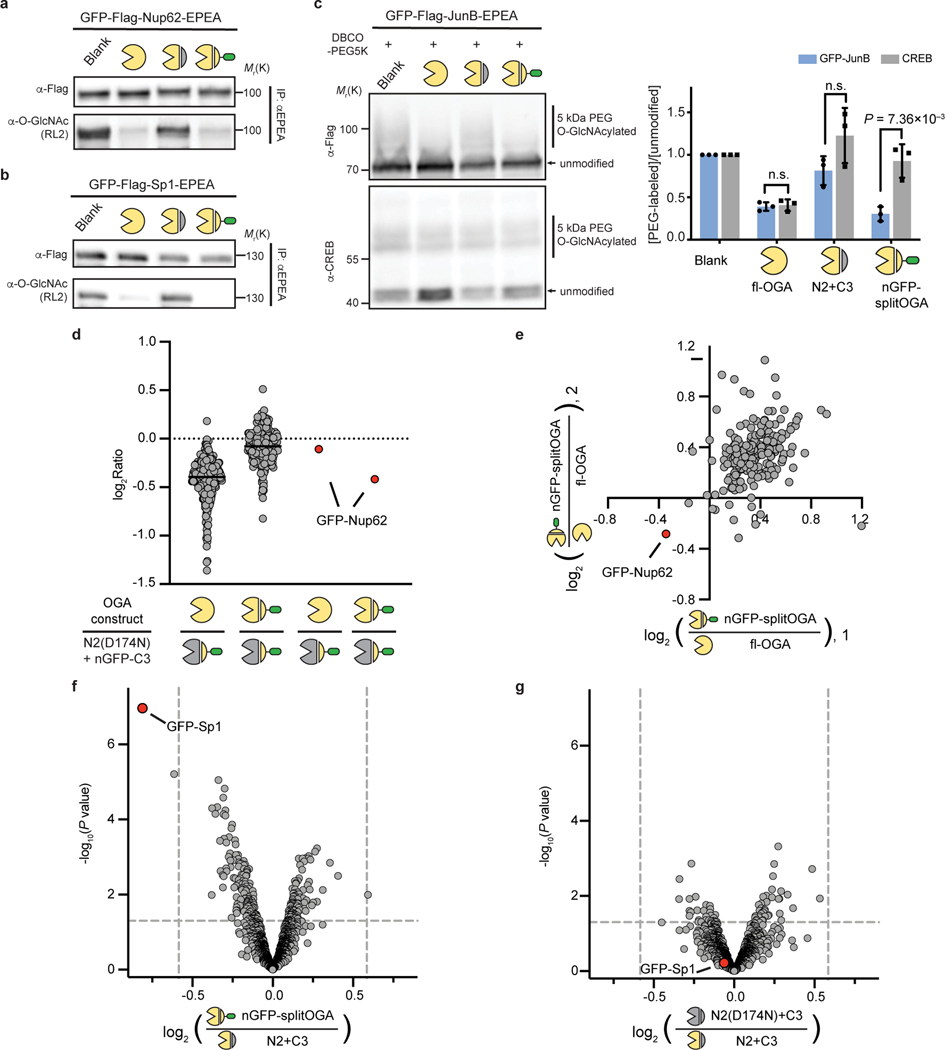
Fig. 3 |. The nanobody-fused split OGA is general for protein-selective deglycosylation on various O-GlcNAcylated proteins.
a–c, nGFP-splitOGA shows deglycosidase activity on GFP-tagged Nup62 (a), Sp1(b), and JunB (c). In a and b, the target protein was enriched and blotted for O-GlcNAc by RL2. In c, samples were subjected to a mass-shift assay with 5kDa DBCO-PEG. O-GlcNAcylation levels were measured by the ratio between the intensity of mass-shifted bands and unmodified bands. The data in a-c are representative of three biological replicates. Quantification is presented as mean ± s.d. of n = 3 independent experiments. d, e, Log2 ratios of changes of enriched O-GlcNAcylated protein abundance of fl-OGA and nGFP-splitOGA versus its inactive form treated cells respectively (d), and nGFP-splitOGA versus fl-OGA-treated cells (e) in the presence of GFP-Nup62. The black solid lines refer to the median of each group. f, g, Volcano plots illustrating the comparison of enriched O-GlcNAcylated proteins of nGFP-splitOGA (f) or of inactive (g) versus split OGA-treated cells co-expression of GFP-Sp1. P = 0.05 and ±1.5-fold change are denoted by gray dashed lines as significance threshold. Each point represents an individual identified protein of two (d, e) or four (f, g) independent biological replicates. GFP-Nup62 or GFP-Sp1 are indicated by the red dot. Symbols represent the corresponding OGA constructs as indicated. A two-tailed, unpaired Student’s t-test was used for statistical analysis in c, f and g. n.s., not significant.
Notably, nGFP-splitOGA selectively deglycosylated GFP-JunB while leaving endogenous CREB24 largely unperturbed, in contrast to the effects of co-expression of fl-OGA (Fig. 3c and Supplementary Figure 4c). The O-GlcNAc levels on CREB were minimally perturbed by nGFP-splitOGA regardless of the target protein (Extended Data Fig. 5a). Global O-GlcNAc levels also showed negligible changes on co-expression of nGFP-splitOGA and GFP-JunB, in contrast to the dramatic reduction caused by the OGT inhibitor OSMI-4b9 (Extended Data Fig. 5b). Furthermore, no perturbation of endogenous OGT or OGA protein levels was observed in the presence of nGFP-splitOGA constructs, though expression levels of nGFP-splitOGA is greater than native expression of OGA (Extended Data Fig. 5c,d). In addition, nGFP-splitOGA did not alter the subcellular localization of the target protein in HEK 293T cells (Extended Data Fig. 6). These data point to the high selectivity, orthogonality, and generality of nGFP-splitOGA in target protein deglycosylation.
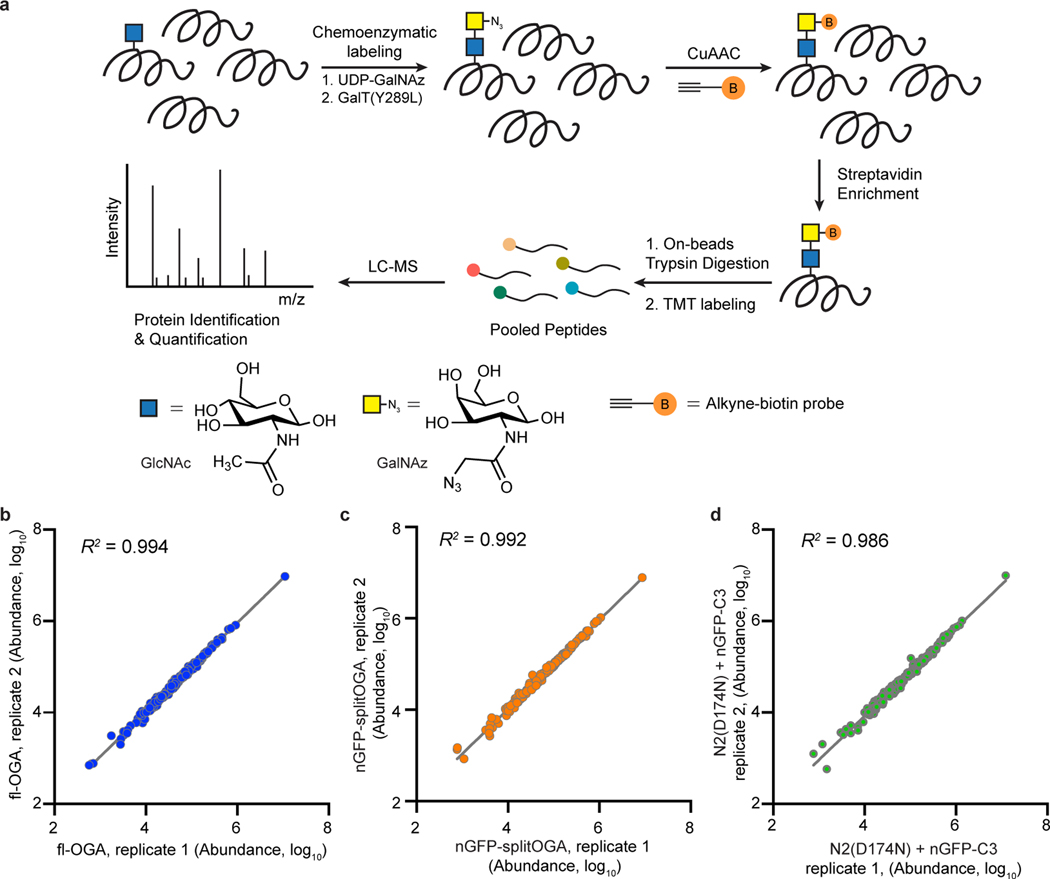
To more quantitatively evaluate the selectivity of nGFP-splitOGA, we performed an unbiased quantitative mass spectrometry (MS) analysis of the O-GlcNAcylated proteome on expression of nGFP-splitOGA (Extended Data Fig. 7a). O-GlcNAcylated proteins from HEK 293T cells co-expressing GFP-Nup62 and (1) fl-OGA, (2) nGFP-splitOGA, or (3) the inactive form of nGFP-splitOGA [N2(D174N) + nGFP-C3] were chemoenzymatically labeled, enriched, and digested for protein identification and quantification by MS using tandem mass tags (TMT). Two independent biological replicates were performed with excellent reproducibility (Extended Data Fig. 7b–d). Normalization to the inactive form of nGFP-splitOGA revealed that fl-OGA expression globally decreased O-GlcNAcylated proteins, while nGFP-splitOGA expression significantly reduced O-GlcNAcylated GFP-Nup62 with negligible perturbation of the O-GlcNAc proteome by comparison (Fig. 3d). The greater reduction of O-GlcNAc on GFP-Nup62 by nGFP-splitOGA than by fl-OGA may reflect the arguably more sensitive measurement by MS. Direct comparison of the O-GlcNAc proteome from nGFP-splitOGA to fl-OGA shows GFP-Nup62 as the only protein that is reproducibly deglycosylated by nGFP-splitOGA more than fl-OGA (Fig. 3e).
To directly compare the nGFP-splitOGA to split OGA alone, we subsequently quantified the global O-GlcNAc proteome from four independent biological replicates with HEK 293T cells co-expressing GFP-Sp1 and (1) nGFP-splitOGA, (2) split OGA (N2+C3), or (3) the inactive form of split OGA [N2(D174N) + C3]. GFP-Sp1 displays the largest O-GlcNAc reduction with the minimal P-value on co-expression of nGFP-splitOGA in comparison to split OGA (Fig. 3f). Notably, the global O-GlcNAc proteome in the presence of split OGA shows no significant difference from that of the inactive form (Fig. 3g). Taken together, both immunoblotting analysis and quantitative proteomics demonstrate that nGFP-splitOGA is a general mechanism to selectively remove O-GlcNAc from target proteins in cells, at efficiency levels comparable to fl-OGA, while minimally affecting the broader O-GlcNAc proteome.
Diverse nanobody-epitope pairs are applicable.
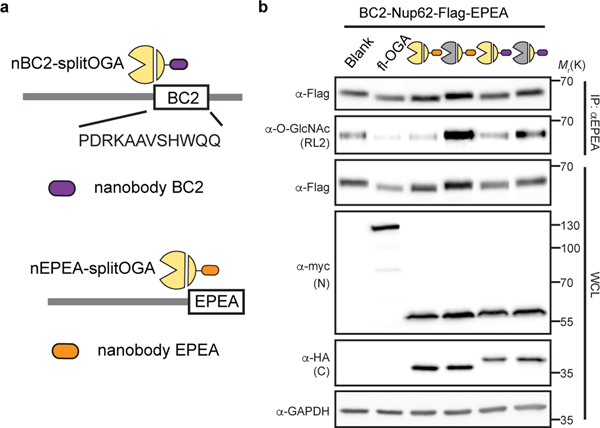
Although we were enthused that nGFP-splitOGA is a highly selective O-GlcNAc eraser of several GFP-fused target proteins, we were cognizant that GFP fusion proteins may not be ideal for the full range of potential biological applications of this system. Hence, we evaluated three additional nanobodies and their epitope tags, with a focus on short peptide epitopes, as additional options for utilizing the protein selective O-GlcNAc eraser. We first evaluated nanobodies against the EPEA-tag and the Ubc tag, which recognize a C-terminal EPEA-tag29, or 14-residue peptide epitope derived from UBC6e30, respectively (Fig. 4a). Application of these nanobodies to the target protein Nup62-Ubc-EPEA revealed that both nanobodies are efficient at reducing O-GlcNAc levels from the target protein (Fig. 4b). The nUbc-splitOGA and the Ubc-tagged substrate were further observed to co-localize by confocal imaging (Supplementary Figure 5). Target protein deglycosylation was also effective with a third nanobody that recognizes the BC2-tag, a 12-residue peptide epitope31 (Extended Data Fig. 8).
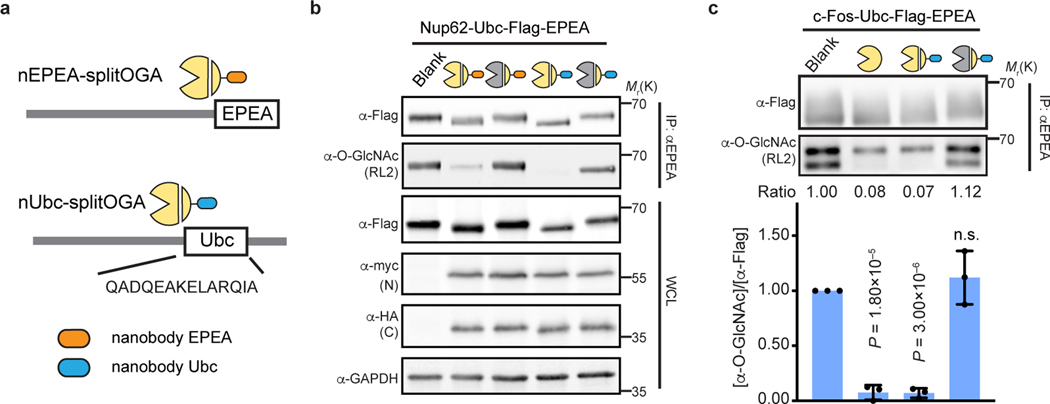
Fig. 4 |. Multiple nanobody-tag pairs are applicable for protein-selective deglycosylation.
a, Schematic of nanobodies against the EPEA and 14-residue Ubc tag adapted to split OGA. b, nEPEA-splitOGA or nUbc-splitOGA remove O-GlcNAc from Nup62 tagged with Ubc and EPEA. Anti-myc and anti-HA blots detect expression of N2 and nanobody-C3, respectively. Results are representative of two biological replicates. c, c-Fos with Ubc tag can be selectively deglycosylated by nUbc-splitOGA. Representative immunoblots are shown. Quantitative results are the mean ± s.d. of n = 3 independent experiments, using unpaired two-tailed Student’s t tests for statistical analysis. n.s., not significant. WCL, whole cell lysate.
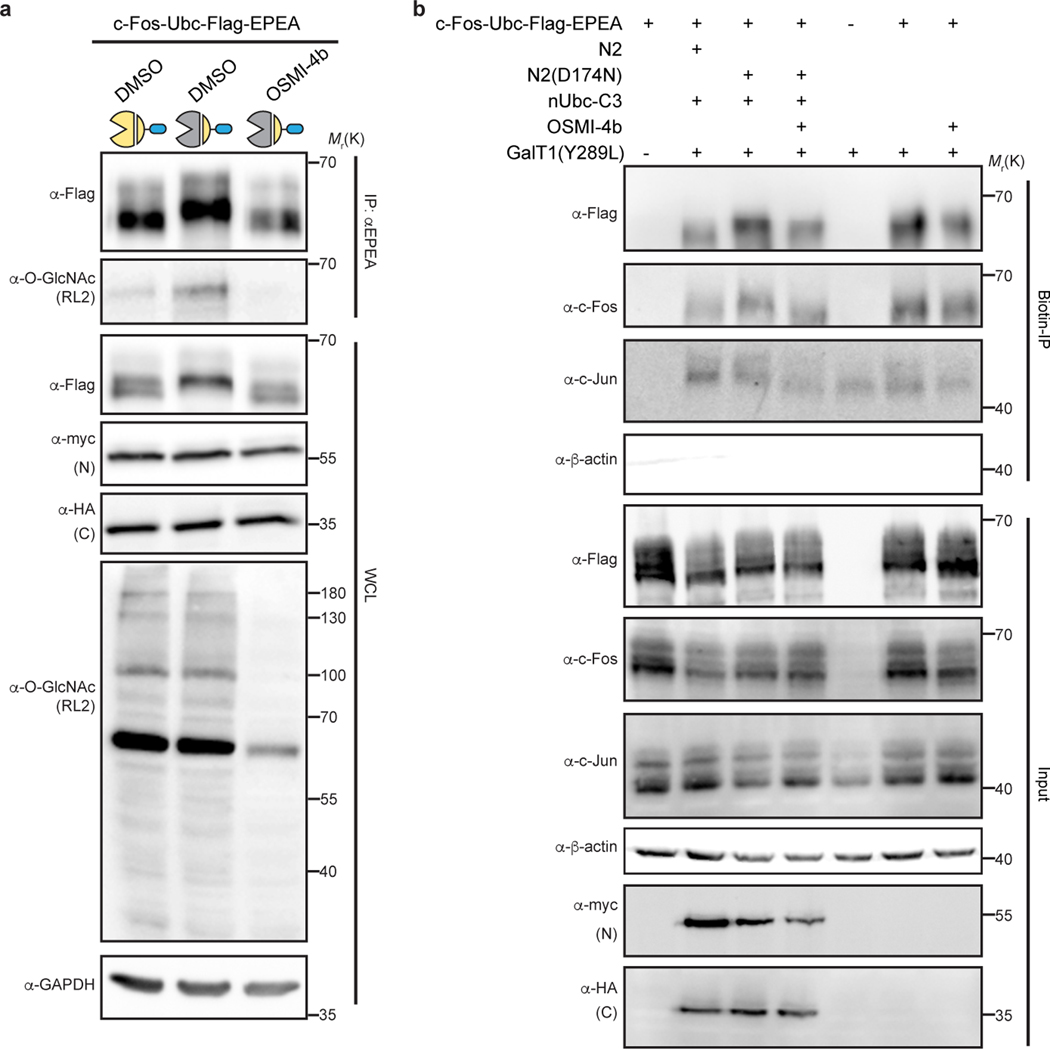
As before, the new nanobody-splitOGA fusions were generalizable to additional target proteins as desired. c-Fos is a core component of AP-1 transcription factor complex32 and reported to be O-GlcNAc modified in mammalian cells33, yet these glycosites await unambiguous assignment by MS. Target deglycosylation of c-Fos tagged with Ubc (c-Fos-Ubc) in HEK 293T cells showed that, as expected, O-GlcNAc was selectively reduced from the target protein by nUbc-splitOGA, and not the inactive mutant [N2(D174N) + nUbc-C3], at levels equivalent to fl-OGA (Fig. 4c and Supplementary Figure 6). Taken together, these results demonstrate that multiple nanobody-tag pairs are readily adapted to split OGA for protein-selective deglycosylation.
Revealing the role of O-GlcNAc on target proteins.
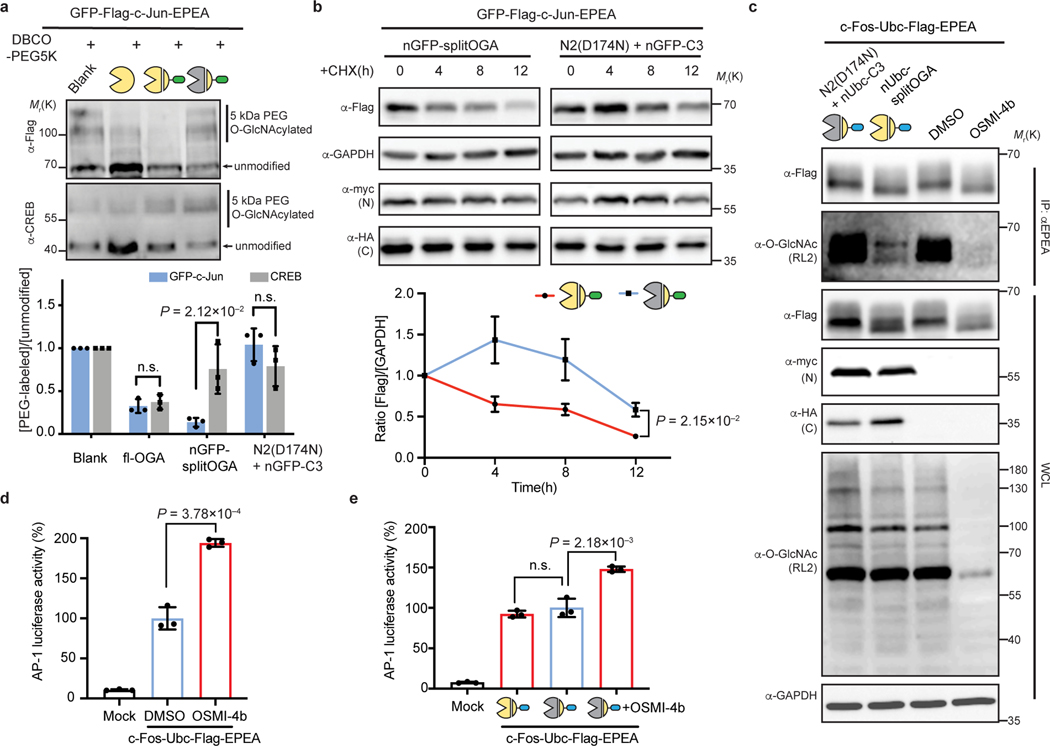
We envisioned that the targeted O-GlcNAc eraser would facilitate engineering and functional analysis of O-GlcNAc on a target protein, and thus evaluated the impact of O-GlcNAc on two members of the AP-1 transcription factor complex, c-Jun and c-Fos. c-Jun carries multiple glycosites34 and is stabilized on global elevation of O-GlcNAc by OGT overexpression in hepatocellular carcinoma (HCC) cells35.However, the direct contribution of O-GlcNAc to stabilization of c-Jun has yet to be established. First, we performed a mass-shift assay on GFP-c-Jun to validate that nGFP-splitOGA can selectively erase O-GlcNAc from O-GlcNAcylated GFP-c-Jun. Similar to GFP-JunB, GFP-c-Jun was selectively deglycosylated by the active nGFP-splitOGA, with minimal disruption of O-GlcNAc levels on endogenous CREB or the global O-GlcNAc proteome (Fig. 5a, Supplementary Figure 7 and Extended Data Fig. 9a,b).
Fig. 5 |. The nanobody-fused split OGA facilitates the functional attribution of O-GlcNAc on c-Jun on protein stability, also O-GlcNAc on c-Fos on AP-1 transcription activity.
a, nGFP-splitOGA can remove O-GlcNAc from GFP-c-Jun selectively. A mass-shift assay was conducted to evaluate the O-GlcNAc level on both GFP-c-Jun and endogenous CREB. b, The stability of GFP-c-Jun is directly related to the extent of O-GlcNAc modification. GFP-c-Jun were co-expressed with indicated constructs in HEK 293T cells. Cells were incubated with 50 μM CHX for up to 12 h and monitored for GFP-c-Jun at different time points. c, nUbc-splitOGA can remove O-GlcNAc from c-Fos-Ubc as OGT inhibitor OSMI-4b, but without causing great reduction of global O-GlcNAc level. Blots are representative of three (a, b) or two (c) biological replicates. d, e, AP-1 luciferase assay showing transcription activity upon OGT inhibition by OSMI-4b (d), or upon the expression of nUbc-splitOGA (e) in c-Fos-Ubc co-transfected HEK 293T cells. Quantitative results are the mean ± s.d. of n = 3 independent experiments, using unpaired two-tailed Student’s t tests for statistical analysis in a, b, d and e. n.s., not significant. WCL, whole cell lysate. Symbols represent corresponding OGA constructs as indicated.
Given that O-GlcNAc may stabilize c-Jun, we monitored the turnover of GFP-c-Jun in HEK 293T cells by adding cycloheximide (CHX) to block protein synthesis. Indeed, degradation of GFP-c-Jun was accelerated upon OGT inhibition with Ac45SGlcNAc and attenuated upon OGA inhibition with Thiamet-G (Extended Data Fig. 9c). However, inhibition of OGT or OGA broadly alters O-GlcNAc on a number of proteins, which may alter protein stability through other mechanisms36. Therefore, we employed nGFP-splitOGA to directly link the O-GlcNAc modification on GFP-c-Jun to protein stability. GFP-c-Jun was co-expressed with nGFP-splitOGA (red line) or the inactive form (blue line) in HEK 293T cells. Upon addition of CHX, the protein level of GFP-c-Jun was monitored over time to reveal that targeted deglycosylation of GFP-c-Jun accelerated degradation (Fig. 5b), implying that the stability of GFP-c-Jun is directly impaired by the loss of O-GlcNAc.
c-Jun forms a heterodimer with c-Fos as part of the AP-1 transcription factor complex. Next, we sought to explore a possible connection between O-GlcNAc and c-Fos on transcriptional activity by comparison of target protein deglycosylation to chemical inhibition of O-GlcNAcylation. We first verified that nUbc-splitOGA selectively removes O-GlcNAc without altering global O-GlcNAc levels, in contrast to the potent OGT inhibitor OSMI-4b9 (Fig. 5c and Supplementary Figure 8). By contrast, the O-GlcNAc modification on endogenous c-Jun was reduced upon OGT inhibition by OSMI-4b, but remained unperturbed under the treatment of nUbc-splitOGA (Extended Data Fig. 10). To evaluate transcriptional activity, c-Fos-Ubc and the AP-1-responsive luciferase reporter were co-transfected in HEK 293T cells and we observed an enhanced AP-1 responsive luciferase signal upon OGT inhibition with OSMI-4b (Fig. 5d). Intriguingly, AP-1 transcriptional activity remained largely unperturbed on selective c-Fos deglycosylation using nUbc-splitOGA in comparison to the inactive mutant, indicating that direct deglycosylation of c-Fos is not promoting AP-1 activity (Fig. 5e). Notably, addition of OSMI-4b again recovered AP-1 transcriptional activity in the presence of the inactive nUbc-splitOGA. Therefore, based on the insight garnered by the target protein deglycosylation approach, we postulate that the enhanced transcriptional activity induced by OSMI-4b may be promoted by removal of O-GlcNAc from other proteins and is not directly linked to removal of O-GlcNAc on c-Fos. For example, AP-1 is composed of members from several protein families32, of which several are O-GlcNAc modified, and other members of transcription machinery are O-GlcNAc modified as well, such as RNA polymerase II37 and TATA-binding protein38. Taken together, the nanobody-fused split OGA can readily translate effects observed on the global O-GlcNAc proteome back to the desired target protein in cells, and will find particular use in the study of target proteins bearing multiple or only partially characterized glycosites.
Discussion
Here we report a method to selectively remove O-GlcNAc and measure the functional contribution on a desired target protein using a nanobody-fused split OGA. The targeted O-GlcNAc eraser enables selective deglycosylation on a protein of interest in cells with minimal perturbation of the global O-GlcNAc proteome, and complements chemical and genetic methods to globally perturb O-GlcNAc levels or target specific glycosites, if known. We demonstrate the generality of nanobody-splitOGA using four nanobodies directed to five protein targets that represent the range of O-GlcNAc substrates and demonstrate selectivity for the target protein through the nanobody by comparison of several split OGA constructs and evaluation of the global O-GlcNAc proteome. Finally, we applied the system to characterize the direct functional contribution of O-GlcNAc on a desired protein, using c-Jun and c-Fos, in the context of protein stabilization and downstream transcriptional activation.
The ability to perform protein-selective deglycosylation within cells bridges an important gap in the translation of phenotypic effects observed from the global perturbation of O-GlcNAc to the contribution of O-GlcNAc on specific target proteins and enables a number of new strategies for the study and engineering of O-GlcNAc. Utilizing inhibitors of OGT or OGA allows for correlation of phenotypic effects with O-GlcNAcylation, yet to link the effect of O-GlcNAc back to a target protein previously required the comprehensive mapping of the glycosites and validation using site-directed mutagenesis. Using the nanobody-splitOGA, candidate proteins can instead be directly evaluated in cells to facilitate the attribution of a phenotype or desired outcome prior to glycosite mapping efforts. We envision that this will be of particular utility for many O-GlcNAc proteins, like c-Jun or c-Fos, which are O-GlcNAc modified at multiple or unmapped glycosites. The approach may further avoid disturbances to other PTM signaling pathways, function, or structure of a protein that may arise from conventional site-directed mutagenesis. Furthermore, unlike mutagenesis, the nanobody-splitOGA is readily complemented with the nanobody-OGT18 to enable bidirectional modulation of the target protein and functional evaluation. Since O-GlcNAc serves as a signal molecule on nucleocytoplasmic proteins, target protein glycosylation and deglycosylation may reveal both native functions for O-GlcNAc and neo-functions that may participate in rewiring related signaling pathways similar to the engineering of other PTMs, like ubiquitylation39 and phosphorylation40, in the long term.
Despite these advantages, the nanobody-splitOGA system contains several limitations in its present form. Although the modularity of nanobody-tag pairs is generalizable, the tagging and expression of a target protein is required for recruitment of the nanobody-splitOGA system. These challenges will be mitigated through the generation of nanobodies that target endogenous proteins 41 or through genetic engineering approaches, like CRISPR, that enable tagging of endogenous proteins with small peptide sequences or GFP42. Nonetheless, binding of the nanobody to the target protein has the potential to influence the functionalities of target proteins, such as by blocking protein-protein interactions, inhibiting enzymatic activity, or interfering with dynamic structural changes43, necessitating the use of appropriate controls (i.e., the catalytically inactive nanobody-splitOGA) and additional validation experiments. In the long term, integration of recent developments in nanobody engineering with this system, such as nanobodies with a wide range of affinities44, or chemo/optogenetically controlled nanobodies45,46 may address these issues and expand the capabilities of this system. As the nanobody-splitOGA system is a target protein deglycosidase, phenotypic effects from modulation of an individual glycosite on a multiply glycosylated target protein may be overlooked. In addition, the protein-selective deglycosylation mediated by nanobody-splitOGA will not differentiate between functions associated with co-translational47 versus post-translational O-GlcNAc modification. Leveraging nanobodies that target unique signal sequences of nascent proteins or relocalization of a nanobody-splitOGA to certain subcellular compartments with more spatiotemporal control may overcome these challenges.
In conclusion, we report a generalizable and flexible strategy for target protein deglycosylation using a nanobody-fused split OGA. The targeted O-GlcNAc eraser enables the direct linkage of phenotypic effects observed in cellular assays to O-GlcNAc on a target protein of interest, thus facilitating characterization of O-GlcNAc functions on a protein level. In combination with additional methods for studying O-GlcNAc, the system reported here will accelerate the discovery of O-GlcNAc functions on the several thousand target proteins bearing O-GlcNAc within different physiological and nutritional contexts.
Methods
Cell culture and transfection.
HEK 293T cells (ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, 11995073) supplemented with penicillin (50 μg/mL) and streptomycin (50 μg/mL) along with 10% (v/v) FBS. Transfections of all plasmids in this study were performed using TransIT-PRO® (Mirus Bio, MIR 5740) according to the manufacturer’s instructions.
Plasmids and subcloning.
Glycoproteins Nup62, Sp1, JunB, c-Jun were subcloned into pcDNA3.1 vector with GFP and a Flag-tag, or BC2 tag in the N terminus and EPEA tag in the C terminus, respectively, unless otherwise noted. For all split OGA constructs with or without the fusion of nanobody, N fragments were tagged with a myc tag and C fragments were tagged with a HA tag, respectively. Human SP1 cDNA ORF plasmid (HG12024-G), human c-Fos cDNA ORF plasmid (HG11279-M) and human OGT cDNA ORF clone with a C-terminal His tag (HG17892-CH) were purchased from Sino Biological. c-Fos was subcloned into pcDNA3.1 vector with a 14-residue Ubc tag (amino acids: QADQEAKELARQIA), Flag tag and EPEA tag in the C terminus. 3xAP1pGL3 was a gift from Alexander Dent (Addgene plasmid # 40342; http://n2t.net/addgene:40342; RRID: Addgene_40342). The full-length OGA plasmid with a myc-tag is a generous gift from Prof. David Vocadlo, Simon Fraser University, Canada. All nanobody DNA fragments were synthesized from IDT. A list of genetic constructs used in this study, with annotated epitope tags, vector name, etc., was provided in Supplementary Table 1.
Antibodies and reagents.
Anti-Flag (F3165) and anti-OGA (HPA036141) were purchased from Sigma-Aldrich. Anti-DYKDDDK Tag (14793), anti-myc (2276), anti-HA (3724), anti-GAPDH (5174), anti-OGT (24083), anti-His-Tag (12698), anti-CREB (9197), anti-c-Jun (60A8), anti-c-Fos (2250) and anti-HA-Tag (Alexa Fluor® 647 Conjugate) (3444) were purchased from Cell Signaling Technology. Anti-O-GlcNAc(RL2) (ab2739) was obtained from Abcam. Anti-Nup62 (610497) was purchased from BD Biosciences. Anti-β-actin (sc-47778) was purchased from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Rockland Immunochemicals. IRDye® secondary antibodies were purchased from LI-COR Biosciences. Alexa Fluor™ 488 anti-rabbit IgG (A11008), Alexa Fluor™ 647 anti-rabbit IgG (A21244), Alex Fluor™ 568 anti-mouse IgG (A11004) and NucBlue™ Fixed Cell Stain ReadyProbes™ reagent (R37606) were purchased from Invitrogen. OSMI-4b, Ac45SGlcNAc and Biotin-Alkyne probe were homemade.
Immunoprecipitation and Immunoblot assays.
After 36~48 h of transfection, cells were harvested and washed with PBS once. Cells were lysed with M-PER lysis buffer (Thermo Scientific, 78501) containing 1× cOmplete™, EDTA-free protease inhibitor cocktail (Sigma-Aldrich, 11873580001) and 10 μM Thiamet-G (Sigma-Aldrich, SML0244) unless otherwise noted. Protein concentrations were determined by the BCA assay kit (G-Biosciences, 786) on a multi-mode microplate reader FilterMax F3 (Molecular Devices LLC, Sunnyvale, CA).
For immunoprecipitation of proteins with the C-terminal EPEA tag, cell lysates with equal amounts of protein were diluted with PBS and incubated with C-tag affinity matrix (Thermo Scientific, 191307005) for 1 h at room temperature, with end-to-end rotation. After washed three times with PBS buffer, the enriched proteins were eluted with SDS sample buffer and subjected to SDS-PAGE.
For immunoprecipitation of proteins with the Flag tag, cells were lysed in a buffer containing 50 mM Tris HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5% glycerol, 1× protease inhibitor cocktail and 10 μM Thiamet-G on ice for 20 min. Cell lysates with equal amounts of protein were diluted with the lysis buffer and incubated with ANTI-FLAG® M2 magnetic beads (Sigma-Aldrich, M8823) for 2 h at 4 °C with rotation. The beads were washed by TBS buffer (50 mM Tris HCl pH 7.4, 150 mM NaCl) for three times. The enriched proteins were eluted with SDS sample buffer and subjected to SDS-PAGE.
For immunoprecipitation of proteins with anti-HA (Pierce, 88836) or anti-c-Myc (Pierce, 88842) magnetic beads, cell lysates with equal amounts of protein were diluted with 1× TBS-T buffer (25 mM Tris HCl pH 7.4, 150 mM NaCl, 0.05% Tween-20) and incubated with pre-washed magnetic beads at room temperature for 30 min with mixing, following the manufacturer’s instructions. Anti-HA magnetic beads were washed three times with TBS-T buffer and once with ultrapure water. Anti-c-Myc magnetic beads were washed three times with 5× TBS-T buffer and once with ultrapure water. The enriched proteins were eluted with SDS sample buffer and subjected to SDS-PAGE.
For immunoprecipitation of proteins with His tag, cells were lysed in a buffer containing 50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 5% glycerol, 1× protease inhibitor cocktail and 10 μM Thiamet-G on ice for 20 min. Cell lysates with equal amounts of protein were diluted with wash buffer (50 mM Tris HCl pH 8.0, 150 mM NaCl, 0.01% Tween-20) and incubated with pre-washed His-Tag Dynabeads (Invitrogen, 10103D) at room temperature for 20 min with mixing, following the manufacturer’s instructions. After washed 4 times with wash buffer, the enriched proteins were eluted with elution buffer (300 mM Imidazole, 50 mM Tris HCl pH 8.0, 150 mM NaCl, 0.01% Tween-20) on a shaker for 10 min at room temperature.
For immunoblotting analysis, proteins were transferred to a nitrocellulose membrane using iBlot (Thermo Scientific). Membranes were blocked with Tris buffered saline containing 0.1% Tween-20 and 5% BSA (Sigma-Aldrich, A9647) and incubated with the primary antibodies (1:1,000 dilution) and the secondary antibodies (1:10,000 dilution) sequentially. Immunoblots images were captured by Azure Imager C600 (Azure Biosystems, Inc., Dublin, CA) and analyzed with Fiji ImageJ. All IR fluorescence western blot images are converted into grayscale images by Fiji ImageJ. The unsaturated exposure of immunoblot images was used for quantification with the appropriate loading controls as standards.
Cycloheximide (CHX) treatment.
HEK 293T cells were transiently transfected with indicated plasmids and treated with DMSO, or Ac45SGlcNAc, or Thiamet-G at the same time if needed. 36~48 h after the transfection, cells were incubated with 50 μM CHX (Sigma-Aldrich, C4859) for up to 12 h. At the indicated time points, cells were harvested and lysed with M-PER lysis buffer. Protein expression and global O-GlcNAc level were determined by immunoblot assays. GAPDH protein level was used as the internal loading control.
Chemoenzymatic labeling of O-GlcNAcylated proteins.
Purification of GalT1 (Y289L) enzyme and labeling of O-GlcNAcylated proteins with GalNAz were performed according to the procedure of Hsieh-Wilson and co-workers48. Briefly, cell samples in 6-well plates or 15-cm dishes were harvested and washed by PBS once. The lysis buffer (PBS with 2% SDS) was added into cell pellets and heated for 5 min at 95 °C, followed by the sonication to sheer DNA. Protein concentrations were determined by BCA assay. Cell lysates were reduced and alkylated with 25 mM DTT (Thermo Scientific, 20290) at 95 °C for 5 min and 50 mM iodoacetamide (Sigma-Aldrich, I1149) at room temperature for 1 h, respectively. Proteins were precipitated by the methanol/chloroform solution (aqueous phase : CH3OH : CHCl3 = 4 : 4 :1) and resuspended in 1% SDS, 20 mM HEPES (pH 7.9) buffer with a concentration of 3.75 mg/mL. For 150 μg proteins, H2O (49 μL), 2.5× GalT labeling buffer (80 μL, final concentrations: 50 mM NaCl, 20 mM HEPES, 2% NP-40, pH 7.9), 100 mM MnCl2 (11 μL), 500 μM UDP-GalNAz (10 μL), 2 mg/mL GalT1 (Y289L) (10 μL) were added into cell lysates orderly. The reaction was gently rotated at 4 °C for at least 20 h and the proteins were precipitated as described above. The proteins were resuspended with 1% SDS, PBS for the further click chemistry. For mass spectrometry analysis, the procedures were scaled up with the starting material of 3 mg input proteins.
Mass shift assay with PEG5K labeling.
For proteins in PBS containing 1% SDS, 10 mM DBCO-PEG-5kDa (Click Chemistry Tools, A118) was added with a final concentration of 1mM. The reaction was conducted at 95 °C for 5 min. The proteins were precipitated as previously described and resuspended in PBS containing 2% SDS. The proteins were mixed with SDS sample buffer and subjected into the immunoblot assay. The relative abundance of O-GlcNAcylated forms and unmodified form of the target protein is obtained by measuring the intensities of mass-shifted bands at higher molecular weights and the bottom band with the original molecular weight, respectively48. The ratio of abundances of O-GlcNAcylated forms versus unmodified form reflects the O-GlcNAcylation level on the protein of interest.
CuAAC and biotin-immunoprecipitation.
For enrichment and identification of the O-GlcNAcylated proteins, experiments were performed based on the procedure of Woo and co-workers49. Briefly, the proteins in PBS containing 1% SDS were diluted with PBS and incubated with 100 μM THPTA (Sigma-Aldrich, 762342), 0.5 mM CuSO4, 2.5 mM fresh sodium ascorbate and 200 μM either Biotin-PEG4-Alkyne (Click Chemistry Tools, TA105) for immunoblotting or Biotin-Alkyne probe for proteomics at 37 °C for 4 h.
For biotin-immunoprecipitation, proteins were precipitated and resuspended into 100 μL PBS containing 1% SDS. The protein solutions were diluted with PBS to lower the final concentration of SDS into 0.2% and incubated with pre-washed 40 μL streptavidin beads slurry at room temperature for 2 h with gentle rotation. Beads were washed sequentially with 0.2 % SDS/PBS three times and PBS three times. Enriched proteins were eluted with SDS sample buffer and subjected to SDS-PAGE.
Quantitative chemical proteomics.
For quantitative proteomics, after reacted with Biotin-Alkyne probe, proteins were precipitated and resuspended into 400 μL PBS containing 2% SDS. The protein solutions were diluted with PBS to lower the final concentration of SDS into 0.2% and incubated with pre-washed 400 μL streptavidin beads slurry. The mixture was incubated at room temperature for 4 h with gentle rotation. The beads were transferred into the Bio-Spin column (Bio-Rad, 7326207) and washed with 1 mL 8 M urea, 5 mL 0.2% SDS/PBS, 5 mL PBS and 5 mL Milli-Q water sequentially with the help of a vacuum manifold. After changing buffer with 500 μL 500 mM urea, 1 mM CaCl2 in PBS, 2 μg trypsin (Promega, V5111) was added, and the resulting mixture was incubated at 37°C for 16 h. The eluant containing trypsin digested peptides were collected as the trypsin fraction for protein identification. The peptides were desalted by C18 Tips (87784) following the manufacturer’s instructions and resuspended in 20 μL 50 mM TEAB buffer. For each sample, 5 μL the corresponding amine-based TMT 10 (90406)/16-plex (A44520) reagents (Thermo Scientific, 10 μg/μL, 11.9 μg/μL, respectively) was added and reacted for 1 h at room temperature. The reactions were quenched with 2 μL 5% hydroxylamine solution and combined. The combined mixture was concentrated using Eppendorf Vacufuge to dryness. For glycoproteomics experiments on GFP-Sp1, the mixture was resuspended and fractionated into 6 samples with High pH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific, 84868), and concentrated to dryness. All samples are stored at –20 °C until analysis.
Mass spectrometry acquisition procedures.
A Thermo Scientific EASY-nLC 1000 system was coupled to an Orbitrap Fusion Tribrid with a nano-electrospray ion source. Mobile phases A and B were water with 0.1% formic acid (v/v) and acetonitrile with 0.1% formic acid (v/v), respectively. For non-fractionated peptides, peptides were separated with a linear gradient from 4 to 32% B within 140 min, followed by an increase to 50% B within 10 min and further to 98% B within 10 min, and re-equilibration. For fractionated peptides, peptides were separated with a linear gradient from 4 to 32% B within 50 min, followed by an increase to 50% B within 10 min and further to 98% B within 10 min, and re-equilibration. The instrument parameters were set as follows: survey scans of peptide precursors were performed at 120K FWHM resolution over a m/z range of 410–1800. HCD fragmentation was performed on the top 10 most abundant precursors exhibiting a charge state from 2 to 5 at a resolving power setting of 50K and fragmentation energy of 37% in the Orbitrap. CID fragmentation was applied with 35% collision energy and resulting fragments detected using the normal scan rate in the ion trap.
Mass spectrometry data analysis.
The raw data was processed using Proteome Discoverer 2.4 (Thermo Fisher Scientific). For the trypsin fraction, the data were searched against the UniProt/SwissProt human (Homo sapiens) protein database (Aug. 19, 2016, 20,156 total entries) and contaminant proteins using Sequest HT algorithm. The database was adjusted by deleting O60502 (OGA) and replacing P37198 (Nup62) with GFP-Nup62 or P08047 (Sp1) with GFP-Sp1 protein sequences, respectively. Searches were performed with the following guidelines: spectra with a signal-to-noise ratio greater than 1.5; trypsin as enzyme, 2 missed cleavages; variable oxidation on methionine residues (15.995 Da); static carboxyamidomethylation of cysteine residues (57.021 Da), static TMT labeling (229.163 Da for TMT 10plex or 304.207 Da for TMT 16-plex) at lysine residues and peptide N-termini; 10 ppm mass error tolerance on precursor ions, and 0.02 Da mass error on fragment ions. Data were filtered with a peptide-to-spectrum match (PSM) of 1% FDR using Percolator. The TMT reporter ions were quantified using the Reporter Ions Quantifier without normalization. For the obtained proteome, the data was further filtered with the following guidelines: protein FDR confidence is high; unique peptides are greater than 2; master protein only; exclude all contaminant proteins. For P-value and fold change calculations, the data was further processed using a custom algorithm as described below. Most of the empty abundances, if any, are filled in with minimum noise level. If all abundances are missing for control and treatment or the variance between existing abundances is above 30%, the PSM is removed. Applied here is a VSN normalization computed on the imputed matrix using a robust variant of the maximum-likelihood estimator for an additive-multiplicative error model and affine calibration50. The model incorporates dependence of the variance on the mean intensity and a variance stabilizing data transformation. A linear model is fitted to the expression data for control and treatment, then t-statistics are computed by empirical Bayes moderation of standard errors towards a common value.
Immunofluorescence microscopy.
Cells were seeded on 22×22 mm glass coverslips No. 1.5 coated with poly-L-lysine (Neuvitro Corporation, H-22–1.5-pll) that had been placed in single wells of a 6-well plate for 24 h prior to transfection. 24~36 h after transfection, cells were washed with PBS twice and fixed in freshly prepared 4% paraformaldehyde in PBS for 20 min at room temperature. After washed with PBS twice, cells were permeabilized and blocked with the blocking buffer (1× PBS / 5% BSA / 0.3% Triton X-100) for 1 h at room temperature. The primary and secondary antibodies were diluted with the dilution buffer (1× PBS / 1% BSA / 0.3% Triton X-100) as the manufacturers recommended on their websites. The cells were incubated with the primary antibodies overnight at 4 °C. The cells were rinsed with PBS three times, followed by 1 h incubation with the secondary antibodies (1:1,000 dilution) at room temperature in the dark. The cells were washed with PBS three times and incubated with extra fluorophore conjugated primary antibodies if needed. After washed with PBS three times, NucBlue™ Fixed Cell Stain ReadyProbes™ reagent (Invitrogen, R37606) was added to stain the nuclei according to the manufacturer’s instructions. Coverslips were washed with PBS and mounted in anti-fade Diamond (Life Technologies, P36961). Images were collected on an OLYMPUS confocal laser scanning microscope (FV3000) and exported to Fiji ImageJ for final processing and assembly.
Luciferase reporter assay.
For luciferase assay in the OGT inhibition experiment, HEK 293T cells co-transfecting with c-Fos-Ubc-Flag-EPEA plasmid and AP-1 responsive luciferase reporter were treated with either DMSO or 25 μM OSMI-4b for 48 h before luciferase activity detection. For luciferase assay in the co-expression of nUbc-splitOGA, HEK 293T cells co-transfecting with c-Fos-Ubc-Flag-EPEA plasmid and AP-1 responsive luciferase reporter with the incubation of either DMSO or 25 μM OSMI-4b were co-expressed with either nUbc-splitOGA or its inactive mutant (D174N) for 48 h before luciferase activity detection. Luciferase reporter assays were performed using Luciferase Assay System (Promega, E1500) according to manufacturer’s protocols. At least three independent biological replicates were done in this assay.
Statistical analysis.
Statistical analyses (unpaired two-tailed Student’s t-test) were performed using GraphPad Prism 8. Data were derived from at least three independent biological replicate experiments and presented as the mean ± s.d., *P < 0.0332, **P < 0.0021, ***P < 0.0002, ****P < 0.0001 and n.s., not significant. For raw data of Fig. 3f, g, the Benjamini-Hochberg adjustment was applied for multiple comparisons.
Data availability
The main data generated or analyzed during this study are included in this published article and its Supplementary Information. The mass spectrometry proteomics data were searched against the UniProt/SwissProt human (Homo sapiens) protein database (Aug. 19, 2016, www.uniprot.org/proteomes/UP000005640) and have been deposited to the ProteomeXchange Consortium via the PRIDE51 partner repository with the dataset identifier PXD022347 (Fig. 3d,e) and PXD018914 (Fig. 3f,g). Source data are provided with this paper.
Code availability
Code used to analyze the mass spectrometry proteomics data has been deposited in GitHub and is available for download at: https://github.com/harvardinformatics/quantproteomics/tree/master/PEA
Extended Data
Extended Data Fig. 1. Schematic representation of OGA and target protein constructs used in this study.
a, Schematic of the structures of human OGA and other truncations. Catalytic domain, stalk domain, HAT domain, and intrinsic disordered regions are shown in pink, cyan, orange and white, respectively. GS linker represents a 15-residue glycine and serine linker. b, Depiction of the strategy to fuse the nanobody on OGAs to achieve protein specificity. nGFP, nanobody against GFP. c, Design of GFP-fused, Ubc tag-fused and BC2 tag-fused proteins of interest used in this study. For GFP-fused proteins, GFP and a Flag tag are placed on the N-terminus and the EPEA tag is in the C-terminus unless otherwise noted. For Ubc tag-fused proteins, the 14-residue peptide tag, Flag tag and EPEA tag are sequentially placed in the C-terminus unless otherwise noted. For BC2 tag-fused proteins, the 12-residue peptide tag is placed on the N-terminus and Flag, EPEA tag are in the C-terminus. Peptide sequences of Ubc and BC2 were shown. d, Symbols used in this manuscript to represent the indicated split OGA constructs.
Extended Data Fig. 2. Identification of the minimal OGA for nanobody-directed deglycosylation on the target protein.
a, Enzymatic activities of OGA and its truncations are evaluated on GFP-Nup62. GFP-Nup62 was co-expressed with indicated constructs, enriched by anti-EPEA beads, and analyzed by immunoblotting to visualize the protein level and O-GlcNAc modification level, respectively. b, Evaluation of enzymatic activities of nGFP-OGA fusion proteins on GFP-Nup62. Expression levels of the indicated proteins and degree of O-GlcNAc modification were quantified by immunoblotting. The ratio equals to the intensity of anti-O-GlcNAc immunoblot normalized by the intensity of anti-Flag immunoblot. WCL, whole cell lysate. The data are representative of two biological replicates.
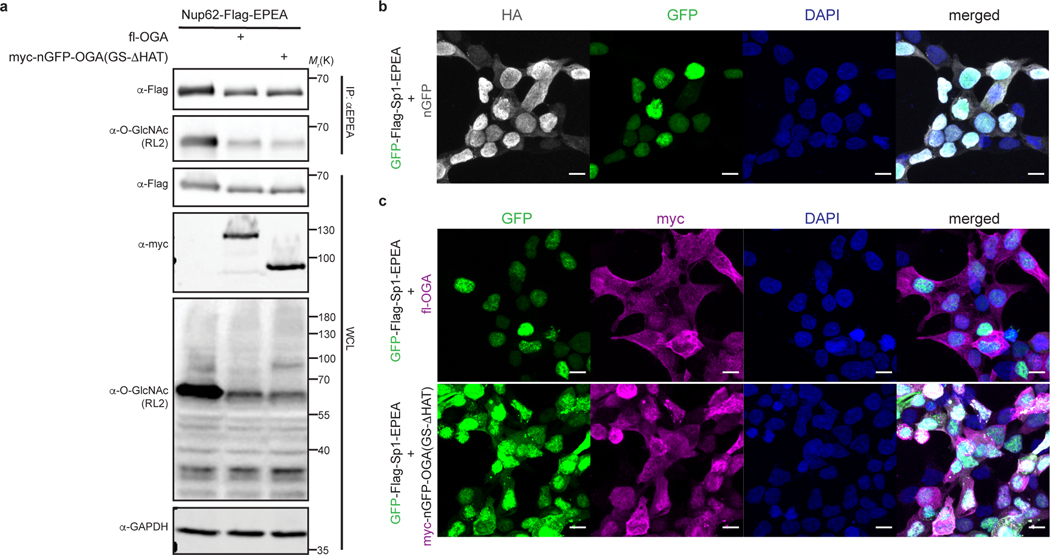
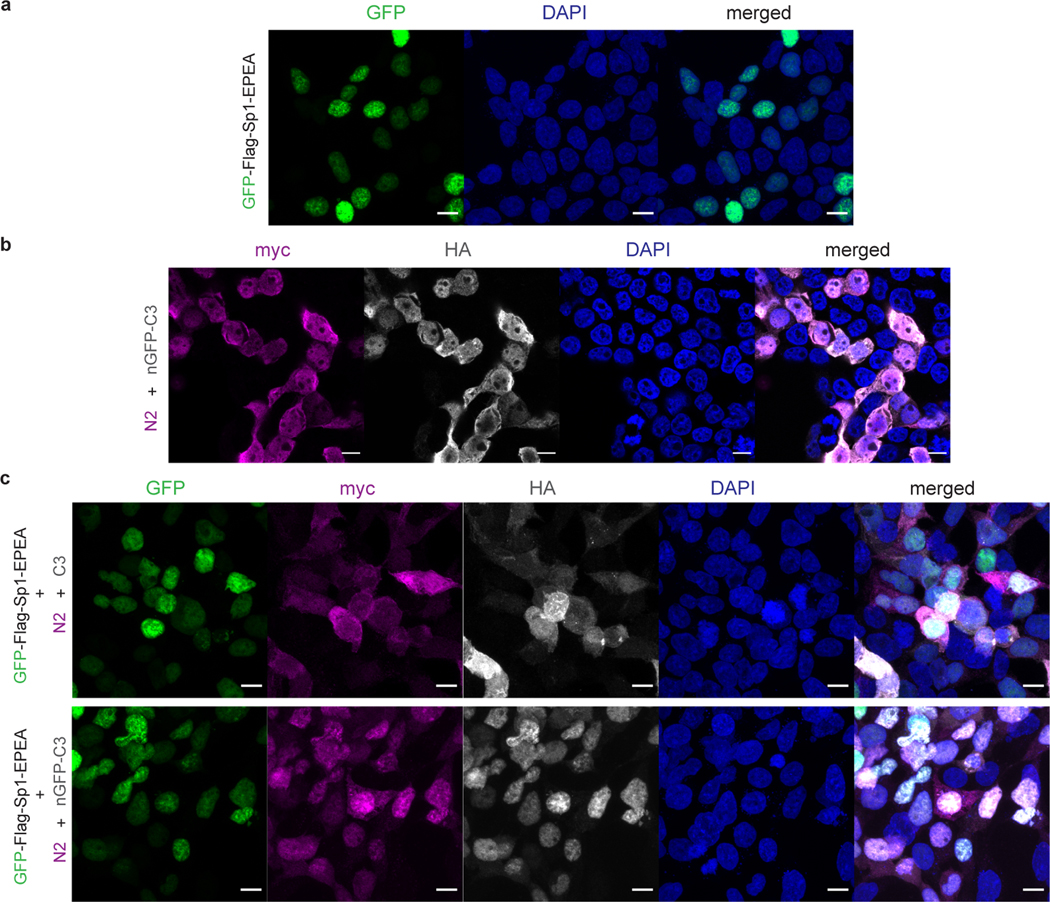
Extended Data Fig. 3. nGFP-OGA(GS-ΔHAT) has limited target protein selectivity and can alter subcellular localization of the target protein.
a, nGFP-OGA(GS-ΔHAT) removes O-GlcNAc from Nup62 without GFP tag similar to the full length OGA (fl-OGA). HEK 293T whole cell lysates (WCL) and immunoprecipitation samples were analyzed by immunoblotting assays using the indicated antibodies. Results are representative of two biological replicates. b, nGFP colocalizes with nuclear transcription factor Sp1 with GFP and does not change the subcellular localization of GFP-Sp1 by immunofluorescence imaging. c, nGFP-OGA(GS-ΔHAT) alters the subcellular localization of GFP-Sp1, but co-expression with fl-OGA does not change nuclear localization of GFP-Sp1 by immunofluorescence imaging. Channels are annotated on the top. Scale bar: 10 μm. Right: merged channel. Proteins co-expressed in each sample were labeled on the left side. Images are representative of at least three randomly selected frames.
Extended Data Fig. 4. Optimization of nGFP-fused split OGA constructs in living cells.
a, Co-expressing N- and C-fragments of OGA reconstitutes deglycosidase activity in HEK 293T cells. b, Split OGA fragments, N2 and C3, instead of C4, associate with each other when co-expressed in HEK 293T cells. The asterisk indicated IgG heavy chain from anti-c-Myc magnetic beads. c, Comparison of nGFP-fused N- and C-terminal OGA fragments on GFP-Nup62 in HEK 293T cells. The pair of N2 and nGFP-fused C3 (N2 + nGFP-C3) shows the best deglycosylation performance. In a and c, activities of fragments alone or pairs with/without nGFP were evaluated on GFP-Nup62, which was enriched by beads against EPEA tag and blotted with RL2 antibody to reveal O-GlcNAc modification level. D174N, a catalytically impaired mutation on OGA. Anti-myc and anti-HA blots detect expression of full-length (fl-OGA) or N-terminal fragment, and C-terminal fragment, respectively. WCL, whole cell lysates. The data in a-c are representative of at least two biological replicates.
Extended Data Fig. 5. nGFP-splitOGA selectively deglycosylates the target protein without affecting the global O-GlcNAc proteome.
a, nGFP-splitOGA has little effect on endogenous glycoprotein CREB. HEK 293T cells co-expressing OGA constructs with GFP-Nup62 were subjected to mass shift assay. The intensities of O-GlcNAcylated and unmodified CREB were quantified. The ratios are shown below the anti-CREB blot. WCL, whole cell lysates. b, Overexpression of selected split OGA constructs with target protein has little effect on global O-GlcNAcylation level. For OGT inhibition, cells were treated with 25 μM OSMI-4b for 30 h. Global O-GlcNAcylation level was evaluated by anti-O-GlcNAc (RL2) antibody. c, nGFP-splitOGA has minimal effect on protein levels of endogenous OGT, OGA and glycoprotein CREB. Anti-myc and anti-HA blots detect expression of N-terminal fragment, and C-terminal fragment, respectively. d, Comparison of overexpressed proteins with the corresponding endogenous proteins. The antibody against OGA recognizes both endogenous OGA and the overexpressed N-terminal fragment of split OGA. Endogenous Nup62(*) and OGA (**) are indicated. The data in a-d are representative of at least two biological replicates.
Extended Data Fig. 6. Confocal imaging of intracellular distributions of GFP-Sp1 and the split OGAs in HEK 293T cells.
a, GFP-Sp1 localized in nucleus. b, Intracellular distributions of N2 and nGFP-C3 fragments when co-expressed in HEK 293T cells. Two fragments of nGFP-splitOGA were distributed on both cytoplasm and nucleus. c, Subcellular localizations of GFP-Sp1, N fragment and C fragment when expressed simultaneously in HEK 293T cells. Two fragments of split OGA without nGFP (c, upper row) were distributed on both cytoplasm and nucleus. C-terminal fragment of nGFP-splitOGA (c, bottom row) reveals better colocalization with nuclear protein GFP-Sp1, showing the binding between nGFP and GFP. Split OGAs do not change the subcellular localization of GFP-Sp1. Channels are annotated on the top. Scale bar: 10 μm. Right: merged channel. Proteins co-expressed in each sample were labeled on the left side. Images are representative of at least three randomly selected frames.
Extended Data Fig. 7. Mass spectrometry analysis on the activity and selectivity of nGFP-splitOGA on GFP-Nup62.
a, Schematic representation of the workflow of O-GlcNAcylated protein enrichment and mass spectrometry-based identification. Proteins with O-GlcNAc modification were labeled with GalNAz by GalT(Y289L)-mediated chemoenzymatic labeling, followed by a click reaction with an alkyne-biotin probe. Biotin-labeled proteome was enriched by streptavidin beads and digested by trypsin. Released peptides were labeled by TMT reagents and compiled into a single pool. Proteins were identified and quantified by LC-MS. b–d, Reproducibility of the TMT experiments of O-GlcNAcylated proteome shown in Fig. 3d,e. The signal abundances of the corresponding TMT channels for each protein were extracted and were log10 transformed for full-length OGA treatment (b, fl-OGA), nGFP-splitOGA treatment (c) and its inactive form [N2(D174N) + nGFP-C3] treatment (d) groups (n = 2 independent biological replicates).
Extended Data Fig. 8. Peptide tag BC2 and its nanobody can be adapted to split OGA to achieve protein-selective deglycosylation.
a, Schematic of nanobodies against BC2 and EPEA tag adapted to split OGA. BC2 tag refers to a 12-residue peptide epitope, which is functional irrespective of its position on the target protein. b, nBC2-splitOGA is able to remove O-GlcNAc from Nup62 tagged with BC2 and EPEA in a similar manner to nEPEA-splitOGA. Symbols represent the corresponding OGA constructs as indicated in Extended Data Fig. 1. Anti-myc and anti-HA blots detect expression of full-length (fl-OGA) or N-terminal and C-terminal fragment, respectively. WCL, whole cell lysate. The data are representative of two biological replicates.
Extended Data Fig. 9. Modulation of O-GlcNAc modification level and validation of its functional contributions on the stability of GFP-c-Jun.
a, O-GlcNAc level on GFP-c-Jun and endogenous CREB were evaluated by the mass-shift assay. GFP-c-Jun was co-expressed with indicated OGA constructs. The intensities of O-GlcNAcylated and unmodified c-Jun and CREB were quantified. Quantification is shown as mean ± s.d. of n = 3 independent biological experiments. All ratios were normalized by the Blank samples. Unpaired two-tailed Student’s t tests were used for statistical analysis. n.s., not significant. b, Whole cell lysates (WCL) were analyzed by immunoblotting assays using the indicated antibodies. Anti-myc and anti-HA blots detect expression of full-length (fl-OGA) or N-terminal fragment, and C-terminal fragment, respectively. The data in a and b are representative of at least three biological replicates. c, The stability of GFP-c-Jun was enhanced by OGA inhibition (Thiamet-G treatment) and impeded by OGT inhibition (Ac45SGlcNAc treatment). HEK 293T cells expressing GFP-c-Jun pre-treated with DMSO or Ac45SGlcNAc or Thiamet-G were incubated with 50 μM CHX for up to 12 h, during which the protein level of GFP-c-Jun and global O-GlcNAcylation level were monitored. Results in c are representative of two biological replicates.
Extended Data Fig. 10. Modulation of O-GlcNAc modification level with nUbc-splitOGA on c-Fos-Ubc in comparison to OGT inhibition.
Immunoblotting analysis of protein expression and O-GlcNAcylation status of c-Fos-Ubc and endogenous c-Jun under the indicated treatments corresponding to Fig. 5d, e. by either enrichment against EPEA-tag (a) or chemoenzymatic labeling followed with Biotin-IP (b). Endogenous c-Jun shows negligible changes on O-GlcNAcylation status with the co-expression of nUbc-splitOGA but shows reduced O-GlcNAc modification upon OGT inhibition with OSMI-4b. No detectable endogenous c-Fos was observed in HEK 293T cells. The data in a and b are representative of two biological replicates.
Supplementary Material
Source Data Fig. 5. Unprocessed Western Blots and/or gels
Source Data Fig. 4. Unprocessed Western Blots and/or gels
Source Data Fig. 6. Unprocessed Western Blots and/or gels
Source Data Fig. 1. Statistical Source Data
Source Data Fig. 2. Statistical Source Data
Source Data Fig. 3. Statistical Source Data
Source Data Fig. 7. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 1. Statistical Source Data
Source Data Extended Data Fig. 2. Statistical Source Data
Source Data Extended Data Fig. 4. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 3. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 7. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 8. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 5. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 6. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 9. Unprocessed Western Blots and/or gels
Acknowledgements
The authors thank P. Schwein and Z. Lin for helpful discussions and B. Budnik, principal scientist of the Harvard University Proteomics Facility. Support from the National Institutes of Health (U01CA242098–01, C.M.W.), the Burroughs Wellcome Fund, a Career Award at the Scientific Interface (C.M.W.), the Sloan Foundation (C.M.W.), the Merck Fellowship Fund, and Harvard University is gratefully acknowledged.
Footnotes
Competing interests
Harvard University has filed a patent application (U.S. Provisional Application No. 63/087,773, filed October 5, 2020) including work described herein. C.M.W and Y.G. are inventors of this patent. All other authors declare no competing financial interest.
References
- 1.Yang X & Qian K Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol 18, 452–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine ZG & Walker S The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells? Annu. Rev. Biochem 85, 631–57 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Alonso J, Schimpl M & van Aalten DM O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J. Biol. Chem 289, 34433–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuzwa SA & Vocadlo DJ O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer’s disease and beyond. Chem. Soc. Rev 43, 6839–58 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Ma J & Hart GW Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics 10, 365–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slawson C & Hart GW O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart GW Nutrient regulation of signaling and transcription. J. Biol. Chem 294, 2211–2231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorelik A & van Aalten DMF Tools for functional dissection of site-specific O-GlcNAcylation. RSC Chem. Biol 1, 98–109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin SES et al. Structure-based evolution of low nanomolar O-GlcNAc transferase inhibitors. J. Am. Chem. Soc 140, 13542–13545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Tan EP, VandenHull NJ, Peterson KR & Slawson C O-GlcNAcase expression is sensitive to changes in o-glcnac homeostasis. Front. Endocrinol. (Lausanne) 5, 206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuzwa SA et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol 4, 483–90 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Alfaro JF et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U. S. A 109, 7280–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vosseller K et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–34 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK & Bertozzi CR Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 12, 561–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorelik A et al. Genetic recoding to dissect the roles of site-specific protein O-GlcNAcylation. Nat. Struct. Mol. Biol 26, 1071–1077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart GW, Slawson C, Ramirez-Correa G & Lagerlof O Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem 80, 825–58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruan HB, Nie Y & Yang X Regulation of protein degradation by O-GlcNAcylation: crosstalk with ubiquitination. Mol. Cell. Proteomics 12, 3489–97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez DH et al. Engineering a Proximity-Directed O-GlcNAc Transferase for Selective Protein O-GlcNAcylation in Cells. ACS Chem. Biol 15, 1059–1066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingram JR, Schmidt FI & Ploegh HL Exploiting nanobodies’ singular traits. Annu. Rev. Immunol 36, 695–715 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Li B, Li H, Lu L & Jiang J Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode. Nat. Struct. Mol. Biol 24, 362–369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth C et al. Structural and functional insight into human O-GlcNAcase. Nat. Chem. Biol 13, 610–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elsen NL et al. Insights into activity and inhibition from the crystal structure of human O-GlcNAcase. Nat. Chem. Biol 13, 613–615 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Gao Y, Wells L, Comer FI, Parker GJ & Hart GW Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem 276, 9838–45 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Rexach JE et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol 6, 645–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchhofer A et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol 17, 133–8 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Butkinaree C et al. Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J. Biol. Chem 283, 23557–66 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cetinbas N, Macauley MS, Stubbs KA, Drapala R & Vocadlo DJ Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 45, 3835–44 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Woo CM et al. Mapping and quantification of over 2000 O-linked glycopeptides in activated human t cells with isotope-targeted glycoproteomics (Isotag). Mol. Cell. Proteomics 17, 764–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Genst EJ et al. Structure and properties of a complex of alpha-synuclein and a single-domain camelid antibody. J. Mol. Biol 402, 326–43 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Ling J et al. A nanobody that recognizes a 14-residue peptide epitope in the E2 ubiquitin-conjugating enzyme UBC6e modulates its activity. Mol. Immunol 114, 513–523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traenkle B et al. Monitoring interactions and dynamics of endogenous beta-catenin with intracellular nanobodies in living cells. Mol. Cell. Proteomics 14, 707–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess J, Angel P & Schorpp-Kistner M AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci 117, 5965–73 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Tai HC, Khidekel N, Ficarro SB, Peters EC & Hsieh-Wilson LC Parallel identification of O-GlcNAc-modified proteins from cell lysates. J. Am. Chem. Soc 126, 10500–1 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Maynard JC, Strickland A, Burlingame AL & Milbrandt J Schwann cell O-GlcNAcylation promotes peripheral nerve remyelination via attenuation of the AP-1 transcription factor JUN. Proc. Natl. Acad. Sci. U. S. A 115, 8019–8024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiao Y et al. High glucose stimulates tumorigenesis in hepatocellular carcinoma cells through AGER-dependent O-GlcNAcylation of c-Jun. Diabetes 65, 619–32 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Zhang F et al. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–25 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Ranuncolo SM, Ghosh S, Hanover JA, Hart GW & Lewis BA Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem 287, 23549–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardiville S et al. TATA-box binding protein O-GlcNAcylation at T114 regulates formation of the B-TFIID complex and is critical for metabolic gene regulation. Mol. Cell 77, 1143–1152 e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu T et al. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol 27, 605–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siriwardena SU et al. Phosphorylation-inducing chimeric small molecules. J. Am. Chem. Soc 142, 14052–14057 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Dong JX et al. A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. Elife 8, e48750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch B et al. Generation and validation of homozygous fluorescent knock-in cells using CRISPR-Cas9 genome editing. Nat. Protoc 13, 1465–1487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuey C, Larocque G, Clarke NI & Royle SJ Unintended perturbation of protein function using GFP nanobodies in human cells. J. Cell Sci 132, jcs234955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fridy PC et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farrants H et al. Chemogenetic control of nanobodies. Nat. Methods 17, 279–282 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Gil AA et al. Optogenetic control of protein binding using light-switchable nanobodies. Nat. Commun 11, 4044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Y et al. O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol 11, 319–25 (2015). [DOI] [PubMed] [Google Scholar]
References
- 48.Thompson JW, Griffin ME & Hsieh-Wilson LC Methods for the detection, dtudy, and dynamic profiling of O-GlcNAc glycosylation. Methods Enzymol 598, 101–135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo CM & Bertozzi CR Isotope targeted glycoproteomics (IsoTaG) to characterize intact, metabolically labeled glycopeptides from complex proteomes. Curr. Protoc. Chem. Biol 8, 59–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huber W, von Heydebreck A, Sultmann H, Poustka A & Vingron M Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1, S96–104 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data Fig. 5. Unprocessed Western Blots and/or gels
Source Data Fig. 4. Unprocessed Western Blots and/or gels
Source Data Fig. 6. Unprocessed Western Blots and/or gels
Source Data Fig. 1. Statistical Source Data
Source Data Fig. 2. Statistical Source Data
Source Data Fig. 3. Statistical Source Data
Source Data Fig. 7. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 1. Statistical Source Data
Source Data Extended Data Fig. 2. Statistical Source Data
Source Data Extended Data Fig. 4. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 3. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 7. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 8. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 5. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 6. Unprocessed Western Blots and/or gels
Source Data Extended Data Fig. 9. Unprocessed Western Blots and/or gels
Data Availability Statement
The main data generated or analyzed during this study are included in this published article and its Supplementary Information. The mass spectrometry proteomics data were searched against the UniProt/SwissProt human (Homo sapiens) protein database (Aug. 19, 2016, www.uniprot.org/proteomes/UP000005640) and have been deposited to the ProteomeXchange Consortium via the PRIDE51 partner repository with the dataset identifier PXD022347 (Fig. 3d,e) and PXD018914 (Fig. 3f,g). Source data are provided with this paper.